Abstract
During protein synthesis, ribosomes encounter many roadblocks, the outcomes of which are largely determined by substrate availability, amino acid features, and reaction kinetics. Prolonged ribosome stalling is likely to be resolved by ribosome rescue [G] or quality control pathways, whereas shorter stalling is likely to be resolved by ongoing productive translation. How ribosome function is affected by such hindrances can therefore have a profound impact on the translational output (yield) of a particular mRNA. In this Review we focus on these roadblocks and the resumption of normal translation elongation, rather than on alternative fates wherein the stalled ribosome triggers degradation of the mRNA and the incomplete protein product. We discuss the fundamental stages of the translation process in eukaryotes, from elongation through ribosome recycling. We pay particular attention to recent discoveries that highlight the complexity of the genetic code and regulatory elements that control gene expression, including ribosome stalling during elongation, the role of mRNA context in translation termination, and mechanisms of ribosome rescue that resemble recycling.
INTRODUCTION
Across all domains of life, translation of the information encoded in mRNA to protein is performed by a large macromolecular machine called the ribosome. The ribosome reads the information one codon (three nucleotides) at a time, translating it into protein through the actions of tRNAs that recognize each codon to insert the appropriate amino acid (Figure 1). The tRNAs are really the central ‘translators’ in the translation process.
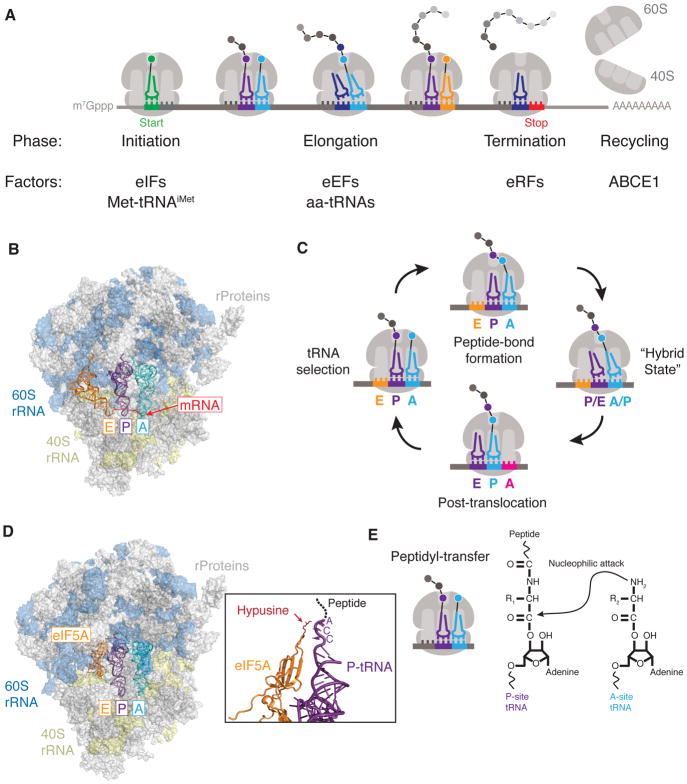
Figure 1. Overview of eukaryotic translation.
(A) Overview of eukaryotic translation. Translation begins with initiation where a complex coordination of many eukaryotic translation initiation factors (eIFs), initiator methionyl-tRNA (Met-tRNAiMet), the ribosomal subunits and mRNA to be translated come together at the AUG start codon of the open reading frame. Next, elongation involves synthesis of the peptide chain through the coordinated actions of eukaryotic elongation factors (eEFs) and aminoacyl-tRNAs (aa-tRNAs) until the ribosome reaches a termination or stop codon. In this termination phase the peptide is released through the actions of eukaryotic peptide chain release factors (eRFs). Finally, the ribosome subunits must be recycled by ATP-binding cassette sub-family E member 1 (ABCE1) for a subsequent round of translation. (B) Structural model of the yeast 80S ribosome depicting three sites for tRNA binding: E (exit; colored orange), P (peptidyl; colored purple) and A (aminoacyl; colored cyan). mRNA is shown in red. The model was created by alignment of the Saccharomyces cerevisiae 80S ribosome (PDB 4V88) and Thermus thermophilus 70S ribosome (PDB 4V9I). (C) Overview of the elongation cycle highlighting tRNA movement through the ribosome. tRNA selection occurs at the A site. Next, peptide-bond formation occurs, which transfers the peptide to the A-site tRNA. Concurrent with peptide bond formation, the tRNAs adopt a ‘hybrid’ state (relative to the large and small ribosomal subunits) and this ribosome complex is the substrate for translocation to allow the decoding of the next codon. (D) Schematic of the peptidyl-transfer reaction that occurs during translation elongation. The amino group of the incoming amino acid (cyan) attacks the ester linkage on the peptidyl-tRNA (purple) in the ribosomal P site to transfer the growing peptide chain to A-site tRNA. (E) Structural model of the S. cerevisiae 80S ribosome bound to eIF5A. eIF5A binds in the ribosomal E site. Zoom-in area shows interactions between the hypusine modification of eIF5A and the CCA nucleotides at the 3′-end of the peptidyl-tRNA. The model was created by alignment of the S. cerevisiae 80S ribosome (PDB 4V88) with coordinates for the 60S subunit bound by eIF5A (PDB 5GAK).
The process of translation can be broken into four main phases: initiation, elongation, termination and ribosome recycling (Figure 1A). Although core aspects of translation are highly conserved between eukaryotes, bacteria, and archaea, there are substantive differences in how each of these four steps is accomplished. In this review, we will focus on eukaryotic translation. In the first step, many eukaryotic translation initiation factors (eIFs) guide the proper assembly of an 80S ribosome at the AUG start codon with an initiator methionyl-tRNA bound in the P site [G]. During elongation, 80S ribosomes move processively along the mRNA, three nucleotides per step, synthesizing the encoded protein one amino acid at a time through the coordinated actions of aminoacyl-tRNAs [G] and eukaryotic elongation factors (eEFs). Throughout this process tRNAs vectorially transition between three sites inside the ribosome, which span both the large ribosomal subunit (60S) and small ribosomal subunit (40S): E site [G], P site and A site [G] (Figures 1B–C). At the end of the open reading frame (ORF) the ribosome encounters a termination codon that is specifically recognized by a set of protein factors called eukaryotic peptide chain release factors (eRFs), which promote the release of the nascent protein from the peptidyl-tRNA [G] (and ultimately the from the ribosome). Finally, in the recycling phase, the post-termination 80S ribosome complex is recycled by ATP-binding cassette sub-family E member 1 (ABCE1) into separate 40S and 60S subunits to begin a new round of translation.
In this Review we discuss our current knowledge of translation elongation, termination and ribosome recycling, including the order and timing (where known) of events that occur at each step. We highlight recent discoveries and regulatory mechanisms that affect each step, including ribosome stalling during elongation, the role of mRNP context in translation termination, and mechanisms of ribosome rescue that resemble recycling. The heart of each step includes a fate decision that is determined by the state of the ribosome, whether to continue translation or invoke quality control mechanisms. For example, some ribosome pauses are resolved by auxiliary factors, such as eIF5A, whereas other pauses lead the ribosome to abandon translation altogether with the help of ribosome rescue factors such as the Dom34–Hbs1 complex (PELO-HBS1L in humans). As with all processes, kinetic events are determined by the cellular concentrations of players in the process and these parameters dictate biological outcome. These decisive moments can have a profound effect on the output from a particular mRNA by determining how much or which products are produced.
Translation elongation
The process of elongation begins immediately after translation initiation has taken place and an 80S ribosome is positioned at an AUG start codon with a methionyl-tRNAiMet in the P site (Figure 2A). Although we do not discuss translation initiation in this review, the details are well described in several excellent reviews1–4. The elongation phase extends from the loading of the first aminoacyl-tRNA at the start of the ORF (after the initiation codon) until the ribosome reaches the termination codon at the end of the ORF, and is thought to be mostly conserved relative to bacterial elongation, as outlined below.
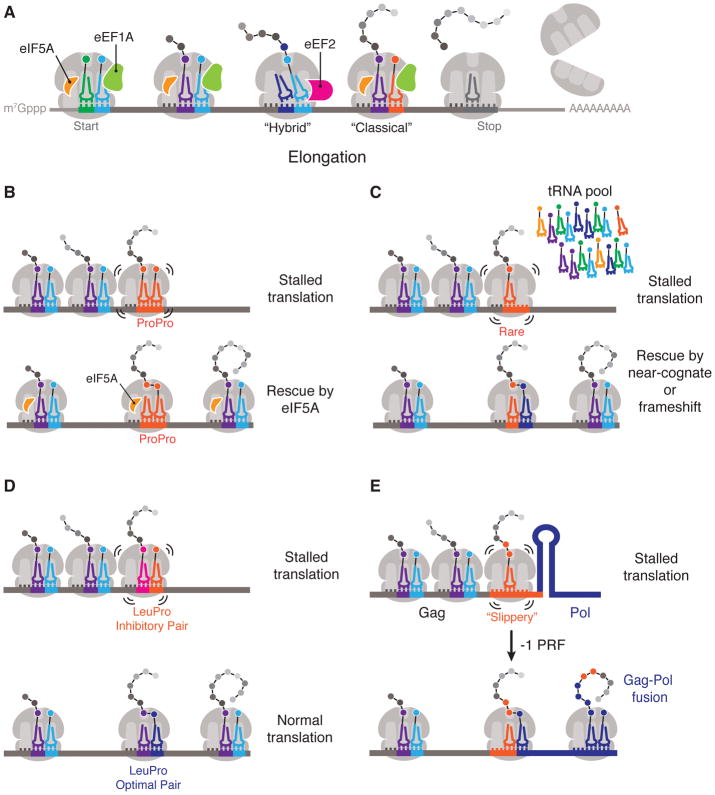
Figure 2. Translation elongation and resolution of ribosome stalling.
(A) Overview of translation elongation. Aminoacyl-tRNAs are delivered to the ribosome in complex with elongation factor 1-alpha (eEF1A) and GTP (not shown). Peptide bond formation occurs and the tRNAs are positioned in a ‘hybrid’ state with respect to the ribosome subunits. Subsequent translocation driven by elongation factor 2 (eEF2) causes tRNA repositioning from a hybrid state to classical state, creating an open A site for the next incoming aminoacyl-tRNA. eIF5A, which is a small protein that binds in the ribosomal E site, stimulates catalysis in the peptidyl transferase center throughout translation elongation. (B) Ribosome stalling due to slow peptidyl-transfer kinetics (such as during the formation of Pro–Pro) is rescued by eIF5A, which promotes peptide-bond formation. (C) Ribosome stalling caused by poor A-site occupancy resulting from poorly-expressed tRNAs (represented in figure) or poor tRNA aminoacylation can be rescued by mis-incorporation of near-cognate tRNAs or by frameshifting (represented as conversion of orange to purple). (D) Ribosome stalling can be caused by certain consecutive tRNA–codon pair orders that are sub-optimal (pink and orange) relative to synonymous pairs (purple and blue). (E) Ribosome stalling caused by mRNA secondary structures can be resolved by programmed ribosomal frameshifting (PRF) at adjacent slippery sequences. The example illustrates the -1 PRF that is required for translation of the Gag-Pol fusion protein of human immunodeficiency virus.
Translation elongation is composed of three basic steps that take place at the incorporation of each amino acid in the elongating peptide chain: tRNA selection (or decoding), peptide-bond formation, and translocation of the mRNA–tRNA complex (Figures 1C and 2A). tRNA selection is the process wherein the aminoacyl-tRNA with the proper anticodon to match the mRNA codon is loaded into the A site of the ribosome. Aminoacyl-tRNAs are delivered to the ribosomal A site by the specialized GTPase eEF1A (elongation factor Tu (EFTu) in bacteria) in a ternary complex with GTP5–7. Once cognate interactions between the codon and anticodon are sensed, eEF1A is activated and hydrolyzes GTP to enable the tRNA to be fully accommodated into the A site (reviewed in 8 for bacteria). In the next step, peptide-bond formation, the amino group of the incoming amino acid attacks the ester linkage on the peptidyl-tRNA in the ribosomal P site and the growing peptide chain is transferred to the tRNA in the A site (a more detailed view of this process is reviewed in 9, again for bacteria) (Figures 1C, 1D). As the peptide bond forms, the ribosomal subunits rotate with respect to one another and the tRNAs adopt an altered conformation referred to as the ‘hybrid’ state, in which the anticodon end of the tRNAs remain positioned essentially in the P and A sites of the small ribosomal subunit, while the acceptor ends of the tRNA are positioned in the E site and P sites of the large subunit (P/E and A/P states, respectively)10–12 (Figures 1C and 2A). This rotated state of the ribosome is then the substrate for the action of another GTPase, eEF2 (elongation factor G (EFG) in bacteria), which translocates the mRNA–tRNA complex relative to the ribosome and returns the tRNAs to their ‘classical’ states (E/E and P/P)13–17. Although no structures exist for translocation intermediates (eEF2 bound with two translocating tRNAs) in eukaryotes, by analogy to structural data from bacteria, translocation is expected to involve the swiveling of the small subunit head and rotation of the small subunit relative to the large subunit to accompany tRNA movement into the classical E/E and P/P states18–20. In addition to eEF1A and eEF2, a third factor, eEF3, is essential for elongation in fungi, potentially by promoting tRNA release from the E site after translocation21,22. This elongation cycle is repeated until each codon has been translated and a complete protein synthesized.
