Abstract
Background
Pulmonary arterial hypertension (PAH) is a rare disease characterized by distinctive changes in pulmonary arterioles that lead to progressive pulmonary arterial pressures, right-sided heart failure, and a high mortality rate. Up to 30% of adult and 75% of pediatric PAH cases are associated with congenital heart disease (PAH-CHD), and the underlying etiology is largely unknown. There are no known major risk genes for PAH-CHD.
Methods
To identify novel genetic causes of PAH-CHD, we performed whole exome sequencing in 256 PAH-CHD patients. We performed a case-control gene-based association test of rare deleterious variants using 7509 gnomAD whole genome sequencing population controls. We then screened a separate cohort of 413 idiopathic and familial PAH patients without CHD for rare deleterious variants in the top association gene.
Results
We identified SOX17 as a novel candidate risk gene (p = 5.5e−7). SOX17 is highly constrained and encodes a transcription factor involved in Wnt/β-catenin and Notch signaling during development. We estimate that rare deleterious variants contribute to approximately 3.2% of PAH-CHD cases. The coding variants identified include likely gene-disrupting (LGD) and deleterious missense, with most of the missense variants occurring in a highly conserved HMG-box protein domain. We further observed an enrichment of rare deleterious variants in putative targets of SOX17, many of which are highly expressed in developing heart and pulmonary vasculature. In the cohort of PAH without CHD, rare deleterious variants of SOX17 were observed in 0.7% of cases.
Conclusions
These data strongly implicate SOX17 as a new risk gene contributing to PAH-CHD as well as idiopathic/familial PAH. Replication in other PAH cohorts and further characterization of the clinical phenotype will be important to confirm the precise role of SOX17 and better estimate the contribution of genes regulated by SOX17.
Electronic supplementary material
The online version of this article (10.1186/s13073-018-0566-x) contains supplementary material, which is available to authorized users.
Keywords: Pulmonary hypertension, Congenital heart disease, Exome sequencing, Genetic association study
Background
Pulmonary arterial hypertension (PAH[MIM:178600]) is a rare disease characterized by distinctive changes in pulmonary arterioles that lead to progressive pulmonary arterial pressures, right-sided heart failure and a high mortality rate. Up to 30% of adult- [1, 2] and 75% of pediatric-onset PAH cases [3] are associated with congenital heart disease (PAH-CHD), and due to improved treatments, the number of adults with PAH-CHD is rising [1, 4]. Congenital heart defects can result in left-to-right (systemic-to-pulmonary) shunts leading to increased pulmonary blood flow and risk of PAH. However, not all patients are exposed to prolonged periods of increased pulmonary flow. PAH may persist following surgical repair of cardiac defects or recur many years after repair. Thus, the underlying etiology is heterogeneous and may include increased pulmonary blood flow, pulmonary vasculature abnormalities, or a combination. In addition to environmental factors, genetic factors likely play an important role in PAH-CHD although no major risk gene has been identified to date [1].
Genetic studies of PAH alone have identified 11 known risk genes for PAH [5–8]. Several of the risk genes encode members of the transforming growth factor beta/bone morphogenetic protein (TGF-β/BMP) signaling pathway, important in both vasculogenesis and embryonic heart development. For example, mutations in bone morphogenetic protein receptor type 2 (BMPR2) are found in approximately 70% of familial and 10–40% of idiopathic PAH cases. Estimates of the frequency of BMPR2 mutations in PAH-CHD are considerably lower than for PAH alone [9–11]. Mutations in other TGFβ family member genes—activin A, receptor type II-like 1 (ACVRL1), endoglin (ENG), BMP receptor type 1A (BMPR1A) and type 1B (BMPR1B)—as well as caveolin-1 (CAV1), eukaryotic initiation translation factor 2 alpha kinase 4 (EIF2AK4), potassium two-pore-domain channel subfamily K member 3 (KCNK3), SMAD family members 4 and 9 (SMAD4 and SMAD9), and T-box4 (TBX4) have all been identified as less frequent or rare causes of PAH [5–8]. The genetics of CHD are complex and no single major risk gene accounts for more than 1% of cases [12, 13]. Aneuploidies and copy number variations underlie up to 23% of CHD cases [14, 15]. Rare, inherited, and de novo variants in hundreds of genes encoding transcription factors, chromatin regulators, signal transduction proteins, and cardiac structural proteins have been implicated in ~ 10% of CHD cases [12, 16–19].
To identify novel genetic causes of PAH-CHD, we performed exome sequencing in a patient cohort of PAH-CHD. Association analysis using population controls identified SOX17, a member of the SRY-related HMG-box family of transcription factors, as a new candidate risk gene.
Methods
An overview of the experimental design and workflow is provided in Additional file 1: Figure S1.
Patients
PAH-CHD patients were recruited from the pulmonary hypertension centers at Columbia University and Children’s Hospital of Colorado (via enrollment in the PAH Biobank at Cincinnati Children's Hospital Medical Center). Patients were diagnosed according to the World Health Organization (WHO) pulmonary hypertension group I classification [20]. The diagnosis of PAH-CHD was confirmed by medical record review including right heart catheterization and echocardiogram to define the cardiac anatomy. The cohort included 15 familial cases, 160 singletons with no family history of PAH, 61 trios (proband and two unaffected biological parents), and 20 duos (proband and one unaffected parent). Written informed consent (and assent when appropriate) was obtained under a protocol approved by the institutional review board at Columbia University Medical Center or Children’s Hospital of Colorado.
Whole exome sequencing (WES)
Familial cases were screened for BMPR2 and ACVRL1 mutations by Sanger sequencing and multiplex ligation-dependent probe amplification (MLPA). Familial cases without mutations in the two risk genes and all other samples were exome sequenced. DNA was extracted from peripheral blood leukocytes using Puregene reagents (Gentra Systems Inc., Minnesota, USA). Exome sequencing was performed in collaboration with the Regeneron Genetics Center (RGC) or at the Children’s Hospital of Cincinnati. In brief, genomic DNA processed at the RGC was prepared with a customized reagent kit from Kapa Biosystems and captured using the SeqCap VCRome 2 exome capture reagent or xGen lockdown probes. Patient DNA samples sequenced at the PAH Biobank/Cincinnati Children’s Hospital Medical Center were prepared with the Clontech Advantage II kit and enriched using the SeqCap EZ exome V2 capture reagent. All samples were sequenced on the Illumina HiSeq 2500 platform, generating 76-bp paired-end reads. Read-depth coverage was ≥ 15× in ≥ 95% of targeted regions for all exome sequencing samples.
WES data analysis
The workflow is outlined in Additional file 1: Figure S1A. We used a previously established bioinformatics procedure [18] to process and analyze exome sequence data. Specifically, we used BWA-MEM (Burrows-Wheeler Aligner) [21] to map and align paired-end reads to the human reference genome (version GRCh37/hg19), Picard MarkDuplicates to identify and flag PCR duplicate reads, GATK HaplotypeCaller (version 3) [22, 23] to call genetic variants, and GATK variant quality score recalibration (VQSR) to estimate accuracy of variant calls. We used heuristic filters to minimize potential technical artifacts, excluding variants that met any of the following conditions: missingness > 10%, minimum read depth ≤ 8 reads, allele balance ≤ 20% [24], genotype quality < 30, mappability < 1 (based on 150 bp fragments), or GATK VQSR < 99.6. Only variants with FILTER “PASS” in gnomAD WGS and restricted to the captured protein coding region were kept.
We used ANNOVAR [25] to annotate the variants and aggregate information about allele frequencies (AF) and in silico predictions of deleteriousness. We used population AF from public databases: Exome Aggregation Consortium (ExAC) [26] and Genome Aggregation Database (gnomAD). Rare variants were defined by AF < 0.01% in both ExAC and gnomAD WES datasets. We employed multiple in silico prediction algorithms including PolyPhen 2, metaSVM [27], Combined Annotation Dependent Depletion (CADD) [28], and REVEL (rare exome variant ensemble learner) [29]. We noted that REVEL outperformed other ensemble methods in pathogenicity prediction in a recent comparison using clinical genetic data [30]. We performed further evaluation of the prediction toolkits using de novo missense variants published in a recent CHD study [19] and published de novo variants of unaffected siblings of Simons Simplex Collection [31] as controls. We observed that REVEL-predicted damaging missense de novo variants reached the highest enrichment rate in cases compared to controls (Additional file 1: Figure S1B). Thus, we ultimately used REVEL to define damaging missense variants (D-mis, REVEL > 0.5) in this study.
