Graphical abstract
Method name: Compound muscle action potential recordings via reconstructed or regenerated facial nerves
Keywords: Facial nerve, Reconstruction, Regeneration, Compound muscle action potential, Retrograde tracer, Rat, Mouse, Analysis, Igor Pro, MATLAB, Version control system
Abstract
Compound muscle action potential (CMAP) recording via reconstructed or regenerated motor axons is a critical examination to evaluate newly developed surgical and regeneration techniques. However, there is currently no documentation on technical aspects of CMAP recordings via reconstructed or regenerated facial nerves. We have studied new techniques of plastic surgery and nerve regeneration using a rat facial nerve defect model for years, standardizing an evaluation pipeline using CMAP recordings. Here we describe our CMAP recording procedure in detail as a package including surgical preparation, data acquisition, analysis and troubleshooting. Each resource is available in public repositories and is maintained as a version control system. In addition, we demonstrate that our analytical pipeline can not only be applied to rats, but also mice. Finally, we show that CMAP recordings can be practically combined with other behavioral and anatomical examinations. For example, retrograde motor neuron labeling provides anatomical evidence for physical routes between the facial motor nucleus and its periphery through reconstructed or regenerated facial nerves, in addition to electrophysiological evidence by CMAP recordings from the same animal.
-
•
Standardized surgical, recording and analytical procedures for the functional evaluation of reconstructed or regenerated facial nerves of rats, extended to mice.
-
•
The functional evaluation can be combined with anatomical evaluations.
-
•
The methods described here are maintained in public repositories as version control systems.
Specifications Table
| Subject area | Medicine and Dentistry |
| More specific subject area | Reconstruction and regeneration of the facial nerves |
| Method name | Compound muscle action potential recordings via reconstructed or regenerated facial nerves |
| Name and reference of original method | Reconstruction or regeneration of the facial nerves of rats. |
| * R. Sasaki, S. Aoki, M. Yamato, H. Uchiyama, K. Wada, T. Okano, H. Ogiuchi, Tubulation with dental pulp cells promotes facial nerve regeneration in rats, Tissue Eng. Part A 14(7) (2008) 1141-7. | |
| * H. Matsumine, R. Sasaki, M. Takeuchi, M. Yamato, H. Sakurai, Surgical procedure for transplanting artificial nerve conduits for peripheral nerve regeneration, Plast. Reconstr. Surg. 128(2) (2011) 95e-97e. | |
| * H. Matsumine, R. Sasaki, Y. Takeuchi, M. Miyata, M. Yamato, T. Okano, H. Sakurai, Vascularized versus nonvascularized island median nerve grafts in the facial nerve regeneration and functional recovery of rats for facial nerve reconstruction study, J. Reconstr. Microsurg. 30(2) (2014) 127-136. | |
| * H. Matsumine, Y. Takeuchi, R. Sasaki, T. Kazama, K. Kano, T. Matsumoto, H. Sakurai, M. Miyata, M. Yamato, Adipocyte-derived and dedifferentiated fat cells promoting facial nerve regeneration in a rat model, Plast. Reconstr. Surg. 134(4) (2014) 686-97. | |
| * Y. Niimi, H. Matsumine, Y. Takeuchi, R. Sasaki, Y. Watanabe, M. Yamato, M. Miyata, H. Sakurai, Effectively axonal-supercharged interpositional jump-graft with an artificial nerve conduit for rat facial nerve paralysis, Plast. Reconstr. Surg. Glob. Open 3 (2015) e416. | |
| Resource availability | CMAPMethods (https://doi.org/10.17632/9g5n35fd3f.1) |
| CMAPAnalysis (https://github.com/yuichi-takeuchi/CMAPAnalysis) | |
| CMAPanalysisMATLAB (https://github.com/hironobu-osaki/CMAPanalysisMATLAB) | |
| RetrogradeMotorNeuronLabeling (https://doi.org/10.6084/m9.figshare.5445199) |
Method details
Animals
LEW/Crl rats (200–300 g; RRID:RGD_737932) have been employed in our research on the reconstruction or regeneration of the buccal branch of facial nerves [[1], [2], [3]]. C57BL/6 mice (20–40 g; RRID:IMSR_JAX:000664) were introduced to our research in this study. All experiments conducted were approved by the Animal Care and Use Committee of the Tokyo Women’s Medical University and performed according to the institutional guidelines.
Surgical procedures for reconstruction or regeneration of the buccal branch of rat facial nerves
Briefly, rats are anesthetized with 4% isoflurane via a nasal mask connected to a Univentor 400 Anesthesia Unit (Univentor, Zejtun, Malta) [4]. A periauricular incision is made on the left side of the face to expose the buccal and marginal mandibular branches of the facial nerves and the parotid gland [1]. The marginal mandibular branch is cut using microsurgical scissors and ligated with 7–0 nylon sutures. After that, the buccal branch is exfoliated from surrounding connective tissues (as shown in Fig. 3D) and a 7 mm-defect is then made on the buccal branch of the facial nerves as previously described [1,4]. This defect could be treated by reconstructing the nerves using an autologous graft from the contralateral buccal branch [2], or by regenerating the nerves using a silicon tube filled with type I collagen solution as a nerve conduit [5]. Further published defect treatments are provided in the Additional information section of the Appendix A., including detailed schematic illustrations and a movie for nerve tube implantations [5,6].
Fig. 3.
Preparation for CMAP recordings.
(A) Introduction of anesthesia with isoflurane. (B) Urethane administration for terminal anesthesia. (C) Shaving. (D) Exposed buccal branch of the facial nerves indicated by white arrows. Black arrows indicate sutures for fixation of the excised skin. (E) Introduction of accessory ear bars. (F) A rat mounted on a stereotaxic apparatus. (G) Introduction of a rectal thermistor for feedback temperature control. (H) A reference electrode fixed on the skull indicated by white arrow heads. (I) A stimulation bipolar electrode hooked to the buccal branch of the facial nerves indicated by magenta arrow heads. Note that anodal and cathodal terminations should be connected to the proximal and distal parts of the nerves, respectively. (J) A small skin incision on the middle of the vibrissal rows C and D of the whisker pad, indicated by a red arrow head. A vibrissal alignment schema is superimposed on the whisker pad. (K) A recording microelectrode inserted into the whisker pad. (L) CMAP recording configuration for mice. The same recording, stimulating and reference electrodes are used. However, the mouthpiece and ear bars are replaced by ones specific to mice.