As the ribosome elongates along an ORF, it can encounter a variety of problematic sequences that slow its progress. First, certain amino acid sequence combinations can stall the ribosome, either because of poor reaction kinetics resulting from the nature of the amino acids themselves or from inhibitory conformations of the nascent peptide in the exit tunnel (Figure 2B). mRNA sequences rich in codons of lowly-expressed cognate tRNAs also pause ribosomes (Figure 2C). In some cases, the order of codons affects how long the ribosome takes to translate them, suggestive of complexity in the interactions of certain tRNAs within the ribosome23 (Figure 2D). Additionally, the ribosome may encounter strong mRNA secondary structures such as stem loops or pseudoknots that can arrest elongation (Figure 2E). In all of these cases, the ribosome can either continue or abort translation. Several of these challenges are corrected simply by waiting – the ribosome waits until the proper factor or tRNA is delivered. In other cases, the ribosome undergoes frameshifting to resume translation in a different frame, where the problematic sequence is no longer translationally relevant. If translation is aborted, ribosomes are recycled by specific machinery (discussed below). Additional emerging evidence suggests that ubiquitylation of ribosomal proteins may affect the fate of ribosome stalling, though the direct mechanistic effects of these modifications remains poorly understood24–27.
Specific amino acid combinations cause ribosome stalling
During elongation the ribosome is faced with the challenge of forming 400 different peptide bonds in its active site, as any of the twenty amino acids can be found on the P- or A-site tRNAs. Unlike many molecular machines that are specific for one substrate, the ribosome must be sufficiently accommodating to allow for reactivity between any of the twenty substrates and to ignore perturbations in peptidyl-tRNA conformation that result from the structure of the newly synthesized peptide. Although the ribosome is capable of making all 400 possible peptide bonds, not all reactions are equally favorable (Figure 2B). For example, proline is unique among the substrates because its reactive amine is found within a five-membered ring, making it a secondary amine with substantially reduced nucleophilicity. This makes Pro-tRNA a poor peptidyl acceptor in the A site28,29. Additionally, due to entropic constraints, proline is not an ideal donor substrate in the P site either. As a result of these different negative contributions to catalysis, the synthesis of Pro–Pro bonds is slow and the addition of a third proline is even more challenging for the ribosome30–33.
Although poly-proline formation is kinetically slow, stretches of proline are found throughout eukaryotic genomes, and the encoded proteins are indeed translated. Initial work in the bacterium Escherichia coli identified the ribosome-interacting protein elongation factor P to be essential for resolving translation stalling at proline stretches30,32,34. In eukaryotes, the homologous factor eIF5A is a highly abundant35 and essential protein36 that is comprised of only 157 amino acids and contains a unique post-translational modification called hypusine [G] 37. Early fractionation experiments showed that eIF5A stimulated a model translation initiation reaction (formation of Met–Puromycin bond)38–40. In retrospect, this biochemical assay more simply reports on peptide-bond formation than true initiation, and multiple studies have suggested that the predominant role of eIF5A in translation is promoting elongation31,41–43. Indeed, following the lead from the work on elongation factor P, eIF5A was shown to be crucial for the eukaryotic ribosome to overcome the challenges of Pro–Pro bond formation31. This activity depends on its unique hypusine modification31 and several high-resolution structures have provided initial mechanistic details of eIF5A binding in the ribosomal E site44,45 (Figure 1E). These structures reveal that eIF5A binding is incompatible with the presence of an E site tRNA, suggesting that the tRNA must be released before eIF5A can bind. One of these studies was able to capture an eIF5A–80S complex with a P-site tRNA present, thereby revealing interactions between the hypusine moiety of eIF5A and the phosphate backbone of the A76 nucleotide at the tRNA 3′ (CCA)-end in the peptidyl-transferase center [G]45. These findings suggested that eIF5A promotes peptide-bond formation by stabilizing the conformation of the peptidyl-tRNA for nucleophilic attack by the aminoacyl-tRNA in the A site (Figures 1D–1E).
Although these studies had clearly identified a crucial role for eIF5A in translation elongation, stretches of three or more consecutive proline codons are found in as few as 10% of yeast genes and thus it was not clear whether this was a complete description of the role of this essential and extremely abundant factor31. More recent approaches to define the in vivo function of eIF5A by unbiased methods including ribosome profiling [G]46 and 5PSeq [G]47 revealed considerable pausing at a wide spectrum of amino acid motifs, including those containing proline, aspartic acid, glycine, alanine, valine, and isoleucine. Moreover, experiments using an in vitro reconstituted translation system showed that eIF5A is crucial for stimulating peptide-bond formation on a broad range of amino acid combinations (seven of eight tested combinations showed stimulation by eIF5A, all except for Phe–Phe)46. Given its high concentration (>273,000 molecules of eIF5A per cell in S. cerevisiae35, which is comparable to 630,000 molecules of eEF1A35, 181,000 molecules of eEF235, and 187,000 ribosomes48 per cell) and high affinity for ribosomes49, eIF5A likely contributes to most (if not all) peptidyl transfer events during translation. These data together argue that eIF5A is a global, core translation factor that functions during the formation of each peptide bond to increase the processivity and efficiency of translation elongation (Figure 2A).
These findings also suggest that the E site has a crucial role as a sensor of ribosome elongation kinetics. Following peptide-bond formation and translocation, the deacylated tRNA moves to the ribosomal E site, the new peptidyl-tRNA moves from the A site to the P site, and the A site is made available for a new aminoacyl-tRNA. It is presumed that upon accommodation of the next aminoacyl-tRNA into the ribosome, the deacylated tRNA is released from the E site, as is the case in in bacterial ribosomes50. Thus, when peptide-bond formation is slow (such as for Pro-Pro), the aminoacyl-tRNA would initially remain unreacted, but the E site would be predicted to be available for eIF5A binding as the E-site tRNA naturally dissociates (Figure 1). Thus, an unoccupied E site effectively functions as a sensor for translation arrest and eIF5A binding as the response to this condition.
Related ideas that were suggested a number of years ago in the so-called allosteric three-site model have argued that E-site occupancy directly affects translation fidelity in the A site51, though this hypothesis has remained controversial (reviewed in8). More recent observations using biochemistry and single-molecule Förster resonance energy transfer [G] (FRET) experiments, mostly in bacteria, have renewed the interest in mechanisms of communication between E-site binding and A-site reactivity13,52,53. In particular, a recent study observed that dissociation of the E-site tRNA is necessary for conformational changes prior to translocation to the next codon52. Taken together, there is good reason to assume that the E site of the ribosome is a crucial modulator of translation elongation kinetics; further investigation will no doubt lead to increased understanding of the precise mechanisms and effects of this communication.
Codon-choice affects ribosome stalling
Ribosome stalling can also occur when the ribosome encounters specific rare or suboptimal codons in the ORF (reviewed in54,55). The idea of rare codons has been discussed in the translation field for many years56 and it has been argued that organisms preferentially use certain codons relative to others to encode amino acids at particular positions or in particular genes54–56. The optimality of a particular codon is a reflection of its usage in the transcriptome and the availability of the corresponding tRNA for use by translating ribosomes57–61. Optimal codons have a pool of tRNAs readily available for elongation; conversely, suboptimal codons have a limited supply of the corresponding tRNA for translation56,62. Over the past decade a correlation was identified between the average optimality of codons over an entire mRNA and its translation rate62–65. A related, but distinct problem can result from inefficient aminoacylation [G] by the corresponding aminoacyl-tRNA synthetase and therefore a reduction in the cognate aminoacyl-tRNA level66.
The molecular signature of stalling owing to decreased levels of either tRNA or aminoacylation is a kinetic barrier to translation elongation resulting from an unoccupied A site (Figure 2C). The ribosome may stall until the proper tRNA is delivered to the A site, until a near-cognate tRNA [G] is delivered (miscoding), or until a non-canonical event such as frameshifting occurs67–70 (Figure 2C). And, if all such events fail to occur, the stalled ribosome may be targeted for quality control71. Other studies have identified correlations between the overall ‘optimality’ of an mRNA and its stability, which are likely related to the more drastic quality control mechanisms that are triggered on problematic mRNAs. A more in-depth discussion of these processes can be found elsewhere72.
An interesting recent study identified a surprisingly subtle difficulty in mRNA coding wherein a particular order of codons can be problematic, even when neither codon is rare. In a screen of a randomized GFP library containing random codons at adjacent positions, 17 different codon pairs that inhibit translation were identified23. In some cases, the inhibition could be explained by the low abundance of a particular tRNA or by likely inefficient wobble decoding [G]. However, in other instances, the authors observed that the particular order of the codons in the pair impacts optimal translation (Figure 2D). For example, the codons pair CUC–CCG (encoding Leu–Pro) strongly inhibited translation compared to the optimally Leu–Pro-encoding UUG–CCA; importantly, however, the same two codons in the reverse order CCG–CUC (Pro–Leu) are no longer inhibitory. These observations are suggestive of subtle coordination between the P- and A-site tRNAs during elongation, and highlight the potential complexity of the genetic code.
Ribosome stalling caused by mRNA structures
Ribosome stalling can also result from the formation of substantial secondary structures in mRNAs. The presence of an RNA stem-loop or pseudoknot structure is widely assumed to cause stalling. In some of these cases the ribosomes stall on top of a repetitive, ‘slippery’ sequence, such as AAAAAAG, that in turn promotes ribosome frameshifting73–78 (Figure 2E). Many such sites are the product of natural selection and are referred to as “programmed” ribosomal frameshifting (PRF) sites79. Although pausing at PRF sites is similar to the pauses discussed above, here it is considered advantageous for the desired gene-expression outcome.
Viruses commonly use PRF sites to more efficiently encode genes in their limited genomes. Often, frameshifting events control the ratio of the viral structural and enzymatic proteins being translated, such as in human immunodeficiency virus 1 (HIV-1), where a -1 PRF adjusts the Gag/Gag–Pol expression ratio needed for proper viral particle replication and assembly80,81. The HIV-1 PRF signal is comprised of two parts: an mRNA stem-loop structure and a U-rich slippery sequence74. The structure leads to ribosome stalling directly over the slippery sequence, where slow reaction kinetics increase the likelihood of frameshifting that would permit translation of the downstream, out-of-frame pol gene (Figure 2E). Again, the ribosome is faced with a barrier that impacts the kinetics of elongation (in this case a stem-loop), but in this case, programmed frameshifting enables the ribosome to get past the barrier and continue translating the downstream gene.
Translation termination
The process of translation termination begins when the ribosome encounters a stop codon in the ribosomal A site (Figure 3A–B)82. In eukaryotes, all three stop codons (UAA, UAG, and UGA) are recognized by a single release factor, eRF1. The overall shape and size of eRF1 is strikingly similar to that of a tRNA, as would be expected of a molecule that binds the A site, and it has two distinct functions. First, eRF1 contains a structural Asn–Ile–Lys–Ser (NIKS) motif and several other conserved elements, including the Gly–Thr–Ser (GTS) and YxCxxxF motifs, which recognize with high specificity the three termination codons83–87. Second, eRF1 has a precisely positioned Gly–Gly–Gln (GGQ) motif that extends into the peptidyl-transferase center to promote the release of the nascent peptide88, similarly to the bacterial release factors RF1 and RF289 (Figure 3B–C). Finally, like tRNA, eRF1 requires a specialized GTPase, eRF390, for proper function; eRF3 is most closely related to eEF1A and EFTu91. eRF3 is known to stimulate termination in a GTPase-dependent manner92–94 and may be required for dissociation of eRF1 following peptide release; as such it is crucial when eRF1 is present at sub-stoichiometric levels95. After the eRF1–eRF3 complex engages the ribosome, eRF3 dissociates following GTP hydrolysis (similarly to EFTu), and the GGQ motif of eRF1 is poised to coordinate a water molecule at the peptidyl-transferase center that hydrolyzes the nascent peptide from the peptidyl-tRNA88 (Figure 3C). Upon release of the peptide, the process of termination is complete.
