Significance
Results presented in this report demonstrate that minor zygotic gene activation (ZGA) must precede major ZGA to execute successfully the maternal-to-zygotic transition, and that the timely occurrence of minor ZGA is crucial for preimplantation development to continue beyond the two-cell stage. In addition, the results show that the gene-expression program proceeds in a step-by-step fashion, and at least initially, is not regulated by a “zygotic clock” (e.g., compaction) or cell cycle progression (e.g., major ZGA that occurs during the two-cell stage).
Keywords: minor zygotic gene activation, preimplantation mouse embryo, gene expression, maternal-to-zygotic transition
Abstract
In mice, transcription initiates at the mid-one-cell stage and transcriptional activity dramatically increases during the two-cell stage, a process called zygotic gene activation (ZGA). Associated with ZGA is a marked change in the pattern of gene expression that occurs after the second round of DNA replication. To distinguish ZGA before and after the second-round DNA replication, the former and latter are called minor and major ZGA, respectively. Although major ZGA are required for development beyond the two-cell stage, the function of minor ZGA is not well understood. Transiently inhibiting minor ZGA with 5, 6-dichloro-1-β-d-ribofuranosyl-benzimidazole (DRB) resulted in the majority of embryos arresting at the two-cell stage and retention of the H3K4me3 mark that normally decreases. After release from DRB, at which time major ZGA normally occurred, transcription initiated with characteristics of minor ZGA but not major ZGA, although degradation of maternal mRNA normally occurred. Thus, ZGA occurs sequentially starting with minor ZGA that is critical for the maternal-to-zygotic transition.
Changes in gene expression underlie development of zygotes into adult multicellular organisms. Mouse oocytes express a set of genes, including oocyte-specific genes (1–3), during the growth phase, but as oocytes approach their final size transcription ceases (4, 5). mRNAs are extremely stable during the growth phase, which permits them to accumulate despite oocytes becoming transcriptionally inactive (6–8). Although resumption of meiosis triggers a transition from mRNA stability to instability, a substantial amount of maternal mRNAs persists in one-cell embryos (9, 10), with the vast majority of these mRNAs degraded by the end of the two-cell stage and replaced with zygotic transcripts. This destruction of maternal mRNAs with expression of new mRNAs is referred to as the maternal-to-zygotic transition (MZT).
Although results of early studies suggested that zygotic gene activation (ZGA) initiates during the two-cell stage, more sensitive assays (e.g., BrUTP incorporation and expression of a transgene from the paternal genome) revealed that zygotic transcription begins during S-phase of one-cell stage (11–13). The changes in gene expression between the one-cell and two-cell stages appear linked to DNA replication because the one-cell pattern of gene expression is still observed in the late two-cell stage when the second round of DNA replication is inhibited (14, 15). The initial ZGA that occurs between S phase of the one-cell embryo and G1 of the two-cell embryo is designated as minor ZGA to discriminate it from the burst of transcription that occurs during the mid-to-late two-cell stage, which is designated as major ZGA.
Major ZGA is essential for development beyond the two-cell stage, because inhibiting major ZGA results in cleavage arrest at the two-cell stage. A role for minor ZGA is ill-defined because α-amanitin–treated one-cell embryos cleave to the two-cell stage and then arrest at the G2 phase of the two-cell stage (16). Nevertheless, whether transcription in one-cell embryos is essential for development beyond the two-cell stage is unresolved because minor ZGA occurs before completion of MZT and it is possible that one-cell embryos complete the first cell cycle using maternally derived proteins and that minor ZGA is required for development beyond the two-cell stage. To identify the biological function of minor ZGA, if any, requires that only minor ZGA, but not major ZGA, is inhibited. To this end, we transiently treated embryos during minor ZGA with 5,6-dichloro-1-β-d-ribofuranosyl-benzimidazole (DRB), which reversibly inhibits RNA polymerase II (POLR2A) (17) by inhibiting protein kinases that target POLR2A (18). We report here that transient inhibition of minor ZGA results in compromised development beyond the two-cell stage.
Results
Minor ZGA Is Critical for Preimplantation Development.
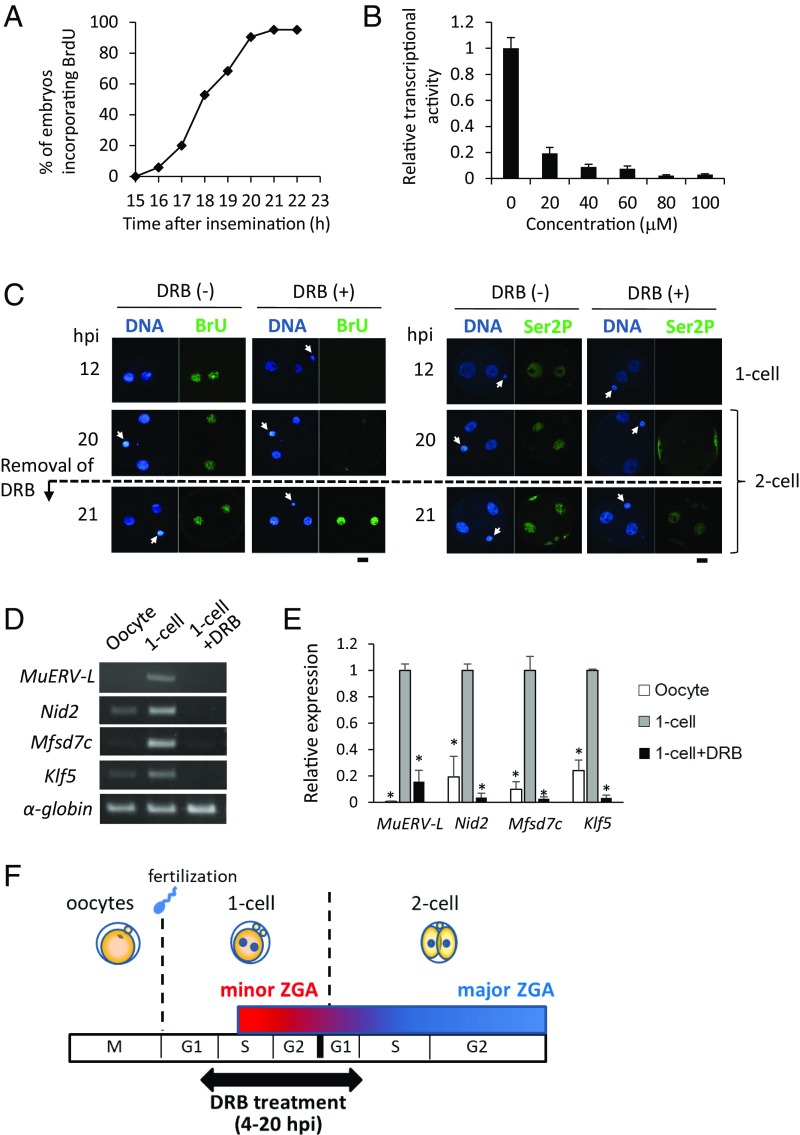
Because DRB is a reversible inhibitor of Pol II-mediated transcription, we used DRB to inhibit transiently transcription during minor ZGA to determine whether minor ZGA is essential for development beyond the two-cell stage. We first determined the period of minor ZGA—the transition from minor to major ZGA occurs during the second round of DNA replication—by ascertaining the time of initiation of DNA replication in two-cell embryos by incorporation of bromodeoxyuridine (BrdU) and found that most embryos reached S phase 20 h postinsemination (hpi) (Fig. 1A). We then determined that 80 µM DRB was the minimum effective concentration to inhibit transcription (Fig. 1B).
