Abstract
Complex I (NADH:ubiquinone oxidoreductase) uses the reducing potential of NADH to drive protons across the energy-transducing inner membrane and power oxidative phosphorylation in mammalian mitochondria. Recently, cryoEM analyses have produced close-to-complete models of all 45 subunits in the bovine, ovine and porcine complexes, and identifed two states relevant to complex I in ischemia-reperfusion injury. Here, we describe the 3.3-Å structure of complex I from mouse heart mitochondria, a biomedically-relevant model system, in the ‘active’ state. We reveal a nucleotide bound in subunit NDUFA10, a nucleoside-kinase homolog, and define mechanistically-critical elements in the mammalian enzyme. By comparisons with a 3.9-Å structure of the ‘deactive’ state and with known bacterial structures we identify differences in helical geometry in the membrane domain that occur upon activation, or that alter the positions of catalytically-important charged residues. Our results demonstrate the capability of cryoEM analyses to challenge and develop mechanistic models for mammalian complex I.
Introduction
Respiratory complex I (NADH:ubiquinone oxidoreductase), one of the largest membrane-bound enzymes in the cell, is a major entry point to the electron transport chain in mammalian mitochondria1. It powers ATP synthesis by capturing the free energy released by electron transfer from NADH to ubiquinone to drive protons across the inner membrane and support the proton-motive force. As an essential metabolic enzyme and an important contributor to cellular reactive oxygen species production, mutations in its subunits and assembly factors cause a wide range of inherited neuromuscular and metabolic diseases2. Mammalian complex I contains 45 subunits, encoded on both the nuclear and mitochondrial genomes1. Fourteen ‘core’ subunits, conserved from bacteria to humans, are sufficient for catalysis, while 31 ‘supernumerary’ subunits are required for assembly, stability or regulation, or fulfil independent roles in mitochondrial metabolism3,4.
Structural knowledge on mammalian complex I (Figure 1) has exploded recently. First, based on data from bacterial complex I5–7, the core subunits were modeled using a 5 Å single-particle electron cryomicroscopy (cryoEM) density map of the enzyme from Bos taurus (bovine) heart mitochondria8. The architecture of the supernumerary ensemble was revealed around the core, and fourteen supernumerary subunits assigned. Crystallographic data on a membrane-domain subcomplex was then used for eight further assignments9. Subsequently, an improved-resolution (4.2 Å) cryoEM map enabled modelling of all 45 subunits, providing the first structure of the entire mammalian complex10. Further structures of complex I from ovine-heart mitochondria11, and of the complex in the respirasome (a supercomplex of complexes I, III and IV) from porcine-heart mitochondria12 and cultured human cells13 have since improved the resolution to 3.6 Å, enabling more accurate models containing greater proportions of the residues and side chains.
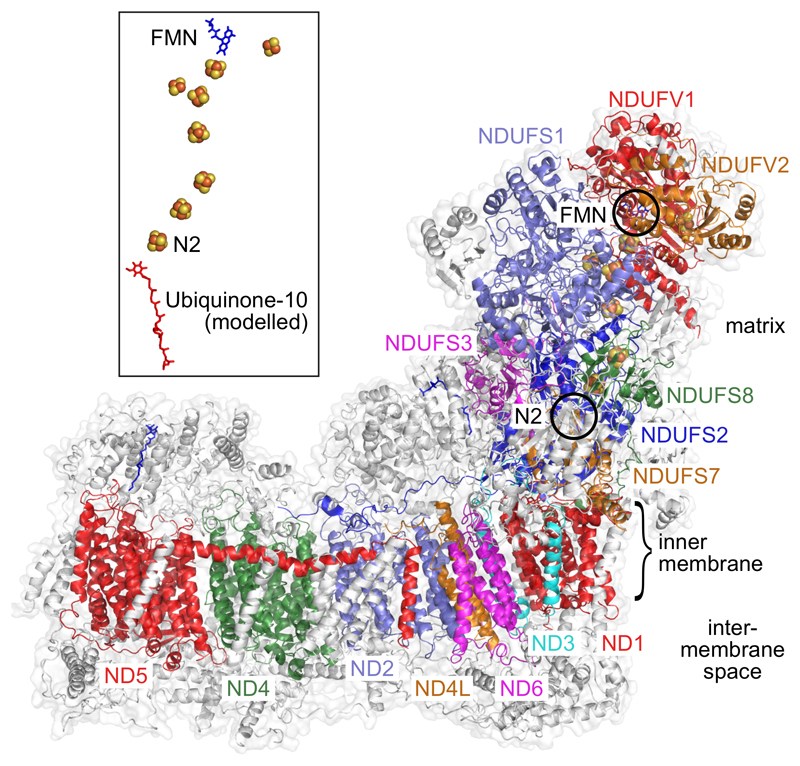
Figure 1. Overview of the structure of mouse complex I.
The fourteen core subunits are in color and labelled accordingly and the 26 supernumerary subunits are in white. The positions of the flavin mononucleotide (FMN), at the NADH binding site, and FeS cluster N2, the immediate electron donor to ubiquinone, are indicated. The structure of the cofactor chain, together with the modeled structure of ubiquinone-1045, is shown separately in the inset also.
In complex I, NADH is oxidized by a flavin mononucleotide in the hydrophilic domain, then electrons are transferred along a chain of seven iron-sulfur (FeS) clusters to ubiquinone (Figure 1), bound in a long channel at the interface between the hydrophilic and membrane domains. As first described in bacterial complex I6,7, then recapitulated for the yeast14 and mammalian complexes8,10,11, the membrane domain contains four antiporter-like units. They contain elements indicative of ion-transport activities, including π-bulges15,16, interrupted transmembrane helices (TMHs), and a series of buried charged residues linking them to the quinone-binding region. Computational simulations have suggested models for how the redox energy is captured, transferred and used to drive proton transfer17–20 but experimental support is lacking so the mechanisms remain undetermined. Solving structures of complex I trapped in different functional states provides a clear strategy to elucidate its mechanisms of catalysis, but focus on obtaining a complete model for the mammalian complex, rather than on understanding the mechanism, have so far left this opportunity little explored.
Three distinct classes of particles were identified during the reconstruction of bovine complex I10. One (class 3) lacked key regions of density and was assigned to enzyme in the first stages of dissociation, whereas classes 2 and 1, which are related by subtle domain movements10, were tentatively assigned to the ‘active’ and ‘deactive’ states, respectively, both of which were present in the imaged preparation. The assignment was subsequently confirmed by imaging enzyme prepared in the deactive state21. The dominant class of ovine complex I11 corresponds to class 3 (partially dissociated), whereas the porcine12 and human13 respirasome complexes resemble the class 2 (active) state21. In the absence of substrates, mammalian complex I relaxes from the ‘active’ state, which is resting but ready to catalyze, into a profound resting state called the ‘deactive’ state22–25. The ubiquinone-binding site in the deactive enzyme is partially disordered10,21, so the slow reactivation process that returns the enzyme to the active state upon addition of NADH and ubiquinone may be initiated by the site reforming around the ubiquinone substrate as it binds and is reduced21. Notably, lack of O2 prevents respiratory chain turnover so the deactive state forms spontaneously during ischemia26, and its slow reactivation has been proposed to protect against ischemia-reperfusion injury by minimizing reactive oxygen species production27.
Here, we present a 3.3 Å resolution structure of complex I from mouse heart mitochondria in the active state (Figure 1). The improved resolution has revealed a nucleotide in the active site of the nucleoside-kinase homolog subunit NDUFA10, and provided structural definition of mechanistically-relevant elements in the mammalian enzyme. Furthermore, comparison with a 3.9 Å resolution structure of the mouse deactive state has uncovered conformational changes in the π-bulge in TMH3 of subunit ND6 that result in residues rotating around the helical axis upon activation. Similarly, comparison with bacterial structures has uncovered conformational changes in the π-bulges in TMH8 of ND2, ND4 and ND5 that alter the positions of charged residues required for proton translocation. Thus, our structures of mammalian complex I in defined states have revealed conformational variations in the membrane domain that may be necessary for catalysis to occur during the catalytic cycle, demonstrating their resolution is now sufficient to challenge and develop mechanistic models.
Results
Structure of mouse complex I
A protocol for isolating pure, monodisperse and catalytically-active complex I from mouse-heart mitochondria was miniaturized from that used to prepare complex I from bovine-heart mitochondria for previous cryoEM analyses8,10,21. The enzyme was frozen onto PEGylated gold grids21,28,29 and imaged using a Titan Krios microscope with a K2 detector at a calibrated pixel size of 1.05 Å. The final cryoEM density map reached a global resolution of 3.3 Å (Supplementary Figure 1 and Table 1), starting from 61K particles. In comparison, the bovine and ovine datasets contained 139K and 241K particles, respectively, for 4.2 and 3.9 Å resolution10,11. Furthermore, the final reconstruction contained only 20K particles in a single major class containing 70% of the 29K particles considered for 3D classification (Supplementary Figure 2), suggesting the mouse preparation is more homogenous than the bovine10 and ovine11 preparations, which each contained three major classes. The 3.3 Å resolution map contains many well-resolved features and clear side-chain densities (Supplementary Figure 3) that enabled confident modelling (with sidechains) of 96% of the residues (Supplementary Table 1). The global resolution and completeness of the model (summarized in Figure 1) now match those of the best resolved thermophilic-bacterial complex I structure7, which, until now, has been the gold-standard structure.
