Abstract
Secondary cell wall (SCW) deposition in Arabidopsis is regulated among others by NAC transcription factors, where SND1 chiefly initiates xylem fibre differentiation while VND6 controls metaxylem vessel SCW development, especially programmed cell death and wall patterning. The translational relevance of Arabidopsis SCW regulation theory and the utility of characterized transcription factors as modular synthetic biology tools for improving commercial fibre crops is unclear. We investigated inter-lineage gene activation dynamics for potential fibre and vessel differentiation regulators from the widely grown hardwood Eucalyptus grandis (Myrtales). EgrNAC26, a VND6 homolog, and EgrNAC61, an SND1 homolog, were transiently expressed in Arabidopsis mesophyll protoplasts in parallel to determine early and late (i.e. 7 and 14 hours post-transfection) gene targets. Surprisingly, across the time series EgrNAC26 activated only a subset of SCW-related transcription factors and biosynthetic genes activated by EgrNAC61, specializing instead in targeting vessel-specific wall pit and programmed cell death markers. Promoters of EgrNAC26 and EgrNAC61 both induced reporter gene expression in vessels of young Arabidopsis plants, with EgrNAC61 also conferring xylem- and cork cambium-preferential expression in Populus. Our results demonstrate partial conservation, with notable exceptions, of SND1 and VND6 homologs in Eucalyptus and a first report of cork cambium expression for EgrNAC61.
Introduction
Secondary cell walls (SCWs) equip plants with pathogen resistance, mechanical support and the ability to effectively transport water from the roots to the aerial organs1–3. Fast-growing angiosperm trees such as Eucalyptus are widely grown as short-rotation lignocellulosic feedstocks for pulp, paper and other renewable biomass products derived from the SCWs within wood4. The deposition of SCWs, which consist primarily of cellulose, hemicelluloses and lignin, is regulated by a complex semi-hierarchical transcriptional network composed mainly of NAC (NAM/ATAF/CUC) and MYELOBLASTOSIS (MYB) transcription factors (TFs)5–8. Among the known master regulators of SCW formation in the herbaceous model Arabidopsis thaliana (Arabidopsis), Secondary Wall NACs (SWNs9) appear to initiate SCW deposition through this regulatory network, occupying the upper network tier and regulating several mid- and lower-tier TFs as well as core SCW biosynthesis genes. The SWNs directly regulate, among others, the expression of several key MYB TF genes, with MYB46 and its functionally redundant homolog MYB83 also being considered master regulators positioned mid-tier, as is the CCCH-type zinc finger C3H1410–14.
Arabidopsis SWNs in the NST clade, among them NST1 (NAC SECONDARY WALL THICKENING PROMOTING FACTOR 1), NST2 and SND1 (SECONDARY WALL-ASSOCIATED NAC DOMAIN THICKENING FACTOR 1), regulate SCW formation in fibres, anther endothecia and silique valves to a large degree of redundancy, while VND (VASCULAR-RELATED NAC DOMAIN) clade TFs encompassing VND1 through VND7 are vessel-specific, with VND6 specifically regulating metaxylem SCW deposition and VND7 regulating both meta- and protoxylem vessel formation15–24. Thus, VND6 is a key regulator of the reticulated and pitted wall patterning observed in secondary xylem vessels, the deposition of which is determined by the bundled microtubule structure of the cytoskeleton25,26. In woody angiosperms such as Populus the expression of NST and VND clade homologs appear to overlap somewhat, with both the SND1 homolog PtrWND1B (a Populus trichocarpa wood-associated NAC domain protein) and the VND7 homolog PtrWND6A being expressed in xylem and phloem fibres, while vessel-specific differentiation appears to be regulated by the exclusive expression of PtrWND6A in vessels27,28. High-resolution spatial transcript profiling in aspen from phloem through the cambium to the lignified xylem zone revealed biphasic expression peaks for SND1 homologs in phloem and early xylem SCW deposition, while VND6 homolog transcripts peaked either during xylem SCW deposition or its cessation29, suggesting a specialization of SND1 homologs in phloem and xylem fibre formation and VND6 homologs in vessel differentiation. However, dominant repression in Populus of either the SND1 homolog PtrWND2B or the VND7 homolog PtrWND6B resulted in significantly reduced xylem SCW deposition in both fibres and vessels30. Although this suggests less distinct roles for SWN-mediated regulation of fibre and vessel SCW formation in woody angiosperms, the question of partially overlapping versus distinct roles in secondary xylem development remains poorly resolved. For example, homologs of SND1 and VND6/VND7 in monocots (a lineage lacking secondary xylem derived from a vascular cambium) appear to be expressed indistinguishably in sclerenchyma fibres and vessels31,32, while in Norway Spruce (a woody gymnosperm lacking fibres and vessels) sufficiently distinct roles could be inferred for VND and NST homologs during xylogenesis33.
Xylem fibre and vessel differentiation is distinguished by differences in SCW thickness and patterning as well as the timing and rate of programmed cell death (PCD) and autolysis, which in the case of water-conducting vessel elements yields hollow lumens shortly after SCW deposition6,34,35. Congruent with their proposed functions in xylem vessel development, VND6 and VND7 strongly activate PCD genes in Arabidopsis9,36. PCD and autolysis, which are distinct biological processes, are initiated by Ca2+ influx signals resulting from extracellular proteolysis by serine proteases37,38. Proteins currently known to be involved in vessel autolysis include XYLEM CYSTEINE PEPTIDASE 1 (XCP1), XCP2 and METACASPASE 9 (MC9) that together cause autolysis of the cytoplasm, the endonuclease ZEN1, XYLEM SERINE PEPTIDASE 1 (XSP1), and possibly ARABIDOPSIS THALIANA SUBTILASE 1.1 (SBT1.1)39–44. The later onset of PCD in fibres may be attributed to the weaker activation of PCD genes by SND1 and slower accumulation of the necessary enzymes to prompt cellular degradation45,46. Compared to fibres, metaxylem vessels exhibit extensive, pronounced pits in their SCWs that allow for the lateral movement of water to living fibres and xylem parenchyma47,48. Pits are established by the recruitment of microtubule disassembly proteins MIDD1 and Kinesin 13 A by pit localization of GTP-bound ROP11, a Rho GTPase that is localized to oval planar domains at the plasma membrane by IQD13, activated by ROPGEF4 and deactivated by ROPGAP349–51.
Despite what is believed to be substantial evolutionary conservation in SCW transcriptional regulation52, the prevalence and significance of lineage-specific adaptations to the SCW transcriptional network architecture among angiosperms are poorly understood. In Populus, for example, numerous co-orthologs of SND1 exist, of which alternative splicing of PtrWND1B in particular produces full-length and intron-retaining, truncated isoforms of the protein that can form non-functional heterodimers27,53–55. Since Arabidopsis does not harbour known SND1 splice variants, this shows that SND1-mediated SCW regulation may differ between plant lineages. In the Eucalyptus woody model, a small number of studies have aided in our understanding of SCW regulation56–61. To date, our knowledge of the functions of SWN master regulators in Eucalyptus is limited to a few gene targets of EgWND1, an E. gunnii homolog of SND130, with some evidence that an intron 2-retaining splice variant may also exist in E. grandis27. In previous work62, we identified putative E. grandis orthologs of SND1 (EgrNAC61, also orthologous to EgWND1) and VND6 (EgrNAC26) which we hypothesised regulate fibre and vessel SCW development, respectively.
While functionally characterized SCW TFs and their promoters are potential tools for the re-engineering of woody biomass, their crude manipulation generally results in undesirable phenotypes and hence refined approaches adopting synthetic and systems biology are favourable63. The development of high-precision multipartite synthetic gene constructs, however, assumes individual parts possess modularity (functional fidelity of discrete parts in different host organisms) and orthogonality (independence from and minimal interference by native endogenous processes), both of which are poorly demonstrated for SCW TFs. For instance, what gene targets might we expect to be activated when SCW TF homologs from diverged lineages are heterologously and ectopically induced, and how is this influenced when their endogenous promoters are exchanged with synthetic promoters? In this study, we functionally characterized E. grandis SCW master regulators through observation of their SCW network targets over time in the context of Arabidopsis mesophyll protoplasts, and their promoter activities in woody and non-woody models. Our results show that EgrNAC61 is a more potent activator of SCW genes while the VND6 homolog EgrNAC26 mainly regulates PCD and autolysis in vessels.
Results
EgrNAC26 and EgrNAC61 transcript splicing, transcriptional activity and spatial expression patterns
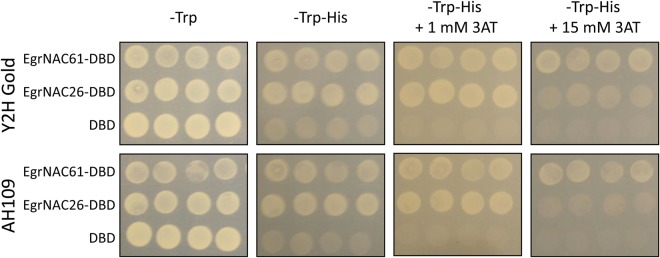
We PCR-amplified the EgrNAC26 and EgrNAC61 coding sequences from E. grandis developing secondary xylem cDNA for cloning. We found no evidence of alternative splice variants among the amplicons (Fig. S1). Similarly, analysis of RNA-seq reads from an independent developing secondary xylem sample64,65 showed no significant coverage across any of the introns (Fig. S2). These results therefore do not support those of the previously observed EgrNAC61 intron 2 retention reported by Zhao et al.27. Since EgrNAC26 and EgrNAC61 are likely master regulators of SCW deposition in E. grandis based on homology, we next tested the ability of these proteins to activate transcription of the His reporter gene in two strains of S. cerevisiae. Both strains expressing either EgrNAC61 or EgrNAC26 grew on histidine-deficient media supplemented with up to 15 mM of the His reporter inhibitor 3AT (Fig. 1), while for the empty-vector control line significantly less growth was observed on media lacking histidine and no growth was observed on media supplemented with 3AT. For both strains, stronger reporter gene activation at 15 mM 3AT was observed for EgrNAC61 than EgrNAC26. These results demonstrate that EgrNAC26 and EgrNAC61 have strong intrinsic transcriptional activation activities.
Figure 1.
Transactivation of the HIS reporter gene in two yeast strains (Y2H Gold and AH109) by EgrNAC61 and EgrNAC26 fused to the GAL4 DNA binding domain (DBD). Yeast transformed with all constructs, including empty vector encoding only the DBD, grew on media lacking Tryptophan (-Trp). Growth of EgrNAC61-DBD and EgrNAC26-DBD fusions on media lacking Tryptophan and Histidine (-Trp-His) demonstrate transcriptional transactivation, while growth on increasing concentrations of 3AT indicate the strength of transcriptional transactivation.