Fig. 1.
DRB reversibly inhibits minor ZGA. (A) Timing of S phase during the two-cell stage. DNA replication was determined by incorporation of BrdU. The percentage of embryos that incorporated BrdU was determined at the indicated times. More than 20 embryos were observed in each time point. (B) Minimum concentration of DRB required for maximal inhibition of transcription during minor ZGA. Embryos were treated with the indicated concentrations of DRB between 4 and 12 hpi and transcriptional activity measured at 12 hpi. Transcriptional activity in control untreated embryos was set as 1 and the relative transcriptional activities were calculated for the other samples. More than 20 pronuclei were analyzed in each sample and the data are presented as the mean ± SEM. (C) Validation of inhibition of transcription and recovery of transcriptional activity (Left) and phosphorylated RNA polymerase II (Right). Embryos were cultured in DRB-containing medium between 4 and 20 hpi and then transferred into the DRB-free medium. The embryos were collected 12, 20, and 21 hpi for determination of transcriptional activity and phosphorylation of RNA polymerase II on the C-terminal Ser2 (Ser2P). Arrowheads indicate polar bodies. More than 20 pronuclei and nuclei were examined in each sample. (Scale bars, 20 µm.) (D and E) DRB inhibits expression of genes transcribed during minor ZGA. Embryos were treated with DRB between 4 and 12 hpi. The MII eggs and one-cell embryos collected at 12 hpi were subjected to RT-PCR assay; rabbit α-globin mRNA was used as an external control. The experiment was performed three times and similar results were obtained. Shown is a representative example (D). The densities of PCR bands were quantified by using image analyzer (E). Data are represented as mean ± SEM. Asterisks indicate a significant difference from the control one-cell embryos (Student’s t test; P < 0.05). (F) Schematic representation of transient inhibition of minor ZGA. The embryos were cultured in the KSOM medium containing 80 µM DRB from 4 to 20 hpi and then transferred to the DRB-free medium.
We next documented that DRB-mediated inhibition of transcription is reversible (Fig. 1C). Because minor ZGA starts around mid-S phase of the one-cell stage (13), we treated embryos with DRB from 4 to 20 hpi and then assessed BrU incorporation following transfer to DRB-free medium. Whereas control embryos were transcriptionally active and embryos continuously exposed to DRB were not at 12 and 20 hpi, BrU incorporation was observed within 1 h after transfer to DRB-free medium. Consistent with DRB reversibly inhibiting transcription during minor ZGA is that Ser2 phosphorylation of the C-terminal domain (Ser2P) of POLR2A was not detected by immunocytochemistry in DRB-treated embryos but was detected within 1 h following transfer to DRB-free medium (Fig. 1C and SI Appendix, Fig. S1). Finally, expression of genes transcribed during minor ZGA (5, 19) was inhibited by DRB treatment (Fig. 1 D and E). Based on these results, we decided to treat embryos with DRB between 4 and 20 hpi to investigate the effect of transient inhibition of transcription during minor ZGA (Fig. 1F).
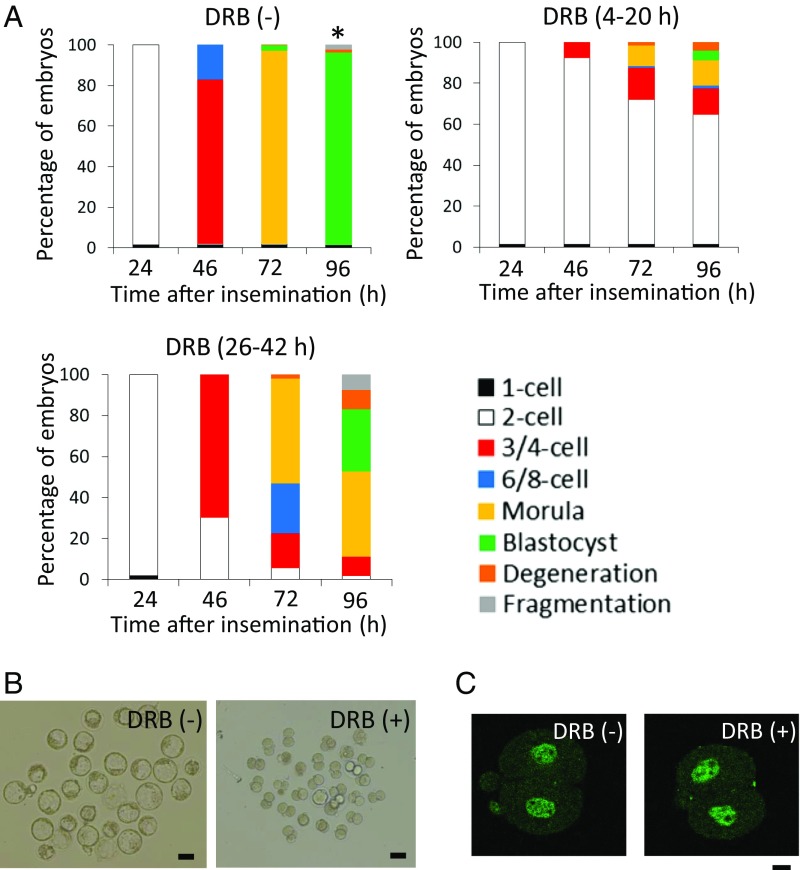
To determine whether minor ZGA is essential for development beyond the two-cell stage, one-cell embryos were transferred to medium containing DRB at 4 hpi, then transferred to DRB-free medium at 20 hpi and cultured until 96 hpi. Control embryos not treated with DRB developed to the blastocyst stage with a high incidence (Fig. 2A), with 98% cleaving to the three- to four-cell or six- to eight-cell stages at 46 hpi. By 72 and 96 hpi, 94% and 92% of the embryos developed to the molura and blastocyst stages, respectively (Fig. 2 A and B). In contrast, development beyond the two-cell stage was highly compromised for DRB-treated embryos (Fig. 2 A and B), with most embryos arresting at the two-cell stage at 46 hpi. By 96 h >60% were still arrested at the two-cell stage and only 10% reached the blastocyst stage. BrdU incorporation studies indicated that the embryos were arrested in G2 because DRB-treated embryos incorporated BrdU at 26 hpi and nuclei were clearly present (Fig. 2C).
Fig. 2.
Effect of transiently inhibiting minor ZGA on preimplantation development. (A) Development of embryos transiently treated with DRB. DRB (−) indicates embryos treated with DMSO, the solvent for DRB, between 4 and 20 hpi. DRB (4–20 h) and DRB (26–42 h) represent embryos that were treated with DRB between 4 and 20 h and between 26 and 42 hpi, respectively. The y axis shows percentages of embryos that developed to the indicated developmental stages. Asterisks represent significant differences by χ2 test (P < 0.05). The experiment was performed three times and >30 embryos were analyzed for each experiment. (B) Morphology of embryos treated with or without DRB at 96 hpi. DRB (−) and (+) represent embryos treated with DMSO and DRB, respectively, between 4 and 20 hpi. (Scale bars, 100 μm.) (C) Detection of the second round of DNA replication in DRB-treated embryos. The embryos were cultured with DRB between 4 and 20 hpi and then transferred to DRB-free medium containing BrdU. After 6 h of incubation, incorporated BrdU were detected by immunocytochemistry with anti-BrdU antibody. The experiments were conducted three times and more than 10 embryos were analyzed in each experiment. Incorporation of BrdU was detected in 96.1% of embryos analyzed. (Scale bars, 20 µm.)
