Significance
Zyxin is a member of the cell–cell adhesion complex and controls cell cytoskeleton and motility. Analysis from clinical samples revealed that Zyxin is highly expressed in colon cancer compared with normal tissue, suggesting an oncogenic role for Zyxin in cancer. Depletion of Zyxin resulted in significantly impaired colon cancer cell proliferation, migration, cellular transformation, and tumor formation in xenograft animal models. We also showed that Zyxin is phosphorylated by CDK1 during mitosis. Mitotic phosphorylation is required for Zyxin activity in promoting colon cancer growth. Zyxin regulates YAP activity through the colon cancer oncogene CDK8. We further showed that CDK8 directly phosphorylates YAP and promotes its activation. These observations identify the Zyxin–CDK8–YAP axis as a potential therapeutic target in cancer.
Keywords: Zyxin, CDK8, Hippo–YAP pathway, mitosis, colon cancer
Abstract
Zyxin is a member of the focal adhesion complex and plays a critical role in actin filament polymerization and cell motility. Several recent studies showed that Zyxin is a positive regulator of Yki/YAP (Yes-associated protein) signaling. However, little is known about the mechanisms by which Zyxin itself is regulated and how Zyxin affects Hippo–YAP activity. We first showed that Zyxin is phosphorylated by CDK1 during mitosis. Depletion of Zyxin resulted in significantly impaired colon cancer cell proliferation, migration, anchorage-independent growth, and tumor formation in xenograft animal models. Mitotic phosphorylation is required for Zyxin activity in promoting growth. Zyxin regulates YAP activity through the colon cancer oncogene CDK8. CDK8 knockout phenocopied Zyxin knockdown in colon cancer cells, while ectopic expression of CDK8 substantially restored the tumorigenic defects of Zyxin-depletion cells. Mechanistically, we showed that CDK8 directly phosphorylated YAP and promoted its activation. Fully activated YAP is required to support the growth in CDK8-knockout colon cancer cells in vitro and in vivo. Together, these observations suggest that Zyxin promotes colon cancer tumorigenesis in a mitotic-phosphorylation-dependent manner and through CDK8-mediated YAP activation.
Zyxin is a LIM domain protein concentrated at sites of cell–cell adhesion, where it is proposed to dock proteins involved in cytoskeletal dynamics and signaling (1). Zyxin has also been implicated in sensing the mechanotransduction signal (2, 3). Of interest, Zyxin contains a nuclear export signal and shuttles between the nucleus and sites of cell adhesion (4), although the underlying mechanisms and function of its localization dynamics are unclear. Several recent studies showed that Zyxin is a positive regulator of tissue growth through Hippo–Yorkie/Yki signaling in Drosophila (5–8). Zyxin regulates Yki-mediated tissue growth by promoting F-actin polymerization independent of Hippo/Warts kinases (5). This mechanism is conserved in mammalian cancer cells (9, 10). Furthermore, in human breast cancer cells, Zyxin promotes growth and tumorigenesis by modulating Hippo–yes-associated protein (YAP) signaling activity (11), which plays an important role in the development and progression of many human malignancies (12–15). Despite the growth-promoting role of Zyxin, however, little is known about the mechanisms by which Zyxin itself is regulated and how Zyxin affects Hippo–YAP (and/or other signaling) activity in cancer cells.
The protein kinase cyclin-dependent kinase 8 (CDK8) is a component of the mediator complex that functions as a bridge between basal transcription machinery and gene-specific transcriptional factors (16). CDK8 is amplified and overexpressed in colon cancer and exerts its oncogenic activity partially through regulating β-catenin activity (17). The precise mechanisms by which CDK8 regulates β-catenin remain obscure. CDK8 mRNA is up-regulated in malignant melanoma by loss of a transcriptional repressor called the histone variant macroH2A, which functions as a tumor suppressor in melanoma (18). In addition, CDK8 protein levels are also controlled by S-phase kinase associated protein 2 (Skp2)-mediated degradation of macroH2A1 protein, and these three proteins work together to regulate G2/M transition and tumorigenesis in breast cancer (19). CDK8 exerts its oncogenic function mainly through phosphorylation of substrates. Several substrates for CDK8 have been identified, including the Notch intracellular domain, SMAD complexes, E2F1, STAT1, and the C-terminal domain of RNA polymerase II (16). These studies highlight an important oncogenic function of CDK8 kinase activity.
A connection between CDK8 and YAP, the critical transcriptional coactivator of Hippo signaling, has not been established. Here, we report that Zyxin promotes colon cancer cell growth, and its oncogenic activity is partially controlled by mitotic phosphorylation. We further showed that Zyxin regulates YAP activity through CDK8 in colon cancer cells. In addition, we identified YAP as a direct substrate of CDK8 and CDK8-mediated phosphorylation that promotes YAP activity in vitro and in vivo.
Results
Zyxin Is Phosphorylated by CDK1 in Vitro During Mitosis.
Others and we have shown that several Hippo–YAP components are regulated and implicated in mitosis (20–26). These studies suggest that the Hippo–YAP pathway controls tumorigenesis through dysregulation during mitosis. Given the connection between Zyxin and Hippo–YAP signaling, we tested the possibility that Zyxin might contribute to tumorigenesis through regulating cell-cycle progression, especially mitosis.
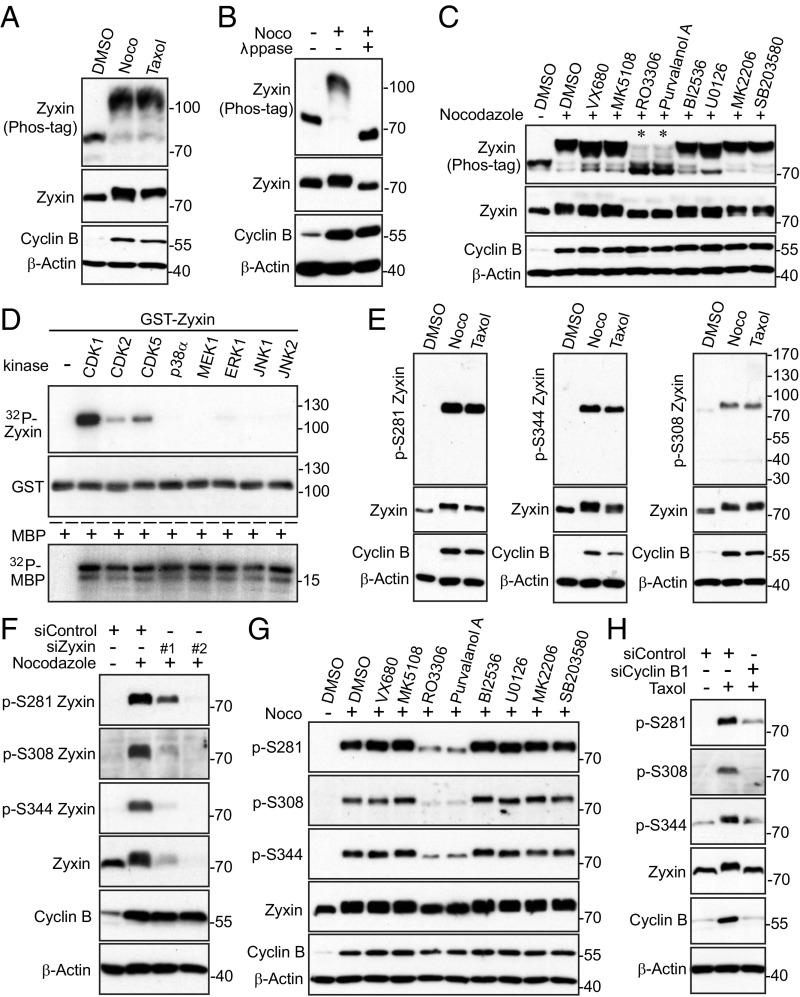
As shown in Fig. 1A, treatment with taxol or nocodazole—both agents that bind to microtubules and arrest cells in mitosis—significantly increased mobility upshift of Zyxin on a Phos-tag gel (Fig. 1A). Lambda phosphatase treatment completely converted all slow-migrating bands to fast-migrating bands, confirming that Zyxin is phosphorylated in mitosis induced by spindle poisons (Fig. 1B).
Fig. 1.
CDK1 phosphorylation of Zyxin during mitotic arrest. (A) Taxol (100 nM for 16 h) or nocodazole (100 ng/mL for 16 h; Noco) treatments in HeLa cells caused mobility upshift of Zyxin on Phos-tag SDS-polyacrylamide gels. (B) HeLa cells were treated with nocodazole (Noco) as indicated and cell lysates were further treated with (+) or without (−) λ phosphatase (ppase). (C) HeLa cells were treated with taxol together with or without various kinase inhibitors as indicated. Inhibitors were added (with MG132 to prevent cyclin B from degradation and cells from exiting from mitosis) 1.5 h before harvesting the cells. (D) In vitro kinase assays with purified kinases using bacterially purified GST–Zyxin (Abnova) and MBP (myelin basic protein) (Sigma) proteins as substrates. MBP proteins were included as positive controls, confirming the kinases are active. (E) Zyxin is phosphorylated at multiple sites during mitotic arrest. HeLa cells were treated as in A and total cell lysates were probed with the indicated antibodies. (F) Validation of phospho-specific antibodies. HeLa cells were transfected with 40 nM scramble (control) or 40 nM siRNA against Zyxin for 48 h and were further treated with nocodazole as indicated. (G) HeLa cells were treated as in C. Total cell lysates were subjected to Western blotting with the indicated antibodies. (H) HeLa cells were transfected with 40 nM scramble (control) or siRNA against Cyclin B1 for 48 h and were further treated with taxol as indicated.
