Significance
Brain aging and neural degenerative diseases are characterized by chronic low-degree inflammation, also known as “inflammaging.” Histone acetylation is a classical mark for active gene expression. Here, using H3K27ac ChIP-seq and RNA-seq of human and mouse brain samples, we found that, during aging, overactivated inflammation-related genes are marked by decreased broad gene-body hyperacetylation. Restoring the gene-body hyper H3K27ac by histone deacetylase inhibitors suppressed such overactivation, implying a potential fine-tuning function of broad gene-body H3K27ac on these inflammaging genes. Our study uncovered a mode of epigenetic regulation of the brain inflammaging genes, suggesting the reversibility of the inflammaging process and a potential angle for intervention of aging-related brain function decline and neural degenerative diseases.
Keywords: inflammation, aging, brain, H3K27ac, gene-body acetylation
Abstract
Brain “inflammaging,” a low-grade and chronic inflammation, is a major hallmark for aging-related neurodegenerative diseases. Here, by profiling H3K27ac and gene expression patterns in human and mouse brains, we found that age-related up-regulated (Age-Up) and down-regulated (Age-Down) genes have distinct H3K27ac patterns. Although both groups show promoter H3K27ac, the Age-Up genes, enriched for inflammation-related functions, are additionally marked by broad H3K27ac distribution over their gene bodies, which is progressively reduced during aging. Age-related gene expression changes can be predicted by gene-body H3K27ac level. Contrary to the presumed transcription activation function of promoter H3K27ac, we found that broad gene-body hyper H3K27ac suppresses overexpression of inflammaging genes. Altogether, our findings revealed opposite regulations by H3K27ac of Age-Up and Age-Down genes and a mode of broad gene-body H3K27ac in repressing transcription.
Aging has been characterized as the progressive loss of function in multiple tissues (1) and is a major risk factor for many diseases, such as diabetes, neurodegenerative diseases, and cancers (2). A low-grade and chronic inflammation, termed “inflammaging” (3), has been found significantly associated with both degenerative disease and death in elderly people (4). Major neurodegenerative diseases, such as Alzheimer’s and Parkinson’s disease, all have chronic nonresolving tissue inflammation as a common hallmark (5). Activation of NF-κB and the NLRP3 inflammasome are the major factors responsible for the production of proinflammatory molecules (6, 7). Loss of IKK-β, the upstream activator of NFκB, can restore the gonadotropin-releasing hormone levels and promote life span in mice (8). However, it is not known whether inflammaging, as a chronic state, is maintained at the epigenetic level, or whether it can be a target for intervention.
Accumulating evidence has revealed epigenetic alterations as a hallmark of aging (9). Genome-wide epigenomics studies have made it possible to identify potential anti-aging targets based on epigenetic mechanisms. For example, in Caenorhabditis elegans, both loss of function of the H3K4 trimethyltransferase complex and H3K27me3 demethylase utx-1 led to global changes in H3K4me3 and H3K27me3, respectively, and extended life span (10–12). In addition, loss of SIRT6, an H3K9ac deacetylase, resulted in hyperactivation of NFκB signaling and accelerated aging (13).
Here, we generated genome-wide profiles of histone H3K27 acetylation (H3K27ac) by ChIP-seq and investigated its association with the age-related up-regulated (Age-Up) and down-regulated (Age-Down) genes from RNA-seq in both the human brain prefrontal cortex (PFC) and the mouse brain. In addition to a global loss of H3K27ac during aging, we found that Age-Up genes are specifically enriched for inflammation-related processes and show broad hyper H3K27ac over gene bodies. In contrast, Age-Down genes are enriched only for H3K27ac around the promoter regions. Using a linear regression model, we inferred that H3K27ac impacted aging-associated gene-expression changes and identified broad gene-body hyper H3K27ac as the top factor predicting a gene to show age-dependent up-regulation. As an implication of the potential causal effect of reduced broad gene-body acetylation to overexpression of Age-Up genes, histone deacetylase (HDAC) inhibition hyperacetylated the gene bodies and down-regulated the expression of Age-Up genes in mouse brain.
Results
Global Transcriptome and H3K27ac Changes in Human and Mouse Brain During Aging.