To assess the potential toxicity of DRB for the observed effects on development, we conducted experiments in which full-grown oocytes were treated with DRB using the same protocol as used for one-cell embryos: the oocytes were treated with DRB during meiotic maturation (16 h), washed from DRB, and then observed for the development after fertilization. Because full-grown oocytes are transcriptionally quiescent, we were therefore able to assess DRB toxicity independent of its effect on transcription. Results of these experiments indicated that DRB treatment did not affect completion of meiosis and that after removal of DRB and fertilization, the embryos developed to the blastocyst stage at a similar rate as controls (SI Appendix, Fig. S2). Thus, no toxic effect of DRB on development was observed.
To assess the effect of inhibiting major ZGA on preimplantation development, embryos were treated with DRB from 26 to 42 hpi. After the embryos were transferred to DRB-containing medium at 26 hpi, transcription ceased rapidly such that Ser2P was not detected within 2 h (SI Appendix, Fig. S3). Although 70% of the embryos cleaved to three to four cells, 30% remained at the two-cell stage at 46 hpi. With additional culture, these two-cell embryos did develop further such that by 96 hpi >40% and 30% reached the morula and blastocyst stage, respectively. Thus, although the rate of development was retarded, developmental potential remained when major ZGA was inhibited, which is in marked contrast to when minor ZGA is inhibited. These results suggest that the timely occurrence of minor ZGA, but not major ZGA, is essential for preimplantation development beyond the two-cell stage.
Inhibiting Minor ZGA Alters the Transcriptome of Two-Cell Embryos and Alters H3K4me3 Level.
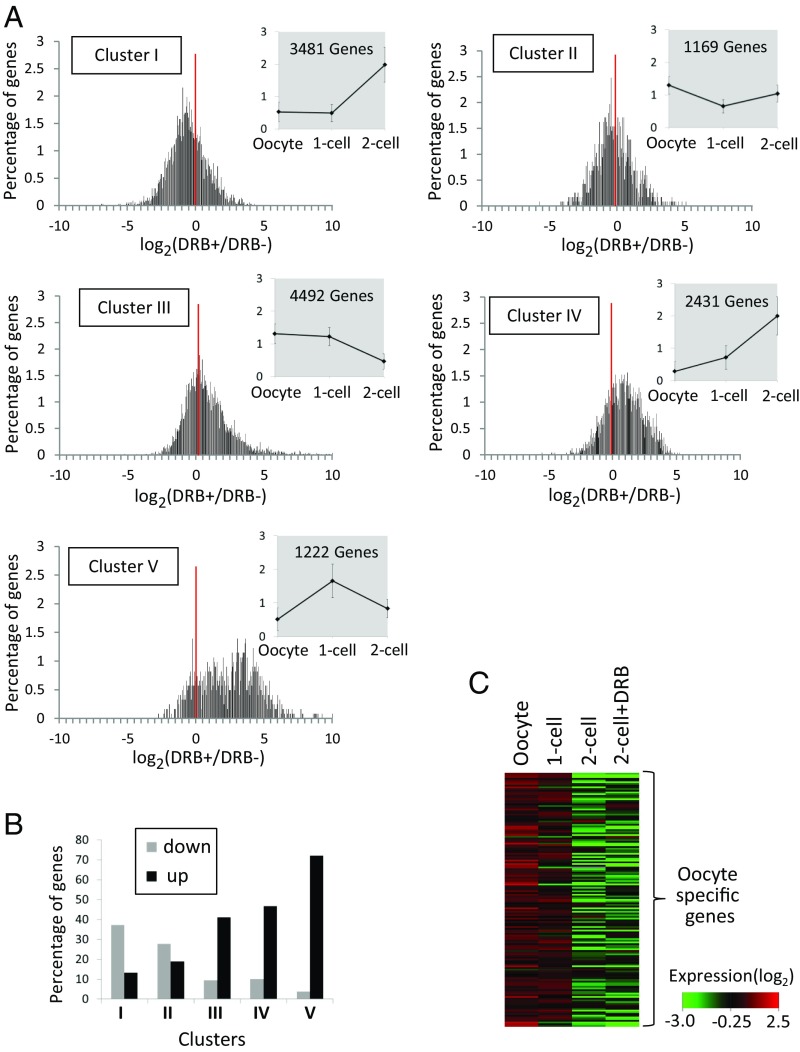
To understand the basis for the two-cell arrest when minor ZGA was inhibited, we examined gene expression in late two-cell embryos following removal of DRB. We first measured transcriptional activity by BrU incorporation and noted that transcriptional activity was drastically increased relative to control embryos up to 29 hpi, after which transcriptional activity was lower than controls (SI Appendix, Fig. S4). These results suggested that Pol II inhibited by DRB paused at promoters of minor ZGA genes, and was rapidly reactivated after removal of DRB. To test this proposal, we conducted RNA-seq on DRB-treated two-cell embryos 12 h after the removal of DRB (32 hpi) and compared the relative transcript abundance between DRB-treated and untreated embryos.
A heatmap of annotated genes shows that a considerable number of transcripts were up- or down-regulated in DRB-treated embryos compared with untreated embryos (SI Appendix, Fig. S5). It was possible that the effect of DRB treatment was different between genes that are transcribed during minor and major ZGA. Therefore, we classified the genes by changes in their relative expression levels at MII, one-cell, and two-cell stages using a k-means nonhierarchical clustering method. Genes were categorized into five classes (Datasets S1–S5). Cluster I consisted of the genes whose expression levels did not change after fertilization (between the MII and one-cell stage) but increased at the two-cell stage, indicating that this cluster predominately contained the genes transcribed during major ZGA but not minor ZGA (Fig. 3A). Cluster II consisted of the genes whose expression decreased after fertilization but increased by the two-cell stage, indicating that this cluster also contains the genes transcribed during major ZGA but not minor ZGA, as well as cluster I. Cluster III consisted of the genes whose expression levels continuously decreased after fertilization and cleavage to the two-cell stage. Genes in this class were unlikely to be activated during minor and major ZGA. Cluster IV mostly contained the genes whose expression levels increased after fertilization and then further increased at the two-cell stage, indicating that they are transcribed during both of minor and major ZGA. Finally, the expression levels in cluster V increased after fertilization and decreased by the two-cell stage, suggesting genes in this cluster were transcribed during minor ZGA but not major ZGA.
Fig. 3.
Effect of transiently inhibiting minor ZGA by DRB on the transcriptome of late-stage two-cell embryos. RNA-seq was conducted for DRB-treated embryos 12 h after removal of DRB (32 hpi). These data and those obtained in the previous study [MII eggs, one- (13 hpi), and two-cell–stage embryos (32 hpi)] (5) were used for analysis of gene expression. (A) Line graphs with gray background: classification of genes by k-means nonhierarchical clustering analysis based on their expression patterns in MII eggs, one-cell, and two-cell embryos treated with DRB. Mapped genes with <11 reads were omitted from the analysis. The average expression levels in the MII eggs, and one- and two-cell–stage embryos were set as 1. Error bars represent SD. Histograms with white background: distribution of up- and down-regulated transcripts in response to DRB in each cluster. The x axis represents the logarithm of the ratio of their expression levels between embryos treated with DRB (DRB+) and those not treated with the DRB (DRB−). The y axis represents the percentage of transcripts with particular DRB+/DRB− ratio in total number belonging to each cluster. The red vertical line represents the position of 0 at which the DRB+/DRB− ratio was 1.0. (B) The percentage of genes up- or down-regulated following transient inhibition of minor ZGA in each cluster. Transcripts whose expression levels were more than two times higher or less than a half, respectively, in the DRB-treated embryos than DRB-untreated ones, were defined as up- or down-regulated, respectively. (C) Analysis of the degradation pattern of 149 oocyte-specific transcripts (5) in MII eggs, one-cell–stage embryos, and two-cell–stage embryos treated with and without DRB.
