Significance
The majority of viral genomic sequences available today are fewer than 50 years old. Parvovirus B19 (B19V) is a ubiquitous human pathogen causing fifth disease in children, as well as other conditions. By isolating B19V DNA from human remains between ∼0.5 and 6.9 thousand years old, we show that B19V has been associated with humans for thousands of years, which is significantly longer than previously thought. We also show that the virus has been evolving at a rate an order of magnitude lower than estimated previously. Access to viral sequences isolated from individuals living thousands of years ago greatly improves our understanding of the timescales of virus evolution, spatiotemporal distribution, and their substitution rates, and can uncover genetic diversity that is now extinct.
Keywords: parvovirus B19, ancient DNA, virus evolution, paleo virology, virology
Abstract
Human parvovirus B19 (B19V) is a ubiquitous human pathogen associated with a number of conditions, such as fifth disease in children and arthritis and arthralgias in adults. B19V is thought to evolve exceptionally rapidly among DNA viruses, with substitution rates previously estimated to be closer to those typical of RNA viruses. On the basis of genetic sequences up to ∼70 years of age, the most recent common ancestor of all B19V has been dated to the early 1800s, and it has been suggested that genotype 1, the most common B19V genotype, only started circulating in the 1960s. Here we present 10 genomes (63.9–99.7% genome coverage) of B19V from dental and skeletal remains of individuals who lived in Eurasia and Greenland from ∼0.5 to ∼6.9 thousand years ago (kya). In a phylogenetic analysis, five of the ancient B19V sequences fall within or basal to the modern genotype 1, and five fall basal to genotype 2, showing a long-term association of B19V with humans. The most recent common ancestor of all B19V is placed ∼12.6 kya, and we find a substitution rate that is an order of magnitude lower than inferred previously. Further, we are able to date the recombination event between genotypes 1 and 3 that formed genotype 2 to ∼5.0–6.8 kya. This study emphasizes the importance of ancient viral sequences for our understanding of virus evolution and phylogenetics.
Infection with human Parvovirus B19 (B19V) can have a number of different outcomes, from asymptomatic or nonspecific symptoms to fifth disease (erythema infectiosum) in children, arthritis and arthralgias in adults, and fetal death (hydrops fetalis) in pregnant women. It can also lead to transient or persistent erythroid aplasia and aplastic crisis in people with underlying hematological disorders (1, 2). Management of B19V infections is generally limited to symptomatic treatment, as there are currently no vaccines or antivirals (1). The virus replicates in erythroid precursor cells in the bone marrow (3). After the initial infection, viral DNA is detectable in multiple human tissues, including kidney, lymph nodes, heart, testes, and synovial tissue, in symptomatic and asymptomatic individuals (4–9).
The persistence of viral DNA in an infected individual is thought to be lifelong, and the persisting virus corresponds to the virus genotype of the initial infection (10). B19V can be transmitted via the respiratory or blood-borne route. The virus can be detected in the saliva and is present in the blood in high titers (1013 virions/mL) during the viraemic phase of the infection (1, 11). In blood donors, seroprevalence of B19V is around 60% on average, and the rate of new individuals infected per year is estimated at between 0.5% and 1% (12).
B19V is a single-stranded DNA (ssDNA) virus with a genome of ∼5,600 nucleotides. The coding region is flanked by terminal repeats of 383 nucleotides and contains three major proteins: the nonstructural protein NS, the capsid proteins VP1 and VP2, and the minor proteins 11 kDa, 9 kDa, and 7.5 kDa (1).
B19V belongs to the Erythrovirus genus in the Parvoviridae family (1). There are three genotypes, with genotype 1 having ∼10% sequence divergence from genotypes 2 and 3 and genotypes 2 and 3 having ∼5% sequence divergence between each other (13). Genotypes 1 and 3 are further split into two subgroups, a and b, with sequence divergence of about 5% (14, 15). All three genotypes form a single serotype (16). The distribution of the three genotypes is not spatially and temporally uniform: genotype 1 has a worldwide distribution (17), genotype 2 is found mainly in elderly adults in northern Europe (10), and genotype 3 is found in Sub-Saharan and West Africa, South America, and France (14). Because genotype 2 is today only found in tissues of people born before the 1970s, it has been hypothesized that it was replaced by genotype 1 and that genotype 1 originated in the second half of the 20th century (18).