Next, we used various kinase inhibitors to identify the candidate kinase for Zyxin phosphorylation. Of interest, treatments with RO3306 (CDK1 inhibitor) or Purvalanol A (CDK1/2/5 inhibitor) significantly inhibited mobility shift and phosphorylation (Fig. 1C, lanes 5 and 6). These data suggest that CDK1, a well-known master mitotic kinase, is likely the relevant kinase for Zyxin phosphorylation. Fig. 1D shows that purified CDK1/cyclin B kinase complex (CDK2 and CDK5/p25 kinases to a lesser extent) directly phosphorylated GST–Zyxin proteins in vitro (Fig. 1D). Zyxin is not a suitable substrate for MAPK-p38α, MEK1, ERK1, and JNK1/2 kinases in vitro (Fig. 1D), although these kinases recognize the same phosphorylation sequence consensus as CDK1.
Zyxin Is Phosphorylated by CDK1 in Cells During Mitosis.
CDK1 phosphorylates an S/TP consensus sequence (27). Database analysis identified three SPs (S281, S308, and S344) as possible main phosphorylation sites in Zyxin during mitosis (28). We generated phospho-specific antibodies against these sites. Nocodazole or taxol treatment significantly increased phosphorylation of S281, S308, and S344 of endogenous Zyxin (Fig. 1E). Phosphopeptide, but not regular nonphosphopeptide, incubation completely blocked the phospho-signal, suggesting that this antibody detects the phosphorylated form of Zyxin (SI Appendix, Fig. S1 A–C). Exogenous Zyxin was also phosphorylated during mitotic arrest, and mutating serines to nonphosphorylatable alanines abolished the phosphorylation (SI Appendix, Fig. S1D). The siRNA-mediated depletion of Zyxin significantly blocked the phospho-signal, confirming the specificity of these phospho-antibodies (Fig. 1F). Phosphorylation of Zyxin occurred in dose-dependent and time course-dependent manners (SI Appendix, Fig. S1 E and F). Using kinase inhibitors, we demonstrated that phosphorylation of Zyxin is CDK1 kinase-dependent (Fig. 1G, lanes 5 and 6). Furthermore, knockdown of Cyclin B1 or CDK1 largely blocked Zyxin phosphorylation (Fig. 1H and SI Appendix, Fig. S1G). Enhanced expression of active CDK1/Cyclin B1 was sufficient to stimulate Zyxin phosphorylation (SI Appendix, Fig. S1H).
Immunofluorescence staining revealed that a strong signal was detected in nocodazole-arrested prometaphase cells for antibodies against Zyxin S281 and S344 (SI Appendix, Fig. S2 A and B, Upper, white arrows). Very low or no signal was detected in interphase cells (SI Appendix, Fig. S2 A and B, yellow arrows). Again, phosphopeptide, but not nonphosphopeptide, incubation completely blocked the signal, suggesting that these antibodies specifically detect phosphorylated Zyxin (SI Appendix, Fig. S2 A and B). The specificity of the antibodies was further confirmed by siRNA knockdown of Zyxin (SI Appendix, Fig. S2 C and D). Addition of RO3306 largely abolished the signals detected by p-Zxyin S281 and S344 antibodies in mitotic cells, further indicating that the phosphorylation is CDK1-dependent (SI Appendix, Fig. S2 A and B, Lower).
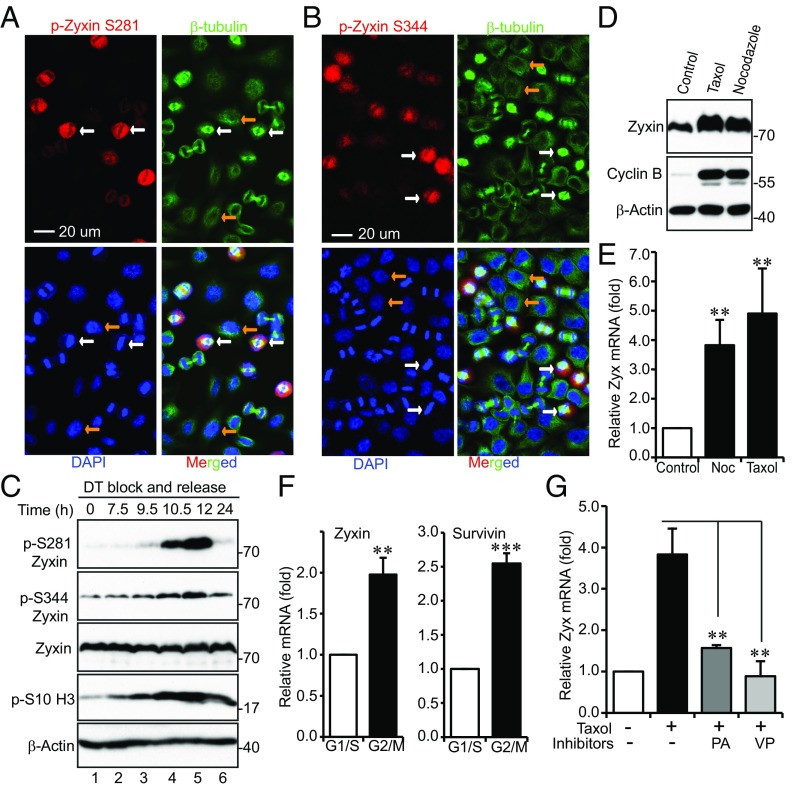
To determine whether mitotic phosphorylation of Zyxin occurs during unperturbed/normal mitosis, we performed immunofluorescence staining on cells collected from a double thymidine block and release (29). Very weak signals were detected in interphase or telophase/cytokinesis cells (Fig. 2 A and B, yellow arrows, and SI Appendix, Fig. S3 A and B). The phospho-signal increased in prometaphase and peaked in metaphase cells (Fig. 2 A and B, white arrows, and SI Appendix, Fig. S3 A and B). After being released from double thymidine block, cells entered into mitosis at 10–12 h (21, 24) (revealed by increased p-S10 H3 levels), and p-Zyxin S281 and S344 signals also increased in these cells (Fig. 2C). Mitotic phosphorylation of Zyxin occurred in freely cycling cells in a CDK1-dependent manner (SI Appendix, Fig. S4 A and B). These observations indicate that Zyxin is phosphorylated by CDK1 during mitosis.
Fig. 2.
Zyxin is phosphorylated and induced during unperturbed mitosis. (A) HeLa cells were synchronized by a double thymidine (DT) block and release method. Cells were stained with antibodies against p-Zyxin S281 or β-tubulin, or with DAPI. A 40× objective lens was used to view various phases of the cells in a field. (B) Experiments were done similarly as in A with p-Zyxin S344 antibodies. White and yellow arrows (in A and B) mark the metaphase and interphase cells, respectively. (C) HeLa cells were synchronized by a DT block and release method. Total cell lysates were harvested at the indicated time points and subjected to Western blotting analysis. (D) Zyxin protein levels increased during mitotic arrest. (E) Zyxin mRNA levels were induced during mitotic arrest. (F) Zyxin mRNA levels were increased during normal mitosis. HeLa cells were treated as in C and harvested at 10 h post release. Survivin serves as a positive control. (G) HeLa cells were treated with taxol and inhibitors were added 2 h before harvesting the cells. qRT-PCR was performed to measure Zyxin mRNA levels. Data were expressed as the mean ± SEM of three independent experiments (E–G). **P < 0.01; ***P < 0.001 (t test).
Zyxin Expression Is Induced During Mitosis.
During our experiments, we noticed that, in addition to mobility shift/phosphorylation, Zyxin protein levels were also increased during taxol or nocodazole-induced mitotic arrest (Fig. 2D) and in normal mitosis (SI Appendix, Fig. S4D). Quantitative RT-PCR (qRT-PCR) confirmed that Zyxin mRNA levels were induced by taxol or nocodazole (Fig. 2E) and in normal mitosis (Fig. 2F). YAP targets survivin and Cyr61, but not YAP itself, were induced in mitosis (Fig. 2F and SI Appendix, Fig. S4 C–E). A previous study showed that Zyxin is a direct target of TAZ (a paralog of YAP)/TEAD (30), and we determined whether Zyxin induction in mitosis is YAP/TAZ-dependent. Indeed, addition of verteporfin [(VP) a YAP/TEAD inhibitor (31)] or knockdown of YAP largely abolished Zyxin mRNA increase induced by taxol or nocodazole treatment (Fig. 2G and SI Appendix, Fig. S4F). Of interest, inhibition of CDK1 by Purvalanol A also blocked Zyxin expression, suggesting that Zyxin expression in mitosis is induced in a CDK1-dependent manner (Fig. 2G).
Zyxin Is Required for Colon Cancer Cell Growth in Vitro and in Vivo.