CMAP recording procedures
Seven to thirteen weeks after reconstruction or regeneration surgery, the buccal branch can be reconnected with sufficient physical strength to become physiologically functional [1,2,7]. CMAPs from the vibrissal muscles are then able to be recorded after stimulation of the reconstructed or regenerated nerves [8,9]. A schematic overview of these CMAP recordings is shown in Fig. 1. The materials for the recordings are listed in Table 1. Note that those materials on the list could be replaced with comparable ones from other manufacturers (see Additional information of the Appendix A.). We provide a hands-on manual for CMAP recordings in the CMAPMethods dataset in Mendeley Data (https://doi.org/10.17632/9g5n35fd3f.1) [10].
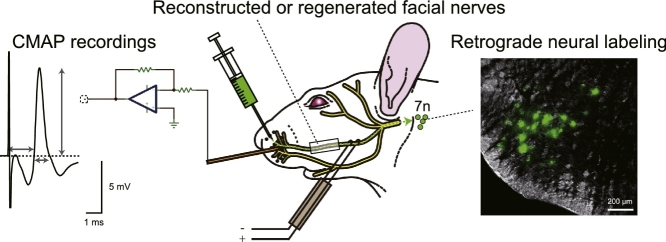
Fig. 1.
Schematic system overview of a compound muscle action potential (CMAP) recording of vibrissal muscles after stimulation of the buccal branch of facial nerves.
A screw-type reference electrode is fixed on the skull and a recording microelectrode is inserted in the whisker pad of an anesthetized rat. Differential potentials between these electrodes are band-pass filtered and amplified with a bio-amplifier and fed to a computer interface. A stimulator delivers TTL signals to trigger data acquisition via the interface. Shortly after beginning the data acquisition process, the stimulator triggers a monopolar square pulse via an isolator to evoke CMAP through the excitation of reconstructed or regenerated facial nerves.
Table 1.
Material list for CMAP recordings. Information on alternatives is provided in the Additional information section of Appendix A.
| Name | Type | Company | Catalog Number | Comments |
|---|---|---|---|---|
| Isoflurane | Reagent | Abbot | Anesthesia | |
| Urethane | Reagent | Wako | 050-05821 | Ethyl Carbamate |
| Physiological saline | Reagent | Otsuka Pharma | OTSUKA NORMAL SALINE | |
| Bio-Amplifier system | Equipment | Nihon Kohden | MEG-6100 | Multi-channel system |
| Amplifier | Equipment | Nihon Kohden | AB-610J | Component |
| Input box | Equipment | Nihon Kohden | JB-610J | Component |
| Stimulator | Equipment | Nihon Kohden | SEN-8203 | 3 ch. |
| Isolator | Equipment | Nihon Kohden | SS-202J | With SEN-8203 |
| DC temperature controller | Equipment | FHC | 40-90-8D | Noise less |
| Rectal thermistor | Supply | FHC | Mini Rectal Thermistor Probe | With 40-90-8D |
| Heating pads | Supply | FHC | Heating Pad | With 40-90-8D |
| Stereotaxic apparatus | Equipment | Narishige | SR-6R | For rats |
| Stereotaxic apparatus | Equipment | Narishige | SR-6M | For mice |
| Accessory ear bar | Supply | Narishige | EB-4N | For rats |
| Accessory ear bar | Supply | Narishige | EB-5N | For mice |
| Micromanipulator | Equipment | Narishige | MM-3 | Holds a recording electrode |
| Magnet stand | Supply | Narishige | GJ-1 | |
| Iron plate with grand termination | Supply | Put below the stereotaxic apparatus | ||
| Operating microscope | Equipment | WILD | Type308795 | |
| Dental drill | Equipment | NSK | Drill Vmax35RV Pack | 120 V |
| Drill bit | Supply | Dentsply | No. 8 | |
| PC | Equipment | Dell | Optiplex910 | Windows 10 |
| Interface | Equipment | ADInstruments | PowerLab4/30 | USB |
| Recording software | Software | ADInstruments | LabChart7 | |
| Hook-shaped bipolar stimulus electrodes | Supply | InterMedical | IMM2-220224 | Custom made |
| Stainless steel microelectrode | Supply | FHC | UESMGCSELNNM | 9–12 MΩ |
| Screw-type reference electrode | Supply | Unique Medical | TN204-089B | Custom made |
| Dental utility wax | Supply | G.C. | UTILITY WAX | |
| 5-0 nylon suture | Supply | Alfresa | EP1105NB45 |
Booting a typical recording system
-
1
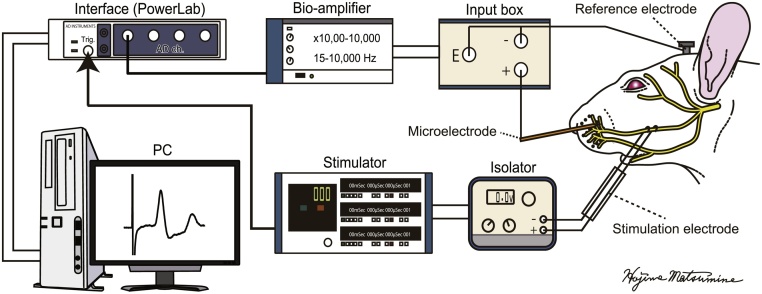
Turn on a bio-amplifier and an interface (PowerLab; ADInstruments), and boot a PC (Fig. 1).
