Abstract
Assessment of genetic diversity is a pre-requisite to broaden the genetic background of cultivated base of sweet corn, an endosperm mutant of field corn that alters starch biosynthesis pathway in endosperm. In the current investigation, genetic divergence among 39 inbred lines was assessed on the basis of 14 agro-morphological traits, two quality parameters and 63 microsatellite markers, selected on the basis of their association with QTLs affecting kernel quality. The cluster analysis based on unweighted pair-group method using arithmetic averages for agro-morphological and quality traits grouped the 39 inbreds into three clusters with 5, 14 and 20 genotypes, respectively. The unweighted neighbor-joining method for microsatellite markers also categorized the inbred lines into three major clusters grouping 10, 9 and 20 genotypes in cluster I, II and III, respectively. The two cluster distribution patterns showed approximately 36 percent similarity. The assay of 30 microsatellite repeats identified 82 alleles with allele size ranging from 80 to 400 bp. The major allele frequency and PIC value of the markers ranged from 0.42 to 0.79 and 0.27 to 0.63, respectively, which suggested the presence of high amount of polymorphism among the inbreds. The average heterozygosity was recorded to be 0.19 which signifies proper maintenance of inbred population. Principle co-ordinate analysis also depicted diverse nature of inbred lines and agreed well with the previously determined clustering pattern. This study has identified several inbreds, having good yield and high sugar content which will not only enhance the genetic background of sweet corn germplasm but will also lead to development of high-yielding hybrids with improved quality.
Keywords: Sweet corn, Endosperm mutant, Microsatellite, Kernel quality, Genetic diversity
Introduction
Sweet corn (Zea mays var. saccharata) among various specialty corns has emerged as one of the popular choice either as fresh vegetable or processed product worldwide. Sweet corn kernels are consumed in green ear/milky stage generally at 20–24 days after pollination and are sold as highly prized fresh or canned vegetables (Khanduri et al. 2011; Mehta et al. 2017). The eating quality of fresh or processed whole kernels, canned or frozen, is determined by its unique combination of flavor, texture and aroma. It is one of the most popular vegetable in the US, Canada and many of the western countries. Brazil, as one of the world’s largest corn producer, also has great potential for sweet corn production. In India, sweet corn occupies good coverage in the states such as Andhra Pradesh, Karnataka and Maharashtra (Dagla et al. 2014). Maharashtra has now become the leading sweet corn hub of India and has maximum contribution in export of sweet corn in all its form fresh, frozen, canned, etc. throughout India and overseas (http://www.cornclub.com).
The distinguishing features of sweet corn kernels are mainly attributed to the presence of several endosperm mutants that increase sugar content and decrease starch content viz., shrunken2 (sh2), brittle1 (bt1), sugary1 (su1), sugary enhancer (se), brittle2 (bt2), dull1 (du1) and waxy1(wx1) (Tracy 2001). Pajic et al. (2004) described these mutants as enzymic “injuries” on the way of starch syntheses that alter the endosperm carbohydrate composition and result, almost in all cases, in decrease of starch content. Among the various mutants, viz., sh2, bt1, su1 and se are the most useful ones in terms of increasing sugar content and decreasing starch content (Lertrat and Pulam 2007). The sh2 and bt1 mutations accumulate sugars at the expense of starch, and are located, respectively, in chromosome number 3 and 5; while su1 and se, located in chromosome 4 and 2, respectively, function in later steps of starch biosynthesis pathway and are involved in changing types and proportion of types of polysaccharides stored in the endosperm (Boyer and Shannon 1984; Tracy 1997). The su1 gene produces three and ten times higher concentration of reducing sugars and water-soluble polysaccharides, respectively, at milky ripening stage compared to field corn and gives a creamy and glossy texture with good flavor to kernels (Creech 1965; James et al. 1995; Feng et al. 2008). At milky ripening stage, sh2 and bt1 genes accumulate about sixfold more reducing sugar and sucrose in kernels than normal maize but the WSP content is same as normal maize (Feng et al. 2008). Moreover, in sh2sh2-based sweet corn types, the depletion of sugar level is much slower and have extended shelf life, hence more suitable for prolonged storage compared to su2su2-based sweet corn types (Lertrat and Pulam 2007; Mehta et al. 2017). The se1 gene on the other hand behaves as carbohydrate modifier that when used in combination with su1 results in increased sugar level equivalent to sh2 and water-soluble polysaccharide (WSP) level similar to unmodified su1 (Tracy 1997).
Sweet corn breeding, while using many of the techniques and theories developed for field corn, is quite different in practice because of the end uses of the variety and the highly perishable nature of the final product. The breeding programs should be emphasized on producing high-yielding cultivars with no compromise in quality. Genetic characterization of sweet corn inbreds assumes great significance in this aspect that not only helps in understanding the population structure but will also serve as a success key to many pre-breeding and conservation programs. The genetic variability present in a sweet corn population can be assessed using agro-morphological traits and/or molecular markers but agro-morphological traits may result in misleading estimates due to higher influence of environment on them. Hence, a comprehensive approach based on both morphological and molecular markers will be more informative. Further, polymorphic marker information will be of great use in genomic structure and evolutionary ecology studies and, marker-assisted selection (Pandey et al. 2018). Use of molecular markers directly associated with quantitative trait loci (QTLs) affecting kernel chemical composition and texture will identify diverse inbreds that confer relevant agronomic and industrial traits for hybrid breeding programs and the consumer market. Hence, with a view to strengthen sweet corn breeding program and to accelerate the pace of progress of sweet corn improvement program, our study has been designed to assess genetic diversity and population structure of sweet corn inbreds using several morphological and biochemical traits in combination with microsatellite markers.