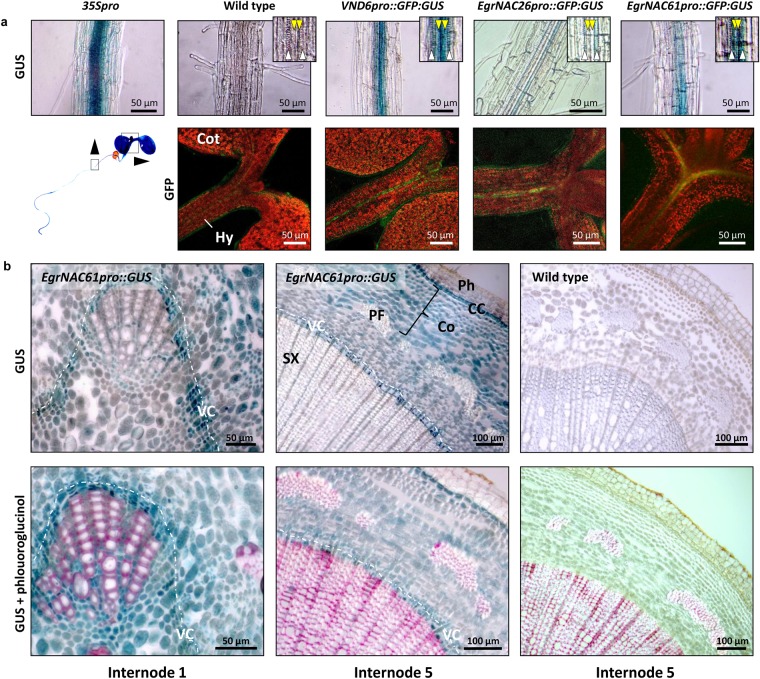
We analysed promoter::GFP:GUS reporter fusions of the EgrNAC26 and EgrNAC61 promoter sequences to determine their heterologous expression patterns. In the vasculature of Arabidopsis seedling roots, which contain only xylem vessels, the metaxylem vessel marker VND6pro::GFP:GUS produced predominant GUS staining in the metaxylem vessels in the centre of the vasculature, with less blue staining in protoxylem vessels (Fig. 2a). In contrast, a 35Spro::GFP:GUS constitutive control showed staining in all root cell types. In most cases, GUS staining in EgrNAC26pro::GFP:GUS lines was not visible; in a few cases, faint expression was observed in the protoxylem vessels. However, GFP expression for this construct was clearly detected in the seedling hypocotyl vasculature (Fig. 2a). Previously, SND1 promoter-directed expression was observed predominantly in interfascicular and xylary fibres of inflorescence stems and hypocotyls of flowering Arabidopsis plants, and also in developing vessels undergoing SCW thickening16,20. In EgrNAC61pro::GFP:GUS plants, GUS staining was evident in proto- and metaxylem vessels in the root vasculature (Fig. 2a). Analysis of GFP expression in the vasculature of seedling hypocotyls for the VND6, EgrNAC26 and EgrNAC61 promoters corroborates our findings of GUS staining in the seedling vessels (Fig. 2a).
Figure 2.
Reporter gene analysis of EgrNAC26 and EgrNAC61 promoters. (a) Promoter analysis in Arabidopsis seedlings. Top panel, GUS histochemical staining in seedling roots. Insets show the positions of protoxylem (white chevrons) and metaxylem (yellow chevrons) vessels. 35 Spro, CaMV 35S promoter (positive control). Bottom panel, corresponding GFP confocal microscopy analysis of seedling shoot tips for each construct. Cot, cotyledon; Hy, hypocotyl. (b) EgrNAC61 promoter::GUS analysis of internodes 1 and 5 in transgenic poplar plants. Bottom panel, phloroglucinol-stained GUS sections. CC, cork cambium; Co, cortex; PF, phloem fibres; Ph, phellem; SX, secondary xylem; VC, vascular cambium.
We also explored the expression patterns of EgrNAC61 during wood formation through promoter::GUS analysis in transgenic poplar plants. GUS signal was strongest in the developing xylem region of the first internode, just before lignification of the primary xylem vessels distinguishable with phloroglucinol staining (Fig. 2b). In secondary xylem in the fifth internode, expression was observed throughout the cortex but was strongest in the xylem expansion zone, sandwiched by the vascular cambium and the lignification zone, as well as in the cork cambial region (phellogen). The latter is a novel report. GUS expression appeared to be visible in developing secondary vessel and fibre cells, forming a near-continuous ring of signal along the expansion zone (Fig. 2b, internode 5). Together, the Arabidopsis and poplar results support a role for EgrNAC26 in the regulation of vessel formation, and EgrNAC61 in the regulation of secondary xylem fibre and vessel deposition.
Identification of early and late transcriptional targets of EgrNAC26 and EgrNAC61 in Arabidopsis protoplasts
In order to identify early (likely direct) and late (likely indirect) gene targets of EgrNAC26 and EgrNAC61 in a heterologous background, we transiently overexpressed these genes in linkage with a dual GFP reporter cassette in Arabidopsis leaf mesophyll protoplasts. Total RNA was isolated 7 and 14 hours post-transfection (hpt) from three independent protoplast isolations per time point. Transfection efficiencies were approximately 30–50% (Fig. S3). Following RNA quality analysis (Fig. S4) and strand-specific RNA sequencing, between 17 and 36 million paired-end reads were obtained per library, most with >90% concordant mapping of read pairs to the Arabidopsis TAIR10 genome (Table S2). After quantification and normalization of the expression values, all transgene FPKM values, including those of GFP for the empty vector (EV) libraries, were within the top percentile of the detected transcript levels and expressed at similar levels across time points and replicates (Fig. S5). Principle Component Analysis (PCA) of transcriptome-wide FPKM values for each library showed stronger differentiation according to time point than construct, while clear clustering by construct was only apparent at 7 hpt (Fig. S6), suggesting that each master regulator affected only a small proportion of the transcriptome uniquely, and most distinguishably for the early targets.
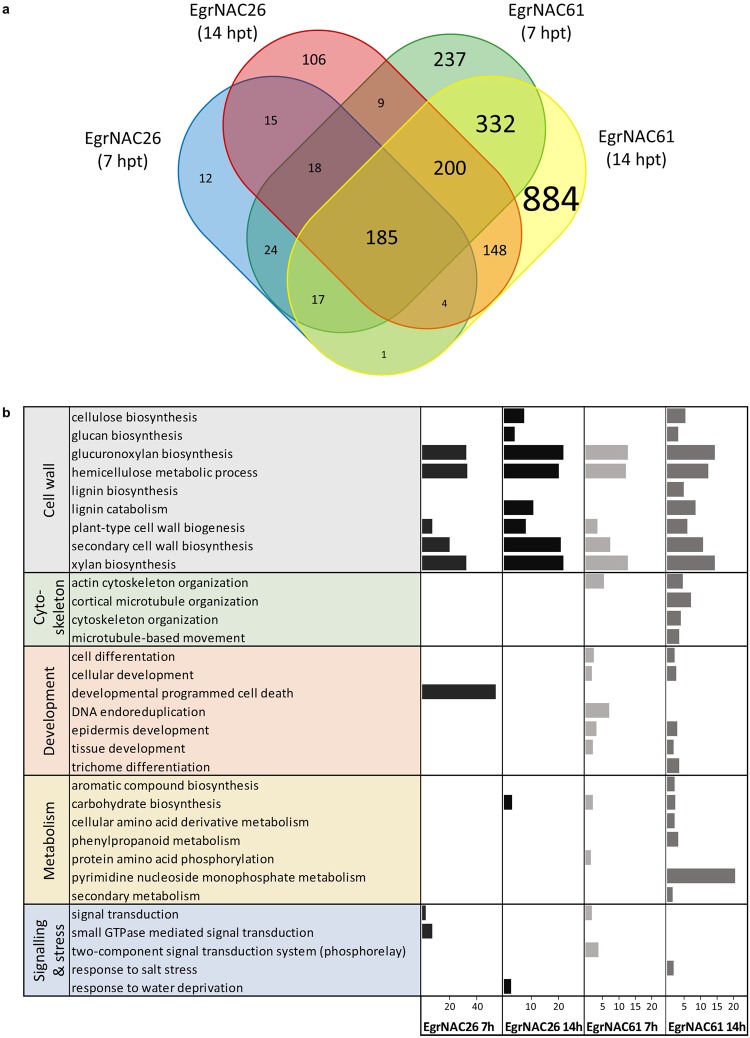
Differential expression analysis of the mesophyll protoplasts induced with EgrNAC26 (EgrNAC26-OX) and EgrNAC61 (EgrNAC61-OX) followed by FPKM and fold-change filtering (FPKM ≥1, fold-change ≥2-fold up or down) identified 276 (EgrNAC26; 7 hpt), 1,022 (EgrNAC61; 7 hpt), 685 (EgrNAC26; 14 hpt) and 1,771 (EgrNAC61; 14 hpt) differentially expressed genes (DEGs), of which the vast majority were upregulated (Figs 3a, S7, Supplementary Dataset 1). The overlap between EgrNAC26-OX and EgrNAC61-OX datasets was highly significant (P < 2.2 × 10−16), only one transcript was alternately up- or down-regulated across time points, and DEGs common to EgrNAC26-OX and EgrNAC61-OX datasets were all up- or down-regulated concordantly, demonstrating robust and consistent activation of a subset of shared genes. Overrepresented biological processes were identified for EgrNAC26-OX and EgrNAC61-OX DEGs through Gene Ontology (GO) enrichment analysis. Significantly overrepresented terms among up-regulated DEGs in the EgrNAC26-OX and EgrNAC61-OX datasets were largely associated with SCW biosynthesis, particularly at 14 hpt. Common terms included secondary cell wall biogenesis, cellulose biosynthesis and (glucurono)xylan biosynthesis (Fig. 3b), supporting the hypothesized roles of these genes as SCW master regulators since the overexpression of the transgenes was sufficient to cause ectopic activation of these genes. Differences between the datasets provided insight into known and novel biological functions that were likely distinctly regulated by EgrNAC26 and EgrNAC61. Specifically, supporting evidence for the role of EgrNAC26 as a vessel-preferential master regulator was the enrichment of genes in the category “developmental PCD” at 7 hpt (Fig. 3b). In contrast, EgrNAC61 induced a good representation of cytoskeleton-related processes, a number of developmental processes such as trichome and epidermal differentiation, and interestingly several primary metabolic processes such as amino acid and pyrimidine metabolism in addition to secondary metabolism. These results are consistent with a role of EgrNAC26 and EgrNAC61 in the regulation of SCW synthesis.