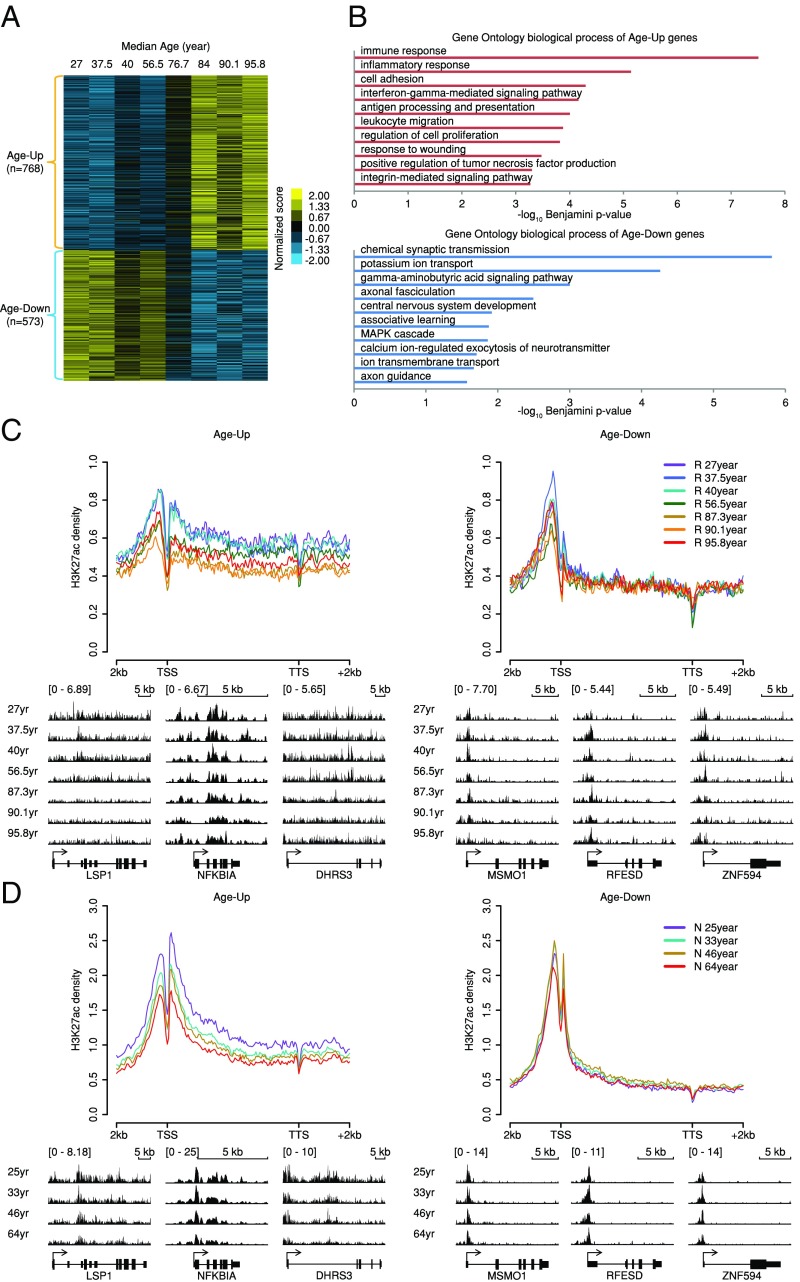
Using RNA-seq, we profiled genome-wide gene expressions in human PFC samples from the Rush Memory and Aging Project and from the Chinese Wuhan collection (R group, SI Appendix, Table S1), which are categorized into eight age groups (a total of 79 of human brain samples). Due to the broad age range of samples mixed in one group and ethnic bias, differential expressed genes (DEGs) were directly compared between old (>60 y old) and young (<60 y old) human brain samples: 768 Age-Up and 573 Age-Down genes were identified (Experimental Procedures and Fig. 1A), which are significantly overlapped with published microarray data (14) (SI Appendix, Fig. S1A). We further found that the Age-Up genes are highly enriched for inflammation-related functions, while the Age-Down genes are enriched for neural-specific functions (Fig. 1B). The differences were also reflected by the enrichment of distinct binding motifs of transcription factors (TFs) between the two sets of genes (TF-binding targets were obtained from ENCODE). For example, NFκB- and BCL3-binding events are enriched only in Age-Up genes, whereas GATA3-binding events are enriched only in Age-Down genes (SI Appendix, Table S2). NFκB and BCL3 are key players in the inflammation process (15, 16), while GATA3 is known to regulate neuron functions (17). We found that inflammatory response genes [Gene Ontology (GO) no. 0006954] are significantly enriched in Age-Up genes (odds ratio = 4.153, Fisher’s exact test P value <2.2e-16) whereas they are depleted in Age-Down genes (odds ratio = 0.667, Fisher’s exact test P value = 0.219), and the functional association network of Age-Up inflammatory response genes is centered around the TNF and NFκB genes with an average degree of 8.312, much higher than that of 2.667 in Age-Down inflammatory response genes (SI Appendix, Fig. S1B). To examine whether epigenetic regulation contributes to the age-associated expression alterations, we profiled the H3K27ac mark, an epigenetic hallmark of active chromatin regions, such as at enhancers and promoters, in the same human brain PFC tissues used for RNA-seq. We found that broad H3K27ac over Age-Up genes is significantly decreased in aged samples compared with the young samples at both promoters and gene bodies [TSS − 2 kb to TTS + 2 kb, Experimental Procedures, Fig. 1C, Left, Kolmogorov–Smirnov (KS) test P value = 6.45e-05 between samples with average age >60 and <60 y]. However, Age-Down genes showed primarily high promoter H3K27ac, which significantly decreased with age, while no obvious change was detected over gene bodies, perhaps due to the relatively low gene-body enrichment of H3K27ac (Fig. 1C, Right, KS test P value = 0.613 between samples with average age >60 and <60 y). To rule out possible ethnic differences (SI Appendix, Table S1), we then investigated PFC H3K27ac patterns in two young and two old Caucasian groups from the National Institute of Child Health and Human Development (N group, SI Appendix, Table S1), and observed similar trends of gene body H3K27ac changes (Fig. 1D, KS test P value = 7.39e-03 and 0.551 between samples with average age >60 and <60 y for Age-Up and Age-Down genes, respectively). The distinctive acetylation changes in Age-Up and Age-Down genes can be similarly observed using Age-Up and Age-Down genes overlapping between a published microarray data (without ethnic confounding factor) (14) and the R group RNA-seq data (SI Appendix, Fig. S1C, KS test P value = 3.70e-03 and 0.912 between samples with average age >60 and <60 y for Age-Up and Age-Down genes, respectively).