-
2Confirm that the recording system is set as follows:
-
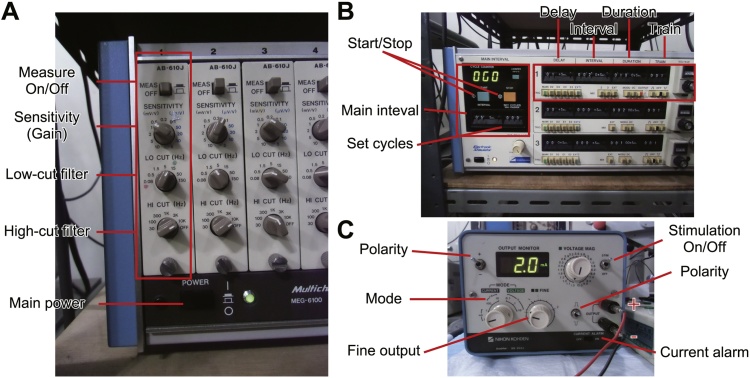
•Bio-amplifier (Fig. 2A)Measure: OnSensitivity (mV/V): 1–10 (1 mV/V in most cases)Low-cut filter (Hz): 15High-cut filter (Hz): 10 k
-
•Stimulator (Fig. 2B)Main interval: 5 sSet cycles: 000 (Free run)1 Ch. Trig: MainDelay: 1 msInterval: 20 msDuration: 100 μsTrain: 1Polarity: +Voltage: over 5000 mV
-
•Isolator (Fig. 2C)Mode: Current 10 mAOutput monitor: +2.0 mAPolarity: +Current alarm: OnStimulation: On
-
•
-
3
Run data acquisition software (e.g. LabChart, RRID:SCR_001620). Confirm that the gain setting of the software is consistent with the sensitivity (gain) setting of the bio-amplifier (e.g. 1000×) (Fig. 2A). Confirm whether the amplitude of the CMAP waveforms is covered by the voltage range of the analog input channel of the interface (e.g. ±10 V). Other settings can be as follows:
Sampling: 40 kHz
Trigger: External
Duration: 20 ms
Sweep: ∞
Fig. 2.
Equipment settings.
(A) Multi-channel bio-amplifier. Recommended setting: 1k times amplification, 15–10,000 Hz band-pass filtering. (B) Stimulator. The main interval circuit triggers the first channel at 0.2 Hz, which determines delay (1 ms), interval (not used), duration (100 μs), and the number of stimuli per trigger (single stimulus). (C) Isolator. This circuit optically isolates the stimulation circuit from the other circuits (e.g. stimulator, interface). It also determines the fine amplitude of constant current stimuli with a fine output volume. If the current alarm switch is turned on and the defined current intensity cannot be achieved due to high resistance between the output terminals, there will be a beeping sound.
Surgical procedures
-
1
Anesthesia: Put a rat into a transparent box with an acrylic lid and introduce 2–3% isoflurane until the rat does not move including its whiskers (Fig. 3A). After that, urethane is intraperitoneally administered with 1.2 g/kg as a final concentration (Fig. 3B). Separate the dose into several administrations to avoid the unexpected death of the rat.
-
2
Shave any hairs on the rat’s face and head using a hair clipper, but leave its whiskers intact (Fig. 3C). Whiskers are required to monitor contractions of the vibrissal muscles after electrical stimulations on the nerves.
-
3
Under an operating microscope, make a preauricular incision on the left side of the face to expose the buccal branch of the facial nerves (Fig. 3D). Incised skin could be stitched using 5–0 nylon sutures to enable a larger field of view. The buccal branch is exfoliated from connective tissues around, resulting in approximately 10 mm-long free nerves.
-
4
Insert accessory ear bars into the rat’s external auditory canals and put its upper incisors into the mouthpiece of the stereotaxic apparatus (Fig. 3E). Pull the mouthpiece toward the rat’s anterior to stably attach its skull (Fig. 3F).
-
5
Put a heater mat under the rat and insert the thermistor probe of a DC temperature controller into its rectum (Fig. 3G).
-
6
Expose the skull on the cerebellum and remove the periosteum on it. Use a dental drill to make a small hole in the skull for a screw-type reference electrode (Fig. 3H). Fix the reference electrode into the skull using a precision driver.
-
7
Lift the reconstructed or regenerated buccal branch of the facial nerves with a hook-shaped bipolar stimulus electrode (Fig. 3I). Properly position the electrode and then fix it to the stereotaxic apparatus using a dental utility wax. If the reconstructed or regenerated nerve seems very thin and fragile, try not to apply excess tension to it. Put several drops of liquid paraffin or mineral oil on the exfoliated facial nerve to prevent it drying.
-
8
Connect the stimulation electrode to an isolator (distal to minus and proximal to plus, respectively), and deliver test pulses via a stimulator (Fig. 2B). Adjust the stimulus intensity to 2.0 mA using a digital isolator (Fig. 2C). After that, make sure that the whiskers and vibrissal muscles are moving after each electrical stimulation.
-
9
Make a skin incision on the middle vibrissal rows C and D of the whisker pad using micro scissors (Fig. 3J) and insert a recording microelectrode into the vibrissal muscles [11]. The microelectrode is then fixed to a micro manipulator (Fig. 3K).
-
10
Connect the reference and recording microelectrodes to an input box (or pre-amplifier). The reference (–) and ground (earth) terminals are shorted.
-
11
Start data acquisition (for example, by pressing the ‘START’ button in LabChart software). The software is then waiting for triggers from the stimulator. Launch your stimulator (Fig. 2B) to start the stimulation and recordings.
-
12
If stable recordings are obtained over ten trials, stop the stimulator and the acquisition software.
-
13
Save the recordings as a data file in your PC (e.g. with an. adicht extension if you are using LabChart software).
-
14
Euthanize the rat with an excess amount of isoflurane or transcardially perfuse it for further anatomical examinations.
CMAP recordings from mice
Almost the same procedures that were used on rats can be applied to CMAP recordings of mice. To ensure stable physical fixation, use accessory ear bars and a mouthpiece specific to mice (Table 1 and Fig. 3L).
Troubleshooting for CMAP recordings
-
•
50 or 60 Hz line noise: First, check whether your recording system is properly grounded. To avoid ground loop, the GND or chassis terminals of all equipment should be connected and ideally only one of them connected to the earth ideally. However, all equipment may be connected to the earth as the second-best option. Practically, this is much easier and hopefully achieves acceptable noise levels. In many cases of line noise troubles, the reference electrode is not properly fixed on the skull. If this is a problem, make another hole on the skull and fix the reference electrode firmly into the new hole. In addition, unplug the power cables of any ungrounded equipment. A small Faraday cage covering the animal might also help to reduce line noises. If these noises are still unmanageable, an on-line or off-line band elimination filter would be useful (e.g. HumBug; DAGAN).
-
•
100 or 120 Hz ripple noise: Make sure that your system does not have any ground loops.
-
•
300–600 Hz pulse-wise noise: It might be the electrocardiogram of your animals. Try not to include their heart between the reference and recording electrodes.
-
•
Continuous DC shift: If you use a DC amplifier, consider an AC differential amplifier instead. Another solution is including a low-cut pre-filter before your DC amplifier.
-
•
The traces after electrical stimulations are out of range and do not come back to the base line immediately: The reference electrode might be loose. Try to make another hole in the skull to secure the reference electrode.