Materials and methods
Plant materials
A set of 39 sweet corn inbred lines procured from different sources and maintained through selfing/sibbing under the All India Co-ordinated Research Project on Maize undergoing at Department of Genetics and Plant Breeding, Institute of Agricultural Sciences, Banaras Hindu University (BHU), Varanasi, were taken for the study (Table 1). All these inbreds were grown in randomized block design with two replications at the Agriculture Research Farm, Institute of Agricultural Sciences, BHU, during rabi 2015–2016. The research farm is situated at 25°18′ North latitude and 83°03′ East longitude and at altitude of 123.23 m from sea level.
Table 1.
List of sweet corn inbred lines
| S.no. | Inbred line | S.no. | Inbred line |
|---|---|---|---|
| 1 | DMSC 1 | 21 | su2su2o2o2Comp(Y)-BBB-28-BBB |
| 2 | DMSC 2 | 22 | WNCDMRSC08R686(A) |
| 3 | DMSC 3 | 23 | WNCDMRSC08R690 |
| 4 | DMSC 4 | 24 | WNCDMRSC08710 |
| 5 | DMSC 6 | 25 | WNCDMRSC08712 |
| 6 | DMSC 8 | 26 | WNCDMRSC08750 |
| 7 | DMSC 9 | 27 | WNCDMRSC08792 |
| 8 | DMSC 19 | 28 | WNCDMRSC08R753 |
| 9 | DMSC 20 | 29 | WNDMRSCY18R715 |
| 10 | DMSC 24 | 30 | WNDMRSCY18R716 |
| 11 | DMSC 27 | 31 | WNDMRSCY18R730 |
| 12 | DMSC 35 | 32 | WNDMRSCY18R736 |
| 13 | DMSC 36 | 33 | WNDMRSCY18R753 |
| 14 | HKI-1827W-1 | 34 | WNDMRSCY18R743 |
| 15 | DulceAmanillo | 35 | SC FEMALE |
| 16 | Win Sweet Corn | 36 | SCF |
| 17 | su2su2o2o2Comp(Y)-BBB-1-BBB4PI | 37 | NSS2W9301A |
| 18 | su2su2o2o2Comp(Y)-BBB-2-BBB | 38 | Phil Super Sweet |
| 19 | su2su2o2o2Comp(Y)-BBB-15-BBB | 39 | SC7-2-1-2-1(N) |
| 20 | su2su2o2o2Comp(Y)-BBB-40-BBB |
Morphological characterization
Observations were recorded on different agro-morphological traits as mentioned below: morphophysiological traits, viz., days to 50% tasseling (DTT), days to 50% silking (DTS), plant height (PH) and ear height (EH): recorded on five random plants in each replication; yield attributing traits, viz., ear index (EI): total number of marketable cobs per plot divided by number of plants per plot, total ear weight (TEW): weight of green cobs with husk, standard ear weight (SEW): weight of cobs disregarding outliers, husk ratio (HR): percentage of husk out of total ear weight, and fodder yield (FY): weight of green and succulent stalks left after harvest of green cobs. Observations on rest of the yield-related traits such as ear length (EL), ear diameter (ED), number of kernels rows per ear (KR/E), number of kernels per row (K/R) and percent of ear filled (PEF): portion of ear completely filled with kernels expressed in percentage were taken by choosing five marketable cobs for each genotype. Two quality parameters, viz., total soluble solids (TSS): recorded in a hand refractometer and reducing sugar (RS): determined in laboratory using Nelson–Somogyi method, were also used in the study.
Molecular characterization
Total genomic DNA was extracted from healthy young leaves using a modified CTAB extraction protocol (Doyle and Doyle 1987). A set of 63 simple sequence repeats distributed throughout the corn genome were chosen based on their association with QTLs governing kernel characteristics and chemical composition (Qi et al. 2009; Park et al. 2013; Hossain et al. 2015; Jha et al. 2016). Primer sequence information of microsatellites was obtained from public domain, i.e., http://www.maizegdb.org. PCR amplifications were performed with a final reaction volume of 15 µl containing ~ 30–40 ng genomic DNA. The amplified products were resolved using 3.5% agarose metaphor gel. The gel was run at a constant voltage of 70 V for about 4 h (until the tracking dye migrated to the end of the gel). The electrophoresed DNA samples were visualized using a UV Trans-illuminator geldoc system and the same was photographed and documented. The allele size was determined by comparing the bands with 50-bp DNA ladder (Bangaluru Genei, India). Out of these 63 SSRs, 30 markers exhibited polymorphism and were further used for analyses. The details of these 30 microsatellites along with QTL information and associated trait are presented in Table 2.
Table 2.
Details of microsatellite markers and associated traits
| S. no. | SSR marker | Trait | References |
|---|---|---|---|
| 1 | umc1165, bnlg1297, umc1465, umc1259, umc1669, bnlg1126, umc1303, phi093, umc1586, bnlg1714, umc1492, bnlg1012, bnlg1079, bnlg1712, umc1605, umc1633, bnlg2323 | Soluble sugar content | Qi et al. (2009) |
| 2 | bnlg1265 | Amylose, sucrose and dextrose content | Park et al. (2013) |
| phi027, umc1634 | Amylose content | ||
| bnlg1867, umc2056 | Dextrose content | ||
| umc2173 | Sucrose content | ||
| umc1130 | |||
| 3 | umc1273 | sh2 locus | Hossain et al. (2015) |
| umc1320 | |||
| umc2276 | |||
| umc1896 | su1 locus | ||
| umc1142 | |||
| umc1031 |
Statistical analysis
The data on agro-morphological traits were subjected to estimation of genetic diversity and clustering of genotypes into different groups using SAS v 9.3. The software computes Euclidian distances, as a measure of (dis)similarity, and performs hierarchical cluster analysis with unweighted pair-group method using arithmetic averages (UPGMA). Major allele frequency, polymorphic information content (PIC) and heterozygosity of different microsatellites were computed using PowerMarker 3.25 (Liu and Muse 2005). The allele with frequency < 0.05 was considered as rare allele. Genetic dissimilarity indices were calculated using simple matching coefficient. The cluster analysis following unweighted neighbor-joining method and principle coordinate analysis (PCoA) was undertaken using DARwin5.0 (Perrier et al. 2003). The analysis was performed at 10,000 bootstraps values and the same has been discussed in “Results and discussion” section.