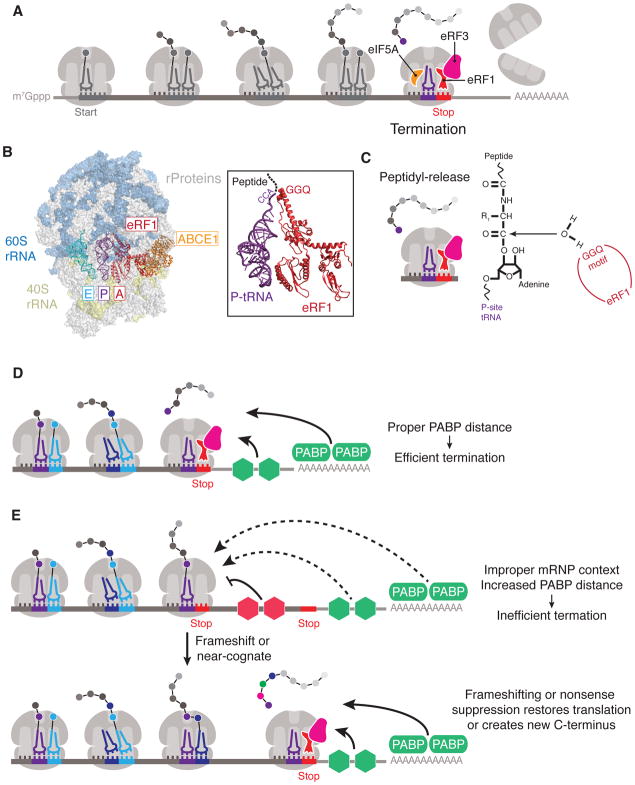
Figure 3. Translation termination and the role of mRNP context.
(A) Overview of translation termination. When the ribosome encounters a termination (stop) codon, eukaryotic peptide chain release factor 1 (eRF1) is delivered by eukaryotic peptide chain release factor 3 (eRF3) to catalyze peptidyl-hydrolysis at the ribosome peptidyl-transferase center. eIF5A binds in the ribosomal E site to stimulate eRF1-mediated hydrolysis. (B) Structure of the 80S–eRF1–ATP-binding cassette sub-family E member 1 (ABCE1) complex (PDB 3JAH) with close-up view showing the GGQ motif of eRF1, which is positioned to coordinate a water molecule for peptidyl-hydrolysis of the P-site peptidyl-tRNA. (C) Schematic of peptidyl-release reaction coordinated by eRF1. (D) Proximity to poly(A)-binding protein (PABP) and other stimulatory RNA-binding proteins (RBPs) can affect translation termination efficiency. (E) Alternatively, if a termination codon is located in a non-ideal mRNP context, far from PABP or stimulatory RBPs, or if certain inhibitory RBPs (colored red) are present near the stop codon, the ribosome may terminate inefficiently. In these cases, the ribosome may undergo frameshifting or incorporate a near-cognate tRNA (orange) to continue translation until a more ideal stop codon is reached.
Although eRF1 is the main catalytic factor of translation termination, other trans-acting factors appear to affect translation termination. ABCE1 (Rli1 in S. cerevisiae) interacts with eRF196–98 to stimulate the catalytic activity (the kcat) of eRF198, and a cryoEM structure of the ABCE1–eRF1–80S complex showed that ABCE1 stabilizes an active eRF1 conformation99 (Figure 3B). We discuss ABCE1 in greater detail below as it is the key catalytic factor for ribosome recycling. Conversely, several components of the multimeric initiation factor eIF3 and its associated factor eIF3j (Hcr1 in S. cerevisiae) promote readthrough at stop codons100,101, which is generally thought to be an indicator of decreased termination efficiency.
Sequence context also affects the efficiency of translation termination. Recent structural studies of 80S–eRF1 complexes revealed interactions between the 18S rRNA and the +4 nucleotide of the termination sequence (the first nucleotide of the 3′ untranslated region (UTR))85,86. These structural observations are consistent with the observation from ribosome profiling that the size of ribosome protected fragments (RPFs) at stop codons is one nucleotide longer than RPFs on sense codons102. Moreover, a role for the +4 nucleotide was suggested years ago using bioinformatics103 and more recently experimentally supported by reporter assays in yeast101,104,105 and mammalian cells106. These studies revealed that particular stop codons and the identity of the +4 nucleotide are more or less likely to promote stop codon readthrough. Weaker termination codons such as UGAC lead to higher levels of readthrough relative to stronger termination codons such as UAAG as measured by readthrough through the incorporation of near-cognate aminoacyl-tRNAs107 or by frameshifting (reviewed in108). Thus, although all three stop codons elicit termination, the +4 nucleotide adds a layer of complexity to the regulation of termination that can affect overall gene expression or create unique carboxy-terminal protein extensions109,110. Emerging evidence suggests that beyond the +4 nucleotide, the sequence context surrounding the termination codon (including in the 3′ UTR)110–114 and elements of the nascent peptide46,115 may also affect termination efficiency.
Translation termination is mechanistically quite similar to translation elongation: in the case of peptidyl-transfer in elongation, a nucleophilic attack on the peptidyl-tRNA occurs from the amino acid conjugated on the incoming A-site tRNA whereas in termination, a nucleophilic attack occurs from the water molecule coordinated by the GGQ motif of eRF1 (Figure 3C). Kinetically, the rate of peptidyl-release is slower than the rate of peptidyl-transfer116, potentially because the water molecule is a worse nucleophile than most amino acids. These in vitro kinetic observations are consistent with the in vivo observation that the accumulation of ribosomes at stop codons in ribosome profiling experiments is typically greater than at an average sense codon (see 46 for example). In a recent study, eIF5A not only strongly enhanced the rate of peptidyl-transfer but also of peptidyl-release46. This is not particularly surprising considering the chemistry of these two events is quite similar (Figures 1D and 3C). Importantly, however, the fact that eIF5A promotes translation termination is an additional evidence for a role for the E site as a sensor of slow ribosome kinetics and as a site through which translation rates can be modulated. In this case, as termination is kinetically slow, the E site is more likely to become unoccupied, and eIF5A could bind and stimulate peptidyl-release.
RNA-binding proteins regulate translation termination
Recent work has begun to shed light on more subtle mechanisms that regulate the termination process. Although the +4 nucleotide involves the direct interaction of the mRNA with the terminating ribosome, the stop codon context could also affect the termination process by recruiting RNA binding proteins that interact with release factors or with the ribosome. Recent work on poly(A)-binding protein (PABP) has suggested that it may influence the efficiency of termination. First, physical interactions between PABP and the amino-terminus of eRF3 have been documented using co-immunoprecipitation and pull-down assays117–119, though the consequence of this interaction has been unclear. And, using an in vitro reconstituted termination system, it was recently shown that PABP can directly promote the recruitment of eRF1–eRF3 to a terminating ribosome120 (Figure 3D). Subsequent biochemical experiments also revealed that PABP stimulated the peptidyl-hydrolysis activity of eRF1–eRF3, though the mechanism is still unknown120.
Additional connections between PABP and translation termination come from investigations of the nonsense-mediated decay (NMD) pathway. NMD is a quality control pathway that selectively degrades mRNAs with premature termination codons (PTCs). This pathway is likely crucial as it minimizes the production of truncated and often deleterious proteins. A reporter-based assay in which PABP was artificially tethered to mRNA at various positions found that the proximity of PABP to a PTC decreased the efficiency of NMD, possibly by enhancing the efficiency of termination or ribosome recycling121. Although the precise connections between termination and NMD remain elusive, these findings suggest that the proximity of PABP or other mRNA-binding proteins could affect termination rates and therefore mRNA decay (Box 1).
Box 1. Connections between nonsense-mediated decay and mRNP context in termination and recycling.
In eukaryotes, the nonsense-mediated decay (NMD) pathway selectively degrades mRNAs that contain what is broadly referred to as a premature termination codon (PTC) dependent on the three conserved UPF proteins: UPF1, UPF2 and UPF3172,173. Such mRNAs are created through multiple routes: genes may carry a mutation that results in a PTC174, inefficient splicing may lead to export of a pre-mRNA with a PTC that is (almost inevitably) encoded in the intron175,176, or the stop codons of upstream open reading frames177,178 and non-coding RNAs179,180 can be sensed as PTCs.
In many eukaryotes, the presence of a termination codon upstream of an exon-junction complex (EJC; a large protein complex deposited at splice junctions) is known to be a strong signal for NMD181. UPF2 interacts with components of the EJC and with UPF1181,182, potentially recruiting UPF1 to a nearby terminating ribosome to signal NMD. However, some organisms that lack an EJC (such as Saccharomyces cerevisiae) have robust NMD, suggesting the NMD machinery can be recruited to PTCs by mechanisms independent of the EJC.
Such a more general mechanism of NMD may in part be mediated by the proximity of poly(A)-binding protein (PABP) to the stop codon. In budding yeast, for example, an experiment tethering PABP to an mRNA at various sites found that the proximity of PABP to a PTC affected the efficiency of NMD, likely by impacting the efficiency of translation termination or ribosome recycling121. As PABP stimulates translation termination directly120, it is possible that the distance between the PTC and PABP can determine whether the mRNA undergoes NMD or not. Indeed, PTCs located near the 3′-end of open-reading frames are weaker substrates for NMD183.
A broad and simple view might be that the efficiency of termination and/or recycling is crucial for distinguishing between authentic and premature stop codons. And, the context of a stop codon is crucial in defining the efficiency of the termination or recycling process. This context may include proximal (or more distal) mRNA sequences or the constellation of RNA binding proteins (both stimulatory and inhibitory) that are recruited to them.

The idea that the mRNA context can affect translation termination has recently been supported by the discovery that some ciliates and trypanosomes can use the three canonical stop codons to encode amino acids (sense) at some sites122–124. In some of these organisms, RNA sequencing readily identified novel tRNAs with anticodons that directly pair with the stop codons (historically known as suppressor tRNAs125). Further work that included ribosome profiling approaches led to a model wherein stop codons are recognized by aminoacyl-tRNA as substrates by default unless they are located sufficiently close to the poly(A) tail, in which case they are recognized by the canonical eRFs (Figures 3D–E)122,123; a termination-stimulatory role for PABP would fit nicely into such a model. Other ciliates appear to contain large numbers of in-frame stop codons that are preceded by a slippery sequence that promotes frameshifting126. Again, in these cases, frameshifting outcompetes relatively inefficient termination when the stop codon is in a relatively upstream position, but termination dominates when the stop codon is sufficiently close to the poly(A) tail. Support for this polyA-proximity-model comes from the observation that mRNAs in these organisms typically contain short (or nonexistent) 3′ UTRs122,123. Because of inherent experimental difficulties in working with these unusual organisms, a direct connection to PABP in vivo has not yet been demonstrated.
In addition to PABP other mRNA-binding proteins might have a role in termination, especially considering the great number of mRNA-binding proteins127 and that many have no determined functions. One such example is heterogeneous nuclear ribonucleoprotein A2/B1, which promotes readthrough of the vascular endothelial growth factor A mRNA to create an isoform with a unique carboxy-terminus128. Additionally, several mRNA-binding proteins that have been implicated in the NMD pathway, including Hrp1129 and Pub1130 in budding yeast and APOBEC1 complementation factor131 and polypyrimidine tract-binding protein 1132 in mammalian cells, are potentially direct regulators of the translation machinery and thus could positively or negatively regulate translation termination.
Ribosome recycling
At the end of translation termination, the 80S complex containing a deacylated-tRNA in the P site must be recycled into its 40S and 60S subunits (Figure 4A). Subunit recycling is accomplished by ABCE1, an essential protein in all eukaryotes that contains two nucleotide-binding domains (NBDs) and an amino-terminal iron–sulfur (Fe–S) cluster133. Using force generated by ATP-binding and hydrolysis, ABCE1 dissociates the post-termination ribosome into the 40S and 60S subunits in collaboration with eRF197,98,134. Findings in S. cerevisiae also suggest that ABCE1–eRF1 interactions couple termination and recycling, since ABCE1 stimulates peptidyl hydrolysis by eRF198. The separated ribosomal subunits are next bound by available initiation factors for a subsequent round of translation initiation135. Following subunit dissociation, the mRNAs and tRNAs must also be removed from the 40S subunit, potentially through the activity of Ligatin (also known as eIF2D) or the related protein complex MCT-1–DENR136,137; we will not further discuss post-subunit dissociation events in this review.
