Abstract
MicroRNAs (miRNAs) are small, single-stranded, noncoding RNAs that can post-transcriptionally regulate the expression of various oncogenes and tumor suppressor genes. Dysregulated expression of many miRNAs have been shown to mediate the signaling pathways critical in the multistep carcinogenesis of colorectal cancer (CRC). MiRNAs are stable and protected from RNase-mediated degradation, thereby enabling its detection in biological fluids and archival tissues for biomarker studies. This review focuses on the role and application of miRNAs in the prognosis and therapy of CRC. While stage II CRC is potentially curable by surgical resection, a significant percentage of stage II CRC patients do develop recurrence. MiRNA biomarkers may be used to stratify such high-risk population for adjuvant chemotherapy to provide better prognoses. Growing evidence also suggests that miRNAs are involved in the metastatic process of CRC. Certain of these miRNAs may thus be used as prognostic biomarkers to identify patients more likely to have micro-metastasis, who could be monitored more closely after surgery and/or given more aggressive adjuvant chemotherapy. Intrinsic and acquired resistance to chemotherapy severely hinders successful chemotherapy in CRC treatment. Predictive miRNA biomarkers for response to chemotherapy may identify patients who will benefit the most from a particular regimen and also spare the patients from unnecessary side effects. Selection of patients to receive the new targeted therapy is becoming possible with the use of predictive miRNA biomarkers. Lastly, forced expression of tumor suppressor miRNA or silencing of oncogenic miRNA in tumors by gene therapy can also be adopted to treat CRC alone or in combination with other chemotherapeutic drugs.
Keywords: MicroRNA, Colorectal cancer, Multidrug resistance, Prognosis, Therapeutic target, Apoptosis, Metastasis, Recurrence, Risk stratification
Core tip: MicroRNAs (miRNAs) are important mediators regulating the initiation, progression, metastasis and recurrence of colorectal cancer (CRC). Numerous studies have reported dysregulation of miRNAs in tumor specimens and body fluids, including serum, plasma and feces. Furthermore, the miRNAs were more recently found in circulating exosomes from CRC patients. Fortunately, this finding suggests potential diagnostic, prognostic and therapeutic applications of miRNAs at different stages of CRC. In this review, we aim to outline the current body of knowledge pertaining to the critical roles played by miRNAs in the molecular pathogenesis of CRC. Our focus is to delineate practical applications of miRNAs as prognostic biomarkers and therapeutic targets in the treatment of CRC.
INTRODUCTION
Colorectal cancer (CRC) is the 3rd most common cancer and the 3rd leading cause of cancer-related deaths worldwide[1]. For 2018, there is an estimated incidence of over 140000 new cases and mortality of over 50000 annually in the United States[2]. It is widely believed that CRC develops in multi-step process, from aberrant crypt foci, through benign precancerous lesions (adenomas), to malignant tumors (adenocarcinomas) over an extended period of time[3]. The majority of CRC is sporadic, though approximately 20%-30% of CRC patients carry inherited mutations[4,5]. Accumulation of numerous genetic mutations and/or epigenetic changes is required to drive the carcinogenic progression through progressive functional disruption of tumor suppressor genes and oncogenes. While the prognosis for advanced CRC remains dismal, the disease is curable in its early stages, thereby highlighting the importance of prevention and early detection.
Treatment of CRC usually involves surgical resection of the primary tumor(s) followed by chemotherapy and/or targeted therapy for the advanced (stage III and IV) disease[6]. New drugs approved for CRC in recent years mainly fall into the class of molecular targeted drugs, including the vascular endothelial growth factor receptor (VEGFR) and epidermal growth factor receptor (EGFR) inhibitors[7].
MicroRNAs (miRNAs) are short (~22 nucleotides in length) endogenous non-coding RNAs that repress gene expression in eukaryotic organisms. There are numerous studies reporting the correlation between dysregulated miRNAs and the aberrant regulation of signaling pathways involved in CRC initiation and progression. Michael et al[8] was the first to report a dramatic downregulation of miR-143 and miR-145 in CRC relative to normal colon epithelial cells, thereby suggesting a role of miRNAs in CRC pathogenesis. Other studies have demonstrated the tumor suppressive or oncogenic functions of miRNAs in CRC. Therefore, miRNAs might have diagnostic and prognostic value for CRC patients. MiRNAs may also represent novel therapeutic targets for gene therapy in the treatment of CRC.
In this review, the dysregulation of miRNAs leading to the initiation, progression and metastasis of CRC will be discussed. We also describe the translation of miRNA research to potential prognostic and therapeutic applications in the management of CRC in clinical settings.
MIRNAS AND DYSREGULATION OF SIGNALING PATHWAYS IN CRC
Gene regulation by miRNAs is mediated by the formation of hybrids with the 3-untranslated region (3’UTR) sequences of the target mRNAs, leading to mRNA degradation and/or translational inhibition[9]. MiRNAs play key roles in numerous cellular processes, including cell proliferation, differentiation, apoptosis, and development. These miRNAs are observed to regulate the expression of approximately one-third of human protein-coding genes[10].
miRNAs and WNT/β-catenin pathway
The dysregulation of the Wnt/β-catenin pathway is one of the earliest events driving CRC carcinogenesis. Constitutively active β-catenin upregulates the expression of Wnt target genes transcriptionally to initiate CRC formation[11]. There is mounting evidence suggesting there is crosstalk between miRNAs and the Wnt/β-catenin pathway in CRC development. While miRNAs were found to activate or inhibit the canonical Wnt pathway, Wnt activation was also shown to increase expression of miRNAs by directly binding to their gene promoters.
MiR-224 was recently shown to activate the Wnt/β-catenin signaling and direct the nuclear translocation of β-catenin in CRC by downregulating GSK3β and SFRP2[12]. Knockdown of miR-224 was found to restore the expression of GSK3β and SFRP2 and inhibit Wnt/β-catenin-mediated cell metastasis and cell proliferation.
Cancer stem cells (CSCs) are widely believed to be the key driving force for the initiation of cancer. CSC subpopulations, capable of self-renewal and having the ability to initiate and sustain tumor growth, metastasis and resistance to chemotherapy, have been identified in CRC[13]. The Wnt pathway plays a key role in the induction of symmetrical cell division of CSCs, which disrupts the homeostasis of normal stem cells and leads to cancer formation[14,15]. To this end, miR-146a was shown to activate the Wnt pathway in colorectal cancer CSCs to stabilize β-catenin, thereby regulating the symmetrical cell division to promote CRC initiation and progression[16]. Interestingly, miR-146a was itself activated transcriptionally by Snail via a β-catenin-TCF4 complex[16]. Therefore, a feedback loop consisting of Snail-miR-146a-β-catenin is operating in CSC of colorectal cancer to maintain Wnt activity.
miRNAs and EGFR pathways
In recent years, anti-EGFR targeted drugs have been approved for treating metastatic CRC. However, treatment response is hindered by dysregulation of the PI3K/AKT and KRAS/RAF/ERK pathways downstream of EGFR.
In the PI3K/AKT pathway, mutation of the PIK3CA gene has been demonstrated in 15%-20% of CRC cases[17]. Interestingly, mutations in the miR-520a and miR-525a binding sites at the 3’UTR of PIK3CA were found to enhance the sensitivity of CRC cell lines to saracatinib, which inhibits the activation of Akt-dependent signaling[18]. Conversely, reduced expression of miR-126 has been shown to mediate amplification of the PI3K/AKT signal in CRC[19].
Activating KRAS mutations constitutes up to 30%-60% of all CRC cases[20], which mediates primary resistance to anti-EGFR targeted therapy. Moreover, even in CRC bearing wild-type KRAS, the response to anti-EGFR targeted therapy is less than 40%[21]. Additional molecular signature(s) are needed for the selection of patients who respond well to anti-EGFR targeted therapy. To this end, miRNAs such as let-7[22], miR-18a*[23], miR-30b[24], miR-143[25], and miR-145[26] have been identified as tumor suppressors that inhibit KRAS expression. These miRNAs may be used as biomarkers to predict favorable response in patients to anti-EGFR therapy.
miRNAs and TGF-β signaling pathway
Transforming growth factor-beta (TGF-β) plays a key role in inhibiting cell proliferation, and it also modulates tumor invasion and tumor microenvironment modification. It has been estimated that 30% of CRC cases are due to mutations in TGF-β type II receptor (TGFβR2)[27,28]. TGF-β binds to its receptors (TGF-βR) and mediates the activation of its downstream pathway through phosphorylation of Smad. The complex is subsequently translocated to the nucleus, and it regulates the expression of transcriptional factors, including Snail, ZEB and Twist. Several miRNAs, including miR-21[29], miR-106a[30], and miR-301a[31], have been reported to induce stemness or promote cancer migration and invasion in CRC by targeting the TGF-β/Smad signaling pathway. Conversely, decreased expression of miR-25 in CRC cell lines was also found to activate SMAD7, which is a negative regulator of TGF-β signaling pathway, to promote cancer proliferation and metastasis[32]. Interestingly, miR-187, a validated downstream effector of the TGFβ pathway, was shown to suppress Smad-mediated epithelial-mesenchymal transition (EMT) in CRC cells[33].
miRNAs and epithelial-to-mesenchymal transition (EMT)
The activation of EMT, a cellular process of converting polarized epithelial cells to mesenchymal cells, enables cancer cells to migrate and invade into metastatic sites[34]. A number of miRNAs have been identified as key regulators of this EMT process in CRC[35]. MiR-29c has been shown to be remarkably downregulated in primary CRC with distant metastasis and it was associated with significantly shorter patient survival[36]. Importantly, forced expression of miR-29c was demonstrated to inhibit cell migration and invasion in vitro and metastasis in vivo[36], which is related to its inhibition of the ERK/GSK3β/β-catenin and AKT/GSK3β/β-catenin pathways. Conversely, liver metastatic tissues were found to express higher level of miR-200c than the primary CRC tumor[37]. This increased expression has been specifically associated with hypomethylation of the promoter of the miRNA gene. The Wnt/β-catenin pathway was also shown to transactivate miR-150, which subsequently promoted EMT of CRC cells by suppressing CREB signaling[38].
MIRNAS AS POTENTIAL CLINICAL BIOMARKERS IN CRC
miRNAS as diagnostic biomarkers for CRC
It is widely believed that a series of sequential genetic changes are needed to drive the conversion of normal colonic epithelium to malignant CRC. If the precancerous adenoma can be detected and treated prior to the development of advanced carcinoma, then patient mortality would be reduced[39]. While colonoscopy remains the gold standard screening test for the diagnosis of CRC[39,40], the application of miRNA as diagnostic markers for CRC has attracted a lot of attention in recent years. Two of the first few miRNAs identified in CRC tumor specimens, miR-143 and miR-145, were, relative to normal colonic mucosa, found to be consistently downregulated in adenoma and other stages of CRC[8,41]. More recently, several other miRNAs (miR-21, miR-29a, miR-92a and miR-135b) have also been shown to, when compared to normal tissues, be upregulated in patients with high-risk adenomas[42]. A few excellent review articles about the development of miRNA-based biomarkers for the diagnosis of CRC can be found in recent literature[43-47]. Our review will focus more on the prognostic application and prediction of therapeutic response by miRNAs in CRC.
miRNA as prognostic biomarkers for CRC
Although tissue-specific signatures of miRNAs have been reported[48,49], more work is still needed to define a set of differentially expressed miRNAs suitable for screening CRC in the clinical setting. Besides using miRNAs as a tool to screen for early adenomas and CRC cases, it may be more useful to use miRNAs as a prognostic tool to guide treatment for CRC patients.
miRNAs in tumor tissues
Since surgical resection of the primary tumor is the treatment of choice in early stages of CRC, tumor tissue can be available as an important source for the identification of CRC-related miRNAs. Numerous miRNAs have been reported to be upregulated or downregulated in CRC cell lines in tumor specimens from patients, suggesting their association with patients’ prognosis and response to anticancer drugs. Table 1 summarizes representative miRNAs of prognostic value in CRC tissues.
Table 1.
A list of representative miRNAs identified in tumor tissues that are of prognostic value in colorectal cancer patients
| miRNA | Method of detection | Patient number | Observations and correlation with clinical outcome | Ref. |
| miR-15a/miR-16 | qRT-PCR | 126 | miR-15a/miR-16 downregulation were significantly associated with advanced TNM staging, poorly histological grade, positive lymph node metastasis. miR-15a/miR-16 combination were identified as independent predictors of unfavorable OS and DFS | [183] |
| miR-17-5p | qRT-PCR | 110 | High expressions were associated with pathological tumor features of poor prognosis. miR-17-5p correlated with DFS only at early stages | [184] |
| miR-21 | In situ hybridization | 84 | High miR-21 expressions were strongly associated with poor survival, more advanced TNM staging and poor therapeutic outcome | [90] |
| miR-29a | miRNA microarray, qRT-PCR | 110 | High expressions were associated with a longer DFS in CRC patients with stage II but not in stage I tumor | [61] |
| miR-34a-5p | qRT-PCR | 205 | The tissue expressions of miR-34a-5p was positively correlated with DFS. Moreover, expression of miR-34a-5p was an independent prognostic factor for CRC recurrence | [185] |
| miR-106a | qRT-PCR | 110 | Downregulation of miR-106a predicted shortened DFS and OS, independent of tumor stage | [184] |
| miR-132 | miRNA microarray, qRT-PCR | 28 (testing); 151 (validation) | Low expressions were associated with poor OS and occurrence of liver metastasis | [186] |
| miR-150 | qRT-PCR, in situ hybridization | 239 | High expressions were associated with longer OS. Low expressions were associated with poor therapeutic outcome in patients treated with 5-FU-based chemotherapy with or without leucovorin, levamisole or cisplatin | [91] |
| miR-181a | qRT-PCR | 162 | High expressions were correlated with poor patient prognosis. Overexpression of miR-181a repressed the expression of the tumor suppressor (PTEN) at mRNA level | [187] |
| miR-181b | qRT-PCR | 345 | High expressions were correlated with poor survival in black patients with stage II CRC | [188] |
| miR-188-3p | Level 3 Illumina miRNASeq data were analyzed from TCGA databasec | 228 | High expressions were associated with lower OS, higher tumor stage and indirectly with BRAF status | [99] |
| miR-195 | qRT-PCR | 85 | Reduced expressions of miR-195 were correlated with occurrence of lymph node metastasis and advanced tumor stage | [189] |
| miR-199b | miRNA microarray and qRT-PCR | 60 | Higher level in metastatic CRC tissue compared with non-metastatic CRC tissue; low expressions were associated with longer OS | [190] |
| miR-203 | microRNA microarray, qRT-PCR | 197 | High expressions were associated with more advanced TNM staging and poor survival | [90] |
| miR-215 | qRT-PCR | 34 | High expressions were closely associated with poor OS | [191] |
| miR-218 | qRT-PCR | 63 | High expressions were significantly associated with higher PFS, OS and response to 5-FU based chemotherapy | [192] |
| miR-320e | miRNA microarray and qRT-PCR | 100 | Elevated expressions were associated with poorer DFS and OS in stage III CRC patients | [93] |
| miR-429 | qRT-PCR | 116 | High levels were correlated with OS; low levels were associated with favorable response to 5-FU-based chemotherapy | [193] |
| miR-494 | qRT-PCR | 104 | High expressions were significantly associated with shorter DFS and OS. When used as a panel with 5 other miRNAs, the signature can distinguish early relapsed from non-early relapsed CRC. | [194] |
| miR-625-3p | qRT-PCR | 94 | High expressions were associated with higher OS, PFS and better response to treatment | [94] |
| 3-miRNA signature (let-7i, miR-10b, miR-30b) | qRT-PCR | 232 | The addition of miR-30b to the 2-miRNA signature allowed the prediction of both distant metastasis and hepatic recurrence in patients with stage I-II CRC who did not receive adjuvant chemotherapy | [195] |
| A multi-RNA-based classifier (consisting of 12 mRNAs, 1 miRNA (miR-27a) and 1 lnc RNA) | mRNA, miRNA and lncRNA data were retrieved from the TCGA data portal | 663 | The classifier can divide patients into high and low risk groups with significantly different OS. Moreover, the classifier is not only independent of clinical features but also with a similar prognostic ability to the well-established TNM stage | [196] |
OS: Overall survival ; DFS: Disease free survival; lnc RNA: Long non-coding RNA; TCGA : The Cancer Genome Atlas Project (http://tcga-data.nci.nih.gov/).
It is known that oncogenic miRNAs (also known as oncomiRs) and tumor suppressor miRNAs are differentially expressed during the development and progression of CRC. Moreover, miRNA expression in CRC is regulated in stage-specific manner (Figure 1). Therefore, tracing the order of miRNA regulation during CRC progression may indicate disease prognosis and predict treatment response. To this end, let-7, miR-21, miR-29a and miR-17-92 cluster were of particular importance.
Figure 1.
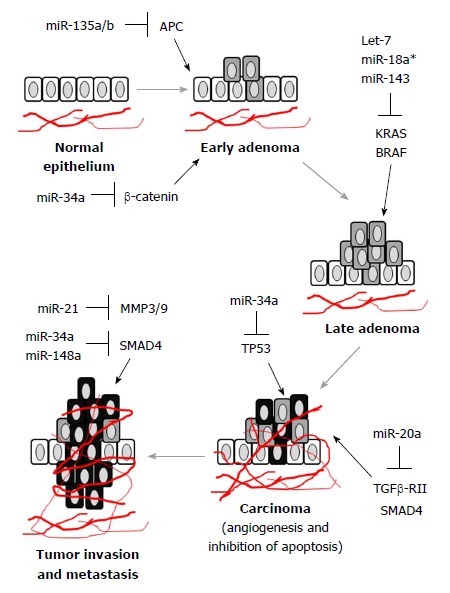
MicroRNA dysregulation and colorectal cancer progression. Altered expression of representative miRNAs are shown in different stages of CRC (in bold: Normal epithelium → early adenoma → late adenoma → carcinoma → metastasis). Upregulation or downregulation of miRNAs can affect signaling pathways and propel progression of CRC, leading to angiogenesis, cell invasion, metastasis and inhibition of apoptosis. APC: adenomatous polyposis coli; CRC: Colorectal cancer; KRAS: Kirsten rat sarcoma viral oncogene homolog; TP53: tumor protein p53.
Let-7 was known to regulate the expression of RAS and MYC genes, both of which are key factors mediating CRC progression and metastasis. Interestingly, the let-7 miRNA single nucleotide polymorphism (SNP) in the K-Ras 3’UTR has been reported to be prognostic in early-stage CRC (see below for more discussion about miRNA binding site SNP)[50]. MiR-17-92 is a highly conserved cluster that gives rise to six mature miRNAs, including miR-17, miR-18a, miR-19a, miR-19b, miR-20a, and miR-92-1. This cluster is frequently amplified in CRC, suggesting an oncogenic role for these miRNAs[51]. Moreover, miR-17 and Tumor, Node, and Metastasis (TNM) staging were shown to be significant but independent prognostic biomarkers in CRC patients[51]. MiR-92a, targeting the anti-apoptotic protein BIM, was found to be expressed at higher level in both adenomas and carcinoma[52]. The other miRNAs within this cluster were shown to regulate multiple targets to drive CRC progression (PTEN – miR-19a and miR-19b-1; BCL2L11 – miR-19a, miR-19b-1, miR-20a, miR-92; E2F1 – miR-17 and miR-20a; TGF-β receptor II – miR-17 and miR-20a[53-56]).