Results and discussion
Cluster analysis based on agro-morphological traits
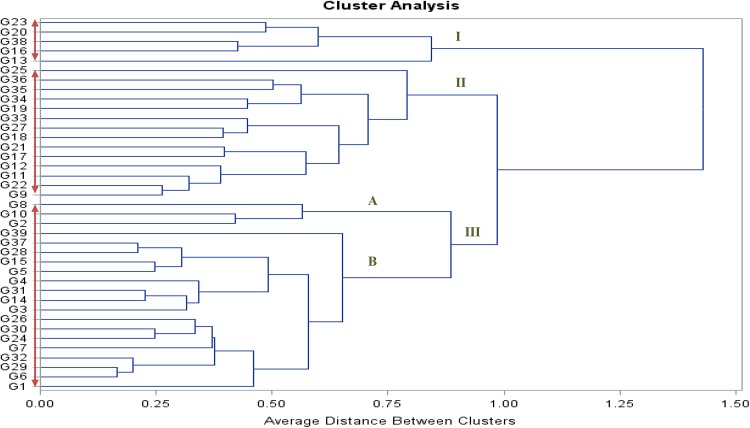
Fourteen morphological and two biochemical traits clustered the 39 sweet corn genotypes into three major clusters. From Fig. 1, it was clear that cluster III was largest, containing 20 genotypes, while clusters I and II had 5 and 14 inbreds, respectively. As shown in Table 3, the first cluster had highest mean values for PH (132.20), EH (50.90), FY (2.39), TEW (1.85) and SEW (1.16) with lower HR (39.52). This cluster was also recorded to have superior mean performance for all yield-attributing traits such as EL, ED, PEF, KR/E and K/R. The highest mean value for TSS (24.83) was recorded for sub-cluster IIIA but the mean value of reducing sugar was only 3.67 which indicated the higher TSS may be attributed to carbohydrates other than sugar. In terms of quality parameters, cluster I was found to be better among all with mean values of TSS and reducing sugar being 14.50 and 3.86, respectively. Hence, cluster I is superior in all aspect with earlier maturity and the genotypes belonging to this cluster will be of great use in sweet corn breeding program.
Fig. 1.
Cluster analysis depicting genetic relationship among inbreds based on agro-morphological traits. Genotypes’ numbers are according to Table 1
Table 3.
Mean values of 14 morphological and two biochemical characters of six clusters revealed by cluster analysis among 39 inbreds of sweet corn
| Cluster | Sub-cluster | PH | EH | DTT | DTS | EI | TEW | SEW | HR | EL | PEF | ED | KR/E | K/R | FY | TSS | RS |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| I | 132.20 | 50.90 | 103.50 | 109.80 | 0.79 | 1.85 | 1.16 | 39.52 | 12.96 | 9.26 | 3.74 | 13.48 | 21.54 | 2.39 | 14.50 | 3.86 | |
| II | 108.13 | 38.51 | 104.39 | 110.93 | 0.82 | 1.42 | 0.87 | 37.41 | 13.08 | 9.28 | 3.68 | 13.61 | 21.77 | 1.84 | 14.93 | 2.30 | |
| III | A | 87.00 | 26.00 | 110.83 | 116.33 | 0.96 | 0.61 | 0.21 | 54.61 | 12.02 | 9.39 | 2.93 | 12.43 | 17.61 | 1.25 | 24.83 | 3.67 |
| B | 84.15 | 28.68 | 100.65 | 108.03 | 0.70 | 1.06 | 0.71 | 35.15 | 12.37 | 9.49 | 3.37 | 12.78 | 21.22 | 1.05 | 15.85 | 1.83 |
PH plant height, EH ear height, DTT days to 50% tasseling, DTS days to 50% silking, EI ear index, TEW total ear weight, SEW standard ear weight, HR Husk ratio, EL ear length, PEF percent of ear filled, ED ear diameter, KR/E kernel rows per ear, K/R kernels per row, FY Fodder yield, PY plot yield, TSS total soluble solids, RS reducing sugar
The per se performance of different inbred lines for all 16 characters is presented in Table 4. The inbred Phil super sweet, belonging to cluster I, was recorded as the best performer for most of the yield-attributing traits. Two inbred lines, viz., HKI-1827W-1 and WNDMRSCY18R730, from cluster IIIB had low per se performance for flowering time and would be of great use in breeding for early maturity hybrids. As far as the two quality traits were concerned, DMSC 27 and DMSC 4, belonging to cluster II and IIIB, respectively, had high per se performance for TSS; whereas, DMSC 36 from cluster I and WNCDMRSC08R686(A) from cluster II showed high per se performance for RS.
Table 4.