Figure 3.
Differentially expressed genes (DEGs) induced by EgrNAC26 and EgrNAC61. (a) Common and unique DEGs for EgrNAC26-OX and EgrNAC61-OX datasets at 7 and 14 hpt with font sizes scaled to the number of genes in each cell. (b) Selected overrepresented biological functions among EgrNAC26-OX and EgrNAC61-OX genes, with fold enrichment indicated on the x-axis.
Temporal regulation of Arabidopsis SCW transcriptional network targets by EgrNAC26 and EgrNAC61
Notable DEGs in the EgrNAC26-OX and EgrNAC61-OX datasets involved in SCW xylem development and transcriptional regulation are shown in Table 1. In total, 151 TFs were differentially expressed, most of them upregulated (Table S3). Many genes were exclusively up-regulated in EgrNAC61-OX protoplasts, among them known SCW transcriptional regulators MYB4, MYB7, MYB42, MYB52, MYB54, MYB83 and MYB103, and lignin regulators MYB58, MYB63 and MYB8511,13,66,67 (Tables 1, S3). Likewise, several lignin-specific genes were upregulated in EgrNAC61-OX at 14 hpt only, including 4CL3, CCoAOMT1, and HCT. Interestingly, far fewer SCW-related DEGs were unique to EgrNAC26-OX; noteworthy examples included LBD30/ASL19, VND2, TED7, a key cytoskeleton-associated protein MAP70-5 and the ROP11 activator ROPGEF4 (Table 1). No activation of the endogenous SND1 gene was observed, suggesting that the self-activation of SND1 observed in plasmid-based promoter transactivation and electrophoretic mobility shift assays68 did occur in the context of native mesophyll chromatin.
Table 1.
Secondary cell wall-associated genes regulated by EgrNAC26 and EgrNAC61.
| Gene ID | Gene name | Linear fold change | |||
|---|---|---|---|---|---|
| EgrNAC26-OX | EgrNAC61-OX | ||||
| 7 hpt | 14 hpt | 7 hpt | 14 hpt | ||
| Transcription factors | |||||
| AT1G75410 | BEL1-LIKE HOMEODOMAIN 3 (BLH3) | — | — | 3.4 | 4.4 |
| AT4G34610 | BEL1-LIKE HOMEODOMAIN 6 (BLH6) | — | 18.5 | 12.9 | 361.7 |
| AT1G19700 | BEL1-LIKE HOMEODOMAIN 10 (BEL10) | — | — | 3.5 | 5.7 |
| AT1G66810 | CCCH ZINC FINGER 14 (C3H14) | — | 5.2 | 7.5 | 34.0 |
| AT1G62990 | KNOTTED-LIKE HOMEOBOX OF ARABIDOPSIS THALIANA 7 (KNAT7) | — | 162.4 | 108.8 | 1139.7 |
| AT2G40470 | LATERAL ORGAN BOUNDARIES 15 (BD15)/ASL11 | — | 26.4 | — | 49.2 |
| AT4G00220 | LATERAL ORGAN BOUNDARIES 30 (LBD30)/ASL19 | — | Inf* | — | — |
| AT1G66230 | MYB20 | 2.2 | — | 3.8 | 2.4 |
| AT4G12350 | MYB42 | — | — | — | 3.5 |
| AT5G12870 | MYB46 | — | 255.9 | — | 1161.7 |
| AT1G17950 | MYB52 | — | — | — | Inf* |
| AT1G73410 | MYB54 | — | — | — | 84.8 |
| AT4G01680 | MYB55 | 9.3 | 3.8 | 22.6 | 4.9 |
| AT1G16490 | MYB58 | — | — | — | 28.3 |
| AT1G79180 | MYB63 | — | — | — | 5.2 |
| AT3G08500 | MYB83 | — | — | Inf* | Inf* |
| AT4G22680 | MYB85 | — | — | 6.9 | 6.8 |
| AT1G63910 | MYB103 | — | — | Inf* | — |
| AT2G38090 | MYB-R | — | 12.3 | 3.5 | 48.4 |
| AT2G34710 | PHABULOSA (PHB) / ATHB14 | — | — | 2.2 | 2.6 |
| AT4G28500 | SECONDARY WALL-ASSOCIATED NAC DOMAIN 2 (SND2) | — | 42.4 | — | 1085.8 |
| AT1G28470 | SECONDARY WALL-ASSOCIATED NAC DOMAIN (SND3) | — | 88.1 | 33.4 | 416.9 |
| AT5G60200 | TARGET OF MONOPTEROS 6 (TMO6) | — | — | 2.8 | 3.4 |
| AT4G36160 | VASCULAR-RELATED NAC-DOMAIN 2 (VND2) | — | 2.1 | — | — |
| AT5G64530 | XYLEM NAC DOMAIN 1 (XND1) | — | — | 2.5 | — |
| Cell wall biosynthesis | |||||
| AT1G65060 | 4-COUMARATE:CoA LIGASE 3 (4CL3) | — | — | — | 61.1 |
| AT4G34050 | CAFFEOYL-CoA 3-O-METHYLTRANSFERASE 1 (CCoAOMT1) | — | — | — | 2.9 |
| AT4G26220 | CAFFEOYL CoA ESTER O-METHYLTRANSFERASE 7 (CCoAOMT7) | — | 9.7 | — | 271.3 |
| AT4G39350 | CELLULOSE SYNTHASE 2 (CESA2) | — | — | — | 2.1 |
| AT5G44030 | CELLULOSE SYNTHASE 4 (CESA4) | — | 35.3 | 29.7 | 649.5 |
| AT5G09870 | CELLULOSE SYNTHASE 5 (CESA5) | — | — | 3.1 | 5.1 |
| AT5G17420 | CELLULOSE SYNTHASE 7 (CESA7) | — | 49.9 | 42.0 | 728.2 |
| AT4G18780 | CELLULOSE SYNTHASE 8 (CESA8) | — | 91.3 | — | 1792.6 |
| AT5G03760 | CELLULOSE SYNTHASE-LIKE A9 (CSLA09) | — | 8.1 | — | 45.1 |
| AT2G32620 | CELLULOSE SYNTHASE-LIKE B2 (CSLB02) | — | 56.2 | Inf* | 1331.2 |
| AT1G15950 | CINNAMOYL COA REDUCTASE 1 (CCR1)/IRREGULAR XYLEM 4 (IRX4) | — | — | — | 2.1 |
| AT5G15630 | COBRA-LIKE4 (COBL4) | — | 44.1 | — | 1083.1 |
| AT2G40890 | COUMARATE 3-HYDROXYLASE 1 (C3H1)/ REF8 | — | — | — | 2.2 |
| AT3G55990 | ESKIMO 1 (ESK1) | — | 220.5 | — | 1676.0 |
| AT5G47820 | FRAGILE FIBER1 (FRA1) | — | — | 2.2 | 2.6 |
| AT2G28110 | FRAGILE FIBER8 (FRA8)/IRREGULAR XYLEM7 (IRX7) | 2.3 | 3.2 | 6.1 | 10.6 |
| AT3G18660 | GLUCURONIC ACID SUBSTITUTION OF XYLAN 1 (GUX1) | — | 411.2 | — | 2763.5 |
| AT4G33330 | GLUCURONIC ACID SUBSTITUTION OF XYLAN 2 (GUX2) | — | 167.0 | Inf* | 1922.8 |
| AT5G48930 | HYDROXYCINNAMOYL-CoA SHIKIMATE/QUINATE HYDROXYCINNAMOYL TRANSFERASE (HCT) | — | — | — | 2.3 |
| AT2G37090 | IRREGULAR XYLEM 9 (IRX9) | — | 103.5 | — | 2449.4 |
| AT1G27440 | IRREGULAR XYLEM 10 (IRX10) | 3.2 | 7.1 | 11.5 | 107.4 |
| AT5G61840 | IRX10-LIKE (IRX10-L) | — | — | — | 4.1 |
| AT4G36890 | IRREGULAR XYLEM 14 (IRX14) | — | — | 2.1 | 7.6 |
| AT5G67230 | IRX14-LIKE (IRX14-L) | — | 8.1 | — | 62.3 |
| AT3G50220 | IRREGULAR XYLEM 15 (IRX15) | — | 173.9 | — | 1575.3 |
| AT5G67210 | IRX15-LIKE (IRX15-L) | — | 12.9 | — | 109.1 |
| AT2G38080 | LACCASE 4 (LAC4) | 19.2 | 206.1 | 71.2 | 1419.8 |
| AT5G60020 | LACCASE 17 (LAC17) | — | 110.9 | — | 423.5 |
| AT1G19300 | PARVUS | 3.0 | 38.6 | 22.5 | 268.9 |
| AT3G47400 | Pectinesterase family protein | Inf* | 555.9 | Inf* | 889.7 |
| AT5G51890 | PEROXIDASE 66 (PER66) | — | 139.6 | 132.1 | 1690.6 |
| AT5G66390 | PEROXIDASE 72 (PER72) | — | 3564.3 | — | 149.3 |
| AT1G70500 | Polygalacturonase | 466.9 | 546.8 | 538.8 | 608.7 |
| AT1G23460 | Polygalacturonase | 3.6 | 22.8 | 3.8 | 18.1 |
| AT2G43880 | Polygalacturonase, putative / pectinase, putative | — | — | — | Inf* |
| AT5G46340 | REDUCED WALL ACETYLATION 1 (RWA1) | — | 2.8 | 7.8 | 33.9 |
| AT2G34410 | REDUCED WALL ACETYLATION 3 (RWA3) | — | — | 3.6 | 12.5 |
| AT1G29890 | REDUCED WALL ACETYLATION 4 (RWA4) | — | 2.6 | 4.1 | 15.1 |
| AT5G01360 | TRICHOME BIREFRINGENCE-LIKE 31 (TBL31) | — | 112.2 | — | 562.8 |
| AT2G40320 | TRICHOME BIREFRINGENCE-LIKE 33 (TBL33) | — | 125.0 | Inf* | 2067.0 |
| AT2G38320 | TRICHOME BIREFRINGENCE-LIKE 34 (TBL34) | — | 761.1 | — | 1268.2 |
| AT1G12780 | UDP-GLC 4-EPIMERASE 1 (UGE1) | 2.6 | 2.7 | 8.2 | 8.6 |
| AT5G59290 | UDP-XYLOSE SYNTHASE 3 (UXS3) | 2.0 | 7.4 | 10.4 | 60.4 |