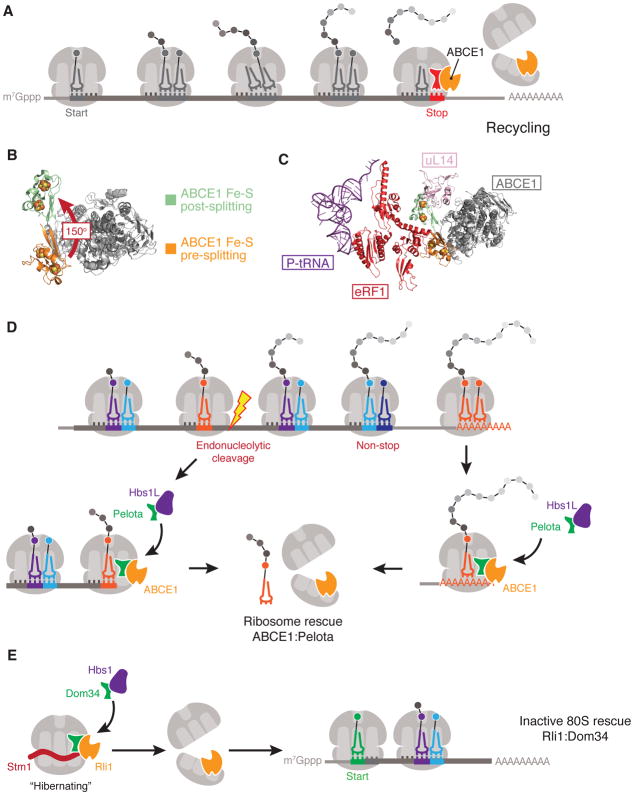
Figure 4. Ribosome recycling and rescue.
(A) Overview of ribosome recycling by ABCE1. ATP-binding cassette sub-family E member 1 (ABCE1) binds to 80S ribosomes loaded with eukaryotic peptide chain release factor subunit 1 (eRF1) and uses the power generated from ATP-binding and hydrolysis to dissociate the ribosomal subunits. ABCE1 remains bound to the 40S subunit to stimulate subsequent translation initiation steps. (B) Superposition of ABCE1 structures in the pre-splitting (PDB 3JAH) and post-splitting (PDB 5LL6) states to highlight the 150-degree rotation of the iron-sulfur (Fe–S) cluster that occurs during ribosomal subunit dissociation. (C) Superposition of ABCE1 structures pre- and post-splitting in the context of eRF1 and ribosomal protein uL14. In the pre-splitting state, the Fe–S cluster of ABCE1 interacts with eRF1. Following splitting, the Fe–S cluster undergoes a dramatic conformational change which drives eRF1 into the inter-subunit space of the ribosome to promote subunit dissociation. In the post-splitting state the Fe–S cluster would clash with 60S ribosomal protein uL14, thereby preventing 60S rejoining and effectively completing the recycling reaction. (D) Dom34–Hbs1 in coordination with ABCE1 rescues stalled ribosomes at the truncated 3′-ends of mRNAs resulting from endo- or exo-nucleolytic cleavage, or those found translating the poly(A) tail. (E) Inactive or ‘hibernating’ Stm1-bound 80S ribosomes in Saccharomyces cerevisiae can be rescued by Dom34–Hbs1 in coordination with ABCE1 to re-enter the cytoplasmic pool of translating ribosomes.
Ribosome profiling experiments in ABCE1-depleted yeast have confirmed its role in ribosome recycling in vivo and highlight the consequences to the cell when recycling is lost138. Upon ABCE1 depletion, increased ribosome occupancy was observed both at the stop codon and in the 3′ UTR, suggestive of defects in termination and/or recycling. Additional experiments using amino acid starvation and reporter constructs revealed that, at least in some cases, the ribosomes in the 3′ UTR were actively translating, having re-initiated translation close to the stop codon of the main ORF.
Before ABCE1 was shown to function in recycling, ABCE1 was initially associated with translation initiation. Work from several labs showed that ABCE1 co-immunoprecipitates with several initiation factors139–141, it was found to sediment with 40S subunits in a sucrose gradient in a manner dependent on its ATPase activity139,141, and genetic depletion of ABCE1 in yeast impaired assembly of pre-initiation complexes as determined by polysome analysis141. As the process of ribosome recycling happens immediately prior to translation initiation in the translation cycle, it is likely that the early and later studies are reconciled by ABCE1 having an important role in connecting the sequential processes of recycling and initiation.
Cryo-EM studies of the structure of ABCE1 bound to 80S ribosomes85,99 and to 40S subunits142 provide a framework for elucidating how ABCE1 may function to stimulate ribosome recycling and recruit translation initiation machinery for the next round of translation. In the 80S–ABCE1 pre-splitting structure, the ABCE1 Fe–S cluster is positioned in the ribosomal A site, making direct contacts with the eRF1 carboxy-terminal domain85,99, whereas in the 40S–ABCE1 post-splitting structure, the Fe–S cluster of ABCE1 adopts a completely different conformation, rotated approximately 150 degrees from the pre-splitting state142 (Figure 4B). Superposition of these structures suggests that, during splitting, movement of the Fe–S domain (coordinated by ATP binding and/or hydrolysis) forces eRF1 deeper into the ribosome inter-subunit space, leading to dissociation of the subunits. After splitting, the Fe–S cluster would clash with the protein uL14 of the 60S subunit, thereby preventing 60S rejoining and ensuring the irreversibility of recycling (Figure 4C).
ABCE1 was also serendipitously visualized in a study reporting the structure of a 48S pre-initiation complex from mammalian cells143–145. Although the ABCE1 density in this structure was originally attributed to eIF3i and eIF3g, this interpretation has since been corrected to ABCE1. Importantly, this structure also includes other members of the pre-initiation complex, including subunits of eIF2 and eIF3, thereby providing some sense as to how ABCE1 might have a dual-role in promoting ribosome recycling and translation initiation through initial interactions with the 80S ribosome post-termination complex and later interactions with the 40S subunit and initiation factors. Further mechanistic and structural studies will be required to fully decipher these connections.
Ribosome rescue
As the ribosome regularly pauses in response to obstacles that it encounters, we suspect that in most cases, the pauses are resolved in a productive manner. We also know, however, that sometimes such mechanisms fail and that in these cases, ribosome rescue may be required as a last resort. In eukaryotes, there is a complex set of machineries that target these aberrant ribosome complexes to degrade the nascent polypeptides and problematic mRNAs, and to rescue the trapped ribosomes146,147. Here we will focus on ribosome rescue and its mechanism, the details of which substantially overlap with ribosome recycling.
Early genetic studies in budding yeast identified Dom34 (Pelota in other eukaryotes) to be crucial for the selective degradation of a stem-loop-containing mRNA and coined the phenomenon no-go decay (NGD)148. NGD is broadly considered to be an mRNA quality control pathway that degrades mRNAs with sequence features that inhibit translation, including mRNA secondary structures, stretches of non-optimal codons or truncated mRNAs148. Dom34 is structurally similar to eRF1149,150 with three important exceptions: it does not contain the NIKS motif-containing domain that recognizes stop codons, it does not contain the GGQ motif that catalyzes peptidyl-hydrolysis, and it is delivered to the ribosome by a distinct GTPase called Hbs1151,152. We note that there are additional isoforms153,154 and homologs155 of Hbs1 in higher eukaryotes that may add further diversity to this system.
The structural similarities with eRF1 suggested a direct role for Dom34 on the ribosome (rather than downstream in mRNA decay148), but likely a somewhat distinct role given the key differences outlined above. Indeed, biochemical work using an in vitro reconstituted translation system showed that Dom34 and its associated GTPase Hbs1 dissociates ribosome subunits independently of peptidyl-release and independently of the identity of the codon positioned in the A site156 (Figure 4D). Additionally, Dom34–Hbs1 appears to preferentially dissociate ribosome complexes carrying mRNA species truncated at their 3′ end98,157. Importantly, whereas Dom34 on its own is sufficient to promote subunit dissociation in vitro156, the recycling factor ABCE1 appears to greatly enhance the rate of subunit dissociation with both eRF1 and Dom3497,98,157.
Though these biochemical studies suggested a role for Dom34 in ribosome rescue, the in vivo targets of this function were unknown. Subsequent experiments using ribosome profiling and reporter constructs in yeast revealed that their natural targets include several distinct ribosome species that accumulate in sometimes unanticipated places. First, Dom34 rescues un-recycled ribosomes stranded in the 3′ UTR138,158 or those that have translated into the poly(A) tail (non-stop decay (NSD)) and have stalled there159,160 (Figure 4D). Additionally, Dom34 appears to rescue stalled ribosomes found at the end of truncated mRNA species generated as a result of endo- (or exo-) nucleolytic cleavage, which can result from a variety of stalling events, including owing to rare codons and stem loops148,160, NSD159,160 or as a part of the unfolded protein response [G]158,161.
Overall, these data suggest a general role for Dom34 in removing ribosomes that translate into problematic regions. Importantly, some internal ribosome-stalling sequences may not depend on Dom34-mediated rescue, including those caused by histidine starvation158, CGA-codon-induced stalling152, or stalling at poly-Arg sequences162. Further exploration with controlled reporter genes in specific genetic backgrounds will be required to completely define the specificities of Dom34 rescue in vivo.
Given its broad function in rescuing ribosomes, Dom34 is crucial in maintaining ribosome homeostasis. The absence of Dom34 was first identified in a genome-wide screen in yeast to exacerbate growth defects arising from ribosomal protein haploinsufficiency163. More recent experiments using both in vivo and in vitro methods showed that Dom34–Hbs1 in concert with ABCE1 recycle inactive, ‘hibernating’ 80S ribosomes bound by Stm1 thereby maintaining the pool of translation-competent ribosomes (Figure 4E)164. In S. cerevisiae Stm1 associates with non-translating 80S ribosomes following nutrient deprivation and has a crucial role in restoring translation after the stress is removed165–167. Structural analysis of a Stm1-bound 80S ribosome revealed that Stm1 binds at the mRNA entry channel where it interacts with both the 40S and 60S subunits to lock this translationally-inhibited state168; the activity of Dom34-Hbs1 is required to dissociate the subunits thereby allowing them to return to translation.
These findings are consistent with recent observations in certain blood cell lineages169,170. In particular, a recent study of human erythroid cells argued that unusual regulation of Dom34 (PELO in humans) expression compensated for cellular defects in ribosome recycling resulting from loss of ABCE1170 (Box 2). This was important in revealing a dynamic regulation of the rescue and recycling factors in order to regulate the availability of ribosomes in this biological system. The implications of these results for the class of diseases known as ribosomopathies [G]171 will be interesting to explore further. What these and other studies suggest is that ribosome rescue and recycling are very important for the maintenance of ribosome homeostasis in all cell types155,163,170.
Box 2. Implications of ribosome recycling defects for human disease.
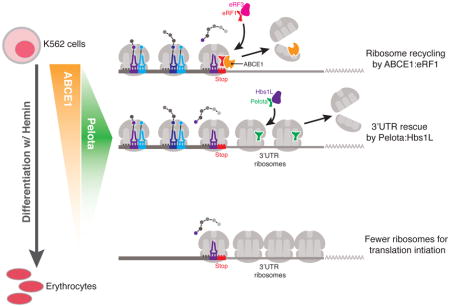
A recent study using a human erythroleukemia cell line discovered a dynamic regulation of the ribosome rescue factors ABCE1 and Dom34-Hbs1 (PELO-HBS1L in humans) that resulted in reprogramming of the cellular translation machinery170. During the hemin–mediated differentiation of K562 cells into erythrocytes, the levels of ABCE1 gradually decline, concomitant with an increase in unrecycled ribosomes at the mRNA 3′ untranslated regions (UTRs) (see the figure). During the initial phase of ABCE1 loss, sharp increase in the levels of the ribosome rescue factor PELO were observed at the exact point of the induction of hemoglobin expression. Following this point, PELO levels decreased and ribosomes began to substantially accumulate at the 3′ UTRs of all the mRNAs in the cell. At this stage, without ABCE1 or PELO to recycle or rescue ribosomes, the cells experienced a global decrease in ribosome availability, which ultimately led to a global translation defect and a trend toward decreased hemoglobin output.
These findings improve our understanding of ribosomopathies, which comprise a heterogeneous set of diseases that result from perturbations in ribosome homeostasis, such as the loss of a ribosomal protein, and in humans most typically present as hematopoietic dysfunction184. Diamond-Blackfan Anemia is a well-documented ribosomopathy that results from the heterozygous loss of any of a number of ribosomal proteins, and is associated with a reduced erythroid progenitor cell population185. The same study that identified a dynamic regulation of ribosome rescue factors in the differentiating K562 cells also explored the connection between ribosome rescue factors and the loss of 40S ribosomal protein S19 (RPS19)170. Importantly, overexpression of PELO–HBS1L was sufficient to rescue defects in hemoglobin synthesis that resulted from RPS19 depletion. Together these results are consistent with the idea that careful regulation of ribosome rescue factors is important for maintaining a cytoplasmic pool of active ribosomes that is suitable for supporting translation homeostasis.