Trait-wise per se performance of different inbred lines
| Trait | Per se performance of superior inbreds | Cluster |
|---|---|---|
| Plant height | su2su2o2o2Comp(Y)-BBB-40-BBB, Phil Super Sweet | I |
| Ear height | WNCDMRSC08R690, Win Sweet Corn | I |
| Days to 50% tasseling | HKI-1827W-1, WNDMRSCY18R730 | IIIB |
| Days to 50% silking | HKI-1827W-1, WNDMRSCY18R730 | IIIB |
| Ear index | DMSC 24 SCF |
IIIA II |
| Total ear weight | Phil Super Sweet su2su2o2o2Comp(Y)-BBB-28-BBB |
I II |
| Standard ear weight | Phil Super Sweet WNDMRSCY18R736 |
I IIIB |
| Husk ratio | Phil Super Sweet WNDMRSCY18R736 |
I IIIB |
| Ear length | Phil Super Sweet su2su2o2o2Comp(Y)-BBB-28-BBB |
I II |
| Percent of ear filled | WNCDMRSC08750, WNDMRSCY18R715 | IIIB |
| Ear diameter | su2su2o2o2Comp(Y)-BBB-15-BBB,WNCDMRSC08712 | II |
| Number of kernel rows per ear | su2su2o2o2Comp(Y)-BBB-2-BBB su2su2o2o2Comp(Y)-BBB-40-BBB |
II I |
| Number of kernels per row | WNCDMRSC08792, su2su2o2o2Comp(Y)-BBB-2-BBB | II |
| Fodder yield | Phil Super Sweet, WNCDMRSC08R690 | I |
| Total soluble solids | DMSC 27 DMSC 4 |
II IIIB |
| Reducing sugar | DMSC 36 WNCDMRSC08R686(A) |
I II |
Genetic dissimilarity between the inbreds varied from 5.71 to 73.69. The range of genetic dissimilarity varied from 14.57 to 32.48, 9.03 to 44.72, 14.38 to 19.54 and 5.71 to 41.89 within cluster I, II, IIIA and IIIB, respectively. This signifies sufficient genetic similarity within the members of a cluster. Based on genetic dissimilarity, DMSC3 and su2su2o2o2Comp(Red)-BBB-40-BBB (79.69) followed by HKI-1827W-1 and Phil Super Sweet (sh2sh2)-1 (79.57) were the most distantly related inbreds. Whereas, lowest genetic dissimilarity was observed for inbred DMSC8 with WNDMRSCY18R730 (5.71) followed by SC FFEMALE (6.60). These results highlighted the efficiency of morphological markers in deciphering genetic diversity and population structure as reported earlier (Choudhary et al. 2017; Babic et al. 2014, 2016; Mazid et al. 2013; Seshu et al. 2015).
Genetic diversity assessment based on microsatellite markers
The study was carried out using 63 simple sequence repeats, of which 30 showed polymorphism and used for characterization of sweet corn inbred lines. The 30 SSRs were distributed on nine linkage groups from one marker on chromosome 1 and 5 to maximum eight markers on chromosome 4. Previously, microsatellite-based diversity has been assessed by utilizing 13 (Rupp et al. 2009), 15 (Lopes et al. 2014), 20 (Solomon et al. 2012), 30 (Lopes et al. 2015), 40 (Srdic et al. 2011) and 56 (Mehta et al. 2017) markers. But, uniqueness of this study is that all the markers used here are directly associated with QTLs affecting kernel chemical composition and texture; thus, it will be helpful in selecting inbreds with good kernel characteristics. These 30 microsatellite repeats identified 82 alleles among the inbreds. The number of alleles detected per loci ranged from 2 to 5 with mean 2.73. Among all 30 loci, 15 were biallelic, 9 were triallelic and 5 loci (bnlg1867, umc2056, bnlg1297, umc1669 and umc1142) produced four alleles while the highest number was recorded in case of umc1165 (Table 5). Here, the presence of higher number of alleles confirms availability of wide genetic diversity among the sweet corn inbreds.
Table 5.
Details of microsatellite markers used for molecular study
| S. no. | Primer | Bin | Repeat | No. of alleles | Major allele frequency | Heterozygosity | PIC |
|---|---|---|---|---|---|---|---|
| 1 | umc1605 | 1.12 | (GGC)4 | 3 | 0.57 | 0.00 | 0.49 |
| 2 | umc1165 | 2.01 | (TA)6 | 5 | 0.51 | 0.34 | 0.59 |
| 3 | bnlg1297 | 2.02 | (AG)32 | 4 | 0.42 | 0.27 | 0.63 |
| 4 | umc1465 | 2.04 | (ACACA)4 | 3 | 0.54 | 0.26 | 0.45 |
| 5 | umc1259 | 2.04 | (GCG)4 | 3 | 0.53 | 0.10 | 0.50 |
| 6 | umc1633 | 2.08 | (GCG)4 | 3 | 0.43 | 0.50 | 0.54 |
| 7 | umc1273 | 3.08 | (AAG)4 | 3 | 0.47 | 0.10 | 0.53 |
| 8 | umc1320 | 3.08 | (GAAC)4 | 2 | 0.62 | 0.41 | 0.36 |
| 9 | umc2276 | 3.08 | (GGC)4 | 2 | 0.51 | 0.26 | 0.37 |