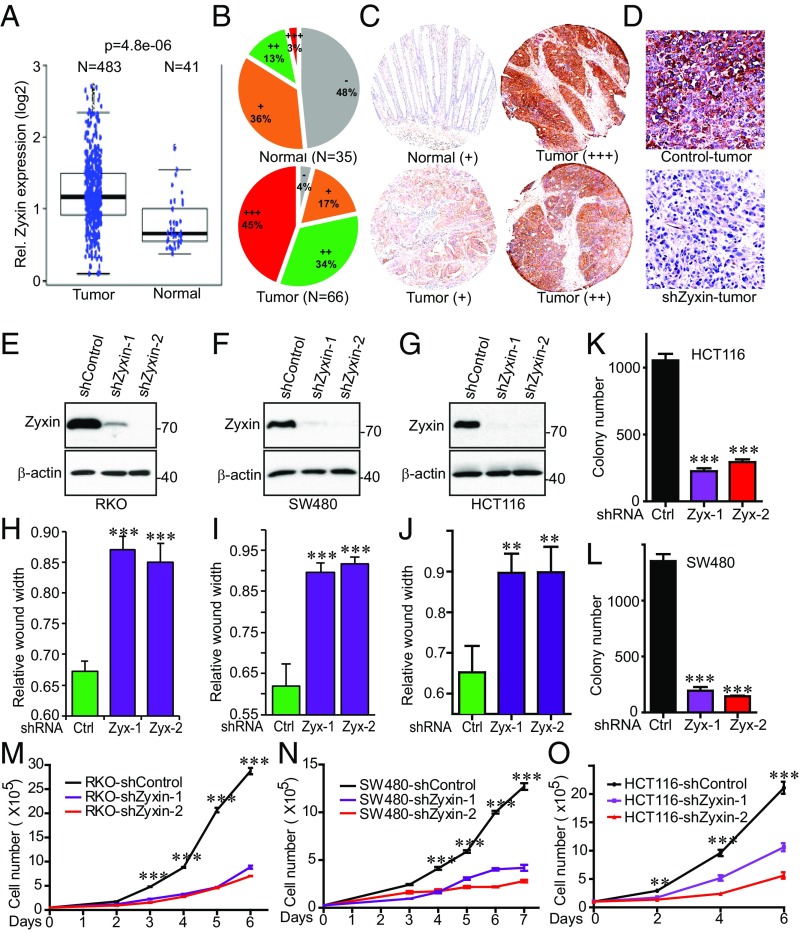
We next explored the biological significance of Zyxin in cancer cell growth. We analyzed the Oncomine and TCGA online databases and found Zyxin to be highly expressed in colon cancer (Fig. 3A). We performed immunohistochemistry (IHC) staining on tissue microarrays (TMAs) (n = 66 cancer/tumor, n = 35 normal) and confirmed that Zyxin protein levels were significantly increased in colon cancer samples compared with normal tissue (Fig. 3 B and C). The specificity of the Zyxin IHC was validated by using Zyxin-knockdown tumor samples (Fig. 3D). These findings highlight a critical role of Zyxin in tumorigenesis in colon cancer.
Fig. 3.
Zyxin levels are increased in colon cancer patients and knockdown of Zyxin inhibits colon cancer cell growth. (A) Zyxin mRNA levels are increased in colon cancer samples. RNA-sequencing data from The Cancer Genome Atlas (TCGA) were downloaded from NCBI’s Gene Expression Omnibus (GEO) with the accession number of GSE62944 (the Student t test was used for statistical evaluation). (B and C) IHC staining analysis of Zyxin on colon cancer TMAs (P < 0.001, Wilcoxon rank sum test). Representative staining images were shown (C). Staining and scoring were done as we described (63). −: negative; +: low expression; ++: moderate expression; +++: strong expression. (D) Validation of the Zyxin antibody in IHC specificity using control and Zyxin-knockdown tumors. (E–G) Establishment of colon cancer cell lines with stable knockdown of Zyxin. (H–J) Cell migration (wound healing) assays of cell lines established in E–G. (K and L) Anchorage-independent growth (colony assays on soft agar) of HCT116 and SW480 cell lines with knockdown of Zyxin (F and G). (M–O) Cell proliferation assays in cell lines established in E–G. Data were expressed as the mean ± SEM of three independent experiments (H–O). **P < 0.01; ***P < 0.001 (t test).
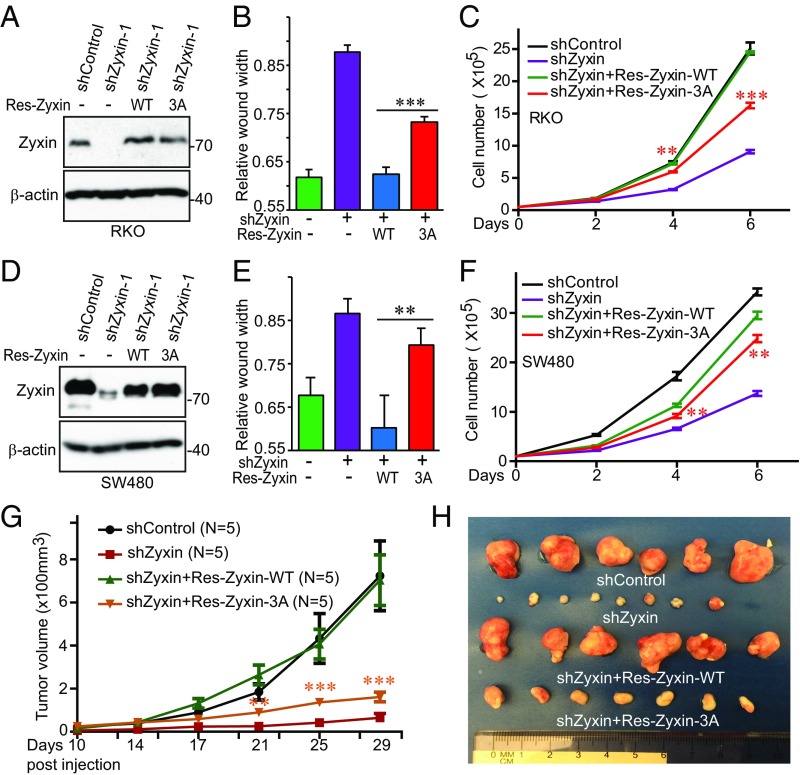
To directly determine the role of Zyxin in colon cancer, we depleted Zyxin (with two independent shRNAs) in various colon cancer cells (Fig. 3 E–G). Knockdown of Zyxin greatly reduced migration (as shown by the wound healing assay) (Fig. 3 H–J and SI Appendix, Fig. S5 A–C), anchorage-independent growth (Fig. 3 K and L and SI Appendix, Fig. S5 D and E), and proliferation (Fig. 3 M–O) in RKO, SW480, and HCT116 cells. Importantly, reexpression of shRNA-resistant wild-type Zyxin completely rescued all these phenotypes in RKO cells (Fig. 4 A–C). However, expression of nonphosphorylatable mutant Zyxin-3A (S281A/S308A/S344A) only partially restored these characteristics (Fig. 4 B and C), suggesting that mitotic phosphorylation is required for Zyxin oncogenic activity in colon cancer cells. Similar results were observed in SW480 cells (Fig. 4 D–F). As expected, Zyxin knockdown also greatly reduced tumor growth in animals, and cells bearing Zyxin-3A formed much smaller tumors compared with cells expressing wild-type Zyxin (Fig. 4 G and H). Knockdown efficiency was confirmed, and reexpression levels between wild-type and mutant Zyxin (Zyxin-3A) were similar in tumors (SI Appendix, Fig. S5F). These results suggest a critical role of Zyxin and its mitotic phosphorylation in colon cancer.
Fig. 4.
Mitotic phosphorylation of Zyxin is required for colon cancer tumorigenesis. (A) Establishment of Zyxin-knockdown cell lines expressing shRNA-resistant (Res) Zyxin or Zyxin-3A in RKO colon cancer cells. 3A: S281A/S308A/S344A. (B and C) Cell migration (wound healing) and proliferation assays in cell lines established in A. (D) Establishment of Zyxin-knockdown cell lines expressing shRNA-resistant (Res) Zyxin or Zyxin-3A in SW480 colon cancer cells. 3A, S281A/S308A/S344A. (E and F) Cell migration (wound healing) and proliferation assays in cell lines established in D. Data were expressed as the mean ± SEM of three independent experiments (B, C, E, and F). **P < 0.01; ***P < 0.001 (Res-Zyxin-WT vs. Res-Zyxin-3A) (t test). (G and H) Zyxin-3A failed to rescue tumor growth in Zyxin-knockdown RKO cells. RKO cell lines from A were s.c. inoculated into athymic nude mice (both left and right sides) and the representative tumors in each group were excised and photographed at the endpoint (H). Yellow asterisks mark the comparisons between Res-Zyxin-WT and Res-Zyxin-3A. **P < 0.01; ***P < 0.001 (t test).
Zyxin Regulates YAP Activity in Colon Cancer Cells.
Zyxin has been shown to be a positive regulator of YAP/Yki activity (5–8, 11). We confirmed that Zyxin knockdown increased p-YAP S127 levels and promoted YAP cytoplasmic localization (SI Appendix, Fig. S6 A and B). Consistent with reduced YAP activity, expression of the target gene CTGF was significantly reduced upon Zyxin knockdown in both RKO and SW480 cells (SI Appendix, Fig. S6C). F-actin cytoskeletal remodeling was shown to control YAP/Yki activity in mammalian cells and in Drosophila (5–9, 32–34). F-actin polymerization was inhibited by cofilin, for which activity is negatively controlled by phosphorylation at Serine3 (35–37). We found that phospho-S3 cofilin levels were reduced and F-actin formation/cytoskeleton was strongly inhibited upon Zyxin knockdown in colon cancer cells (SI Appendix, Fig. S6 D and E), suggesting that Zyxin might control YAP activity and subsequent cell migration and proliferation through F-actin remodeling.