We identified de novo variants in a set of 60 PAH-CHD trios using methods described previously [18, 32], and manually inspected all candidate de novo variants using the Integrative Genomics Viewer (IGV) [33] to exclude potential false positives.
Identification of rare, deleterious variants in established risk genes
We screened for variants in 11 known risk genes for PAH [5–8]: ACVRL1, BMPR1A, BMPR1B, BMPR2, CAV1, EIF2AK4, ENG, KCNK3, SMAD4, SMAD9, and TBX4. We also screened for variants in the recently curated list of 253 candidate risk genes for CHD [19]. Variants identified in the PAH-CHD cohort were compared to mutations reported in the literature and in genetic databases (Online Mendelian Inheritance in Man database, Human Genome Mutation Database [34] and ClinVar [35]). We defined deleterious variants as likely gene-disrupting (LGD) (including premature stopgain, frameshift indels, canonical splicing variants, and deletion of exons) or damaging missense with REVEL score > 0.5 (D-mis). Insertion/deletion variants in known risk genes were confirmed with Sanger sequencing and tested for disease segregation when family DNA samples were available.
Statistical analysis
To identify novel candidate risk genes, we performed a case-control association test comparing frequency of rare deleterious variants in each gene in PAH-CHD cases with gnomAD whole genome sequencing (WGS) subjects as population controls. To control for ethnicity, we selected cases of European ancestry (n = 144) using principal components analysis (PCA) (Peddy software package) [36] (Additional file 1: Figure S1C) and gnomAD subjects of non-Finnish European (NFE) ancestry (n = 7509). Since cases and controls were sequenced using different platforms, we assessed the batch effect based on the burden of rare synonymous variants, a variant class that is mostly neutral with respect to disease status. We observed that the frequency of rare synonymous variants in cases and controls was virtually identical (enrichment rate = 1.01, p value = 0.4) (Additional file 1: Table S3a). The analysis of disease-associated genes was confined to gene-specific enrichment of rare, deleterious variants (AF < 0.01%, LGD or D-mis). We assumed that under the null model, the number of rare deleterious variants observed in cases should follow a binomial distribution, given the total number of such variants in cases and controls, and a rate determined by fraction of cases in total number of subjects (cases and controls). The enrichment rate was then determined by the average number of variants in cases over the sum of average number of variants in cases and controls. The statistical significance of enrichment was tested using binom.test in R. We defined the threshold for genome-wide significance by Bonferroni correction for multiple testing (n = 17,701, threshold p value = 2.8e−6). We used the Benjamini-Hochberg procedure to estimate false discovery rate (FDR) by p.adjust in R. All SOX17 variants reported herein were confirmed with Sanger sequencing and inheritance determined when parental DNA samples were available.
To guard against spurious association results due to population differences or batch effects inherent to the use of publicly available gnomAD data, we repeated the association analysis using a set of 1319 European control subjects with individual level data obtained from the same analytical pipeline and called jointly with the PAH-CHD cases. These controls were comprised of unrelated, unaffected European parents from the Pediatric Cardiac Genomics Consortium [18]. The data were captured using NimbleGen V2.0. We performed principle components analysis of ethnicity with cases and controls together.
To estimate the burden of de novo variants in cases, we calculated the background mutation rate using a previously published tri-nucleotide change table [32, 37] and calculated the rate in protein-coding regions that are uniquely mappable. We assumed that the number of de novo variants of various types (e.g., synonymous, missense, LGD) expected by chance in gene sets or all genes followed a Poisson distribution [32]. For a given type of de novo variant in a gene set, we set the observed number of cases to m1, the expected number to m0, estimated the enrichment rate by (m1/m0), and tested for significance using an exact Poisson test (poisson.test in R) with m0 as the expectation.
Results
Characteristics of the PAH-CHD cohort are shown in Table 1. The cohort included 15 familial and 241 sporadic cases, including 61 parent-child trios and 20 duos. The majority of cases (56%) had an age of PAH onset < 18 years (pediatric-onset). There were more females among both pediatric-onset (n = 91/53, 1.7:1 female-to-male ratio) and adult-onset (n = 88/24, 3.7:1) patients, with a significant ~ 2-fold enrichment of females for adult- compared to pediatric-onset PAH (p = 0.009) (Table 1). Fifty-six percent of the patients were of European ancestry, 26% Hispanic, and 5–7% each of African, East Asian, or South Asian. The most common cardiac defects were atrial and ventricular septum defects; however, more severe defects were more frequent in pediatric-onset cases.
Table 1.
PAH-CHD patient population
| Pediatric | Adult | |
|---|---|---|
| Male, n (%) | 53 (36.8) | 24 (21.4) |
| Female, n (%) | 91 (63.2) | 88 (78.6) |
| Total, n (%) | 144 (56.3) | 112 (43.7) |
| Female-to-male ratio | 1.7:1 | 3.7:1a |
| Ancestry, n (%) | ||
| East Asian | 7 (4.9) | 7 (6.3) |
| Hispanic | 30 (20.8) | 27 (24.1) |
| African | 13 (9) | 6 (5.4) |
| South Asian | 10 (6.9) | 7 (6.3) |
| European | 81 (56.3) | 63 (56.3) |
| Unknown | 3 (2.1) | 2 (1.8) |
| Primary cardiac defect, % | ||
| Atrial septal defect (ASD) | 33.8 | 55.7 |
| Ventricular septal defect (VSD) | 22.5 | 17.7 |
| ASD + VSD | 13.8 | 7.6 |
| Atrioventricular canal defect | 7.5 | 6.3 |
| Tetralogy of Fallot | 5.6 | 1.3 |
| Transposition of the great vessels | 3.8 | 3.8 |
| Hypoplastic left heart syndrome | 1.3 | 0 |
| Coarctation of the artery | 0.6 | 0 |
| Other/complex | 11.3 | 7.6 |
aFisher’s exact test, p = 0.009, indicating a higher female-to-male ratio in adult-onset cases compared to pediatric-onset cases
Rare deleterious variants in known PAH and CHD risk genes
We screened for rare, predicted deleterious variants in 11 known risk genes for PAH and 253 candidate risk genes for CHD (Additional file 1: Table S1). PAH risk gene variants were identified in only 6.4% (16/250) of sporadic PAH-CHD cases and four of 15 familial cases (Additional file 1: Table S2). Of these cases, the majority had pediatric-onset disease (17/144 pediatric vs 3/112 adult, p = 0.0085 Fisher’s exact test). Most of the rare deleterious variants were identified in BMPR2 (n = 7, 6 pediatric) and TBX4 (n = 7, all pediatric) with a few variants in BMPR1A (n=1), BMPR1B (1), CAV1 (1), ENG (1), and SMAD9 (2). Parental DNA samples were available for a subset of the cases and three TBX4 variants were confirmed to be de novo: c.C293G:p.P98R, c.537_546del:p.1801 fs*45, and c.669_671del:p.223_224delF. We performed enrichment analysis for the PAH gene set in all PAH-CHD individuals of European ancestry (n = 143), using NFE gnomAD WGS subjects (n = 7509) as population controls. Similar frequencies of synonymous variants in cases and controls indicated that potential batch effects were minimal between the two independent datasets (Additional file 1: Table S3a). For the known PAH gene set, we observed a 5.7-fold enrichment of rare deleterious (LGD or D-mis) variants in PAH-CHD (P = 0.001) (Additional file 1: Table S3b). In contrast, there was no enrichment of rare deleterious variants in CHD risk genes in cases compared to controls (Additional file 1: Table S3b; Additional file 2: Table S4), indicating that overall these variants contribute little to PAH-CHD risk.