The remarkable upregulation of miR-21 in CRC has advocated its function as a prognostic marker[57]. In a small cohort of 29 CRC patients, high level of miR-21 in the tumor specimens was found to be associated with lymph node metastasis and also distal metastasis[58]. In a bigger cohort of 156 CRC cases, elevated level of miR-21 was also found to be correlated with venous invasion, liver metastasis and advanced Dukes’ stage[59]. A differentially expressed microRNA signature was recently reported in American CRC patients with liver metastasis[60]. miR-21, together with miR-93 and miR-103, were found to be increased in advanced CRC with liver metastasis.
The risk of CRC recurrence was found to be associated with high expression of miR-29a in tumor tissues in patients with stage II CRC[61]. miR-29a was later identified as a novel metastasis-promoting factor through upregulation of matrix metalloproteinase 2 and downregulation of E-cadherin via targeting the tumor suppressor gene KLF4[62].
LIMITATION WITH MEASURING MIRNAS IN ARCHIVAL TUMOR TISSUES
The major limitation of detecting miRNAs in archival tumor tissues is heterogeneity (from within the same primary tumor and also between different metastatic sites). Therefore, evaluating miRNA expression from serum/plasma (i.e. circulating miRNAs) may be potentially preferable for predicting prognosis in the clinical setting.
Circulating miRNAs
For CRC, the traditionally used blood-based biomarkers, including carcinoembryonic antigen (CEA) and carbohydrate antigen 19-9 (CA19-9), suffer from low sensitivity, particularly for early stage CRC[63]. In recent years, miRNA-based biomarkers have shown promise as CRC-specific prognostic tests due to their high sensitivity and specificity. MiRNAs are remarkably stable in blood despite the high level of ribonuclease that rapidly degrade RNA[64]. MiRNAs are protected from enzyme degradation because they are stored in exosomes or vesicles, or bound to lipoproteins[65,66].
Several miRNAs isolated from serum have been evaluated for their use as prognostic markers in CRC (Table 2). A recent report revealed a panel of miRNAs (miR-21, let-7g, miR-31, miR-92a, miR-181b, and miR-203) that can be reliably used as prognostic marker for CRC, with 93% sensitivity and 91% specificity in comparison with the traditional markers (CEA and CA19-9)[67]. Furthermore, miR-21 has also been shown to be able to differentiate both adenoma and CRC from healthy controls[68,69]. The detection of miR-141 in plasma samples has been used as a supplementary prognostic marker besides CEA for the detection of CRC patients with distant metastasis[70]. Moreover, the changes in plasma levels of miR-24, miR-320a and miR423-5p can be evaluated to predict the risk of post-surgery metastasis in CRC patients[71].
Table 2.
A list of representative circulating miRNAs of prognostic value in colorectal cancer patients
| miRNA | Method of detection | Patient number | Observations and correlation with clinical outcome | Ref. |
| miR-21 | qRT-PCR | 102 | Lower serum expressions were associated with higher local recurrence and mortality | [197] |
| miR-23b | qRT-PCR | 96 | CRC patients with low miR-23b expression in plasma exhibited a shorter recurrence-free survival and poorer overall survival rate | [198] |
| miR-96 | TaqMan miRNA microarray | 50 (screening); 234 (validation) | Elevated plasma levels were strongly correlated with shorter OS, especially in stage II and III CRC patients | [199] |
| miR-124-5p | qRT-PCR | 71 | Higher plasma levels were correlated with longer OS | [200] |
| miR-141 | qRT-PCR | 102 | High plasma levels were significantly associated with stage IV colon cancer and poor prognosis | [70] |
| miR-148a | qRT-PCR | 55 | Lower levels were associated significantly with shorter DFS and poorer OS rates | [201] |
| miR-155 | qRT-PCR | 146 | High serum levels correlated with poor PFS and OS | [202] |
| miR-183 | qRT-PCR | 118 | High plasma levels were significantly associated with lymph node metastasis, distant metastasis, higher TNM staging, and tumor recurrence | [203] |
| miR-200b | TaqMan miRNA microarray | 50 (screening); 234 (validation) | Elevated plasma levels were correlated with shorter OS, especially in stage II and III CRC patients | [199] |
| miR-200c | qRT-PCR | 182 CRC patients and 24 controls | High serum expressions were strongly correlated with lymph node, distant metastasis, tumor recurrence and poor prognosis. | [204] |
| miR-203 | qRT-PCR | 144 (validation) | High serum levels were associated with poor survival and metastasis; | [205] |
| miR-218 | qRT-PCR | 189 | Serum levels were significantly associated with TNM stage, lymph node metastasis and differentiation. Patients with low miR-218 serum level had shorter survival. | [206] |
| miR-221 | qRT-PCR | 103 | Elevated plasma level is a significant prognostic factor for poor overall survival in CRC patients | [207] |
| miR-345 | TaqMan miRNA microarray | 138 | High plasma levels were significantly associated with shorter PFS and lack of response to treatment with cetuximab and irinotecan | [108] |
| miR-885-5p | miRNA microarray, qRT-PCR | 169 | High serum expressions were highly correlated with poor prognosis, lymph node metastasis and distant metastasis | [208] |
| miR-1290 | miRNA microarray, qRT-PCR | 324 | High serum levels were associated with lower OS, lower DFS and more advanced tumor stage | [209] |
| 2-miRNA prognostic panel (miR-23a-3p & miR-376c-3p) | miRNA microarray | 427 | A prognostic panel consisting of miR-23a-3p and miR-376c-3p, independent of TNM stage, was established | [210] |
| miR-200 & miR-141 | qRT-PCR | 380 | High serum levels of miR-200 and miR-141 were associated with higher propensity of CRC patients to develop liver metastasis | [211] |
| miR-122 & miR-200 family | miRNA microarray | 543 | Increased plasma miR-122 levels were associated with a “bad” prognostic subtype in metastatic CRC and a shorter relapse-free survival and overall survival for non-metastatic and metastatic CRC patients. Several members of the miR-200 family were associated with patients’ prognosis and clinicopathological characteristics | [212] |
OS : Overall survival; DFS: Disease free survival; PFS : Progression free survival; CRC: Colorectal cancer; TNM: Tumor, node, and metastasis.
Fecal-based miRNAs
MiRNAs are known to be sufficiently stable for detection in stool samples because they are protected in exosomes[72]. Moreover, fecal matter comes into direct contact with the lumen of colon and may include cells exfoliated from cancerous colonocytes. Molecular changes in CRC are more readily detected in the stool rather than in the blood[73]. Although fecal miRNAs were less extensively studied than circulatory miRNAs, the expression of numerous fecal miRNAs have been reported to be dysregulated in patients with CRC or advanced adenomas (Table 3). Recently, a panel of miRNAs isolated from the stool of CRC patients were found to differentiate not only CRC cases from healthy subjects but also differentiate TNM stages with high sensitivity and specificity[74]. Additionally, miR-135b was found to differentiate among different stages of CRC[75].
Table 3.
A list of representative miRNAs identified in fecal samples from colorectal cancer patients that are of prognostic value
| miRNA | Method of detection | Patient number | Observations and correlation with clinical outcome | Ref. |
| miR-135b | qRT-PCR | 424 | miR-135b showed a significant increasing trend across the adenoma to cancer sequence. miR-135b level may be used to differentiate between different stages of CRC. Stool miR-135b level dropped significantly upon removal of CRC or advanced adenoma. | [75] |
| miR-19-b-3p, miR-20a-5p, miR-21-3p, miR-92a-3p, miR-141 | qRT-PCR | 20 | Expression levels of three out of the five overexpressing miRNAs returned to values comparable to normal controls after curative surgery; this was correlated with the | [213] |
| 12 upregulated and 8 downregulated miRNA panel | miRNA microarray, qRT-PCR | 60 | A panel of 12 upregulated miRNAs (miR-7, miR-17, miR-20a, miR-21, miR-92a, miR-96, miR-106a, miR-134, miR-183, miR-196a, miR-199a-3p, miR-214) and 8 downregulated miRNAs (miR-9, miR-29b, miR-127-5p, miR-138, miR-143, miR-146a, miR-222, miR-938) were found to differentiate not only CRC cases from healthy subjects but also differentiate TNM stages with high sensitivity and specificity. | [74] |
miRNAs in CRC-derived exosomes
Exosomes are a unique forms of extracellular vesicles, ranging from 30 nm to 100 nm in diameter, which are released into the extracellular space upon the fusion of multivesicular bodies with plasma membranes from diverse cell types. Tumor-derived exosomes are emerging as local and systemic intercellular mediators of oncogenic information through the horizontal transfer of mRNAs, miRNAs, and protein during tumorigenesis[76]. In a recent proteome profiling study of exosomes derived from human primary and metastatic CRC cells, a selective enrichment of metastatic factors and signaling pathway components was observed[77]. High expression of exosomal miR-17-92a cluster was found to be associated with recurrence in late stage CRC patients[78]. Moreover, an elevated exosomal level of miR-19a was reported in the sera samples of CRC patients with poor prognosis[78]. Interestingly, a recent report has demonstrated an enrichment of miR-328 from CRC patients’ plasma samples collected from colonic veins (i.e., mesenteric veins) when compared to peripheral vein plasma samples and that it was significantly correlated with the development of liver metastasis[79]. Table 4 summarizes a short list of miRNAs of prognostic value identified in exosomes released from CRC cells.
Table 4.
A list of representative miRNAs isolated from exosomes in biological samples from colorectal cancer patients or cell lines that are of prognostic value
| miRNA | Method of detection | Type of samples | Tumor stage | Ref. |
| miR-17-92a | miRNA microarray | Tumor specimens | IV | [78] |
| miR-19a | miRNA microarray | Tumor specimens | IV | [78] |
| let-7a | qRT-PCR | Serum | I, II, IIIa, IIIb, IV | [214] |
| miR-21 | qRT-PCR and miRNA microarray | Serum; plasma | I, II, IIIa, IIIb, IV | [214] |
| miR-23a | qRT-PCR | Serum | I, II, IIIa, IIIb, IV | [214] |
| miR-150 | qRT-PCR | Serum | I, II, IIIa, IIIb, IV | [214] |
| miR-203 | qRT-PCR | Serum | I, II, III, IV | [215] |
| miR-223 | qRT-PCR | Serum | I, II, IIIa, IIIb, IV | [214] |
| miR-1246 | qRT-PCR | Serum | I, II, IIIa, IIIb, IV | [214] |
| miR-1229 | qRT-PCR | Serum | I, II, IIIa, IIIb, IV | [214] |
| miR-548c-5p | miRNA microarray and qRT-PCR | Serum (downregulated) | I, II, IIIa, IIIb, IV | [216] |
| miR-638 | miRNA microarray and qRT-PCR | Serum (downregulated) | I, II, IIIa, IIIb, IV | [216] |
| miR-5787 | miRNA microarray and qRT-PCR | Serum (downregulated) | I, II, IIIa, IIIb, IV | [216] |
| miR-8075 | miRNA microarray and qRT-PCR | Serum (downregulated) | I, II, IIIa, IIIb, IV | [216] |
| miR-6869-5p | miRNA microarray and qRT-PCR | Serum (downregulated) | I, II, IIIa, IIIb, IV | [216] |
| miR-486-5p | miRNA microarray and qRT-PCR | Serum (upregulated) | I, II, IIIa, IIIb, IV | [216] |
| miR-3180-5p | miRNA microarray and qRT-PCR | Serum (upregulated) | I, II, IIIa, IIIb, IV | [216] |
| miR-96-5p and miR-149 | qRT-PCR | Plasma | III | [217] |
| miR-100 | qRT-PCR | Cell lines (DKO-1, Dks-8, DLD-1) | [218] | |
| miR-192 | qRT-PCR | Cell lines (HCT-15, SW480, WiDr) | [219] | |
| miR-210 | qRT-PCR | Cell line (HCT-8) | [220] | |
| miR-221 | qRT-PCR | Cell lines (HCT-15, SW480, WiDr) | [219] | |
| miR-379 | qRT-PCR | Cell lines (HCT-116, HT-29) | [221] |
MIRNAS FOR STRATIFING RISK OF RECURRENCE IN STAGE II CRC PATIENTS
Adjuvant chemotherapy following surgery is adopted as the standard treatment to improve survival in stage III CRC patients[80]. Conversely, in stage II CRC patients, surgical resection of the primary tumor is highly effective without other concomitant treatment. Therefore, the use of adjuvant chemotherapy in all stage II CRC patients is debatable[81-83]. However, a notable sub-group of stage II CRC patients (~30%) develop tumor recurrence and have poor prognoses. At present, there are a few clinical parameters, including poorly differentiated histology, extramural venous invasion, intestinal obstruction and perforation, that are used as risk factors to identify high-risk patients for adjuvant chemotherapy. Since these clinical parameters are not specific, more reliable biomarkers are needed to identify the high-risk stage II CRC patients and initiate adjuvant chemotherapy to improve their survival. Likewise, other Stage II CRC patients with lower risk can be spared from the toxicity of conventional cytotoxic chemotherapeutic drugs. To this end, clinical trials have been initiated to investigate the use of miRNA to identify stage II CRC patients who may benefit from chemotherapy (including an on-going study being conducted in China (NCT02635087).
Schetter et al[84] showed that the combination of miR-21 expression in primary tumor tissues and inflammatory risk score was a better predictor of prognosis than either biomarker alone. Data from a large population-based study conducted on 520 stage II CRC patients showed that elevated miR-21 expression in tumor specimens correlated significantly with poor cancer-free survival[85]. Another six-miRNAs based classifiers (miR-20a-5p, miR-21-5p, miR-103a-3p, miR-106a-5p, miR-143-5p, and miR-215) were reported to be a promising predictive markers for tumor recurrence in patients with stage II CRC[86]. More recently, the up-regulation of miR-181c in CRC specimens was also found to be associated with higher recurrence[87].
miRNAs for predicting response to chemotherapy
There has been intense research interest in the development of miRNA signature to predict response to anticancer treatment, thus allowing a more personalized approach for treating CRC with better efficacy and fewer adverse effects. To this end, a genome-wide miRNA profiling in twelve CRC cell lines has been performed to derive an in vitro signature of chemosensitivity[88]. Numerous studies have also reported the potential use of miRNAs as biomarkers in tissues and blood to predict response to the most common drug therapies (5-fluorouracil (5-FU)/oxaliplatin cytotoxic chemotherapy and EGFR targeted therapy) in CRC patients (Table 5).
Table 5.
MiRNAs in tumor specimens and plasma/serum samples reported to predict therapeutic response in colorectal cancer patients
| miRNA | Treatment regimen | Detection method | Expression that suggests inadequate response | Patient number | Ref. |
| Tumor specimens | |||||
| Let-7 | Irinotecan, cetuximab | qRT-PCR | Low | 59 | [102] |
| miR-7 | Cetuximab | qRT-PCR | Low | 105 | [97] |
| miR-21 | 5-FU | qRT-PCR | High | 84 | [90] |
| miR-21 | 5-FU | qRT-PCR | High | 67 | [89] |
| miR-21-5p | 5-FU + radiation | Microarray | Low | 27 | [222] |
| miR-31-3p | Anti-EGFR | Microarray, qRT-PCR | High | 33 | [98] |
| miR-31-5p | Anti-EGFR | qRT-PCR | High | 102 | [100] |
| miR-126 | Capecitabine, oxaliplatin | qRT-PCR, ISH | Low | 89 | [104] |
| miR-143 | Capecitabine, oxaliplatin, anti-EGFR | Microarray, qRT-PCR | High | 34 | [103] |
| miR-146b-3p | Cetuximab | qRT-PCR | High | 25 | [223] |
| miR-148a | 5-FU | qRT-PCR, ISH | Low | 273 | [92] |
| miR-150 | 5-FU | qRT-PCR, ISH | Low | 239 | [91] |
| miR-181a | Anti-EGFR | qRT-PCR | Low | 80 | [99] |
| miR-200 family | Fluoropyrimidine | qRT-PCR | Low | 127 | [103] |
| miR-200b | Capecitabine, oxaliplatin, anti-EGFR | Microarray, qRT-PCR | Low | 34 | [103] |
| miR-320e | 5-FU, oxaliplatin | Microarray, qRT-PCR | High | 100 | [93] |
| miR-455-5p | Capecitabine, oxaliplatin, bevacizumab | qRT-PCR, ISH | High | 212 | [224] |
| miR-486-5p | Cetuximab | qRT-PCR | High | 25 | [223] |
| miR-519c | 5-FU, irinotecan | qRT-PCR | Low | 26 | [153] |
| miR-592 | Anti-EGFR | Microarray | Low | 33 | [98] |
| miR-625-3p | Capecitabine, oxaliplatin | Microarray, qRT-PCR | High | 94 | [94] |
| miR-664-3p | Capecitabine, oxaliplatin, bevacizumab | qRT-PCR, ISH | Low | 212 | [224] |
| Let-7c, miR-99a, miR-125b | Anti-EGFR | Microarray, qRT-PCR | Low | 74 | [101] |
| miR-1274b, miR-720 | Capecitabine, oxaliplatin, radiation | Microarray, qRT-PCR | High | 38 | [96] |
| miR-107, miR-99a-3p | Fluoropyrimidine | Microarray, qRT-PCR | Low | 39 | [225] |
| miR-215, miR-190b, miR-29b-2 | 5-FU, radiotherapy | Microarray, qRT-PCR | High | 20 | [95] |
| Let-7e, miR-196, miR-450a, miR-450b-5p, miR-99a | 5-FU, radiotherapy | Microarray, qRT-PCR | Low | 20 | [95] |
| miR-17-3p, miR-193b-5p, miR-204-5p, miR-501-5p, miR-545-3p, miR-592, miR-644-3p, miR-15a-5p, miR-196b-5p, miR-552 | First-line capecitabine and oxaliplatin with or without bevacizumab | qRT-PCR, ISH | Low | 212 (screening); 121 (validation) | [224] |
| miR-1183, miR-622. miR-765, miR-1471, miR-125a-3p, miR-1224-5p, miR-188-5p, miR-483-5p, miR-671-5p, miR-1909 | Capectabine, oxaliplatin, radiation | Microarray, qRT-PCR | Low | 38 | [96] |
| Blood | |||||
| miR-19a | FOLFOXa | Microarray, qRT-PCR | High | 72 | [226] |
| miR-126 | Bevacizumab | High | [104] | ||
| miR-143 | Oxaliplatin | qRT-PCR | Low | 62 | [227] |
| miR-155 | Leucovorin, cetuximab, 5-FU | qRT-PCR | High | 15 | [107] |
| miR-345 | Cetuximab, irinotecan | Microarray, qRT-PCR | High | 138 | [108] |
| miR-1914* | Capecitabine + oxaliplatin | qRT-PCR | Low | 49 | [228] |
| miR-106a, miR-130b, miR-484 | 5-FU, oxaliplatin | qRT-PCR | High | 150 | [105] |
| miR-20a, miR-130, miR-145, miR-216, miR-372 | FOLFOX | Microarray, qRT-PCR | High | 40 | [106] |
FOLFOX: 5-FU + oxaliplatin + leucovorin; 5-FU: 5-fluorouracil; ISH: In situ hybridization; Anti-EGFR: Anti-EGFR targeted monoclonal antibody therapy; EGFR: Epidermal growth factor receptor.