-
•
No physiological response with an intact animal: First, confirm that your amplifier is turned on and a recording microelectrode is connected to a signal terminal of your preamplifier (input box), as shown in Fig. 1. After that, check whether there is a stimulus artifact and if you can see a muscular twitch on the whisker pad at each trial. If stimuli do not seem to be delivered properly and your isolator is power-supplied by batteries, check whether the compliance voltage is high enough to achieve the intended current magnitude.
-
•
No physiological response with a facial nerve-reconstructed or -regenerated animal: In addition to the above-mentioned possibilities, the absence of responses may stem from poor functional recovery after treatments. Check whether there are whisker or whisker pad movements just after each stimulus delivery. If there is still nothing, retract the recording electrode and test another locus on the whisker pad. You might find a small CMAP with a long latency and duration [8].
Safety
Always make sure that your recording system is properly grounded. Avoid inflammable anesthetics. For example, diethyl ether.
Representative results
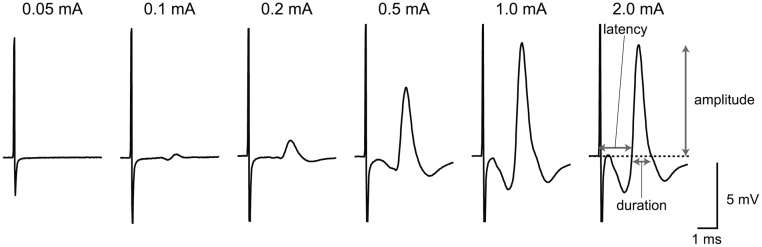
Representative CMAP recordings from an intact mouse are shown in Fig. 4. Because CMAPs are extracellular field potentials originated from many muscle action potentials, their amplitude is graded with an upper limit in response to increasing stimulus intensity, rather than it being in an all-or-none fashion. In most cases of the buccal branch stimulation of both rats and mice, 2.0 mA is the supramaximal intensity. In addition, because the holistic physiological function of reconstructed or regenerated facial nerves can be evaluated, we always employ 2.0 mA supramaximal stimulation for quantitative evaluations [6,8,9,12,13]. Amplitude, latency and duration of the evoked potentials have been measured as CMAP parameters (Fig. 4). The means ± the standard deviations of these parameters for six intact rats and mice are summarized as Table 2. There were no significant differences between the rats and the mice (p > 0.05, two-sample t-test; the raw dataset and the entire statistical results are available as supplemental materials).
Fig. 4.
Representative CMAP traces and measurable parameters.
Representative CMAP traces recorded from an intact mouse. The numbers above the traces represent stimuli intensities. Note that the amplitude of CMAP is graded until it reaches an upper limit before 1.0 mA. The amplitude is the differential voltage between the baseline (zero) and the positive peak potentials. Latency is the time between the beginning of stimuli artifacts and when the CMAP traces rise over the zero line. Duration is the time between the zero crossing points of the rising and falling trace directions. Recorded potentials relative to the reference electrode is inverted according to the traditional convention.
Table 2.
Statistical summary of the CMAP parameters of rats and mice.
| Group | Amplitude (mV) | Duration (ms) | Latency (ms) |
|---|---|---|---|
| Rat (n = 6) | 4.12 ± 2.13 | 1.17 ± 0.33 | 1.41 ± 0.37 |
| Mouse (n = 6) | 5.02 ± 1.43 | 0.97 ± 0.17 | 1.11 ± 0.34 |
Data were obtained from six intact rats and mice. Values are presented as the mean ± sd. There was no significant difference between rats and mice on each parameter. P > 0.05, non-paired t-test; two-tailed.
Analysis
Data saved as LabChart extension (.adicht) can be exported or converted to various formats, including Igor Pro experiments (.pxp) (WaveMetrics, RRID:SCR_000325), MATLAB data files (.mat) (MathWorks, RRID:SCR_001622), Axon binary files (.abf) (Molecular Devices, RRID:SCR_011323), comma-separated value files (.csv) and general ASCII text files. We utilize a custom-made program for Igor Pro: CMAPAnalysis [14], or MATLAB: CMAPanalysisMATLAB [15] for routine analyses.
How to analyze CMAP recording data using the CMAP analysis program
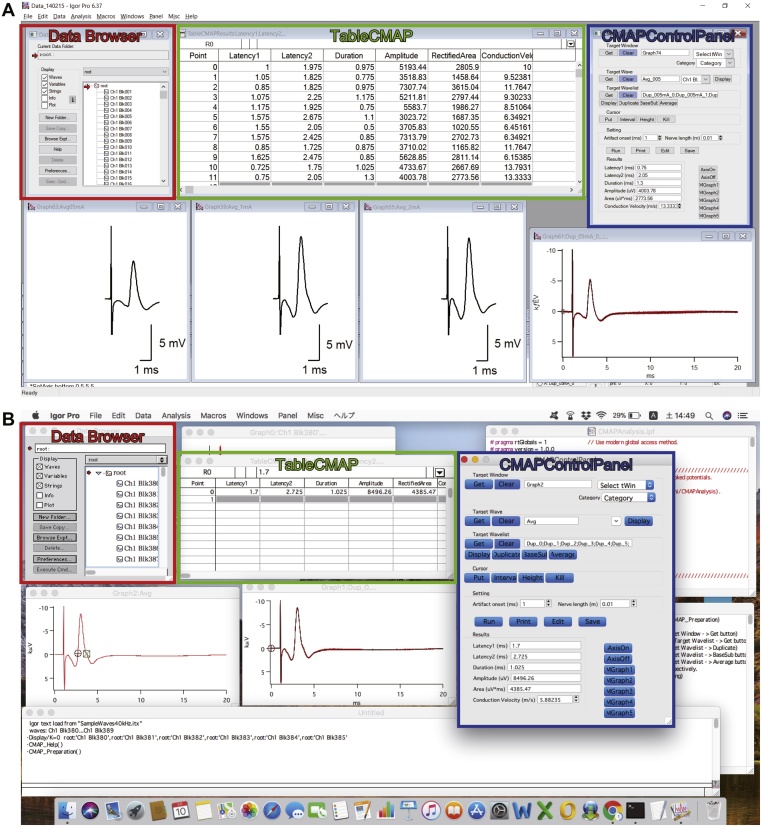
CMAPAnalysis is a custom Igor Pro program developed by Y.T. It offers a user-friendly graphical interface to analyze CMAP recordings and runs on both Windows and Mac OS (Fig. 5). The use of CMAPAnalysis does not require programming skills, experimenters only need to do mouse and cursor work. Information on system requirements and installation procedures is available on the GitHub website (https://github.com/yuichi-takeuchi/CMAPAnalysis). Sample raw (Data_140215_Raw.pxp) and analyzed data (Data_140215_Analyzed.pxp) are available in the CMAPMethods dataset (https://doi.org/10.17632/9g5n35fd3f.1) [10]. To analyze your data, follow the steps below.