Association analysis identifies transcription factor SOX17 as a new candidate PAH-CHD risk gene
To identify novel risk genes for PAH-CHD, we performed an association analysis comparing per-gene rate of rare deleterious variants in European cases and NFE gnomAD WGS controls. We used a binomial test to assess the significance in 17,701 genes and found SOX17 to be associated with PAH-CHD with genome-wide significance (5/143, 3.3% of cases vs 5/7509, 0.07% of controls; enrichment rate = 52, p value = 5.5e−07) (Fig. 1). Analysis of the depth of coverage in the targeted SOX17 region indicated nearly 100% of gnomAD samples and a slightly lower percentage of PAH-CHD samples attained read depths of at least 10 (Additional file 1: Figure S2), excluding the possibility that the association is driven by coverage difference between cases and population data. No other genes reached the threshold for genome-wide significance. The top associations with a Benjamini-Hochberg FDR < 1.0 are listed in Fig. 1b. Notably, three of these genes (BZW2, FTSJ3, BAZ1B) encode putative SOX17 downstream targets [38] and two have been implicated in CHD (BAZ1B [39]) or cardiac defects associated with syndromic intellectual ability (THOC3 [40]). Similar results were obtained using a smaller cohort of European controls with individual-level data, called and annotated together with the PAH-CHD cases (Additional file 1: Figure S3). Based on the different frequencies between cases and population controls, we estimate that rare deleterious variants in SOX17 contribute to about 3.2% of European PAH-CHD patients.
Fig. 1.
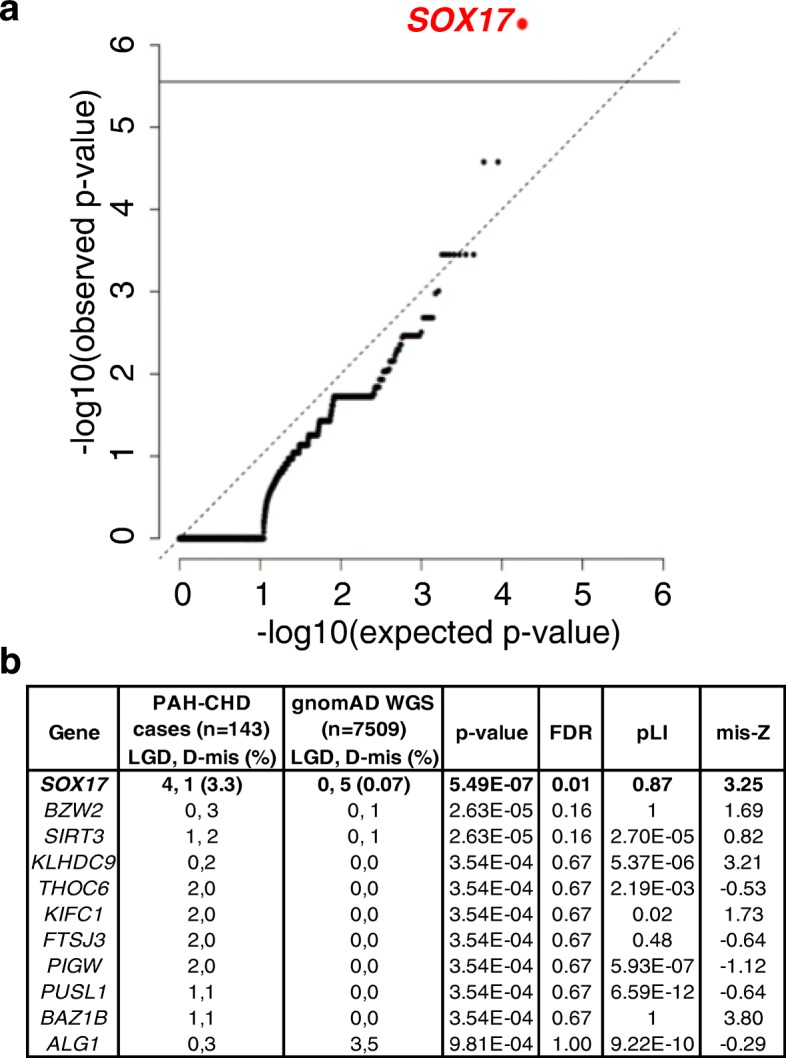
Significant association of SOX17 with PAH-CHD. a Quantile-quantile plot showing results of test of rare variant association in 17,701 genes, using 143 cases of European ancestry and 7509 gnomAD whole genome sequencing subjects of non-Finnish European ancestry. The association of SOX17 is genome-wide significant following Bonferroni correction for multiple testing. b Table of all genes with p value < 0.001 in the association tests. False discovery rate (FDR) was estimated using Benjamini-Hochberg procedure. LGD, likely gene-disrupting; D-mis, damaging missense defined as REVEL score > 0.5
We then searched for SOX17 variants in the non-European cases in the PAH-CHD cohort, and an additional cohort of 413 idiopathic and familial PAH patients without CHD (IPAH/HPAH) [41]. We identified two additional rare LGD and three additional rare D-mis variants in the PAH-CHD cohort, and one additional rare LGD (Table 2) and two rare D-mis variants in the IPAH/HPAH cohort. Variant c.C398T:p.133L, from a European patient, was not included in the initial association analysis due to in silico quality control failure but was later confirmed by Sanger sequencing. Frameshift variant c.489_510del/ p.Q163fs was observed in three unrelated patients of European or Hispanic ancestry. Closer examination of the sequence revealed a 10-bp repeat, once at the start of the deletion and once just downstream (data not shown), suggesting that a replication error may explain the recurrence. Among these three c.489_510del/p.Q163fs mutations, one was a de novo variant and another inherited from an asymptomatic parent (Table 2). Five of the six missense mutations occur within a highly conserved DNA-binding HMG-box domain (Fig. 2a). Three-dimensional modeling indicates that three of these mutations (M76V, N95S, W106L) localize within the DNA binding pocket (Fig. 2b). Comparative sequence analysis shows that all six of the missense variants are in sites highly conserved between species, including vertebrates and invertebrates (Fig. 2c).
Table 2.
Rare deleterious SOX17 variants identified in 258 PAH-CHD and 413 IPAH/HPAH samples
| Proband ID | Gender | Age at dx (years) | Disease class | Heart defecta | Ancestry | SOX17 exonb | Nucleotide change | AA change | Inheritance | Allele frequency (gnomAD) | CADD | REVEL scorec |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| JM0016 | M | 5 | PAH-CHD | ASD | European | 2 | c.C398T | p.P133L | Paternal | – | 32.0 | 0.91 |
| JM0025 | M | 7 months | PAH-CHD | VSD | European | 2 | c.489_510del | p.Q163fs | De novo | – | 33 | N/A |
| JM1277 | F | 30 | PAH-CHD | ASD | Asian | 2 | c.1203delC | p.D401fs | Unknown | – | 24.1 | N/A |
| JM1417 | F | 3 | PAH-CHD | ASD | European | 2 | c.489_510del | p.Q163fs | Paternal or de novo | – | 33 | N/A |
| JM174 | F | 14 | PAH-CHD | ASD | European | 2 | c.344delG | p.R115fs | Maternal | – | 35 | N/A |
| JM654 | M | 1 | PAH-CHD | PDA | Hispanic | 1 | c.A284G | p.N95S | Unknown | – | 24.7 | 0.93 |
| JM673 | M | 34 | PAH-CHD | ASD | European | 2 | c.C388T | p.Q130X | Unknown | – | 39.0 | N/A |
| JM887 | F | 3 | PAH-CHD | PDA | European | 1 | c.A226G | p.M76V | Unknown | – | 28.7 | 0.97 |
| JM951 | M | 9 | PAH-CHD | ASD, VSD, AV canal defect, sinus inversus, mitral cleft | Hispanic | 2 | c.C664G | p.P222A | Unknown | – | 26.1 | 0.57 |
| SPH1070EW5480 | F | 38 | PAH-CHD | Unknown | Hispanic | 2 | c.A392G | p.D131G | Unknown | – | 22.4 | 0.89 |
| SPH831KB5173 | F | 32 | IPAH | N/A | European | 2 | c.G317T | p.W106L | Unknown | – | 28.4 | 0.9 |
| JM1363 | F | 5 | IPAH | N/A | Hispanic | 2 | c.489_510del | p.Q163fs | Maternal | – | 33 | N/A |
| FPPH126-01 | M | 3 | HPAH | N/A | European | 1 | c.72_76del | p.M24fs | Unknown | – | 33 | N/A |
aASD, atrial septal defect; PDA, patent ductus arteriosus; VSD, ventricular septal defect; AV, atrioventricular
bSOX17 variants identified from transcript NM_022454
cRare, deleterious variants defined as gnomAD AF < 0.01% and REVEL > 0.5
Fig. 2.