| AT2G28760 | UDP-XYLOSE SYNTHASE 6 (UXS6) | 9.9 | 40.8 | 60.1 | 242.3 |
| AT2G14620 | XYLOGLUCAN ENDOTRANSGLYCOSYLASE/HYDROLASE 10 (XTH10) | 6.0 | 8.0 | 36.1 | 9.4 |
| AT3G48580 | XYLOGLUCAN ENDOTRANSGLYCOSYLASE/HYDROLASE 11 (XTH11) | — | 3.0 | 2.4 | 2.8 |
| Cytoskeleton | |||||
| AT1G14840 | MICROTUBULE-ASSOCIATED PROTEIN 70-5 (MAP70-5) | — | 63.4 | — | — |
| AT3G53350 | MICROTUBULE DEPLETION DOMAIN 1 (MIDD1) | — | 2.6 | 5.7 | 11.3 |
| AT1G50010 | TUBULIN ALPHA-2 CHAIN (TUA2) | — | 5.1 | 4.3 | 21.4 |
| AT1G04820 | TUBULIN ALPHA-4 CHAIN (TUA4) | — | — | — | 3.6 |
| AT4G14960 | TUBULIN ALPHA-6 CHAIN (TUA6) | — | — | — | 3.8 |
| AT5G23860 | TUBULIN BETA-8 (TUB8) | 2.3 | 3.6 | 6.3 | 19.8 |
| AT3G16630 | Kinesin-13A | — | — | — | 2.5 |
| Signaling | |||||
| AT4G13195 | CLE44 | — | — | 0.4 | 0.3 |
| AT4G17220 | IQ-DOMAIN 10 (IQD10) | — | — | — | Inf* |
| AT2G45890 | RHO GUANYL-NUCLEOTIDE EXCHANGE FACTOR 4 (ROPGEF4) | — | 39.2 | — | — |
| AT2G46710 | RHO GTPASE-ACTIVATING PROTEIN 3 (ROPGAP3) | 2.7 | 6.4 | 29.7 | 59.0 |
| AT5G62880 | RHO-RELATED PROTEIN FROM PLANTS 11 (ROP11) | 2.7 | 3.3 | 6.2 | 7.2 |
| AT1G08340 | RAC GTPase | — | 55.7 | — | 1020.4 |
| Programmed cell death and autolysis | |||||
| AT1G11190 | BIFUNCTIONAL NUCLEASE 1 (BFN1) | — | 359.3 | — | 147.9 |
| AT5G04200 | METACASPASE 9 (MC9) | 90.3 | 112.3 | 87.1 | 44.5 |
| AT1G26820 | RIBONUCLEASE 3 (RNS3) | Inf* | 678.7 | Inf* | 926.7 |
| AT4G35350 | XYLEM CYSTEINE PEPTIDASE 1 (XCP1) | 423.0 | 1371.1 | 117.7 | 119.9 |
| AT1G20850 | XYLEM CYSTEINE PEPTIDASE 2 (XCP2) | 83.9 | 1208.3 | 45.0 | 126.4 |
| Miscellaneous genes linked to xylem development | |||||
| AT3G16920 | CHITINASE-LIKE PROTEIN 2 (CTL2) | 6.7 | 62.6 | 96.0 | 890.8 |
| AT3G62020 | Germin-like protein 10 (GLP10) | 4.2 | 48.1 | 16.0 | 348.2 |
| AT1G33800 | Glucuronoxylan(GX)-specific 4-O-methyltransferase | — | 97.3 | 167.6 | 1165.0 |
| AT1G31720 | MODIFYING WALL LIGNIN 1 (MWL1) | — | Inf* | — | Inf* |
| AT4G27435 | MODIFYING WALL LIGNIN 2 (MWL2) | — | 68.3 | — | 1626.9 |
| AT5G18460 | Protein of Unknown Function (DUF239) | 242.1 | 295.1 | 651.0 | 453.4 |
| AT4G09990 | Protein of Unknown Function (DUF579) | — | 8.6 | 8.1 | 113.8 |
| AT4G18425 | Protein of Unknown Function (DUF679) | Inf* | — | Inf* | — |
| AT1G43790 | TRACHEARY ELEMENT DIFFERENTIATION-RELATED 6 (TED6) | — | 432.0 | — | 624.6 |
| AT5G48920 | TRACHEARY ELEMENT DIFFERENTIATION-RELATED 7 (TED7) | 18.2 | — | — | — |
*Inf refers to instances where the FPKM was 0 in the empty-vector control, but was larger than 1 in the test sample.
hpt, hours post transfection.
Several SCW-related genes and TFs were upregulated in at least one time point for both EgrNAC26-OX and EgrNAC61-OX. However, we noted that apart from the abovementioned SCW-related genes and TFs being exclusively activated by EgrNAC61, this TF activated several key TFs and many SCW biosynthetic genes five- to ten-fold more strongly than EgrNAC26 by 14 hpt. These included TFs BLH6, C3H14, KNAT7, MYB46, SND2 and SND3, cellulose-associated CesA4, CesA7, CesA8 and COBL4, xylan-associated genes such as IRX9, IRX10, IRX15, GUX1, GUS2 and PARVUS, and lignin-associated genes such as CCoAOMT7, LAC4 and LAC17 (Table 1). This, despite a lower transfection efficiency for the EgrNAC61-OX construct (Fig. S3b). In contrast, although PCD genes were expressed in all datasets, BFN1, MC9, XCP1 and XCP2 were upregulated by as much as an order of magnitude more in EgrNAC26-OX compared to EgrNAC61-OX at 14 h (Table 1). Supported by the apparently stronger transactivation ability of EgrNAC61 in yeast (demonstrated by its persistent growth at 15 mM 3AT, Fig. 1), our results demonstrate that EgrNAC61 is a more potent transcriptional activator of the Arabidopsis SCW pathway compared to EgrNAC26, while the latter shows a similar trend for PCD-associated genes in particular.
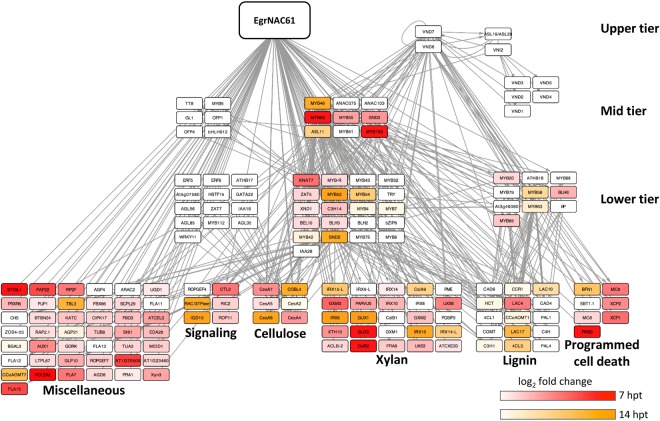
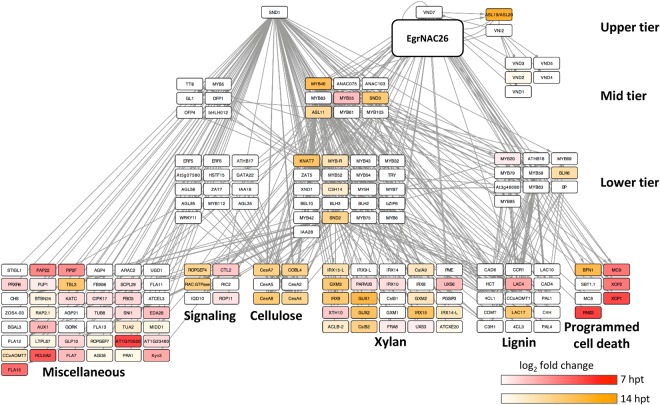
To understand the temporal activation of SCW transcriptional network nodes following EgrNAC26 and EgrNAC61 induction in Arabidopsis protoplasts, we mapped the DEGs to a curated Arabidopsis SCW transcriptional network compiled from the literature52. Time-lapse activation of the known SCW transcriptional network nodes by EgrNAC61 and EgrNAC26 is represented in Figs 4 and 5, respectively. EgrNAC61 induced strong, early upregulation of second and third tier TFs as well as structural genes associated with cellulose, hemicellulose and lignin biosynthesis by 7 hpt and a few additional targets in all tiers by 14 hpt (Fig. 4). The effect of EgrNAC26 induction clearly differed from that of EgrNAC61, where strong activation of PCD-associated genes but weak activation of only a few SCW structural genes and second- or third-tier TFs were apparent at 7 hpt, with a better representation of mid-tier SCW-related TFs and biosynthetic genes by 14 hpt, although weakly activated compared to EgrNAC61-OX (Fig. 5). Given that EgrNAC26 and EgrNAC61 transfections were performed in parallel on a common preparation of protoplasts, these results demonstrate a weaker and possibly delayed onset of SCW structural gene activation for EgrNAC26 in comparison. Finally, we noted that two clusters of mid- and lower-tier TFs implicated in the SCW network52 were unaffected by induction of either EgrNAC26 or EgrNAC61 (Figs 4 and 5, left). These clusters are not known to be activated by SWNs, consistent with our results and suggesting an independent route of regulation.
Figure 4.
Time-course activation of the Arabidopsis SCW transcriptional network following EgrNAC61 induction. The red heatmap indicates the fold change (log2) of transcripts activated at 7 hpt (hours post-transfection). The orange heatmap indicates the fold change (log2) of nodes that were exclusively upregulated at 14 hpt. Structural genes (bottom tier) are grouped according to biological function. The transcriptional network scaffold was adapted from Hussey et al.52.
Figure 5.
Time-course activation of the Arabidopsis SCW transcriptional network following EgrNAC26 induction. The red heatmap indicates the fold change (log2) of transcripts activated at 7 hpt (hours post-transfection). The orange heatmap indicates the fold change (log2) of nodes that were exclusively upregulated at 14 hpt. Structural genes (bottom tier) are grouped according to biological function. The transcriptional network scaffold was adapted from Hussey et al.52.