-
1
Export your data from LabChart as an Igor Pro experiment (e.g. blahblah.pxp).
-
2
Open the exported experiment file in Igor Pro.
-
3
Click ‘CMAP_Preparation’ in the macro menu and the ‘CMAPControlPanel’ window will appear (Fig. 5). Global variables are stored in the root/Packages/CMAP folder in the current Igor Pro experiment.
-
4
Select multiple waves to be analyzed in ‘Data Browser’ and click ‘Display’ in the short-cut menu after right clicking. Selected waves will be plotted in a new graph window (Fig. 5).
-
5
Specify the graph as a target window by using the ‘Get’ button in the Target Window group on the control panel (CMAPControlPanel) (Fig. 5).
-
6
Get a list of source waves by using the ‘Get’ button in the Target Wave group on the control panel.
-
7
Duplicate the source waves by using the ‘Duplicate’ button in the Target Wavelist group on the control panel.
-
8
Subtract baseline DC shifts from the source waves by using the ‘BaseSub’ button in the Target Wavelist group on the control panel.
-
9
Average the duplicated waves by using the ‘Average’ button. The averaged wave will be plotted in a graph window.
-
10
Place the ‘A’ and ‘B’ cursors on the beginning and ending of the averaged CMAP wave on the graph window, respectively. This should indicate the rising and falling crossings with the zero line (Fig. 4).
-
11
Set the stimulus artifact onset time (e.g. 1 ms) in the edit box on the control panel.
-
12
Click the ‘Run’, ‘Print’, and ‘Edit’ buttons on the control panel and you will have summary table results (‘TableCMAP’) and edit boxes on the control panel (Fig. 5). Use the ‘Save’ button on the control panel to export the summary table as a. csv file. A prompt window will appear for this purpose.
-
13
Prepare publish quality graphs of the averaged traces by using the ‘MGraph1–5’ buttons on the control panel (Fig. 5). The graphs can be exported as. eps files suitable for preparing a figure panel with other vector graphics software (e.g. Adobe Illustrator, RRID:SCR_010279).
Fig. 5.
Analysis of CMAP recordings with Igor Pro software.
A screenshot of the Igor Pro analytical environment with the CMAPAnalysis add-in program on (A) Windows and (B) Mac OS [14]. CMAPAnalysis offers an integrated control panel (CMAPControlPanel) and TableCMAP, which enables a graphical user interface-based analysis without any programming skills. Results collected on the summary table (TableCMAP) via buttons on the control panel are exported as a csv file and the averaged CMAP traces are automatically built into publication-quality (vectorized) graph windows with scale bars.
Troubleshooting for CMAPAnalysis
-
•
Help information is available via the ‘CMAP_Help’ function in the macro menu, or in the ‘CMAPAnalysis’ topic of the ‘Help Browser’ of Igor Pro.
-
•
‘CMAP_Preparation’ does not appear: The CMAPAnalysis.ipf may not be compiled properly. Follow the installation instructions for CMAPAnalysis (https://github.com/yuichi-takeuchi/CMAPAnalysis) [14].
-
•
‘CMAPAnalysis’ is not found as a topic in the ‘Help Browser’: The CMAPAnalysis Help.ihf may not be compiled properly. Follow the installation instructions for CMAPAnalysis [14].
Statistical analysis
Any statistical packages can be employed for statistical tests. We have utilized commercially available packages including GraphPad Prism (RRID:SCR_002798), Igor Pro and MATLAB. R is widely used for statistical computing software (RRID:SCR_001905) as it is freely available, and it runs on Windows, MacOS and a wide variety of Unix platforms. We have developed a practical R script library for basic biostatistics: RStatisticalTests [16]. In this library, TwoSampleTest.R is used for parametric and non-parametric comparisons between two independent groups. OneWayANOVA.R is used for parametric and non-parametric comparisons between multiple groups, followed by post-hoc comparisons.
How to use RStatisticalTests for CMAP parameters
System requirements and installation procedures are described on the GitHub website (https://github.com/yuichi-takeuchi/RStatisticalTests). To analyze your data, follow the steps below.
-
1
The input data file must be a csv file (e.g. AmplitudeData.csv) that has two columns for two independent groups. The first row must include the names of the groups.
-
2
Make a working directory in your computer (e.g. D:RWD for Windows or/User/<user>/Desktop for Macintosh).
-
3
To indicate the working directory, edit the first line of a TwoSampleTest.R script file [e.g. AmplitudeTwoSampleTestR.R) as 'setwd("D:RWD")' for Windows, setwd("/User/<user>/Desktop" for Macintosh)], for example.
-
4
Place the input csv file in the same working directory.
-
5
Launch the R software.
-
6
Open the TwoSampleTest.R file (File -> Open Script…).
-
7
Run the script (Edit -> Run all, or Ctrl + A > Ctrl + R for Windows; Edit > Cmd + A > Cmd + Return for Macintosh).
-
8
The results will be exported as a text file in the same directory (e.g. AmplitudeResults_TwoSampleTest.txt).
-
9
Template csv and R files prepared for CMAP recordings (TemplateData.csv.zip and TwoSampleTest.R) are available in the CMAPMethods dataset (https://doi.org/10.17632/9g5n35fd3f.1) [10].
How to combine CMAP recordings with other examinations
CMAP recordings of facial nerve reconstruction or regeneration can be combined with other behavioral and anatomical examinations. For example, the facial palsy score (obtained by the visual inspection of the symmetry of the vibrissae at rest, the motion of the vibrissae, the symmetry of the nose at rest, and the motion of the nose) [7] can be longitudinally examined during the 7–13 week-long survival time before the CMAP recordings.
During the CMAP recordings, movements of the whiskers can be monitored using a high-speed camera (e.g. 500 Hz frame rate) [17] after each stimulation of the reconstructed or regenerated facial nerves. This provides a correlation between electrophysiological and behavioral observations. Monitoring of whisker movements during CMAP recordings is also useful for detecting a very weak CMAP with a long latency.