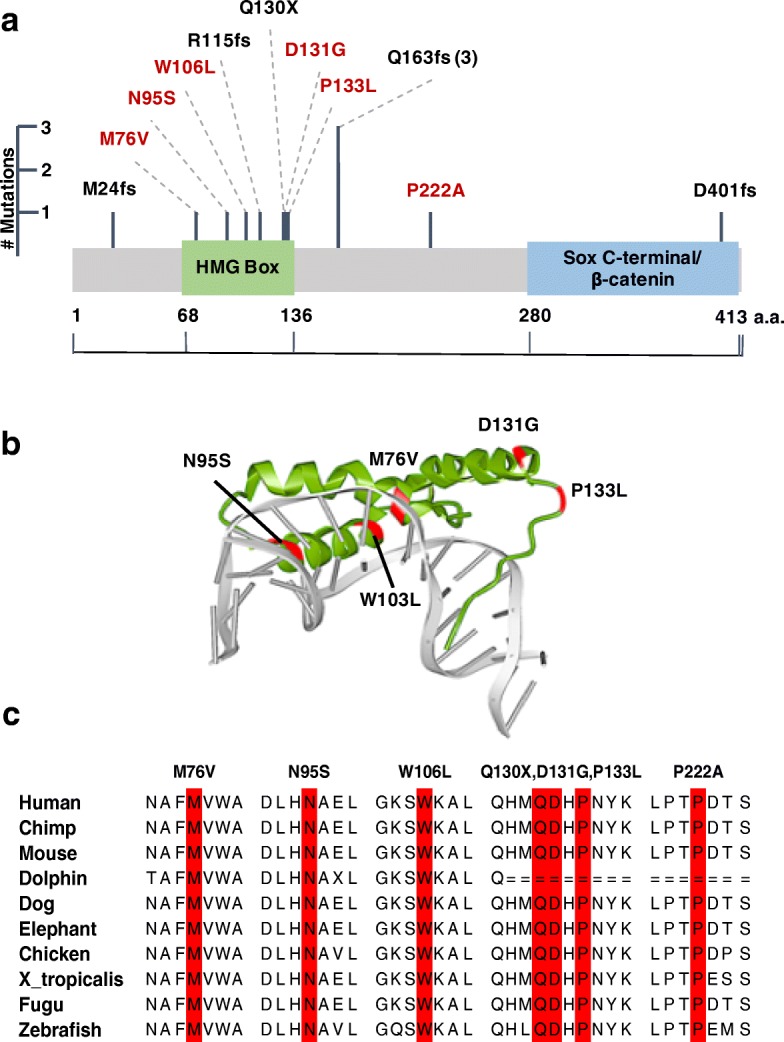
Rare deleterious variants in SOX17. a Linear schematic of the SOX17 encoded protein and location of genetic variants identified by WES. LGD variants are in black, D-mis variants in red. b Three-dimensional structure of the SOX17 HMG box domain, comprised of three alpha-helices, bound to the minor groove of DNA (Protein Data Bank 3F27). Localization of the five patient D-mis variants (red) indicates that three reside within the DNA binding pocket. c Multiple sequence alignment indicating a high degree of sequence conservation across species at the locations of SOX17 missense variants
We hypothesized that deleterious variants in SOX17 confer PAH-CHD risk through dysregulation of SOX17 target genes and some of these genes may contribute to PAH-CHD risk directly, independent of SOX17. Therefore, we tested for enrichment of rare variants in 1947 putative SOX17 target genes identified by genome-wide ChIP-X experiments [38] in European cases compared to NFE gnomAD WGS subjects. We observed a moderate but significant enrichment of rare missense variants (enrichment rate = 1.16, p value = 3.4e−4) (Additional file 1: Table S5). Since there are 618 rare missense variants in these genes in 143 cases, even a moderate enrichment suggests a large number of rare variants in SOX17-regulated genes may contribute to PAH-CHD risk. Using publicly available gene expression data for developing heart [17] and adult pulmonary artery endothelial cells (ENCODE RNA-seq data, ENCBS024RNA), we found that the majority of the SOX17 target genes with rare deleterious variants are expressed in one or both of these tissue/cell types, with 28% (42/149) having top quartile expression in both tissue/cell types (Additional file 1: Table S6 and Fig. S4a). We assessed the statistical significance of this expression pattern by building a background distribution with randomly selected sets of 149 genes that carry at least one rare LGD or D-mis variant in cases and counted the number of genes with top quartile ranked expression in both tissues. Based on 100,000 simulations, the number of observed genes in the top quartile of developing heart and PAEC expression in the SOX17 targets (42 out of 149) is significantly larger than expectation by chance (p ≤ 10–5) (Additional file 1: Figure S4b), supporting functional relevance of these SOX17 target genes. Pathway enrichment analysis using Reactome 2016 [42, 43] through Enrichr (amp.pharm.mssm.edu/Enrichr/enrich) showed that the SOX17 target genes with deleterious variants are over-represented (FDR-adjusted p value < 0.05) in (1) developmental processes, (2) transmembrane transport of small molecules and ion homeostasis, and (3) extracellular matrix interactions (Additional file 1: Table S7).
Contribution of de novo mutations to PAH-CHD
We have previously reported an enrichment of de novo predicted deleterious variants in a CHD cohort ascertained without considering PAH [17, 18]. We tested for a role of de novo mutations in PAH-CHD in 60 cases with WES data of biological parents (“trios”). The complete list of 60 rare de novo variants is provided in Additional file 1: Table S8. As mentioned previously, three de novo variants were identified in PAH risk gene TBX4 and one variant each in CHD risk genes NOTCH1 and PTPN11. However, testing for enrichment of all rare de novo variants in PAH-CHD trio probands compared to an estimated background mutation rate indicated no overall enrichment, likely due to the small sample size.
Discussion
Exome sequencing in our cohort of 256 PAH-CHD patients indicated that the genetic contribution of known/candidate risk genes for PAH or CHD alone is minimal. An unbiased, gene-based association analysis of rare deleterious variants identified SOX17 as a novel PAH-CHD candidate risk gene, explaining up to 3.2% of cases. A recent study of 1038 PAH cases (not including PAH-CHD) also found an association of SOX17 with IPAH but with a smaller effect size (relative risk ~ 2.9) [44]. The observed frequency of rare variants was ~ 0.9% of PAH cases [44], similar to our observation of SOX17 variants in ~ 0.7% of IPAH/HPAH patients without CHD. Of note, no rare deleterious SOX17 variants were identified in a recently published cohort of 1200 patients with CHD [18]. Additionally, we observed an enrichment of rare variants in putative target genes of SOX17. There was no enrichment of de novo mutations in this cohort, possibly due to the relatively small number of available trios.
SOX17 is a member of the conserved SOX family of transcription factors widely expressed in development, and the subgroup of SOXF genes (including SOX7, SOX17, and SOX18) participate in vasculogenesis and remodeling [45]. In the embryonic vasculature, SOX17 is selectively expressed in arterial endothelial cells [46–48]. Early studies of Sox17 knock-out mice did not find obvious abnormalities in embryonic vasculature [49, 50], at least partially explained by functional redundancy and compensatory roles of Sox17 and Sox18 [50, 51]. Subsequent genetic studies revealed that gene compensation and phenotypic effects were dependent on strain background [52]. Recent endothelial-specific inactivation of Sox17 in murine embryo or postnatal retina led to impaired arterial specification and embryonic death or arterial-venous malformations, respectively [46]. SOX17 has also been associated with intracranial aneurysms in genome-wide association studies [53–55], and endothelial-specific Sox17 deficiency was subsequently shown to induce intracranial aneurysm pathology in an angiotensin II infusion mouse model [56]. Finally, conditional deletion of Sox17 in mesenchymal progenitor cells demonstrated that SOX17 is required for normal pulmonary vasculature morphogenesis in utero and deficiency results in postnatal cardiac defects [57].