To explore the functional interaction between Zyxin and YAP, we reexpressed YAP–S127A in Zyxin knockdown cells (SI Appendix, Fig. S6 F and G). Enhanced expression of YAP–S127A completely rescued the effects (migration and proliferation) of Zyxin knockdown in both SW480 and HCT116 cells (SI Appendix, Fig. S6 H–K), suggesting that Zyxin promotes colon cancer cell growth through YAP.
Zyxin Regulates CDK8 to Promote Migration and Proliferation in Colon Cancer Cells.
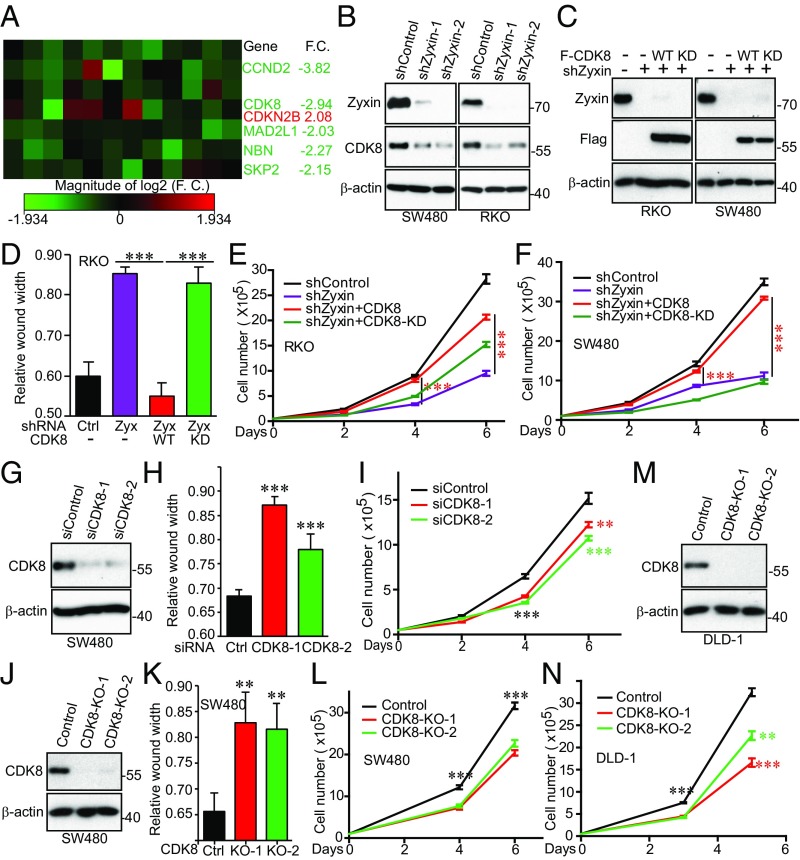
To elucidate the underlying mechanisms in which Zyxin regulates YAP activity and controls colon cancer cell growth, we performed an array analysis involving 84 cell-cycle regulators in control and Zyxin-knockdown cells. Five genes were down-regulated and one gene up-regulated (CDKN2B) in Zyxin-depleted cells compared with control cells (Fig. 5A). Most (except for NBN) were confirmed by qRT-PCR in an independent Zyxin-knockdown cell line (SI Appendix, Fig. S7A). Among these genes, CDK8 (cyclin-dependent kinase 8) attracted us, since it functions as an oncogene in colon cancer (17) and in melanoma (18). We confirmed that CDK8 protein levels were also reduced in Zyxin-knockdown RKO and SW480 cells (Fig. 5B) and restored by reexpression of exogenous Zyxin (SI Appendix, Fig. S7B). The Zyxin-3A mutant had similar activity to wild-type Zyxin in rescuing CDK8 levels, suggesting that mitotic phosphorylation of Zyxin is dispensable in controlling CDK8 expression (SI Appendix, Fig. S7B). Importantly, ectopic expression of CDK8, but not the kinase-dead form (CDK8-KD), substantially restored the migratory (assayed by wound healing) and proliferating ability of Zyxin-depleted RKO and SW480 cells (Fig. 5 C–F). Moreover, siRNA-mediated transient knockdown (two independent siRNAs) or CRISPR/double nickase-mediated knockout (KO; two independent gRNAs) (38) of CDK8 significantly impaired cell migration, proliferation, and anchorage-independent growth in SW480 cells (Fig. 5 G–L). CDK8 KO also reduced proliferation in DLD-1 cells (Fig. 5 M and N). Of note, both mRNA and protein levels of CDK8 were up-regulated in colon tumors compared with normal tissue (SI Appendix, Fig. S7 C and D). We found that CDK8 and Zyxin protein levels were positively correlated in metastatic colon cancer samples (SI Appendix, Fig. S7 E and F). Together, these data suggest that Zyxin promotes colon cancer cell growth via CDK8.
Fig. 5.
Zyxin regulates migration and proliferation through CDK8 in colon cancer cells. (A) Expression profiling of cell cycle regulators identified multiple genes including CDK8 that are regulated by Zyxin. The RT2 cell cycle arrays (Qiagen) were used and experiments were performed following manufacturer’s instructions. F.C., fold change. (B) CDK8 protein levels were decreased in Zyxin-knockdown cells. (C) Establishment of Zyxin-knockdown cells expressing Flag-tagged CDK8 or CDK8-KD (kinase dead, D173A). (D–F) Migration (wound healing) and proliferation assays in cell lines from C. Ectopic expression of CDK8, but not CDK8-KD, rescued growth defects of Zyxin-knockdown cells. (G–I) Transient knockdown (two independent siRNAs) of CDK8 inhibited SW480 cell migration (wound healing) and proliferation. (J) Establishment of CDK8 KO cell lines. Two independent gRNAs were used. (K and L) Cell migration (wound healing) assays and proliferation curves of cell lines from J. (M and N) CDK8 KO inhibited proliferation in DLD-1 cells. Data were expressed as the mean ± SEM of three independent experiments (D, F, H, I, K, L, and N). **P < 0.01; ***P < 0.001 (t test).
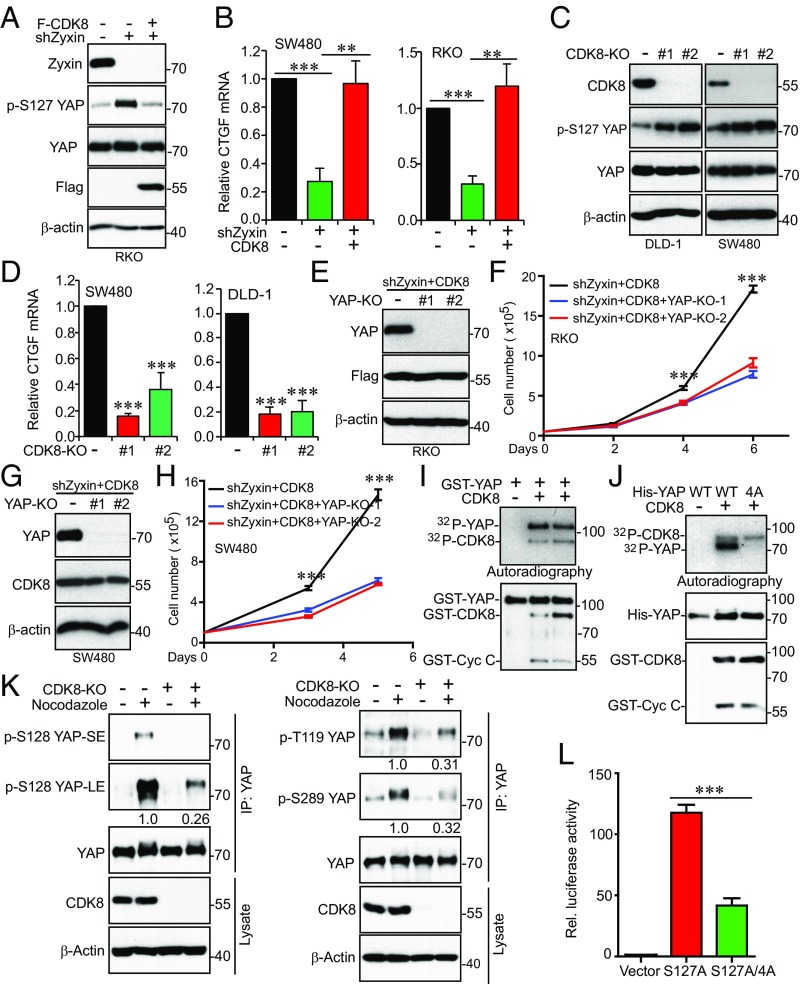
In line with the above functional studies, reexpression of CDK8 largely restored the YAP phosphorylation and target gene (CTGF) expression in Zyxin knockdown cells (Fig. 6 A and B), suggesting that Zyxin regulates YAP activity through CDK8.
Fig. 6.