Response to classical cytotoxic chemotherapy (5-FU/oxaliplatin) overexpression or downregulation of single miRNA in CRC tumor specimens
In 5-FU-based neoadjuvant chemotherapy setting to shrink the tumor size before surgery, low tumoral expression of miR-21 was found to be predictive of pathological response to the drug in locally advanced CRC patients[89]. In another study performing miRNA expression profiling of CRC, along with their paired non-cancerous tissues, high miR-21 expression in the tumors were found to associate with poor therapeutic response to 5-FU-based adjuvant chemotherapy as an add-on treatment[90]. Also predictively, low miR-148a, low miR-150 or high miR-320e level in tumor specimens were found to be associated with poor response to adjuvant 5-FU/oxaliplatin (FOLFOX) regimen in several other studies[91-93]. Interestingly, in a recent study on responsive and non-responsive metastatic CRC patients receiving XELOX/FOLFOX regimen, high expression of mR-625-3p in the tumor was found to be associated with poor response to the drug but was not related to disease recurrence[94].
Signature of miRNAs panel in tumor specimens
While the increased/decreased expression of a single miRNA may predict response to chemotherapy as described above, the use of a panel of miRNAs demonstrating a signature may be more reliable and specific. In a recent study on fluoropyrimidine-based chemotherapy, the responders were found to exhibit high tumoral levels of let-7e, miR-99a*, miR196b, miR450a and miR450b-5p whereas the non-responders were found to have high expression of miR-29b-2*, miR-196b and miR-215[95]. Another set of thirteen miRNAs, exhibiting a unique signature with 11 upregulated miRNAs (miR-125a-3p, miR-188-5p, miR-483-5p, miR-622, miR-630, miR-671-5p, miR-765; miR-1183, miR-1224-5p, miR-1471, and miR-1909*) and 2 downregulated miRNAs (miR-720 and miR-1274b) were shown to be strongly associated with complete pathological response to neoadjuvant capecitabine and oxaliplatin (XELOX) regimen in CRC patients[96].
Response to anti-EGFR/anti-VEGF targeted therapy
There has been intense research effort to identify miRNAs predictive of a favorable response to anti-EGFR monoclonal antibody. Consistent with the fact that miR-7 targets EGFR, reduced miR-7 expression was linked to poor prognosis of anti-EGFR therapy[97]. The efficacy of anti-EGFR therapy is related to the mutation status of KRAS/BRAF. Patients bearing wild-type KRAS/BRAF are normally considered candidates for anti-EGFR therapy. Therefore, numerous studies have been carried out with an aim to identify miRNAs that predict success with EGFR inhibition with wild-type KRAS. In metastatic CRC patients bearing wild-type KRAS/BRAF, high expression of miR-31[98], low expression of miR-592[98], or low expression of miR-181a[99] were found to be strongly associated with disease progression and short progression-free survival after treatment with anti-EGFR therapy (cetuximab or panitumab). To this end, miR-31-5p has been reported to regulate BRAF activation and thus the downstream signaling pathway of EGFR[100]. Conversely, miR-181a has been reported to regulate β-catenin expression[99]. Apart from individual miRNAs, the high-intensity signature of the cluster let-7c/miR-99a/miR-125b was found to be associated with favorable response (longer progression-free survival and overall survival) to cetuximab or panitumumab in metastatic CRC patients bearing wild-type KRAS[101].
Drug resistance mediated by KRAS mutations is severely hindering the clinical efficacy of anti-EGFR therapy in CRC because KRAS mutations drive the downstream MAPK and Akt signaling independent of EGFR. However, treatment with anti-EGFR therapy is still feasible, and a few miRNAs have been reported to predict favorable responses in metastatic CRC patients bearing KRAS mutations. To this end, let-7 is the most extensively studied miRNA as predictive biomarker for anti-EGFR therapy in patients with KRAS mutations[102]. Let-7 downregulates KRAS by directly binding to its mRNA 3’UTR. Interestingly, in patients with KRAS mutations, high levels of let-7 were shown to be significantly associated with better survival following cetuximab treatment[102]. Thus, let-7 level may be used to select subgroup of CRC patients with KRAS mutations to benefit from anti-EGFR therapy. In another study investigating KRAS-mutated metastatic CRC tumors, low miR-143 and high miR-200b expression in the tumor were found to be associated with better progression-free survival after treatment with combination of XELOX, cetuximab and bevacizumab[103].
The combination of anti-VEGF monoclonal antibody (bevacizumab) and cytotoxic chemotherapy has been approved for treating metastatic CRC. The predictive value of circulating miR-126 in treatment response to first-line chemotherapy combined with bevacizumab has been demonstrated in patients with metastatic CRC[104]. While non-responding patients had a remarkable increase in circulating miR-126 level, responding patients exhibited a mild decrease in circulating miR-126.
CIRCULATING MIRNAS AS NON-INVASIVE BIOMARKERS OF DRUG RESPONSE IN CRC
Besides serving as prognostic markers, circulating miRNAs can also be used as non-invasive and convenient biomarkers of drug response to classical cytotoxic drugs and EGFR-targeted therapy in CRC patients.
Pre-treatment level of plasma miRNAs have been shown to predict treatment response to 5-FU/oxaliplatin in metastatic colorectal cancer patients[105]. By measuring the expression profile of plasma miRNAs in metastatic CRC patients before and after four cycles of 5-FU/oxaliplatin chemotherapy, a set of three miRNAs (miR-106a, miR-130b, and miR-484) were found to be significantly upregulated in non-responders[105]. In another study using Taqman low-density array to evaluate the expression profile of serum miRNAs in chemotherapy responsive versus resistant CRC patients by unsupervised cluster analysis, a set of five serum miRNAs (miR-20a, miR-130, miR-145, miR-216, and miR-372) was found to predict response to classical chemotherapy and it may guide the selection of the chemotherapy regimen[106]. Recent studies have also shown that elevated levels of serum miR-155[107] and miR-345[108], respectively, were associated with inadequate response to 5-FU/leucovorin/cetuximab and irinotecan/cetuximab regimens in metastatic CRC patients.
POLYMORPHISMS IN MIRNA BINDING SITES PREDICT CRC PROGNOSIS AND TREATMENT RESPONSE
Single nucleotide polymorphisms (SNPs) located in the 3’UTR of miRNA target genes have been shown to affect miRNA-mRNA interaction and thus expression of target genes[109,110]. While several studies have reported the correlation between miRNA SNPs and risk of developing CRC[110,111], only a few studies have been conducted to investigate the use of miRNA SNPs to predict clinical outcomes of cancer therapy.
Using genome-wide approach, two SNPs rs1051424 and rs11704 were identified in the putative miRNA target sites of the 3’UTR of RPS6KB1 and ZNF839[112]. Importantly, overall survival was found to be inversely proportional to the occurrence of the two SNPs in CRC patients[112]. The SNPs resulted in the loss of the miRNA binding sites on both RPS6KB1 and ZNF839, thereby enabling the upregulation of the two genes. Activated RPS6KB1 is known to phosphorylate ribosomal protein S6 to increase protein synthesis and to promote cancer cell survival[113]. On the other hand, ZNF839 is not well characterized but reportedly involved in humoral immune response to cancer[114].
Similar miRNA SNP-mRNA interaction has also been shown to predict therapeutic response to cancer treatment in CRC. While it is well known that KRAS mutations predict resistance to anti-EGFR monoclonal antibodies treatment in CRC, the predictive marker for therapeutic response in CRC patients bearing wild-type KRAS is less studied. In a phase II study, the occurrence of a SNP in the let-7 miRNA complementary site of wild-type KRAS 3’UTR was found to correlate with the objective response rate to cetuximab monotherapy in metastatic CRC patients[115].
MIRNAS DEMONSTRATED TO BE THERAPEUTIC TAGETS IN CRC
The development of miRNA-based therapy for cancer treatment is based on the premise that aberrantly expressed miRNAs play crucial role in the development of cancer and therapeutic response to anticancer drugs. Therefore, correcting the miRNA deficiency or restoring the miRNA function could be used as novel strategy in cancer treatment[116,117]. Key miRNAs that have been studied are described below.
miR-21 and tumor invasion
Overexpression of miR-21 was associated with tumor invasion of CRC by promoting nuclear translocation of β-catenin[118]. Interestingly, CRC patients bearing adenomatous polyposis coli (APC) mutated tumor who have high levels of serum miR-21 were found to have poor prognosis, but this correlation was not observed in CRC patients bearing APC-wild type tumor. Mechanistically, miR-21 targets PDCD4, which inhibits cancer transformation and invasion[119].
Furthermore, miR-21 was also shown to confer resistance to 5-FU chemotherapy by downregulation of human MutS homolog 2 (MSH2)[120]. Overexpression of miR-21 was found to suppress the G2/M arrest and apoptosis induced by 5-FU. To this end, a miR-21 antisense inhibitor against hepatocellular carcinoma is under development by Regulus Therapeutics[121].
miR-34a and tumor suppressor p53
miR-34a is one of the most important downstream molecules of p53 that is transcriptionally activated to exert the tumor suppressive effect through cell cycle arrest and induction of apoptosis[122,123]. However, p53 mutation is observed in 50%-75% of CRC[3]. Downregulation of miR-34a is commonly observed in CRC. It has been reported that miR-34a expression was significantly decreased in CRC patients versus healthy individuals, suggesting that miR-34a could be a useful biomarker and therapeutic target in CRC[124,125]. Recently, it has been shown that p53-dependent expression of miR-34a blocks the IL-6R/STAT3/miR-34 feedback loop and subsequently inhibit tumor progression in CRC[126]. Since activation of IL-6R and STAT3 promote cancer proliferation, the overexpression/restoration of miR-34a may develop into a new treatment strategy for CRC.
miR-101 and WNT/β-catenin pathway
More than 60% of CRC cases are known to carry inactivating APC gene mutations, which lead to stimulation of the Wnt/β-catenin pathway to drive tumor initiation and recurrence[3]. Besides this extensively studied APC gene-mediated mechanism, miRNAs have also been shown to regulate the Wnt pathway in CRC. Downregulation of miR-101 was associated with activation of Wnt/β-catenin pathway[127]. Interestingly, overexpression of miR-101 in CRC was found to impair β-catenin nuclear localization and inhibit cancer stem cells-related genes[127]. Therefore, pharmacological restoration of miR-101 may represent a novel strategy to prevent recurrence of CRC.
miR-135B and downregulation of APC in CRC
miR-135a/b has been shown to be upregulated in colorectal adenomas and carcinomas, which correlates well with low APC expression in the tumor[128]. MiR-135a/b was found to target the 3’UTR of APC to suppress the expression of APC gene, and thus activating the downstream Wnt pathway to promote tumor transformation and progression[129]. To this end, miR-135b up-regulation was also found to be common in sporadic and inflammatory bowel disease-associated human CRCs; similarly, increased miR-135b expression was correlated with advanced tumor stages and poor clinical prognosis[129]. Importantly, inhibition of miR-135b, in CRC mouse xenograft models, was able to inhibit tumor growth by regulating genes involved in proliferation, invasion and apoptosis.
miR-143/145 as tumor suppressors in CRC
miR-143 and miR-145 were first identified as potential tumor suppressors in an analysis investigating the alteration of microRNAs expression from adenomatous to carcinoma stages in CRC[8]. Consistent with this finding, the upregulation of miR143/145 was observed in tumor tissues from advanced rectal cancer patients demonstrating favorable response to neoadjuvant chemoradiotherapy[130]. Mechanistically, miR-143 was found to bind directly to and suppress KRAS, DNMT3A and ERK5 whereas miR-145 was shown to target IRS-1, c-Myc, FLI1, IRS-1, STAT1 and YES1 to suppress tumor growth[131]. Forced expression of miR-143 and miR-145 have been shown to inhibit proliferation of CRC cell lines in vitro and in vivo[132].
MIRNAS REGULATING THE EGFR SIGNALING PATHWAY
The crucial role played by EGFR signaling in the survival of numerous cancer types, including CRC, has led to the approval of anti-EGFR targeted therapy, including tyrosine kinase inhibitors and monoclonal antibodies for treating CRC. However, drug resistance to these targeted agents is severely hindering their clinical efficacy. To this end, the activation of PI3K and mutation of KRAS, two key signaling pathways downstream of EGFR, have been extensively studied as mechanisms leading to the drug resistance to anti-EGFR therapy[133]. In recent years, increasing evidence has suggested that aberrant expression of miRNAs are affecting EGFR signaling and are causing drug resistance to anti-EGFR therapy.
Low expression of miR-30b was observed in CRC tumor specimens, which was significantly associated with advanced TNM staging and poor prognosis[24]. Forced expression of miR-30b was found to promote G1 arrest and induce apoptosis by targeting PIK3CD and BCL2, thereby inhibiting CRC cell growth in vitro and in vivo. On the other hand, miR-126 has been shown to inhibit PI3K signaling by targeting the p85b regulatory subunit responsible for PI3K stabilization[19].
Activating mutation of KRAS is known to drive resistance to anti-EGFR therapy by bypassing the inhibited EGFR. To this end, let-7 has been shown to post-transcriptionally downregulate KRAS and administration of let-7 to CRC animal models expressing activating KRAS mutations was shown to inhibit tumor growth[134]. More importantly, in CRC patients bearing mutant KRAS tumor who received salvage cetuximab plus irinotecan, higher let-7 level was found to be significantly associated with better survival outcomes[102]. miR-4689 was found to directly target KRAS and AKT1, therefore activation of the KRAS pathway may thus be mediated by downregulation of miR-4689. Forced expression of miR-4689 has been reported to inhibit CRC cancer growth and induce apoptosis both in vitro and in vivo[135].
The tumor suppressor PTEN (phosphatase and tensin homolog) plays an important role in regulating the PI3K/Akt pathway. Overexpression of miR-17-5p has been shown to promote drug resistance and tumor metastasis of CRC by downregulating PTEN[136]. Similarly, miR-32 was found to directly repress PTEN to regulate tumorigenesis of CRC[137].
MIRNAS REGULATING EMT
EMT is a complex process controlled by a number of transcriptional regulators and signaling pathways. In CRC, EMT is associated with a more malignant phenotype and metastasis. Recently, miRNAs have been suggested to be involved in the acquisition of cancer stem cell-like properties by regulating EMT. To this end, TGF-β is a major inducer of EMT by activating Smad signaling pathway. The miR-200 family have been reported to target TGF-β2. In particular, miR-200c was found to be significantly upregulated in tumor samples from patients with metastatic colon cancer and also colon cancer cell lines[37], which has been correlated with reduced expression of its direct target (ZEB1) to inhibit EMT[37,138,139].
miR-130a/301a/454 family of microRNAs have also been shown to regulate TGF-β signaling by inhibiting SMAD4[140]. Overexpression of these miRNAs was reported to increase cell proliferation and migration in colon cancer cell lines (HCT116 and SW480)[140]. Similarly, miR-1269a was also found to promote EMT and metastasis in CRC cells in vitro and in vivo[141]. Importantly, the overexpression of miR-1269a in tumor specimens from patients with late-stage CRC was associated with relapse and metastasis[141].
MIRNAS AS REGULATORS OF DRUG RESISTANCE IN CRC
While surgical resection of the primary tumor is the treatment of choice for most CRC patients, chemotherapy is commonly used as both neoadjuvant and adjuvant therapy to reduce the tumor load and to prevent recurrence and metastasis, respectively. Commonly used chemotherapeutic agents include 5-FU, oxaliplatin, irinotecan, and the anti-EGFR/VEGF monoclonal antibodies[142]. Drug resistance to these anticancer drugs pose a major obstacle for effective chemotherapy[143]. To this end, miRNAs have been found to play an important role in the induction of chemoresistance and they also represent emerging prognostic markers to indicate treatment success[144-146] (Table 6).
Table 6.
A list of representative miRNAs involved in drug resistance to colorectal cancer therapy
| miRNA | Drug(s) affected | Known molecular target(s) in CRC | Ref. |
| Overexpression of miRNA causing drug resistance to conventional chemotherapeutic drugs | |||
| Let-7g | S-1 (Tegafur/gimeracil/oteracil) | Cyclin D, c-myc, E2F, RAS | [229] |
| miR-10b | 5-FU | BIM | [230] |
| miR-19b | 5-FU | MYBL2 | [231] |
| miR-20a | 5-FU, oxaliplatin | BNIP2 | [170] |
| miR-21 | 5-FU | hMSH2 and hMSH6 | [120] |
| miR-23a | 5-FU | APAF-1 | [149] |
| miR-31 | 5-FU | - | [232] |
| miR-140 | 5-FU | HDAC4 | [233] |
| miR-148a | 5-FU, oxaliplatin | - | [92] |
| miR-181b | S-1, 5-FU | Cyclin D, c-myc, E2F, RAS | [229] |
| miR-192/miR-215 | 5-FU | DHFR | [234,235] |
| miR-195 | 5-FU | WEE1 | [236] |
| miR-203 | Oxaliplatin | ATM | [154] |
| miR-224 | 5-FU | - | [237] |
| miR-520g | 5-FU, oxaliplatin | p21 | [238] |
| miR-587 | 5-FU | PPP2R1B | [151] |
| miR-625-3p | Oxaliplatin | - | [94] |
| Downregulation of miRNA causing drug resistance to conventional chemotherapeutic drugs | |||
| miR-34a | 5-FU, oxaliplatin | SIRT1, KIT, LDHA, TGF-β, SMAD4 | [239-241] |
| miR-139-5p | 5-FU | NOTCH-1 | [242] |
| miR-153 | Oxaliplatin | FOXO3 | [243] |
| miR-194 | Oxaliplatin, irinotecan | HMGA2 | [244] |
| miR-200 family | 5-FU | EMT-related genes | [245] |
| miR-200b-3p | Oxaliplatin | PRDX2 | [246] |
| miR-203 | 5-FU | TYMS | [247] |
| miR-218 | FOLFOX | EZH2 | [248] |
| miR-761 | 5-FU | FOXM1 | [249] |
| miR-1915 | Oxaliplatin | BCL2 | [155] |
| miR-141/miR-200c | Oxaliplatin | ZEB1 | [250] |
| miR-18a*/miR-4802 | 5-FU, capecitabine | ATG7, ULK1 | [251] |
| Overexpression of miRNA causing drug resistance to molecular targeted drugs | |||
| miR-31 | Cetuximab | - | [252] |
| miR-126 | Bevacizumab | VEGF | [104] |
| miR-100/miR-125b | Cetuximab | Wnt/β-catenin negative regulators | [253] |
| Downregulation of miRNA causing drug resistance to molecular targeted drugs | |||
| Let-7 family | Cetuximab, panitumumab | KRAS | [115,223] |
| miR-7 | Cetuximab | EGFR, ERK1/2, AKT | [97] |
EGFR: Epidermal growth factor receptor; CRC: Colorectal cancer.
miRNAs mediating resistance to 5-FU
5-FU is the most commonly used first-line chemotherapeutic drug for CRC. Hypoxia inside CRC tumors has been shown to reduce drug sensitivity to 5-FU by upregulating two hypoxia-responsive miR-21 and miR-30d, which altered amino acid metabolism. Importantly, treatment with miR-21 and miR-30d antagonists were able to sensitize hypoxic and resistant CRC cells to 5-FU[147]. Reduced miR-34a expression in CRC cell line was also shown to mediate resistance to 5-FU. Ectopic expression of miR-34a was able to reverse the resistance by downregulating Sirt1 and E2F3[148]. It has been demonstrated that high expression of miR-23a could trigger 5-FU resistance by inhibiting the APAF-1/caspase-9 apoptotic pathways in CRC cell lines[149]. While miR-494 expression was found to be reduced in drug-resistant cell lines, forced expression of miR-494 was shown to potentiate the anticancer effect of 5-FU by targeting DPYD[150]. On the other hand, overexpression of miR-587 has been observed in CRC cells resistant to 5-FU in vitro and in vivo[151], which is mediated by downregulation of PPP2R1B and thus increased Akt phosphorylation and elevated XIAP expression to inhibit apoptosis.