When expression levels were compared at 32 hpi (12 h after removal of DRB) between embryos treated with or without DRB, more than half of the genes in clusters I and II, which are transcribed during only major ZGA but not minor ZGA, displayed lower levels of expression in DRB-treated embryos than controls (Fig. 3A). In contrast, a large portion of the genes in clusters IV and V, which are transcribed during minor ZGA, showed higher expression in DRB-treated embryos (Fig. 3A). Similar results were obtained when the percentages of highly up-regulated and down-regulated genes were examined in each cluster (Fig. 3B). In clusters I and II, the percentages of down-regulated genes were higher than those of up-regulated ones, whereas in clusters IV and V the percentages of up-regulated genes were much higher. These results indicated that a large portion of genes presumed to be transcribed during minor ZGA were activated after removal of DRB, but that those transcribed during major ZGA were not sufficiently transcribed at the time (32 hpi) when major ZGA normally occurs. In cluster I, the transcripts of Ccna2, Cdk1, Cdc25c, whose encoding proteins are essential components of the M-phase promoting factor and its activator (20–22), were significantly decreased (SI Appendix, Fig. S6). Therefore, it is possible that the compromised expression of genes (e.g., Ccna2, Cdk1, Cdc25c) during major ZGA caused cell-cycle arrest before entry into M phase in DRB-treated two-cell embryos.
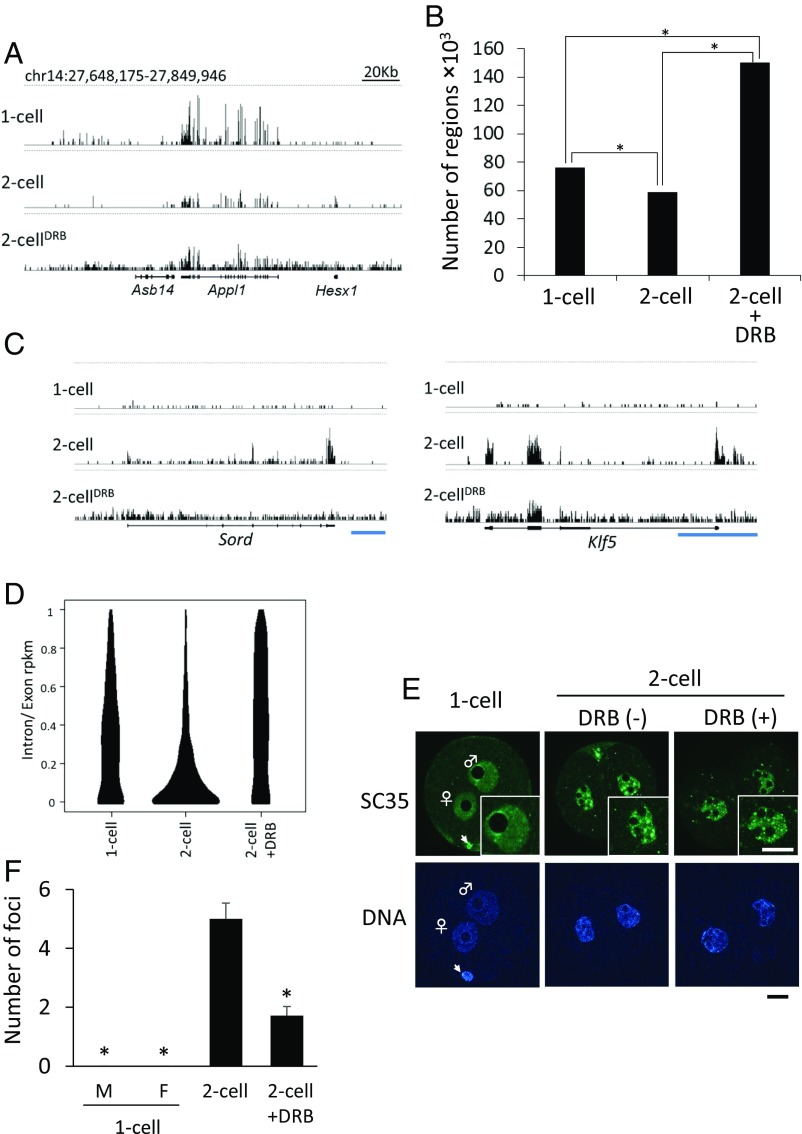
To confirm further whether minor ZGA occurred after removal of DRB, we examined transcription from intergenic regions, because widespread transcription from intergenic regions is a unique characteristic of minor ZGA (5). As expected, the transcripts originated from all over the genome, including intergenic regions in DRB-treated embryos (Fig. 4A). Quantitative analysis for intergenic expression, as we previously described (5), also demonstrated extensive expression from intergenic regions in these embryos. We divided the intergenic regions across the entire genome into 1-kb segments and enumerated divided segments in which at least a single read was uniquely mapped. The number of mapped regions was around threefold higher in DRB-treated two-cell embryos than the control two-cell embryos (Fig. 4B). Thus, the transcriptome of these late two-cell embryos following removal of DRB was similar to that of minor ZGA but not major ZGA.
Fig. 4.
Transcription with the characteristics of minor ZGA in DRB-treated two-cell stage embryos. Embryos were cultured with DRB from 4 to 20 hpi and transferred to DRB-free medium. (A) Screenshots from the University of California, Santa Cruz (UCSC) genome browser (51) of RNA-seq data in one-cell embryos (13 hpi) and two-cell embryos (32 hpi) treated with and without DRB. The vertical scale was trimmed to 0–20 reads; trimming is indicated by horizontal dashed lines. (B) Quantification of the number of transcribed intergenic regions in one-cell embryos and two-cell embryos treated with or without DRB. Intergenic regions were divided into 1-kb regions across the genome and then the number of regions that were uniquely mapped by at least a single read was counted. Asterisks indicate significant differences by χ2 test (P < 0.05). (C) Distribution of the mapped reads of Sord and Klf5 that are transcribed during both of minor and major ZGA but not in oocytes. Shown are RNA sequencing data from one-cell embryos and two-cell embryos treated with or without DRB visualized using the UCSC genome browser. The vertical scale was trimmed to 0–20 reads. Dashed lines indicate trimming at 20 reads. An exon–intron structure of each gene is described under the screenshot. (Blue scale bars, 5 kb.) (D) Violin plot representing intron RPKM/exon RPKM (intron and exon read counts normalized to 1-kb length) ratios for transcripts expressed in embryos but not in MII eggs. (E) Images of serine/arginine-rich splicing factor 2 (SC35) immunocytochemical staining in pronuclei of one-cell embryos and nuclei of two-cell embryos treated with [DRB(+)] and without DRB [DRB(−)]. Insets represent the magnified images. The experiment was conducted two times and six nuclei were examined for each experiment. (Scale bars, 20 μm.) Arrowheads indicate polar bodies. (F) The number of SC35 foci in the nucleus/pronucleus in the immunofluorescence images of E. The foci whose size occupied more than 0.03% of the area of whole embryos were recognized as those of SC35. Data are represented as mean ± SEM. Asterisks indicate a significant difference from the control two-cell embryos (Student’s t test; P < 0.05).