Cardiogenesis occurs in a highly conserved and regulated manner in the developing embryo [58]. Precise temporal and spatial control of gene expression is controlled by master transcription factors such as GATA4, MEF2C, TBX5, and NKX2–5 [59], In addition, signaling pathways, including canonical and non-canonical WNT/β-catenin [60, 61] and NOTCH [62] signaling cascades, drive cardiac morphogenesis and differentiation. SOX17 is a direct transcriptional target of GATA4, giving rise to SOX17-positive endoderm from embryonic stem cells [63] and the two proteins co-localize in the primitive endoderm [64, 65]. SOX17 induction inhibits WNT/β-catenin signaling by direct protein interaction with β-catenin through a carboxyl terminal domain of SOX17 required for transactivation of target genes [66, 67]. NOTCH1 has recently been shown to be a direct transcriptional target of SOX17 in early arterial development [68]. Thus, it is possible that impaired functional interactions between these molecules during embryogenesis could provide an underlying mechanism for the development of CHD in some PAH-CHD patients.
SOX17 is a highly constrained gene depleted of LGD and missense variants in a large population data set (ExAC pLI = 0.87, missense Z-score = 3.25) [26]. About half of the observed rare, deleterious variants in cases are LGD variants, and most of the missense variants are located in a conserved HMG box domain. The HMG box is a 79-amino acid domain that binds in a sequence-specific manner within the minor groove of DNA causing bending and facilitating assembly of nucleoprotein complexes [45]. Localization of the five HMG box missense variants within a three-dimensional model of the protein domain interacting with DNA indicated that three of the patient missense mutations (M76V, N95S, W106L) localize to the DNA binding pocket (Fig. 2b). Previously reported site-directed mutagenesis studies indicate that similar point mutations within this region (M76A, G103R) can impair both direct DNA binding [69] and complex nucleoprotein interactions, including SOX17/β-catenin protein complexes, at target gene promoters [70, 71]. This suggests that haploinsufficiency with loss of function alleles is the likely mechanism of SOX17 risk in PAH-CHD.
Some variants in SOX17 downstream target genes may be predicted to mimic some of the consequences of SOX17 loss of function mutations or haploinsufficiency. We identified 163 rare deleterious variants (131 D-mis and 32 LGD) in 149 putative target genes. Using published gene expression data, we found that most of these genes are expressed in developing heart and/or pulmonary artery endothelial cells, with significant enrichment of top quartile expression in both tissue/cell types compared to randomly selected sets of genes carrying deleterious variants in European PAH-CHD cases. Additionally, we showed that these target genes are overrepresented in pathways related to developmental biology, ion transport/homeostasis, and extracellular matrix interactions. A wide range of transmembrane small molecule transporters/channels/pumps are expressed in developing heart and pulmonary vasculature, and some have been shown to be differentially expressed in lung tissue from PAH patients compared to non-disease controls or PH with interstitial fibrosis [72]. As key regulators of vascular tone, some of these molecules function as targets of vasodilatory pharmacotherapy [73]. We recently identified the potassium channel gene, KCNK3, as a risk gene for PAH using exome sequencing [74]. Extracellular matrix proteins, including laminins, play key roles in embryonic development of both pulmonary vasculature and heart [75]. Thus, it is likely that mutations in SOX17, and possibly downstream target genes, may increase risk for PAH-CHD via multiple pathways.
The striking clinical finding was that nine out of 13 patients had pediatric-onset disease. The mean age of PAH onset for all patients with rare SOX17 variants was 14.2 years. Most of the congenital heart defects were simple (i.e., atrial septal defect, ventricular septal defect, or patent ductus arteriosus). However, most of the patients had severe PAH with systemic or supersystemic resting pulmonary arterial pressures, right ventricular hypertrophy with diminished right ventricular function, and requiring chronic intravenous vasodilator treatment. Severe PAH was observed in all patients carrying variants in the HMG-box domain or the recurrent c.489_510del/ p.Q163fs variant.
Conclusions
Together, these data strongly implicate SOX17 as a new risk gene contributing to ~ 3% of PAH-CHD cases and suggest that rare variants in genes regulated by SOX17 also contribute to PAH-CHD. Expansion of the number of PAH-CHD patients assessed and characterization of the clinical phenotypes will be important to confirm the role of SOX17 in PAH-CHD and IPAH, and more precisely estimate the contribution of genes regulated by SOX17 and de novo mutations.
Additional files
Figure S1. Study overview. Figure S2. Depth of sequencing coverage for SOX17. Figure S3. Gene-based association analysis using in-house controls. Figure S4. SOX17 target gene expression in murine E14.5 developing heart and human adult pulmonary aortic endothelial cells. Figure S5. Gene ontology analysis of SOX17 target genes harboring PAH-CHD patient-derived rare deleterious variants. Table S1. List of known PAH and CHD candidate risk genes. Table S2. Variants in known PAH risk genes. Table S3. Enrichment analyses in European cases and controls. Table S5. Enrichment analysis for SOX17 target genes. Table S6. SOX17 target gene variants and gene expression rank. Table S7. De novo variants. Table S8. List of all rare de novo variants in pediatric-onset PAH-CHD trios (n=60). (DOCX 2.05 mb)
Table S4. Variants in known CHD risk genes. (XLSX 46 kb)
Acknowledgements
We thank the patients and their families for their generous contribution. Robyn Barst and Jane Morse were critical members of the team to enroll and clinically characterize patients. Patricia Lanzano provided oversight of the Columbia biorepository. Hongjian Qi provided helpful discussions on bioinformatics analysis of WES data.
Funding
Funding support was provided by NHLBI HL060056 (to WKC), NIH/NCATS Colorado Clinical and Translational Science Award UL1 TR001082 (DDI), and The Jayden de Luca Foundation (DDI). Funding for the PAH Biobank was provided by NHLBI R24HL105333 (WCN). Y.S. was partly supported by NIH grant R01GM120609.
Availability of data and materials
The datasets used and/or analyzed in the current study are available from the corresponding author upon request. The variants in known PAH risk genes (included in Additional file 1) are deposited in ClinVar, accession numbers SCV000784722-SCV000784741.
Abbreviations
- ACVRL1
Activin A receptor-like 1
- AF
Allele frequency
- BMPR1A
Bone morphogenetic protein receptor type 1A
- BMPR1B
Bone morphogenetic protein receptor type 1B
- BMPR2
Bone morphogenetic protein receptor type 2
- bp
Base pair
- BWA-MEM
Burrows-Wheeler Aligner
- CADD
Combined Annotation Dependent Depletion
- CAV1
Caveolin-1
- CHD
Congenital heart disease
- D-mis
Damaging missense variants
- EIF2AK4
Eukaryotic initiation translation factor 2 alpha kinase 4
- ENG
Endoglin
- ExAC
Exome Aggregation Consortium
- FDR
False discovery rate
- gnomAD
Genome Aggregation Database
- IGV
Integrative Genomics Viewer
- IPAH
Idiopathic pulmonary arterial hypertension
- KCNK3
Potassium two-pore-domain channel subfamily K member 3
- LGD
Likely gene-disrupting
- MLPA
Multiplex ligation-dependent probe amplification
- NFE
Non-Finnish Europeans
- NOTCH1
Notch (Drosophila) homolog 1
- PAH
Pulmonary arterial hypertension
- PAH-CHD
Pulmonary arterial hypertension associated with congenital heart disease
- PCA
Principal components analysis
- PTPN11
Protein tyrosine phosphatase non-receptor type 11
- REVEL
Rare exome variant ensemble learner
- RGC
Regeneron Genetics Center
- SMAD4
SMAD family member 4
- SMAD9
SMAD family member 9
- SOX17
SRY-related HMG-box family member 17
- TBX4
T-box 4
- TGF-β/BMP
Transforming growth factor beta/bone morphogenetic protein
- VQSR
Variant quality score recalibration
- WES
Whole exome sequencing
- WGS
Whole genome sequencing
- WHO
World Health Organization
Authors’ contributions
WKC conceived and designed the study. NZ, YS, WKC, CW, JW, and PMA analyzed and interpreted the data. CW, YS, WKC, NZ, CG-J, and FED wrote the manuscript. LM, UK, and EBR collected the samples and clinical information. DDI, EDA, RH, WCN, MWP, and KAL collected the samples and provided the WES data and clinical information. JDO, AKK, JGR, and AB provided the WES data. All authors contributed to and discussed the results and critically reviewed the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Written informed consent (and assent when appropriate) was obtained from patients or parents/legal guardians under a protocol approved by the Institutional Review Board at Columbia University Medical Center or Children’s Hospital of Colorado. The research complied with the principles of the Declaration of Helsinki.