After the CMAP recordings, toluidine blue staining and transmission electron microscopy examinations of cross-sections of the reconstructed or regenerated nerves can be conducted to investigate the number of myelinated fibers in the nerve, the diameter of each axon and the thickness of myelin sheath [2,7,12]. Furthermore, retrograde motor neuron labeling via reconstructed or regenerated nerves from the peripheral tissue can be performed to examine whether the reconstructed or regenerated facial nerves are parts of physical route between the facial motor nucleus in the brain stem and its periphery [1,6,9,13,18].
In the following sections, we describe the practical procedures of retrograde tracer experiments along with the CMAP recordings for both rats and mice. Hands-on manuals are available and maintained in the RetrogradeMotorNeuronLabeling dataset (https://doi.org/10.6084/m9.figshare.5445199) [19]. We have employed lipophilic carbocyanine dyes and cholera toxin B subunit (CTB) for rats and mice, respectively [9]. CTB is highly sensitive [20,21] and works with the rat peripheral nervous system as well [22,23]. Furthermore, CTBs labeled with distinct Alexa fluorophores (Thermo Fisher Scientific) could be employed for labeling from different peripheral loci [24]. However, we have not tried CTB for rats so far as CTB and their Alexa conjugates are very expensive; subcutaneous injections of these tracers into rats are also not cost-effective. We have had scientifically sufficient results with carbocyanine dyes for rats [6,9,13,25].
For rats
If carbocyanine dyes with different fluorescent spectra are injected separately into different peripheral loci (e.g. DiO, DiI and DiD), motor neurons with different origins are distinctly labeled according to their axonal targets [6,13]. The materials are listed in Table 3. The procedure is outlined below.
-
1
Dyes are dissolved and stored at 4 °C as a 10% solution in N,N-dimethylformamide (DMF).
-
2
Two weeks before the CMAP recordings, the rats are anesthetized with 4% isoflurane and placed under a dissection microscope.
-
3
For DiI and DiD, inject 100 μl 1% dye solution into the whisker pads subcutaneously using a Hamilton syringe (Fig. 6). For DiO, inject 200 μl 0.5% solution.
-
4
After a two-week incubation, record the CMAPs as described above.
-
5
Under deep urethane anesthesia (1.5 g/kg, i.p.), perfuse the rat transcardially with physiological saline followed by 4% paraformaldehyde in 0.1 M PB (pH 7.4). Use the same fixative overnight.
-
6
Prepare 50 μm-thick coronal brain stem sections with a vibrating blade microtome and harvest them into 12-well culture plates containing 0.1 M PB.
-
7
Mount the sections on gelatin-coated glass slides and coverslip with an aqueous mounting medium (e.g. PermaFluorTM).
-
8
Observe the sections with an epifluorescence or a confocal microscopy (Fig. 6).
-
9
Note that brain sections with carbocyanine dyes should not be prepared as frozen sections because freeze-throw cycles injure the plasma membrane of cells and disrupt the integrity of the labeled motor neurons.
-
10
Avoid mounting media containing glycerol, which can extract membrane-bound dye (e.g. ProLong Gold, VectaShield HardSetTM, Aqua-Poly/Mount).
-
11
Abbreviated solution: PB, phosphate buffer.
Table 3.
Material list for retrograde motor neuron labeling with carbocyanine dyes for rats.
| Name | Type | Company | Catalog Number | Comments |
|---|---|---|---|---|
| Isoflurane | Reagent | Abbot | Anesthesia | |
| DiO | Reagent | Thermo Fisher Scientific | D275 | Retrograde tracer (Green) |
| DiI | Reagent | Thermo Fisher Scientific | D282 | Retrograde tracer (Red) |
| DiD | Reagent | Thermo Fisher Scientific | D307 | Retrograde tracer (Far red) |
| DMF | Reagent | Wako | 050-05821 | Solvent |
| EtOH | Reagent | Wako | 057-00451 | Solvent |
| PermaFluor | Reagent | Thermo Fisher Scientific | TA-030-FM | Aqueous mounting medium |
| 25 μl Hamilton syringe | Supply | Hamilton | 80401 | Luer Tip |
| Vibratome | Equipment | Leica Microsystems | VT1000S | Vibrating blade microtome |
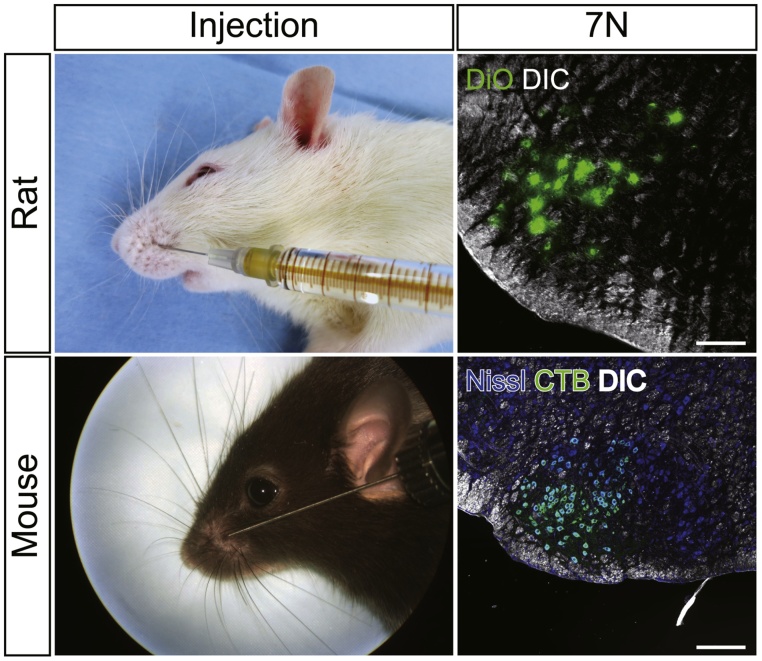
Fig. 6.
Retrograde motor neuron labeling along CMAP recordings.
Lipophilic carbocyanine dyes (e.g. DiO, DiI and DiD) are used to label motor neurons in the facial nerve nucleus from the whisker pad for rats, whereas cholera toxin B subunit is used for mice. Scale bars: 200 μm. Detailed protocols are documented in the text and available in figshare in the RetrogradeMotorNeuronLabeling dataset [19].