Given that promiscuous transcription was observed in DRB-treated embryos in late two-cell embryos, the other unique characteristic of minor ZGA, inefficient splicing (5), was explored. On the genome browser, DRB-treated embryos showed little enrichment of sequence reads mapped to exons compared with introns, indicating that introns are not sufficiently removed (Fig. 4C). We determined the reads per kilobase and million mapped reads (RPKMs) using the sequence reads mapped to exons and introns, and calculated their ratio (intron RPKM/exon RPKM) for each gene. Violin plots of these ratios are very narrow—above 0.3 in the control two-cell embryos—but much wider in DRB-treated embryos (Fig. 4D), indicating that splicing does not occur efficiently in DRB-treated embryos.
To confirm further inefficient splicing in DRB-treated embryos, we examined the presence of nuclear speckles, which store splicing factors required for pre-mRNA splicing (23), because few nuclear speckles are observed in one-cell–stage embryos (5). Although at 32 hpi several distinct foci of serine/arginine-rich splicing factor 2 (SC35), which is a component of nuclear speckles (23, 24), were observed in control two-cell embryos, only a few small foci were detected in DRB-treated embryos (Fig. 4 E and F). Therefore, functional proteins would not be synthesized from the unspliced pre-mRNA in these embryos. These results strongly suggest the pattern of gene expression in DRB-treated embryos at the late two-cell stage was quite similar to that during minor ZGA.
It is possible that an aberrant zygotic program caused by DRB treatment affected the maternal program (i.e., degradation of maternal transcripts and/or proteins) and lead to the observed developmental arrest. Therefore, we determined whether DRB treatment affected degradation of maternal mRNAs. Because some of genes expressed in oocytes are also transcribed during ZGA, we examined transcript abundance of oocyte-specific genes (i.e., not transcribed during ZGA). A heatmap of 149 oocyte-specific transcripts (5) (Dataset S6) in MII eggs, one-cell embryos, and two-cell embryos treated with and without DRB, indicated that DRB treatment did not affect degradation of these transcripts (Fig. 3C).
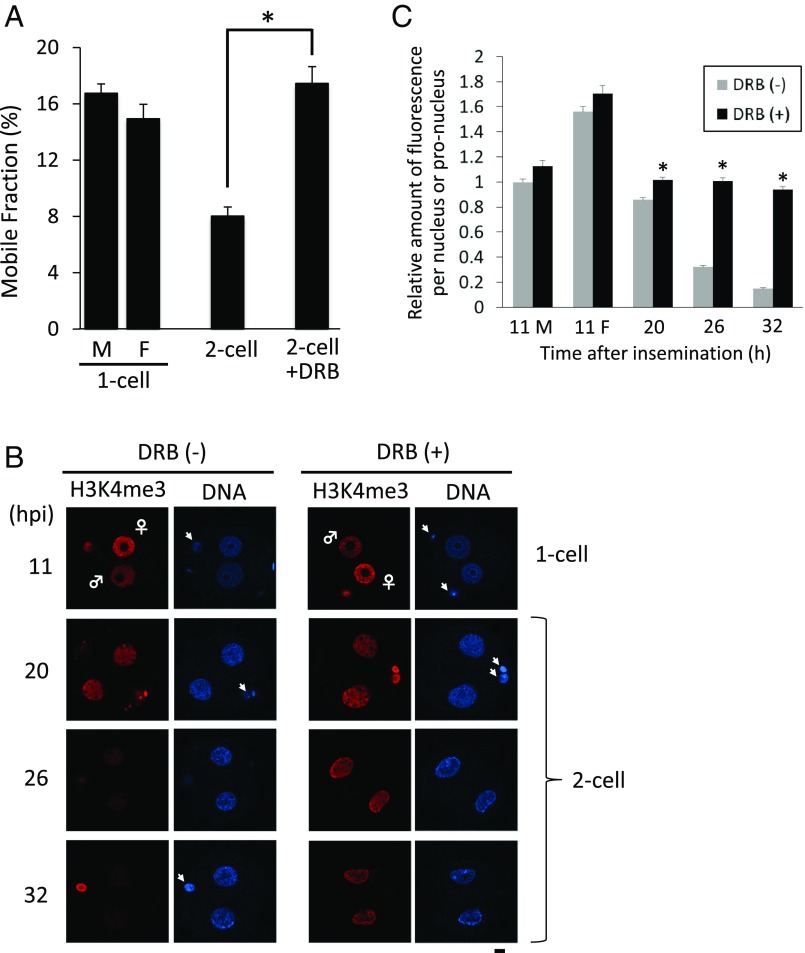
Chromatin structure plays an important role in regulating transcription and it has been suggested that a loosened chromatin structure is involved in the promiscuous transcription that occurs during minor ZGA (5, 25). We demonstrated by fluorescence recovery after photobleaching (FRAP) analysis that chromatin structure is extremely loosened in one-cell embryos and that it become tighter during the two-cell stage (26). FRAP analysis using an eGFP-tagged core histone estimates the mobility of core histones that reflects the looseness of chromatin structure (27, 28). Because inhibition of minor ZGA led to the persistence of promiscuous transcription in two-cell embryos (Fig. 4), minor ZGA could induce the change of chromatin structure to a tighter one. Consistent with this proposal is that the mobile fraction, an index of the looseness of chromatin structure, remained high in DRB-treated embryos (Fig. 5A).
Fig. 5.
Deficiency in the formation of tight chromatin structure and demethylation of H3K4me3 in DRB-treated two-cell–stage embryos. Embryos were cultured with DRB from 4 to 20 hpi and transferred to DRB-free medium. (A) Embryos at the one- and two-cell stages and DRB-treated two-cell–stage embryos were subjected to FRAP for analysis of chromatin structure. The embryos were collected at 11 and 26 hpi for one- and two-cell–stage embryos, respectively. M and F represent male and female pronuclei, respectively, in one-cell embryos. The experiment was reproducible at three times and more than seven embryos were examined. Asterisks indicate a significant difference from the control two-cell embryos (Student’s t test; P < 0.05). Data are represented as mean ± SEM. (B) Embryos at the one- and two-cell stages and DRB-treated two-cell–stage embryos were subjected to immunocytochemistry with an anti-H3K4me3 antibody at 11, 20, 26, and 32 hpi. (Scale bar, 10 μm.) Arrowheads indicate polar bodies. (C) Quantification of H3K4me3 signal in the images of immunocytochemistry in B. Three independent experiments were performed and 8–12 embryos were analyzed in total at each time point. Data are represented as mean ± SEM. An asterisk represents significant differences (by Student’s t test; P < 0.01).