| 10 | umc1669 | 4.01 | (AGA)4 | 4 | 0.53 | 0.15 | 0.55 |
| 11 | bnlg1126 | 4.03 | (AG)20 | 3 | 0.57 | 0.54 | 0.45 |
| 12 | umc1303 | 4.03 | (CCG)4 | 2 | 0.67 | 0.00 | 0.35 |
| 13 | bnlg1265 | 4.05 | (AG)33 | 3 | 0.53 | 0.11 | 0.54 |
| 14 | umc1896 | 4.05 | (CA)8 | 2 | 0.74 | 0.23 | 0.31 |
| 15 | umc1142 | 4.05 | (TGGA)5 | 4 | 0.51 | 0.06 | 0.51 |
| 16 | umc1031 | 4.05 | (CT)6AT(CT)9 | 3 | 0.53 | 0.23 | 0.49 |
| 17 | phi093 | 4.08 | AGCT | 2 | 0.74 | 0.41 | 0.31 |
| 18 | bnlg2323 | 5.04 | (AG)25 | 2 | 0.54 | 0.92 | 0.37 |
| 19 | bnlg1867 | 6.01 | (AG)17 | 4 | 0.42 | 0.03 | 0.63 |
| 20 | umc2056 | 6.01 | (ATC)5 | 4 | 0.75 | 0.14 | 0.36 |
| 21 | umc2173 | 8.03 | (CGT)5 | 2 | 0.74 | 0.00 | 0.31 |
| 22 | umc1130 | 8.05 | (TAA)4 | 2 | 0.79 | 0.03 | 0.28 |
| 23 | phi027 | 9.03 | GCGCT | 2 | 0.68 | 0.08 | 0.34 |
| 24 | umc1634 | 9.03 | (AG)7 | 2 | 0.79 | 0.00 | 0.27 |
| 25 | umc1586 | 9.03 | (ATC)5 | 2 | 0.58 | 0.23 | 0.37 |
| 26 | bnlg1714 | 9.04 | (AG)25 | 3 | 0.59 | 0.05 | 0.48 |
| 27 | umc1492 | 9.04 | (GCT)4 | 2 | 0.73 | 0.13 | 0.32 |
| 28 | bnlg1012 | 9.04 | (AG)16 | 2 | 0.58 | 0.03 | 0.37 |
| 29 | bnlg1079 | 10.03 | (AG)14 | 2 | 0.59 | 0.00 | 0.37 |
| 30 | bnlg1712 | 10.03 | (AG)20 | 2 | 0.51 | 0.00 | 0.37 |
| Mean | 2.73 | 0.43 | 0.19 | 0.59 | |||
The allele size across 30 markers ranged from 80 to 400. As evident from Table 5, the major allele frequency for the markers ranged from 0.42 to 0.79 with minimum frequency in bnlg1297 for 150 bp and maximum in case of umc1634 and umc2056 for 120 and 150 bp, respectively. The PIC value of markers ranged from 0.27 in umc1634 to 0.63 in bnlg1297 and bnlg1867 (Table 5). The mean PIC value was recorded to be 0.59 suggesting that the markers contained sufficiently high amount of polymorphism that enabled efficient grouping of genotypes into different groups. The 30 marker assay identified 3 rare alleles out of 82. Marker umc2056 generated two rare alleles, one of 130 bp in genotypes HKI-1827W-1 and DMSC20, and another of 120 bp in DMSC27, DMSC35 and su2su2o2o2Comp(Y)-BBB-1-BBB4PI. The third rare allele of size 160 bp was produced by marker umc1142 in genotypes, WNDMRSCY18R736 and SC FEMALE.
SSR markers being co-dominant in nature are of great use in determining population structure of inbred lines. Estimation of heterozygosity among inbreds at molecular level will not only determine the purity of seed lot but will also assess the various cycles of maintenance which the inbreds had gone through during the course of development. In the present study, the mean heterozygosity was recorded to be 0.19 which signifies proper maintenance of most of these inbreds in breeding program (Table 5). However, being a highly cross-pollinated crop, some amount of heterozygosity remain inherent in many inbred lines during the course of maintaining morphological uniformity. That is the reason some of the markers showed heterozygosity up to 0.50. Marker bnlg2323 showed maximum heterozygosity of 0.92 which may be attributed to the residual heterozygosity retained among the inbreds for that particular locus during process of development and maintenance of inbreds. Mehta et al. (2017) also reported high heterozygosity up to 0.35 for some of the markers.
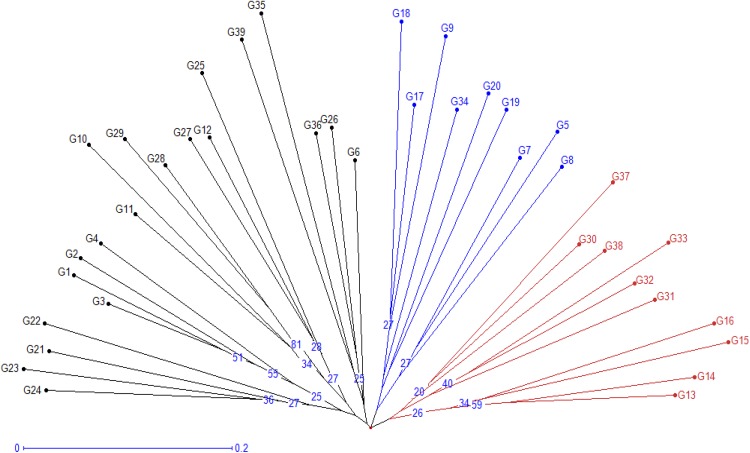
The pair-wise genetic dissimilarity indices varied from 0.28 to 0.79 with mean 0.60 indicating considerable diversity among the inbreds. This result is comparable with other estimates obtained using various markers in sweet corn (Gerdes and Tracy 1994; Amorim et al. 2003; Rupp et al. 2009). The cluster analysis based on molecular diversity classified the 39 inbreds into three major clusters (Fig. 2). Cluster I contained 10 inbreds, cluster II had 9 and cluster III, being the largest, included 20 inbred lines. Cluster III was further grouped into two sub clusters containing 5 and 15 inbred lines. The polymorphism among sweet corn inbred lines based on two microsatellites has been presented in Fig 3.
Fig. 2.