B19V is deemed unique among the DNA viruses because its substitution rate, inferred from modern heterochronous sequences, is unusually high, in the range of 1.0–4.0 × 10−4 nucleotide substitutions per site per year (s/s/y) (18–20), which is more similar to substitution rates of RNA viruses than either single-stranded or double-stranded DNA viruses (21).
Recent advances in sequencing ancient DNA (aDNA) have provided important insights into past human population dynamics (22) and the evolution of bacterial human pathogens (23, 24), but have only recently been applied to viruses (25, 26). Viral sequences recovered from aDNA samples have provided important reference points for the calibration of molecular clocks (25, 26). These samples can also provide insight into the spatiotemporal distribution of past viral variants and strains (25). B19V has characteristics that should favor molecular preservation, including a DNA genome, high viraemia, and a stable nonenveloped virion (27), and indeed the persistence of B19V DNA in bones from individuals deceased ∼70 years ago has been established (20). Here, we extend our knowledge of the virus further back in time and present 10 B19V coding region sequences (63.9–99.7% genome coverage) recovered from humans living in Eurasia and Greenland between ∼0.5 and 6.9 kya, leading to an improved understanding of the timescale of B19V evolution.
Results
Identification and Authentication.
We screened shotgun DNA sequencing data representing dental or skeletal remains of 1,578 ancient human individuals (∼0.2–24.0 kya) recovered from across Eurasia, Southeast Asia, and Greenland for the presence of reads matching B19V. A total of 20 samples had reads covering >30% of the coding region of B19V, 10 of which had coverage >50% (Table 1 and SI Appendix, Table S1). The samples with coverage >50% were between ∼0.5 and ∼6.9 thousand years old and from different archaeologically defined cultures (four Viking Age Scandinavians, three Early Neolithic to Early Bronze Age Baikal Hunter-Gatherers, one early Slav from the Czech Republic, and one Tian Shan Hun) (Table 1). The individuals came from a wide geographic range, spanning Europe, Central Asia, and Greenland (Fig. 1 and Table 1). The samples with coverage >50% showed DNA damage patterns typical for aDNA (28) when at least 100 reads were available (8 of 10 samples; SI Appendix, Fig. S1). Clear damage patterns could not be definitively identified when there were fewer than 100 reads (2 of 10 samples); there was no evidence for this being for any reason other than a low number of reads. Authenticity of the ancient B19V sequences presented here is further supported by our compliance with standard precautions for working with aDNA (29) and because B19V sequences were found in only 20 of 1,578 ancient human individuals, which would not be expected in case of ubiquitous laboratory contaminant. Finally, the ancient sequences mostly occupy basal positions in the phylogenetic tree, with short branch lengths from the trunk, for which the only explanation is that they are ancestors of modern strains.
Table 1.