Conclusions and future perspective
Although the ribosome was discovered more than sixty years ago and the genetic code deciphered in the 1960s, a host of new data underscores the complexity of translation and the possible layers of regulation of gene expression that exist beyond the simple three-letter codon table. As the ribosome proceeds through the phases of translation (Figure 1), it encounters many roadblocks that can have a profound impact on the translational output of a particular mRNA. Although these problematic encounters are diverse in nature, in each case the subsequent steps of translation are affected and so the ribosome must either continue or abort translation. The different outcomes can have a dramatic effect on global gene expression and thus on the physiology of the organism.
During translation elongation (Figure 2), the ribosome’s task is to synthesize the mRNA-encoded polypeptide, even when particular sequences of amino acids, codons, or interactions between tRNAs inhibit elongation kinetics. In some cases, the cell has evolved intricate machinery to promote faster translation kinetics (eIF5A, for example) or to permit ribosome frameshifting to resume proper translation, but in other cases the cellular mechanism that resolves these kinetic defects remains unknown. For instance, the discovery of specific tRNA–codon pairs that inhibit translation elongation remains quite puzzling. What about these particular pairs elicits a translation arrest by the ribosome? And why does the order of the tRNA–codon pair matter? Moving forward it will be important to improve our understanding of the communication between the ribosomal P and A sites during elongation, as mediated by the codons, tRNAs, and active site of the ribosome. Additionally, the role of the ribosomal E site remains to be fully defined. The general stimulatory factor eIF5A binds the unoccupied ribosome E site to stimulate peptide-bond formation when peptidyl-transfer kinetics are slow between the P- and A-site tRNAs. Thus, the occupancy of the E site is likely to be a key indicator of the translation elongation rate at a particular site in the coding sequence. Further investigation into the coordination of the E, P, and A sites using careful biochemical or single-molecule approaches should provide a deeper understanding of the dynamics of ribosome pauses during translation elongation and their influence on gene expression.
Translation termination (Figure 3) is also a rich area for continued exploration. What are the determinants of stop codon recognition beyond the codons themselves? Recent studies in organisms that use termination codons for both coding and termination functions (sense and nonsense) increased the interest in models where termination is guided by the mRNP context near the termination codon. One potential candidate for directly stimulating termination is PABP120. However, although we have begun to characterize the diverse collection of mRNA binding proteins using computational and biochemical approaches127, we still have little idea of the composition of a single mRNP and how this composition varies on mRNAs encoding different genes (or even the same gene). More relevant to our interests here, we have no idea how this mRNP context might influence the actions of the ribosome and translation factors. A more thorough investigation using both proteomic and single-molecule approaches to define an mRNP-code will provide insight into the communication between the ribosome, translation factors, and mRNPs as they relate to translation.
Finally, although the last phase of translation, ribosome recycling (Figure 4), is reasonably well-determined biochemically and structurally, potential connections between recycling and translation initiation through ABCE1 remain particularly intriguing. Although there is some evidence to suggest a role for ABCE1 in connecting these phases of translation, how ABCE1 works to recruit initiation factors and promote pre-initiation complex assembly remains to be determined. Additionally, further dissection of the connections between the activity of recycling factors, the availability of translation-competent ribosomes and gene expression patterns (that is, how ribosome concentration affects gene-specific expression) will be of great importance to better understand the etiology of ribosomopathies. Given all these areas of further investigation and the constant discovery of new regulatory factors and sequence elements that modulate translation, the field will continue to be an exciting area for scientific exploration.
Glossary
- Ribosome rescue
Mechanism of ribosomal subunit dissociation when translation stalls cannot be resolved to return to productive translation.
- P site
‘Peptidyl’ site of the ribosome, which binds the tRNA attached to the elongating peptide chain.
- Aminoacyl-tRNAs
tRNA molecules charged with their corresponding amino acids.
- E site
‘Exit’ site of the ribosome, which binds the de-acylated (uncharged) tRNA after translocation and before this tRNA dissociates from the ribosome.
- A site
‘Aminoacyl’ site of the ribosome, which binds the incoming aminoacyl-tRNA for peptide-bond formation.
- Peptidyl-tRNA
tRNA molecule covalently attached to the nascent polypeptide.
- Hypusine
A unique lysine post-translational modification of eIF5A that is crucial for its translation elongation and termination functions.
- Peptidyl-transferase center
The active site of the ribosome where peptide-bond formation and peptide release occur during translation.
- Ribosome profiling
High-throughput sequencing method that provides a global snapshot of translating ribosomes in the cell by mapping mRNA fragments protected by ribosomes.
- 5PSeq
Technique to identify mRNAs that contain phosphorylated 5′-ends resulting from exonucleolytic (5′ to 3′) mRNA degradation pathways.
- Förster resonance energy transfer (FRET)
Biophysical technique to monitor molecular dynamics that relies on energy transfer between two fluorescent molecules located close in space.
- Aminoacylation
Chemical reaction whereby a tRNA molecule is ‘charged’ with its corresponding amino acid.
- Near-cognate tRNA
tRNA molecule that is not a perfect match to the codon; usually unmatched at one position of the codon.
- Wobble decoding
Codon–anticodon interactions typically at the third (3′) position of the codon that do not follow strict Watson–Crick base pairing rules.
- Unfolded protein response
Cellular stress response that is activated upon the accumulation of unfolded or misfolded proteins in the endoplasmic reticulum.
- Ribosomopathies
Broad group of human disorders resulting from ribosomal protein haploinsufficiency or defects in ribosome biogenesis.
References
- 1.Hinnebusch AG. The scanning mechanism of eukaryotic translation initiation. Annual review of biochemistry. 2014;83:779–812. doi: 10.1146/annurev-biochem-060713-035802. [DOI] [PubMed] [Google Scholar]
- 2.Hinnebusch AG, Lorsch JR. The mechanism of eukaryotic translation initiation: new insights and challenges. Cold Spring Harbor perspectives in biology. 2012;4:a011544. doi: 10.1101/cshperspect.a011544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jackson RJ, Hellen CU, Pestova TV. The mechanism of eukaryotic translation initiation and principles of its regulation. Nature reviews. Molecular cell biology. 2010;11:113–127. doi: 10.1038/nrm2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009;136:731–745. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carvalho MD, Carvalho JF, Merrick WC. Biological characterization of various forms of elongation factor 1 from rabbit reticulocytes. Archives of biochemistry and biophysics. 1984;234:603–611. doi: 10.1016/0003-9861(84)90310-2. [DOI] [PubMed] [Google Scholar]
- 6.Fischer N, et al. Structure of the E. coli ribosome-EF-Tu complex at <3 A resolution by Cs-corrected cryo-EM. Nature. 2015;520:567–570. doi: 10.1038/nature14275. [DOI] [PubMed] [Google Scholar]
- 7.Shao S, et al. Decoding Mammalian Ribosome-mRNA States by Translational GTPase Complexes. Cell. 2016;167:1229–1240. doi: 10.1016/j.cell.2016.10.046. High-resolution cryoEM structures of mammalian ribosomes bound by factors involved in translation elongation, termination, and ribosome rescue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zaher HS, Green R. Fidelity at the molecular level: lessons from protein synthesis. Cell. 2009;136:746–762. doi: 10.1016/j.cell.2009.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beringer M, Rodnina MV. The ribosomal peptidyl transferase. Molecular cell. 2007;26:311–321. doi: 10.1016/j.molcel.2007.03.015. [DOI] [PubMed] [Google Scholar]
- 10.Behrmann E, et al. Structural snapshots of actively translating human ribosomes. Cell. 2015;161:845–857. doi: 10.1016/j.cell.2015.03.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Budkevich T, et al. Structure and dynamics of the mammalian ribosomal pretranslocation complex. Molecular cell. 2011;44:214–224. doi: 10.1016/j.molcel.2011.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moazed D, Noller HF. Intermediate states in the movement of transfer RNA in the ribosome. Nature. 1989;342:142–148. doi: 10.1038/342142a0. [DOI] [PubMed] [Google Scholar]
- 13.Ferguson A, et al. Functional Dynamics within the Human Ribosome Regulate the Rate of Active Protein Synthesis. Molecular cell. 2015;60:475–486. doi: 10.1016/j.molcel.2015.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Spahn CM, et al. Domain movements of elongation factor eEF2 and the eukaryotic 80S ribosome facilitate tRNA translocation. The EMBO journal. 2004;23:1008–1019. doi: 10.1038/sj.emboj.7600102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Taylor DJ, et al. Structures of modified eEF2 80S ribosome complexes reveal the role of GTP hydrolysis in translocation. The EMBO journal. 2007;26:2421–2431. doi: 10.1038/sj.emboj.7601677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ling C, Ermolenko DN. Structural insights into ribosome translocation. Wiley interdisciplinary reviews. RNA. 2016;7:620–636. doi: 10.1002/wrna.1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shoji S, Walker SE, Fredrick K. Ribosomal translocation: one step closer to the molecular mechanism. ACS chemical biology. 2009;4:93–107. doi: 10.1021/cb8002946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ermolenko DN, Noller HF. mRNA translocation occurs during the second step of ribosomal intersubunit rotation. Nature structural & molecular biology. 2011;18:457–462. doi: 10.1038/nsmb.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ramrath DJ, et al. Visualization of two transfer RNAs trapped in transit during elongation factor G-mediated translocation. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:20964–20969. doi: 10.1073/pnas.1320387110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ratje AH, et al. Head swivel on the ribosome facilitates translocation by means of intra-subunit tRNA hybrid sites. Nature. 2010;468:713–716. doi: 10.1038/nature09547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Andersen CB, et al. Structure of eEF3 and the mechanism of transfer RNA release from the E-site. Nature. 2006;443:663–668. doi: 10.1038/nature05126. [DOI] [PubMed] [Google Scholar]
- 22.Triana-Alonso FJ, Chakraburtty K, Nierhaus KH. The elongation factor 3 unique in higher fungi and essential for protein biosynthesis is an E site factor. The Journal of biological chemistry. 1995;270:20473–20478. doi: 10.1074/jbc.270.35.20473. [DOI] [PubMed] [Google Scholar]
- 23.Gamble CE, Brule CE, Dean KM, Fields S, Grayhack EJ. Adjacent Codons Act in Concert to Modulate Translation Efficiency in Yeast. Cell. 2016;166:679–690. doi: 10.1016/j.cell.2016.05.070. Study finding that the order of certain codon pairs inhibits translation, suggesting potential communication between tRNAs inside the ribosome during elongation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Juszkiewicz S, Hegde RS. Initiation of Quality Control during Poly(A) Translation Requires Site-Specific Ribosome Ubiquitination. Molecular cell. 2017;65:743–750e744. doi: 10.1016/j.molcel.2016.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Matsuo Y, et al. Ubiquitination of stalled ribosome triggers ribosome-associated quality control. Nat Commun. 2017;8:159. doi: 10.1038/s41467-017-00188-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sundaramoorthy E, et al. ZNF598 and RACK1 Regulate Mammalian Ribosome-Associated Quality Control Function by Mediating Regulatory 40S Ribosomal Ubiquitylation. Molecular cell. 2017;65:751–760. doi: 10.1016/j.molcel.2016.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Silva GM, Finley D, Vogel C. K63 polyubiquitination is a new modulator of the oxidative stress response. Nature structural & molecular biology. 2015;22:116–123. doi: 10.1038/nsmb.2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pavlov MY, et al. Slow peptide bond formation by proline and other N-alkylamino acids in translation. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:50–54. doi: 10.1073/pnas.0809211106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wohlgemuth I, Brenner S, Beringer M, Rodnina MV. Modulation of the rate of peptidyl transfer on the ribosome by the nature of substrates. The Journal of biological chemistry. 2008;283:32229–32235. doi: 10.1074/jbc.M805316200. [DOI] [PubMed] [Google Scholar]