Histone modifications play crucial roles in regulating transcription. The global level of H3 lysine 4 trimethylation (H3K4me3), a hallmark of active genes, decreases during the two-cell stage (29, 30). Consistent with loss of H3K4me3 from nucleosomes being associated with the transition from minor to major ZGA is that this decrease was not observed in DRB-treated embryos (Fig. 5 B and C). We also examined the effect of DRB treatment on nuclear deposition of H3K9ac, H3K27me3, and H3K36me3, whose levels change between the one- and two-cell stages (31–33), and no obvious difference was detected for these modified histones between DRB-treated and the control embryos (SI Appendix, Fig. S7)
Taken together, these results demonstrate that the zygotic program, including epigenetic changes, is compromised at the two-cell stage by transiently inhibiting minor ZGA, whereas the maternal program (i.e., degradation of maternal mRNA) occurs with the expected timing
Discussion
Genome activation in preimplantation embryos occurs in two phases—namely, minor and major—the timing being species-dependent. For example, in mouse minor ZGA occurs during the one-cell stage with major ZGA occurring during the two-cell stage, whereas in bovine, minor ZGA occurs during the two/four-cell stage with major ZGA initiating during the eight-cell stage. Major ZGA and the reprogramming of gene expression that occurs during major ZGA are critical for further development (16, 34). The role of minor ZGA in development, however, has remained enigmatic (see introductory section) but inroads have been made. For example, in mouse the products of minor ZGA transcription were characterized, with the unexpected finding that transcription is relatively promiscuous, low-level, genome-wide, and produces transcripts from thousands of protein-coding genes that are inefficiently spliced and polyadenylated (5, 35). By transiently inhibiting minor ZGA with DRB, we report here that minor ZGA is indispensable for preimplantation development.
Transient inhibition of minor ZGA (i.e., from the mid one-cell to the early two-cell stage) results in developmental arrest mostly at the two-cell stage, with many minor ZGA genes, but not major ones, being expressed. Furthermore, the transcription characteristics of minor ZGA (i.e., pervasive transcription from intergenic regions and inefficient splicing) are observed in late two-cell embryos. The transcriptome similarity of one-cell and transiently inhibited late two-cell embryos suggests minor ZGA must precede major ZGA for continued development (i.e., the program of gene expression is not driven by a zygotic clock) (36). It is also unlikely that failure to clear maternal mRNAs causes developmental arrest because degradation of maternal mRNAs appears unaffected. Further support for an essential role of minor ZGA for development is that the effect on development of transiently inhibiting major ZGA is much less pronounced.
Although >90% of the embryos arrested at the two-cell stage after removal of DRB at 46 hpi, 40% of the embryos developed beyond the two-cell stage and arrested at various stages at 96 hpi (Fig. 2A). This difference of arrested stage could be due to asynchronized embryo development. Alternatively, it could arise from differences in expression levels of RNAs and proteins that play important roles for development. Indeed, some genes transcribed during major ZGA were still expressed or up-regulated after removal of DRB (Fig. 3 A and B).
DRB inhibits Pol I-mediated transcription as well as a Pol II-mediated one (37). However, it is unlikely that the developmental arrest in the DRB-treated embryos was due to the inhibition of Pol I-mediated transcription. Inhibition of Pol I caused developmental arrest at the one-cell stage (38), whereas most of the DRB-treated embryos developed to and arrested at the two-cell stage in our study (Fig. 2A). In addition, although DRB inhibits Pol I-mediated transcription, the degree of inhibition is lower in Pol I-mediated transcription than Pol II-mediated transcription (37). Because we used the minimum concentration of DRB to inhibit the Pol II-mediated transcription (Fig. 1B), Pol I-mediated transcription would not be efficiently inhibited in our experimental conditions.
Chromatin structure becomes “tighter” during MZT (26, 39). The lack of a requirement for an enhancer—enhancers function by overcoming chromatin-mediated transcriptional repression—for efficient expression of plasmid-borne reporter genes and promiscuous genome-wide transcription during minor ZGA likely reflect a loosened chromatin structure (5, 25, 40). Reciprocally, the requirement for an enhancer for efficient expression of plasmid-borne reporter genes and a decrease in intergenic transcription during major ZGA presumably reflects chromatin structure becoming “tighter” (5, 25, 26, 40). Indeed, the absence of minor ZGA caused the persistence of loosened state of chromatin structure at the two-cell stage (Fig. 5A).
H3K4me3 is a hallmark of active genes and facilitates a loosened chromatin structure by recruiting CHD1, a chromatin remodeler (41); H3K4me3 typically maps to a highly defined narrow region over a promoter of an active gene. Results from recent chromatin immunoprecipitation experiments revealed that broad peaks of H3K4me3 are observed on the maternal genome of zygotes and that this distribution transforms into a sharp peak localized at the promoter region of active genes in two-cell embryos (27, 42, 43). Interestingly, a broad H3K4me3 distribution is still observed at the late two-cell stage when transcription is inhibited by treatment with α-amanitin during the one- and early two-cell stages. Thus, minor ZGA and transcription per se could drive this change. A possible mechanism is that transcriptional elongation erases H3K4 methylation through H3K36 methylation, which is regulated by phosphorylation of S2 of the carboxyl-terminal domain (44). In addition, the transcriptome of DRB-treated two-cell embryos indicates that functional proteins are not produced because transcripts are inefficiently spliced. A consequence would be reduced amounts of KDM5A and -B, and H3K4me3 demethylases (29) in late two-cell embryos, which would be an additional factor contributing to maintaining high levels of H3K4me3 in these treated embryos.
The picture that emerges regarding control of early preimplantation development in mouse is that maternally inherited transcripts and proteins are responsible for development to the two-cell stage because zygotes treated with α-amanitin cleave but then arrest at G2 phase after completing DNA replication (45). Minor ZGA, which is characterized by promiscuous transcription that produces nonfunctional transcripts, except for transcripts encoded by intronless genes, provides a mechanism to remodel chromatin into a mature structure that supports major ZGA and expression of genes necessary for further development.
Materials and Methods
Collection and Culture of Oocytes and Embryos.
The methods of collecting fully grown oocytes and MII stage eggs, and fertilization and culturing embryos were described previously (5, 46) and are detailed in SI Appendix. All of the procedures using animals were reviewed and approved by the University of Tokyo Institutional Animal Care and Use Committee and were performed in accordance with the Guiding Principles for the Care and Use of Laboratory Animals.
Treatment with DRB.
To transiently inhibit minor ZGA, one-cell embryos were cultured in potassium simplex optimized medium (KSOM) containing 80 μM DRB (Sigma-Aldrich) between 4 and 20 hpi, and the treated embryos were then washed six times in KSOM without DRB and then cultured in KSOM without DRB. For transient inhibition of major ZGA, two-cell embryos were cultured in KSOM containing 80 μM DRB between 26 and 42 hpi. Control embryos were cultured in DRB-free medium that contained the appropriate amount of dimethyl sulfoxide (DMSO), the solvent used to prepare the DRB stock solution.
In Vitro Transcription Assay.
Transcriptional activity was measured by incorporation of BrU into nascent RNA, as described previously (13), and is detailed in SI Appendix.
Detection of DNA Replication.
The embryos were transferred into KSOM containing 10 μM BrdU (Roche Diagonostic) at 14 hpi and DNA synthesis was detected at 1-h intervals by BrdU incorporation, as previously described (47).
Immunocytochemistry.
The methods of immunocytochemistry for phosphorylated Ser2 of the carboxyl-terminal domain, SC35, H3K4me3, H3K9ac, H3K27me3, and H3K36me3 are described in SI Appendix.
FRAP.