Cluster dendrogram depicting genetic divergence among 39 inbreds based on microsatellite markers. Genotypes’ numbers are according to Table 1
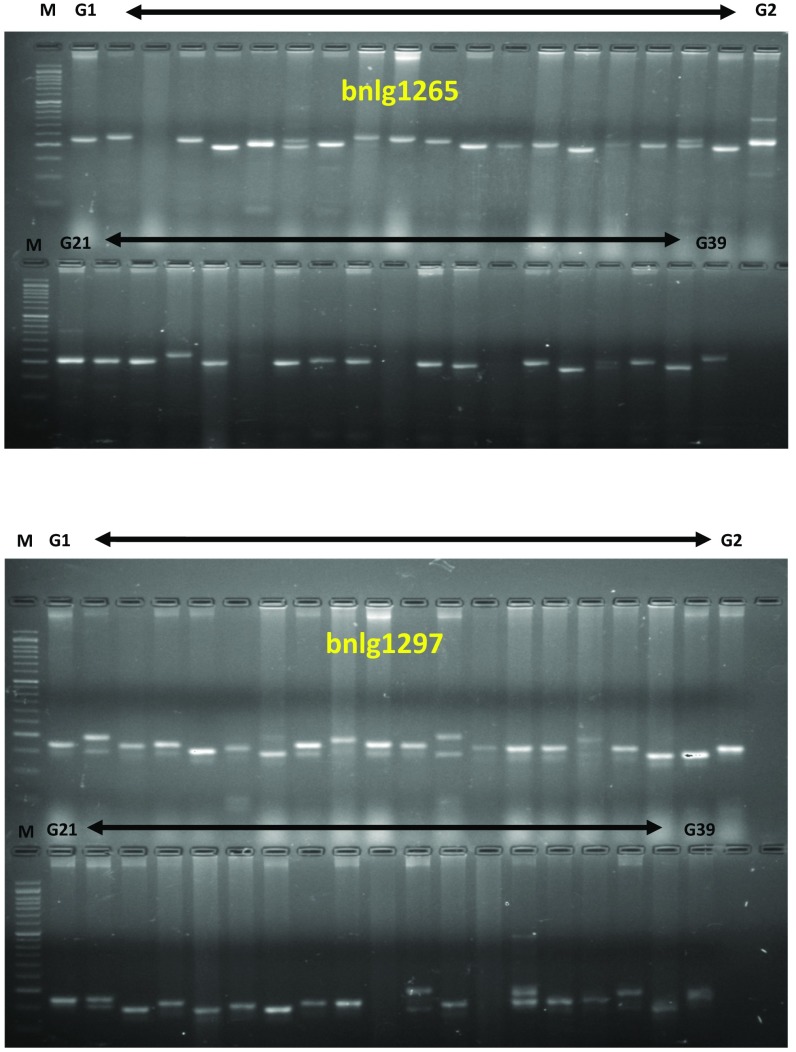
Fig. 3.
Microsatellite polymorphism among sweet corn inbreds. M: 50-bp ladder
This clustering pattern was quite similar with the pattern obtained on the basis of morphological and biochemical parameters. Out of 39, nearly 36% inbreds showed similar distribution pattern.
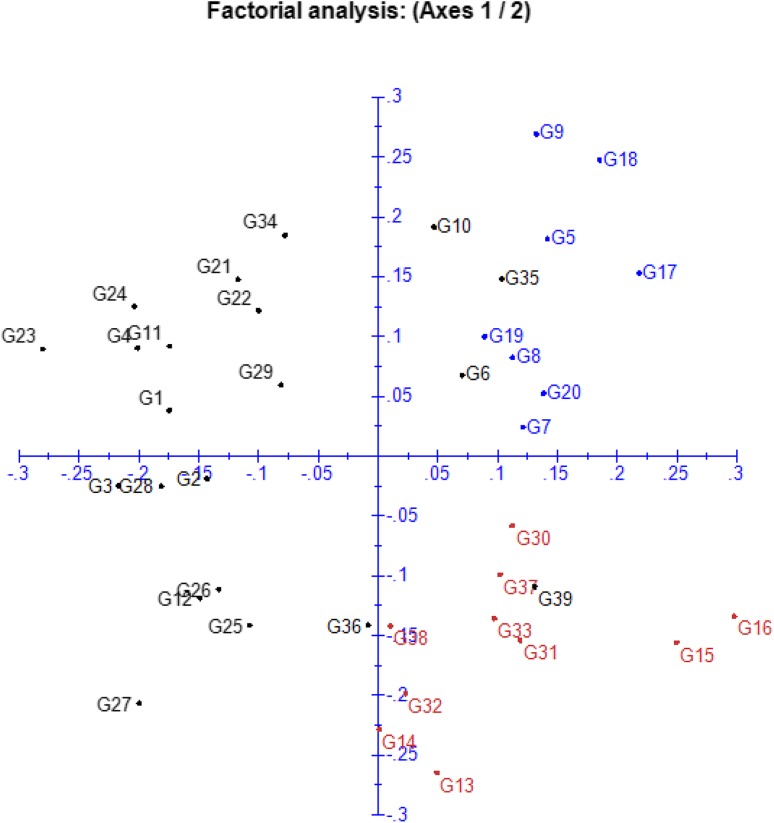
The result of Principle coordinate analysis also depicts the diverse nature of inbreds used in the study and agreed well with their clustering pattern (Figs. 2, 4). All ten inbreds belonging to cluster I were distributed in the right lower quadrangle and that of nine inbreds of cluster II were present in right upper quadrangle except inbred SCF which fell in left upper quadrangle. The third and largest cluster mostly occupied left quadrangle except four inbreds. The diverse nature of inbreds has already been proven to be source of superior heterotic hybrid combinations by Mehta et al. (2017), Solomon et al. (2012) and Babic et al. (2014).
Fig. 4.