Fig. 1.
H3K27ac changes of age-dependently expressed genes in human brain prefrontal cortex. (A) Age-dependently expressed genes in human brain. Cuffdiff was used to define the age-dependent gene sets between young (<60 y old) and old (>60 y old) groups (q-value <0.05). The z-score normalized expression values are visualized in the heatmap. (B) Top 10 enriched GO biological processes among human brain Age-Up and Age-Down genes determined by DAVID. (C) R-group H3K27ac profiles on Age-Up and Age-Down genes. Integrative Genomics Viewer (IGV) views of exemplary genes are shown below. Ranges of profiles and heatmaps are from −2 kb upstream to +2 kb downstream unless noted otherwise. (D) N-group H3K27ac profiles on Age-Up and Age-Down genes. IGV views of exemplary genes are shown below.
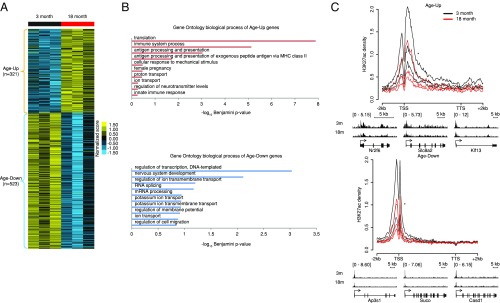
To further confirm the age-related changes of H3K27ac, we verified our findings in two groups of 3- and 18-mo-old mice using a similar approach (Fig. 2A and SI Appendix, Table S3). Age-Up (n = 321) genes in mouse brain are also related to immune functions, e.g., “immune system process” and “antigen processing and presentation,” and the Age-Down (n = 523) genes are enriched for neural functions, e.g., “nervous system development” (Fig. 2B). Importantly, mouse Age-Up and Age-Down genes also showed similar age-dependent gene-body and promoter changes of H3K27ac, respectively, as in human brains (Fig. 2C).
Fig. 2.
H3K27ac changes of age-dependently expressed genes in mouse brain. (A) Age-dependently expressed genes in mouse brain. Cuffdiff was used to define the Age-Up and Age-Down gene sets (q-value <0.05). The z-score normalized expression values are visualized in the heatmap. (B) Top 10 enriched GO biological process terms among mouse brain Age-Up and Age-Down genes determined by DAVID. (C) Average H3K27ac profiles of Age-Up and Age-Down genes in individual 3- and 18-mo-old mice brains (n = 3 for each group). Merged IGV views of exemplary genes are shown below. The significance of difference between 3 and 18 mo was determined by KS test. *P value <0.0001.
Overall, the broad gene-body H3K27ac of the Age-Up genes was a feature distinguishing the young (high H3K27ac) from the old (low H3K27ac) samples (Figs. 1 C and D and 2C).
Aging-Associated Gene Expression Changes Can Be Predicted by the Gene-Body H3K27ac Pattern.