Table 1. Cryo-EM data collection, refinement and validation statistics.
| Active state (EMDB 4345, PDB 6G2J) |
Deactive state (EMDB 4356, PDB 6G72) |
|
|---|---|---|
| Data collection and processing | ||
| Magnification | 47,600 | 37,600 |
| Voltage (kV) | 300 | 300 |
| Electron exposure (e–/Å2) | 50 | 50 |
| Defocus range (μm) | -1.5 to -3.1 | -2.2 to -3.4 |
| Pixel size (Å) | 1.05 | 1.33 |
| Symmetry imposed | None | None |
| Initial particle images (no.) | 60,851 | 40,131 |
| Final particle images (no.) | 20,370 | 17,066 |
| Map resolution (Å) | 3.3 | 3.9 |
| FSC threshold | 0.143 | 0.143 |
| Map resolution range (Å) | 472 to 3.3 | 484 to 3.9 |
| Refinement | ||
| Initial model used | PDB 5LC5 | PDB 6G2J |
| Model resolution (Å) | 3.6 | 4.1 |
| FSC threshold | 0.5 | 0.5 |
| Model resolution range (Å) | 472 to 3.3 | 484 to 3.9 |
| Map sharpening B factor (Å2) | -61 | -67 |
| Model composition | ||
| Nonhydrogen atoms | 66,811 | 64,923 |
| Protein residues | 8,164 | 8,072 |
| Ligands | 34 | 14 |
| B factors (Å2) | ||
| Protein | 47 | 71 |
| Ligand | 45 | 67 |
| R.m.s. deviations | ||
| Bond lengths (Å) | 0.009 | 0.012 |
| Bond angles (°) | 1.33 | 1.33 |
| Validation | ||
| MolProbity score | 2.03 | 2.18 |
| Clashscore | 6.63 | 9.10 |
| Poor rotamers (%) | 0.06 | 0.23 |
| Ramachandran plot | ||
| Favored (%) | 84.7 | 82.7 |
| Allowed (%) | 14.9 | 16.9 |
| Disallowed (%) | 0.4 | 0.4 |
Local resolution as an indicator of rigidity
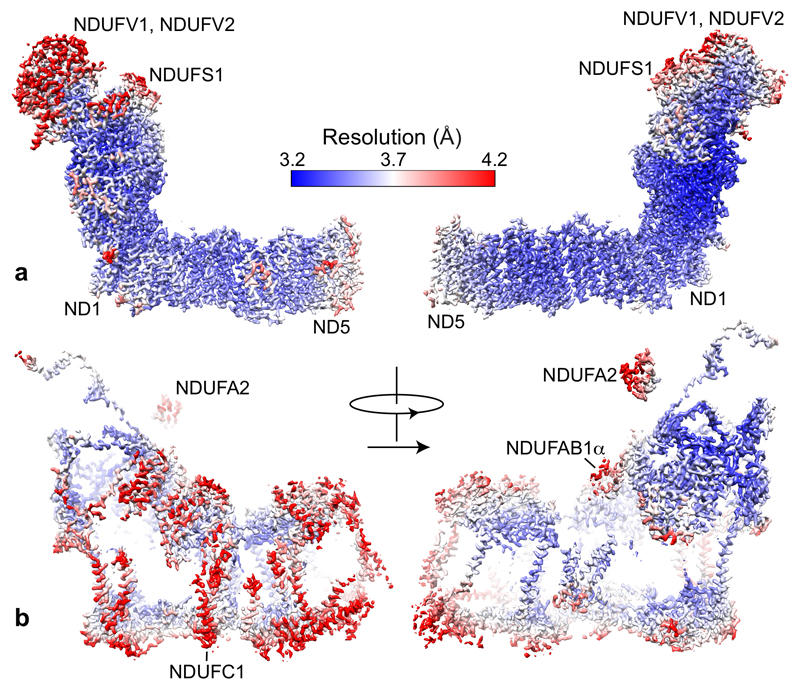
Figure 2 shows how the local resolution of the cryoEM map varies in the core and supernumerary subunits, and Supplementary Table 2 gives the local resolution estimate for each subunit. The central core subunits are highly resolved, but the resolution decreases towards the end of each core domain (Figure 2a), for ND5 in the membrane domain, and for NDUFV1, NDUFV2 and the large domain of NDUFS1 at the top of the hydrophilic domain (see Supplementary Table 1 for alternative subunit nomenclatures). Notably, the relatively low resolutions of these subunits coincide with the smaller surface-area contacts they make with the rest of the complex (49, 33, 36 and 30%, respectively, vs. 69 ± 6% for the other core subunits, Supplementary Table 2). Figure 2 also shows that the supernumerary subunits are lower resolution than the core subunits, and they also have lower contact areas (42 ± 12%) than the core subunits. Strikingly, the resolution is asymmetric between the two ‘sides’ of the complex (Figure 2b), arguing against imperfect particle alignment (which would affect the extremities most) as a substantial effect. The origin of the asymmetry is unclear, although the better-resolved side contains the ND5 transverse helix and contacts complex III in the respirasome12,30. Finally, supernumerary subunits distant from the core stand out in Figure 2b: NDUFA2 and NDUFAB1α contact only the large domain of NDUFS1 and NDUFA6, respectively (contact areas 25 and 17%), and NDUFC1 is bound peripherally only to supernumerary NDUFC2. The concept of low surface area contacts producing decreased resolution through increased structural dispersion or flexibility may thus help to explain how the resolution varies through the complex, particularly for the supernumerary subunits, which rely less on tight and specific interactions with the complex for their structure and function than the core subunits.
Figure 2. The resolution of densities for the core and supernumerary subunits in the 3.3 Å resolution map of active mouse complex I.
Local resolutions were estimated using the Local Resolution function in RELION with default parameters. The figure was created using UCSF Chimera, using the model to separate the core (a) and supernumerary (b) subunit densities.
Assignment of the 3.3 Å resolution structure to the active state, and comparison with a deactive structure
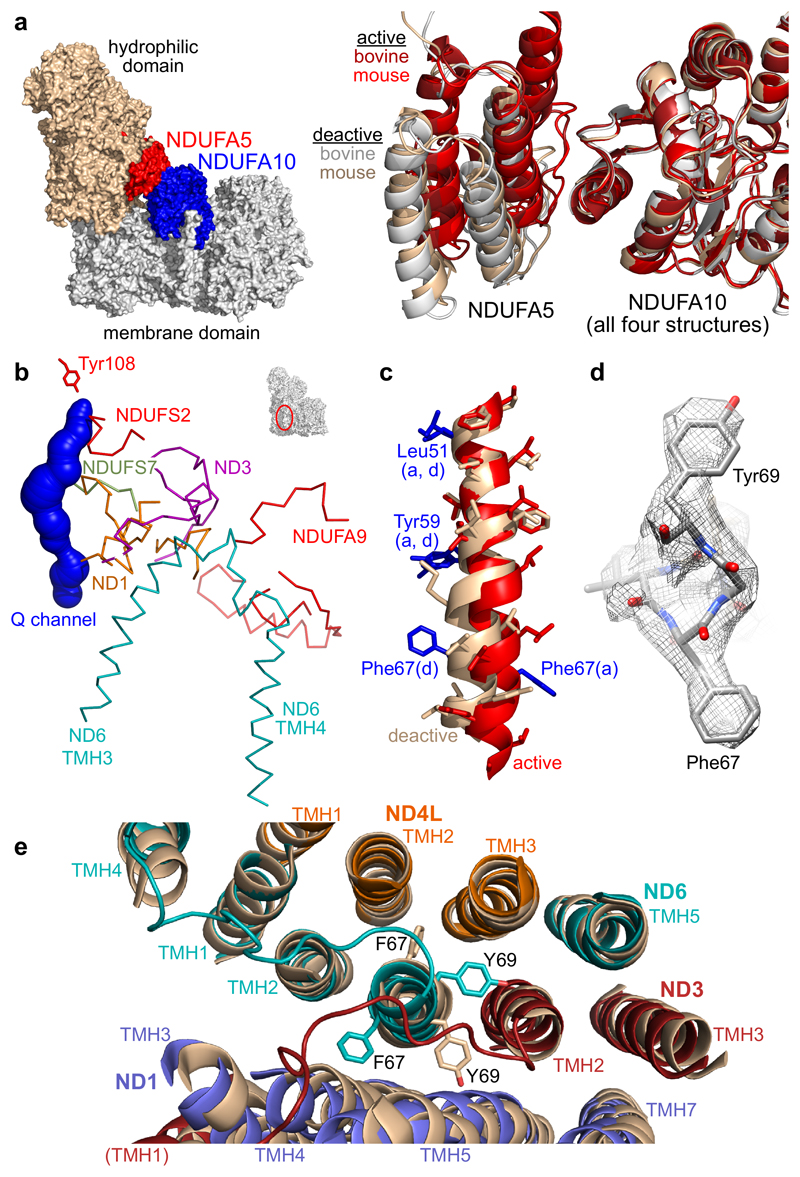
The 3.3 Å resolution structure of mouse complex I was assigned to the active state as follows. First, global RMSD values calculated in Pymol were much lower for the active (class 2) bovine model than the deactive (class 1) or broken (class 3) models (1.6, 3.2 and 4.2 Å, respectively). Second, the different orientations of the hydrophilic and membrane domains in the active and deactive states, captured visually by the different positions of NDUFA5 (hydrophilic domain) and NDUFA10 (membrane domain), show clearly that the mouse structure matches the active bovine structure (Figure 3a). Third, densities for all the loops that become disordered in the deactive state10,21 are clearly visible in the mouse map, and remained so even when its resolution was decreased to 4.3 Å, to match the original bovine structures.
Figure 3. Differences in structure between the active and deactive states of mouse complex I.
a) Left - NDUFA5, on the hydrophilic domain, and NDUFA10, on the membrane domain, form a second contact between the two domains. Right – the different arrangements of NDUFA5 and NDUFA10 are illustrated by superimposing the NDUFA10 subunit in the NDUFA5-NDUFA10 pairs from the bovine and mouse deactive and active states. NDUFA5 is present in markedly different positions in the active (red) and deactive (wheat and grey) states. b) Structural elements that become disordered or change between the active and deactive states. The ubiquinone binding channel determined by CAVER 3.054 is shown for reference. c) The different structures of ND6-TMH3 in the active (red) and deactive (wheat) states, superimposed on the N-terminal section, and the positions of key sidechains. The π-bulge is present only in the deactive state. d) Densities for key sidechains in the active state. e) Rotation of the bulky sidechains of Phe67 and Tyr69 between the TMHs of ND3, ND4L and ND1. The structures were superimposed on ND4L with the active structure colored by subunit and the deactive in wheat. Only the TMHs are shown, plus the loops between ND3-TMHs 1 and 2 and ND6-TMHs 3 and 4.