- 30.Doerfel LK, et al. EF-P is essential for rapid synthesis of proteins containing consecutive proline residues. Science. 2013;339:85–88. doi: 10.1126/science.1229017. [DOI] [PubMed] [Google Scholar]
- 31.Gutierrez E, et al. eIF5A promotes translation of polyproline motifs. Molecular cell. 2013;51:35–45. doi: 10.1016/j.molcel.2013.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ude S, et al. Translation Elongation Factor EF-P Alleviates Ribosome Stalling at Polyproline Stretches. Science. 2013;339:82–85. doi: 10.1126/science.1228985. [DOI] [PubMed] [Google Scholar]
- 33.Woolstenhulme CJ, et al. Nascent peptides that block protein synthesis in bacteria. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:E878–887. doi: 10.1073/pnas.1219536110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Woolstenhulme CJ, Guydosh NR, Green R, Buskirk AR. High-precision analysis of translational pausing by ribosome profiling in bacteria lacking EFP. Cell reports. 2015;11:13–21. doi: 10.1016/j.celrep.2015.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kulak NA, Pichler G, Paron I, Nagaraj N, Mann M. Minimal, encapsulated proteomic-sample processing applied to copy-number estimation in eukaryotic cells. Nature methods. 2014;11:319–324. doi: 10.1038/nmeth.2834. [DOI] [PubMed] [Google Scholar]
- 36.Schnier J, Schwelberger HG, Smit-McBride Z, Kang HA, Hershey JW. Translation initiation factor 5A and its hypusine modification are essential for cell viability in the yeast Saccharomyces cerevisiae. Molecular and cellular biology. 1991;11:3105–3114. doi: 10.1128/mcb.11.6.3105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Park MH, Cooper HL, Folk JE. Identification of hypusine, an unusual amino acid, in a protein from human lymphocytes and of spermidine as its biosynthetic precursor. Proceedings of the National Academy of Sciences of the United States of America. 1981;78:2869–2873. doi: 10.1073/pnas.78.5.2869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Benne R, Hershey JW. The mechanism of action of protein synthesis initiation factors from rabbit reticulocytes. The Journal of biological chemistry. 1978;253:3078–3087. [PubMed] [Google Scholar]
- 39.Kemper WM, Berry KW, Merrick WC. Purification and properties of rabbit reticulocyte protein synthesis initiation factors M2Balpha and M2Bbeta. The Journal of biological chemistry. 1976;251:5551–5557. [PubMed] [Google Scholar]
- 40.Schreier MH, Erni B, Staehelin T. Initiation of mammalian protein synthesis. I. Purification and characterization of seven initiation factors. Journal of molecular biology. 1977;116:727–753. doi: 10.1016/0022-2836(77)90268-6. [DOI] [PubMed] [Google Scholar]
- 41.Gregio AP, Cano VP, Avaca JS, Valentini SR, Zanelli CF. eIF5A has a function in the elongation step of translation in yeast. Biochemical and biophysical research communications. 2009;380:785–790. doi: 10.1016/j.bbrc.2009.01.148. [DOI] [PubMed] [Google Scholar]
- 42.Henderson A, Hershey JW. Eukaryotic translation initiation factor (eIF) 5A stimulates protein synthesis in Saccharomyces cerevisiae. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:6415–6419. doi: 10.1073/pnas.1008150108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Saini P, Eyler DE, Green R, Dever TE. Hypusine-containing protein eIF5A promotes translation elongation. Nature. 2009;459:118–121. doi: 10.1038/nature08034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Melnikov S, et al. Crystal Structure of Hypusine-Containing Translation Factor eIF5A Bound to a Rotated Eukaryotic Ribosome. Journal of molecular biology. 2016;428:3570–3576. doi: 10.1016/j.jmb.2016.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schmidt C, et al. Structure of the hypusinylated eukaryotic translation factor eIF-5A bound to the ribosome. Nucleic acids research. 2016;44:1944–1951. doi: 10.1093/nar/gkv1517. High-resolution cryoEM structure of yeast 80S ribosome bound by eIF5A reveals mechanistic information regarding the relevance of eIF5A in global translation elongation and termination. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schuller AP, Wu CC, Dever TE, Buskirk AR, Green R. eIF5A Functions Globally in Translation Elongation and Termination. Molecular cell. 2017;66:194–205. doi: 10.1016/j.molcel.2017.03.003. Characterization of eIF5A as a general translation factor involved in both elongation and termination using a combination of ribosome profiling and in vitro reconstituted biochemistry. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pelechano V, Alepuz P. eIF5A facilitates translation termination globally and promotes the elongation of many non polyproline-specific tripeptide sequences. Nucleic acids research. 2017;45:7326–7338. doi: 10.1093/nar/gkx479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.von der Haar T. A quantitative estimation of the global translational activity in logarithmically growing yeast cells. BMC Syst Biol. 2008;2:87. doi: 10.1186/1752-0509-2-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rossi D, et al. Evidence for a Negative Cooperativity between eIF5A and eEF2 on Binding to the Ribosome. PloS one. 2016;11:e0154205. doi: 10.1371/journal.pone.0154205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gnirke A, Geigenmuller U, Rheinberger HJ, Nierhaus LH. The allosteric three-site model for the ribosomal elongation cycle. Analysis with a heteropolymeric mRNA. The Journal of biological chemistry. 1989;264:7291–7301. [PubMed] [Google Scholar]
- 51.Nierhaus KH. The allosteric three-site model for the ribosomal elongation cycle: features and future. Biochemistry. 1990;29:4997–5008. doi: 10.1021/bi00473a001. [DOI] [PubMed] [Google Scholar]
- 52.Choi J, Puglisi JD. Three tRNAs on the ribosome slow translation elongation. Proceedings of the National Academy of Sciences of the United States of America. 2017;114:13691–13696. doi: 10.1073/pnas.1719592115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zaher HS, Green R. Quality control by the ribosome following peptide bond formation. Nature. 2009;457:161–166. doi: 10.1038/nature07582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Plotkin JB, Kudla G. Synonymous but not the same: the causes and consequences of codon bias. Nature reviews. Genetics. 2011;12:32–42. doi: 10.1038/nrg2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Quax TE, Claassens NJ, Soll D, van der Oost J. Codon Bias as a Means to Fine-Tune Gene Expression. Molecular cell. 2015;59:149–161. doi: 10.1016/j.molcel.2015.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ikemura T, Ozeki H. Codon usage and transfer RNA contents: organism-specific codon-choice patterns in reference to the isoacceptor contents. Cold Spring Harbor symposia on quantitative biology. 1983;47(Pt 2):1087–1097. doi: 10.1101/sqb.1983.047.01.123. [DOI] [PubMed] [Google Scholar]
- 57.dos Reis M, Savva R, Wernisch L. Solving the riddle of codon usage preferences: a test for translational selection. Nucleic acids research. 2004;32:5036–5044. doi: 10.1093/nar/gkh834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pechmann S, Frydman J. Evolutionary conservation of codon optimality reveals hidden signatures of cotranslational folding. Nature structural & molecular biology. 2013;20:237–243. doi: 10.1038/nsmb.2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sharp PM, Li WH. The codon Adaptation Index--a measure of directional synonymous codon usage bias, and its potential applications. Nucleic acids research. 1987;15:1281–1295. doi: 10.1093/nar/15.3.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sabi R, Tuller T. Modelling the efficiency of codon-tRNA interactions based on codon usage bias. DNA Res. 2014;21:511–526. doi: 10.1093/dnares/dsu017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sabi R, Volvovitch Daniel R, Tuller T. stAIcalc: tRNA adaptation index calculator based on species-specific weights. Bioinformatics. 2017;33:589–591. doi: 10.1093/bioinformatics/btw647. [DOI] [PubMed] [Google Scholar]
- 62.Dana A, Tuller T. The effect of tRNA levels on decoding times of mRNA codons. Nucleic acids research. 2014;42:9171–9181. doi: 10.1093/nar/gku646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hussmann JA, Patchett S, Johnson A, Sawyer S, Press WH. Understanding Biases in Ribosome Profiling Experiments Reveals Signatures of Translation Dynamics in Yeast. PLoS genetics. 2015;11:e1005732. doi: 10.1371/journal.pgen.1005732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yu CH, et al. Codon Usage Influences the Local Rate of Translation Elongation to Regulate Co-translational Protein Folding. Molecular cell. 2015;59:744–754. doi: 10.1016/j.molcel.2015.07.018. Study finding that codon choice affects translation elongation kinetics and co-translational folding using a combination of biochemistry and ribosome profiling. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gardin J, et al. Measurement of average decoding rates of the 61 sense codons in vivo. eLife. 2014;3:e03735. doi: 10.7554/eLife.03735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Elf J, Nilsson D, Tenson T, Ehrenberg M. Selective charging of tRNA isoacceptors explains patterns of codon usage. Science. 2003;300:1718–1722. doi: 10.1126/science.1083811. [DOI] [PubMed] [Google Scholar]
- 67.Gallant JA, Lindsley D. Ribosomes can slide over and beyond “hungry” codons, resuming protein chain elongation many nucleotides downstream. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:13771–13776. doi: 10.1073/pnas.95.23.13771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gurvich OL, Baranov PV, Gesteland RF, Atkins JF. Expression levels influence ribosomal frameshifting at the tandem rare arginine codons AGG_AGG and AGA_AGA in Escherichia coli. Journal of bacteriology. 2005;187:4023–4032. doi: 10.1128/JB.187.12.4023-4032.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kane JF. Effects of rare codon clusters on high-level expression of heterologous proteins in Escherichia coli. Current opinion in biotechnology. 1995;6:494–500. doi: 10.1016/0958-1669(95)80082-4. [DOI] [PubMed] [Google Scholar]
- 70.Temperley R, Richter R, Dennerlein S, Lightowlers RN, Chrzanowska-Lightowlers ZM. Hungry codons promote frameshifting in human mitochondrial ribosomes. Science. 2010;327:301. doi: 10.1126/science.1180674. [DOI] [PubMed] [Google Scholar]
- 71.Graille M, Seraphin B. Surveillance pathways rescuing eukaryotic ribosomes lost in translation. Nature reviews. Molecular cell biology. 2012;13:727–735. doi: 10.1038/nrm3457. [DOI] [PubMed] [Google Scholar]
- 72.Hanson G, Coller J. Codon optimality, bias and usage in translation and mRNA decay. Nature reviews. Molecular cell biology. 2018;19:20–30. doi: 10.1038/nrm.2017.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Belew AT, et al. Ribosomal frameshifting in the CCR5 mRNA is regulated by miRNAs and the NMD pathway. Nature. 2014;512:265–269. doi: 10.1038/nature13429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Biswas P, Jiang X, Pacchia AL, Dougherty JP, Peltz SW. The human immunodeficiency virus type 1 ribosomal frameshifting site is an invariant sequence determinant and an important target for antiviral therapy. Journal of virology. 2004;78:2082–2087. doi: 10.1128/JVI.78.4.2082-2087.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Caliskan N, et al. Conditional Switch between Frameshifting Regimes upon Translation of dnaX mRNA. Molecular cell. 2017;66:558–567e554. doi: 10.1016/j.molcel.2017.04.023. [DOI] [PubMed] [Google Scholar]
- 76.Namy O, Moran SJ, Stuart DI, Gilbert RJ, Brierley I. A mechanical explanation of RNA pseudoknot function in programmed ribosomal frameshifting. Nature. 2006;441:244–247. doi: 10.1038/nature04735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yan S, Wen JD, Bustamante C, Tinoco I., Jr Ribosome excursions during mRNA translocation mediate broad branching of frameshift pathways. Cell. 2015;160:870–881. doi: 10.1016/j.cell.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Koutmou KS, et al. Ribosomes slide on lysine-encoding homopolymeric A stretches. eLife. 2015;4:e05534. doi: 10.7554/eLife.05534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Dinman JD. Mechanisms and implications of programmed translational frameshifting. Wiley interdisciplinary reviews. RNA. 2012;3:661–673. doi: 10.1002/wrna.1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jacks T, et al. Characterization of ribosomal frameshifting in HIV-1 gag-pol expression. Nature. 1988;331:280–283. doi: 10.1038/331280a0. [DOI] [PubMed] [Google Scholar]
- 81.Wilson W, et al. HIV expression strategies: ribosomal frameshifting is directed by a short sequence in both mammalian and yeast systems. Cell. 1988;55:1159–1169. doi: 10.1016/0092-8674(88)90260-7. [DOI] [PubMed] [Google Scholar]