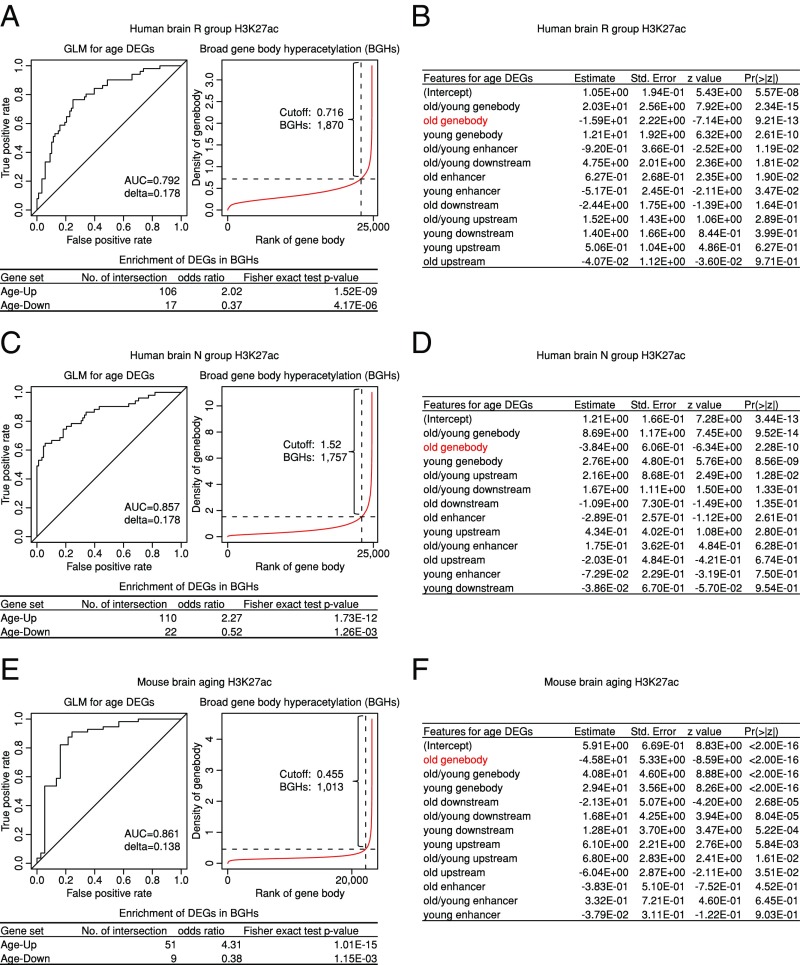
To quantitatively determine the contribution of histone acetylation in different genomic regions to age-associated gene-expression changes in human or mouse brains, we used a generalized linear model (GLM) (Experimental Procedures) to predict the DEGs showing Age-Up/-Down patterns. We found that H3K27ac density over a gene and its closest enhancer could predict the effect of aging using both human and mouse datasets [Fig. 3 with area under the Receiver Operating Characteristic (ROC) curves; area under the curve (AUC): 0.792 ∼ 0.861]. Importantly, change in gene-body H3K27ac density is ranked as the most significant predictive feature in all three datasets to distinguish Age-Up versus Age-Down genes (Fig. 3 B, D, and F). These findings suggest that broad gene-body acetylation is likely functionally involved in transcription regulation in these processes.
Fig. 3.
Loss of broad gene-body acetylation is predictive of aging-induced gene expression activation. (A) ROC curve for a GLM using R-group human brain oldest and youngest acetylation features to discriminate Age-Up (value = 0) and Age-Down (value = 1) genes in R-group human brain. AUC and delta from cross-validation are shown. BGHs are defined by the inflection point on acetylation density rank versus density plot, similar to super enhancers. Features used to define BGHs are marked by a red typeface in the corresponding GLM feature table in B, with fold of enrichment and Fisher’s exact test P values of the defined BGHs for Age-Up and Age-Down genes shown in the table below. (B) Related to A, features used in GLM; red typeface indicates the feature used to identify BGHs. (C) Same as A, but using N-group human brain oldest and youngest acetylation features. (D) Related to C, features used in GLM; red typeface indicates the feature used to identify BGHs. (E) Same as A, but using 3- and 18-mo-old mouse brain acetylation features to discriminate Age-Up and Age-Down genes. (F) Related to E, features used in GLM, red typeface indicates the feature used to identify BGHs.
Interestingly, using a definition similar to that of “super enhancers” (18, 19), broad gene-body hyperacetylations (BGHs) could be readily defined by the inflection point on the rank versus intensity plot (Fig. 3 A, C, and E). We found that BGHs are 2.02- to 4.31- fold enriched for Age-Up genes, and 0.37- to 0.52-fold depleted for Age-Down genes compared with genome background (Fig. 3 A, C, and E), suggesting that BGHs could be used to predict the Age-Up genes.
HDAC Inhibitors Suppress the Overexpression of Age-Up Genes.