To confirm that the active and deactive states observed for bovine complex I apply equally to the mouse enzyme, mouse complex I was prepared in the deactivate state21 and its structure determined to 3.9 Å resolution (Supplementary Figure 1), following 3D particle classification into a major (69%) and a minor (25%) class (Supplementary Figure 2). Comparisons of the minor class with the three classes of bovine complex I10 suggested it represents the active state. The model for the major class exhibited the lowest global RMSD value with deactive bovine complex I (1.6, 2.4 and 2.7 Å for classes 1, 2 and 3, respectively), and Figure 3a reveals a close match in the relative positions of NDUFA5 and NDUFA10. Consistent with deactive bovine complex I10,21, no clear densities are observed for the ND1 TMH5-6, ND3 TMH1-2 and NDUFS2 β1-β2 loops, or several loops in NDUFA9 (Supplementary Table 1), whereas key features not resolved in class 310, notably the ND5 transverse helix, are clearly visible. Therefore, the structural changes accompanying deactivation are common to the bovine and mouse enzymes, providing a robust basis for understanding the active-deactive transition in mammalian complex I.
Figure 3a shows that the interface between NDUFA5 and NDUFA10 (the only contact between the hydrophilic and membrane domains outside the main interface) changes upon deactivation. In the active state the buried interface of 400 Å2 includes three salt-bridges and three hydrogen bonds. In contrast, in the deactive state the interface is decreased to 200 Å2 and includes only one salt-bridge and one hydrogen bond. The implication that interactions between NDUFA5 and NDUFA10 stabilize the active state is supported by coarse-grained simulations that illustrated how NDUFA5 and NDUFA10 restrict global vibrational modes in the mammalian complex that resemble the changes required to interconvert the active and deactive states31.
In addition to the loops in ND1, ND3, NDUFS2 and NDUFA9 known to become disordered upon deactivation10,21, comparison of the higher-resolution active and deactive mouse structures reveals subtle differences in ND1, ND6 and NDUFS7. Figure 3b shows the structural elements that become disordered or change between the two states, illustrating how the perturbation propagates from the ubiquinone-binding site. In ND6 the TMH3-4 loop is altered, and in the deactive map TMH4 is poorly resolved. Furthermore, a striking difference between the active and deactive states is revealed in ND6-TMH3. In all existing structures6–8,10–14, a π-bulge (stretch of π-helix) has been modeled in ND6-TMH3, following its identification in bacterial complex I6. The π-bulge is present in deactive mouse complex I, but not in the active enzyme. Consequently, in the active state TMH3 is extended, and the residues in its C-terminal section are rotated around the helical axis (Figure 3c). Although ND6-Tyr59, which may form a functionally-important interaction with ND4L-Glu34, is at the start of the π-bulge and little affected, two aromatic residues, Phe67 and Tyr69 (Figure 3d), undergo large changes (Figure 3e). They rotate, respectively, from between ND4L-TMH2, 3 and ND1-TMH5, ND3-TMH2 in the deactive state to between ND1-TMH4, 5 and ND3-TMH2, ND4L-TMH3 in the active state. The rotation locks the enzyme into different conformations, explaining why ND6 moves relative to ND1 and ND3 between the active and deactive states10, and confirming the importance of the ND6 π-bulge for catalysis.
Phospholipid densities
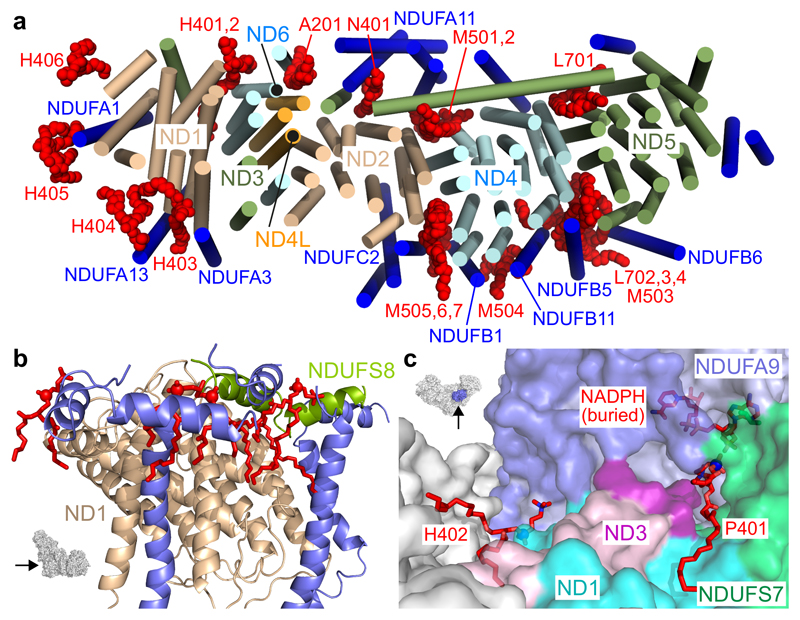
Densities for twenty phospholipids were identified in the map for active mouse complex I (Figure 4a and Supplementary Table 3, see Supplementary Figure 4 for example densities), with many of their headgroup phosphates stabilized by arginines, histidines or tyrosines32. In four cases, four acyl chains were observed so they were assigned as cardiolipins. All other phospholipids were modelled as phosphatidylethanolamines, unless density features indicated phosphatidylcholine is more likely (12 and four cases, respectively). Eight phospholipids overlap with the 12 phospholipids modeled previously around ovine complex I11, and nine and 11 of them with the 11 and 18 phospholipids modeled around complex I in the porcine12 and human13 respirasomes, respectively (Supplementary Table 3). Altogether nine cardiolipins have been modeled, corresponding to the ~10 cardiolipins that copurify with bovine complex I, along with a similar number of interchangeable phosphatidylethanolamines and phosphatidylcholines33.
Figure 4. Phospholipids in the 3.3 Å resolution structure of mouse complex I.
a) Membrane-bound helices in the complex I viewed from the matrix side, with phospholipids in red and labelled with the nomenclature in Supplementary Table 3. b) Phospholipids stabilizing amphipathic helices on the matrix side of the heel of the complex. The structure shown in blue is from supernumerary subunits NDUFA1, NDUFA3, NDUFA7, NDUFA12 and NDUFA13 and phosphorus atoms are marked with spheres. c) Two phospholipids that interact with NDUFA9 may stabilize loops in ND3 and NDUFA9 that become disordered in the deactive state (shown in darker colors than the rest of the two subunits). The arrows on the insets to b) and c) indicate the directions of view.
The locations of the phospholipids (Figure 4a and Supplementary Table 3) suggest they stabilize intraprotein and subunit interactions. Four stabilize the C-terminus of ND5, one at its interface with NDUFA11 and ND2 and two ‘behind’ its transverse helix, and on the heel they stabilize hydrophilic-domain amphipathic helices and supernumerary TMHs against ND1 (Figure 4b). Many phospholipids are packed between the core membrane subunits. Notably, the contact areas between ND2, ND4 and ND5 are low (less than 20% of the surface of ND4 is buried at interfaces with ND2 and ND5) so, along with the ‘cage’ of supernumerary subunits, phospholipids may be crucial for the integrity of the membrane domain. Finally, two phospholipids (Figure 4c) interact with NDUFA9 residues relevant to the active-deactive transition. In particular, one of them extends from close to the NAD(P)H ribose-phosphate, along an interface between NDUFS7 and two regions (a loop in NDUFA9 and the start of the ND3-TMH1-2 loop) that become disordered in the deactive state. Therefore, protein-phospholipid interactions may stabilize loops that are inherently unstable and susceptible to becoming disordered.
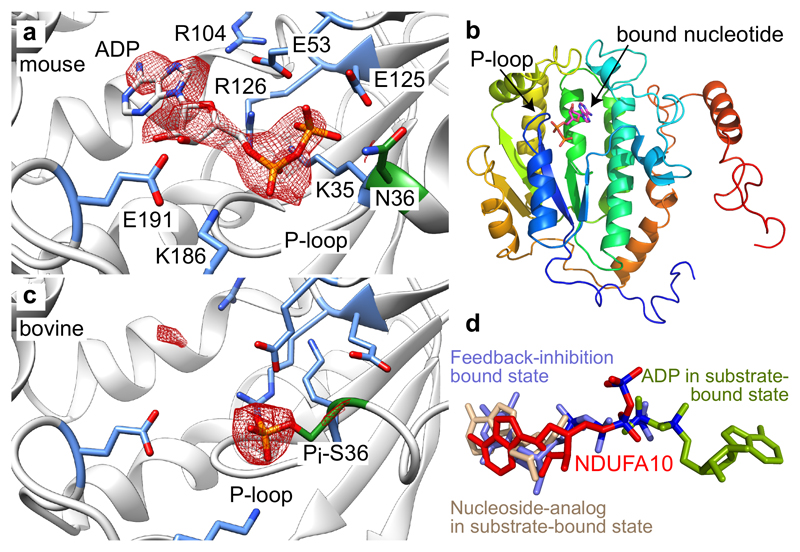
Nucleotide binding to NDUFA10
The improved resolution of the active mouse structure has allowed the tetrahedral coordination of the Zn in NDUFS6 and the acyl chains in NDUFAB1α and NDUFAB1β to be clearly visualized, revealed the coordination of the NAD(P)H in NDUFA9 by conserved charged residues, confirmed 11 disulfide bonds on the intermembrane space side of the complex — and showed that no disulfide is present in the thioredoxin-like subunit NDUFA2 (located in the reducing matrix environment). Most strikingly, however, there is clear density for a bound nucleotide in subunit NDUFA10, a member of the nucleoside-kinase family (Figures 5a and 5b) that transfer a phosphate from a nucleotide triphosphate to a nucleoside34. The density has been tentatively modeled as ADP, because it resembles a purine nucleoside plus two phosphates, and ADP is abundant in the matrix. The putative ADP is coordinated by the sidechains of widely conserved residues: the adenosine by Tyr83, Gln97, Arg104 (which interacts with Glu53 and Arg126), Phe134, Glu191 and Tyr198, and the phosphates by Lys35 (P-loop), Glu125 and Lys186. However, comparison with structural information from canonical nucleoside kinases, such as ADP-bound human deoxycytidine kinase35, showed that the ADP is bound with its adenosine in the nucleoside-binding site and its two phosphates towards the P-loop nucleotide-phosphates binding site (Figure 5d). Therefore, it is bound in a configuration resembling the feedback-inhibition mode characterized for deoxythymidine triphosphate bound to Drosophila melanogaster deoxyribonucleoside kinase36.