Discussion
In this study, we explored the temporal transactivation dynamics of Arabidopsis genes (Brassicales) by homologs of SCW master regulators of fibre and metaxylem vessel formation from E. grandis, a commercially important hardwood. Our results provide (1) insight into the possible roles of EgrNAC26 and EgrNAC61 SWNs in fibre and vessel SCW regulation in eucalypts, (2) evidence of considerable conservation in SWN gene targets between Eucalyptus as a representative of the basal eudicot lineage the Myrtales which divergence from the Brassicales over 100 million years ago69, and (3) a demonstration of the regulatory dynamics and specific targets of orthologous master regulators associated with fibre and vessel differentiation in a heterologous context typical of a synthetic biology strategy.
Few studies have explored the genome-wide gene targets of Arabidopsis SND1 and VND6/VND79,24,36,70. These previous investigations revealed preferential activation of PCD-associated genes such as XCP1 and XCP2 during vessel differentiation by VND6 and VND7. In woody angiosperms, only one study has reported the genomic targets of a fibre-associated SCW master regulator, in this case a time-course experiment of poplar stem differentiating xylem protoplasts transfected with Ptr-SND1-B1, a homolog of SND1 from P. trichocarpa71. Ours is the first study to probe the global transcriptional targets of EgrNAC26 and EgrNAC61, albeit in a heterologous system (Arabidopsis). One advantage of our approach is that the transfections for each construct were performed in parallel from a single protoplast preparation, allowing for a direct comparison of the gene targets and temporal activation dynamics of each SWN candidate. Two limitations should be noted: first, mesophyll protoplasts may not reflect the accessible chromatin landscape or contain the same co-factors as xylem mother cells. However, SWNs and their homologs are known to be particularly powerful SCW master regulators, sufficient for ectopic tracheary element transdifferentiation in many cell types20,72–74. The inherent transcriptional transactivation properties of EgrNAC26 and especially EgrNAC61 in yeast (Fig. 1) and the extensive and significant enrichment of SCW-associated biological processes in all EgrNAC26-OX and EgrNAC61-OX DEG datasets (Fig. 3b) support this statement. Thus, we can infer that EgrNAC26 and EgrNAC61 activate homologs of genes similarly in protoplasts as they do in developing xylem. Second, since the use of a heterologous system may yield Arabidopsis-specific targets and cannot detect novel gene targets unique to E. grandis, validation of the Eucalyptus orthologs of the target genes in this study is recommended.
Given the predominant fibre- and vessel-preferential SCW regulatory functions of SND1 and VND6 respectively in Arabidopsis, we expected to observe similar cell type associations for their putative E. grandis orthologs EgrNAC61 and EgrNAC26. In contrast to this hypothesis, we observed a substantial overlap in promoter activity between EgrNAC61pro and EgrNAC26pro in during Arabidopsis vascular development, namely in seedling vessels (Fig. 2). The EgrNAC26 promoter-driven reporter expression in Arabidopsis primary xylem vessels was substantially weaker, however. It is possible that the EgrNAC26 promoter construct, which included only 1,223 bp upstream of the predicted transcription start site due to a large 2,422 bp 5′ UTR region containing an embedded intron, lacked a critical cis-element for stronger expression, or that the promoter is regulated differently in E. grandis.
In comparison with our promoter analysis, expression of the aspen SND1 homolog peaked in phloem and early-onset SCW deposition in the xylem from cryosectioned xylogenesis developmental series29, consistent with our observation for developing xylem although it is unclear why no EgrNAC61pro activity was observed in phloem fibres (Fig. 2b). The aspen series29 did not include cork cambium, but we here report strong GUS induction by EgrNAC61pro in this tissue layer, while EgrNAC26pro was not assessed. Cork (phellem) consists of lignified and suberized cells that undergo PCD and have cell walls in which a guaiacyl-rich lignin backbone predominates75,76. Phloroglucinol staining revealed small amounts of lignin in the phellem of the poplar transgenics (Fig. 2b, bottom panel). Since EgrNAC61 activates the phenylpropanoid pathway (Figs 3, 5), we postulate that it has been co-opted to activate phellem lignification in the cork cambium too.
In addition to its homology to VND6, several lines of evidence point to a vessel-specific SCW regulatory role for EgrNAC26 in the transient expression experiment: (i) EgrNAC26 transfection uniquely induced vessel-specific regulators of SCW deposition, among them LATERAL ORGAN BOUNDARIES 30 (LBD30/ASL19), VND2 and TRACHEARY ELEMENT DIFFERENTION-RELATED 7 (TED7). LBD30/ASL19 and LBD18/ASL20 are targets of VND6 that participate in a positive feedback loop during vessel differentiation77, and VND2 is a vessel-specific SCW master regulator that interacts with VND715,23,74. TED7 is a vessel-specific protein with an unknown role in tracheary element SCW formation, acting with TED6 and interacting with the SCW-related CesA complex78. While TED6 was significantly upregulated by both EgrNAC26 and EgrNAC61 at 14 hpt (Table 1), the exclusive activation of TED7 at the earliest time point by EgrNAC26 suggests a vessel-related specialization. (ii) EgrNAC26 activated proteins associated with SCW pit formation in vessels, uniquely among them RHO GUANYL-NUCLEOTIDE EXCHANGE FACTOR 4 (ROPGEF4) and MICROTUBULE-ASSOCIATED PROTEIN 70-5 (MAP70-5) (Table 1). ROPGEF4 is essential for pit formation through its highly localized activation of the small GTPase ROP11, which was a common target of EgrNAC26 and EgrNAC61 in addition to the microtubule-associated protein MIDD1 and the ROP11-deactivating protein ROPGAP379. MAP70-5 is a tracheary element-specific microtubule-implicated protein, the abundance of which has a profound effect on the frequency of pitted, reticulated or smooth wall patterns observed in transdifferentiated cells80. Thus, its strong upregulation at 14 hpt in EgrNAC26-OX may re-structure the cytoskeleton for vessel-specific microtubule-directed SCW deposition. (iii) EgrNAC26 transfection induced stronger activation of the chief genes associated with autolysis of tracheary elements during PCD, namely XCP1, XCP2 and MC946, as well as BIFUNCTIONAL NUCLEASE 1 (BFN1), the closest homolog of the Zinnia endonuclease ZEN1 known to degrade nuclei in tracheary elements41 (Table 1). The exclusive enrichment of “programmed cell death” as a biological function term in the early induction (7 hpt) of EgrNAC26 (Fig. 3b) strengthens this observation. A ribonuclease with a currently unknown role in xylem PCD, RNS3, was rapidly induced following transfection by EgrNAC26 and EgrNAC61 (Table 1), while expression of suspected but non-validated xylem PCD genes XSP1 and SBT1.1 were not affected in this experiment.
We also observed what appears to be a much weaker and possibly delayed response in SCW target activation by EgrNAC26 compared to EgrNAC61. Interestingly, the expression of EgrNAC26 in protoplasts was substantially lower than that of EgrNAC61, despite the use of the same constitutive promoter and the fact that the transfection efficiency for EgrNAC26 was somewhat higher (Figs S3b, S5a). We cannot currently explain this phenomenon. However, several promoter transactivation experiments in Populus and Arabidopsis showed that NST clade SWNs activate mid-level regulators PtrMYB021 (i.e. MYB46), PtrMYB020 (MYB83), PtrMYB128 (MYB103), PtrNAC156 (SND2), PtrKNAT7 (KNAT7) and PtrMYB28 (MYB58/MYB63) more strongly than VND clade SWNs13,28,30,55, a trend that also emerged in our time-course experiment where SCW structural genes were preferentially activated by EgrNAC61 much earlier and to a stronger level than for EgrNAC26 (Figs 4, 5). Since fibre cells exhibit thicker SCWs than vessels, stronger activation of SCW structural genes in fibres is developmentally plausible, and transcript levels of SCW cellulose, xylan and lignin genes appear higher in laser micro-dissected fibres compared to vessels in Populus in support of this81. An enrichment for primary metabolic processes in EgrNAC61-OX (Fig. 3b) may reflect a metabolic shift toward carbon investment in this carbon sink which is in agreement with previous observations of several amino acid pathways being upregulated during in vitro tracheary element transdifferentiation82. Fibre-specific mutant phenotypes for EgrNAC61-preferential targets such as SND2 and SND313,83,84 suggest that EgrNAC61 is an important fibre SCW regulator, but the clear reporter gene expression conferred by its promoter in Arabidopsis vessels and all cell types in the developing xylem in poplar plants (Fig. 2) suggests a dual role in SCW formation in fibres and vessels, and possible other xylem cells.
Given the combined evidence of vessel-specific specialization of EgrNAC26 and preferential PCD activation, versus vessel- and fibre-preferential expression of EgrNAC61 and strong activation of SCW structural genes and TFs following induction in Arabidopsis protoplasts, we favour the hypothesis that EgrNAC61 is the chief fibre SCW master regulator that plays a supporting role in vessel development, while EgrNAC26 complements EgrNAC61 in vessels with PCD and SCW pit-related regulatory functions. The potential role of EgrNAC61 in regulating cell wall deposition in cork cells is an interesting topic for further investigation.
Methods
Construct preparation
Promoter fragments of 2,009 bp (EgrNAC61 promoter; Eucgr. E01053), 3,660 bp (EgrNAC26 promoter; Eucgr. A02887) and 1,115 bp (VND6 promoter; AT5G62380) directly upstream of the translational start site were PCR-amplified from genomic DNA (Table S1), cloned into pCR8/GW/TOPO® (EgrNAC61) or pENTR/D-TOPO® (EgrNAC26 and VND6) entry vectors (Invitrogen, Carlsbad, CA) and transferred to pBGWFS785 using the Gateway LR ClonaseII Enzyme Mix (Invitrogen). For Populus GUS constructs, the promoters were transferred to pMDC16286. Developing secondary xylem RT-PCR products of EgrNAC26 and EgrNAC61 coding regions were cloned into pCR8/GW/TOPO® and transferred to the pUC-35S-Rfa-35S-GFP protoplast expression vector71 and pDEST-GBKT7 yeast expression vector87 similarly using LR recombination. Empty vector controls of the expression vectors were produced through LR recombination with a self-ligated pCR8/GW/TOPO® vector. Large-scale plasmid isolations for protoplast transfections were performed using the PureLink HiPure Plasmid FP Maxiprep Kit (Invitrogen), or caesium chloride purification88. The pHBT::sGFP(S65T)-NOS vector89 was used as a fluorescent marker to assess the effect of different plasmid purification methods on protoplast transfection efficiency.