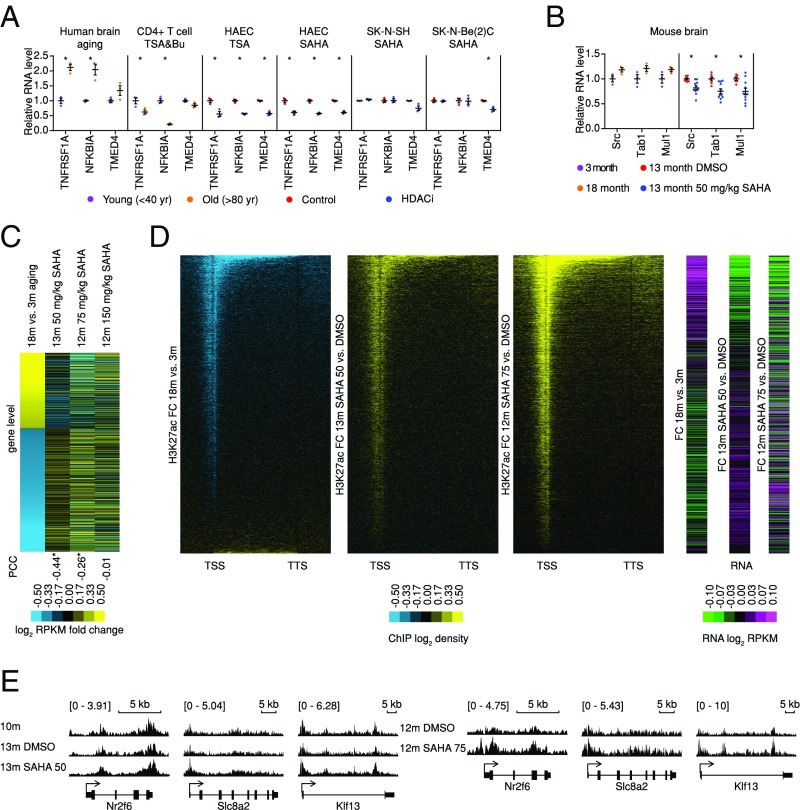
Finding aging-associated H3K27ac loss in Age-Up genes is surprising, as it suggests that broad gene-body H3K27ac might play a suppressive role in transcription, in contrast to a commonly accepted activation function. However, our findings are consistent with previously reported HDAC inhibitor (HDACi)-induced transcriptional suppression of a large number of genes through interference of transcription elongation occurring mainly within gene bodies (20). To investigate whether H3K27ac changes are causative to the gene expression changes or merely a consequence, we first examined whether HDACi down-regulated Age-Up genes in published datasets (21, 22). We found that trichostatin A (TSA), together with sodium butyrate treatment of human CD4+ T cells [Gene Expression Omnibus (GSE) no. 15735], TSA or suberanilohydroxamic acid (SAHA) treatments of human aortic epithelial cells (GSE37378), and SAHA treatment of neuroblastoma (GSE47670) indeed induced opposite expression changes in both the age up-regulated and down-regulated pathways (SI Appendix, Fig. S2A and Table S4). Despite differences across different cell lines, NFκB pathway regulators, such as TNFRSF1A, NFKBIA, and TMED4, which are up-regulated during human brain aging, were commonly suppressed by HDACi (Fig. 4A).
Fig. 4.
Overexpression of inflammation-related genes can be repressed by restoring gene-body acetylation. (A) Relative RNA levels of NFκB pathway regulator changes during human brain aging and with HDACi treatment in different cell lines (n = 3 for each group). Mean value and SEM are shown, two-tail Student’s t test is used, *P value <0.05. (B) Relative RNA levels of NFκB pathway regulators changed during mouse brain aging and with SAHA treatment, respectively (n = 3 for 3 and 18 mo, n = 6 for 13 mo DMSO, n = 11 for 13 mo SAHA). Mean value and SEM are shown, two-tail Student’s t test is used, *P value <0.05. (C) SAHA attenuates age-dependent transcriptome changes in mouse brain. The mouse brain age-dependent genes are sorted by their log2 fold change of expression between old (18 mo) and young (3 mo) mice (first column) and then are compared with those under SAHA for 3 mo with different concentrations (in columns two to four). Yellow/blue color in the first column indicates Age-Up/Down genes. The rest of the columns show SAHA effects in different groups for the same genes. *P value of PCC <0.0001. (D) Heatmaps of log2 fold changes of average acetylation and expression in 18 vs. 3 mo, 13 mo 50 mg/kg SAHA vs. DMSO, and 12 mo 75 mg/kg SAHA vs. DMSO treatment. Genes are ranked by average ChIP signal from upstream 2 kb to downstream 2 kb in 18 mo vs. 3 mo from low to high. (E) Merged IGV views of H3K27ac profiles of exemplary Age-Up genes.
Because mice of middle age already display age-related changes at the phenotype level that are similar to those of 18-mo-old mice but to a lesser extent (23), to further validate such findings, we next treated middle age (10 mo) mice with DMSO or 50 mg/kg SAHA every other day for 3 mo (Experimental Procedures and SI Appendix, Table S3) and then compared brain gene-expression profiles to the pretreatment mice (10 mo). We found that the SAHA-treated mouse brains (13 mo old) displayed profiles more similar to the 10-mo-old pretreated mouse brains than the vehicle-treated controls (SI Appendix, Fig. S2B). Similar to the results in human cell lines, SAHA treatment in mice repressed the NFκB pathway regulators Src, Tab1, and Mul1, which were up-regulated during mouse brain aging (Fig. 4B). Importantly, SAHA treatment was able to induce a significant reversal of age-associated expression changes of both Age-Up and Age-Down genes, [Fig. 4C, first two columns, Pearson correlation coefficient (PCC): −0.47, P value <2.2e-16].
We also treated mice at two higher dosages of SAHA (75 and 150 mg/kg every other day) (Experimental Procedures and SI Appendix, Table S3). At both dosages, reversals of aging effects were observed at the gene-expression level, although the high 150 mg/kg dosage seems to be less pronounced, perhaps due to toxicity (Fig. 4C, columns three and four). Phenotypically, and consistent with a previous report (24), we also observed that memory improved in mice after 75 mg/kg SAHA treatment by water maze test, while the 150 mg/kg dosage did not produce such results (SI Appendix, Fig. S2C).