Figure 5. Nucleotide/nucleoside binding to NDUFA10.
a) Density for the bound nucleotide/nucleoside (modeled as ADP) in NDUFA10 in active mouse complex I. The density has been carved around the modeled ADP and the phosphate in the overlaid model from panel c (2 Å carve radius, threshold level of 0.075). Key residues that interact with the nucleotide/nucleoside are indicated, and Asn36, the homolog of Ser36 is in green. b) Overview of the typical nucleoside-kinase fold of NDUFA10 with nucleotide bound and the P-loop adjacent to the bound phosphates. c) The nucleotide/nucleoside-binding site in bovine complex (taken from PDB 5O3121) with a phosphate added to the side-chain of Ser36 (green) on the basis of mass spectrometry evidence. The density has been carved around the phosphate and the ADP in the overlaid model from panel a (2 Å carve radius, threshold level of 0.16). d) Comparison of the binding mode of the ADP modeled in NDUFA10 (red) with deoxythymidine triphosphate bound in the feedback inhibition mode in Drosophila melanogaster deoxyribonucleoside kinase (PDB 1OE036, blue) and with ADP (green) and a nucleoside analog (wheat) in the substrate-binding site of human deoxycytidine kinase (PDB 2ZI435). The structures were overlaid using the whole nucleoside kinase fold structures.
Similar, less well-defined densities are present (but were not interpreted) in NDUFA10 in the maps for the human13 and porcine12 respirasomes, but not in the maps for ovine11 or bovine complex I10. In the bovine maps increased density in the ADP phosphate-binding region can be attributed to phosphorylation of Ser3637 (Figure 5c), which has now been confirmed in the structurally-characterized (bovine) preparation (Supplementary Figure 5). Ser36 is conserved in ovine complex I, but substituted by Asn or Gly in the mouse, porcine and human homologs, suggesting phosphorylation blocks nucleotide binding. Other nucleoside kinases are present in mitochondria34, and as the NDUFA10 active site is accessible to solution and key residues are present38, phosphorylation of Ser36 and inhibitory substrate binding may together explain why no nucleoside kinase activity has yet been reported for mammalian complex I.
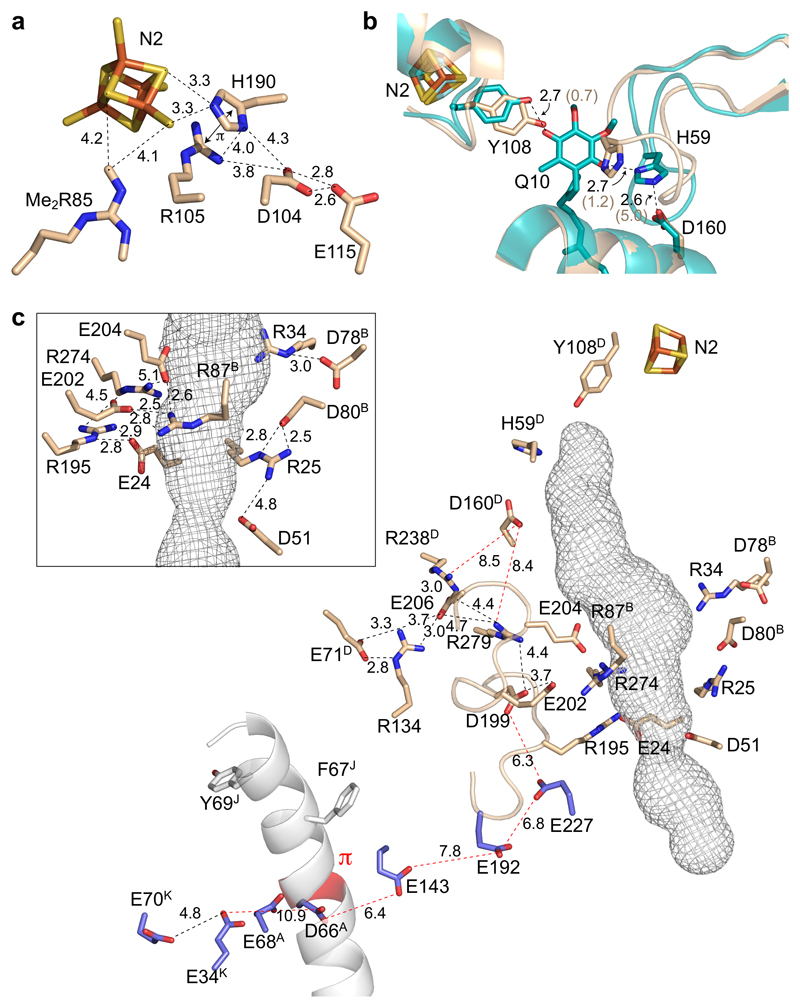
The second coordination sphere of cluster N2
The residues surrounding cluster N2, the final cluster in the FeS chain that donates electrons to ubiquinone (Figure 1), are clearly resolved in the active mouse complex I map (see, for example, Arg85 in Supplementary Figure 3), defining its environment in the mammalian enzyme (Figure 6a). Three basic residues, Arg85, Arg105 and His190 in NDUFS2, are close to N2 and their positive charges may explain its high reduction potential39. Arg85 is widely conserved, but absent from Thermus thermophilus, in which N2 has a lower reduction potential39. Furthermore, it has been shown to be symmetrically dimethylated in the mammalian and yeast enzymes40, but not in Escherichia coli, in which the N2 reduction potential is also unusually low39. Protonation of His190 upon N2 reduction explains why N2 has a weakly pH-dependent reduction potential41,42. Although the His190Met variant of Yarrowia lipolytica complex I remains catalytically competent41,42, molecular simulations have suggested His190 deprotonation is required for electron transfer from N2 to semiquinone, and that both His190 and Arg105 (which form a cation π-stacking arrangement) contribute to the pH dependence43. Notably, no channels from the His190 sidechain to the outside could be detected in Caver analyses (even with a 0.9 Å probe radius). Conversely, similar analyses revealed channels from both NDUFS2-Asp104 (within hydrogen-bonding distance of His190) and Glu115 (hydrogen bonded to Asp104). Unusually for cryoEM-density maps Asp104 and Glu115 both have well resolved sidechains, so their negative charges may be neutralized by protonation44. Therefore, they may act as a proton reservoir, for rapid proton delivery to His190 upon N2 reduction and/or rapid proton capture upon N2 oxidation.
Figure 6. The environment of cluster N2, the ubiquinone-binding channel and the E-channel in active mouse complex I.
a) Cluster N2 and nearby key residues in NDUFS2. Dimethyl-Arg85 and Arg105 are positively charged, His190 is the redox-Bohr group that changes protonation state upon N2 reduction41,42, and Asp104 and Glu115 may transfer protons to/from His190. b) The structure around the ubiquinone-headgroup binding site with key residues in NDUFS2. The structure of active mouse complex I (wheat) is overlaid on the modeled structure of bovine complex I with ubiquinone-10 bound (teal)45 in which hydrogen bonds are present between the headgroup and Tyr108 and His59, and His59 and Asp160. Distances in brackets are for the active mouse structure. c) The ubiquinone-binding channel (mesh) in active mouse complex I was predicted using CAVER 3.054 with a 1.4 Å probe radius. The key residues from panel b are shown with charged residues in the vicinity of the site (inset, charged residues around the center of the channel and their interactions). The main figure highlights the carboxylate-rich ND1 TMH 5-6 loop and the E channel residues (blue) that connect the ubiquinone-binding site to the π-helix present in ND6 in the deactive enzyme, and to charged residues in the central membrane plane7. Long distances of interest are in red, and all residues are in ND1 unless otherwise indicated (B - NDUFS7, D - NDUFS2, J - ND6 and K - ND4L). All distances are in Å.
The ubiquinone binding site and E-channel
Tyr108 and His59 in NDUFS2 are generally considered to bind the ubiquinone headgroup in complex I by forming hydrogen bonds to its redox-active C=O groups. Both residues are well resolved in the mouse map, but positioned too close for ubiquinone to fit between them (5.9 Å O-to-N distance). Figure 6b compares the structure with a modeled structure for ubiquinone bound to bovine complex I (7.8 Å O-to-N distance)45, showing the substantial change in the His59 loop required for binding. Compared to previous structures, the mouse and porcine ubiquinone-binding sites match closely12, but differ from the ovine11 and Y. lipolytica14 structures, which have markedly different His59-loop conformations. The variation attests to the flexibility of the site, suggesting it is configured by substrate or inhibitor binding. Finally, in the mouse structure the His59 to NDUFS2-Asp160 (O-to-N) distance is 5.0 Å, whereas in the modeled ubiquinone-bound bovine45 and T. thermophilus7 structures the two are within hydrogen-bonding distance. Although Asp160 has been proposed to undergo large movements upon ubiquinone reduction to initiate proton translocation18, its position on a nearby helix is conserved in all structures reported so far.
Figure 6c shows how networks of charged residues surround the central section (isoprenoids ~4-7) of the predicted ubiquinone-binding channel, with the regions before and after dominated by hydrophobic residues45. As expected for cryoEM maps, the Arg sidechains in Figure 6c are better defined than the (negatively-charged) carboxylates44. Nevertheless, tight interactions are observed within the networks (Figure 6c, inset) that, by neutralizing individual charges and restricting hydration, may assist the hydrophobic isoprenoids to bind. The charged networks may also be involved in coupling ubiquinone reduction to proton translocation. Carboxylate residues on the ND1-TMH5-6 loop (that becomes disordered in the deactive state, but exhibits similar conformations in the active mouse and bovine10, porcine12, human13 and T. thermophilus7 structures) contribute to the ‘E-channel’7 (Figure 6c), which connects them (and thus the ubiquinone-binding site) to the π-bulge in ND6-TMH3 and two glutamates in ND4L, presenting a route for energy transfer to the chain of charged residues through ND2, ND4 and ND5.