Plant transformation and GUS analysis
The pBGWFS7 constructs (VND6pro::GFP:GUS, EgrNAC26pro::GFP:GUS and EgrNAC61pro::GFP:GUS) were transformed into Arabidopsis thaliana using the Agrobacterium-mediated floral dip method90 and the transformed lines were selected on half-strength MS agar medium using Glufosinate-ammonium (20 mg/l). Transgenic Arabidopsis seeds were surface-sterilized, plated on nonselective nitrate-enriched media and allowed to germinate for 2–3 weeks. For poplar transformation, the EgrNAC61pro::GUS construct in the pMDC162 backbone was introduced into poplar hybrid Populus alba × P. tremula 717-1B4 by co-cultivation of Agrobacterium solution with leaf discs followed by shoot regeneration on selective media containing 30 mg/l Hygromycin. Transgenic poplar plants were propagated using greenwood stem cuttings and young developing stems (internodes 1 and 5) were harvested. GUS staining was performed as described by Spokevicius et al.91, excluding acetone treatment, but adding 0.5 mM potassium ferrocyanide and 0.5 mM potassium ferricyanide. After incubation at 37 °C overnight, seedlings were destained in 70% ethanol and visualized with differential interference contrast and brightfield microscopy.
Yeast transactivation assays
The pDEST-GBKT7 constructs containing EgrNAC61 or EgrNAC26 were used to transform Saccharomyces cerevisiae strains AH109 and Y2HGold (Clontech™, CA, USA) through lithium acetate/polyethylene glycol-mediated transformation as previously described92. Transformed colonies were screened on media lacking tryptophan (Trp-), media lacking tryptophan and histidine (Trp- His-), as well as Trp- His- media supplemented with 1 mM or 15 mM 3-amino-1,2,4-triazole (3AT, Sigma-Aldrich®, South Africa). Trp- His- media was prepared using -Trp-Leu-His drop-out supplement (Clontech™) and adding the appropriate amount of leucine supplement (Sigma-Aldrich®). Colonies were incubated for 3 days at 30 °C.
Isolation and transfection of Arabidopsis leaf mesophyll protoplasts
A. thaliana ecotype Columbia (Col-0) seedlings were grown for four weeks at 22 °C under a 10 h photoperiod prior to mesophyll protoplast isolation from leaves as previously described93. Multiple plants were pooled for protoplast isolations and de-bulked to form three independent biological replicates. Large-scale transfections with pUC-35S-EgrNAC26-35S-GFP, pUC-35S-EgrNAC61-35S-GFP, and pUC-35S-NOS-35S-GFP (empty vector) constructs were conducted using 4 × 105 protoplasts for each of three technical replicates per biological replicate (n = 3) to account for transfection variation and incubated for 7 h and 14 h at room temperature in the dark. Technical replicates were bulked and harvested through centrifugation at 500 × g. Of the harvested cells, 10 μl was used to determine transfection efficiency (at 14 hpt only) and the remaining cells were flash-frozen and stored at −80 °C.
RNA-seq analysis
Total RNA was isolated using the Qiagen RNeasy Mini Kit (Sigma-Aldrich®, South Africa) and RNA quality was determined using an Experion instrument (Bio-Rad). Stranded 100 nt paired-end (PE100) mRNA sequencing was performed at Beijing Genomics Institute (BGI, Hong Kong) on the Illumina HiSeq4000 platform using fragment sizes of up to 200 bp. Raw RNA-seq reads passing initial quality filtering by BGI were further assessed for quality with FastQC94. The RNA-seq reads were formatted with FastQGroomer95 prior to mapping to the A. thaliana genome (TAIR10) using TopHat 296. The mapped RNA-seq reads were assigned to gene models and the fragments per kilobase of transcript per million reads mapped (FPKM) values for each mapped transcript was calculated in Cufflinks97 using the A. thaliana TAIR10 annotation. The list of genes generated by Cufflinks was compiled into a single list with all nine libraries and Principle Component Analysis (PCA) was applied in R98 using the prcomp function99. Differentially expressed genes (DEG) were identified using the mapped RNA-seq reads in Cuffdiff97, with FPKM ≥1, false discovery rate (FDR) >0.05 and fold change ≥2 or ≤0.5.
The BiNGO plugin in Cytoscape100,101 was used to determine overrepresented Gene Ontology (GO) terms in the up- and down-regulated EgrNAC26 and EgrNAC61 DEG datasets. A hypergeometric test with Benjamini and Hochberg FDR correction with a P-value cut-off of 0.05 was applied with the whole A. thaliana annotation as a reference set.
Data availability
The RNA-seq data reported in this study is freely available at the NCBI Sequence Read Archive (https://www.ncbi.nlm.nih.gov/sra), accession SRP117192.
Electronic supplementary material
Acknowledgements
We thank Alan Hall and Eudri Venter (UP) for confocal microscopy assistance, Vincent Chiang (University of North Carolina State University) for providing the pUC-35S-Rfa-35S-GFP vector, Mamadile Kgaphu (UP) for assistance with GUS transgenic plants, Adri Veale (UP) for assistance with the transgenic poplar trial, Karen van der Merwe (UP) for bioinformatics input and Jonathan Botha (UP) for cloning the EgrNAC26 and EgrNAC61 coding sequences. This work was supported by a strategic (Eucalyptus Genomics Platform) grant from the Department of Science and Technology (DST) and the National Research Foundation of South Africa (NRF Bioinformatics and Functional Genomics Programme grant UID 97911). Seed funding was provided by the Genomics Research Institute (University of Pretoria). Finally, the work was supported by Sappi and Mondi through the Forest Molecular Genetics (FMG) Programme and matching funding from the Technology and Human Resources for Industry Programme (THRIP grant UID 96413).
Author Contributions
M.L. prepared the constructs, performed the protoplast RNA-seq analyses and yeast transactivation experiments, and co-wrote the manuscript with S.H. K.B., L.T. and S.H. conducted promoter::GUS analyses and drafted these sections of the manuscript. E.M. and A.M. edited the manuscript, co-supervised the study with S.H. and assisted with funding.
Competing Interests
The authors declare no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-29278-w.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Cano-Delgado A, Lee JY, Demura T. Regulatory mechanisms for specification and patterning of plant vascular tissues. Annual Review of Cell Developmental Biology. 2010;26:605–637. doi: 10.1146/annurev-cellbio-100109-104107. [DOI] [PubMed] [Google Scholar]
- 2.Demura T, Ye ZH. Regulation of plant biomass production. Current opinion in plant biology. 2010;13:299–304. doi: 10.1016/j.pbi.2010.03.002. [DOI] [PubMed] [Google Scholar]
- 3.Plomion C, Leprovost G, Stokes A. Wood Formation in Trees. Plant Physiology. 2001;127:1513–1523. doi: 10.1104/pp.010816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rockwood DL, Rudie AW, Ralph SA, Zhu JY, Winandy JE. Energy product options for Eucalyptus species grown as short rotation woody crops. International journal of molecular sciences. 2008;9:1361–1378. doi: 10.3390/ijms9081361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pimrote K, Tian Y, Lu X. Transcriptional regulatory network controlling secondary cell wall biosynthesis and biomass production in vascular plants. African Journal of Biotechnology. 2012;11:13928–13937. doi: 10.5897/AJB12.1928. [DOI] [Google Scholar]
- 6.Schuetz M, Smith R, Ellis B. Xylem tissue specification, patterning, and differentiation mechanisms. Journal of Experimental Botany. 2013;64:11–31. doi: 10.1093/jxb/ers287. [DOI] [PubMed] [Google Scholar]
- 7.Ko JH, Jeon HW, Kim WC, Kim JY, Han KH. The MYB46/MYB83-mediated transcriptional regulatory programme is a gatekeeper of secondary wall biosynthesis. Ann Bot. 2014;114:1099–1107. doi: 10.1093/aob/mcu126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ye ZH, Zhong R. Molecular control of wood formation in trees. J Exp Bot. 2015;66:4119–4131. doi: 10.1093/jxb/erv081. [DOI] [PubMed] [Google Scholar]
- 9.Zhong R, Lee C, Ye Z-H. Global analysis of direct targets of secondary wall NAC master switches in Arabidopsis. Molecular Plant. 2010;3:1087–1103. doi: 10.1093/mp/ssq062. [DOI] [PubMed] [Google Scholar]
- 10.Ko J-H, Kim W-C, Han K-H. Ectopic expression of MYB46 identifies transcriptional regulatory genes involved in secondary wall biosynthesis in Arabidopsis. The Plant Journal. 2009;60:649–665. doi: 10.1111/j.1365-313X.2009.03989.x. [DOI] [PubMed] [Google Scholar]
- 11.McCarthy RL, Zhong R, Ye Z-H. MYB83 is a direct target of SND1 and acts redundantly with MYB46 in the regulation of secondary cell wall biosynthesis in Arabidopsis. Plant and Cell Physiology. 2009;50:1950–1964. doi: 10.1093/pcp/pcp139. [DOI] [PubMed] [Google Scholar]
- 12.Kim W-C, et al. MYB46 directly regulates the gene expression of secondary wall-associated cellulose synthases in Arabidopsis. The Plant Journal. 2012;73:26–36. doi: 10.1111/j.1365-313x.2012.05124.x. [DOI] [PubMed] [Google Scholar]
- 13.Zhong R, Lee C, Zhou J, McCarthy RL, Ye Z-H. A battery of transcription factors involved in the regulation of secondary cell wall biosynthesis in Arabidopsis. The Plant Cell. 2008;20:2763–2782. doi: 10.1105/tpc.108.061325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhong R, Richardson EA, Y Z-H. The MYB46 transcription factor is a direct target of SND1 and regulates secondary wall biosynthesis in Arabidopsis. The Plant Cell. 2007;19:2776–2792. doi: 10.1105/tpc.107.053678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou J, Zhong R, Ye ZH. Arabidopsis NAC domain proteins, VND1 to VND5, are transcriptional regulators of secondary wall biosynthesis in vessels. PLoS One. 2014;9:e105726. doi: 10.1371/journal.pone.0105726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mitsuda N, et al. NAC transcription factors, NST1 and NST3, are key regulators of the formation of secondary walls in woody tissues of Arabidopsis. The Plant Cell. 2007;19:270–280. doi: 10.1105/tpc.106.047043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mitsuda N, Ohme-Takagi M. NAC transcription factors NST1 and NST3 regulate pod shattering in a partially redundant manner by promoting secondary wall formation after the establishment of tissue identity. The Plant Journal. 2008;56:768–778. doi: 10.1111/j.1365-313X.2008.03633.x. [DOI] [PubMed] [Google Scholar]