Principle coordinate analysis based on molecular data depicting genetic divergence of 39 inbred lines. Genotypes’ numbers are according to Table 1. The proportion of total variation by axis 1: 10.84, axis 2: 9.99, axis 3: 8.56 and axis 4: 7.65
The present study implied both morphological and molecular markers for discriminating the inbred lines into divergent groups. Although information obtained from morphological data gives significant results and classifies the inbreds into distinct groups, it is a well-known fact that a major portion of phenotypic variability is probably a consequence of environment that affects expression of a trait to a great extent. Babic et al. (2012, 2014) pointed out morphological markers, based on UPOV (International Union for the Protection of New Varieties Plant) descriptor, could produce sufficient level of discrimination in divergent groups as well as the fact that obtained information could be useful in maize breeding. Babic et al. (2016) also reported the use of adequate statistical methods as well as scale of measurement could significantly increase quality and utility of morphological markers. In addition, use of molecular markers not only presents validatory proof of diversity assessed through morphological markers, but is also informative and robust tools for selection of elite inbreds without any biasness of environment.
As we discussed earlier, use of microsatellite markers associated with kernel sugar and eating quality is the uniqueness of this study. Hence, the molecular markers used here will be informative in terms of selecting inbreds with high sugar content. Out of 30 microsatellite loci, 7 loci, associated with QTLs affecting kernel sugar content, showed polymorphism among the inbreds used in this study. A biallelic locus umc2276, mapped as nearest marker from sh2 by Hossain et al. (2015), showed major allele frequency up to 0.51 with PIC 0.37. This study will be of great use in selecting inbreds for developing high-yielding hybrids with good commercial standards.
Genetic dissimilarity index of > 0.5 between the clusters indicated that inbreds with higher mean performance for yield and yield-attributing traits could be selected from any of the cluster for use in hybridization program. Based on average performance, inbred SC FEMALE and Phil Super Sweet (sh2sh2)-1 from cluster I; su2su2o2o2Comp(Y)-BBB-1-BBB4PI and su2su2o2o2Comp(Y)-BBB-2-BBB from cluster II; DMSC8 and SC7-2-1-2-1(N) from cluster IIIA and; Dulce Amallino, WNCDMRSC08R686(A) and WNCDMRSC08710 belonging to cluster IIIB can be recommended for further use in breeding program.
Genetic improvement is the most viable approach to any breeding program. In case of sweet corn, a relatively narrow genetic background with limited sources of germplasm and poorly defined heterotic groups are the reasons for modest improvement of sweet corn compared to field corn. In conclusion, the present study, based on comprehensive use of morphological and molecular markers, has generated many useful information regarding genetic diversity and population structure of sweet inbred lines that will definitely help in germplasm enhancement and hybrid breeding.
Acknowledgements
The corresponding author is grateful to Banaras Hindu University for providing UGC Research Fellowship for pursuing Ph.D. Authors are thankful to All India Co-ordinated Research Project on Maize lead by Indian Institute of Maize Research, Ludhiana, for providing sweet corn inbred lines used in the study. The help of Dr. Rajkumar Zunjare, Division of Genetics, IARI, New Delhi, is duly acknowledged.
Compliance with ethical standards
Conflict of interest
The authors declared that they have no conflict of interest in the publication.
References
- Amorim EP, Almeida CCD, Sereno M, Bered F, Neto JFB. Genetic variability in sweet corn using molecular markers. Maydica. 2003;48:177–181. [Google Scholar]
- Babic M, Babic V, Prodanovic S, Filipovic M, Andjelkovic V. Comparison of morphological and molecular genetic distances of maize inbreds. Genetika. 2012;44(1):119–128. doi: 10.2298/GENSR1201119B. [DOI] [Google Scholar]
- Babic V, Srdic J, Pajic Z, Grcic N, Filipovic M. The prediction of heterosis based on the phenotypic distance of sweet maize parental lines. Ratar Povrt. 2014;51(1):23–28. doi: 10.5937/ratpov51-5915. [DOI] [Google Scholar]
- Babic V, Nikolic A, Andjelkovic V, Kovacevic D, Filipovic M, Vasic V, Mladenovic-Drinic S. AUPOV morphological versus molecular markers for maize inbred lines variability determination. Chilean J Agric Res. 2016;76(4):417–426. doi: 10.4067/S0718-58392016000400004. [DOI] [Google Scholar]
- Boyer CD, Shannon JC. The use of endosperm genes for sweet corn improvement. In: Janick J, editor. Plant breeding reviews. Hoboken: Wiley; 1984. pp. 139–161. [Google Scholar]
- Choudhary SB, Sharma HK, Kumar AA, Maruthi RT, Mitra J, Chowdhury I, Singh BK, Karmakar PG. SSR and morphological trait based population structure analysis of 130 diverse flax (Linum usitatissimum L.) accessions. CR Biol. 2017;340:65–75. doi: 10.1016/j.crvi.2016.12.002. [DOI] [PubMed] [Google Scholar]
- Creech RG. Genetic control of carbohydrate synthesis in maize. Genetics. 1965;52:1175–1186. doi: 10.1093/genetics/52.6.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagla MC, Gadak RN, Kumar N, Ajay BC, Ram C. A potential scope of sweet corn for peri-urban farmers in India. Popul Kheti. 2014;2:69–73. [Google Scholar]