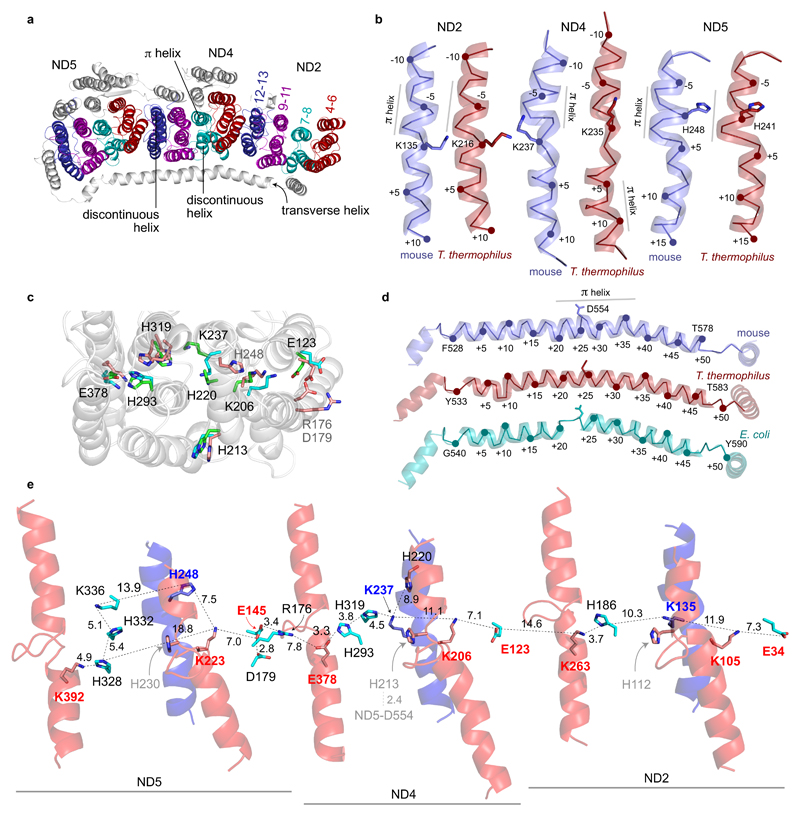
π-helices in ND2, ND4 and ND5
The antiporter-like subunits ND2, ND4 and ND5 contain a common core structure of ten TMHs, TMHs 4–13 in ND4 and ND5. In higher metazoans three TMHs have been lost from the N-terminus of ND2 during evolution46, so ND2 TMHs 1–10 correspond to ND4 and ND5 TMHs 4–13. For simplicity, we use the numbering system for ND4 and ND5 throughout. TMHs 4–8 and 9–13 comprise two symmetry-related domains6 that face opposite sides of the membrane in a front-to-back arrangement related by rotation in an axis along the membrane domain. As first observed in E. coli complex I6, symmetry-related TMHs 7 and 12 are discontinuous (interrupted in the center by substantial loops), and TMH 8 (but not the symmetry-related TMH13) is distorted. The six domains of ND2, ND4 and ND5 thus form alternating ‘layers’ of interrupted or distorted (7-8 and 12) and continuous-α-helical (4-6 and 9-11) TMHs, along the membrane domain (Figure 7a).
Figure 7. Charged residues and discontinuous and π-helices in subunits ND2, ND4 and ND5.
a) Overview of subunits ND2, ND4 and ND5 showing the layered arrangement of core TMHs 4 to 13. TMHs 4 to 6 (red) and 9 to 11 (magenta) are α helical whereas TMH7 is discontinuous, TMH8 contains π-helical structure (cyan) and TMH12 (blue) is also discontinuous. The transverse helix from ND5 is also shown. b) TMH8 in ND2, ND4 and ND5 orientated with the mitochondrial matrix at the top. c) Overlay of conserved charged residues in the central membrane plane in ND2, ND4 and ND5. A cartoon representation of ND4 is shown for reference, and conserved residues1,7 are colored cyan for ND2, green for ND4 and pink for ND5. Numbers refer to ND4, except for residues only in ND5 numbered in grey. d) Comparison of structures of the transverse helix in ND5 with TMH16 (the C-terminus) on the right and the conserved aspartate residue indicated. Groups of structures have been aligned using the whole subunit structures. The ribbons join the Cα atoms together, and each fifth residue is numbered. The approximate positions of stretches of π-helix are indicated. e) The distribution of charged residues and helical elements along the membrane arm with key residues labelled and the distances between ionizable groups shown in Å. TMH7 and 12 are in red and TMH8 is in blue. Residues labeled in red are on TMH5, TMH7 and TMH12. The mouse structure is for the active state, the structure of T. thermophilus complex I is from PDB 4HEA7 and the structure of the membrane domain of complex I from E. coli is from PDB 3RKO6.
The distortions in TMH8 of ND2, ND4 and ND5 have been described as π-bulges, although the formal (i + 5 → i) hydrogen-bonding pattern is not always fulfilled in current structures, and they are clearly distorted outside the π-bulges also (Figure 7b and Supplementary Figure 6). Formal π-bulges are present in ND2-TMH8 only in the T. thermophilus7 and human respirasome13 structures, although the mouse and human structures match closely. The π-bulge is centered on Leu131, close to Lys135, a conserved charged residue in the central membrane plane (Figures 7c and 8a). Comparison of all available structures, particularly the higher-resolution (≤3.3 Å) structures from mouse, T. thermophilus7 and E. coli6, revealed no substantial differences in ND2-TMH8 or Lys135. In contrast, in ND4-TMH8, formal π-bulges are present in a common position in the E. coli6, human13 and ovine11 structures (the human and mouse structures again match closely) but in a different position in T. thermophilus complex I7 (Figure 7b). Strikingly, the two π-bulges border a central unwound region that contains conserved Lys237 in different positions, rotated around the helical axis. In ND5-TMH8, formal π-bulges were detected centered on His248, a further conserved residue, in the mouse, porcine12, E. coli6 and T. thermophilus7 structures (Figure 7b). The position of His248 varies, although the positions of Pro242 and Phe258 are conserved to each side.
Figure 7c compares the positions of the conserved charged residues in ND2, ND4 and ND5, and Figure 7e shows how they form a chain along the membrane domain. In each subunit a His residue on the loop in interrupted-TMH7 points outside the subunit (Figure 7e, grey labels) [ND4-His213 interacts with ND5-Asp554 on the transverse helix, clasping it against ND4]. Glutamate residues on TMH5 may be stabilized electrostatically by charged residues on interrupted-TMH7 and -TMH12 (Figure 7e, red labels). ND2-Lys135 and ND4-Lys237 (Figure 7e, blue labels) are on TMH8 but not conserved in ND5. Instead, ND5-His248 on TMH8 is positioned, like ND4-His220, closer to the matrix side of the membrane, suggesting a role in proton uptake. Further residues, not conserved in all three subunits, contribute additional stepping stones along the domain (Figure 7e, black labels).
Finally, the ND5 transverse helix, which runs alongside ND5, ND4 and ND2 (Figure 1), was originally proposed to consistute an important coupling element for energy transfer6 — although this proposal was subsequently challenged by alterations to the helix that do not substantially affect catalysis47,48. The transverse helix contains a substantial stretch of π-helix in the mouse structure (Figure 7d), and also in the porcine and E. coli structures (in which the C-terminus of ND5 is clearly resolved)6,12. Conversely, in T. thermophilus complex I7 the transverse helix is α-helical throughout, except for a short distortion at the start. Despite these differences in structure, the position of ND5-Asp554, which interacts with ND4-His213 and has been proposed to be important for catalysis49, is conserved, and the whole transverse helix extends the same distance in each case — due to its different structures it contains three more residues in mouse than in T. thermophilus, and five more than in E. coli.
Discussion
A central question for complex I research is the mechanism of energy transduction: how energy from the redox reaction is captured, transferred through the protein, and consumed in proton transport. Here, we have provided the first experimental evidence for conformational changes in helical structures in the membrane domain. First, we reveal that ND6-TMH3 contains a π-bulge in the deactive state, but not the active state. Thus, it may be held in an energized conformation for catalysis, or the π-bulge may be formed and destroyed during each cycle to induce conformational changes. Second, we reveal differences in TMH8 in ND2, ND4 and ND5, resulting from different helical geometries and π-bulges in different subunits and species, which may simply reflect how they have come to rest following catalysis. Crucially, ND4-Lys237, a conserved charged residue, takes up different rotational positions around the helical axis, such that when the mouse residue is placed into the position observed in T. thermophilus the Lys-Nζ comes into hydrogen-bonding distance of ND4-His220 (Figure 7e). We therefore propose that geometrical changes in TMH8 may form and destroy interactions within the chain of charged residues, suggesting a mechanism for energy propagation. Specific water molecules have also been proposed, on the basis of computational simulations, to be crucial for forming functional connections within the chain17,19,20. Similar rotations of residues around a helical axis due to π-helix geometry changes have been observed previously in methane monooxygenase hydroxylase upon substrate binding15,50. Interestingly, comparable changes are not observed in TMH13, the symmetry-related partner of TMH8. TMH8 contributes to the matrix half-channel7, so we may speculate that the energy required for proton translocation is consumed mostly in proton uptake (rather than expulsion). Finally, the transverse helix in the mammalian enzyme also contains π-helical structure, which, if it is catalytically-relevant, could move along the helix during catalysis but not be formed or destroyed, because the helix ends are anchored. Alternatively, the compressed π-helix geometry may contribute to the pronounced curvature of the mammalian membrane domain and, consistent with increased resolution on this side of the structure (Figure 2b), ‘pull’ the otherwise-loosely-associated ND2, ND4 and ND5 subunits together into a tightly-bound functional assembly.