FRAP analysis was conducted as described previously with slight modification (26). Briefly, after photobleaching images were taken at 5-s intervals for 60 s. The mobile fraction (MF), an index of the rate of fluorescence recovery, was calculated using the following equation (38–40): MF = (Fend − Fpost)/(Fpre − Fpost), where Fend is the relative intensity of fluorescence at the endpoint, Fpost is immediately after photo-bleaching, and Fpre is before photo-bleaching.
RNA Extraction and Preparation of the RNA-Seq Library.
The methods of RNA extraction and preparation of the RNA-seq library are described in SI Appendix.
Heatmap, k-Means Nonhierarchical Clustering, Hierarchical Clustering.
Relative expression levels were calculated by RPKM values in MII eggs, one- and two-cell embryos treated with or without DRB. The average RPKM value of these four samples was calculated and set as 1 for each gene. Relative expression values were converted into log2 values and represented in a heatmap using TIGR Multiple Experiment Viewer (MeV) (48).
To conduct analysis of k-means nonhierarchical clustering, RPKM values in oocytes were set as 1 and then relative expression values were transformed into log10 values and subjected to k-means nonhierarchical clustering using MeV.
To compare transcript levels between two-cell embryos treated with and without DRB, average RPKM values of two samples were set as 1 and then relative expression values were transformed into log2 values and subjected to hierarchical clustering using Cluster 3.0 (49). These data were visualized by Java Treeview (50).
Reverse Transcription and PCR.
Reverse transcription and PCR was conducted as described in SI Appendix.
Supplementary Material
Acknowledgments
This work was supported in part by Grants-in-Aid Grants 20062002, 2525205, 416H01215, and 17K19318 from the Ministry of Education, Culture, Sports, Science and Technology of Japan (to F.A.). R.M.S. was supported by NIH Grant HD022681.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequence reported in this paper has been deposited in the DDBJ Sequence Read Archive, trace.ddbj.nig.ac.jp/dra/index.html (accession no. DRA006557).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1804309115/-/DCSupplemental.
References
- 1.Bleil JD, Wassarman PM. Mammalian sperm-egg interaction: Identification of a glycoprotein in mouse egg zonae pellucidae possessing receptor activity for sperm. Cell. 1980;20:873–882. doi: 10.1016/0092-8674(80)90334-7. [DOI] [PubMed] [Google Scholar]
- 2.McGrath SA, Esquela AF, Lee SJ. Oocyte-specific expression of growth/differentiation factor-9. Mol Endocrinol. 1995;9:131–136. doi: 10.1210/mend.9.1.7760846. [DOI] [PubMed] [Google Scholar]
- 3.Rajkovic A, Matzuk MM. Functional analysis of oocyte-expressed genes using transgenic models. Mol Cell Endocrinol. 2002;187:5–9. doi: 10.1016/s0303-7207(01)00710-9. [DOI] [PubMed] [Google Scholar]
- 4.Moore GPM, Lintern-Moore S. A correlation between growth and RNA synthesis in the mouse oocyte. J Reprod Fertil. 1974;39:163–166. doi: 10.1530/jrf.0.0390163. [DOI] [PubMed] [Google Scholar]
- 5.Abe K, et al. The first murine zygotic transcription is promiscuous and uncoupled from splicing and 3′ processing. EMBO J. 2015;34:1523–1537. doi: 10.15252/embj.201490648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Paynton BV, Bachvarova R. Polyadenylation and deadenylation of maternal mRNAs during oocyte growth and maturation in the mouse. Mol Reprod Dev. 1994;37:172–180. doi: 10.1002/mrd.1080370208. [DOI] [PubMed] [Google Scholar]
- 7.Ma J, Flemr M, Strnad H, Svoboda P, Schultz RM. Maternally recruited DCP1A and DCP2 contribute to messenger RNA degradation during oocyte maturation and genome activation in mouse. Biol Reprod. 2013;88:11. doi: 10.1095/biolreprod.112.105312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yu C, et al. BTG4 is a meiotic cell cycle-coupled maternal-zygotic-transition licensing factor in oocytes. Nat Struct Mol Biol. 2016;23:387–394. doi: 10.1038/nsmb.3204. [DOI] [PubMed] [Google Scholar]
- 9.Pikó L, Clegg KB. Quantitative changes in total RNA, total poly(A), and ribosomes in early mouse embryos. Dev Biol. 1982;89:362–378. doi: 10.1016/0012-1606(82)90325-6. [DOI] [PubMed] [Google Scholar]
- 10.Alizadeh Z, Kageyama S, Aoki F. Degradation of maternal mRNA in mouse embryos: Selective degradation of specific mRNAs after fertilization. Mol Reprod Dev. 2005;72:281–290. doi: 10.1002/mrd.20340. [DOI] [PubMed] [Google Scholar]
- 11.Matsumoto K, et al. Onset of paternal gene activation in early mouse embryos fertilized with transgenic mouse sperm. Mol Reprod Dev. 1994;39:136–140. doi: 10.1002/mrd.1080390203. [DOI] [PubMed] [Google Scholar]
- 12.Bouniol C, Nguyen E, Debey P. Endogenous transcription occurs at the 1-cell stage in the mouse embryo. Exp Cell Res. 1995;218:57–62. doi: 10.1006/excr.1995.1130. [DOI] [PubMed] [Google Scholar]
- 13.Aoki F, Worrad DM, Schultz RM. Regulation of transcriptional activity during the first and second cell cycles in the preimplantation mouse embryo. Dev Biol. 1997;181:296–307. doi: 10.1006/dbio.1996.8466. [DOI] [PubMed] [Google Scholar]
- 14.Christians E, Campion E, Thompson EM, Renard JP. Expression of the HSP 70.1 gene, a landmark of early zygotic activity in the mouse embryo, is restricted to the first burst of transcription. Development. 1995;121:113–122. doi: 10.1242/dev.121.1.113. [DOI] [PubMed] [Google Scholar]
- 15.Davis W, Jr, De Sousa PA, Schultz RM. Transient expression of translation initiation factor eIF-4C during the 2-cell stage of the preimplantation mouse embryo: Identification by mRNA differential display and the role of DNA replication in zygotic gene activation. Dev Biol. 1996;174:190–201. doi: 10.1006/dbio.1996.0065. [DOI] [PubMed] [Google Scholar]
- 16.Warner CM, Versteegh LR. In vivo and in vitro effect of α-amanitin on preimplantation mouse embryo RNA polymerase. Nature. 1974;248:678–680. doi: 10.1038/248678a0. [DOI] [PubMed] [Google Scholar]
- 17.Tamm I, Hand R, Caliguiri LA. Action of dichlorobenzimidazole riboside on RNA synthesis in L-929 and HeLa cells. J Cell Biol. 1976;69:229–240. doi: 10.1083/jcb.69.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zandomeni R, Zandomeni MC, Shugar D, Weinmann R. Casein kinase type II is involved in the inhibition by 5,6-dichloro-1-beta-D-ribofuranosylbenzimidazole of specific RNA polymerase II transcription. J Biol Chem. 1986;261:3414–3419. [PubMed] [Google Scholar]
- 19.Kigami D, Minami N, Takayama H, Imai H. MuERV-L is one of the earliest transcribed genes in mouse one-cell embryos. Biol Reprod. 2003;68:651–654. doi: 10.1095/biolreprod.102.007906. [DOI] [PubMed] [Google Scholar]
- 20.Hutchins JR, Clarke PR. Many fingers on the mitotic trigger: Post-translational regulation of the Cdc25C phosphatase. Cell Cycle. 2004;3:41–45. [PubMed] [Google Scholar]