CDK8 phosphorylates YAP and promotes its activation. (A and B) Zyxin regulates YAP activity through CDK8. Ectopic expression of CDK8 inhibited p-YAP S127 (A) and rescued CTGF (YAP target gene) expression (B) in Zyxin-knockdown cells. (C and D) CDK8 regulates YAP activity. CDK8 KO increased YAP phosphorylation (S127) (C) and inhibited CTGF expression (D). (E–H) YAP is required for CDK8-driven proliferation in colon cancer cells. (E and G) Establishment of YAP KO cell lines. CRISPR/double nickase-mediated (two independent gRNAs were used) KO of YAP in RKO and SW480 cells where Zyxin was depleted and CDK8 reexpressed. (F and H) Cell proliferation assays for cell lines from E and G. (I) In vitro kinase assays using GST-YAP as substrates. Two batches of CDK8 kinases (SignalChem) were used. (J) In vitro kinase assays with His-YAP-WT and -YAP-4A (T119A/S128A/S289A/S367A) proteins as substrates. (K) CDK8 phosphorylates YAP in mitosis. SW480 control and CDK8-KO cells were treated with DMSO or nocodazole for 16 h. Endogenous YAP proteins were immunoprecipitated and probed with phospho-antibodies against YAP T119, S128, S289 and total YAP antibodies. Relative phosphorylation (fold changes in signal intensity) was quantified by Image J and shown below the blots from three independent experiments. LE, long exposure; SE, short exposure. (L) CDK8-mediated phosphorylation stimulates YAP transcriptional activity. Luciferase reporter assays were done as we described (60, 64). Data were expressed as the mean ± SEM of three biological repeats (B, D, F, H, and L). **P < 0.01; ***P < 0.001 (t test).
CDK8 Requires YAP to Promote Cell Growth in Colon Cancer.
Based on our observations that Zyxin regulates YAP through CDK8 (Figs. 5 and 6 A and B), we reasoned that CDK8 also controls YAP activity. Indeed, we found that S127 phosphorylation of YAP was increased in CDK8-KO cells (Fig. 6C). Consistent with decreased activity of YAP in CDK8-KO cells, the expression of YAP target gene CTGF was inhibited upon CDK8 deletion (Fig. 6D). Furthermore, CRISPR-mediated KO (two independent gRNAs) of YAP strongly inhibited migration and proliferation in RKO and SW480 cells in which Zyxin was depleted and CDK8 reexpressed (Fig. 6 E–H), suggesting that YAP is essential for CDK8-driven tumorigenesis in colon cancer. Thus, we identified CDK8 as a positive regulator of YAP activity.
CDK8 Phosphorylates and Promotes YAP Activity.
CDK8 is a member of the mediator complex, which couples transcriptional regulators to control basal transcription (16). Since CDK8 is a kinase and regulates YAP transcriptional activity, we tested whether CDK8 directly phosphorylates YAP. In vitro kinase assays showed that purified CDK8/Cyclin C complex readily phosphorylated GST- or His-tagged YAP proteins (Fig. 6 I and J). CDK8 could also undergo autophosphorylation under these conditions (SI Appendix, Fig. S8). CDKs phosphorylate at S/T-P minimal consensus sequences. YAP contains seven conserved S/TPs (T119, S128, S138, S217, S289, S367, and T412; the T143, T154, and S340 sites do not exist in mouse and were excluded from further analysis). Of interest, mutating four amino acid positions (T119, S128, S289, and S367) to alanines (YAP-4A) completely blocked phosphorylation (Fig. 6J), suggesting that CDK8 phosphorylates these sites in YAP in vitro.
Given that both Zyxin (Figs. 1 and 2) and YAP are phosphorylated during mitosis (21, 39), we next explored whether CDK8 phosphorylates YAP in mitosis. Interestingly, S128 phosphorylation was significantly increased in nocodazole-arrested mitotic cells (Fig. 6K). Consistent with our previous study (21), phosphorylation of T119 and S289 was also increased in mitotic cells (Fig. 6K). Importantly, CDK8 deletion largely blocked the phosphorylation of all three sites induced by mitotic arrest, suggesting that CDK8 is a relevant YAP kinase in vivo, specifically in mitosis (Fig. 6K). Using reporter assays, we next examined whether phosphorylation of YAP affects its activity. Mutating these four sites to alanines greatly suppressed YAP–S127A activity (Fig. 6L). These observations suggest that CDK8 directly phosphorylates and promotes YAP transcriptional activity, identifying YAP as a direct substrate for CDK8.
YAP Phosphorylation Is Required for CDK8-Driven Migration and Proliferation.
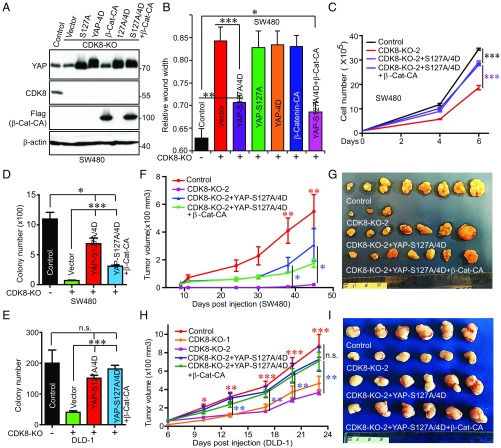
To further explore the functional relationship between CDK8 and YAP, we performed complementary experiments. Ectopic expression of the constitutively active form of YAP (YAP–S127A) had little impact on proliferation and migration in SW480 and DLD-1 CDK8-KO cells (SI Appendix, Fig. S10 A and B). Expression of phospho-mimetic mutant YAP-4D failed to rescue proliferation and migration in CDK8-KO cells (Fig. 7 A and B). However, expression of YAP-S127A/4D (highest activity) substantially (but not completely) restored CDK8-KO–mediated migration, proliferation, and anchorage-independent growth, suggesting that CDK8-mediated phosphorylation is essential for full activation of YAP (Fig. 7 B–E and SI Appendix, Fig. S10 C–E), which is consistent with our earlier observations (Fig. 6). CDK8 KO significantly inhibited tumor growth in vivo in both SW480 and DLD-1 cells (Fig. 7 F–I). Ectopic expression of YAP–S127A/4D also largely restored tumor growth of CDK8-KO cells in animals (Fig. 7 F–I).
Fig. 7.
YAP phosphorylation is required for CDK8-driven tumorigenesis in colon cancer cells. (A) Establishment of cell lines expressing various YAP mutants and/or β-catenin in CDK8 KO SW480 cells. CA, constitutive active form (S33Y mutant). (B–D) Functional assays of cell lines established in A. Cell migration (wound healing) assays in various cell lines as indicated (B). Cell proliferation curves of cell lines with various gene expression status (C). Anchorage-independent growth (colony assays in soft agar) in cell lines as indicated (D). Data (B–D) are from three to four independent experiments and expressed as the mean ± SEM. *P < 0.05; **P < 0.01; ***P < 0.001 (t test). (E) Anchorage-independent growth in various DLD-1 cells lines as indicated. Data are from three independent experiments and expressed as the mean ± SEM. ***P < 0.001. n.s., not significant. (F and G) Tumor growth curves and images of all tumors (at the endpoint) from SW480 cell lines. 2.5 × 106 cells (1:1 mixed with Matrigel) were s.c. inoculated into athymic nude mice on both flanks. n = 5 mice and 10 injections per group. Only three tumors were formed at the last two time points in the CDK8-KO group and the other three groups formed seven to eight tumors (G). Red asterisks mark the comparisons between control and CDK8-KO-2; Blue asterisks mark the comparisons between YAP-S127A/4D and CDK8-KO-2. *P < 0.05; **P < 0.01 (t test). (H and I) Tumor growth curves and representative images of tumors from DLD-1 cell lines. 1.0 × 106 cells (1:1 mixed with Matrigel) were s.c. inoculated into athymic nude mice on both flanks (5 mice and 10 injections/group, 10 tumors were averaged for each group at each time point). Red asterisks mark the comparisons between control and CDK8-KO-2; Blue asterisks mark the comparisons between YAP-S127A/4D and CDK8-KO-2. *P < 0.05; **P < 0.01; ***P < 0.001 (t test). n.s., not significant.
A previous report showed that CDK8 functions as a colon oncogene through regulating β-catenin (17). Consistent with this study, the inhibitory phosphorylation of β-catenin was increased, and target gene (LEF1) expression was inhibited in CDK8-KO cells (SI Appendix, Fig. S9 A and B). Interestingly, KO of β-catenin also greatly impaired migration and proliferation in RKO cells with Zyxin depletion and CDK8 reexpression (SI Appendix, Fig. S9 C–E). These findings suggest that CDK8 controls YAP and β-catenin activity, both of which are critical regulators in colon cancer.
Like YAP–S127A or YAP-4D, enhanced expression of β-catenin–S33Y (active form) had little effect on CDK8-KO cell growth (Fig. 7B and SI Appendix, Fig. S10 A, B, and D), suggesting that active β-catenin alone is not sufficient to rescue CDK8-KO–mediated cell growth. Coexpression of β-catenin-S33Y with YAP–S127A/4D did not further increase growth compared with YAP–S127A/4D alone in CDK8-KO cells (Fig. 7 B–I and SI Appendix, Fig. S10 D and E). Our data suggest that CDK8 promotes YAP activation through a twofold mechanism: canonical S127 phosphorylation and direct phosphorylation. Both YAP and β-catenin are required for CDK8-driven tumorigenesis in colon cancer. Altogether, our findings indicate that Zyxin promotes colon tumorigenesis in a mitotic phosphorylation-dependent manner and through CDK8-mediated YAP activation.