Consent for publication
Written informed consent for publication was obtained at enrollment.
Competing interests
CG-J, AKK, JGR, JDO, AB, and FD are full time employees of Regeneron Pharmaceuticals Inc. and receive stock options as part of compensation. The remaining authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Na Zhu and Carrie L. Welch contributed equally to this work.
Electronic supplementary material
The online version of this article (10.1186/s13073-018-0566-x) contains supplementary material, which is available to authorized users.
Contributor Information
Na Zhu, Email: nz2274@cumc.columbia.edu.
Carrie L. Welch, Email: cbw13@cumc.columbia.edu
Jiayao Wang, Email: jw3514@cumc.columbia.edu.
Philip M. Allen, Email: pma2130@cumc.columbia.edu
Claudia Gonzaga-Jauregui, Email: clau.gonzagajauregui@regeneron.com.
Lijiang Ma, Email: malijiang@yahoo.com.
Alejandra K. King, Email: alenjandra.king@regeneron.com
Usha Krishnan, Email: usk1@cumc.columbia.edu.
Erika B. Rosenzweig, Email: esb14@cumc.columbia.edu
D. Dunbar Ivy, Email: Dunbar.Ivy@childrenscolorado.org.
Eric D. Austin, Email: eric.austin@Vanderbilt.edu
Rizwan Hamid, Email: rizwan.hamid@Vanderbilt.edu.
Michael W. Pauciulo, Email: Mike.Pauciulo@cchmc.org
Katie A. Lutz, Email: Katie.Lutz@cchmc.org
William C. Nichols, Email: Bill.Nichols@cchmc.org
Jeffrey G. Reid, Email: jeffrey.reid@regeneron.com
John D. Overton, Email: john.overton@regeneron.com
Aris Baras, Email: aris.baris@regeneron.com.
Frederick E. Dewey, Email: frederick.dewey@regeneron.com
Yufeng Shen, Email: ys2411@cumc.columbia.edu.
Wendy K. Chung, Phone: 212-851-5313, Email: wkc15@cumc.columbia.edu, Email: wkc15@columbia.edu
References
- 1.van Dissel AC, Mulder BJ, Bouma BJ. The changing landscape of pulmonary arterial hypertension in the adult with congenital heart disease. J Clin Med. 2017;6(4) [DOI] [PMC free article] [PubMed]
- 2.Dimopoulos K, Wort SJ, Gatzoulis MA. Pulmonary hypertension related to congenital heart disease: a call for action. Eur Heart J. 2014;35(11):691–700. doi: 10.1093/eurheartj/eht437. [DOI] [PubMed] [Google Scholar]
- 3.Li L, Jick S, Breitenstein S, Hernandez G, Michel A, Vizcaya D. Pulmonary arterial hypertension in the USA: an epidemiological study in a large insured pediatric population. Pulm Circ. 2017;7(1):126–136. doi: 10.1086/690007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marelli AJ, Ionescu-Ittu R, Mackie AS, Guo L, Dendukuri N, Kaouache M. Lifetime prevalence of congenital heart disease in the general population from 2000 to 2010. Circulation. 2014;130(9):749–756. doi: 10.1161/CIRCULATIONAHA.113.008396. [DOI] [PubMed] [Google Scholar]
- 5.Best DH, Austin ED, Chung WK, Elliott CG. Genetics of pulmonary hypertension. Curr Opin Cardiol. 2014;29(6):520–527. doi: 10.1097/HCO.0000000000000105. [DOI] [PubMed] [Google Scholar]
- 6.Chida A, Shintani M, Nakayama T, Furutani Y, Hayama E, Inai K, et al. Missense mutations of the BMPR1B (ALK6) gene in childhood idiopathic pulmonary arterial hypertension. Circ J. 2012;76(6):1501–1508. doi: 10.1253/circj.CJ-11-1281. [DOI] [PubMed] [Google Scholar]
- 7.Nasim MT, Ogo T, Ahmed M, Randall R, Chowdhury HM, Snape KM, et al. Molecular genetic characterization of SMAD signaling molecules in pulmonary arterial hypertension. Hum Mutat. 2011;32(12):1385–1389. doi: 10.1002/humu.21605. [DOI] [PubMed] [Google Scholar]
- 8.Kerstjens-Frederikse WS, Bongers EMHF, Roofthooft MTR, Leter EM, Douwes JM, Van Dijk A, et al. TBX4 mutations (small patella syndrome) are associated with childhood-onset pulmonary arterial hypertension. J Med Genet. 2013;50(8):500–506. doi: 10.1136/jmedgenet-2012-101152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roberts KE, McElroy JJ, Wong WP, Yen E, Widlitz A, Barst RJ, et al. BMPR2 mutations in pulmonary arterial hypertension with congenital heart disease. Eur Respir J. 2004;24(3):371–374. doi: 10.1183/09031936.04.00018604. [DOI] [PubMed] [Google Scholar]
- 10.Pfarr N, Fischer C, Ehlken N, Becker-Grunig T, Lopez-Gonzalez V, Gorenflo M, et al. Hemodynamic and genetic analysis in children with idiopathic, heritable, and congenital heart disease associated pulmonary arterial hypertension. Respir Res. 2013;14:3. doi: 10.1186/1465-9921-14-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Levy M, Eyries M, Szezepanski I, Ladouceur M, Nadaud S, Bonnet D, et al. Genetic analyses in a cohort of children with pulmonary hypertension. Eur Respir J. 2016;48(4):1118–1126. doi: 10.1183/13993003.00211-2016. [DOI] [PubMed] [Google Scholar]
- 12.Vecoli C, Pulignani S, Foffa I, Andreassi MG. Congenital heart disease: the crossroads of genetics, epigenetics and environment. Curr Genomics. 2014;15(5):390–399. doi: 10.2174/1389202915666140716175634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zaidi S, Brueckner M. Genetics and genomics of congenital heart disease. Circ Res. 2017;120(6):923–940. doi: 10.1161/CIRCRESAHA.116.309140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Soemedi R, Wilson IJ, Bentham J, Darlay R, Topf A, Zelenika D, et al. Contribution of global rare copy-number variants to the risk of sporadic congenital heart disease. Am J Hum Genet. 2012;91(3):489–501. doi: 10.1016/j.ajhg.2012.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Glessner JT, Bick AG, Ito K, Homsy J, Rodriguez-Murillo L, Fromer M, et al. Increased frequency of de novo copy number variants in congenital heart disease by integrative analysis of single nucleotide polymorphism array and exome sequence data. Circ Res. 2014;115(10):884–896. doi: 10.1161/CIRCRESAHA.115.304458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fahed AC, Gelb BD, Seidman JG, Seidman CE. Genetics of congenital heart disease: the glass half empty. Circ Res. 2013;112(4):707–720. doi: 10.1161/CIRCRESAHA.112.300853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zaidi S, Choi M, Wakimoto H, Ma L, Jiang J, Overton JD, et al. De novo mutations in histone-modifying genes in congenital heart disease. Nature. 2013;498(7453):220–223. doi: 10.1038/nature12141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Homsy J, Zaidi S, Shen Y, Ware JS, Samocha KE, Karczewski KJ, et al. De novo mutations in congenital heart disease with neurodevelopmental and other congenital anomalies. Science. 2015;350(6265):1262–1266. doi: 10.1126/science.aac9396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jin SC, Homsy J, Zaidi S, Lu Q, Morton S, DePalma SR, et al. Contribution of rare inherited and de novo variants in 2,871 congenital heart disease probands. Nat Genet. 2017;49(11):1593–1601. doi: 10.1038/ng.3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Simonneau G, Robbins IM, Beghetti M, Channick RN, Delcroix M, Denton CP, et al. Updated clinical classification of pulmonary hypertension. J Am Coll Cardiol. 2009;54(1 Suppl):S43–S54. doi: 10.1016/j.jacc.2009.04.012. [DOI] [PubMed] [Google Scholar]