- 82.Dever TE, Green R. The elongation, termination, and recycling phases of translation in eukaryotes. Cold Spring Harbor perspectives in biology. 2012;4:a013706. doi: 10.1101/cshperspect.a013706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Song H, et al. The crystal structure of human eukaryotic release factor eRF1--mechanism of stop codon recognition and peptidyl-tRNA hydrolysis. Cell. 2000;100:311–321. doi: 10.1016/s0092-8674(00)80667-4. [DOI] [PubMed] [Google Scholar]
- 84.Frolova L, et al. A highly conserved eukaryotic protein family possessing properties of polypeptide chain release factor. Nature. 1994;372:701–703. doi: 10.1038/372701a0. [DOI] [PubMed] [Google Scholar]
- 85.Brown A, Shao S, Murray J, Hegde RS, Ramakrishnan V. Structural basis for stop codon recognition in eukaryotes. Nature. 2015;524:493–496. doi: 10.1038/nature14896. High-resolution cryoEM structure of a mammalian terminating ribosome reveals the molecular basis of stop codon recognition by eRF1 and the role of the +4 nucleotide. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Matheisl S, Berninghausen O, Becker T, Beckmann R. Structure of a human translation termination complex. Nucleic acids research. 2015;43:8615–8626. doi: 10.1093/nar/gkv909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.des Georges A, et al. Structure of the mammalian ribosomal pre-termination complex associated with eRF1.eRF3.GDPNP. Nucleic acids research. 2014;42:3409–3418. doi: 10.1093/nar/gkt1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Frolova LY, et al. Mutations in the highly conserved GGQ motif of class 1 polypeptide release factors abolish ability of human eRF1 to trigger peptidyl-tRNA hydrolysis. Rna. 1999;5:1014–1020. doi: 10.1017/s135583829999043x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Korostelev AA. Structural aspects of translation termination on the ribosome. Rna. 2011;17:1409–1421. doi: 10.1261/rna.2733411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Frolova L, et al. Eukaryotic polypeptide chain release factor eRF3 is an eRF1- and ribosome-dependent guanosine triphosphatase. Rna. 1996;2:334–341. [PMC free article] [PubMed] [Google Scholar]
- 91.Kong C, et al. Crystal structure and functional analysis of the eukaryotic class II release factor eRF3 from S. pombe. Molecular cell. 2004;14:233–245. doi: 10.1016/s1097-2765(04)00206-0. [DOI] [PubMed] [Google Scholar]
- 92.Alkalaeva EZ, Pisarev AV, Frolova LY, Kisselev LL, Pestova TV. In vitro reconstitution of eukaryotic translation reveals cooperativity between release factors eRF1 and eRF3. Cell. 2006;125:1125–1136. doi: 10.1016/j.cell.2006.04.035. [DOI] [PubMed] [Google Scholar]
- 93.Eyler DE, Green R. Distinct response of yeast ribosomes to a miscoding event during translation. Rna. 2011;17:925–932. doi: 10.1261/rna.2623711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Salas-Marco J, Bedwell DM. GTP hydrolysis by eRF3 facilitates stop codon decoding during eukaryotic translation termination. Molecular and cellular biology. 2004;24:7769–7778. doi: 10.1128/MCB.24.17.7769-7778.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Eyler DE, Wehner KA, Green R. Eukaryotic release factor 3 is required for multiple turnovers of peptide release catalysis by eukaryotic release factor 1. The Journal of biological chemistry. 2013;288:29530–29538. doi: 10.1074/jbc.M113.487090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Khoshnevis S, et al. The iron-sulphur protein RNase L inhibitor functions in translation termination. EMBO Rep. 2010;11:214–219. doi: 10.1038/embor.2009.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Pisarev AV, et al. The role of ABCE1 in eukaryotic posttermination ribosomal recycling. Molecular cell. 2010;37:196–210. doi: 10.1016/j.molcel.2009.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Shoemaker CJ, Green R. Kinetic analysis reveals the ordered coupling of translation termination and ribosome recycling in yeast. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:E1392–1398. doi: 10.1073/pnas.1113956108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Preis A, et al. Cryoelectron microscopic structures of eukaryotic translation termination complexes containing eRF1–eRF3 or eRF1-ABCE1. Cell reports. 2014;8:59–65. doi: 10.1016/j.celrep.2014.04.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Beznoskova P, et al. Translation initiation factors eIF3 and HCR1 control translation termination and stop codon read-through in yeast cells. PLoS genetics. 2013;9:e1003962. doi: 10.1371/journal.pgen.1003962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Beznoskova P, Wagner S, Jansen ME, von der Haar T, Valasek LS. Translation initiation factor eIF3 promotes programmed stop codon readthrough. Nucleic acids research. 2015;43:5099–5111. doi: 10.1093/nar/gkv421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ingolia NT, Lareau LF, Weissman JS. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell. 2011;147:789–802. doi: 10.1016/j.cell.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Brown CM, Stockwell PA, Trotman CN, Tate WP. Sequence analysis suggests that tetra-nucleotides signal the termination of protein synthesis in eukaryotes. Nucleic acids research. 1990;18:6339–6345. doi: 10.1093/nar/18.21.6339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Bonetti B, Fu L, Moon J, Bedwell DM. The efficiency of translation termination is determined by a synergistic interplay between upstream and downstream sequences in Saccharomyces cerevisiae. Journal of molecular biology. 1995;251:334–345. doi: 10.1006/jmbi.1995.0438. [DOI] [PubMed] [Google Scholar]
- 105.Beznoskova P, Gunisova S, Valasek LS. Rules of UGA-N decoding by near-cognate tRNAs and analysis of readthrough on short uORFs in yeast. Rna. 2016;22:456–466. doi: 10.1261/rna.054452.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Floquet C, Hatin I, Rousset JP, Bidou L. Statistical analysis of readthrough levels for nonsense mutations in mammalian cells reveals a major determinant of response to gentamicin. PLoS genetics. 2012;8:e1002608. doi: 10.1371/journal.pgen.1002608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Roy B, Leszyk JD, Mangus DA, Jacobson A. Nonsense suppression by near-cognate tRNAs employs alternative base pairing at codon positions 1 and 3. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:3038–3043. doi: 10.1073/pnas.1424127112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Baranov PV, Atkins JF, Yordanova MM. Augmented genetic decoding: global, local and temporal alterations of decoding processes and codon meaning. Nature reviews. Genetics. 2015;16:517–529. doi: 10.1038/nrg3963. [DOI] [PubMed] [Google Scholar]
- 109.Dunn JG, Foo CK, Belletier NG, Gavis ER, Weissman JS. Ribosome profiling reveals pervasive and regulated stop codon readthrough in Drosophila melanogaster. eLife. 2013;2:e01179. doi: 10.7554/eLife.01179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Schueren F, Thoms S. Functional Translational Readthrough: A Systems Biology Perspective. PLoS genetics. 2016;12:e1006196. doi: 10.1371/journal.pgen.1006196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Mottagui-Tabar S, Tuite MF, Isaksson LA. The influence of 5′ codon context on translation termination in Saccharomyces cerevisiae. European journal of biochemistry. 1998;257:249–254. doi: 10.1046/j.1432-1327.1998.2570249.x. [DOI] [PubMed] [Google Scholar]
- 112.Harrell L, Melcher U, Atkins JF. Predominance of six different hexanucleotide recoding signals 3′ of read-through stop codons. Nucleic acids research. 2002;30:2011–2017. doi: 10.1093/nar/30.9.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Namy O, Hatin I, Rousset JP. Impact of the six nucleotides downstream of the stop codon on translation termination. EMBO Rep. 2001;2:787–793. doi: 10.1093/embo-reports/kve176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Yordanova MM, et al. AMD1 mRNA employs ribosome stalling as a mechanism for molecular memory formation. Nature. 2018;553:356–360. doi: 10.1038/nature25174. [DOI] [PubMed] [Google Scholar]
- 115.Janzen DM, Frolova L, Geballe AP. Inhibition of translation termination mediated by an interaction of eukaryotic release factor 1 with a nascent peptidyl-tRNA. Molecular and cellular biology. 2002;22:8562–8570. doi: 10.1128/MCB.22.24.8562-8570.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.McClary B, et al. Inhibition of Eukaryotic Translation by the Antitumor Natural Product Agelastatin A. Cell Chem Biol. 2017;24:605–613e605. doi: 10.1016/j.chembiol.2017.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Hoshino S, Imai M, Kobayashi T, Uchida N, Katada T. The eukaryotic polypeptide chain releasing factor (eRF3/GSPT) carrying the translation termination signal to the 3′-Poly(A) tail of mRNA. Direct association of erf3/GSPT with polyadenylate-binding protein. The Journal of biological chemistry. 1999;274:16677–16680. doi: 10.1074/jbc.274.24.16677. [DOI] [PubMed] [Google Scholar]
- 118.Ivanov PV, Gehring NH, Kunz JB, Hentze MW, Kulozik AE. Interactions between UPF1, eRFs, PABP and the exon junction complex suggest an integrated model for mammalian NMD pathways. The EMBO journal. 2008;27:736–747. doi: 10.1038/emboj.2008.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Uchida N, Hoshino S, Imataka H, Sonenberg N, Katada T. A novel role of the mammalian GSPT/eRF3 associating with poly(A)-binding protein in Cap/Poly(A)-dependent translation. The Journal of biological chemistry. 2002;277:50286–50292. doi: 10.1074/jbc.M203029200. [DOI] [PubMed] [Google Scholar]
- 120.Ivanov A, et al. PABP enhances release factor recruitment and stop codon recognition during translation termination. Nucleic acids research. 2016;44:7766–7776. doi: 10.1093/nar/gkw635. Identification of a direct role for PABP in stimulating translation termination by increasing the efficiency of recruitment of eRF1–eRF3 to the ribosome in a reconstituted biochemical system. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Amrani N, et al. A faux 3′-UTR promotes aberrant termination and triggers nonsense-mediated mRNA decay. Nature. 2004;432:112–118. doi: 10.1038/nature03060. [DOI] [PubMed] [Google Scholar]
- 122.Heaphy SM, Mariotti M, Gladyshev VN, Atkins JF, Baranov PV. Novel Ciliate Genetic Code Variants Including the Reassignment of All Three Stop Codons to Sense Codons in Condylostoma magnum. Molecular biology and evolution. 2016;33:2885–2889. doi: 10.1093/molbev/msw166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Swart EC, Serra V, Petroni G, Nowacki M. Genetic Codes with No Dedicated Stop Codon: Context-Dependent Translation Termination. Cell. 2016;166:691–702. doi: 10.1016/j.cell.2016.06.020. Study establishing a link between stop codon context, PABP proximity, and the efficiency of translation termination in organisms where all 64 codons can code for amino acids. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Zahonova K, Kostygov AY, Sevcikova T, Yurchenko V, Elias M. An Unprecedented Non-canonical Nuclear Genetic Code with All Three Termination Codons Reassigned as Sense Codons. Current biology. 2016;26:2364–2369. doi: 10.1016/j.cub.2016.06.064. [DOI] [PubMed] [Google Scholar]
- 125.Capecchi MR, Hughes SH, Wahl GM. Yeast super-suppressors are altered tRNAs capable of translating a nonsense codon in vitro. Cell. 1975;6:269–277. doi: 10.1016/0092-8674(75)90178-6. [DOI] [PubMed] [Google Scholar]
- 126.Lobanov AV, et al. Position-dependent termination and widespread obligatory frameshifting in Euplotes translation. Nature structural & molecular biology. 2017;24:61–68. doi: 10.1038/nsmb.3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Castello A, et al. Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell. 2012;149:1393–1406. doi: 10.1016/j.cell.2012.04.031. [DOI] [PubMed] [Google Scholar]
- 128.Eswarappa SM, et al. Programmed translational readthrough generates antiangiogenic VEGF-Ax. Cell. 2014;157:1605–1618. doi: 10.1016/j.cell.2014.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Gonzalez CI, Ruiz-Echevarria MJ, Vasudevan S, Henry MF, Peltz SW. The yeast hnRNP-like protein Hrp1/Nab4 marks a transcript for nonsense-mediated mRNA decay. Molecular cell. 2000;5:489–499. doi: 10.1016/s1097-2765(00)80443-8. [DOI] [PubMed] [Google Scholar]
- 130.Ruiz-Echevarria MJ, Peltz SW. The RNA binding protein Pub1 modulates the stability of transcripts containing upstream open reading frames. Cell. 2000;101:741–751. doi: 10.1016/s0092-8674(00)80886-7. [DOI] [PubMed] [Google Scholar]
- 131.Chester A, et al. The apolipoprotein B mRNA editing complex performs a multifunctional cycle and suppresses nonsense-mediated decay. The EMBO journal. 2003;22:3971–3982. doi: 10.1093/emboj/cdg369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Ge Z, Quek BL, Beemon KL, Hogg JR. Polypyrimidine tract binding protein 1 protects mRNAs from recognition by the nonsense-mediated mRNA decay pathway. eLife. 2016;5:e11155. doi: 10.7554/eLife.11155. Study detailing the role of PTBP1 in protecting mRNAs from nonsense-mediated decay, potentially by affecting translation termination. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Karcher A, Schele A, Hopfner KP. X-ray structure of the complete ABC enzyme ABCE1 from Pyrococcus abyssi. The Journal of biological chemistry. 2008;283:7962–7971. doi: 10.1074/jbc.M707347200. [DOI] [PubMed] [Google Scholar]