Discussion
Our study revealed that the cell-adhesion protein Zyxin promotes colon cancer cell growth by controlling CDK8 expression. It is currently not known how Zyxin regulates CDK8 expression. CDK8 mRNA expression is negatively regulated by the histone variant macroH2A (18). A recent study demonstrated that the E3 ligase, Skp2, promotes macroH2A ubiquitination and degradation (19). Interestingly, in our array analysis, Skp2 is one of the five genes down-regulated by Zyxin depletion (Fig. 5 and SI Appendix, Fig. S7A). Therefore, it is possible that Zyxin controls CDK8 expression through the Skp2–macroH2A axis. Since both Zyxin and CDK8 levels are increased in colon cancer patients (Fig. 3 and SI Appendix, Fig. S7 C and D) and possess oncogenic roles in colon cancer development (17), it will be interesting to explore the biological significance of Skp2 and macroH2A in colon cancer and their functional interactions with Zyxin/CDK8.
CDK8 positively regulates β-catenin transcriptional activity in colon cancer cells (17), although the precise mechanism remains obscure. It may involve the phosphorylation of E2F1 and relieve E2F1-mediated repression on β-catenin/TCF signaling activity (17, 40, 41). The kinase activity of CDK8 is necessary for β-catenin–driven transcription (17). Furthermore, a dominant-negative TCF construct, previously shown to inhibit β-catenin–induced cellular transformation, also only partially abrogated CDK8 activity (17). This notion is supported by the observations that several profiling experiments failed to identify the broad deregulation of β-catenin signaling in CDK8-disturbed cells (16). These results strongly indicate that CDK8 controls additional factor(s) also outside the β-catenin signaling pathway to promote colon cancer cell growth (i.e., other factors in addition to β-catenin exist downstream of CDK8). Although our data (SI Appendix, Fig. S9 D and E) and previous studies (17) showed that β-catenin–KO suppressed the CDK8-driven cell migration and proliferation, restoration of β-catenin in CDK8-KO cells was not sufficient to support colon cancer cell growth (SI Appendix, Fig. S10), further suggesting that β-catenin signaling is not the main (or only) effector of CDK8. Our study highlights the importance of YAP in mediating CDK8-driven tumorigenesis in colon cancer (Figs. 6 and 7). CDK8 regulates YAP activity through a twofold mechanism: phosphorylation on S127/localization and direct phosphorylation at T119/S128/S289/S367 (Figs. 6 and 7). CDK8-mediated direct phosphorylation promotes YAP further activation and is required for CDK8-KO cell growth (Fig. 7). These observations not only added additional layers of YAP regulation, but also raised complex questions. For example, S128 has been shown to be phosphorylated by Nemo-like kinase (42, 43), and T119/S289/S367 are phosphorylated by CDK1 (21). How these kinases and phosphorylation are coordinated in cells requires further investigation. Another question is: How does CDK8 regulate YAP S127 phosphorylation and localization? YAP S127 is largely mediated by Lats1/2 kinases in the Hippo pathway; however, we did not observe significant changes of Lats1/2 kinase activity (revealed by phosphorylation levels of their activation loop) in CDK8-KO cells and -intact cells, suggesting a Hippo-independent regulation of YAP activity.
Our study showed that the active β-catenin mutant (β-catenin–S33Y) had little effect on rescuing the phenotypes of CDK8 KO in colon cancer cells (SI Appendix, Fig. S10). This implies that additional target(s) exists or, alternatively, that β-catenin–S33Y (like YAP–S127A) is not fully active in CDK8-deleted cells. The similarity between YAP and β-catenin prompted us to reason that β-catenin is a direct substrate for CDK8. Indeed, β-catenin sequence analysis revealed three S/TP phospho-consensus (S191/S246/S605) as potential sites for CDK8 phosphorylation, and these sites have been shown to be important for β-catenin cellular localization and activity (44).
The global burden of colon cancer is rising steadily. Although much progress has been made in therapeutic approaches, for patients with locally advanced primary or recurrent colon cancer, often unresectable for cure, survival remains poor (45). Furthermore, available treatment options for colon cancer are very limited due to lack of validated targets. Our study has demonstrated the critical role and regulatory mechanisms of the Zyxin–CDK8–YAP/β-catenin signaling in colon cancer. These studies suggest that this axis is a potential therapeutic target and provide several options for targeted therapy using small-molecule inhibitors in colon cancer. For example, CDK8 function requires its kinase activity, and several selective inhibitors have been developed (16, 46, 47). Furthermore, animal studies confirmed the potential efficacy of CDK8 inhibitors (48), although interpretation of CDK8 inhibitor data requires caution since CDK8 may also have a tumor-suppressive function during early stages of colon tumorigenesis (49). VP was recently identified as an effective compound for preventing YAP activity (31). Interestingly, VP (without light activation) significantly inhibited YAP activity in vitro and suppressed YAP-driven tumorigenesis in the mouse liver (31). Subsequent studies further confirmed that VP is also effective in inhibiting tumor growth in prostate cancer (50), uveal melanoma (51, 52), and colon cancer in vivo (53), suggesting that inhibiting YAP is a viable strategy in cancer treatment. Cyclin D1 is frequently deregulated in colon cancer and is a known target of β-catenin signaling (17, 54). Interestingly, Cyclin D1 was also induced by YAP activation in the intestine (55) and was identified as a direct target of YAP/TEAD in mesothelioma cells (56). Therefore, Cyclin D1 is a common target of YAP and β-catenin. Cyclin D1 is up-regulated (probably through YAP/β-catenin activation) in the ApcMin/+ mice, and deletion of Cyclin D1 blocks/reduces ApcMin/+-driven adenoma formation (57). Acute deletion of Cyclin D1 in adult mice does not affect the animal’s health (58). These observations suggest that targeting Cyclin D1 (e.g., with Palbociclib/PD-0332991, which is a Food and Drug Administration-approved selective inhibitor of Cyclin D1–CDK4/6) would be a viable strategy in colon cancer, especially in CDK8 and YAP/β-catenin–activated subtypes.
Materials and Methods
CRISPR/Double Nickase-Mediated KO.
To construct the all-in-one CRISPR/Cas9n (double nickase) plasmid targeting β-catenin, the sense and antisense oligonucleotides from Table 1 were synthesized, annealed, and Golden Gate assembled into the pX330A_D10A-1 × 2-EGFP (38) and pX330S-2 vectors as described (59). After the two vectors were generated, a final Golden Gate assembly was performed to generate the all-in-one vector as described (59). The resulting pX330A_D10A-1 × 2-EGFP–β-catenin-1 or -2 construct was transfected into cells, and GFP-positive clones were selected by flow cytometry-based cell sorting. CDK8 and YAP CRISPR/double nickase KO plasmids were purchased from Santa Cruz Biotechnology, and transfection and clone selection were done as instructed by the manufacturer.
Table 1.
β-catenin guide sequences
| Oligo name | Sequence |
| β-catenin-1A-Fwd | CACCGGACGGTCGGACTCCCGCGG |
| β-catenin-1A-Rev | aaacCCGCGGGAGTCCGACCGTCC |
| β-catenin-1B-Fwd | CACCGCGACCGTCCTCGACCTGCGG |
| β-catenin-1B-Rev | aaacCCGCAGGTCGAGGACGGTCGC |
| β-catenin-2A-Fwd | CACCGCGAGGACGGTCGGACTCCCG |
| β-catenin-2A-Rev | aaacCGGGAGTCCGACCGTCCTCGC |
| β-catenin-2B-Fwd | CACCGACCTGCGGTGGCGGCTCGG |
| β-catenin-2B-Rev | aaacCCGAGCCGCCACCGCAGGTC |
Fwd, forward; Rev, reverse.
Phos-Tag and Western Blot Analysis.
Phos-tag was obtained from Wako Pure Chemical Industries, Ltd. (catalog no. 304-93521) and used at a concentration of 20 µM (with 100 µM MnCl2) in 8% SDS-acrylamide gels. Before transferring, the gels were equilibrated in transfer buffer containing 10 mM EDTA two times, each for 10 min. The gels were then soaked in transfer buffer (without EDTA) for another 10 min. Western blotting and lambda phosphatase treatment assays were done as described (20).
Immunofluorescence Staining and Confocal Microscopy.
Cell fixation, permeabilization, fluorescence staining, and microscopy were done as described (29). The stained cells were mounted with Fluoromount (Vector Laboratories) and visualized on an LSM710 Zeiss fluorescence microscope (Carl Zeiss). The Slidebook 4.2 software (Intelligent Imaging Innovations) was used for analyzing and processing all immunofluorescence images. F-actin was stained by using rhodamine-labeled phalloidin (Cytoskeleton). YAP cellular localization was visualized by YAP immunofluorescence staining with an Alexa Fluor 647-conjugated YAP antibody (Cell Signaling Technology). For peptide blocking, the phospho-Zyxin antibodies were first neutralized by an excess of immunizing (phosphorylated) peptides (1 µg/mL for 1 h at room temperature). The antibody (containing the phospho-peptide) was then used for staining in parallel with staining using antibodies with no peptide or non-phospho-peptide.