- 21.Li H, Ruan J, Durbin R. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 2008;18(11):1851–1858. doi: 10.1101/gr.078212.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43(5):491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Van der Auwera GA, Carneiro MO, Hartl C, Poplin R, Del Angel G, Levy-Moonshine A, et al. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinformatics. 2013;43:11 0 1–11 033. doi: 10.1002/0471250953.bi1110s43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krumm N, Turner TN, Baker C, Vives L, Mohajeri K, Witherspoon K, et al. Excess of rare, inherited truncating mutations in autism. Nat Genet. 2015;47(6):582–588. doi: 10.1038/ng.3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38(16):e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536(7616):285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dong C, Wei P, Jian X, Gibbs R, Boerwinkle E, Wang K, et al. Comparison and integration of deleteriousness prediction methods for nonsynonymous SNVs in whole exome sequencing studies. Hum Mol Genet. 2015;24(8):2125–2137. doi: 10.1093/hmg/ddu733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kircher M, Witten DM, Jain P, O'Roak BJ, Cooper GM, Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46(3):310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ioannidis NM, Rothstein JH, Pejaver V, Middha S, McDonnell SK, Baheti S, et al. REVEL: an ensemble method for predicting the pathogenicity of rare missense variants. Am J Hum Genet. 2016;99(4):877–885. doi: 10.1016/j.ajhg.2016.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ghosh RO, Oak N, Plon SE. Evaluation of in silico algorithms for use with ACMG/AMP clinical variant interpretation guidelines. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Iossifov I, O'Roak BJ, Sanders SJ, Ronemus M, Krumm N, Levy D, et al. The contribution of de novo coding mutations to autism spectrum disorder. Nature. 2014;515(7526):216–221. doi: 10.1038/nature13908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Samocha KE, Robinson EB, Sanders SJ, Stevens C, Sabo A, McGrath LM, et al. A framework for the interpretation of de novo mutation in human disease. Nat Genet. 2014;46(9):944–950. doi: 10.1038/ng.3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thorvaldsdottir H, Robinson JT, Mesirov JP. Integrative genomics viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 2013;14(2):178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stenson PD, Ball EV, Mort M, Phillips AD, Shiel JA, Thomas NS, et al. Human Gene Mutation Database (HGMD): 2003 update. Hum Mutat. 2003;21(6):577–581. doi: 10.1002/humu.10212. [DOI] [PubMed] [Google Scholar]
- 35.Landrum MJ, Lee JM, Riley GR, Jang W, Rubinstein WS, Church DM, et al. ClinVar: public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res. 2014;42(Database issue):D980–D985. doi: 10.1093/nar/gkt1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pedersen BS, Quinlan AR. Who's who? Detecting and resolving sample anomalies in human DNA sequencing studies with Peddy. Am J Hum Genet. 2017;100(3):406–413. doi: 10.1016/j.ajhg.2017.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ware JS, Samocha KE, Homsy J, Daly MJ. Interpreting de novo variation in human disease using denovolyzeR. Curr Protoc Hum Genet. 2015;87:7 25 1–7 7 15. doi: 10.1002/0471142905.hg0725s87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lachmann A, Xu H, Krishnan J, Berger SI, Mazloom AR, Ma'ayan A. ChEA: transcription factor regulation inferred from integrating genome-wide ChIP-X experiments. Bioinformatics. 2010;26(19):2438–2444. doi: 10.1093/bioinformatics/btq466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Andersen TA, Troelsen Kde L, Larsen LA. Of mice and men: molecular genetics of congenital heart disease. Cell Mol Life Sci. 2014;71(8):1327–1352. doi: 10.1007/s00018-013-1430-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Amos JS, Huang L, Thevenon J, Kariminedjad A, Beaulieu CL, Masurel-Paulet A, et al. Autosomal recessive mutations in THOC6 cause intellectual disability: syndrome delineation requiring forward and reverse phenotyping. Clin Genet. 2017;91(1):92–99. doi: 10.1111/cge.12793. [DOI] [PubMed] [Google Scholar]
- 41.Zhu N, Gonzaga-Jauregui C, Welch CL, Ma L, Qi H, King AK, et al. Exome sequencing in children with pulmonary arterial hypertension demonstrates differences compared with adults. Circ Genom Precis Med. 2018;11(4):e001887. doi: 10.1161/CIRCGEN.117.001887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Croft D, Mundo AF, Haw R, Milacic M, Weiser J, Wu G, et al. The Reactome pathway knowledgebase. Nucleic Acids Res. 2014;42(Database issue):D472–D477. doi: 10.1093/nar/gkt1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fabregat A, Sidiropoulos K, Viteri G, Forner O, Marin-Garcia P, Arnau V, et al. Reactome pathway analysis: a high-performance in-memory approach. BMC Bioinformatics. 2017;18(1):142. doi: 10.1186/s12859-017-1559-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Graf S, Haimel M, Bleda M, Hadinnapola C, Southgate L, Li W, et al. Identification of rare sequence variation underlying heritable pulmonary arterial hypertension. Nat Commun. 9(1):2018, 1416. [DOI] [PMC free article] [PubMed]
- 45.Francois M, Koopman P, Beltrame M. SoxF genes: key players in the development of the cardio-vascular system. Int J Biochem Cell Biol. 2010;42(3):445–448. doi: 10.1016/j.biocel.2009.08.017. [DOI] [PubMed] [Google Scholar]
- 46.Corada M, Orsenigo F, Morini MF, Pitulescu ME, Bhat G, Nyqvist D, et al. Sox17 is indispensable for acquisition and maintenance of arterial identity. Nat Commun. 2013;4:2609. doi: 10.1038/ncomms3609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liao WP, Uetzmann L, Burtscher I, Lickert H. Generation of a mouse line expressing Sox17-driven Cre recombinase with specific activity in arteries. Genesis. 2009;47(7):476–483. doi: 10.1002/dvg.20520. [DOI] [PubMed] [Google Scholar]
- 48.Sacilotto N, Monteiro R, Fritzsche M, Becker PW, Sanchez-Del-Campo L, Liu K, et al. Analysis of Dll4 regulation reveals a combinatorial role for sox and notch in arterial development. Proc Natl Acad Sci U S A. 2013;110(29):11893–11898. doi: 10.1073/pnas.1300805110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kanai-Azuma M, Kanai Y, Gad JM, Tajima Y, Taya C, Kurohmaru M, et al. Depletion of definitive gut endoderm in Sox17-null mutant mice. Development. 2002;129(10):2367–2379. doi: 10.1242/dev.129.10.2367. [DOI] [PubMed] [Google Scholar]
- 50.Sakamoto Y, Hara K, Kanai-Azuma M, Matsui T, Miura Y, Tsunekawa N, et al. Redundant roles of Sox17 and Sox18 in early cardiovascular development of mouse embryos. Biochem Biophys Res Commun. 2007;360(3):539–544. doi: 10.1016/j.bbrc.2007.06.093. [DOI] [PubMed] [Google Scholar]
- 51.Matsui T, Kanai-Azuma M, Hara K, Matoba S, Hiramatsu R, Kawakami H, et al. Redundant roles of Sox17 and Sox18 in postnatal angiogenesis in mice. J Cell Sci. 2006;119(Pt 17):3513–3526. doi: 10.1242/jcs.03081. [DOI] [PubMed] [Google Scholar]
- 52.Hosking B, Francois M, Wilhelm D, Orsenigo F, Caprini A, Svingen T, et al. Sox7 and Sox17 are strain-specific modifiers of the lymphangiogenic defects caused by Sox18 dysfunction in mice. Development. 2009;136(14):2385–2391. doi: 10.1242/dev.034827. [DOI] [PubMed] [Google Scholar]