Originally, the driving force for cryoEM studies on mouse complex I was to enable molecular-level characterization of complex I-linked mitochondrial disease models. For example, the physiological effects of deleting subunit NDUFS4 have been studied extensively in mice51, but a molecular description of the compromised assembly, stability or reactivity of the complex remains obscure. However, despite technical challenges from the small scale of the preparation, the mouse enzyme led to a much higher resolution map, from far fewer particles, than the large-scale bovine preparation characterized previously. Although changes have been made to our data collection strategies (use of a K2 instead of a Falcon II detector, at higher magnification, and use of PEGylated-gold instead of carbon-Quantfoil grids21), and we have used updated data analysis software52,53, the fact that the mouse particles gave a single major class of the active state, rather than a mixture of active, deactive and broken states, is likely crucial for the improvement. We propose the increased homogeneity results from the much shorter time required to extract and cool the mouse-heart tissue. During slaughterhouse procedures and transport to the laboratory the large bovine heart maintains its temperature and becomes ischemic, promoting both degradation and deactivation. In any case, the resolution available from cryoEM studies of mammalian complex I is now sufficient to visualize individual sidechains and access mechanistically-relevant information. Structures of the enzyme trapped in different intermediate states are required next, for further elucidation of its mechanism of catalysis.
Online Methods
Preparation of mouse complex I
C57BL/6 mice were sacrificed by cervical dislocation in accord with the UK Animals (Scientific Procedures) Act, 1986 (PPL: 70/7538, approved by the local ethics committee and the UK Home Office). Hearts (typically 0.17 g) were excised, then immersed immediately in cold solution containing 10 mM Tris-HCl (pH 7.4 at 4 °C), 75 mM sucrose, 225 mM sorbitol, 1 mM EGTA, and 0.1% (w/v) fatty acid-free bovine serum albumin (Sigma-Aldrich). By coordinating with other researchers, most of the hearts used were harvested from animals also required for other purposes. All the following procedures were at 4 °C. Mitochondria were isolated from heart tissue as described previously55. Briefly, the hearts were cut into 1 mm sized pieces, washed, then homogenized (10 mL buffer (g tissue)-1) by 7-10 strokes of a Potter-Elvehjem homogenizer fitted with a Teflon pestle at ~1000 rpm. The homogenate was centrifuged (1,000 x g, 5 min.), then the supernatant centrifuged (9,000 x g, 10 min.) to collect the crude mitochondria. They were washed several times by resuspension and centrifugation then collected by centrifugation at 15,000 x g for 2 min., resuspended in 20 mM Tris-HCl (pH 7.4 at 4 °C), 1 mM EDTA and 10% (v/v) glycerol to 10-20 mg protein mL-1 and frozen for storage. The total time required from extracting the first heart to freezing the mitochondria was less than two hours. Mitochondria were thawed, diluted to 5 mg protein mL-1, then ruptured by three 5 s bursts of sonication (with 30 s intervals on ice) using a Q700 Sonicator (Qsonica) at 65% amplitude setting. The membranes were collected by centrifugation (75,000 x g, 1 hour), resuspended to 5 mg protein mL-1 and frozen for storage. The active enzyme preparation used 27.3 mg of membrane protein, isolated from 26 hearts that had been harvested from two batches of 10 and 16 mice. The deactive enzyme preparation used 17.2 mg of membrane protein, isolated from a single batch of 20 hearts.
Complex I was prepared using a method adapted from that for bovine complex I33. Mitochondrial membranes, typically containing ~15 mg protein, were thawed, then 1% n-dodecyl β-D-maltoside (DDM, Glycon) and 0.005% (w/v) PMSF were added, the mixture stirred for 30 min. to solubilize the complexes and centrifuged (48,000 x g, 30 min.). The supernatant was loaded onto a 1 mL Hi-Trap Q HP anion exchange column (GE Healthcare) pre-equilibrated with buffer A (20 mM Tris-HCl (pH 7.14 at 20 °C), 1 mM EDTA, 0.1% DDM, 10% (v/v) ethylene glycol, 0.005% asolectin (Avanti) and 0.005% CHAPS (Calbiochem)). The column was washed at 0.3 mL min-1 with buffer A for 2 min. then with 20% buffer B (buffer A + 1 M NaCl) for 7 min., then complex I was eluted in 35% buffer B for 10 min. Complex I-containing fractions (typically 1 mL total) were pooled and concentrated to ~100 µL using a 100 kDa cut-off Vivaspin 500 concentrator (Sartorius), loaded onto a Superose 6™ Increase 5/150 column (GE Healthcare) and eluted in 20 mM Tris-HCl (pH 7.14 at 20 °C), 150 mM NaCl and 0.05% DDM at 0.03 mL min-1. Complex I eluted at ~1.65 mL, and the highest concentration fraction (3 – 5 mg mL-1) was collected and used directly for cryoEM grid preparation. Typically, the NADH:decylubiquinone oxidoreductase activity was 10-12 µmol min-1 mg-1. For the deactive enzyme the membranes were incubated at 37 °C for 30 min. with the ratio of active and deactive complexes monitored by comparing the rates of NADH oxidation following a 5 min treatment with or without 2.5 mM N-ethyl maleimide, as described previously21. The proportion of active complex I decreased from 94 ± 13 to 39 ± 7% during the incubation.
Electrospray ionization-MS was performed on purified bovine complex I as described previously56 on a Sciex 4000QTRAP spectrometer, and data analyzed using Peakview 2.1. Tryptic digests of the NDUFA10-containing region were extracted from SDS-PAGE gels of the iodoacetamide-treated enzyme and analyzed by LTQ Orbitrap XL as described previously57, except using the ETD fragmentation mode.
CryoEM grid preparation and imaging
UltrAuFoil® gold grids (0.6/1, Quantifoil Micro Tools GmbH)28 were prepared as described previously21. Briefly, they were glow discharged (20 mA, 60 s), incubated in a solution of 5 mM 11-mercaptoundecyl hexaethyleneglycol (SPT-0011P6, SensoPath Technologies) in ethanol29 for two days in an anaerobic glovebox, then washed with ethanol and air-dried just before use. Using an FEI Vitrobot Mark IV, 2.5 μL of complex I solution (3 – 5 mg mL-1) were applied to each grid at 4 °C in 100% relative humidity, and blotted for 8 – 16 s at force setting -10, before the grid was frozen by plunging it into liquid ethane.
Grids were imaged using a Gatan K2 summit camera and GIF Quantum energy filter mounted on an FEI 300 keV Titan Krios microscope with a 100 μm objective aperture and EPU-1.9 software. Data for the active complex were recorded at the Astbury Centre for Structural Molecular Biology, University of Leeds, and data for the deactive complex at the UK National electron Bio-Imaging Centre (eBIC) at the Diamond Light Source. The detectors were operated in counting mode, and the energy filters in zero-energy-loss mode with a slit width of 20 eV. The active complex was imaged at 1.05 Å/pixel (47,600× actual magnification) and the deactive complex at 1.33 Å/pixel (37,600× actual magnification) with defocus ranges -1.5 to -3.1 μm and -2.2 to -3.4 μm, respectively, and the autofocus routine run every 10 μm. For the active complex, the dose rate was 5 electrons Å-2 s-1 with 10 s exposures captured in 25 frames; for the deactive complex it was 3.25 electrons Å-2 s-1 with 15 s exposures captured in 30 frames. The total dose was thus ~50 electrons Å-2 in each case.
Data processing
All analyses were carried out using the RELION 2.1 (patch b1) pipeline52,53. First, MotionCor2-1.0.5 (with dose weighting) was applied to both datasets to account for beam-induced movement58 and Gctf-1.06 was used for CTF estimation59. Then, for the active complex, 60,851 particles were extracted following autopicking and manual curation and subjected to 2D classification; 28,935 particles were retained. Following an initial movie refinement with a three frame running average, the particles were subjected to ‘particle polishing’, using default parameters for B-factor weighting; the highest resolution reconstruction was obtained using frames 1–12. The particles were then subjected to 3D classification into five classes, with the angular sampling increased to 3.7°. The two major classes (containing a total of 20,370 particles, 70%) were indistinguishable and combined for the final 3D autorefinement. The final resolution of the map is 3.3 Å. For the deactive complex, 40,131 particles were picked manually. Following one round of 2D classification (leaving 30,378 particles), a preliminary 3D autorefinement was performed and the particles re-centered and re-extracted. Then 24,644 particles were retained from 3D classification with a coarse angular sampling of 7.5°. Following an initial movie refinement with a five frame running average, particle polishing was performed using the default parameters for B-factor weighting; the highest resolution reconstruction was obtained using all 30 frames. The polished particles were then subjected to high-resolution 3D classification. The resolution was limited to 8 Å, 5 classes were provided and the angular sampling was increased gradually from 7.5° to 0.9°. Two major classes were obtained. The highest resolution class, which was attributed to the deactive enzyme, contained 69% (17,066) of the particles and was taken forward to 3D autorefinement, leading to a 3.9 Å resolution map. The second class, containing 25% of the particles, matched the structure of the active enzyme and the remaining three classes contained negligible numbers of particles. In all cases, the final 3D autorefinements used solvent-flattened FSC curves with a soft mask to minimize the effects of the large solvent contribution; this increases the accuracy of the resolution estimate after each iteration and gave slight increases in resolution. Finally, post-processing was performed with default parameters using masks created from fitted coordinate files (using the Molmap command in UCSF Chimera (1.11.2)60 and mask creation tools in RELION) in order to exclude interference from the detergent belt. Resolution estimates were based on the gold standard FSC = 0.143 criterion61.
Global structure resolutions were estimated from FSC curves in RELION61, as shown in Supplementary Figure 1. Local resolutions were estimated using the Local Resolution function in RELION (similar to the ‘blocres’ function in Bsoft62), with default parameters (sphere of radius 12.5 Å with a soft edge of 25 Å). Using the local resolution function to estimate the overall resolution of the active state structure gave 3.4 Å, close to the globally-calculated value of 3.3 Å. For individual subunits, UCSF Chimera was used to extract the resolution at each atomic coordinate and the values were averaged. Values calculated by masking individual subunit volumes in the local resolution map gave equivalent results.