Cell Proliferation, Cell Migration (Wound Healing), and Colony Formation Assays.
Colony formation assays in soft agar were performed as described (60). Cells (5,000 per well) were seeded in six-well plates, and colonies were counted by ImageJ online. For cell proliferation assays, cells (SW480 and HCT116: 100,000 per well; RKO and DLD-1: 50,000 per well) were seeded in a six-well plate in triplicate. Cells were counted by the automated cell counter (Invitrogen) at indicated time points and proliferation curves were made based on the cell number in each well from at least three independent experiments. For wound healing (migration) assays, the cell monolayer was scratched by a P200 pipet tip when cells were confluent. Cells were washed three times with PBS and incubated with 2 mL of serum-free medium for an additional 24 h. The wound width was measured by QCapture Software (QImaging).
qRT-PCR.
Total RNA isolation, RNA reverse transcription, and qRT-PCR were done as we have described (61).
IHC Staining and Scoring.
The TMA slides consisted of 66 colon adenocarcinomas and 35 normal adjacent tissues collected from colon cancer patients at the University of Nebraska Medical Center treated during 2008–2009. Slide deparaffinization, antigen retrieving, and blocking were performed as we have described (60). Tumors from RKO xenografts (control and Zyxin knockdown) were also used for Zyxin antibody validation. The sections were then stained with anti-Zyxin antibody (at 1:100 dilutions; Cell Signaling Technology) by using a Histostain-Plus IHC kit following the manufacturer’s instructions (Invitrogen). The TMAs were also stained with anti-CDK8 antibody (IHC-00704, at 1:300 dilutions; Bethyl Laboratory). Cell nuclei were stained with hematoxylin. Ventana iScan HT (Roche) was used for slide scanning with a 20× objective lens. The staining results were independently evaluated by three researchers, including one pathologist (S.M.L.). The staining intensity (a scale of 0–3 was used: 0, negative; 1, weak; 2, moderate; and 3, strong) was scored (62).
Animal Studies.
For in vivo xenograft studies, RKO cells (shControl, shZyxin, shZyxin with Res-Zyxin-WT, or Res-Zyxin-3A) (1.0 × 106 cells each injection, mixed with Matrigel at 1:1 ratio) were s.c. injected into the left or right flank of 6-wk-old male athymic nude mice (Nude-Foxn1nu; Envigo). Five animals (10 injections) were used per group. Similar approaches were used for DLD-1 (1.0 × 106 cells per injection) and SW480 (2.5 × 106 cells per injection) cells in Fig. 7. Tumor sizes were measured at the indicated time points. Tumor volume (V) was calculated by the formula: V = 0.5 × length × width2 (60). Mice were euthanized at the end of the experiments, and the tumors were excised for subsequent analysis. The animals were housed in pathogen-free facilities. All animal experiments were approved by the University of Nebraska Medical Center Institutional Animal Care and Use Committee.
Statistical Analysis.
Statistical significance was evaluated by using a two-tailed, unpaired Student’s t test. The Wilcoxon rank sum test was used to compare the IHC staining data between groups. A P value of <0.05 was considered as indicating statistical significance.
Supplementary Material
Acknowledgments
We thank Dr. Shian Wu (Nankai University, China) for the Zyxin shRNA constructs, Dr. Eek-hoon Jho (University of Seoul, Korea) for the p-S128 YAP antibody, and Xiaolong Yang (Queen’s University, Canada) for the TetOn-shCDK1 cell line. We thank Dr. Wayne Xu (University of Manitoba, Canada) for analyzing the mRNA expression levels of Zyxin and CDK8 from TCGA. We also thank Dr. Joyce Solheim for critical reading and comments on the manuscript. All fluorescence images were acquired by Zeiss LSM 710 or LSM 800 confocal microscopes at the Advanced Microscopy Core at the University of Nebraska Medical Center. The core is supported in part by National Institutes of Health (NIH) Grant P30 GM106397. Research in the J.D. laboratory is supported by Fred and Pamela Buffett Cancer Center Support Grant P30 CA036727; NIH Grants P30 GM106397 and R01 GM109066; and Department of Defense Health Program Grant W81XWH-14-1-0150.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. K.-L.G. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1800621115/-/DCSupplemental.
References
- 1.Beckerle MC. Zyxin: Zinc fingers at sites of cell adhesion. BioEssays. 1997;19:949–957. doi: 10.1002/bies.950191104. [DOI] [PubMed] [Google Scholar]
- 2.Hirata H, Tatsumi H, Sokabe M. Zyxin emerges as a key player in the mechanotransduction at cell adhesive structures. Commun Integr Biol. 2008;1:192–195. doi: 10.4161/cib.1.2.7001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hirata H, Tatsumi H, Sokabe M. Mechanical forces facilitate actin polymerization at focal adhesions in a zyxin-dependent manner. J Cell Sci. 2008;121:2795–2804. doi: 10.1242/jcs.030320. [DOI] [PubMed] [Google Scholar]
- 4.Nix DA, Beckerle MC. Nuclear-cytoplasmic shuttling of the focal contact protein, zyxin: A potential mechanism for communication between sites of cell adhesion and the nucleus. J Cell Biol. 1997;138:1139–1147. doi: 10.1083/jcb.138.5.1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gaspar P, Holder MV, Aerne BL, Janody F, Tapon N. Zyxin antagonizes the FERM protein expanded to couple F-actin and Yorkie-dependent organ growth. Curr Biol. 2015;25:679–689. doi: 10.1016/j.cub.2015.01.010. [DOI] [PubMed] [Google Scholar]
- 6.Fernández BG, et al. Actin-capping protein and the Hippo pathway regulate F-actin and tissue growth in Drosophila. Development. 2011;138:2337–2346. doi: 10.1242/dev.063545. [DOI] [PubMed] [Google Scholar]
- 7.Rauskolb C, Pan G, Reddy BV, Oh H, Irvine KD. Zyxin links fat signaling to the hippo pathway. PLoS Biol. 2011;9:e1000624. doi: 10.1371/journal.pbio.1000624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sansores-Garcia L, et al. Modulating F-actin organization induces organ growth by affecting the Hippo pathway. EMBO J. 2011;30:2325–2335. doi: 10.1038/emboj.2011.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aragona M, et al. A mechanical checkpoint controls multicellular growth through YAP/TAZ regulation by actin-processing factors. Cell. 2013;154:1047–1059. doi: 10.1016/j.cell.2013.07.042. [DOI] [PubMed] [Google Scholar]
- 10.Feng X, et al. Hippo-independent activation of YAP by the GNAQ uveal melanoma oncogene through a trio-regulated rho GTPase signaling circuitry. Cancer Cell. 2014;25:831–845. doi: 10.1016/j.ccr.2014.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ma B, et al. Zyxin-Siah2-Lats2 axis mediates cooperation between Hippo and TGF-β signalling pathways. Nat Commun. 2016;7:11123. doi: 10.1038/ncomms11123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pan D. The hippo signaling pathway in development and cancer. Dev Cell. 2010;19:491–505. doi: 10.1016/j.devcel.2010.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yu FX, Zhao B, Guan KL. Hippo pathway in organ size control, tissue homeostasis, and cancer. Cell. 2015;163:811–828. doi: 10.1016/j.cell.2015.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hong AW, Meng Z, Guan KL. The Hippo pathway in intestinal regeneration and disease. Nat Rev Gastroenterol Hepatol. 2016;13:324–337. doi: 10.1038/nrgastro.2016.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meng Z, Moroishi T, Guan KL. Mechanisms of Hippo pathway regulation. Genes Dev. 2016;30:1–17. doi: 10.1101/gad.274027.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rzymski T, Mikula M, Wiklik K, Brzózka K. CDK8 kinase—An emerging target in targeted cancer therapy. Biochim Biophys Acta. 2015;1854:1617–1629. doi: 10.1016/j.bbapap.2015.05.011. [DOI] [PubMed] [Google Scholar]
- 17.Firestein R, et al. CDK8 is a colorectal cancer oncogene that regulates beta-catenin activity. Nature. 2008;455:547–551. doi: 10.1038/nature07179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kapoor A, et al. The histone variant macroH2A suppresses melanoma progression through regulation of CDK8. Nature. 2010;468:1105–1109. doi: 10.1038/nature09590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu D, et al. Skp2-macroH2A1-CDK8 axis orchestrates G2/M transition and tumorigenesis. Nat Commun. 2015;6:6641. doi: 10.1038/ncomms7641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xiao L, et al. KIBRA protein phosphorylation is regulated by mitotic kinase aurora and protein phosphatase 1. J Biol Chem. 2011;286:36304–36315. doi: 10.1074/jbc.M111.246850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang S, et al. CDK1 phosphorylation of YAP promotes mitotic defects and cell motility and is essential for neoplastic transformation. Cancer Res. 2013;73:6722–6733. doi: 10.1158/0008-5472.CAN-13-2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang L, et al. CDK1 phosphorylation of TAZ in mitosis inhibits its oncogenic activity. Oncotarget. 2015;6:31399–31412. doi: 10.18632/oncotarget.5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen X, Chen Y, Dong J. MST2 phosphorylation at serine 385 in mitosis inhibits its tumor suppressing activity. Cell Signal. 2016;28:1826–1832. doi: 10.1016/j.cellsig.2016.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen X, Stauffer S, Chen Y, Dong J. Ajuba phosphorylation by CDK1 promotes cell proliferation and tumorigenesis. J Biol Chem. 2016;291:14761–14772. doi: 10.1074/jbc.M116.722751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hergovich A, Hemmings BA. Hippo signalling in the G2/M cell cycle phase: Lessons learned from the yeast MEN and SIN pathways. Semin Cell Dev Biol. 2012;23:794–802. doi: 10.1016/j.semcdb.2012.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zeng Y, et al. Cyclin-dependent kinase 1 (CDK1)-mediated mitotic phosphorylation of the transcriptional co-repressor Vgll4 inhibits its tumor-suppressing activity. J Biol Chem. 2017;292:15028–15038. doi: 10.1074/jbc.M117.796284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nigg EA. Cellular substrates of p34(cdc2) and its companion cyclin-dependent kinases. Trends Cell Biol. 1993;3:296–301. doi: 10.1016/0962-8924(93)90011-o. [DOI] [PubMed] [Google Scholar]