- 53.Bilguvar K, Yasuno K, Niemela M, Ruigrok YM, von Und Zu Fraunberg M, van Duijn CM, et al. Susceptibility loci for intracranial aneurysm in European and Japanese populations. Nat Genet. 2008;40(12):1472–1477. doi: 10.1038/ng.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yasuno K, Bilguvar K, Bijlenga P, Low SK, Krischek B, Auburger G, et al. Genome-wide association study of intracranial aneurysm identifies three new risk loci. Nat Genet. 2010;42(5):420–425. doi: 10.1038/ng.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Foroud T, Koller DL, Lai D, Sauerbeck L, Anderson C, Ko N, et al. Genome-wide association study of intracranial aneurysms confirms role of Anril and SOX17 in disease risk. Stroke. 2012;43(11):2846–2852. doi: 10.1161/STROKEAHA.112.656397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lee S, Kim IK, Ahn JS, Woo DC, Kim ST, Song S, et al. Deficiency of endothelium-specific transcription factor Sox17 induces intracranial aneurysm. Circulation. 2015;131(11):995–1005. doi: 10.1161/CIRCULATIONAHA.114.012568. [DOI] [PubMed] [Google Scholar]
- 57.Lange AW, Haitchi HM, LeCras TD, Sridharan A, Xu Y, Wert SE, et al. Sox17 is required for normal pulmonary vascular morphogenesis. Dev Biol. 2014;387(1):109–120. doi: 10.1016/j.ydbio.2013.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li X, Martinez-Fernandez A, Hartjes KA, Kocher JP, Olson TM, Terzic A, et al. Transcriptional atlas of cardiogenesis maps congenital heart disease interactome. Physiol Genomics. 2014;46(13):482–495. doi: 10.1152/physiolgenomics.00015.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.McCulley DJ, Black BL. Transcription factor pathways and congenital heart disease. Curr Top Dev Biol. 2012;100:253–277. doi: 10.1016/B978-0-12-387786-4.00008-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gillers BS, Chiplunkar A, Aly H, Valenta T, Basler K, Christoffels VM, et al. Canonical wnt signaling regulates atrioventricular junction programming and electrophysiological properties. Circ Res. 2015;116(3):398–406. doi: 10.1161/CIRCRESAHA.116.304731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Klaus A, Muller M, Schulz H, Saga Y, Martin JF, Birchmeier W. Wnt/beta-catenin and Bmp signals control distinct sets of transcription factors in cardiac progenitor cells. Proc Natl Acad Sci U S A. 2012;109(27):10921–10926. doi: 10.1073/pnas.1121236109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Luxan G, D'Amato G, MacGrogan D, de la Pompa JL. Endocardial notch signaling in cardiac development and disease. Circ Res. 2016;118(1):e1–e18. doi: 10.1161/CIRCRESAHA.115.305350. [DOI] [PubMed] [Google Scholar]
- 63.Holtzinger A, Rosenfeld GE, Evans T. Gata4 directs development of cardiac-inducing endoderm from ES cells. Dev Biol. 2010;337(1):63–73. doi: 10.1016/j.ydbio.2009.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Artus J, Piliszek A, Hadjantonakis AK. The primitive endoderm lineage of the mouse blastocyst: sequential transcription factor activation and regulation of differentiation by Sox17. Dev Biol. 2011;350(2):393–404. doi: 10.1016/j.ydbio.2010.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Viotti M, Nowotschin S, Hadjantonakis AK. SOX17 links gut endoderm morphogenesis and germ layer segregation. Nat Cell Biol. 2014;16(12):1146–1156. doi: 10.1038/ncb3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Morrison G, Scognamiglio R, Trumpp A, Smith A. Convergence of cMyc and beta-catenin on Tcf7l1 enables endoderm specification. EMBO J. 2016;35(3):356–368. doi: 10.15252/embj.201592116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zorn AM, Barish GD, Williams BO, Lavender P, Klymkowsky MW, Varmus HE. Regulation of Wnt signaling by Sox proteins: XSox17 alpha/beta and XSox3 physically interact with beta-catenin. Mol Cell. 1999;4(4):487–498. doi: 10.1016/S1097-2765(00)80200-2. [DOI] [PubMed] [Google Scholar]
- 68.Chiang IK, Fritzsche M, Pichol-Thievend C, Neal A, Holmes K, Lagendijk A, et al. SoxF factors induce Notch1 expression via direct transcriptional regulation during early arterial development. Development. 2017;144(14):2629–2639. doi: 10.1242/dev.146241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sinner D, Kordich JJ, Spence JR, Opoka R, Rankin S, Lin SC, et al. Sox17 and Sox4 differentially regulate beta-catenin/T-cell factor activity and proliferation of colon carcinoma cells. Mol Cell Biol. 2007;27(22):7802–7815. doi: 10.1128/MCB.02179-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Liu X, Luo M, Xie W, Wells JM, Goodheart MJ, Engelhardt JF. Sox17 modulates Wnt3A/beta-catenin-mediated transcriptional activation of the Lef-1 promoter. Am J Physiol Lung Cell Mol Physiol. 2010;299(5):L694–L710. doi: 10.1152/ajplung.00140.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Banerjee A, Ray S. Structural insight, mutation and interactions in human beta-catenin and SOX17 protein: a molecular-level outlook for organogenesis. Gene. 2017;610:118–126. doi: 10.1016/j.gene.2017.01.026. [DOI] [PubMed] [Google Scholar]
- 72.Rajkumar R, Konishi K, Richards TJ, Ishizawar DC, Wiechert AC, Kaminski N, et al. Genomewide RNA expression profiling in lung identifies distinct signatures in idiopathic pulmonary arterial hypertension and secondary pulmonary hypertension. Am J Physiol Heart Circ Physiol. 2010;298(4):H1235–H1248. doi: 10.1152/ajpheart.00254.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Olschewski A, Papp R, Nagaraj C, Olschewski H. Ion channels and transporters as therapeutic targets in the pulmonary circulation. Pharmacol Ther. 2014;144(3):349–368. doi: 10.1016/j.pharmthera.2014.08.001. [DOI] [PubMed] [Google Scholar]
- 74.Piovan E, Yu J, Tosello V, Herranz D, Ambesi-Impiombato A, Da Silva AC, et al. Direct reversal of glucocorticoid resistance by AKT inhibition in acute lymphoblastic leukemia. Cancer Cell. 2013;24(6):766–776. doi: 10.1016/j.ccr.2013.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Durbeej M. Laminins. Cell Tissue Res. 2010;339(1):259–268. doi: 10.1007/s00441-009-0838-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Study overview. Figure S2. Depth of sequencing coverage for SOX17. Figure S3. Gene-based association analysis using in-house controls. Figure S4. SOX17 target gene expression in murine E14.5 developing heart and human adult pulmonary aortic endothelial cells. Figure S5. Gene ontology analysis of SOX17 target genes harboring PAH-CHD patient-derived rare deleterious variants. Table S1. List of known PAH and CHD candidate risk genes. Table S2. Variants in known PAH risk genes. Table S3. Enrichment analyses in European cases and controls. Table S5. Enrichment analysis for SOX17 target genes. Table S6. SOX17 target gene variants and gene expression rank. Table S7. De novo variants. Table S8. List of all rare de novo variants in pediatric-onset PAH-CHD trios (n=60). (DOCX 2.05 mb)
Table S4. Variants in known CHD risk genes. (XLSX 46 kb)
Data Availability Statement
The datasets used and/or analyzed in the current study are available from the corresponding author upon request. The variants in known PAH risk genes (included in Additional file 1) are deposited in ClinVar, accession numbers SCV000784722-SCV000784741.