Given the distinct H3K27ac patterns over the promoter and gene-body regions associated with Age-Up and Age-Down genes the expression of which could be reversed by SAHA, we hypothesized that H3K27ac patterns are also differentially reversed by SAHA treatment. After 3 mo of 50 mg/kg SAHA treatment, we found a global increase of H3K27ac in mouse brain (SI Appendix, Fig. S2D). Consistent with our hypothesis, decreasing H3K27ac on promoter or gene body is reversed to the young mode, in line with their gene-expression changes, 75 mg/kg SAHA also had a similar impact on the H3K27ac profiles (see Fig. 4 D and E and SI Appendix, Fig. S2E for examples). Overall, upon SAHA treatment, the H3K27ac levels of Age-Up and SAHA-Down genes increased both at promoters and broadly in gene bodies, which were accompanied by a decrease of expression. In contrast, the Age-Down and SAHA-Up genes showed increases of H3K27ac primarily at promoter regions, which were accompanied by an increase of expression (Fig. 4 and SI Appendix, Fig. S2). As the Age-Up genes were overexpressed in aged brains, and attenuated by HDAC inhibition (SI Appendix, Fig. S3), this suggests that, while hyperacetylation at promoters activates gene expression, hyperacetylation at gene bodies represses overactivation of genes.
Discussion
Through ChIP-seq experiments using human brain PFC and mouse brain tissues, we have shown a progressive loss of histone H3K27 acetylation and dysregulated gene expression during aging in both human and mouse brains. Two sets of genes, namely Age-Up and Age-Down, were identified by RNA-seq. Interestingly, although both sets of genes display promoter H3K27ac enrichment, the Age-Up genes also have robust broad gene-body H3K27ac. Importantly, and contrary to the well-established transcription activation role of H3K27ac, we found that broad gene-body hyper H3K27ac is a different mode of transcription regulation in that it tends to suppress gene expression, rather than activate gene expression; more precisely, broad gene body hyper acetylation may prevent overactivation of highly expressed genes. HDAC inhibition restores H3K27ac levels in both sets of genes.
It has been established that, during aging, there is a general loss of repressive epigenetic marks, such as H3K27me3, and a gain of active marks such as H3K4me2/3 (25). However, we found in human and mouse brain, there is a general loss of H3K27ac during aging. This occurs in the promoters of both Age-Up and Age-Down genes, but is only broadly distributed over the gene bodies of the Age-Up genes. Such drastic age-associated H3K27ac loss, especially in the broad distribution of gene bodies of the Age-Up genes, has never been reported. Although we focus mainly on the enhancer mark H3K27ac, the age-related change in histone acetylation might not be restricted to this one site, and reanalysis of a published dataset (GSE63945) (20) from mouse brain finds that another histone acetylation mark, H4K12ac, also shows a similar aging-related decline as H3K27ac in the gene bodies of Age-Up genes (SI Appendix, Fig. S4). Altogether, considering the high correlations between acetylation sites on histones (26) and low specificity of HATs and HDACs (27–30), H3K27ac may be just one of the marks and contributors indicating the acetylation states at a genomic region.
An increase of HDAC2 activity has been demonstrated to cause cognitive decline in mice, which can be rescued by HDACi treatment (24, 31, 32). In the same study, the restored expression of neural genes was considered partially responsible for HDACi effect (31, 32). Consistently, here we also found that HDACi treatment could reverse the age-related decline of neural gene expression in addition to our observation of the repression of inflammaging genes. Here we used HDACi only to probe whether the gene-body acetylation changes might cause gene-expression changes. Due to their nonspecificity and toxicity, HDACi may not be ideal for exploring functional mechanisms or for using as anti-aging drugs. However, the fact that age-related functions were restored by the HDACi SAHA suggests that these functions are histone-acetylation–dependent, although not necessarily exclusive at the H3K27 site, since SAHA also promotes histone acetylation at other sites. Consistent with the dependency of the expression changes on the histone acetylation changes, by examining the overlap of mouse brain age or SAHA DEGs with different HATs and HDAC KO-induced DEGs, we found that Hdac6 KO could mimic SAHA and anti-aging transcriptome profiles, while Kat2A/B and Kat6 KO induced aging-like transcriptome changes (SI Appendix, Fig. S5A), which hints at a direct or indirect role of these enzymes in regulating Age-Up genes. Interestingly, HDAC6 shows slightly increased expression during aging in both human and mouse brain and prefers to localize on gene bodies compared with other HDACs as mentioned above (21).
Due to the limitations of human sample collection, not all samples were from the same ethnic group in this study. However, we have shown that age-dependent expression and acetylation changes are applicable to both Caucasian and Chinese groups. More importantly, our mouse data indicate that such alterations are likely to be evolutionally conserved in mammals. Our in vivo human and mouse brain sample analysis established a global H3K27ac decrease during brain aging, its prominent consequence on gene expression, and, in particular, the link between broad gene-body hyperacetylation of inflammaging gene repression.