- 18.Mitsuda N, Seki M, Shinozaki K, Ohme-Takagi M. The NAC transcription factors NST1 and NST2 of Arabidopsis regulate secondary wall thickenings and are required for anther dehiscence. Plant Cell. 2005;17:2993–3006. doi: 10.1105/tpc.105.036004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhong R, Ye ZH. The Arabidopsis NAC transcription factor NST2 functions together with SND1 and NST1 to regulate secondary wall biosynthesis in fibers of inflorescence stems. Plant Signal Behav. 2015;10:e989746. doi: 10.4161/15592324.2014.989746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhong R, Demura T, Ye ZH. SND1, a NAC domain transcription factor, is a key regulator of secondary wall synthesis in fibers of Arabidopsis. Plant Cell. 2006;18:3158–3170. doi: 10.1105/tpc.106.047399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhong R, Richardson EA, Ye Z-H. Two NAC domain transcription factors, SND1 and NST1, function redundantly in regulation of secondary wall synthesis in fibers of Arabidopsis. Planta. 2007;225:1603–1611. doi: 10.1007/s00425-007-0498-y. [DOI] [PubMed] [Google Scholar]
- 22.Kubo M, et al. Transcription switches for protoxylem and metaxylem vessel formation. Genes and Development. 2005;19:1855–1860. doi: 10.1101/gad.1331305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yamaguchi M, Kubo M, Fukuda H, Demura T. VASCULAR-RELATED NAC-DOMAIN7 is involved in the differentiation of all types of xylem vessels in Arabidopsis roots and shoots. The Plant Journal. 2008;55:652–664. doi: 10.1111/j.1365-313X.2008.03533.x. [DOI] [PubMed] [Google Scholar]
- 24.Yamaguchi M, et al. VASCULAR-RELATED NAC-DOMAIN7 directly regulates the expression of a broad range of genes for xylem vessel formation. The Plant Journal. 2011;66:579–590. doi: 10.1111/j.1365-313X.2011.04514.x. [DOI] [PubMed] [Google Scholar]
- 25.Hepler PK, Newcomb EH. Microtubules and fibrils in the cytoplasm of Coleus cells undergoing secondary wall deposition. J Cell Biol. 1964;20:529–533. doi: 10.1083/jcb.20.3.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Demura T, Fukuda H. Transcriptional regulation in wood formation. Trends in Plant Science. 2007;12:64–70. doi: 10.1016/j.tplants.2006.12.006. [DOI] [PubMed] [Google Scholar]
- 27.Zhao Y, Sun J, Xu P, Zhang R, Li L. Intron-mediated alternative splicing of WOOD-ASSOCIATED NAC TRANSCRIPTION FACTOR1B regulates cell wall thickening during fiber development in Populus species. Plant Physiology. 2014;164:765–776. doi: 10.1104/pp.113.231134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ohtani M, et al. A NAC domain protein family contributing to the regulation of wood formation in poplar. Plant J. 2011;67:499–512. doi: 10.1111/j.1365-313X.2011.04614.x. [DOI] [PubMed] [Google Scholar]
- 29.Sundell D, et al. AspWood: High-spatial-resolution transcriptome profiles reveal uncharacterized modularity of wood formation in Populus tremula. Plant Cell. 2017;29:1585–1604. doi: 10.1105/tpc.17.00153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhong R, McCarthy RL, Lee C, Ye Z-H. Dissection of the transcriptional program regulating secondary wall biosynthesis during wood formation in poplar. Plant Physiology. 2011;157:1452–1468. doi: 10.1104/pp.111.181354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xiao W, Yang Y, Yu J. ZmNST3 and ZmNST4 are master switches for secondary wall deposition in maize (Zea mays L.) Plant Sci. 2018;266:83–94. doi: 10.1016/j.plantsci.2017.03.012. [DOI] [PubMed] [Google Scholar]
- 32.Zhong R, et al. Transcriptional activation of secondary wall biosynthesis by rice and maize NAC and MYB transcription factors. Plant and Cell Physiology. 2011;52:1856–1871. doi: 10.1093/pcp/pcr123. [DOI] [PubMed] [Google Scholar]
- 33.Jokipii-Lukkari S, et al. NorWood: a gene expression resource for evo-devo studies of conifer wood development. New Phytol. 2017;216:482–494. doi: 10.1111/nph.14458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pesquet E, et al. Non-cell-autonomous postmortem lignification of tracheary elements in Zinnia elegans. The Plant cell. 2013;25:1314–1328. doi: 10.1105/tpc.113.110593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smith RA, et al. Neighboring parenchyma cells contribute to Arabidopsis xylem lignification, while lignification of interfascicular fibers is cell autonomous. The Plant cell. 2013;25:3988–3999. doi: 10.1105/tpc.113.117176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ohashi-Ito K, Oda Y, Fukuda H. Arabidopsis VASCULAR-RELATED NAC-DOMAIN6 directly regulates the genes that govern programmed cell death and secondary wall formation during xylem differentiation. Plant Cell. 2010;22:3461–3473. doi: 10.1105/tpc.110.075036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Groover A, Jones AM. Tracheary Element Differentiation Uses a Novel Mechanism Coordinating Programmed Cell Death and Secondary Cell Wall Synthesis. Plant Physiology. 1999;119:375–384. doi: 10.1104/pp.119.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Böllhoner B, Prestele J, Tuominen H. Xylem cell death: emerging understanding of regulation and function. J Exp Bot. 2012;63:1081–1094. doi: 10.1093/jxb/err438. [DOI] [PubMed] [Google Scholar]
- 39.Avci U, Petzold HE, Ismail IO, Beers EP, Haigler CH. Cysteine proteases XCP1 and XCP2 aid micro-autolysis within the intact central vacuole during xylogenesis in Arabidopsis roots. The Plant Journal. 2008;56:303–315. doi: 10.1111/j.1365-313X.2008.03592.x. [DOI] [PubMed] [Google Scholar]
- 40.Petzold HE, Zhao M, Beers EP. Expression and functions of proteases in vascular tissues. Physiologia plantarum. 2012;145:121–129. doi: 10.1111/j.1399-3054.2011.01538.x. [DOI] [PubMed] [Google Scholar]
- 41.Ito J, Fukuda H. ZEN1 Is a Key Enzyme in the Degradation of Nuclear DNA during Programmed Cell Death of Tracheary Elements. The Plant Cell Online. 2002;14:3201–3211. doi: 10.1105/tpc.006411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Funk V, Kositsup B, Zhao C, Beers EP. The Arabidopsis Xylem Peptidase XCP1 Is a Tracheary Element Vacuolar Protein That May Be a Papain Ortholog. Plant Physiology. 2002;128:84–94. doi: 10.1104/pp.010514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Böllhoner B, et al. Post mortem function of AtMC9 in xylem vessel elements. New Phytol. 2013;200:498–510. doi: 10.1111/nph.12387. [DOI] [PubMed] [Google Scholar]
- 44.Zhao C, Johnson BJ, Kositsup B, Beers EP. Exploiting Secondary Growth in Arabidopsis. Construction of Xylem and Bark cDNA Libraries and Cloning of Three Xylem Endopeptidases. Plant Physiology. 2000;123:1185–1196. doi: 10.1104/pp.123.3.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ohashi-Ito K, Fukuda H. Transcriptional regulation of vascular cell fates. Curr Opin Plant Biol. 2010;13:670–676. doi: 10.1016/j.pbi.2010.08.011. [DOI] [PubMed] [Google Scholar]
- 46.Escamez S, Tuominen H. Programmes of cell death and autolysis in tracheary elements: when a suicidal cell arranges its own corpse removal. Journal of Experimental Botany. 2014;65:1313–1321. doi: 10.1093/jxb/eru057. [DOI] [PubMed] [Google Scholar]
- 47.Ruzicka K, Ursache R, Hejatko J, Helariutta Y. Xylem development - from the cradle to the grave. New Phytol. 2015;207:519–535. doi: 10.1111/nph.13383. [DOI] [PubMed] [Google Scholar]
- 48.Spicer R. Symplasmic networks in secondary vascular tissues: parenchyma distribution and activity supporting long-distance transport. J Exp Bot. 2014;65:1829–1848. doi: 10.1093/jxb/ert459. [DOI] [PubMed] [Google Scholar]
- 49.Oda Y, Fukuda H. Initiation of cell wall pattern by a Rho- and microtubule-driven symmetry breaking. Science. 2012;337:1333–1336. doi: 10.1126/science.1222597. [DOI] [PubMed] [Google Scholar]
- 50.Oda Y, Fukuda H. Rho of plant GTPase signaling regulates the behavior of Arabidopsis kinesin-13A to establish secondary cell wall patterns. Plant Cell. 2013;25:4439–4450. doi: 10.1105/tpc.113.117853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sugiyama Y, Wakazaki M, Toyooka K, Fukuda H, Oda Y. A Novel Plasma Membrane-Anchored Protein Regulates Xylem Cell-Wall Deposition through Microtubule-Dependent Lateral Inhibition of Rho GTPase Domains. Current Biology. 2017;27:2522–2528.e2524. doi: 10.1016/j.cub.2017.06.059. [DOI] [PubMed] [Google Scholar]
- 52.Hussey SG, Mizrachi E, Creux NM, Myburg AA. Navigating the transcriptional roadmap regulating plant secondary cell wall deposition. Front Plant Sci. 2013;4:325. doi: 10.3389/fpls.2013.00325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang L, et al. Identification and characterization of a wood-associated NAC domain transcription factor PtoVNS11 from Populus tomentosa Carr. Trees. 2015;29:1091–1101. doi: 10.1007/s00468-015-1188-1. [DOI] [Google Scholar]
- 54.Li Q, et al. Splice variant of the SND1 transcription factor is a dominant negative of SND1 members and their regulation in Populus trichocarpa. Proc Natl Acad Sci USA. 2012;109:14699–14704. doi: 10.1073/pnas.1212977109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhong R, Ye ZH. The poplar PtrWNDs are transcriptional activators of secondary cell wall biosynthesis. Plant Signal Behav. 2010;5:469–472. doi: 10.4161/psb.5.4.11400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Creux NM, Bossinger G, Myburg AA, Spokevicius AV. Induced somatic sector analysis of cellulose synthase (CesA) promoter regions in woody stem tissues. Planta. 2013;237:799–812. doi: 10.1007/s00425-012-1792-x. [DOI] [PubMed] [Google Scholar]