Overexpression of the multidrug resistance transporter ABCG2 has been shown to cause resistance to 5-FU. Our research team has previously demonstrated the specific targeting of ABCG2 by miR-519c[152]. Interestingly, we revealed the shortening of the ABCG2 3’UTR in several ABCG2-overexpressing resistant cell lines, which removes the miR-519c binding site and has a repressive effect on mRNA stability and translocation blockadeallowing ABCG2 overexpression without a change in miR-519c expression to mediate drug resistance[152]. We further validated the effect of ABCG2 dysregulation on CRC sensitivity to 5-FU-based chemotherapy using pairs of archival tissue blocks of CRC tumors and their matched non-cancerous colon epithelial cells from CRC patients[153]. In the unresponsive tumor, high ABCG2 expression was found to be associated with the concomitant overexpression of a mRNA binding protein HuR but a low expression of miR-519c because miR-519c was known to target both HuR and ABCG2[153].
miRNAs mediating resistance to oxaliplatin
Oxaliplatin is a key platinum-based anticancer drug in combination regimens for CRC treatment. High expression of miR-203 has been shown to mediate oxaliplatin resistance by suppressing the key DNA double-strand break sensor ATM[154]. Likewise, high expression of miR-625-3p was associated with poor response to first-line treatment with oxaliplatin in metastatic CRC[94]. Moreover, forced expression of miR-1915 in an oxaliplatin-selected multidrug resistant cell line HCT116/L-OHP was found to induce apoptosis by inhibiting Bcl-2[155].
miRNAs mediating resistance to irinotecan
Irinotecan is another key cytotoxic chemotherapeutic drug in the FOLFIRI regimen (5-FU + leucovorin + irinotecan) for metastatic CRC. In colonospheres exhibiting cancer stem cell properties and chemoresistance from CRC cell lines, miR-451 was found to be remarkably downregulated[156]. Restoration of miR-451 in the colonospheres was found to specifically reverse drug resistance to irinotecan by decreasing the expression of the drug efflux ATP-binding cassette drug transporter ABCB1[156].
miRNAs mediating resistance to targeted drugs for CRC
Clinical efficacy with the anti-EGFR or anti-VEGF monoclonal antibodies in CRC is known to be severely hindered by drug resistance due to numerous mechanisms. Downregulation of miR-216b has been reported to mediate adapted autophagy (through upregulation of Beclin-1) in CRC cell lines to mediate resistance to anti-EGFR therapy[157]. Strategies to restore miR-216b or inhibit autophagy are thus proposed to improve the efficacy of anti-EGFR therapy in CRC. Conversely, the reduced expression of a tumor suppressor miR-7, which targets EGFR and RAF-1, was observed in CRC tumor specimens from patients unresponsive to cetuximab[97]. Reduced expression of miR-181a in tumor specimens from CRC patients was also found to correlate with poor response to EGFR antibodies[99].
MODULATING MIRNAS FOR TREATMENT OF CRC
Modulation of miRNA expressions and/or miRNAs-regulated pathways may be used as a therapeutic strategy to potentiate the cytotoxic effect from other anticancer drugs in CRC[158]. Guidelines have been proposed for the investigation and evaluation of potential microRNA-based therapeutics for cancer therapy[159]. Briefly, the up- or downregulation of miRNAs have to be established in appropriate cancer models. Gain/loss of function studies are then performed in vitro and in animal models. After the potential miRNA targets have been identified, an efficient delivery system has to be available to administer the miRNA modulating modalities for pharmacological, pharmacokinetic and pharmacodynamic studies. Finally, clinical trials will be conducted to evaluate the efficacy and safety of the potential miRNA therapeutics in patients. Only a few miRNA modulating systems have been investigated in vivo, which are summarized in Table 7.
Table 7.
miRNA therapeutics investigated in animal models of colorectal cancer
| miRNA | Animal model | Restoring or suppressing miRNA function | Therapeutic outcome | Ref. |
| miR-34a | Transgenic mice | Restoring | Anti-tumor | [126] |
| miR-135b | Xenotranplantation of tumor-derived organoids to mice | Suppression (antisense) | Anti-tumor | [129] |
| miR-143 | Mice xenograft | Restoring | Anti-tumor | [132] |
| miR-145 | Mice xenograft | Restoring | Negative effect | [132] |
| miR-4689 | Mice xenograft | Restoring (mimic) | Anti-tumor | [135] |
RESTORING TUMOR SUPPRESSIVE MIRNAS
Downregulation of tumor-suppressive miRNAs are known to contribute to cancer development. Strategies have been devised to effectively restore the function of these “lost” miRNAs using synthetic miRNA-like molecules, including miRNA mimics and miRNAs encoded in expression vectors. MiRNA mimics are short and chemically modified (2’-O-methoxy) RNA duplexes, which are processed into single-strand form intracellularly and loaded into RNA-induced silencing complex (RISC) to mediate the target mRNA degradation and/or post-translational blockade[160,161]. This approach has been investigated in cell models and orthotopic mice models of colorectal and hepatocellular carcinomas and hepatic metastasis from primary CRC to restore the expression of downregulated miR-26a, miR-33a, miR-34a and miR-145[159,162-164]. The restoration of these tumor suppressor miRNAs was found to inhibit cancer cell proliferation, induce apoptosis, shrink tumor size and prolong animal survival.
miR-34 is remarkably downregulated in a number of different cancer types including CRC[165]. As discussed above, miR-34 has been shown to regulate multiple oncogenic pathways. Thus, the restoration of miR-34 may represent a novel therapeutic approach to treat cancer. To this end, a first-in-human phase I clinical trial (NCT01829971) has been conducted to investigate the safety, pharmacokinetics and clinical activity of a liposomal formulation of miR-34 mimic (known as MRX34) in patients with advanced solid tumors including CRC[166]. While MRX34 was tolerated and displayed antitumor activity in a subset of patients with refractory advanced solid tumors[166], the sponsoring company (Mirna Therapeutic, Inc.) has voluntarily halted the enrollment and dosing of MRX34 in clinical study in September 2016 following multiple immune-related severe adverse events reported in patients dosed with MRX34.
INHIBITING ONCOGENIC MIRNAS
Different approaches have been developed to inhibit mature miRNAs, which include antisense oligonucleotides (also known as ASOs, anti-miRs, or antagomirs), miRNA masking, miRNA sponges, and anti-miR peptides. ASOs are single-stranded oligonucleotides that are complementary to the target miRNA[167]. The miRNAs bind to the RISC complexes, which subsequently block the interaction of miRNAs with their endogenous mRNA targets. This strategy has been evaluated in cell culture and mice models of CRC by targeting overexpressed oncogenic miRNAs. Specific inhibition of miR-135b has been achieved in human colon cancer SW480 cells to inhibit cell proliferation and to induce apoptosis[129] and in HCT116 cells to suppress cell migration[168]. Other oncogenic miRNAs, including miR-20a, miR-21, miR-95, and miR-675, have also been inhibited by similar approaches in human CRC cell lines[120,169-171]. ASOs are often chemically modified by conjugation to cholesterol to avoid cleavage by RISC nuclease and to increase their stability. Locked nucleic acids (LNAs) are nucleic acid analogs in which the ribose ring is “locked” with a methylene bridge connecting the 2’-O atom with the 4’-C atom[172]. Modification by LNAs is another approach used to enhance stability of ASOs, which also increases the binding affinity to the target miRNA[173]. LNA-containing oligonucleotides also have excellent mismatch discrimination capabilities[174], thus avoiding potential off-target secondary effects. LNA-anti-miR-21 was an ASO developed by the LNA technology[175]. It has been shown to inhibit cancer growth and invasion in a colon cancer cell line LS174T[175]. On the other hand, miRNA masking refers to the use of single-stranded 2’-O-methyl modified oligonucleotides complementary to miRNA binding sites in the 3’UTR of the target mRNAs to interfere with specific miRNA-mRNA interaction[176]. A miR-34a sponge construct, encoding a decoy mRNA containing multiple tandem miR-34a binding sites, has been shown to inhibit self-renewal of colon cancer stem cells in vitro[177]. Anti-miR peptides are peptide nucleic acids (PNAs), modified from DNA or RNA, with the sugar backbone replaced by a peptide-like backbone consisting of N-(2-aminoethyl)glycine units. Aside from being more stable and able to be administered systematically with low toxicity, PNAs have been shown to bind to target miRNA more tightly than equivalent oligonucleotides[178].
USE OF SMALL MOLECULE MODULATORS OF MIRNA EXPRESSION
Besides making use of pharmaceutical carriers to deliver the miRNA mimics/inhibitors to the tumor, attempts have also been made in preclinical studies to screen for small molecule drugs that modulate miRNA expression and function by luciferase-based reporter assay[179]. These compounds do not inhibit target mRNA recognition by the miRNAs, but they instead modulate the expression of the targeted miRNAs. With potential application in treating CRC, diazobenzene and its derivatives were identified to specifically inhibit miR-21 expression[179].
Most recently, aberrant expression of the lipid metabolic enzymes, acyl-CoA synthetase/stearoyl-CoA desaturase (ACSL/SCD) have been shown in CRC patients to promote cancer migration and invasion[180]. miR-19b-1 was found to a major regulator of ACSL/SCD expression. Importantly, lower miR-19b-1 expression and higher ACSL/SCD levels in tumor specimens from CRC patients were strongly associated with shorter disease-free survival[180]. Therefore, miRNA replacement therapy was proposed to increase the expression level of miR-19b-1 in colorectal tissue to improve the prognosis. Interestingly, since induction of miR-19b-1 expression has been achieved by a low protein diet in piglets[181], dietary modulation of this particular miRNA may also be feasible. More in vivo investigation is needed to test the efficacy and safety before we might benefit from miR-19b-1 replacement therapy in the management of CRC.
HURDLES IN MODULATING MIRNAS TO TREAT CRC
Since miRNAs only require partial sequence complementarity to target mRNA, non-specific targeting of bystander mRNAs may lead to off-target effects, toxicity and/or unfavorable immune response. Moreover, rapid degradation of miRNAs or anti-miRNAs by cellular nucleases and poor cellular uptake are also preventing the development of successful miRNA-based gene therapies. Furthermore, the administration of anti-miRs in vivo with an aim to block oncogenic miRNAs may adversely affect physiological functions normally regulated by these miRNAs. Thus, specific site or cell targeting will be needed to avoid the potential adverse effects. The lowest optimum concentration of miRNAs and appropriate delivery systems, such as viral and/or non-viral vectors, are critical factors required to achieve the desired therapeutic goal while minimizing adverse effects.
CONCLUSION
Accumulating evidence suggests that miRNAs play a crucial role in the regulation of genes driving CRC initiation, progression and metastasis. Differential expression of miRNAs in CRC has provided opportunities for their applications in diagnosis, prognostic prediction and stratification of risk groups. While numerous miRNA biomarkers have been reported to predict prognosis and drug response in CRC patients, it is interesting to note that the miRNA signature identified in different studies usually do not overlap and, in certain cases, they were even found to be contradictory[182]. The different selection criteria for patients, collection method and processing of biological samples, and detection methods may contribute to the different miRNA signatures obtained. To obtain reliable results, precaution should be taken to prevent the degradation of miRNAs during sample processing. It may be helpful to use these novel miRNA signatures together with the conventional biomarkers, such as CEA, to increase the sensitivity and specificity. As novel therapeutic targets, underexpressed miRNAs can be restored, whereas overexpressed miRNAs can be inhibited. Different delivery systems of these miRNA modulating moieties have been extensively studied in cell culture and animal models. Despite adverse effects having yielded a discouraging initial clinical experience with miR-34 replacement therapy, the correction of miRNA dysregulation is a promising therapeutic approach for the treatment of CRC; still, it is necessary to resolve the bystander effects and address toxicity concerns prior to large-scale clinical use.
Footnotes
Manuscript source: Invited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: China
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): D, D
Grade E (Poor): 0
Conflict-of-interest statement: No potential conflicts of interest. No financial support.
Peer-review started: May 4, 2018
First decision: June 6, 2018
Article in press: June 30, 2018
P- Reviewer: Kato J, Nakayama Y, Tomazic A S- Editor: Gong ZM L- Editor: A E- Editor: Huang Y
Contributor Information
Kenneth KW To, School of Pharmacy, Faculty of Medicine, the Chinese University of Hong Kong, Hong Kong, China. kennethto@cuhk.edu.hk.
Christy WS Tong, School of Pharmacy, Faculty of Medicine, the Chinese University of Hong Kong, Hong Kong, China.
Mingxia Wu, School of Pharmacy, Faculty of Medicine, the Chinese University of Hong Kong, Hong Kong, China.
William CS Cho, Department of Clinical Oncology, Queen Elizabeth Hospital, Hong Kong, China.