- Doyle JJ, Doyle JL. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Photochem Bull. 1987;19:11–15. [Google Scholar]
- Feng ZL, Liu J, Fu FL, Li WC. Molecular mechanism of sweet and waxy in maize. Int J Plant Breed Genet. 2008;2:93–100. doi: 10.3923/ijpbg.2008.93.100. [DOI] [Google Scholar]
- Gerdes JT, Tracy WF. Diversity of historically important sweet corn inbreds as estimated by RFLPs, morphology, isozymes, and pedigree. Crop Sci. 1994;34:26–33. doi: 10.2135/cropsci1994.0011183X003400010004x. [DOI] [Google Scholar]
- Hossain F, Nepolean T, Vishwakarma AK, Pandey N, Prasanna BM, Gupta HS. Mapping and validation of microsatellite markers linked to sugary1 and shrunken2 genes in maize (Zea mays L.) J Plant Biochem Biotechnol. 2015;24(2):135–142. doi: 10.1007/s13562-013-0245-3. [DOI] [Google Scholar]
- James MG, Robertson DS, Myers AM. Characterization of the maize gene sugary1, a determinant of starch composition in kernels. Plant Cell. 1995;7:417–429. doi: 10.1105/tpc.7.4.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jha S, Singh NK, Agrawal PK. Complementation of sweet corn mutants: a method for grouping sweet corn genotypes. J Genet. 2016;95(1):183–187. doi: 10.1007/s12041-015-0608-8. [DOI] [PubMed] [Google Scholar]
- Khanduri A, Hossain F, Lakhera PC, Prasanna BM. Effect of harvest time on kernel sugar concentration in sweet corn. Indian J Genet Plant Breed. 2011;71:231–234. [Google Scholar]
- Lertrat K, Pulam T. Breeding for increased sweetness in sweet corn. Int J Plant Breed. 2007;1(1):27–30. [Google Scholar]
- Liu KJ, Muse SV. Power marker: an integrated analysis environment for genetic marker analysis. Bioinformatics. 2005;21:2128–2129. doi: 10.1093/bioinformatics/bti282. [DOI] [PubMed] [Google Scholar]
- Lopes AD, Scapim CA, Mangolin CA, Machado MFPS. Genetic divergence among sweet corn lines estimated by microsatellite markers. Genet Mol Res. 2014;13(4):10415–10426. doi: 10.4238/2014.December.12.3. [DOI] [PubMed] [Google Scholar]
- Lopes AD, Scapim CA, da Silva Machado MFP, Mangolin CA, Silva TA, Cantagali LB, Teixeira FF, Mora F. Genetic diversity assessed by microsatellite markers in sweet corn cultivars. Scientia Agricola. 2015;72(6):513–519. doi: 10.1590/0103-9016-2014-0307. [DOI] [Google Scholar]
- Mazidf MS, Rafii MY, Hanafi MM, Rahim HA, Shabanimofrad M, Latif MA. Agro-morphological characterization and assessment of variability, heritability, genetic advance and divergence in bacterial blight resistant rice genotypes. S Afr J Bot. 2013;86:15–22. doi: 10.1016/j.sajb.2013.01.004. [DOI] [Google Scholar]
- Mehta B, Hossain F, Muthusamy V, Baveja A, Zunjare R, Jha SK, Gupta HS. Microsatellite-based genetic diversity analyses of sugary1-, shrunken2- and double mutant- sweet corn inbreds for their utilization in breeding programme. Physiol Mol Biol Plants. 2017;23(2):411–426. doi: 10.1007/s12298-017-0431-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monsoon Agro Bio Ltd (2017) Sweet corn exchange in India. http://www.cornclub.com. Accessed 9 Aug 2017
- Pajic Z, Dukanovic L, Eric U. Effects of endosperm mutants on maize seed germination. Genetika. 2004;36(3):265–270. doi: 10.2298/GENSR0403265P. [DOI] [Google Scholar]
- Pandey S, Ansari WA, Choudhary BR, Pandey M, Jena SN, Singh AK, Dubey RK, Singh B. Microsatellite analysis of genetic diversity and population structure of hermaphrodite ridge gourd (Luffa hermaphrodita) 3 Biotech. 2018;8:17. doi: 10.1007/s13205-017-1030-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park KJ, Sa KJ, Koh H, Lee JK. QTL analysis for eating quality-related traits in an F2:3 population derived from waxy corn×sweet corn cross. Breed Sci. 2013;63:325–332. doi: 10.1270/jsbbs.63.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrier X, Flori A, Bonnot F. Data analysis methods. In: Hamon P, Seguin M, Perrier X, Glaszmann JC, editors. Genetic diversity of cultivated tropical plants. Enfield: Science Publishers Montpellier; 2003. pp. 43–76. [Google Scholar]
- Qi X, Zhao Y, Jiang L, Cui Y, Wang Y, Liu B. QTL analysis of kernel soluble sugar content in supersweet corn. Afr J Biotechnol. 2009;8(24):6913–6917. [Google Scholar]
- Rupp JV, Mangolin CA, Scapim CA, Machado MFPS. Genetic structure and diversity among sweet corn (su1-germplasm) progenies using SSR markers. Maydica. 2009;54:125–132. [Google Scholar]
- Seshu G, Rao MVB, Sudarshan MR. Combining ability in diallel crosses of elite inbred lines of sweet corn (Zea mays L. saccharata) Green Farming. 2015;6(2):222–226. [Google Scholar]
- Solomon KF, Martin I, Zeppa A. Genetic effects and genetic relationships among shrunken (sh2) sweet corn lines and F1 hybrids. Euphytica. 2012;185:385–394. doi: 10.1007/s10681-011-0555-2. [DOI] [Google Scholar]
- Srdic J, Nikolic A, Pajic Z, Drinic SM, Filipovic M. Genetic similarity of sweet corn inbred lines in correlation with heterosis. Maydica. 2011;56(3):251–256. [Google Scholar]
- Tracy WF. History, Genetics and Breeding of super sweet (shrunken2) sweet corn. In: Janick J, editor. Plant breeding reviews. Hoboken: Wiley; 1997. pp. 189–236. [Google Scholar]
- Tracy WF. Sweet corn. In: Hallauer AR, editor. Specialty corns. 2. Boca Raton: CRC Press; 2001. pp. 155–197. [Google Scholar]