For mice
We have developed highly sensitive retrograde labeling of facial motor neurons for mice along with CMAP recordings. This method not only labels motor neurons, but also sensory afferent fibers to the trigeminal nuclei [21]. Materials are listed in Table 4.
Table 4.
Material list for retrograde motor neuron labeling with cholera toxin B subunit for mice.
| Name | Type | Company | Catalog Number | Comments |
|---|---|---|---|---|
| Ketamine | Reagent | Daiichi-Sankyo | KETALAR | Anesthesia: Strictly regulated. |
| Xylazine | Reagent | Sigma-Aldrich | X1251 | Sedating drug |
| Cholera Toxin B subunit | Reagent | Sigma-Aldrich | C9905 | Retrograde tracer. Need documentation to purchase. |
| Microneedle syringe | Supply | WPI | NANOFIL | Comes with 26 gauge needle. |
| Cryo-blocking medium | Supply | Sakura Finetek | Tissue-TekTM O.C.T. compoundTM | Transparent |
| Cryomold | Supply | Sakura Finetek | Tissue-TekTM CryomoldTM | 10 mm × 10 mm × 5 mm |
| Blocking serum | Reagent | Vector Lab | S-5000 | Normal rabbit serum |
| 1st antibody | Reagent | List Biological Lab | 703 | Go anti-CTB |
| RRID:AB_10013220 | ||||
| 2nd antibody | Reagent | Vector Lab | BA-5000 | Rb anti-Go IgG, biotinylated |
| Streptavidin | Reagent | Thermo Fisher Scientific | S21375 | Alexa Fluor 633-conjugated |
| Fluoro Nissl solution | Reagent | Thermo Fisher Scientific | N-21470 | Blue |
| Cryostat | Equipment | Leica Microsystems | CM1860 | Freezing microtome |
Injection
-
1
Two to four days before the CMAP recordings, the mice are anesthetized with a ketamine/xylazine cocktail (80/10 mg/kg, i.p) and placed under a dissecting microscope.
-
2
Fill a microneedle syringe (NANOFIL; WPI) with 1% CTB solution in 50 mM PBS and inject 5 μl of the solution into the whisker pads subcutaneously (Fig. 6).
-
3
After two to four days of incubation, record the CMAPs as described above.
Perfusion, cryo-embedding, sectioning
-
1
Under deep pentobarbital anesthesia (120 mg/kg, i.p.), perfuse the mice transcardially with ice-cold saline, followed by 4% paraformaldehyde and 0.2% picric acid in 0.1 M PB (pH 7.2–7.3). Use the same fixative overnight.
-
2
Infiltrate the brain with sucrose gradient (10–30%).
-
3
Cryo-embed the brain into a cryomold filled with O.C.T. compound on −80 °C acetone solution with dry ice.
-
4
Prepare 20-μm-thick coronal brain stem sections using a freezing microtome and harvest them into 24-well culture plates containing an anti-freeze solution (30% glycerol, 30% ethylene glycol, 40% PBS).
-
5
PBS wash.
Staining in free-floating configuration (room temperature, light shielded)
-
1
10% NRS in PBS-XCR, 30 min on shaker.
-
2
1:10,000 dil. Go anti-CTB (703, List) in PBS- XCR, overnight on shaker.
-
3
PBS-X wash (quick × 1, 10 min × 2).
-
4
1:400 dil. biotinylated Rb anti-Go IgG (Vector) in PBS-XCR, 2 h on shaker.
-
5
PBS-X wash (quick × 1, 10 min × 2).
-
6
1:400 dil. Alexa 633-conjugated streptavidin in PBS-X, 2 h on shaker
-
7
PBS wash (quick × 1, 10 min × 2).
Counterstaining with fluorescent Nissl
-
•
1:150 dil. fluorescent Nissl solution (blue), 40–60 min.
-
•
PBS wash (quick × 1, 10 min × 2).
Mounting, cover-slipping, observation
-
•
Mount the sections on gelatin-coated glass slides and air-dry for 30 min.
-
•
Coverslip with 50% (v/v) glycerol and 2.5% (w/v) DABCO (1,4-diazabicyclo [2.2.2] octane) in PBS.
-
•
Observe the sections with an epifluorescence or a confocal microscopy (Fig. 6).
-
•
Abbreviated solutions: PB, phosphate buffer; PBS, 0.1 M phosphate buffered saline; PBS-X, 0.3% Triton-X 100 in PBS; PBS-XCR, 0.12% λ-carrageenan, 1% normal rabbit serum, 0.02% sodium azide in PBS-X.
Conflicts of interest
The authors declare no conflicts of interest.
Acknowledgements
We thank E. Naraba for technical assistance. This study was supported by the Grants-in-Aid for Scientific Research (25870757 and 17H05752), the Keiko Watanabe Award and the Hiroto Yoshioka Memorial Fund for Medical Research.
Footnotes
Supplementary material related to this article can be found, in the online version, at doi:10.1016/j.mex.2018.03.007.