In this study, we found that brain Age-Up genes include many genes related to inflammation, such as genes belonging to the NFκB pathway that are commonly up-regulated in many tissues during aging (33, 34). Our findings thus show that such an inflammaging process is not only regulated at the epigenomic level, but also, more importantly, is reversible.
Experimental Procedures
Human and Mouse Samples.
Mouse experiments were approved by ethics committee of Shanghai Model Organisms Center; informed consent was obtained for the use of the human tissues.
RNA-Seq Data Processing.
Reads from RNA-seq were mapped to human genome hg19 and mouse genome mm10, respectively, by using STAR (v2.5.2a) (35) with 2% maximum mismatch. Cuffdiff (v2.2.1) (36) with default parameters was used to generate RPKM and define DEGs (q-value < 0.05).
ChIP-Seq Data Processing.
Reads from ChIP-seq were mapped to human genome hg19 and mouse genome mm10, respectively, by using Bowtie (v1.1.1) (37) (reads shorter than 150 bp) or Bowtie2 (v2.2.4) (38) (read length is 150 bp). When comparing different batches, data are trimmed to the same length as the shortest samples, and reads are mapped by Bowtie1 with single-end strategy. Heatmaps and profiles for ChIP-seq data were generated and normalized with Danpos2 (v2.2.2) (39), total reads were normalized to 1 × 107, and genes shorter than 2,000 bp were excluded to avoid spillover signal from promoters to gene bodies. The KS test was performed by R (v3.1.0) on young (<60 y old) and old (>60 y old) human brain groups.
Data Availability.
RNA-seq and ChIP-seq data that support the findings of this study have been deposited in GEO with the GSE106670 accession code. Data are available in SI Appendix.
Supplementary Material
Acknowledgments
This work was supported by China Ministry of Science and Technology Grants 2015CB964803 and 2016YFE0108700; National Natural Science Foundation of China Grants 91749205, 31210103916, and 91519330; Chinese Academy of Sciences Grants XDB19020301 and XDA01010303 (to J.-D.J.H.); National Institutes of Health Grants P30AG10161, R01AG15819, and R01AG17917 (to D.A.B.); China Ministry of Science and Technology Grant 2016YFA0101800; and the Science and Technology Commission of Shanghai Municipality Grant 16XD1400500 (to F.L.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: RNA-seq and ChIP-seq data that support the findings of this study have been deposited in the Gene Expression Omnibus database (accession no. GSE106670).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1800656115/-/DCSupplemental.
References
- 1.Campisi J. Aging, cellular senescence, and cancer. Annu Rev Physiol. 2013;75:685–705. doi: 10.1146/annurev-physiol-030212-183653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guarente L. Aging research: Where do we stand and where are we going? Cell. 2014;159:15–19. doi: 10.1016/j.cell.2014.08.041. [DOI] [PubMed] [Google Scholar]
- 3.Franceschi C, et al. Inflamm-aging: An evolutionary perspective on immunosenescence. Ann N Y Acad Sci. 2000;908:244–254. doi: 10.1111/j.1749-6632.2000.tb06651.x. [DOI] [PubMed] [Google Scholar]
- 4.Franceschi C, Campisi J. Chronic inflammation (inflammaging) and its potential contribution to age-associated diseases. J Gerontol A Biol Sci Med Sci. 2014;69(Suppl 1):S4–S9. doi: 10.1093/gerona/glu057. [DOI] [PubMed] [Google Scholar]
- 5.Glass CK, Saijo K, Winner B, Marchetto MC, Gage FH. Mechanisms underlying inflammation in neurodegeneration. Cell. 2010;140:918–934. doi: 10.1016/j.cell.2010.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bollrath J, Greten FR. IKK/NF-kappaB and STAT3 pathways: Central signalling hubs in inflammation-mediated tumour promotion and metastasis. EMBO Rep. 2009;10:1314–1319. doi: 10.1038/embor.2009.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Iyer SS, et al. Mitochondrial cardiolipin is required for Nlrp3 inflammasome activation. Immunity. 2013;39:311–323. doi: 10.1016/j.immuni.2013.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang G, et al. Hypothalamic programming of systemic ageing involving IKK-β, NF-κB and GnRH. Nature. 2013;497:211–216. doi: 10.1038/nature12143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.López-Otín C, Blasco MA, Partridge L, Serrano M, Kroemer G. The hallmarks of aging. Cell. 2013;153:1194–1217. doi: 10.1016/j.cell.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Greer EL, et al. Members of the H3K4 trimethylation complex regulate lifespan in a germline-dependent manner in C. elegans. Nature. 2010;466:383–387. doi: 10.1038/nature09195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maures TJ, Greer EL, Hauswirth AG, Brunet A. The H3K27 demethylase UTX-1 regulates C. elegans lifespan in a germline-independent, insulin-dependent manner. Aging Cell. 2011;10:980–990. doi: 10.1111/j.1474-9726.2011.00738.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jin C, et al. Histone demethylase UTX-1 regulates C. elegans life span by targeting the insulin/IGF-1 signaling pathway. Cell Metab. 2011;14:161–172. doi: 10.1016/j.cmet.2011.07.001. [DOI] [PubMed] [Google Scholar]