References
- 1.Arnold M, Sierra MS, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global patterns and trends in colorectal cancer incidence and mortality. Gut. 2017;66:683–691. doi: 10.1136/gutjnl-2015-310912. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 3.Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759–767. doi: 10.1016/0092-8674(90)90186-i. [DOI] [PubMed] [Google Scholar]
- 4.DA Silva FC, Wernhoff P, Dominguez-Barrera C, Dominguez-Valentin M. Update on Hereditary Colorectal Cancer. Anticancer Res. 2016;36:4399–4405. doi: 10.21873/anticanres.10983. [DOI] [PubMed] [Google Scholar]
- 5.Peters U, Bien S, Zubair N. Genetic architecture of colorectal cancer. Gut. 2015;64:1623–1636. doi: 10.1136/gutjnl-2013-306705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Graham JS, Cassidy J. Adjuvant therapy in colon cancer. Expert Rev Anticancer Ther. 2012;12:99–109. doi: 10.1586/era.11.189. [DOI] [PubMed] [Google Scholar]
- 7.Yalcin S, Trad D, Kader YA, Halawani H, Demir OG, Mall R, Meshcheryakov A, Nasr F, Nosworthy A, Osinsky D, et al. Personalized treatment is better than one treatment fits all in the management of patients with mCRC: a consensus statement. Future Oncol. 2014;10:2643–2657. doi: 10.2217/fon.14.203. [DOI] [PubMed] [Google Scholar]
- 8.Michael MZ, O’ Connor SM, van Holst Pellekaan NG, Young GP, James RJ. Reduced accumulation of specific microRNAs in colorectal neoplasia. Mol Cancer Res. 2003;1:882–891. [PubMed] [Google Scholar]
- 9.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 11.Segditsas S, Tomlinson I. Colorectal cancer and genetic alterations in the Wnt pathway. Oncogene. 2006;25:7531–7537. doi: 10.1038/sj.onc.1210059. [DOI] [PubMed] [Google Scholar]
- 12.Li T, Lai Q, Wang S, Cai J, Xiao Z, Deng D, He L, Jiao H, Ye Y, Liang L, et al. MicroRNA-224 sustains Wnt/β-catenin signaling and promotes aggressive phenotype of colorectal cancer. J Exp Clin Cancer Res. 2016;35:21. doi: 10.1186/s13046-016-0287-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Medema JP. Targeting the Colorectal Cancer Stem Cell. N Engl J Med. 2017;377:888–890. doi: 10.1056/NEJMcibr1706541. [DOI] [PubMed] [Google Scholar]
- 14.Vermeulen L, De Sousa E Melo F, van der Heijden M, Cameron K, de Jong JH, Borovski T, Tuynman JB, Todaro M, Merz C, Rodermond H, et al. Wnt activity defines colon cancer stem cells and is regulated by the microenvironment. Nat Cell Biol. 2010;12:468–476. doi: 10.1038/ncb2048. [DOI] [PubMed] [Google Scholar]
- 15.Le Grand F, Jones AE, Seale V, Scimè A, Rudnicki MA. Wnt7a activates the planar cell polarity pathway to drive the symmetric expansion of satellite stem cells. Cell Stem Cell. 2009;4:535–547. doi: 10.1016/j.stem.2009.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hwang WL, Jiang JK, Yang SH, Huang TS, Lan HY, Teng HW, Yang CY, Tsai YP, Lin CH, Wang HW, et al. MicroRNA-146a directs the symmetric division of Snail-dominant colorectal cancer stem cells. Nat Cell Biol. 2014;16:268–280. doi: 10.1038/ncb2910. [DOI] [PubMed] [Google Scholar]
- 17.Velho S, Oliveira C, Ferreira A, Ferreira AC, Suriano G, Schwartz S Jr, Duval A, Carneiro F, Machado JC, Hamelin R, Seruca R. The prevalence of PIK3CA mutations in gastric and colon cancer. Eur J Cancer. 2005;41:1649–1654. doi: 10.1016/j.ejca.2005.04.022. [DOI] [PubMed] [Google Scholar]
- 18.Arcaroli JJ, Quackenbush KS, Powell RW, Pitts TM, Spreafico A, Varella-Garcia M, Bemis L, Tan AC, Reinemann JM, Touban BM, et al. Common PIK3CA mutants and a novel 3’ UTR mutation are associated with increased sensitivity to saracatinib. Clin Cancer Res. 2012;18:2704–2714. doi: 10.1158/1078-0432.CCR-11-3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guo C, Sah JF, Beard L, Willson JK, Markowitz SD, Guda K. The noncoding RNA, miR-126, suppresses the growth of neoplastic cells by targeting phosphatidylinositol 3-kinase signaling and is frequently lost in colon cancers. Genes Chromosomes Cancer. 2008;47:939–946. doi: 10.1002/gcc.20596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cekaite L, Eide PW, Lind GE, Skotheim RI, Lothe RA. MicroRNAs as growth regulators, their function and biomarker status in colorectal cancer. Oncotarget. 2016;7:6476–6505. doi: 10.18632/oncotarget.6390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lièvre A, Bachet JB, Boige V, Cayre A, Le Corre D, Buc E, Ychou M, Bouché O, Landi B, Louvet C, et al. KRAS mutations as an independent prognostic factor in patients with advanced colorectal cancer treated with cetuximab. J Clin Oncol. 2008;26:374–379. doi: 10.1200/JCO.2007.12.5906. [DOI] [PubMed] [Google Scholar]
- 22.Sebio A, Paré L, Páez D, Salazar J, González A, Sala N, del Río E, Martín-Richard M, Tobeña M, Barnadas A, et al. The LCS6 polymorphism in the binding site of let-7 microRNA to the KRAS 3’-untranslated region: its role in the efficacy of anti-EGFR-based therapy in metastatic colorectal cancer patients. Pharmacogenet Genomics. 2013;23:142–147. doi: 10.1097/FPC.0b013e32835d9b0b. [DOI] [PubMed] [Google Scholar]
- 23.Tsang WP, Kwok TT. The miR-18a* microRNA functions as a potential tumor suppressor by targeting on K-Ras. Carcinogenesis. 2009;30:953–959. doi: 10.1093/carcin/bgp094. [DOI] [PubMed] [Google Scholar]
- 24.Liao WT, Ye YP, Zhang NJ, Li TT, Wang SY, Cui YM, Qi L, Wu P, Jiao HL, Xie YJ, et al. MicroRNA-30b functions as a tumour suppressor in human colorectal cancer by targeting KRAS, PIK3CD and BCL2. J Pathol. 2014;232:415–427. doi: 10.1002/path.4309. [DOI] [PubMed] [Google Scholar]
- 25.Chen X, Guo X, Zhang H, Xiang Y, Chen J, Yin Y, Cai X, Wang K, Wang G, Ba Y, et al. Role of miR-143 targeting KRAS in colorectal tumorigenesis. Oncogene. 2009;28:1385–1392. doi: 10.1038/onc.2008.474. [DOI] [PubMed] [Google Scholar]
- 26.Pagliuca A, Valvo C, Fabrizi E, di Martino S, Biffoni M, Runci D, Forte S, De Maria R, Ricci-Vitiani L. Analysis of the combined action of miR-143 and miR-145 on oncogenic pathways in colorectal cancer cells reveals a coordinate program of gene repression. Oncogene. 2013;32:4806–4813. doi: 10.1038/onc.2012.495. [DOI] [PubMed] [Google Scholar]
- 27.Grady WM, Myeroff LL, Swinler SE, Rajput A, Thiagalingam S, Lutterbaugh JD, Neumann A, Brattain MG, Chang J, Kim SJ, et al. Mutational inactivation of transforming growth factor beta receptor type II in microsatellite stable colon cancers. Cancer Res. 1999;59:320–324. [PubMed] [Google Scholar]
- 28.Grady WM, Markowitz SD. Genetic and epigenetic alterations in colon cancer. Annu Rev Genomics Hum Genet. 2002;3:101–128. doi: 10.1146/annurev.genom.3.022502.103043. [DOI] [PubMed] [Google Scholar]
- 29.Yu Y, Kanwar SS, Patel BB, Oh PS, Nautiyal J, Sarkar FH, Majumdar AP. MicroRNA-21 induces stemness by downregulating transforming growth factor beta receptor 2 (TGFβR2) in colon cancer cells. Carcinogenesis. 2012;33:68–76. doi: 10.1093/carcin/bgr246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Feng B, Dong TT, Wang LL, Zhou HM, Zhao HC, Dong F, Zheng MH. Colorectal cancer migration and invasion initiated by microRNA-106a. PLoS One. 2012;7:e43452. doi: 10.1371/journal.pone.0043452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang W, Zhang T, Jin R, Zhao H, Hu J, Feng B, Zang L, Zheng M, Wang M. MicroRNA-301a promotes migration and invasion by targeting TGFBR2 in human colorectal cancer. J Exp Clin Cancer Res. 2014;33:113. doi: 10.1186/s13046-014-0113-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li Q, Zou C, Zou C, Han Z, Xiao H, Wei H, Wang W, Zhang L, Zhang X, Tang Q, et al. MicroRNA-25 functions as a potential tumor suppressor in colon cancer by targeting Smad7. Cancer Lett. 2013;335:168–174. doi: 10.1016/j.canlet.2013.02.029. [DOI] [PubMed] [Google Scholar]
- 33.Zhang F, Luo Y, Shao Z, Xu L, Liu X, Niu Y, Shi J, Sun X, Liu Y, Ding Y, et al. MicroRNA-187, a downstream effector of TGFβ pathway, suppresses Smad-mediated epithelial-mesenchymal transition in colorectal cancer. Cancer Lett. 2016;373:203–213. doi: 10.1016/j.canlet.2016.01.037. [DOI] [PubMed] [Google Scholar]
- 34.Singh A, Settleman J. EMT, cancer stem cells and drug resistance: an emerging axis of evil in the war on cancer. Oncogene. 2010;29:4741–4751. doi: 10.1038/onc.2010.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Carstens JL, Lovisa S, Kalluri R. Microenvironment-dependent cues trigger miRNA-regulated feedback loop to facilitate the EMT/MET switch. J Clin Invest. 2014;124:1458–1460. doi: 10.1172/JCI75239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang JX, Mai SJ, Huang XX, Wang FW, Liao YJ, Lin MC, Kung HF, Zeng YX, Xie D. MiR-29c mediates epithelial-to-mesenchymal transition in human colorectal carcinoma metastasis via PTP4A and GNA13 regulation of β-catenin signaling. Ann Oncol. 2014;25:2196–2204. doi: 10.1093/annonc/mdu439. [DOI] [PubMed] [Google Scholar]
- 37.Hur K, Toiyama Y, Takahashi M, Balaguer F, Nagasaka T, Koike J, Hemmi H, Koi M, Boland CR, Goel A. MicroRNA-200c modulates epithelial-to-mesenchymal transition (EMT) in human colorectal cancer metastasis. Gut. 2013;62:1315–1326. doi: 10.1136/gutjnl-2011-301846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guo YH, Wang LQ, Li B, Xu H, Yang JH, Zheng LS, Yu P, Zhou AD, Zhang Y, Xie SJ, et al. Wnt/β-catenin pathway transactivates microRNA-150 that promotes EMT of colorectal cancer cells by suppressing CREB signaling. Oncotarget. 2016;7:42513–42526. doi: 10.18632/oncotarget.9893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zauber AG, Winawer SJ, O’Brien MJ, Lansdorp-Vogelaar I, van Ballegooijen M, Hankey BF, Shi W, Bond JH, Schapiro M, Panish JF, et al. Colonoscopic polypectomy and long-term prevention of colorectal-cancer deaths. N Engl J Med. 2012;366:687–696. doi: 10.1056/NEJMoa1100370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nishihara R, Wu K, Lochhead P, Morikawa T, Liao X, Qian ZR, Inamura K, Kim SA, Kuchiba A, Yamauchi M, et al. Long-term colorectal-cancer incidence and mortality after lower endoscopy. N Engl J Med. 2013;369:1095–1105. doi: 10.1056/NEJMoa1301969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Akao Y, Nakagawa Y, Naoe T. MicroRNAs 143 and 145 are possible common onco-microRNAs in human cancers. Oncol Rep. 2006;16:845–850. [PubMed] [Google Scholar]
- 42.Uratani R, Toiyama Y, Kitajima T, Kawamura M, Hiro J, Kobayashi M, Tanaka K, Inoue Y, Mohri Y, Mori T, et al. Diagnostic Potential of Cell-Free and Exosomal MicroRNAs in the Identification of Patients with High-Risk Colorectal Adenomas. PLoS One. 2016;11:e0160722. doi: 10.1371/journal.pone.0160722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moridikia A, Mirzaei H, Sahebkar A, Salimian J. MicroRNAs: Potential candidates for diagnosis and treatment of colorectal cancer. J Cell Physiol. 2018;233:901–913. doi: 10.1002/jcp.25801. [DOI] [PubMed] [Google Scholar]
- 44.Carter JV, Galbraith NJ, Yang D, Burton JF, Walker SP, Galandiuk S. Blood-based microRNAs as biomarkers for the diagnosis of colorectal cancer: a systematic review and meta-analysis. Br J Cancer. 2017;116:762–774. doi: 10.1038/bjc.2017.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pan C, Yan X, Li H, Huang L, Yin M, Yang Y, Gao R, Hong L, Ma Y, Shi C, et al. Systematic literature review and clinical validation of circulating microRNAs as diagnostic biomarkers for colorectal cancer. Oncotarget. 2017;8:68317–68328. doi: 10.18632/oncotarget.19344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vatandoost N, Ghanbari J, Mojaver M, Avan A, Ghayour-Mobarhan M, Nedaeinia R, Salehi R. Early detection of colorectal cancer: from conventional methods to novel biomarkers. J Cancer Res Clin Oncol. 2016;142:341–351. doi: 10.1007/s00432-015-1928-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Clancy C, Joyce MR, Kerin MJ. The use of circulating microRNAs as diagnostic biomarkers in colorectal cancer. Cancer Biomark. 2015;15:103–113. doi: 10.3233/CBM-140456. [DOI] [PubMed] [Google Scholar]
- 48.Sood P, Krek A, Zavolan M, Macino G, Rajewsky N. Cell-type-specific signatures of microRNAs on target mRNA expression. Proc Natl Acad Sci USA. 2006;103:2746–2751. doi: 10.1073/pnas.0511045103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ludwig N, Leidinger P, Becker K, Backes C, Fehlmann T, Pallasch C, Rheinheimer S, Meder B, Stähler C, Meese E, et al. Distribution of miRNA expression across human tissues. Nucleic Acids Res. 2016;44:3865–3877. doi: 10.1093/nar/gkw116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sha D, Lee AM, Shi Q, Alberts SR, Sargent DJ, Sinicrope FA, Diasio RB. Association study of the let-7 miRNA-complementary site variant in the 3’ untranslated region of the KRAS gene in stage III colon cancer (NCCTG N0147 Clinical Trial) Clin Cancer Res. 2014;20:3319–3327. doi: 10.1158/1078-0432.CCR-14-0069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yu G, Tang JQ, Tian ML, Li H, Wang X, Wu T, Zhu J, Huang SJ, Wan YL. Prognostic values of the miR-17-92 cluster and its paralogs in colon cancer. J Surg Oncol. 2012;106:232–237. doi: 10.1002/jso.22138. [DOI] [PubMed] [Google Scholar]
- 52.Tsuchida A, Ohno S, Wu W, Borjigin N, Fujita K, Aoki T, Ueda S, Takanashi M, Kuroda M. miR-92 is a key oncogenic component of the miR-17-92 cluster in colon cancer. Cancer Sci. 2011;102:2264–2271. doi: 10.1111/j.1349-7006.2011.02081.x. [DOI] [PubMed] [Google Scholar]
- 53.Peric D, Chvalova K, Rousselet G. Identification of microprocessor-dependent cancer cells allows screening for growth-sustaining micro-RNAs. Oncogene. 2012;31:2039–2048. doi: 10.1038/onc.2011.391. [DOI] [PubMed] [Google Scholar]
- 54.Liang Z, Li Y, Huang K, Wagar N, Shim H. Regulation of miR-19 to breast cancer chemoresistance through targeting PTEN. Pharm Res. 2011;28:3091–3100. doi: 10.1007/s11095-011-0570-y. [DOI] [PubMed] [Google Scholar]
- 55.Olive V, Bennett MJ, Walker JC, Ma C, Jiang I, Cordon-Cardo C, Li QJ, Lowe SW, Hannon GJ, He L. miR-19 is a key oncogenic component of mir-17-92. Genes Dev. 2009;23:2839–2849. doi: 10.1101/gad.1861409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tili E, Michaille JJ, Liu CG, Alder H, Taccioli C, Volinia S, Calin GA, Croce CM. GAM/ZFp/ZNF512B is central to a gene sensor circuitry involving cell-cycle regulators, TGF{beta} effectors, Drosha and microRNAs with opposite oncogenic potentials. Nucleic Acids Res. 2010;38:7673–7688. doi: 10.1093/nar/gkq637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ellermeier C, Vang S, Cleveland K, Durand W, Resnick MB, Brodsky AS. Prognostic microRNA expression signature from examination of colorectal primary and metastatic tumors. Anticancer Res. 2014;34:3957–3967. [PubMed] [Google Scholar]
- 58.Slaby O, Svoboda M, Fabian P, Smerdova T, Knoflickova D, Bednarikova M, Nenutil R, Vyzula R. Altered expression of miR-21, miR-31, miR-143 and miR-145 is related to clinicopathologic features of colorectal cancer. Oncology. 2007;72:397–402. doi: 10.1159/000113489. [DOI] [PubMed] [Google Scholar]
- 59.Shibuya H, Iinuma H, Shimada R, Horiuchi A, Watanabe T. Clinicopathological and prognostic value of microRNA-21 and microRNA-155 in colorectal cancer. Oncology. 2010;79:313–320. doi: 10.1159/000323283. [DOI] [PubMed] [Google Scholar]
- 60.Drusco A, Nuovo GJ, Zanesi N, Di Leva G, Pichiorri F, Volinia S, Fernandez C, Antenucci A, Costinean S, Bottoni A, et al. MicroRNA profiles discriminate among colon cancer metastasis. PLoS One. 2014;9:e96670. doi: 10.1371/journal.pone.0096670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Weissmann-Brenner A, Kushnir M, Lithwick Yanai G, Aharonov R, Gibori H, Purim O, Kundel Y, Morgenstern S, Halperin M, Niv Y, et al. Tumor microRNA-29a expression and the risk of recurrence in stage II colon cancer. Int J Oncol. 2012;40:2097–2103. doi: 10.3892/ijo.2012.1403. [DOI] [PubMed] [Google Scholar]
- 62.Tang W, Zhu Y, Gao J, Fu J, Liu C, Liu Y, Song C, Zhu S, Leng Y, Wang G, et al. MicroRNA-29a promotes colorectal cancer metastasis by regulating matrix metalloproteinase 2 and E-cadherin via KLF4. Br J Cancer. 2014;110:450–458. doi: 10.1038/bjc.2013.724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Locker GY, Hamilton S, Harris J, Jessup JM, Kemeny N, Macdonald JS, Somerfield MR, Hayes DF, Bast RC Jr; ASCO. ASCO 2006 update of recommendations for the use of tumor markers in gastrointestinal cancer. J Clin Oncol. 2006;24:5313–5327. doi: 10.1200/JCO.2006.08.2644. [DOI] [PubMed] [Google Scholar]
- 64.Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O’Briant KC, Allen A, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci USA. 2008;105:10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Turchinovich A, Weiz L, Langheinz A, Burwinkel B. Characterization of extracellular circulating microRNA. Nucleic Acids Res. 2011;39:7223–7233. doi: 10.1093/nar/gkr254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ, Lötvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 67.Wang J, Huang SK, Zhao M, Yang M, Zhong JL, Gu YY, Peng H, Che YQ, Huang CZ. Identification of a circulating microRNA signature for colorectal cancer detection. PLoS One. 2014;9:e87451. doi: 10.1371/journal.pone.0087451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Toiyama Y, Takahashi M, Hur K, Nagasaka T, Tanaka K, Inoue Y, Kusunoki M, Boland CR, Goel A. Serum miR-21 as a diagnostic and prognostic biomarker in colorectal cancer. J Natl Cancer Inst. 2013;105:849–859. doi: 10.1093/jnci/djt101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hollis M, Nair K, Vyas A, Chaturvedi LS, Gambhir S, Vyas D. MicroRNAs potential utility in colon cancer: Early detection, prognosis, and chemosensitivity. World J Gastroenterol. 2015;21:8284–8292. doi: 10.3748/wjg.v21.i27.8284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cheng H, Zhang L, Cogdell DE, Zheng H, Schetter AJ, Nykter M, Harris CC, Chen K, Hamilton SR, Zhang W. Circulating plasma MiR-141 is a novel biomarker for metastatic colon cancer and predicts poor prognosis. PLoS One. 2011;6:e17745. doi: 10.1371/journal.pone.0017745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Fang Z, Tang J, Bai Y, Lin H, You H, Jin H, Lin L, You P, Li J, Dai Z, et al. Plasma levels of microRNA-24, microRNA-320a, and microRNA-423-5p are potential biomarkers for colorectal carcinoma. J Exp Clin Cancer Res. 2015;34:86. doi: 10.1186/s13046-015-0198-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ahmed FE, Jeffries CD, Vos PW, Flake G, Nuovo GJ, Sinar DR, Naziri W, Marcuard SP. Diagnostic microRNA markers for screening sporadic human colon cancer and active ulcerative colitis in stool and tissue. Cancer Genomics Proteomics. 2009;6:281–295. [PubMed] [Google Scholar]