- 21.Wang Q, et al. Cyclin dependent kinase 1 inhibitors: A review of recent progress. Curr Med Chem. 2011;18:2025–2043. doi: 10.2174/092986711795590110. [DOI] [PubMed] [Google Scholar]
- 22.Wolgemuth DJ. Function of the A-type cyclins during gametogenesis and early embryogenesis. Results Probl Cell Differ. 2011;53:391–413. doi: 10.1007/978-3-642-19065-0_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang S, Spector DL. U1 and U2 small nuclear RNAs are present in nuclear speckles. Proc Natl Acad Sci USA. 1992;89:305–308. doi: 10.1073/pnas.89.1.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim YD, et al. NSrp70 is a novel nuclear speckle-related protein that modulates alternative pre-mRNA splicing in vivo. Nucleic Acids Res. 2011;39:4300–4314. doi: 10.1093/nar/gkq1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Majumder S, Miranda M, DePamphilis ML. Analysis of gene expression in mouse preimplantation embryos demonstrates that the primary role of enhancers is to relieve repression of promoters. EMBO J. 1993;12:1131–1140. doi: 10.1002/j.1460-2075.1993.tb05754.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ooga M, Fulka H, Hashimoto S, Suzuki MG, Aoki F. Analysis of chromatin structure in mouse preimplantation embryos by fluorescent recovery after photobleaching. Epigenetics. 2016;11:85–94. doi: 10.1080/15592294.2015.1136774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chalut KJ, et al. Chromatin decondensation and nuclear softening accompany Nanog downregulation in embryonic stem cells. Biophys J. 2012;103:2060–2070. doi: 10.1016/j.bpj.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martin RM, Cardoso MC. Chromatin condensation modulates access and binding of nuclear proteins. FASEB J. 2010;24:1066–1072. doi: 10.1096/fj.08-128959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dahl JA, et al. Broad histone H3K4me3 domains in mouse oocytes modulate maternal-to-zygotic transition. Nature. 2016;537:548–552. doi: 10.1038/nature19360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yu C, et al. CFP1 regulates histone H3K4 trimethylation and developmental potential in mouse oocytes. Cell Rep. 2017;20:1161–1172. doi: 10.1016/j.celrep.2017.07.011. [DOI] [PubMed] [Google Scholar]
- 31.Santenard A, et al. Heterochromatin formation in the mouse embryo requires critical residues of the histone variant H3.3. Nat Cell Biol. 2010;12:853–862. doi: 10.1038/ncb2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hayashi-Takanaka Y, et al. Tracking epigenetic histone modifications in single cells using Fab-based live endogenous modification labeling. Nucleic Acids Res. 2011;39:6475–6488. doi: 10.1093/nar/gkr343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bošković A, et al. Analysis of active chromatin modifications in early mammalian embryos reveals uncoupling of H2A.Z acetylation and H3K36 trimethylation from embryonic genome activation. Epigenetics. 2012;7:747–757. doi: 10.4161/epi.20584. [DOI] [PubMed] [Google Scholar]
- 34.Schultz RM. The molecular foundations of the maternal to zygotic transition in the preimplantation embryo. Hum Reprod Update. 2002;8:323–331. doi: 10.1093/humupd/8.4.323. [DOI] [PubMed] [Google Scholar]
- 35.Park SJ, et al. Inferring the choreography of parental genomes during fertilization from ultralarge-scale whole-transcriptome analysis. Genes Dev. 2013;27:2736–2748. doi: 10.1101/gad.227926.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nothias JY, Majumder S, Kaneko KJ, DePamphilis ML. Regulation of gene expression at the beginning of mammalian development. J Biol Chem. 1995;270:22077–22080. doi: 10.1074/jbc.270.38.22077. [DOI] [PubMed] [Google Scholar]
- 37.Hellung-Larsen P, Jensen EG, Frederiksen S. Effect of 5,6-dichloro-1-beta-D-ribofuranosylbenzimidazole on the synthesis of low molecular weight, RNA components. Biochem Biophys Res Commun. 1981;99:1303–1310. doi: 10.1016/0006-291x(81)90761-0. [DOI] [PubMed] [Google Scholar]
- 38.Lin CJ, Koh FM, Wong P, Conti M, Ramalho-Santos M. Hira-mediated H3.3 incorporation is required for DNA replication and ribosomal RNA transcription in the mouse zygote. Dev Cell. 2014;30:268–279. doi: 10.1016/j.devcel.2014.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cho T, Sakai S, Nagata M, Aoki F. Involvement of chromatin structure in the regulation of mouse zygotic gene activation. Anim Sci J. 2002;73:113–122. [Google Scholar]
- 40.Wiekowski M, Miranda M, DePamphilis ML. Regulation of gene expression in preimplantation mouse embryos: Effects of the zygotic clock and the first mitosis on promoter and enhancer activities. Dev Biol. 1991;147:403–414. doi: 10.1016/0012-1606(91)90298-h. [DOI] [PubMed] [Google Scholar]
- 41.Sims RJ, 3rd, Reinberg D. Stem cells: Escaping fates with open states. Nature. 2009;460:802–803. doi: 10.1038/460802a. [DOI] [PubMed] [Google Scholar]
- 42.Liu X, et al. Distinct features of H3K4me3 and H3K27me3 chromatin domains in pre-implantation embryos. Nature. 2016;537:558–562. doi: 10.1038/nature19362. [DOI] [PubMed] [Google Scholar]
- 43.Zhang B, et al. Allelic reprogramming of the histone modification H3K4me3 in early mammalian development. Nature. 2016;537:553–557. doi: 10.1038/nature19361. [DOI] [PubMed] [Google Scholar]
- 44.Mbogning J, et al. Functional interaction of Rpb1 and Spt5 C-terminal domains in co-transcriptional histone modification. Nucleic Acids Res. 2015;43:9766–9775. doi: 10.1093/nar/gkv837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Goddard MJ, Pratt HP. Control of events during early cleavage of the mouse embryo: An analysis of the ‘2-cell block’. J Embryol Exp Morphol. 1983;73:111–133. [PubMed] [Google Scholar]
- 46.Abe K, Inoue A, Suzuki MG, Aoki F. Global gene silencing is caused by the dissociation of RNA polymerase II from DNA in mouse oocytes. J Reprod Dev. 2010;56:502–507. doi: 10.1262/jrd.10-068a. [DOI] [PubMed] [Google Scholar]
- 47.Aoki E, Schultz RM. DNA replication in the 1-cell mouse embryo: Stimulatory effect of histone acetylation. Zygote. 1999;7:165–172. doi: 10.1017/s0967199499000532. [DOI] [PubMed] [Google Scholar]
- 48.Saeed AI, et al. TM4: A free, open-source system for microarray data management and analysis. Biotechniques. 2003;34:374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- 49.de Hoon MJ, Imoto S, Nolan J, Miyano S. Open source clustering software. Bioinformatics. 2004;20:1453–1454. doi: 10.1093/bioinformatics/bth078. [DOI] [PubMed] [Google Scholar]
- 50.Saldanha AJ. Java Treeview—Extensible visualization of microarray data. Bioinformatics. 2004;20:3246–3248. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
- 51.Karolchik D, Hinrichs AS, Kent WJ. The UCSC genome browser. Curr Protoc Bioinformatics. 2012;Chapter 1:Unit1.4. doi: 10.1002/0471250953.bi0104s28. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.