- 134.Barthelme D, et al. Ribosome recycling depends on a mechanistic link between the FeS cluster domain and a conformational switch of the twin-ATPase ABCE1. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:3228–3233. doi: 10.1073/pnas.1015953108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Pisarev AV, Hellen CU, Pestova TV. Recycling of eukaryotic posttermination ribosomal complexes. Cell. 2007;131:286–299. doi: 10.1016/j.cell.2007.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Dmitriev SE, et al. GTP-independent tRNA delivery to the ribosomal P-site by a novel eukaryotic translation factor. The Journal of biological chemistry. 2010;285:26779–26787. doi: 10.1074/jbc.M110.119693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Skabkin MA, et al. Activities of Ligatin and MCT-1/DENR in eukaryotic translation initiation and ribosomal recycling. Genes & development. 2010;24:1787–1801. doi: 10.1101/gad.1957510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Young DJ, Guydosh NR, Zhang F, Hinnebusch AG, Green R. Rli1/ABCE1 Recycles Terminating Ribosomes and Controls Translation Reinitiation in 3′UTRs In Vivo. Cell. 2015;162:872–884. doi: 10.1016/j.cell.2015.07.041. Study detailing the in vivo role of ABCE1 in ribosome recycling and the role of Dom34 in rescuing inefficiently-recycled ribosomes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Andersen DS, Leevers SJ. The essential Drosophila ATP-binding cassette domain protein, pixie, binds the 40 S ribosome in an ATP-dependent manner and is required for translation initiation. The Journal of biological chemistry. 2007;282:14752–14760. doi: 10.1074/jbc.M701361200. [DOI] [PubMed] [Google Scholar]
- 140.Chen ZQ, et al. The essential vertebrate ABCE1 protein interacts with eukaryotic initiation factors. The Journal of biological chemistry. 2006;281:7452–7457. doi: 10.1074/jbc.M510603200. [DOI] [PubMed] [Google Scholar]
- 141.Dong J, et al. The essential ATP-binding cassette protein RLI1 functions in translation by promoting preinitiation complex assembly. The Journal of biological chemistry. 2004;279:42157–42168. doi: 10.1074/jbc.M404502200. [DOI] [PubMed] [Google Scholar]
- 142.Heuer A, et al. Structure of the 40S-ABCE1 post-splitting complex in ribosome recycling and translation initiation. Nature structural & molecular biology. 2017;24:453–460. doi: 10.1038/nsmb.3396. High-resolution cryoEM structure of 40S–ABCE1 complex provides mechanistic details into a potential dual role for ABCE1 in both ribosome recycling and translation initiation. [DOI] [PubMed] [Google Scholar]
- 143.Simonetti A, et al. eIF3 Peripheral Subunits Rearrangement after mRNA Binding and Start-Codon Recognition. Molecular cell. 2016;63:206–217. doi: 10.1016/j.molcel.2016.05.033. [DOI] [PubMed] [Google Scholar]
- 144.Mancera-Martinez E, Brito Querido J, Valasek LS, Simonetti A, Hashem Y. ABCE1: A special factor that orchestrates translation at the crossroad between recycling and initiation. RNA biology. 2017;14:1279–1285. doi: 10.1080/15476286.2016.1269993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Schuller AP, Green R. The ABC(E1)s of Ribosome Recycling and Reinitiation. Molecular cell. 2017;66:578–580. doi: 10.1016/j.molcel.2017.05.017. [DOI] [PubMed] [Google Scholar]
- 146.Brandman O, Hegde RS. Ribosome-associated protein quality control. Nature structural & molecular biology. 2016;23:7–15. doi: 10.1038/nsmb.3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Joazeiro CAP. Ribosomal Stalling During Translation: Providing Substrates for Ribosome-Associated Protein Quality Control. Annu Rev Cell Dev Biol. 2017;33:343–368. doi: 10.1146/annurev-cellbio-111315-125249. [DOI] [PubMed] [Google Scholar]
- 148.Doma MK, Parker R. Endonucleolytic cleavage of eukaryotic mRNAs with stalls in translation elongation. Nature. 2006;440:561–564. doi: 10.1038/nature04530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Graille M, Chaillet M, van Tilbeurgh H. Structure of yeast Dom34: a protein related to translation termination factor Erf1 and involved in No-Go decay. The Journal of biological chemistry. 2008;283:7145–7154. doi: 10.1074/jbc.M708224200. [DOI] [PubMed] [Google Scholar]
- 150.Lee HH, et al. Structural and functional insights into Dom34, a key component of no-go mRNA decay. Molecular cell. 2007;27:938–950. doi: 10.1016/j.molcel.2007.07.019. [DOI] [PubMed] [Google Scholar]
- 151.Carr-Schmid A, Pfund C, Craig EA, Kinzy TG. Novel G-protein complex whose requirement is linked to the translational status of the cell. Molecular and cellular biology. 2002;22:2564–2574. doi: 10.1128/MCB.22.8.2564-2574.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Chen L, et al. Structure of the Dom34-Hbs1 complex and implications for no-go decay. Nature structural & molecular biology. 2010;17:1233–1240. doi: 10.1038/nsmb.1922. [DOI] [PubMed] [Google Scholar]
- 153.Kalisiak K, et al. A short splicing isoform of HBS1L links the cytoplasmic exosome and SKI complexes in humans. Nucleic acids research. 2017;45:2068–2080. doi: 10.1093/nar/gkw862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Kowalinski E, et al. Structure of a Cytoplasmic 11-Subunit RNA Exosome Complex. Molecular cell. 2016;63:125–134. doi: 10.1016/j.molcel.2016.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Ishimura R, et al. RNA function. Ribosome stalling induced by mutation of a CNS-specific tRNA causes neurodegeneration. Science. 2014;345:455–459. doi: 10.1126/science.1249749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Shoemaker CJ, Eyler DE, Green R. Dom34:Hbs1 promotes subunit dissociation and peptidyl-tRNA drop-off to initiate no-go decay. Science. 2010;330:369–372. doi: 10.1126/science.1192430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Pisareva VP, Skabkin MA, Hellen CU, Pestova TV, Pisarev AV. Dissociation by Pelota, Hbs1 and ABCE1 of mammalian vacant 80S ribosomes and stalled elongation complexes. The EMBO journal. 2011;30:1804–1817. doi: 10.1038/emboj.2011.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Guydosh NR, Green R. Dom34 rescues ribosomes in 3′ untranslated regions. Cell. 2014;156:950–962. doi: 10.1016/j.cell.2014.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Guydosh NR, Green R. Translation of poly(A) tails leads to precise mRNA cleavage. Rna. 2017;23:749–761. doi: 10.1261/rna.060418.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Tsuboi T, et al. Dom34:hbs1 plays a general role in quality-control systems by dissociation of a stalled ribosome at the 3′ end of aberrant mRNA. Molecular cell. 2012;46:518–529. doi: 10.1016/j.molcel.2012.03.013. [DOI] [PubMed] [Google Scholar]
- 161.Guydosh NR, Kimmig P, Walter P, Green R. Regulated Ire1-dependent mRNA decay requires no-go mRNA degradation to maintain endoplasmic reticulum homeostasis in S. pombe. eLife. 6:e29216–2017. doi: 10.7554/eLife.29216. Study using yeast genetics and ribosome profiling to define a role for Dom34–Hbs1 in the Ire1-mediated unfolded protein response. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Brandman O, et al. A ribosome-bound quality control complex triggers degradation of nascent peptides and signals translation stress. Cell. 2012;151:1042–1054. doi: 10.1016/j.cell.2012.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Bhattacharya A, McIntosh KB, Willis IM, Warner JR. Why Dom34 stimulates growth of cells with defects of 40S ribosomal subunit biosynthesis. Molecular and cellular biology. 2010;30:5562–5571. doi: 10.1128/MCB.00618-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.van den Elzen AM, Schuller A, Green R, Seraphin B. Dom34-Hbs1 mediated dissociation of inactive 80S ribosomes promotes restart of translation after stress. The EMBO journal. 2014;33:265–276. doi: 10.1002/embj.201386123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Balagopal V, Parker R. Stm1 modulates translation after 80S formation in Saccharomyces cerevisiae. Rna. 2011;17:835–842. doi: 10.1261/rna.2677311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Van Dyke N, Baby J, Van Dyke MW. Stm1p, a ribosome-associated protein, is important for protein synthesis in Saccharomyces cerevisiae under nutritional stress conditions. Journal of molecular biology. 2006;358:1023–1031. doi: 10.1016/j.jmb.2006.03.018. [DOI] [PubMed] [Google Scholar]
- 167.Van Dyke N, Chanchorn E, Van Dyke MW. The Saccharomyces cerevisiae protein Stm1p facilitates ribosome preservation during quiescence. Biochemical and biophysical research communications. 2013;430:745–750. doi: 10.1016/j.bbrc.2012.11.078. [DOI] [PubMed] [Google Scholar]
- 168.Ben-Shem A, et al. The structure of the eukaryotic ribosome at 3.0 A resolution. Science. 2011;334:1524–1529. doi: 10.1126/science.1212642. [DOI] [PubMed] [Google Scholar]
- 169.Mills EW, Green R, Ingolia NT. Slowed decay of mRNAs enhances platelet specific translation. Blood. 2017;129:e38–e48. doi: 10.1182/blood-2016-08-736108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Mills EW, Wangen J, Green R, Ingolia NT. Dynamic Regulation of a Ribosome Rescue Pathway in Erythroid Cells and Platelets. Cell reports. 2016;17:1–10. doi: 10.1016/j.celrep.2016.08.088. Identification of unusual ribosome distributions on 3′ UTRs in erythroid cells and platelets and a link to defects in ribosome recycling and rescue in these tissues. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Mills EW, Green R. Ribosomopathies: There’s strength in numbers. Science. 2017;358 doi: 10.1126/science.aan2755. [DOI] [PubMed] [Google Scholar]
- 172.Popp MW, Maquat LE. Organizing principles of mammalian nonsense-mediated mRNA decay. Annual review of genetics. 2013;47:139–165. doi: 10.1146/annurev-genet-111212-133424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Kervestin S, Jacobson A. NMD: a multifaceted response to premature translational termination. Nature reviews. Molecular cell biology. 2012;13:700–712. doi: 10.1038/nrm3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Holbrook JA, Neu-Yilik G, Hentze MW, Kulozik AE. Nonsense-mediated decay approaches the clinic. Nature genetics. 2004;36:801–808. doi: 10.1038/ng1403. [DOI] [PubMed] [Google Scholar]
- 175.He F, Peltz SW, Donahue JL, Rosbash M, Jacobson A. Stabilization and ribosome association of unspliced pre-mRNAs in a yeast upf1- mutant. Proceedings of the National Academy of Sciences of the United States of America. 1993;90:7034–7038. doi: 10.1073/pnas.90.15.7034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Mitrovich QM, Anderson P. Unproductively spliced ribosomal protein mRNAs are natural targets of mRNA surveillance in C. elegans. Genes & development. 2000;14:2173–2184. doi: 10.1101/gad.819900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Mendell JT, Sharifi NA, Meyers JL, Martinez-Murillo F, Dietz HC. Nonsense surveillance regulates expression of diverse classes of mammalian transcripts and mutes genomic noise. Nature genetics. 2004;36:1073–1078. doi: 10.1038/ng1429. [DOI] [PubMed] [Google Scholar]
- 178.Welch EM, Jacobson A. An internal open reading frame triggers nonsense-mediated decay of the yeast SPT10 mRNA. The EMBO journal. 1999;18:6134–6145. doi: 10.1093/emboj/18.21.6134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Marquardt S, Hazelbaker DZ, Buratowski S. Distinct RNA degradation pathways and 3′ extensions of yeast non-coding RNA species. Transcription. 2011;2:145–154. doi: 10.4161/trns.2.3.16298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Smith JE, et al. Translation of small open reading frames within unannotated RNA transcripts in Saccharomyces cerevisiae. Cell reports. 2014;7:1858–1866. doi: 10.1016/j.celrep.2014.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Le Hir H, Gatfield D, Izaurralde E, Moore MJ. The exon-exon junction complex provides a binding platform for factors involved in mRNA export and nonsense-mediated mRNA decay. The EMBO journal. 2001;20:4987–4997. doi: 10.1093/emboj/20.17.4987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Melero R, et al. The cryo-EM structure of the UPF-EJC complex shows UPF1 poised toward the RNA 3′ end. Nature structural & molecular biology. 2012;19:498–505. doi: 10.1038/nsmb.2287. [DOI] [PubMed] [Google Scholar]
- 183.Peltz SW, Brown AH, Jacobson A. mRNA destabilization triggered by premature translational termination depends on at least three cis-acting sequence elements and one trans-acting factor. Genes & development. 1993;7:1737–1754. doi: 10.1101/gad.7.9.1737. [DOI] [PubMed] [Google Scholar]
- 184.Narla A, Ebert BL. Ribosomopathies: human disorders of ribosome dysfunction. Blood. 2010;115:3196–3205. doi: 10.1182/blood-2009-10-178129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Draptchinskaia N, et al. The gene encoding ribosomal protein S19 is mutated in Diamond-Blackfan anaemia. Nature genetics. 1999;21:169–175. doi: 10.1038/5951. [DOI] [PubMed] [Google Scholar]