- 73.Ahlquist DA. Molecular detection of colorectal neoplasia. Gastroenterology. 2010;138:2127–2139. doi: 10.1053/j.gastro.2010.01.055. [DOI] [PubMed] [Google Scholar]
- 74.Ahmed FE, Ahmed NC, Vos PW, Bonnerup C, Atkins JN, Casey M, Nuovo GJ, Naziri W, Wiley JE, Mota H, et al. Diagnostic microRNA markers to screen for sporadic human colon cancer in stool: I. Proof of principle. Cancer Genomics Proteomics. 2013;10:93–113. [PubMed] [Google Scholar]
- 75.Wu CW, Ng SC, Dong Y, Tian L, Ng SS, Leung WW, Law WT, Yau TO, Chan FK, Sung JJ, et al. Identification of microRNA-135b in stool as a potential noninvasive biomarker for colorectal cancer and adenoma. Clin Cancer Res. 2014;20:2994–3002. doi: 10.1158/1078-0432.CCR-13-1750. [DOI] [PubMed] [Google Scholar]
- 76.Braicu C, Tomuleasa C, Monroig P, Cucuianu A, Berindan-Neagoe I, Calin GA. Exosomes as divine messengers: are they the Hermes of modern molecular oncology? Cell Death Differ. 2015;22:34–45. doi: 10.1038/cdd.2014.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ji H, Greening DW, Barnes TW, Lim JW, Tauro BJ, Rai A, Xu R, Adda C, Mathivanan S, Zhao W, et al. Proteome profiling of exosomes derived from human primary and metastatic colorectal cancer cells reveal differential expression of key metastatic factors and signal transduction components. Proteomics. 2013;13:1672–1686. doi: 10.1002/pmic.201200562. [DOI] [PubMed] [Google Scholar]
- 78.Matsumura T, Sugimachi K, Iinuma H, Takahashi Y, Kurashige J, Sawada G, Ueda M, Uchi R, Ueo H, Takano Y, et al. Exosomal microRNA in serum is a novel biomarker of recurrence in human colorectal cancer. Br J Cancer. 2015;113:275–281. doi: 10.1038/bjc.2015.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Monzo M, Santasusagna S, Moreno I, Martinez F, Hernández R, Muñoz C, Castellano JJ, Moreno J, Navarro A. Exosomal microRNAs isolated from plasma of mesenteric veins linked to liver metastases in resected patients with colon cancer. Oncotarget. 2017;8:30859–30869. doi: 10.18632/oncotarget.16103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.NIH consensus conference. Adjuvant therapy for patients with colon and rectal cancer. JAMA. 1990;264:1444–1450. [PubMed] [Google Scholar]
- 81.Quasar Collaborative Group. Gray R, Barnwell J, McConkey C, Hills RK, Williams NS, Kerr DJ. Adjuvant chemotherapy versus observation in patients with colorectal cancer: a randomised study. Lancet. 2007;370:2020–2029. doi: 10.1016/S0140-6736(07)61866-2. [DOI] [PubMed] [Google Scholar]
- 82.Labianca R, Nordlinger B, Beretta GD, Brouquet A, Cervantes A; ESMO Guidelines Working Group. Primary colon cancer: ESMO Clinical Practice Guidelines for diagnosis, adjuvant treatment and follow-up. Ann Oncol. 2010;21 Suppl 5:v70–v77. doi: 10.1093/annonc/mdq168. [DOI] [PubMed] [Google Scholar]
- 83.Agesen TH, Sveen A, Merok MA, Lind GE, Nesbakken A, Skotheim RI, Lothe RA. ColoGuideEx: a robust gene classifier specific for stage II colorectal cancer prognosis. Gut. 2012;61:1560–1567. doi: 10.1136/gutjnl-2011-301179. [DOI] [PubMed] [Google Scholar]
- 84.Schetter AJ, Nguyen GH, Bowman ED, Mathé EA, Yuen ST, Hawkes JE, Croce CM, Leung SY, Harris CC. Association of inflammation-related and microRNA gene expression with cancer-specific mortality of colon adenocarcinoma. Clin Cancer Res. 2009;15:5878–5887. doi: 10.1158/1078-0432.CCR-09-0627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kjaer-Frifeldt S, Hansen TF, Nielsen BS, Joergensen S, Lindebjerg J, Soerensen FB, dePont Christensen R, Jakobsen A; Danish Colorectal Cancer Group. The prognostic importance of miR-21 in stage II colon cancer: a population-based study. Br J Cancer. 2012;107:1169–1174. doi: 10.1038/bjc.2012.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhang JX, Song W, Chen ZH, Wei JH, Liao YJ, Lei J, Hu M, Chen GZ, Liao B, Lu J, et al. Prognostic and predictive value of a microRNA signature in stage II colon cancer: a microRNA expression analysis. Lancet Oncol. 2013;14:1295–1306. doi: 10.1016/S1470-2045(13)70491-1. [DOI] [PubMed] [Google Scholar]
- 87.Yamazaki N, Koga Y, Taniguchi H, Kojima M, Kanemitsu Y, Saito N, Matsumura Y. High expression of miR-181c as a predictive marker of recurrence in stage II colorectal cancer. Oncotarget. 2017;8:6970–6983. doi: 10.18632/oncotarget.14344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Salendo J, Spitzner M, Kramer F, Zhang X, Jo P, Wolff HA, Kitz J, Kaulfuß S, Beißbarth T, Dobbelstein M, et al. Identification of a microRNA expression signature for chemoradiosensitivity of colorectal cancer cells, involving miRNAs-320a, -224, -132 and let7g. Radiother Oncol. 2013;108:451–457. doi: 10.1016/j.radonc.2013.06.032. [DOI] [PubMed] [Google Scholar]
- 89.Caramés C, Cristóbal I, Moreno V, del Puerto L, Moreno I, Rodriguez M, Marín JP, Correa AV, Hernández R, Zenzola V, et al. MicroRNA-21 predicts response to preoperative chemoradiotherapy in locally advanced rectal cancer. Int J Colorectal Dis. 2015;30:899–906. doi: 10.1007/s00384-015-2231-9. [DOI] [PubMed] [Google Scholar]
- 90.Schetter AJ, Leung SY, Sohn JJ, Zanetti KA, Bowman ED, Yanaihara N, Yuen ST, Chan TL, Kwong DL, Au GK, et al. MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. JAMA. 2008;299:425–436. doi: 10.1001/jama.299.4.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ma Y, Zhang P, Wang F, Zhang H, Yang J, Peng J, Liu W, Qin H. miR-150 as a potential biomarker associated with prognosis and therapeutic outcome in colorectal cancer. Gut. 2012;61:1447–1453. doi: 10.1136/gutjnl-2011-301122. [DOI] [PubMed] [Google Scholar]
- 92.Takahashi M, Cuatrecasas M, Balaguer F, Hur K, Toiyama Y, Castells A, Boland CR, Goel A. The clinical significance of MiR-148a as a predictive biomarker in patients with advanced colorectal cancer. PLoS One. 2012;7:e46684. doi: 10.1371/journal.pone.0046684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Perez-Carbonell L, Sinicrope FA, Alberts SR, Oberg AL, Balaguer F, Castells A, Boland CR, Goel A. MiR-320e is a novel prognostic biomarker in colorectal cancer. Br J Cancer. 2015;113:83–90. doi: 10.1038/bjc.2015.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Rasmussen MH, Jensen NF, Tarpgaard LS, Qvortrup C, Rømer MU, Stenvang J, Hansen TP, Christensen LL, Lindebjerg J, Hansen F, et al. High expression of microRNA-625-3p is associated with poor response to first-line oxaliplatin based treatment of metastatic colorectal cancer. Mol Oncol. 2013;7:637–646. doi: 10.1016/j.molonc.2013.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Svoboda M, Sana J, Fabian P, Kocakova I, Gombosova J, Nekvindova J, Radova L, Vyzula R, Slaby O. MicroRNA expression profile associated with response to neoadjuvant chemoradiotherapy in locally advanced rectal cancer patients. Radiat Oncol. 2012;7:195. doi: 10.1186/1748-717X-7-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Della Vittoria Scarpati G, Falcetta F, Carlomagno C, Ubezio P, Marchini S, De Stefano A, Singh VK, D’Incalci M, De Placido S, Pepe S. A specific miRNA signature correlates with complete pathological response to neoadjuvant chemoradiotherapy in locally advanced rectal cancer. Int J Radiat Oncol Biol Phys. 2012;83:1113–1119. doi: 10.1016/j.ijrobp.2011.09.030. [DOI] [PubMed] [Google Scholar]
- 97.Suto T, Yokobori T, Yajima R, Morita H, Fujii T, Yamaguchi S, Altan B, Tsutsumi S, Asao T, Kuwano H. MicroRNA-7 expression in colorectal cancer is associated with poor prognosis and regulates cetuximab sensitivity via EGFR regulation. Carcinogenesis. 2015;36:338–345. doi: 10.1093/carcin/bgu242. [DOI] [PubMed] [Google Scholar]
- 98.Mosakhani N, Lahti L, Borze I, Karjalainen-Lindsberg ML, Sundström J, Ristamäki R, Osterlund P, Knuutila S, Sarhadi VK. MicroRNA profiling predicts survival in anti-EGFR treated chemorefractory metastatic colorectal cancer patients with wild-type KRAS and BRAF. Cancer Genet. 2012;205:545–551. doi: 10.1016/j.cancergen.2012.08.003. [DOI] [PubMed] [Google Scholar]
- 99.Pichler M, Winter E, Ress AL, Bauernhofer T, Gerger A, Kiesslich T, Lax S, Samonigg H, Hoefler G. miR-181a is associated with poor clinical outcome in patients with colorectal cancer treated with EGFR inhibitor. J Clin Pathol. 2014;67:198–203. doi: 10.1136/jclinpath-2013-201904. [DOI] [PubMed] [Google Scholar]
- 100.Igarashi H, Kurihara H, Mitsuhashi K, Ito M, Okuda H, Kanno S, Naito T, Yoshii S, Takahashi H, Kusumi T, et al. Association of MicroRNA-31-5p with Clinical Efficacy of Anti-EGFR Therapy in Patients with Metastatic Colorectal Cancer. Ann Surg Oncol. 2015;22:2640–2648. doi: 10.1245/s10434-014-4264-7. [DOI] [PubMed] [Google Scholar]
- 101.Cappuzzo F, Sacconi A, Landi L, Ludovini V, Biagioni F, D’Incecco A, Capodanno A, Salvini J, Corgna E, Cupini S, et al. MicroRNA signature in metastatic colorectal cancer patients treated with anti-EGFR monoclonal antibodies. Clin Colorectal Cancer. 2014;13:37–45.e4. doi: 10.1016/j.clcc.2013.11.006. [DOI] [PubMed] [Google Scholar]
- 102.Ruzzo A, Graziano F, Vincenzi B, Canestrari E, Perrone G, Galluccio N, Catalano V, Loupakis F, Rabitti C, Santini D, et al. High let-7a microRNA levels in KRAS-mutated colorectal carcinomas may rescue anti-EGFR therapy effects in patients with chemotherapy-refractory metastatic disease. Oncologist. 2012;17:823–829. doi: 10.1634/theoncologist.2012-0081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Mekenkamp LJ, Tol J, Dijkstra JR, de Krijger I, Vink-Börger ME, van Vliet S, Teerenstra S, Kamping E, Verwiel E, Koopman M, et al. Beyond KRAS mutation status: influence of KRAS copy number status and microRNAs on clinical outcome to cetuximab in metastatic colorectal cancer patients. BMC Cancer. 2012;12:292. doi: 10.1186/1471-2407-12-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Hansen TF, Carlsen AL, Heegaard NH, Sørensen FB, Jakobsen A. Changes in circulating microRNA-126 during treatment with chemotherapy and bevacizumab predicts treatment response in patients with metastatic colorectal cancer. Br J Cancer. 2015;112:624–629. doi: 10.1038/bjc.2014.652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kjersem JB, Ikdahl T, Lingjaerde OC, Guren T, Tveit KM, Kure EH. Plasma microRNAs predicting clinical outcome in metastatic colorectal cancer patients receiving first-line oxaliplatin-based treatment. Mol Oncol. 2014;8:59–67. doi: 10.1016/j.molonc.2013.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zhang J, Zhang K, Bi M, Jiao X, Zhang D, Dong Q. Circulating microRNA expressions in colorectal cancer as predictors of response to chemotherapy. Anticancer Drugs. 2014;25:346–352. doi: 10.1097/CAD.0000000000000049. [DOI] [PubMed] [Google Scholar]
- 107.Chen J, Wang W, Zhang Y, Chen Y, Hu T. Predicting distant metastasis and chemoresistance using plasma miRNAs. Med Oncol. 2014;31:799. doi: 10.1007/s12032-013-0799-x. [DOI] [PubMed] [Google Scholar]
- 108.Schou JV, Rossi S, Jensen BV, Nielsen DL, Pfeiffer P, Høgdall E, Yilmaz M, Tejpar S, Delorenzi M, Kruhøffer M, et al. miR-345 in metastatic colorectal cancer: a non-invasive biomarker for clinical outcome in non-KRAS mutant patients treated with 3rd line cetuximab and irinotecan. PLoS One. 2014;9:e99886. doi: 10.1371/journal.pone.0099886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Landi D, Gemignani F, Landi S. Role of variations within microRNA-binding sites in cancer. Mutagenesis. 2012;27:205–210. doi: 10.1093/mutage/ger055. [DOI] [PubMed] [Google Scholar]
- 110.Ryan BM, Robles AI, Harris CC. Genetic variation in microRNA networks: the implications for cancer research. Nat Rev Cancer. 2010;10:389–402. doi: 10.1038/nrc2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Vodicka P, Pardini B, Vymetalkova V, Naccarati A. “Polymorphisms in non-coding RNA genes and their targets sites as risk factors of sporadic colorectal cancer,” in Non-Coding RNAs in Colorectal Cancer, vol.937 of Advances in Experimental Medicine and Biology, pp. 123-149, Springer International, Cham, Switzerland, 2016. doi: 10.1007/978-3-319-42059-2_7. [DOI] [PubMed] [Google Scholar]
- 112.Yang YP, Ting WC, Chen LM, Lu TL, Bao BY. Polymorphisms in MicroRNA Binding Sites Predict Colorectal Cancer Survival. Int J Med Sci. 2017;14:53–57. doi: 10.7150/ijms.17027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Bahrami-B F, Ataie-Kachoie P, Pourgholami MH, Morris DL. p70 Ribosomal protein S6 kinase (Rps6kb1): an update. J Clin Pathol. 2014;67:1019–1025. doi: 10.1136/jclinpath-2014-202560. [DOI] [PubMed] [Google Scholar]
- 114.Scanlan MJ, Gordan JD, Williamson B, Stockert E, Bander NH, Jongeneel V, Gure AO, Jäger D, Jäger E, Knuth A, et al. Antigens recognized by autologous antibody in patients with renal-cell carcinoma. Int J Cancer. 1999;83:456–464. doi: 10.1002/(sici)1097-0215(19991112)83:4<456::aid-ijc4>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- 115.Zhang W, Winder T, Ning Y, Pohl A, Yang D, Kahn M, Lurje G, Labonte MJ, Wilson PM, Gordon MA, et al. A let-7 microRNA-binding site polymorphism in 3’-untranslated region of KRAS gene predicts response in wild-type KRAS patients with metastatic colorectal cancer treated with cetuximab monotherapy. Ann Oncol. 2011;22:104–109. doi: 10.1093/annonc/mdq315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Aslam MI, Patel M, Singh B, Jameson JS, Pringle JH. MicroRNA manipulation in colorectal cancer cells: from laboratory to clinical application. J Transl Med. 2012;10:128. doi: 10.1186/1479-5876-10-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Shah MY, Calin GA. MicroRNAs as therapeutic targets in human cancers. Wiley Interdiscip Rev RNA. 2014;5:537–548. doi: 10.1002/wrna.1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Lin PL, Wu DW, Huang CC, He TY, Chou MC, Sheu GT, Lee H. MicroRNA-21 promotes tumour malignancy via increased nuclear translocation of β-catenin and predicts poor outcome in APC-mutated but not in APC-wild-type colorectal cancer. Carcinogenesis. 2014;35:2175–2182. doi: 10.1093/carcin/bgu110. [DOI] [PubMed] [Google Scholar]
- 119.Asangani IA, Rasheed SA, Nikolova DA, Leupold JH, Colburn NH, Post S, Allgayer H. MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene. 2008;27:2128–2136. doi: 10.1038/sj.onc.1210856. [DOI] [PubMed] [Google Scholar]
- 120.Valeri N, Gasparini P, Braconi C, Paone A, Lovat F, Fabbri M, Sumani KM, Alder H, Amadori D, Patel T, et al. MicroRNA-21 induces resistance to 5-fluorouracil by down-regulating human DNA MutS homolog 2 (hMSH2) Proc Natl Acad Sci USA. 2010;107:21098–21103. doi: 10.1073/pnas.1015541107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Wagenaar TR, Zabludoff S, Ahn SM, Allerson C, Arlt H, Baffa R, Cao H, Davis S, Garcia-Echeverria C, Gaur R, et al. Anti-miR-21 Suppresses Hepatocellular Carcinoma Growth via Broad Transcriptional Network Deregulation. Mol Cancer Res. 2015;13:1009–1021. doi: 10.1158/1541-7786.MCR-14-0703. [DOI] [PubMed] [Google Scholar]
- 122.Hermeking H. MicroRNAs in the p53 network: micromanagement of tumour suppression. Nat Rev Cancer. 2012;12:613–626. doi: 10.1038/nrc3318. [DOI] [PubMed] [Google Scholar]
- 123.Chang TC, Wentzel EA, Kent OA, Ramachandran K, Mullendore M, Lee KH, Feldmann G, Yamakuchi M, Ferlito M, Lowenstein CJ, et al. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol Cell. 2007;26:745–752. doi: 10.1016/j.molcel.2007.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Nugent M, Miller N, Kerin MJ. Circulating miR-34a levels are reduced in colorectal cancer. J Surg Oncol. 2012;106:947–952. doi: 10.1002/jso.23174. [DOI] [PubMed] [Google Scholar]
- 125.Hahn S, Jackstadt R, Siemens H, Hünten S, Hermeking H. SNAIL and miR-34a feed-forward regulation of ZNF281/ZBP99 promotes epithelial-mesenchymal transition. EMBO J. 2013;32:3079–3095. doi: 10.1038/emboj.2013.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Rokavec M, Öner MG, Li H, Jackstadt R, Jiang L, Lodygin D, Kaller M, Horst D, Ziegler PK, Schwitalla S, et al. IL-6R/STAT3/miR-34a feedback loop promotes EMT-mediated colorectal cancer invasion and metastasis. J Clin Invest. 2014;124:1853–1867. doi: 10.1172/JCI73531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Strillacci A, Valerii MC, Sansone P, Caggiano C, Sgromo A, Vittori L, Fiorentino M, Poggioli G, Rizzello F, Campieri M, et al. Loss of miR-101 expression promotes Wnt/β-catenin signalling pathway activation and malignancy in colon cancer cells. J Pathol. 2013;229:379–389. doi: 10.1002/path.4097. [DOI] [PubMed] [Google Scholar]