- 28.Hornbeck PV, et al. PhosphoSitePlus, 2014: Mutations, PTMs and recalibrations. Nucleic Acids Res. 2015;43:D512–D520. doi: 10.1093/nar/gku1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang L, et al. KIBRA regulates aurora kinase activity and is required for precise chromosome alignment during mitosis. J Biol Chem. 2012;287:34069–34077. doi: 10.1074/jbc.M112.385518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Diepenbruck M, et al. Tead2 expression levels control the subcellular distribution of Yap and Taz, zyxin expression and epithelial-mesenchymal transition. J Cell Sci. 2014;127:1523–1536. doi: 10.1242/jcs.139865. [DOI] [PubMed] [Google Scholar]
- 31.Liu-Chittenden Y, et al. Genetic and pharmacological disruption of the TEAD-YAP complex suppresses the oncogenic activity of YAP. Genes Dev. 2012;26:1300–1305. doi: 10.1101/gad.192856.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dai X, et al. Phosphorylation of angiomotin by Lats1/2 kinases inhibits F-actin binding, cell migration, and angiogenesis. J Biol Chem. 2013;288:34041–34051. doi: 10.1074/jbc.M113.518019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhao B, et al. Cell detachment activates the Hippo pathway via cytoskeleton reorganization to induce anoikis. Genes Dev. 2012;26:54–68. doi: 10.1101/gad.173435.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mana-Capelli S, Paramasivam M, Dutta S, McCollum D. Angiomotins link F-actin architecture to Hippo pathway signaling. Mol Biol Cell. 2014;25:1676–1685. doi: 10.1091/mbc.E13-11-0701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ostrowska Z, Moraczewska J. Cofilin—A protein controlling dynamics of actin filaments. Postepy Hig Med Dosw. 2017;71:339–351. doi: 10.5604/01.3001.0010.3818. [DOI] [PubMed] [Google Scholar]
- 36.Mizuno K. Signaling mechanisms and functional roles of cofilin phosphorylation and dephosphorylation. Cell Signal. 2013;25:457–469. doi: 10.1016/j.cellsig.2012.11.001. [DOI] [PubMed] [Google Scholar]
- 37.Prunier C, Prudent R, Kapur R, Sadoul K, Lafanechère L. LIM kinases: Cofilin and beyond. Oncotarget. 2017;8:41749–41763. doi: 10.18632/oncotarget.16978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stauffer S, et al. CDK1-mediated mitotic phosphorylation of PBK is involved in cytokinesis and inhibits its oncogenic activity. Cell Signal. 2017;39:74–83. doi: 10.1016/j.cellsig.2017.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bui DA, et al. Cytokinesis involves a nontranscriptional function of the Hippo pathway effector YAP. Sci Signal. 2016;9:ra23. doi: 10.1126/scisignal.aaa9227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhao J, Ramos R, Demma M. CDK8 regulates E2F1 transcriptional activity through S375 phosphorylation. Oncogene. 2013;32:3520–3530. doi: 10.1038/onc.2012.364. [DOI] [PubMed] [Google Scholar]
- 41.Morris EJ, et al. E2F1 represses beta-catenin transcription and is antagonized by both pRB and CDK8. Nature. 2008;455:552–556. doi: 10.1038/nature07310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hong AW, et al. Osmotic stress-induced phosphorylation by NLK at Ser128 activates YAP. EMBO Rep. 2017;18:72–86. doi: 10.15252/embr.201642681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moon S, et al. Phosphorylation by NLK inhibits YAP-14-3-3-interactions and induces its nuclear localization. EMBO Rep. 2017;18:61–71. doi: 10.15252/embr.201642683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu X, et al. Rac1 activation controls nuclear localization of beta-catenin during canonical Wnt signaling. Cell. 2008;133:340–353. doi: 10.1016/j.cell.2008.01.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Haddock MG. Intraoperative radiation therapy for colon and rectal cancers: A clinical review. Radiat Oncol. 2017;12:11. doi: 10.1186/s13014-016-0752-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Koehler MF, et al. Development of a potent, specific CDK8 kinase inhibitor which phenocopies CDK8/19 knockout cells. ACS Med Chem Lett. 2016;7:223–228. doi: 10.1021/acsmedchemlett.5b00278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mallinger A, et al. Discovery of potent, selective, and orally bioavailable small-molecule modulators of the mediator complex-associated kinases CDK8 and CDK19. J Med Chem. 2016;59:1078–1101. doi: 10.1021/acs.jmedchem.5b01685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Clarke PA, et al. Assessing the mechanism and therapeutic potential of modulators of the human mediator complex-associated protein kinases. eLife. 2016;5:e20722. doi: 10.7554/eLife.20722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McCleland ML, et al. Cdk8 deletion in the Apc(Min) murine tumour model represses EZH2 activity and accelerates tumourigenesis. J Pathol. 2015;237:508–519. doi: 10.1002/path.4596. [DOI] [PubMed] [Google Scholar]
- 50.Nguyen LT, et al. ERG activates the YAP1 transcriptional program and induces the development of age-related prostate tumors. Cancer Cell. 2015;27:797–808. doi: 10.1016/j.ccell.2015.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lyubasyuk V, Ouyang H, Yu FX, Guan KL, Zhang K. YAP inhibition blocks uveal melanogenesis driven by GNAQ or GNA11 mutations. Mol Cell Oncol. 2014;2:e970957. doi: 10.4161/23723548.2014.970957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yu FX, et al. Mutant Gq/11 promote uveal melanoma tumorigenesis by activating YAP. Cancer Cell. 2014;25:822–830. doi: 10.1016/j.ccr.2014.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang H, et al. Tumor-selective proteotoxicity of verteporfin inhibits colon cancer progression independently of YAP1. Sci Signal. 2015;8:ra98. doi: 10.1126/scisignal.aac5418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tetsu O, McCormick F. Beta-catenin regulates expression of cyclin D1 in colon carcinoma cells. Nature. 1999;398:422–426. doi: 10.1038/18884. [DOI] [PubMed] [Google Scholar]
- 55.Camargo FD, et al. YAP1 increases organ size and expands undifferentiated progenitor cells. Curr Biol. 2007;17:2054–2060. doi: 10.1016/j.cub.2007.10.039. [DOI] [PubMed] [Google Scholar]
- 56.Mizuno T, et al. YAP induces malignant mesothelioma cell proliferation by upregulating transcription of cell cycle-promoting genes. Oncogene. 2012;31:5117–5122. doi: 10.1038/onc.2012.5. [DOI] [PubMed] [Google Scholar]
- 57.Hulit J, et al. Cyclin D1 genetic heterozygosity regulates colonic epithelial cell differentiation and tumor number in ApcMin mice. Mol Cell Biol. 2004;24:7598–7611. doi: 10.1128/MCB.24.17.7598-7611.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Choi YJ, et al. The requirement for cyclin D function in tumor maintenance. Cancer Cell. 2012;22:438–451. doi: 10.1016/j.ccr.2012.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sakuma T, Nishikawa A, Kume S, Chayama K, Yamamoto T. Multiplex genome engineering in human cells using all-in-one CRISPR/Cas9 vector system. Sci Rep. 2014;4:5400. doi: 10.1038/srep05400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dong J, et al. Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell. 2007;130:1120–1133. doi: 10.1016/j.cell.2007.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xiao L, Chen Y, Ji M, Dong J. KIBRA regulates Hippo signaling activity via interactions with large tumor suppressor kinases. J Biol Chem. 2011;286:7788–7796. doi: 10.1074/jbc.M110.173468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Song S, et al. Hippo coactivator YAP1 upregulates SOX9 and endows esophageal cancer cells with stem-like properties. Cancer Res. 2014;74:4170–4182. doi: 10.1158/0008-5472.CAN-13-3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang L, et al. The hippo pathway effector YAP regulates motility, invasion, and castration-resistant growth of prostate cancer cells. Mol Cell Biol. 2015;35:1350–1362. doi: 10.1128/MCB.00102-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yang S, et al. Active YAP promotes pancreatic cancer cell motility, invasion and tumorigenesis in a mitotic phosphorylation-dependent manner through LPAR3. Oncotarget. 2015;6:36019–36031. doi: 10.18632/oncotarget.5935. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.