- 57.Goicoechea M, et al. EgMYB2, a new transcriptional activator from Eucalyptus xylem, regulates secondary cell wall formation and lignin biosynthesis. The Plant Journal. 2005;43:553–567. doi: 10.1111/j.1365-313X.2005.02480.x. [DOI] [PubMed] [Google Scholar]
- 58.Legay S, et al. Molecular characterization of EgMYB1, a putative transcriptional repressor of the lignin biosynthetic pathway. Plant Science. 2007;173:542–549. doi: 10.1016/j.plantsci.2007.08.007. [DOI] [Google Scholar]
- 59.Legay S, et al. EgMYB1, an R2R3 MYB transcription factor from eucalyptus negatively regulates secondary cell wall formation in Arabidopsis and poplar. New Phytologist. 2010;188:774–786. doi: 10.1111/j.1469-8137.2010.03432.x. [DOI] [PubMed] [Google Scholar]
- 60.Creux NM, Ranik M, Berger DK, Myburg AA. Comparative analysis of orthologous cellulose synthase promoters from Arabidopsis, Populus and Eucalyptus: evidence of conserved regulatory elements in angiosperms. New Phytologist. 2008;179:722–737. doi: 10.1111/j.1469-8137.2008.02517.x. [DOI] [PubMed] [Google Scholar]
- 61.Soler M, et al. The Eucalyptus linker histone variant EgH1.3 cooperates with the transcription factor EgMYB1 to control lignin biosynthesis during wood formation. New Phytol. 2017;213:287–299. doi: 10.1111/nph.14129. [DOI] [PubMed] [Google Scholar]
- 62.Hussey SG, Saidi MN, Hefer CA, Myburg AA, Grima-Pettenati J. Structural, evolutionary and functional analysis of the NAC domain protein family in Eucalyptus. New Phytol. 2015;206:1337–1350. doi: 10.1111/nph.13139. [DOI] [PubMed] [Google Scholar]
- 63.Zuo J, Niu Q-W, Chua N-H. An estrogen receptor-based transactivator XVE mediates highly inducible gene expression in transgenic plants. The Plant Journal. 2000;24:265–273. doi: 10.1046/j.1365-313x.2000.00868.x. [DOI] [PubMed] [Google Scholar]
- 64.Hefer C, Mizrachi E, Joubert F, Myburg A. The Eucalyptus genome integrative explorer (EucGenIE): a resource for Eucalyptus genomics and transcriptomics. BMC Proceedings. 2011;5:O49. doi: 10.1186/1753-6561-5-S7-O49. [DOI] [Google Scholar]
- 65.Vining KJ, et al. The floral transcriptome of Eucalyptus grandis. New Phytologist. 2014;206:1406–1422. doi: 10.1111/nph.13077. [DOI] [PubMed] [Google Scholar]
- 66.Öhman D, et al. MYB103 is required for FERULATE-5-HYDROXYLASE expression and syringyl lignin biosynthesis in Arabidopsis stems. Plant J. 2013;73:63–76. doi: 10.1111/tpj.12018. [DOI] [PubMed] [Google Scholar]
- 67.Zhou J, Lee C, Zhong R, Ye Z-H. MYB58 and MYB63 are transcriptional activators of the lignin biosynthetic pathway during secondary cell wall formation in Arabidopsis. The Plant Cell. 2009;21:248–266. doi: 10.1105/tpc.108.063321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang H, Zhao Q, Chen F, Wang M, Dixon RA. NAC domain function and transcriptional control of a secondary cell wall master switch. The Plant Journal. 2011;68:1104–1114. doi: 10.1111/j.1365-313X.2011.04764.x. [DOI] [PubMed] [Google Scholar]
- 69.Myburg AA, et al. The genome of Eucalyptus grandis. Nature. 2014;510:356–362. doi: 10.1038/nature13308. [DOI] [PubMed] [Google Scholar]
- 70.Ko J-H, Yang SH, Park AH, Lerouxel O, Han K-H. ANAC012, a member of the plant-specific NAC transcription factor family, negatively regulates xylary fiber development in Arabidopsis thaliana. The Plant Journal. 2007;50:1035–1048. doi: 10.1111/j.1365-313X.2007.03109.x. [DOI] [PubMed] [Google Scholar]
- 71.Lin Y-C, et al. SND1 transcription factor–directed quantitative functional hierarchical genetic regulatory network in wood formation in Populus trichocarpa. The Plant Cell. 2013;25:4324–4341. doi: 10.1105/tpc.113.117697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Valdivia ER, et al. Regulation of secondary wall synthesis and cell death by NAC transcription factors in the monocot Brachypodium distachyon. Journal of Experimental Botany. 2013;64:1333–1343. doi: 10.1093/jxb/ers394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yamaguchi M, et al. VASCULAR-RELATED NAC-DOMAIN6 (VND6) and VND7 effectively induce transdifferentiation into xylem vessel elements under control of an induction system. Plant Physiology. 2010;153:906–914. doi: 10.1104/pp.110.154013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Endo H, et al. Multiple classes of transcription factors regulate the expression of VASCULAR-RELATED NAC-DOMAIN7, a master switch of xylem vessel differentiation. Plant Cell Physiol. 2015;56:242–254. doi: 10.1093/pcp/pcu134. [DOI] [PubMed] [Google Scholar]
- 75.Marques, A. V., Pereira, H., Meier, D. & Faix, O. Structural Characterization of Cork Lignin by Thioacidolysis and Permanganate Oxidation. Holzforschung53, 10.1515/hf.1999.028 (1999).
- 76.Jones, R., Ougham, H., Thomas, H. & Waaland, S. The Molecular Life of Plants. (John Wiley & Sons, Ltd, 2013).
- 77.Soyano T, Thitamadee S, Machida Y, Chua N-H. Asymmetric Leaves2-Like19/Lateral Organ Boundaries Domain30 and ASL20/LBD18 regulate tracheary element differentiation in Arabidopsis. The Plant Cell. 2008;20:3359–3373. doi: 10.1105/tpc.108.061796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Endo S, et al. Identifying New Components Participating in the Secondary Cell Wall Formation of Vessel Elements in Zinnia and Arabidopsis. The Plant Cell. 2009;21:1155–1165. doi: 10.1105/tpc.108.059154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Oda Y, Fukuda H. Emerging roles of small GTPases in secondary cell wall development. Front Plant Sci. 2014;5:428. doi: 10.3389/fpls.2014.00428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Pesquet E, Korolev AV, Calder G, Lloyd CW. The microtubule-associated protein AtMAP70-5 regulates secondary wall patterning in Arabidopsis wood cells. Curr Biol. 2010;20:744–749. doi: 10.1016/j.cub.2010.02.057. [DOI] [PubMed] [Google Scholar]
- 81.Shi R, et al. Tissue and cell-type co-expression networks of transcription factors and wood component genes in Populus trichocarpa. Planta. 2017;245:927–938. doi: 10.1007/s00425-016-2640-1. [DOI] [PubMed] [Google Scholar]
- 82.Li Z, et al. A Transcriptional and Metabolic Framework for Secondary Wall Formation in Arabidopsis. Plant Physiol. 2016;172:1334–1351. doi: 10.1104/pp.16.01100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hussey SG, et al. SND2, a NAC transcription factor gene, regulates genes involved in secondary cell wall development in Arabidopsis fibres and increases fibre cell area in Eucalyptus. BMC Plant Biology. 2011;11:173. doi: 10.1186/1471-2229-11-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wang HH, et al. Chimeric repressor of PtSND2 severely affects wood formation in transgenic Populus. Tree Physiology. 2013;33:878–886. doi: 10.1093/treephys/tpt058. [DOI] [PubMed] [Google Scholar]
- 85.Karimi M, Inzé D, Depicker A. GATEWAY™ vectors for Agrobacterium-mediated plant transformation. Trends in Plant Science. 2002;7:193–195. doi: 10.1016/S1360-1385(02)02251-3. [DOI] [PubMed] [Google Scholar]
- 86.Curtis MD, Grossniklaus U. A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiology. 2003;133:462–469. doi: 10.1104/pp.103.027979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Rossignol P, Collier S, Bush M, Shaw P, Doonan JH. Arabidopsis POT1A interacts with TERT-V(I8), an N-terminal splicing variant of telomerase. J Cell Sci. 2007;120:3678–3687. doi: 10.1242/jcs.004119. [DOI] [PubMed] [Google Scholar]
- 88.Lin Y-C, et al. A simple improved-throughput xylem protoplast system for studying wood formation. Nature Protocols. 2014;9:2194–2205. doi: 10.1038/nprot.2014.147. [DOI] [PubMed] [Google Scholar]
- 89.Chiu W-L, et al. Engineered GFP as a vital reporter in plants. Current Biology. 1996;6:325–330. doi: 10.1016/S0960-9822(02)00483-9. [DOI] [PubMed] [Google Scholar]
- 90.Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. The Plant Journal. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 91.Spokevicius AV, Van Beveren K, Leitch MA, Bossinger G. Agrobacterium-mediated in vitro transformation of wood-producing stem segments in eucalypts. Plant Cell Rep. 2005;23:617–624. doi: 10.1007/s00299-004-0856-1. [DOI] [PubMed] [Google Scholar]
- 92.Maritz-Olivier, C. The mechanisms regulating exocytosis of the salivary glands of the soft tick, Ornithodoros savignyl PhD thesis. (University of Pretoria, 2005).
- 93.Yoo SD, Cho YH, Sheen J. Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nature protocols. 2007;2:1565–1572. doi: 10.1038/nprot.2007.199. [DOI] [PubMed] [Google Scholar]
- 94.Andrews, S. FastQC: A quality control tool for high throughput sequence data. http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (2010).
- 95.Blankenberg D, et al. Manipulation of FASTQ data with Galaxy. Bioinformatics. 2010;26:1783–1785. doi: 10.1093/bioinformatics/btq281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kim D, et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biology. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Trapnell C, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nature biotechnology. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.R Core Team. R: A language and environment for statistical computing. (R Foundation for Statistical Computing, 2016).
- 99.Venables, W. N. & Ripley, B. D. Modern Applied Statistics with S. Springer-Verlag (2002).
- 100.Maere S, Heymans K, Kuiper M. BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics. 2005;21:3448–3449. doi: 10.1093/bioinformatics/bti551. [DOI] [PubMed] [Google Scholar]
- 101.Smoot ME, Ono K, Ruscheinski J, Wang PL, Ideker T. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics. 2011;27:431–432. doi: 10.1093/bioinformatics/btq675. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-seq data reported in this study is freely available at the NCBI Sequence Read Archive (https://www.ncbi.nlm.nih.gov/sra), accession SRP117192.