Model building
The model for the active complex (Supplementary Table 1) was built first using Coot (0.8.9.1)63 and existing models for bovine complex I (PDB 5LC510 and PDB 5O3121) as templates, with the residues converted to the mouse sequences. The core subunit models were easily fit into the density map, with good agreement between the map and initial models; the only significant extension was to the C-terminus of NDUFV2. In total 4,408 core residues were modeled out of 4,504 (97.9%). The supernumerary subunits were less well resolved in the bovine maps, so the template models were less accurate. However, the higher resolution of the current map revealed clear side-chain densities for the majority of residues, allowing all 31 supernumerary subunits to be confidently modeled. The structure on the intermembrane-space side of the tip of the membrane domain is relatively poorly resolved; although we were able to extend NDUFB2 (C-terminus), NDUFB6 (C-terminus), NDUFB7 (N-terminus) and NDUFB8 (C-terminus) around the already-built C-terminal helix of NDUFB7, the model is less confident in this region. A number of minor extensions were added to other supernumerary subunits. In total 3,756 supernumerary residues were modeled out of 4,014 (93.6%). Thus, overall 95.9% of the residues were modeled, 95.7% with sidechains. Stretches of more than 10 unresolved residues comprise the likely flexible amino and carboxy termini of NDUFS1, NDUFS3, NDUFS7 (34 residues at the N-terminus), NDUFA2, NDUFA6, NDUFB3, NDUFB7, NDUFB11 and NDUFV3, as well as 13 residues in the middle of NDUFA7 and 26 residues in the middle of NDUFB6 (Supplementary Table 1).
The densities observed for the phosphopantetheine and acyl groups attached to both copies of NDUFAB1 were modeled using 11 and 12 carbon chains for chains T and U, respectively, matching the lengths of density observed. The unknown nucleotide bound to NDUFA10 was modeled as ADP. Arg-85 of NDUFS2 was modeled as symmetrically dimethylated based on the results from mass spectrometry analyses40 and the bulky density observed for its headgroup. The densities for possible disulfide bonds were inspected individually and bonds formed where appropriate: in the four CHCH domains in NDUFA8, NDUFB7 and NDUFS5 and in NDUFB10 (Cys76-Cys89 and Cys112-Cys123) which are all on the intermembrane space side, and at the membrane-intermembrane space interface in NDUFA11 (Cys20-Cys77). No density was observed for the disulfide between Cys94 and Cys114 in NDUFA11 observed previously in the porcine respirasome12. Post-translational modifications to peptide termini detected in bovine complex I3, notably N-formylation and N-acetylation, were assumed to be identical if the residues are conserved in mouse, and built where possible. Densities for twenty phospholipid molecules were identified and modeled as cardiolipin (PDB code CDL) in cases where four acyl chains were observed, or as phosphatidylethanolamine (3PE), unless density features indicated that phosphatidylcholine (PC1) is more likely.
The model was refined into the map by real-space refinement in Phenix 1.1364 using default parameters without secondary structure restraints. Cycles of manual adjustment and Phenix refinement were used to obtain the final model. The results were assessed by MolProbity 4.465 (see Table 1) and EMringer66 (score 3.37).
To build the deactive model, the model for the active complex was fitted roughly into the deactive map using the ‘fit-in-map’ function in UCSF Chimera, then rigid body refinement in Phenix was used to optimize the fit for each chain. Loops in ND1, ND3, NDUFS2 and NDUFA9 that are not observed in the bovine deactive structure were not observed in the deactive mouse enzyme either so were deleted from the model (Supplementary Table 1) giving a total of 94.8% of the residues modeled. Phospholipids were also removed from the deactive model since their densities were not sufficiently clearly defined. The final model was obtained by cycles of manual adjustment and Phenix refinement and assessed by MolProbity 4.465 (see Table 1) and EMringer66 (score 2.10).
Secondary structures were analyzed using DSSP (http://swift.cmbi.ru.nl/gv/dssp/)67. Surface areas of contacts between subunits were calculated using the PDBePISA server (http://www.ebi.ac.uk/pdbe/pisa/)68. The ubiquinone-binding channel was predicted using CAVER 3.054 with a with a 1.4 Å probe radius, starting from the sidechain O atom of NDUFS2-Tyr108. Protein structure and density figures were created using MacPymol or UCSF Chimera.
Supplementary Material
Acknowledgements
We thank the staff at the Astbury Biostructure Laboratory, University of Leeds (funded by the University of Leeds and Wellcome Trust 108466/Z/15/Z), and at the UK National electron Bio-Imaging Centre (eBIC) at the Diamond Light Source for assistance with cryoEM data collection, the staff at Phenomics Animal Care Facility, S. Ding and I. Fearnley (Cambridge) for mass spectrometry analyses, and M. Hartley and A. Raine (Cambridge) for IT support. This work was supported by The Medical Research Council grant numbers MC_U105663141 (J.H.), MC_UU_00015/2 (J.H.) and MC_UU_00015/5 (C.V.).
Footnotes
Data Availability Statement
The electron microscopy maps and masks and the models for the active and deactive states have been deposited in the Electron Microscopy Data Bank (EMDB) with accession numbers EMD-4345 (active) and EMD-4356 (deactive), and in the Protein Data Bank with accession codes 6G2J (active) and 6G72 (deactive).
Author Contributions
A.N.A.A. prepared and characterized mouse complex I assisted by H.R.B. and C.V.; J.N.B. prepared cryoEM grids and collected data, assisted by S.R. and S.P.M.; A.N.A.A. and J.N.B. processed cryoEM data assisted by H.R.B., S.R. and S.P.M.; J.N.B. built the models assisted by A.N.A.A.; J.H. analyzed and interpreted the models assisted by A.N.A.A., J.N.B. and H.R.B; J.H. directed the project and wrote the manuscript with input from all authors.
Competing Financial Interests statement
The authors declare no competing interests.
References
- 1.Hirst J. Mitochondrial complex I. Annu Rev Biochem. 2013;82:551–575. doi: 10.1146/annurev-biochem-070511-103700. [DOI] [PubMed] [Google Scholar]
- 2.Fassone E, Rahman S. Complex I deficiency: clinical features, biochemistry and molecular genetics. J Med Genet. 2012;49:578–590. doi: 10.1136/jmedgenet-2012-101159. [DOI] [PubMed] [Google Scholar]
- 3.Hirst J, Carroll J, Fearnley IM, Shannon RJ, Walker JE. The nuclear encoded subunits of complex I from bovine heart mitochondria. Biochim Biophys Acta. 2003;1604:135–150. doi: 10.1016/s0005-2728(03)00059-8. [DOI] [PubMed] [Google Scholar]
- 4.Stroud DA, et al. Accessory subunits are integral for assembly and function of human mitochondrial complex I. Nature. 2016;538:123–126. doi: 10.1038/nature19754. [DOI] [PubMed] [Google Scholar]
- 5.Sazanov LA, Hinchliffe P. Structure of the hydrophilic domain of respiratory complex I from Thermus thermophilus. Science. 2006;311:1430–1436. doi: 10.1126/science.1123809. [DOI] [PubMed] [Google Scholar]
- 6.Efremov RG, Sazanov LA. Structure of the membrane domain of respiratory complex I. Nature. 2011;476:414–420. doi: 10.1038/nature10330. [DOI] [PubMed] [Google Scholar]
- 7.Baradaran R, Berrisford JM, Minhas GS, Sazanov LA. Crystal structure of the entire respiratory complex I. Nature. 2013;494:443–448. doi: 10.1038/nature11871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vinothkumar KR, Zhu J, Hirst J. Architecture of mammalian complex I. Nature. 2014;515:80–84. doi: 10.1038/nature13686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhu J, et al. Structure of subcomplex Iβ of mammalian respiratory complex I leads to new supernumerary subunit assignments. Proc Natl Acad Sci U S A. 2015;112:12087–12092. doi: 10.1073/pnas.1510577112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhu J, Vinothkumar KR, Hirst J. Structure of mammalian respiratory complex I. Nature. 2016;536:354–358. doi: 10.1038/nature19095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fiedorczuk K, et al. Atomic structure of the entire mammalian mitochondrial complex I. Nature. 2016;538:406–410. doi: 10.1038/nature19794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu M, Gu J, Guo R, Huang Y, Yang M. Structure of mammalian respiratory supercomplex I1III2IV1. Cell. 2016;167:1598–1609. doi: 10.1016/j.cell.2016.11.012. [DOI] [PubMed] [Google Scholar]
- 13.Guo R, Zong S, Wu M, Gu J, Yang M. Architecture of human mitochondrial respiratory megacomplex I2III2IV2. Cell. 2017;170:1247–1257. doi: 10.1016/j.cell.2017.07.050. [DOI] [PubMed] [Google Scholar]
- 14.Zickermann V, et al. Mechanistic insight from the crystal structure of mitochondrial complex I. Science. 2015;347:44–49. doi: 10.1126/science.1259859. [DOI] [PubMed] [Google Scholar]
- 15.Cooley RB, Arp DJ, Karplus PA. Evolutionary origin of a secondary structure: π-helices as cryptic but widespread insertional variations of α-helices that enhance protein functionality. J Mol Biol. 2010;404:232–246. doi: 10.1016/j.jmb.2010.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kumar P, Bansal M. Dissecting π-helices: sequence, structure and function. FEBS J. 2015;282:4415–4432. doi: 10.1111/febs.13507. [DOI] [PubMed] [Google Scholar]
- 17.Kaila VRI, Wikström M, Hummer G. Electrostatics, hydration, and proton transfer dynamics in the membrane domain of respiratory complex I. Proc Natl Acad Sci U S A. 2014;111:6988–6993. doi: 10.1073/pnas.1319156111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sharma V, et al. Redox-induced activation of the proton pump in the respiratory complex I. Proc Natl Acad Sci U S A. 2015;112:11571–11576. doi: 10.1073/pnas.1503761112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Di Luca A, Gamiz-Hernandez AP, Kaila VRI. Symmetry-related proton transfer pathways in respiratory complex I. Proc Natl Acad Sci U S A. 2017;114:E6314–E6321. doi: 10.1073/pnas.1706278114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haapanen O, Sharma V. Role of water and protein dynamics in proton pumping by respiratory complex I. Sci Rep. 2017;7:7747. doi: 10.1038/s41598-017-07930-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Blaza JN, Vinothkumar KR, Hirst J. Structure of the deactive state of mammalian respiratory complex I. Structure. 2018;26:312–319. doi: 10.1016/j.str.2017.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kotlyar AB, Vinogradov AD. Slow active/inactive transition of the mitochondrial NADH-ubiquinone reductase. Biochim Biophys Acta. 1990;1019:151–158. doi: 10.1016/0005-2728(90)90137-s. [DOI] [PubMed] [Google Scholar]