- 13.Kawahara TL, et al. SIRT6 links histone H3 lysine 9 deacetylation to NF-kappaB-dependent gene expression and organismal life span. Cell. 2009;136:62–74. doi: 10.1016/j.cell.2008.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lu T, et al. Gene regulation and DNA damage in the ageing human brain. Nature. 2004;429:883–891. doi: 10.1038/nature02661. [DOI] [PubMed] [Google Scholar]
- 15.Lawrence T. The nuclear factor NF-kappaB pathway in inflammation. Cold Spring Harb Perspect Biol. 2009;1:a001651. doi: 10.1101/cshperspect.a001651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chang TP, Vancurova I. Bcl3 regulates pro-survival and pro-inflammatory gene expression in cutaneous T-cell lymphoma. Biochim Biophys Acta. 2014;1843:2620–2630. doi: 10.1016/j.bbamcr.2014.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kizil C, et al. Regenerative neurogenesis from neural progenitor cells requires injury-induced expression of Gata3. Dev Cell. 2012;23:1230–1237. doi: 10.1016/j.devcel.2012.10.014. [DOI] [PubMed] [Google Scholar]
- 18.Whyte WA, et al. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153:307–319. doi: 10.1016/j.cell.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lovén J, et al. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell. 2013;153:320–334. doi: 10.1016/j.cell.2013.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Greer CB, et al. Histone deacetylases positively regulate transcription through the elongation machinery. Cell Rep. 2015;13:1444–1455. doi: 10.1016/j.celrep.2015.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Z, et al. Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell. 2009;138:1019–1031. doi: 10.1016/j.cell.2009.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rafehi H, et al. Vascular histone deacetylation by pharmacological HDAC inhibition. Genome Res. 2014;24:1271–1284. doi: 10.1101/gr.168781.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shoji H, Takao K, Hattori S, Miyakawa T. Age-related changes in behavior in C57BL/6J mice from young adulthood to middle age. Mol Brain. 2016;9:11. doi: 10.1186/s13041-016-0191-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Benito E, et al. HDAC inhibitor-dependent transcriptome and memory reinstatement in cognitive decline models. J Clin Invest. 2015;125:3572–3584. doi: 10.1172/JCI79942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brunet A, Berger SL. Epigenetics of aging and aging-related disease. J Gerontol A Biol Sci Med Sci. 2014;69(Suppl 1):S17–S20. doi: 10.1093/gerona/glu042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang Z, et al. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet. 2008;40:897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Furdas SD, Kannan S, Sippl W, Jung M. Small molecule inhibitors of histone acetyltransferases as epigenetic tools and drug candidates. Arch Pharm (Weinheim) 2012;345:7–21. doi: 10.1002/ardp.201100209. [DOI] [PubMed] [Google Scholar]
- 28.Sterner DE, Berger SL. Acetylation of histones and transcription-related factors. Microbiol Mol Biol Rev. 2000;64:435–459. doi: 10.1128/mmbr.64.2.435-459.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marmorstein R, Zhou MM. Writers and readers of histone acetylation: Structure, mechanism, and inhibition. Cold Spring Harb Perspect Biol. 2014;6:a018762. doi: 10.1101/cshperspect.a018762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Seto E, Yoshida M. Erasers of histone acetylation: The histone deacetylase enzymes. Cold Spring Harb Perspect Biol. 2014;6:a018713. doi: 10.1101/cshperspect.a018713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Penney J, Tsai LH. Histone deacetylases in memory and cognition. Sci Signal. 2014;7:re12. doi: 10.1126/scisignal.aaa0069. [DOI] [PubMed] [Google Scholar]
- 32.Shimazu T, et al. Suppression of oxidative stress by β-hydroxybutyrate, an endogenous histone deacetylase inhibitor. Science. 2013;339:211–214. doi: 10.1126/science.1227166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Adler AS, et al. Motif module map reveals enforcement of aging by continual NF-kappaB activity. Genes Dev. 2007;21:3244–3257. doi: 10.1101/gad.1588507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.O’Brown ZK, Van Nostrand EL, Higgins JP, Kim SK. The inflammatory transcription factors NFκB, STAT1 and STAT3 drive age-associated transcriptional changes in the human kidney. PLoS Genet. 2015;11:e1005734. doi: 10.1371/journal.pgen.1005734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dobin A, et al. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Trapnell C, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen K, et al. Broad H3K4me3 is associated with increased transcription elongation and enhancer activity at tumor-suppressor genes. Nat Genet. 2015;47:1149–1157. doi: 10.1038/ng.3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq and ChIP-seq data that support the findings of this study have been deposited in GEO with the GSE106670 accession code. Data are available in SI Appendix.