- 128.Nagel R, le Sage C, Diosdado B, van der Waal M, Oude Vrielink JA, Bolijn A, Meijer GA, Agami R. Regulation of the adenomatous polyposis coli gene by the miR-135 family in colorectal cancer. Cancer Res. 2008;68:5795–5802. doi: 10.1158/0008-5472.CAN-08-0951. [DOI] [PubMed] [Google Scholar]
- 129.Valeri N, Braconi C, Gasparini P, Murgia C, Lampis A, Paulus-Hock V, Hart JR, Ueno L, Grivennikov SI, Lovat F, et al. MicroRNA-135b promotes cancer progression by acting as a downstream effector of oncogenic pathways in colon cancer. Cancer Cell. 2014;25:469–483. doi: 10.1016/j.ccr.2014.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Drebber U, Lay M, Wedemeyer I, Vallböhmer D, Bollschweiler E, Brabender J, Mönig SP, Hölscher AH, Dienes HP, Odenthal M. Altered levels of the onco-microRNA 21 and the tumor-supressor microRNAs 143 and 145 in advanced rectal cancer indicate successful neoadjuvant chemoradiotherapy. Int J Oncol. 2011;39:409–415. doi: 10.3892/ijo.2011.1036. [DOI] [PubMed] [Google Scholar]
- 131.Schetter AJ, Okayama H, Harris CC. The role of microRNAs in colorectal cancer. Cancer J. 2012;18:244–252. doi: 10.1097/PPO.0b013e318258b78f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Akao Y, Nakagawa Y, Hirata I, Iio A, Itoh T, Kojima K, Nakashima R, Kitade Y, Naoe T. Role of anti-oncomirs miR-143 and -145 in human colorectal tumors. Cancer Gene Ther. 2010;17:398–408. doi: 10.1038/cgt.2009.88. [DOI] [PubMed] [Google Scholar]
- 133.Huang L, Fu L. Mechanisms of resistance to EGFR tyrosine kinase inhibitors. Acta Pharm Sin B. 2015;5:390–401. doi: 10.1016/j.apsb.2015.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, Labourier E, Reinert KL, Brown D, Slack FJ. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 135.Hiraki M, Nishimura J, Takahashi H, Wu X, Takahashi Y, Miyo M, Nishida N, Uemura M, Hata T, Takemasa I, et al. Concurrent Targeting of KRAS and AKT by MiR-4689 Is a Novel Treatment Against Mutant KRAS Colorectal Cancer. Mol Ther Nucleic Acids. 2015;4:e231. doi: 10.1038/mtna.2015.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Fang L, Li H, Wang L, Hu J, Jin T, Wang J, Yang BB. MicroRNA-17-5p promotes chemotherapeutic drug resistance and tumour metastasis of colorectal cancer by repressing PTEN expression. Oncotarget. 2014;5:2974–2987. doi: 10.18632/oncotarget.1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Wu W, Yang J, Feng X, Wang H, Ye S, Yang P, Tan W, Wei G, Zhou Y. MicroRNA-32 (miR-32) regulates phosphatase and tensin homologue (PTEN) expression and promotes growth, migration, and invasion in colorectal carcinoma cells. Mol Cancer. 2013;12:30. doi: 10.1186/1476-4598-12-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Chen ML, Liang LS, Wang XK. miR-200c inhibits invasion and migration in human colon cancer cells SW480/620 by targeting ZEB1. Clin Exp Metastasis. 2012;29:457–469. doi: 10.1007/s10585-012-9463-7. [DOI] [PubMed] [Google Scholar]
- 139.Zaravinos A. The Regulatory Role of MicroRNAs in EMT and Cancer. J Oncol. 2015;2015:865816. doi: 10.1155/2015/865816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Wang J, Du Y, Liu X, Cho WC, Yang Y. MicroRNAs as Regulator of Signaling Networks in Metastatic Colon Cancer. Biomed Res Int. 2015;2015:823620. doi: 10.1155/2015/823620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Bu P, Wang L, Chen KY, Rakhilin N, Sun J, Closa A, Tung KL, King S, Kristine Varanko A, Xu Y, et al. miR-1269 promotes metastasis and forms a positive feedback loop with TGF-β. Nat Commun. 2015;6:6879. doi: 10.1038/ncomms7879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Loupakis F, Cremolini C, Masi G, Lonardi S, Zagonel V, Salvatore L, Cortesi E, Tomasello G, Ronzoni M, Spadi R, et al. Initial therapy with FOLFOXIRI and bevacizumab for metastatic colorectal cancer. N Engl J Med. 2014;371:1609–1618. doi: 10.1056/NEJMoa1403108. [DOI] [PubMed] [Google Scholar]
- 143.Hu T, Li Z, Gao CY, Cho CH. Mechanisms of drug resistance in colon cancer and its therapeutic strategies. World J Gastroenterol. 2016;22:6876–6889. doi: 10.3748/wjg.v22.i30.6876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Amirkhah R, Farazmand A, Irfan-Maqsood M, Wolkenhauer O, Schmitz U. The role of microRNAs in the resistance to colorectal cancer treatments. Cell Mol Biol (Noisy-le-grand) 2015;61:17–23. [PubMed] [Google Scholar]
- 145.Xie T, Huang M, Wang Y, Wang L, Chen C, Chu X. MicroRNAs as Regulators, Biomarkers and Therapeutic Targets in the Drug Resistance of Colorectal Cancer. Cell Physiol Biochem. 2016;40:62–76. doi: 10.1159/000452525. [DOI] [PubMed] [Google Scholar]
- 146.Wu QB, Sheng X, Zhang N, Yang MW, Wang F. Role of microRNAs in the resistance of colorectal cancer to chemoradiotherapy. Mol Clin Oncol. 2018;8:528–532. doi: 10.3892/mco.2018.1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Nijhuis A, Thompson H, Adam J, Parker A, Gammon L, Lewis A, Bundy JG, Soga T, Jalaly A, Propper D, et al. Remodelling of microRNAs in colorectal cancer by hypoxia alters metabolism profiles and 5-fluorouracil resistance. Hum Mol Genet. 2017;26:1552–1564. doi: 10.1093/hmg/ddx059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Akao Y, Noguchi S, Iio A, Kojima K, Takagi T, Naoe T. Dysregulation of microRNA-34a expression causes drug-resistance to 5-FU in human colon cancer DLD-1 cells. Cancer Lett. 2011;300:197–204. doi: 10.1016/j.canlet.2010.10.006. [DOI] [PubMed] [Google Scholar]
- 149.Shang J, Yang F, Wang Y, Wang Y, Xue G, Mei Q, Wang F, Sun S. MicroRNA-23a antisense enhances 5-fluorouracil chemosensitivity through APAF-1/caspase-9 apoptotic pathway in colorectal cancer cells. J Cell Biochem. 2014;115:772–784. doi: 10.1002/jcb.24721. [DOI] [PubMed] [Google Scholar]
- 150.Chai J, Dong W, Xie C, Wang L, Han DL, Wang S, Guo HL, Zhang ZL. MicroRNA-494 sensitizes colon cancer cells to fluorouracil through regulation of DPYD. IUBMB Life. 2015;67:191–201. doi: 10.1002/iub.1361. [DOI] [PubMed] [Google Scholar]
- 151.Zhang Y, Talmon G, Wang J. MicroRNA-587 antagonizes 5-FU-induced apoptosis and confers drug resistance by regulating PPP2R1B expression in colorectal cancer. Cell Death Dis. 2015;6:e1845. doi: 10.1038/cddis.2015.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.To KK, Robey RW, Knutsen T, Zhan Z, Ried T, Bates SE. Escape from hsa-miR-519c enables drug-resistant cells to maintain high expression of ABCG2. Mol Cancer Ther. 2009;8:2959–2968. doi: 10.1158/1535-7163.MCT-09-0292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.To KK, Leung WW, Ng SS. Exploiting a novel miR-519c-HuR-ABCG2 regulatory pathway to overcome chemoresistance in colorectal cancer. Exp Cell Res. 2015;338:222–231. doi: 10.1016/j.yexcr.2015.09.011. [DOI] [PubMed] [Google Scholar]
- 154.Zhou Y, Wan G, Spizzo R, Ivan C, Mathur R, Hu X, Ye X, Lu J, Fan F, Xia L, et al. miR-203 induces oxaliplatin resistance in colorectal cancer cells by negatively regulating ATM kinase. Mol Oncol. 2014;8:83–92. doi: 10.1016/j.molonc.2013.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Xu K, Liang X, Cui D, Wu Y, Shi W, Liu J. miR-1915 inhibits Bcl-2 to modulate multidrug resistance by increasing drug-sensitivity in human colorectal carcinoma cells. Mol Carcinog. 2013;52:70–78. doi: 10.1002/mc.21832. [DOI] [PubMed] [Google Scholar]
- 156.Bitarte N, Bandres E, Boni V, Zarate R, Rodriguez J, Gonzalez-Huarriz M, Lopez I, Javier Sola J, Alonso MM, Fortes P, et al. MicroRNA-451 is involved in the self-renewal, tumorigenicity, and chemoresistance of colorectal cancer stem cells. Stem Cells. 2011;29:1661–1671. doi: 10.1002/stem.741. [DOI] [PubMed] [Google Scholar]
- 157.Chen Z, Gao S, Wang D, Song D, Feng Y. Colorectal cancer cells are resistant to anti-EGFR monoclonal antibody through adapted autophagy. Am J Transl Res. 2016;8:1190–1196. [PMC free article] [PubMed] [Google Scholar]
- 158.Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov. 2017;16:203–222. doi: 10.1038/nrd.2016.246. [DOI] [PubMed] [Google Scholar]
- 159.Christopher AF, Kaur RP, Kaur G, Kaur A, Gupta V, Bansal P. MicroRNA therapeutics: Discovering novel targets and developing specific therapy. Perspect Clin Res. 2016;7:68–74. doi: 10.4103/2229-3485.179431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.van Rooij E, Kauppinen S. Development of microRNA therapeutics is coming of age. EMBO Mol Med. 2014;6:851–864. doi: 10.15252/emmm.201100899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Thorsen SB, Obad S, Jensen NF, Stenvang J, Kauppinen S. The therapeutic potential of microRNAs in cancer. Cancer J. 2012;18:275–284. doi: 10.1097/PPO.0b013e318258b5d6. [DOI] [PubMed] [Google Scholar]
- 162.Bader AG. miR-34 - a microRNA replacement therapy is headed to the clinic. Front Genet. 2012;3:120. doi: 10.3389/fgene.2012.00120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Ibrahim AF, Weirauch U, Thomas M, Grünweller A, Hartmann RK, Aigner A. MicroRNA replacement therapy for miR-145 and miR-33a is efficacious in a model of colon carcinoma. Cancer Res. 2011;71:5214–5224. doi: 10.1158/0008-5472.CAN-10-4645. [DOI] [PubMed] [Google Scholar]
- 164.Liang G, Zhu Y, Jing A, Wang J, Hu F, Feng W, Xiao Z, Chen B. Cationic microRNA-delivering nanocarriers for efficient treatment of colon carcinoma in xenograft model. Gene Ther. 2016;23:829–838. doi: 10.1038/gt.2016.60. [DOI] [PubMed] [Google Scholar]
- 165.Adams BD, Parsons C, Slack FJ. The tumor-suppressive and potential therapeutic functions of miR-34a in epithelial carcinomas. Expert Opin Ther Targets. 2016;20:737–753. doi: 10.1517/14728222.2016.1114102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Beg MS, Brenner AJ, Sachdev J, Borad M, Kang YK, Stoudemire J, Smith S, Bader AG, Kim S, Hong DS. Phase I study of MRX34, a liposomal miR-34a mimic, administered twice weekly in patients with advanced solid tumors. Invest New Drugs. 2017;35:180–188. doi: 10.1007/s10637-016-0407-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Garzon R, Marcucci G, Croce CM. Targeting microRNAs in cancer: rationale, strategies and challenges. Nat Rev Drug Discov. 2010;9:775–789. doi: 10.1038/nrd3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Wu W, Wang Z, Yang P, Yang J, Liang J, Chen Y, Wang H, Wei G, Ye S, Zhou Y. MicroRNA-135b regulates metastasis suppressor 1 expression and promotes migration and invasion in colorectal cancer. Mol Cell Biochem. 2014;388:249–259. doi: 10.1007/s11010-013-1916-z. [DOI] [PubMed] [Google Scholar]
- 169.Huang Z, Huang S, Wang Q, Liang L, Ni S, Wang L, Sheng W, He X, Du X. MicroRNA-95 promotes cell proliferation and targets sorting Nexin 1 in human colorectal carcinoma. Cancer Res. 2011;71:2582–2589. doi: 10.1158/0008-5472.CAN-10-3032. [DOI] [PubMed] [Google Scholar]
- 170.Chai H, Liu M, Tian R, Li X, Tang H. miR-20a targets BNIP2 and contributes chemotherapeutic resistance in colorectal adenocarcinoma SW480 and SW620 cell lines. Acta Biochim Biophys Sin (Shanghai) 2011;43:217–225. doi: 10.1093/abbs/gmq125. [DOI] [PubMed] [Google Scholar]
- 171.Tsang WP, Ng EK, Ng SS, Jin H, Yu J, Sung JJ, Kwok TT. Oncofetal H19-derived miR-675 regulates tumor suppressor RB in human colorectal cancer. Carcinogenesis. 2010;31:350–358. doi: 10.1093/carcin/bgp181. [DOI] [PubMed] [Google Scholar]
- 172.Hagedorn PH, Persson R, Funder ED, Albæk N, Diemer SL, Hansen DJ, Møller MR, Papargyri N, Christiansen H, Hansen BR, et al. Locked nucleic acid: modality, diversity, and drug discovery. Drug Discov Today. 2018;23:101–114. doi: 10.1016/j.drudis.2017.09.018. [DOI] [PubMed] [Google Scholar]
- 173.Vester B, Wengel J. LNA (locked nucleic acid): high-affinity targeting of complementary RNA and DNA. Biochemistry. 2004;43:13233–13241. doi: 10.1021/bi0485732. [DOI] [PubMed] [Google Scholar]
- 174.You Y, Moreira BG, Behlke MA, Owczarzy R. Design of LNA probes that improve mismatch discrimination. Nucleic Acids Res. 2006;34:e60. doi: 10.1093/nar/gkl175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Nedaeinia R, Sharifi M, Avan A, Kazemi M, Rafiee L, Ghayour-Mobarhan M, Salehi R. Locked nucleic acid anti-miR-21 inhibits cell growth and invasive behaviors of a colorectal adenocarcinoma cell line: LNA-anti-miR as a novel approach. Cancer Gene Ther. 2016;23:246–253. doi: 10.1038/cgt.2016.25. [DOI] [PubMed] [Google Scholar]
- 176.Choi WY, Giraldez AJ, Schier AF. Target protectors reveal dampening and balancing of Nodal agonist and antagonist by miR-430. Science. 2007;318:271–274. doi: 10.1126/science.1147535. [DOI] [PubMed] [Google Scholar]
- 177.Bu P, Chen KY, Chen JH, Wang L, Walters J, Shin YJ, Goerger JP, Sun J, Witherspoon M, Rakhilin N, et al. A microRNA miR-34a-regulated bimodal switch targets Notch in colon cancer stem cells. Cell Stem Cell. 2013;12:602–615. doi: 10.1016/j.stem.2013.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Avitabile C, Fabbri E, Bianchi N, Gambari R, Romanelli A. Inhibition of miRNA maturation by peptide nucleic acids. Methods Mol Biol. 2014;1095:157–164. doi: 10.1007/978-1-62703-703-7_13. [DOI] [PubMed] [Google Scholar]
- 179.Gumireddy K, Young DD, Xiong X, Hogenesch JB, Huang Q, Deiters A. Small-molecule inhibitors of microrna miR-21 function. Angew Chem Int Ed Engl. 2008;47:7482–7484. doi: 10.1002/anie.200801555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Cruz-Gil S, Sanchez-Martinez R, Gomez de Cedron M, Martin-Hernandez R, Vargas T, Molina S, Herranz J, Davalos A, Reglero G, Ramirez de Molina A. Targeting the lipid metabolic axis <i>ACSL/SCD</i> in colorectal cancer progression by therapeutic miRNAs: miR-19b-1 role. J Lipid Res. 2018;59:14–24. doi: 10.1194/jlr.M076752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Sun RP, Xi QY, Sun JJ, Cheng X, Zhu YL, Ye DZ, Chen T, Wei LM, Ye RS, Jiang QY, et al. In low protein diets, microRNA-19b regulates urea synthesis by targeting SIRT5. Sci Rep. 2016;6:33291. doi: 10.1038/srep33291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Jarry J, Schadendorf D, Greenwood C, Spatz A, van Kempen LC. The validity of circulating microRNAs in oncology: five years of challenges and contradictions. Mol Oncol. 2014;8:819–829. doi: 10.1016/j.molonc.2014.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Xiao G, Tang H, Wei W, Li J, Ji L, Ge J. Aberrant Expression of MicroRNA-15a and MicroRNA-16 Synergistically Associates with Tumor Progression and Prognosis in Patients with Colorectal Cancer. Gastroenterol Res Pract. 2014;2014:364549. doi: 10.1155/2014/364549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Díaz R, Silva J, García JM, Lorenzo Y, García V, Peña C, Rodríguez R, Muñoz C, García F, Bonilla F, et al. Deregulated expression of miR-106a predicts survival in human colon cancer patients. Genes Chromosomes Cancer. 2008;47:794–802. doi: 10.1002/gcc.20580. [DOI] [PubMed] [Google Scholar]
- 185.Gao J, Li N, Dong Y, Li S, Xu L, Li X, Li Y, Li Z, Ng SS, Sung JJ, et al. miR-34a-5p suppresses colorectal cancer metastasis and predicts recurrence in patients with stage II/III colorectal cancer. Oncogene. 2015;34:4142–4152. doi: 10.1038/onc.2014.348. [DOI] [PubMed] [Google Scholar]
- 186.Mokutani Y, Uemura M, Munakata K, Okuzaki D, Haraguchi N, Takahashi H, Nishimura J, Hata T, Murata K, Takemasa I, et al. Down-Regulation of microRNA-132 is Associated with Poor Prognosis of Colorectal Cancer. Ann Surg Oncol. 2016;23:599–608. doi: 10.1245/s10434-016-5133-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Nishimura J, Handa R, Yamamoto H, Tanaka F, Shibata K, Mimori K, Takemasa I, Mizushima T, Ikeda M, Sekimoto M, et al. microRNA-181a is associated with poor prognosis of colorectal cancer. Oncol Rep. 2012;28:2221–2226. doi: 10.3892/or.2012.2059. [DOI] [PubMed] [Google Scholar]
- 188.Bovell LC, Shanmugam C, Putcha BD, Katkoori VR, Zhang B, Bae S, Singh KP, Grizzle WE, Manne U. The prognostic value of microRNAs varies with patient race/ethnicity and stage of colorectal cancer. Clin Cancer Res. 2013;19:3955–3965. doi: 10.1158/1078-0432.CCR-12-3302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Wang X, Wang J, Ma H, Zhang J, Zhou X. Downregulation of miR-195 correlates with lymph node metastasis and poor prognosis in colorectal cancer. Med Oncol. 2012;29:919–927. doi: 10.1007/s12032-011-9880-5. [DOI] [PubMed] [Google Scholar]
- 190.Shen ZL, Wang B, Jiang KW, Ye CX, Cheng C, Yan YC, Zhang JZ, Yang Y, Gao ZD, Ye YJ, et al. Downregulation of miR-199b is associated with distant metastasis in colorectal cancer via activation of SIRT1 and inhibition of CREB/KISS1 signaling. Oncotarget. 2016;7:35092–35105. doi: 10.18632/oncotarget.9042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Karaayvaz M, Pal T, Song B, Zhang C, Georgakopoulos P, Mehmood S, Burke S, Shroyer K, Ju J. Prognostic significance of miR-215 in colon cancer. Clin Colorectal Cancer. 2011;10:340–347. doi: 10.1016/j.clcc.2011.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Li PL, Zhang X, Wang LL, Du LT, Yang YM, Li J, Wang CX. MicroRNA-218 is a prognostic indicator in colorectal cancer and enhances 5-fluorouracil-induced apoptosis by targeting BIRC5. Carcinogenesis. 2015;36:1484–1493. doi: 10.1093/carcin/bgv145. [DOI] [PubMed] [Google Scholar]