Appendix A. Supplementary data
The following are Supplementary data to this article:
References
- 1.Sasaki R., Aoki S., Yamato M., Uchiyama H., Wada K., Okano T., Ogiuchi H. Tubulation with dental pulp cells promotes facial nerve regeneration in rats. Tissue Eng. Part A. 2008;14(7):1141–1147. doi: 10.1089/ten.tea.2007.0157. [DOI] [PubMed] [Google Scholar]
- 2.Matsumine H., Sasaki R., Yamato M., Okano T., Sakurai H. A polylactic acid non-woven nerve conduit for facial nerve regeneration in rats. J. Tissue Eng. Regener. Med. 2012;8(6):454–462. doi: 10.1002/term.1540. [DOI] [PubMed] [Google Scholar]
- 3.Sasaki R., Aoki S., Yamato M., Uchiyama H., Wada K., Ogiuchi H., Okano T., Ando T. PLGA artificial nerve conduits with dental pulp cells promote facial nerve regeneration. J. Tissue Eng. Regener. Med. 2011;5(10):823–830. doi: 10.1002/term.387. [DOI] [PubMed] [Google Scholar]
- 4.Sasaki R., Matsumine H., Watanabe Y., Yamato M., Ando T. Anesthesia for research on reconstructive facial surgery in rats. J. Reconstr. Microsurg. 2013;29(3):209–210. doi: 10.1055/s-0032-1331147. [DOI] [PubMed] [Google Scholar]
- 5.Matsumine H., Sasaki R., Takeuchi M., Yamato M., Sakurai H. Surgical procedure for transplanting artificial nerve conduits for peripheral nerve regeneration. Plast. Reconstr. Surg. 2011;128(2):95e–97e. doi: 10.1097/PRS.0b013e31821ef380. [DOI] [PubMed] [Google Scholar]
- 6.Niimi Y., Matsumine H., Takeuchi Y., Sasaki R., Watanabe Y., Yamato M., Miyata M., Sakurai H. Effectively axonal-supercharged interpositional jump-graft with an artificial nerve conduit for rat facial nerve paralysis. Plast. Reconstr. Surg. Glob. Open. 2015;3:e416. doi: 10.1097/GOX.0000000000000397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Watanabe Y., Sasaki R., Matsumine H., Yamato M., Okano T. Undifferentiated and differentiated adipose-derived stem cells improve nerve regeneration in a rat model of facial nerve defect. J. Tissue Eng. Regener. Med. 2014;11(2):362–374. doi: 10.1002/term.1919. [DOI] [PubMed] [Google Scholar]
- 8.Matsumine H., Sasaki R., Takeuchi Y., Miyata M., Yamato M., Okano T., Sakurai H. Vascularized versus nonvascularized island median nerve grafts in the facial nerve regeneration and functional recovery of rats for facial nerve reconstruction study. J. Reconstr. Microsurg. 2014;30(2):127–136. doi: 10.1055/s-0033-1357500. [DOI] [PubMed] [Google Scholar]
- 9.Matsumine H., Takeuchi Y., Sasaki R., Kazama T., Kano K., Matsumoto T., Sakurai H., Miyata M., Yamato M. Adipocyte-derived and dedifferentiated fat cells promoting facial nerve regeneration in a rat model. Plast. Reconstr. Surg. 2014;134(4):686–697. doi: 10.1097/PRS.0000000000000537. [DOI] [PubMed] [Google Scholar]
- 10.Takeuchi Y. 2018. CMAPMethods, Mendeley Data, v1.0.0. [Google Scholar]
- 11.Dorfl J. The musculature of the mystacial vibrissae of the white mouse. J. Anat. 1982;135(Pt. 1):147–154. [PMC free article] [PubMed] [Google Scholar]
- 12.Sasaki R., Matsumine H., Watanabe Y., Takeuchi Y., Yamato M., Okano T., Miyata M., Ando T. Electrophysiologic and functional evaluations of regenerated facial nerve defects with a tube containing dental pulp cells in rats. Plast. Reconstr. Surg. 2014;134(5):970–978. doi: 10.1097/PRS.0000000000000602. [DOI] [PubMed] [Google Scholar]
- 13.Matsumine H., Sasaki R., Takeuchi Y., Watanabe Y., Niimi Y., Sakurai H., Miyata M., Yamato M. Unilateral multiple facial nerve branch reconstruction using “end-to-side loop graft” supercharged by hypoglossal nerve. Plast. Reconstr. Surg. Glob. Open. 2014;2:e240. doi: 10.1097/GOX.0000000000000206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Y. Takeuchi, CMAPAnalysis, Zenodo, v1.0.0, 2017. 10.5281/zenodo.800747. [DOI]
- 15.H. Osaki, CMAPanalysisMATLAB, GitHub, 2017. https://github.com/hironobu-osaki/CMAPanalysisMATLAB.
- 16.Y. Takeuchi, RStatisticalTests, Zenodo, v1.0.0, 2017. 10.5281/zenodo.817654. [DOI]
- 17.Ferezou I., Haiss F., Gentet L.J., Aronoff R., Weber B., Petersen C.C. Spatiotemporal dynamics of cortical sensorimotor integration in behaving mice. Neuron. 2007;56(5):907–923. doi: 10.1016/j.neuron.2007.10.007. [DOI] [PubMed] [Google Scholar]
- 18.Furukawa H., Saito A., Mol W., Sekido M., Sasaki S., Yamamoto Y. Double innervation occurs in the facial mimetic muscles after facial-hypoglossal end-to-side neural repair: rat model for neural supercharge concept. J. Plast. Reconstr. Aesthet. Surg. 2008;61(3):257–264. doi: 10.1016/j.bjps.2007.01.075. [DOI] [PubMed] [Google Scholar]
- 19.Takeuchi Y. 2018. RetrogradeMotorNeuronLabeling, Figshare, v1.0.0. [Google Scholar]
- 20.Takeuchi Y., Asano H., Katayama Y., Muragaki Y., Imoto K., Miyata M. Large-scale somatotopic refinement via functional synapse elimination in the sensory thalamus of developing mice. J. Neurosci. 2014;34(4):1258–1270. doi: 10.1523/JNEUROSCI.3865-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Takeuchi Y., Osaki H., Yagasaki Y., Katayama Y., Miyata M. Afferent fiber remodeling in the somatosensory thalamus of mice as a neural basis of somatotopic reorganization in the brain and ectopic mechanical hypersensitivity after peripheral sensory nerve injury. eNeuro. 2017;4(2):e0345–e0346. doi: 10.1523/ENEURO.0345-16.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee H., White J.M., Chung J., Tansey K.E. Peripheral and central anatomical organization of cutaneous afferent subtypes in a rat nociceptive intersegmental spinal reflex. J. Comp. Neurol. 2017;525(9):2216–2234. doi: 10.1002/cne.24201. [DOI] [PubMed] [Google Scholar]
- 23.Fernández-Montoya J., Martin Y.B., Negredo P., Avendaño C. Changes in the axon terminals of primary afferents from a single vibrissa in the rat trigeminal nuclei after active touch deprivation or exposure to an enriched environment. Brain Struct. Funct. 2018;223(1):47–61. doi: 10.1007/s00429-017-1472-5. [DOI] [PubMed] [Google Scholar]
- 24.Wang S., Su D., Li J., Li D., Wan H., Schumacher M., Liu S. Donor nerve axotomy and axonal regeneration after end-to-side neurorrhaphy in a rodent model. J. Neurosurg. 2018 doi: 10.3171/2017.8.JNS17739. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 25.Niimi Y., Matsumine H., Takeuchi Y., Osaki H., Tsunoda S., Miyata M., Yamato M., Sakurai H. A collagen-coated PGA conduit for interpositional-jump graft with end-to-side neurorrhaphy for treating facial nerve paralysis in rat. Microsurgery. 2018 doi: 10.1002/micr.30291. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.