- 23.Vinogradov AD. Catalytic properties of the mitochondrial NADH-ubiquinone oxidoreductase (complex I) and the pseudo-reversible active/inactive enzyme transition. Biochim Biophys Acta. 1998;1364:169–185. doi: 10.1016/s0005-2728(98)00026-7. [DOI] [PubMed] [Google Scholar]
- 24.Babot M, Birch A, Labarbuta P, Galkin A. Characterisation of the active/deactive transition of mitochondrial complex I. Biochim Biophys Acta. 2014;1837:1083–1092. doi: 10.1016/j.bbabio.2014.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Galkin A, Moncada S. Modulation of the conformational state of mitochondrial complex I as a target for therapeutic intervention. Interface Focus. 2017;7 doi: 10.1098/rsfs.2016.0104. 20160104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Galkin A, Abramov AY, Frakich N, Duchen MR, Moncada S. Lack of oxygen deactivates mitochondrial complex I: implications for ischemic injury? J Biol Chem. 2009;284:36055–36061. doi: 10.1074/jbc.M109.054346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chouchani ET, et al. Cardioprotection by S-nitrosation of a cysteine switch on mitochondrial complex I. Nat Med. 2013;19:753–759. doi: 10.1038/nm.3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Russo CJ, Passmore LA. Ultrastable gold substrates for electron cryomicroscopy. Science. 2014;346:1377–1380. doi: 10.1126/science.1259530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meyerson JR, et al. Self-assembled monolayers improve protein distribution on holey carbon cryo-EM supports. Sci Rep. 2014;4:7084. doi: 10.1038/srep07084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Letts JA, Fiedorczuk K, Sazanov LA. The architecture of respiratory supercomplexes. Nature. 2016;537:644–648. doi: 10.1038/nature19774. [DOI] [PubMed] [Google Scholar]
- 31.Di Luca A, Kaila VRI. Global collective motions in the mammalian and bacterial respiratory complex I. Biochim Biophys Acta. 2018;1859:326–332. doi: 10.1016/j.bbabio.2018.02.001. [DOI] [PubMed] [Google Scholar]
- 32.Hunte C. Specific protein–lipid interactions in membrane proteins. Biochem Soc Trans. 2005;33:938–942. doi: 10.1042/BST20050938. [DOI] [PubMed] [Google Scholar]
- 33.Sharpley MS, Shannon RJ, Draghi F, Hirst J. Interactions between phospholipids and NADH:ubiquinone oxidoreductase (complex I) from bovine mitochondria. Biochemistry. 2006;45:241–248. doi: 10.1021/bi051809x. [DOI] [PubMed] [Google Scholar]
- 34.Eriksson S, Munch-Petersen B, Johansson K, Eklund H. Structure and function of cellular deoxyribonucleoside kinases. Cell Mol Life Sci. 2002;59:1327–1346. doi: 10.1007/s00018-002-8511-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sabini E, Hazra S, Ort S, Konrad M, Lavie A. Structural basis for substrate promiscuity of dCK. J Mol Biol. 2008;378:607–621. doi: 10.1016/j.jmb.2008.02.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mikkelsen NE, et al. Structural basis for feedback inhibition of the deoxyribonucleoside salvage pathway: studies of the Drosophila deoxyribonucleoside kinase. Biochemistry. 2003;42:5706–5712. doi: 10.1021/bi0340043. [DOI] [PubMed] [Google Scholar]
- 37.Schilling B, et al. Mass spectrometric identification of a novel phosphorylation site in subunit NDUFA10 of bovine mitochondrial complex I. FEBS Lett. 2005;579:2485–2490. doi: 10.1016/j.febslet.2005.03.061. [DOI] [PubMed] [Google Scholar]
- 38.Elurbe DM, Huynen MA. The origin of the supernumerary subunits and assembly factors of complex I: A treasure trove of pathway evolution. Biochim Biophys Acta. 2016;1857:971–979. doi: 10.1016/j.bbabio.2016.03.027. [DOI] [PubMed] [Google Scholar]
- 39.Hirst J, Roessler MM. Energy conversion, redox catalysis and generation of reactive oxygen species by respiratory complex I. Biochim Biophys Acta. 2016;1857:872–883. doi: 10.1016/j.bbabio.2015.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Carroll J, Ding S, Fearnley IM, Walker JE. Post-translational modifications near the quinone binding site of mammalian complex I. J Biol Chem. 2013;288:24799–24808. doi: 10.1074/jbc.M113.488106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zwicker K, et al. The redox-Bohr group associated with iron-sulfur cluster N2 of complex I. J Biol Chem. 2006;281:23013–23017. doi: 10.1074/jbc.M603442200. [DOI] [PubMed] [Google Scholar]
- 42.Le Breton N, et al. Using hyperfine electron paramagnetic resonance spectroscopy to define the proton-coupled electron transfer reaction at Fe-S cluster N2 in respiratory complex I. J Am Chem Soc. 2017;139:16319–16326. doi: 10.1021/jacs.7b09261. [DOI] [PubMed] [Google Scholar]
- 43.Gamiz-Hernandez AP, Jussupow A, Johansson MP, Kaila VRI. Terminal electron–proton transfer dynamics in the quinone reduction of respiratory complex I. J Am Chem Soc. 2017;139:16282–16288. doi: 10.1021/jacs.7b08486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hryc CF, et al. Accurate model annotation of a near-atomic resolution cryo-EM map. Proc Nat Acad Sci U S A. 2017;114:3103–3108. doi: 10.1073/pnas.1621152114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fedor JG, Jones AJY, Di Luca A, Kaila VRI, Hirst J. Correlating kinetic and structural data on ubiquinone binding and reduction by respiratory complex I. Proc Nat Acad Sci USA. 2017;114:12737–12742. doi: 10.1073/pnas.1714074114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Birrell JA, Hirst J. Truncation of subunit ND2 disrupts the threefold symmetry of the antiporter-like subunits in complex I from higher metazoans. FEBS Lett. 2010;584:4247–4252. doi: 10.1016/j.febslet.2010.09.017. [DOI] [PubMed] [Google Scholar]
- 47.Belevich G, Knuuti J, Verkhovsky MI, Wikström M, Verkhovskaya M. Probing the mechanistic role of the long α-helix in subunit L of respiratory complex I from Escherichia coli by site-directed mutagenesis. Mol Microbiol. 2011;82:1086–1095. doi: 10.1111/j.1365-2958.2011.07883.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhu S, Vik SB. Constraining the lateral helix of respiratory complex I by cross-linking does not impair enzyme activity or proton translocation. J Biol Chem. 2015;290:20761–20773. doi: 10.1074/jbc.M115.660381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Steimle S, et al. Asp563 of the horizontal helix of subunit NuoL is involved in proton translocation by the respiratory complex I. FEBS Lett. 2012;586:699–704. doi: 10.1016/j.febslet.2012.01.056. [DOI] [PubMed] [Google Scholar]
- 50.Sazinsky MH, Lippard SJ. Product bound structures of the soluble methane monooxygenase hydroxylase from Methylococcus capsulatus (Bath): protein motion in the α-subunit. J Am Chem Soc. 2005;127:5814–5825. doi: 10.1021/ja044099b. [DOI] [PubMed] [Google Scholar]
- 51.Calvaruso MA, et al. Mitochondrial complex III stabilizes complex I in the absence of NDUFS4 to provide partial activity. Hum Mol Genet. 2012;21:115–120. doi: 10.1093/hmg/ddr446. [DOI] [PubMed] [Google Scholar]
- 52.Kimanius D, Forsberg BO, Scheres SH, Lindahl E. Accelerated cryo-EM structure determination with parallelisation using GPUs in RELION-2. Elife. 2016;5:e18722. doi: 10.7554/eLife.18722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fernandez-Leiro R, Scheres SHW. A pipeline approach to single-particle processing in RELION. Acta Crystallogr. 2017;D73:496–502. doi: 10.1107/S2059798316019276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chovancova E, et al. CAVER 3.0: a tool for the analysis of transport pathways in dynamic protein structures. PLoS Comput Biol. 2012;8:e1002708. doi: 10.1371/journal.pcbi.1002708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fernández-Vizarra E, et al. Isolation of mitochondria for biogenetical studies: an update. Mitochondrion. 2010;10:253–262. doi: 10.1016/j.mito.2009.12.148. [DOI] [PubMed] [Google Scholar]
- 56.Carroll J, Altman MC, Fearnley IM, Walker JE. Identification of membrane proteins by tandem mass spectrometry of protein ions. Proc Natl Acad Sci U S A. 2007;104:14330–14335. doi: 10.1073/pnas.0706817104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bridges HR, Mohammed K, Harbour ME, Hirst J. Subunit NDUFV3 is present in two distinct isoforms in mammalian complex I. Biochim Biophys Acta. 2017;1858:197–207. doi: 10.1016/j.bbabio.2016.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zheng SQ, et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods. 2017;14:331–332. doi: 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang K. Gctf: real-time CTF determination and correction. J Struct Biol. 2016;193:1–12. doi: 10.1016/j.jsb.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pettersen EF, et al. UCSF Chimera - a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 61.Scheres SHW, Chen S. Prevention of overfitting in cryo-EM structure determination. Nat Methods. 2012;9:853–854. doi: 10.1038/nmeth.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Heymann JB, Belnap DM. Bsoft: image processing and molecular modeling for electron microscopy. J Struct Biol. 2007;157:3–18. doi: 10.1016/j.jsb.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 63.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Cryst. 2010;D66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Cryst. 2010;D66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Williams CJ, et al. MolProbity: more and better reference data for improved all-atom structure validation. Protein Sci. 2018;27:293–315. doi: 10.1002/pro.3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Barad BA, et al. EMringer: side chain-directed model and map validation for 3D cryo-electron microscopy. Nat Methods. 2015;12:943–946. doi: 10.1038/nmeth.3541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Touw WG, et al. A series of PDB related databases for everyday needs. Nucleic Acids Res. 2015;43:D364–D368. doi: 10.1093/nar/gku1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.