- 193.Dong SJ, Cai XJ, Li SJ. The Clinical Significance of MiR-429 as a Predictive Biomarker in Colorectal Cancer Patients Receiving 5-Fluorouracil Treatment. Med Sci Monit. 2016;22:3352–3361. doi: 10.12659/MSM.900674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Yang IP, Tsai HL, Miao ZF, Huang CW, Kuo CH, Wu JY, Wang WM, Juo SH, Wang JY. Development of a deregulating microRNA panel for the detection of early relapse in postoperative colorectal cancer patients. J Transl Med. 2016;14:108. doi: 10.1186/s12967-016-0856-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Coebergh van den Braak RRJ, Sieuwerts AM, Lalmahomed ZS, Smid M, Wilting SM, Bril SI, Xiang S, van der Vlugt-Daane M, de Weerd V, van Galen A, et al. Confirmation of a metastasis-specific microRNA signature in primary colon cancer. Sci Rep. 2018;8:5242. doi: 10.1038/s41598-018-22532-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Xiong Y, Wang R, Peng L, You W, Wei J, Zhang S, Wu X, Guo J, Xu J, Lv Z, et al. An integrated lncRNA, microRNA and mRNA signature to improve prognosis prediction of colorectal cancer. Oncotarget. 2017;8:85463–85478. doi: 10.18632/oncotarget.20013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Menéndez P, Padilla D, Villarejo P, Palomino T, Nieto P, Menéndez JM, Rodríguez-Montes JA. Prognostic implications of serum microRNA-21 in colorectal cancer. J Surg Oncol. 2013;108:369–373. doi: 10.1002/jso.23415. [DOI] [PubMed] [Google Scholar]
- 198.Kou CH, Zhou T, Han XL, Zhuang HJ, Qian HX. Downregulation of mir-23b in plasma is associated with poor prognosis in patients with colorectal cancer. Oncol Lett. 2016;12:4838–4844. doi: 10.3892/ol.2016.5265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Sun Y, Liu Y, Cogdell D, Calin GA, Sun B, Kopetz S, Hamilton SR, Zhang W. Examining plasma microRNA markers for colorectal cancer at different stages. Oncotarget. 2016;7:11434–11449. doi: 10.18632/oncotarget.7196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Jinushi T, Shibayama Y, Kinoshita I, Oizumi S, Jinushi M, Aota T, Takahashi T, Horita S, Dosaka-Akita H, Iseki K. Low expression levels of microRNA-124-5p correlated with poor prognosis in colorectal cancer via targeting of SMC4. Cancer Med. 2014;3:1544–1552. doi: 10.1002/cam4.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Tsai HL, Yang IP, Huang CW, Ma CJ, Kuo CH, Lu CY, Juo SH, Wang JY. Clinical significance of microRNA-148a in patients with early relapse of stage II stage and III colorectal cancer after curative resection. Transl Res. 2013;162:258–268. doi: 10.1016/j.trsl.2013.07.009. [DOI] [PubMed] [Google Scholar]
- 202.Lv ZC, Fan YS, Chen HB, Zhao DW. Investigation of microRNA-155 as a serum diagnostic and prognostic biomarker for colorectal cancer. Tumour Biol. 2015;36:1619–1625. doi: 10.1007/s13277-014-2760-9. [DOI] [PubMed] [Google Scholar]
- 203.Yuan D, Li K, Zhu K, Yan R, Dang C. Plasma miR-183 predicts recurrence and prognosis in patients with colorectal cancer. Cancer Biol Ther. 2015;16:268–275. doi: 10.1080/15384047.2014.1002327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Toiyama Y, Hur K, Tanaka K, Inoue Y, Kusunoki M, Boland CR, Goel A. Serum miR-200c is a novel prognostic and metastasis-predictive biomarker in patients with colorectal cancer. Ann Surg. 2014;259:735–743. doi: 10.1097/SLA.0b013e3182a6909d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Hur K, Toiyama Y, Okugawa Y, Ide S, Imaoka H, Boland CR, Goel A. Circulating microRNA-203 predicts prognosis and metastasis in human colorectal cancer. Gut. 2017;66:654–665. doi: 10.1136/gutjnl-2014-308737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Yu H, Gao G, Jiang L, Guo L, Lin M, Jiao X, Jia W, Huang J. Decreased expression of miR-218 is associated with poor prognosis in patients with colorectal cancer. Int J Clin Exp Pathol. 2013;6:2904–2911. [PMC free article] [PubMed] [Google Scholar]
- 207.Pu XX, Huang GL, Guo HQ, Guo CC, Li H, Ye S, Ling S, Jiang L, Tian Y, Lin TY. Circulating miR-221 directly amplified from plasma is a potential diagnostic and prognostic marker of colorectal cancer and is correlated with p53 expression. J Gastroenterol Hepatol. 2010;25:1674–1680. doi: 10.1111/j.1440-1746.2010.06417.x. [DOI] [PubMed] [Google Scholar]
- 208.Hur K, Toiyama Y, Schetter AJ, Okugawa Y, Harris CC, Boland CR, Goel A. Identification of a metastasis-specific MicroRNA signature in human colorectal cancer. J Natl Cancer Inst. 2015;107:pii: dju492. doi: 10.1093/jnci/dju492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Imaoka H, Toiyama Y, Fujikawa H, Hiro J, Saigusa S, Tanaka K, Inoue Y, Mohri Y, Mori T, Kato T, et al. Circulating microRNA-1290 as a novel diagnostic and prognostic biomarker in human colorectal cancer. Ann Oncol. 2016;27:1879–1886. doi: 10.1093/annonc/mdw279. [DOI] [PubMed] [Google Scholar]
- 210.Vychytilova-Faltejskova P, Radova L, Sachlova M, Kosarova Z, Slaba K, Fabian P, Grolich T, Prochazka V, Kala Z, Svoboda M, et al. Serum-based microRNA signatures in early diagnosis and prognosis prediction of colon cancer. Carcinogenesis. 2016;37:941–950. doi: 10.1093/carcin/bgw078. [DOI] [PubMed] [Google Scholar]
- 211.Ding M, Zhang T, Li S, Zhang Y, Qiu Y, Zhang B. Correlation analysis between liver metastasis and serum levels of miR200 and miR141 in patients with colorectal cancer. Mol Med Rep. 2017;16:7791–7795. doi: 10.3892/mmr.2017.7538. [DOI] [PubMed] [Google Scholar]
- 212.Maierthaler M, Benner A, Hoffmeister M, Surowy H, Jansen L, Knebel P, Chang-Claude J, Brenner H, Burwinkel B. Plasma miR-122 and miR-200 family are prognostic markers in colorectal cancer. Int J Cancer. 2017;140:176–187. doi: 10.1002/ijc.30433. [DOI] [PubMed] [Google Scholar]
- 213.Rotelli MT, Di Lena M, Cavallini A, Lippolis C, Bonfrate L, Chetta N, Portincasa P, Altomare DF. Fecal microRNA profile in patients with colorectal carcinoma before and after curative surgery. Int J Colorectal Dis. 2015;30:891–898. doi: 10.1007/s00384-015-2248-0. [DOI] [PubMed] [Google Scholar]
- 214.Ogata-Kawata H, Izumiya M, Kurioka D, Honma Y, Yamada Y, Furuta K, Gunji T, Ohta H, Okamoto H, Sonoda H, et al. Circulating exosomal microRNAs as biomarkers of colon cancer. PLoS One. 2014;9:e92921. doi: 10.1371/journal.pone.0092921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Takano Y, Masuda T, Iinuma H, Yamaguchi R, Sato K, Tobo T, Hirata H, Kuroda Y, Nambara S, Hayashi N, et al. Circulating exosomal microRNA-203 is associated with metastasis possibly via inducing tumor-associated macrophages in colorectal cancer. Oncotarget. 2017;8:78598–78613. doi: 10.18632/oncotarget.20009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Yan S, Han B, Gao S, Wang X, Wang Z, Wang F, Zhang J, Xu D, Sun B. Exosome-encapsulated microRNAs as circulating biomarkers for colorectal cancer. Oncotarget. 2017;8:60149–60158. doi: 10.18632/oncotarget.18557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Li J, Li B, Ren C, Chen Y, Guo X, Zhou L, Peng Z, Tang Y, Chen Y, Liu W, et al. The clinical significance of circulating GPC1 positive exosomes and its regulative miRNAs in colon cancer patients. Oncotarget. 2017;8:101189–101202. doi: 10.18632/oncotarget.20516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Cha DJ, Franklin JL, Dou Y, Liu Q, Higginbotham JN, Demory Beckler M, Weaver AM, Vickers K, Prasad N, Levy S, et al. KRAS-dependent sorting of miRNA to exosomes. Elife. 2015;4:e07197. doi: 10.7554/eLife.07197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.219 Chiba M, Kimura M, Asari S. Exosomes secreted from human colorectal cancer cell lines contain mRNAs, microRNAs and natural antisense RNAs, that can transfer into the human hepatoma HepG2 and lung cancer A549 cell lines. Oncol Rep. 2012;28:1551–1558. doi: 10.3892/or.2012.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Bigagli E, Luceri C, Guasti D, Cinci L. Exosomes secreted from human colon cancer cells influence the adhesion of neighboring metastatic cells: Role of microRNA-210. Cancer Biol Ther. 2016:1–8. doi: 10.1080/15384047.2016.1219815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Clancy C, Khan S, Glynn CL, Holian E, Dockery P, Lalor P, Brown JA, Joyce MR, Kerin MJ, Dwyer RM. Screening of exosomal microRNAs from colorectal cancer cells. Cancer Biomark. 2016;17:427–435. doi: 10.3233/CBM-160659. [DOI] [PubMed] [Google Scholar]
- 222.Lopes-Ramos CM, Habr-Gama A, Quevedo Bde S, Felício NM, Bettoni F, Koyama FC, Asprino PF, Galante PA, Gama-Rodrigues J, Camargo AA, et al. Overexpression of miR-21-5p as a predictive marker for complete tumor regression to neoadjuvant chemoradiotherapy in rectal cancer patients. BMC Med Genomics. 2014;7:68. doi: 10.1186/s12920-014-0068-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Ragusa M, Majorana A, Statello L, Maugeri M, Salito L, Barbagallo D, Guglielmino MR, Duro LR, Angelica R, Caltabiano R, et al. Specific alterations of microRNA transcriptome and global network structure in colorectal carcinoma after cetuximab treatment. Mol Cancer Ther. 2010;9:3396–3409. doi: 10.1158/1535-7163.MCT-10-0137. [DOI] [PubMed] [Google Scholar]
- 224.Boisen MK, Dehlendorff C, Linnemann D, Nielsen BS, Larsen JS, Osterlind K, Nielsen SE, Tarpgaard LS, Qvortrup C, Pfeiffer P, et al. Tissue microRNAs as predictors of outcome in patients with metastatic colorectal cancer treated with first line Capecitabine and Oxaliplatin with or without Bevacizumab. PLoS One. 2014;9:e109430. doi: 10.1371/journal.pone.0109430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Molina-Pinelo S, Carnero A, Rivera F, Estevez-Garcia P, Bozada JM, Limon ML, Benavent M, Gomez J, Pastor MD, Chaves M, et al. MiR-107 and miR-99a-3p predict chemotherapy response in patients with advanced colorectal cancer. BMC Cancer. 2014;14:656. doi: 10.1186/1471-2407-14-656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Chen Q, Xia HW, Ge XJ, Zhang YC, Tang QL, Bi F. Serum miR-19a predicts resistance to FOLFOX chemotherapy in advanced colorectal cancer cases. Asian Pac J Cancer Prev. 2013;14:7421–7426. doi: 10.7314/apjcp.2013.14.12.7421. [DOI] [PubMed] [Google Scholar]
- 227.Qian X, Yu J, Yin Y, He J, Wang L, Li Q, Zhang LQ, Li CY, Shi ZM, Xu Q, et al. MicroRNA-143 inhibits tumor growth and angiogenesis and sensitizes chemosensitivity to oxaliplatin in colorectal cancers. Cell Cycle. 2013;12:1385–1394. doi: 10.4161/cc.24477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Hu J, Cai G, Xu Y, Cai S. The Plasma microRNA miR-1914* and -1915 Suppresses Chemoresistant in Colorectal Cancer Patients by Down-regulating NFIX. Curr Mol Med. 2016;16:70–82. doi: 10.2174/1566524016666151222144656. [DOI] [PubMed] [Google Scholar]
- 229.Nakajima G, Hayashi K, Xi Y, Kudo K, Uchida K, Takasaki K, Yamamoto M, Ju J. Non-coding MicroRNAs hsa-let-7g and hsa-miR-181b are Associated with Chemoresponse to S-1 in Colon Cancer. Cancer Genomics Proteomics. 2006;3:317–324. [PMC free article] [PubMed] [Google Scholar]
- 230.Nishida N, Yamashita S, Mimori K, Sudo T, Tanaka F, Shibata K, Yamamoto H, Ishii H, Doki Y, Mori M. MicroRNA-10b is a prognostic indicator in colorectal cancer and confers resistance to the chemotherapeutic agent 5-fluorouracil in colorectal cancer cells. Ann Surg Oncol. 2012;19:3065–3071. doi: 10.1245/s10434-012-2246-1. [DOI] [PubMed] [Google Scholar]
- 231.Kurokawa K, Tanahashi T, Iima T, Yamamoto Y, Akaike Y, Nishida K, Masuda K, Kuwano Y, Murakami Y, Fukushima M, et al. Role of miR-19b and its target mRNAs in 5-fluorouracil resistance in colon cancer cells. J Gastroenterol. 2012;47:883–895. doi: 10.1007/s00535-012-0547-6. [DOI] [PubMed] [Google Scholar]
- 232.Wang CJ, Stratmann J, Zhou ZG, Sun XF. Suppression of microRNA-31 increases sensitivity to 5-FU at an early stage, and affects cell migration and invasion in HCT-116 colon cancer cells. BMC Cancer. 2010;10:616. doi: 10.1186/1471-2407-10-616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Song B, Wang Y, Xi Y, Kudo K, Bruheim S, Botchkina GI, Gavin E, Wan Y, Formentini A, Kornmann M, et al. Mechanism of chemoresistance mediated by miR-140 in human osteosarcoma and colon cancer cells. Oncogene. 2009;28:4065–4074. doi: 10.1038/onc.2009.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Song B, Wang Y, Titmus MA, Botchkina G, Formentini A, Kornmann M, Ju J. Molecular mechanism of chemoresistance by miR-215 in osteosarcoma and colon cancer cells. Mol Cancer. 2010;9:96. doi: 10.1186/1476-4598-9-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Boni V, Bitarte N, Cristobal I, Zarate R, Rodriguez J, Maiello E, Garcia-Foncillas J, Bandres E. miR-192/miR-215 influence 5-fluorouracil resistance through cell cycle-mediated mechanisms complementary to its post-transcriptional thymidilate synthase regulation. Mol Cancer Ther. 2010;9:2265–2275. doi: 10.1158/1535-7163.MCT-10-0061. [DOI] [PubMed] [Google Scholar]
- 236.Kim C, Hong Y, Lee H, Kang H, Lee EK. MicroRNA-195 desensitizes HCT116 human colon cancer cells to 5-fluorouracil. Cancer Lett. 2018;412:264–271. doi: 10.1016/j.canlet.2017.10.022. [DOI] [PubMed] [Google Scholar]
- 237.Amankwatia EB, Chakravarty P, Carey FA, Weidlich S, Steele RJ, Munro AJ, Wolf CR, Smith G. MicroRNA-224 is associated with colorectal cancer progression and response to 5-fluorouracil-based chemotherapy by KRAS-dependent and -independent mechanisms. Br J Cancer. 2015;112:1480–1490. doi: 10.1038/bjc.2015.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Zhang Y, Geng L, Talmon G, Wang J. MicroRNA-520g confers drug resistance by regulating p21 expression in colorectal cancer. J Biol Chem. 2015;290:6215–6225. doi: 10.1074/jbc.M114.620252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Li X, Zhao H, Zhou X, Song L. Inhibition of lactate dehydrogenase A by microRNA-34a resensitizes colon cancer cells to 5-fluorouracil. Mol Med Rep. 2015;11:577–582. doi: 10.3892/mmr.2014.2726. [DOI] [PubMed] [Google Scholar]
- 240.Akao Y, Khoo F, Kumazaki M, Shinohara H, Miki K, Yamada N. Extracellular disposal of tumor-suppressor miRs-145 and -34a via microvesicles and 5-FU resistance of human colon cancer cells. Int J Mol Sci. 2014;15:1392–1401. doi: 10.3390/ijms15011392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Sun C, Wang FJ, Zhang HG, Xu XZ, Jia RC, Yao L, Qiao PF. miR-34a mediates oxaliplatin resistance of colorectal cancer cells by inhibiting macroautophagy via transforming growth factor-β/Smad4 pathway. World J Gastroenterol. 2017;23:1816–1827. doi: 10.3748/wjg.v23.i10.1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Liu H, Yin Y, Hu Y, Feng Y, Bian Z, Yao S, Li M, You Q, Huang Z. miR-139-5p sensitizes colorectal cancer cells to 5-fluorouracil by targeting NOTCH-1. Pathol Res Pract. 2016;212:643–649. doi: 10.1016/j.prp.2016.04.011. [DOI] [PubMed] [Google Scholar]
- 243.Zhang L, Pickard K, Jenei V, Bullock MD, Bruce A, Mitter R, Kelly G, Paraskeva C, Strefford J, Primrose J, et al. miR-153 supports colorectal cancer progression via pleiotropic effects that enhance invasion and chemotherapeutic resistance. Cancer Res. 2013;73:6435–6447. doi: 10.1158/0008-5472.CAN-12-3308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Chang HY, Ye SP, Pan SL, Kuo TT, Liu BC, Chen YL, Huang TC. Overexpression of miR-194 Reverses HMGA2-driven Signatures in Colorectal Cancer. Theranostics. 2017;7:3889–3900. doi: 10.7150/thno.20041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Senfter D, Holzner S, Kalipciyan M, Staribacher A, Walzl A, Huttary N, Krieger S, Brenner S, Jäger W, Krupitza G, et al. Loss of miR-200 family in 5-fluorouracil resistant colon cancer drives lymphendothelial invasiveness in vitro. Hum Mol Genet. 2015;24:3689–3698. doi: 10.1093/hmg/ddv113. [DOI] [PubMed] [Google Scholar]
- 246.Lv Z, Wei J, You W, Wang R, Shang J, Xiong Y, Yang H, Yang X, Fu Z. Disruption of the c-Myc/miR-200b-3p/PRDX2 regulatory loop enhances tumor metastasis and chemotherapeutic resistance in colorectal cancer. J Transl Med. 2017;15:257. doi: 10.1186/s12967-017-1357-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Li T, Gao F, Zhang XP. miR-203 enhances chemosensitivity to 5-fluorouracil by targeting thymidylate synthase in colorectal cancer. Oncol Rep. 2015;33:607–614. doi: 10.3892/or.2014.3646. [DOI] [PubMed] [Google Scholar]
- 248.Li P, Zhang X, Wang H, Wang L, Liu T, Du L, Yang Y, Wang C. MALAT1 Is Associated with Poor Response to Oxaliplatin-Based Chemotherapy in Colorectal Cancer Patients and Promotes Chemoresistance through EZH2. Mol Cancer Ther. 2017;16:739–751. doi: 10.1158/1535-7163.MCT-16-0591. [DOI] [PubMed] [Google Scholar]
- 249.Cao S, Lin L, Xia X, Wu H. MicroRNA-761 promotes the sensitivity of colorectal cancer cells to 5-Fluorouracil through targeting FOXM1. Oncotarget. 2017;9:321–331. doi: 10.18632/oncotarget.20109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Tanaka S, Hosokawa M, Yonezawa T, Hayashi W, Ueda K, Iwakawa S. Induction of epithelial-mesenchymal transition and down-regulation of miR-200c and miR-141 in oxaliplatin-resistant colorectal cancer cells. Biol Pharm Bull. 2015;38:435–440. doi: 10.1248/bpb.b14-00695. [DOI] [PubMed] [Google Scholar]
- 251.Yu T, Guo F, Yu Y, Sun T, Ma D, Han J, Qian Y, Kryczek I, Sun D, Nagarsheth N, et al. Fusobacterium nucleatum Promotes Chemoresistance to Colorectal Cancer by Modulating Autophagy. Cell. 2017;170:548–563.e16. doi: 10.1016/j.cell.2017.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Mlcochova J, Faltejskova-Vychytilova P, Ferracin M, Zagatti B, Radova L, Svoboda M, Nemecek R, John S, Kiss I, Vyzula R, et al. MicroRNA expression profiling identifies miR-31-5p/3p as associated with time to progression in wild-type RAS metastatic colorectal cancer treated with cetuximab. Oncotarget. 2015;6:38695–38704. doi: 10.18632/oncotarget.5735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Lu Y, Zhao X, Liu Q, Li C, Graves-Deal R, Cao Z, Singh B, Franklin JL, Wang J, Hu H, et al. lncRNA MIR100HG-derived miR-100 and miR-125b mediate cetuximab resistance via Wnt/β-catenin signaling. Nat Med. 2017;23:1331–1341. doi: 10.1038/nm.4424. [DOI] [PMC free article] [PubMed] [Google Scholar]