Overview of samples included in phylogenetic analyses
| Sample | 14C date BP, SD | Mean cal BP, SD | Estimated age, years | Sample age, years | Site | Period/culture | Reads included in consensus | Coverage of consensus | Coverage depth |
| DA66 | 1,546 (33) | 1,451 (47) | N/A | 1,518 | Tian Shan, Kyrgyzstan | Hun | 535 | 99.70% | 7.8× |
| DA337 | 3,593 (67) | 3,900 (99) | N/A | 3,967 | Lake Baikal (Shamanka II), Russia | Early Bronze Age (EBA) | 883 | 98.40% | 13.5× |
| VK477 | N/D | N/D | ∼1,000 | 1,000 | Gotland, Sweden | Viking | 242 | 87.70% | 3.1× |
| RISE569 | 1,300 (30) | 1,237 (34) | N/A | 1,304 | Brandysek, Czech Republic | Early Slav | 285 | 84.10% | 3.4× |
| NEO105 | 415 (25) | 477 (42) | N/A | 544 | Bazaiha, Krasnoyarsk, Russia | Historical period | 219 | 83.60% | 2.4× |
| DA251 | 5,955 (72) | 6,795 (90) | N/A | 6,862 | Lake Baikal (Shamanka II), Russia | Early Neolithic | 228 | 82.80% | 3.2× |
| VK6 | N/D | N/D | ∼1,000 | 1,000 | Eastern Settlement, Greenland | Viking | 301 | 82.50% | 4.0× |
| VK154 | N/D | N/D | 980/990–1030–1035 AD | 1,000 | Bodzia, Poland | Viking | 134 | 71.40% | 1.6× |
| VK143 | N/D | N/D | 880–1000 AD | 1,077 | Oxford, UK | Viking | 120 | 67.30% | 1.6× |
| DA336 | 3,723 (70) | 4,079 (107) | N/A | 4,146 | Lake Baikal (Shamanka II), Russia | EBA | 92 | 63.90% | 1.3× |
Samples included in phylogenetic analysis, sorted by decreasing genome coverage. From left to right, the columns denote the sample name, the 14C age in years before the present (BP) and SD, the mean calibrated age BP and SD, the archaeological age estimated from the sample context, the sample age used in the phylogenetic analyses corresponding to the mean calibrated age BP relative to 2017, the site where the sample was found, the period or culture that the sample is from, the number of reads included in the consensus sequence, the coverage of the consensus sequence, and the coverage depth.
Fig. 1.
Geographic locations of the samples with reads matching B19V. Samples are colored by age. Squares indicate genotype 1, circles genotype 2. Empty symbols indicate samples with 30–50% coverage of the coding region, filled symbols samples with >50% coverage of the coding region that were included for further analysis. Samples that were negative for B19V are shown as small gray symbols (samples outside Eurasia are omitted for clarity).
Similarity to Modern Genotypes.
We compared the ancient sequences with all published nonartificial, complete B19V genomes in GenBank (SI Appendix, Fig. S2A). Five ancient sequences are most similar to genotype 1 (DA66, DA251, DA336, DA337, NEO105), and five to genotype 2 (RISE569, VK6, VK143, VK154, VK477). The mean pairwise sequence identity between the ancient and the modern sequences is below the mean within-genotype pairwise sequence identity of the modern sequences (blue and red dashed lines in SI Appendix, Fig. S2A). None of the ancient sequences are more diverged from any modern sequence than the modern genotypes are from each other (dashed black lines in SI Appendix, Fig. S2A); hence, we consider them part of the current genotypes and do not propose that they should be classified as new genotypes. We did not find any differences, such as insertions or deletions, between the ancient and the modern sequences. Of the samples with a genome coverage between 30% and 50% that were not included in the phylogenetic analysis, three had the highest number of reads matching genotype 1, and seven best matched genotype 2 (SI Appendix, Fig. S2B).
Recombination Analysis.
We performed recombination analyses using RDP4 (30) on the 10 ancient sequences with coverage >50% and a selection of 13 modern genotype 1–3 sequences, selected to represent the modern diversity. All seven recombination detection algorithms used by RDP4 identified all eight genotype 2 sequences included in the analysis (three modern and five ancient sequences) as potential recombinants, with very high probabilities (SI Appendix, Table S2). Recombination plots for the RDP and MaxChi algorithms of the RDP4 package for all eight genotype 2 sequences are shown in SI Appendix, Figs. S3 and S4. In all cases, the major parent was found to be most similar to one of the modern genotype 3 sequences included in the analysis, and the minor parent to resemble an ancient genotype 1 sequence most similar to DA337. To determine the timing of the recombination, we subsequently removed DA337, DA66, and a modern sequence (AB126262). Only after the removal of these three sequences was DA251, our oldest sequence, found to be the minor parent, indicating that the recombination event most likely took place between DA251 and DA337. Because of the age of the samples, the major parent clearly cannot be a modern sequence, suggesting that a genotype 3 ancestor and a genotype 1 virus that existed sometime during the evolutionary span from DA251 (6.9 kya) to DA337 (4.0 kya) recombined to form genotype 2. The identification of genotype 2 as a recombinant and the inferred recombination breakpoints closely correspond to the results of Shen et al. (31) in their analysis of modern B19V sequences (SI Appendix, Figs. S3 and S4).
Phylogenetic Analysis.
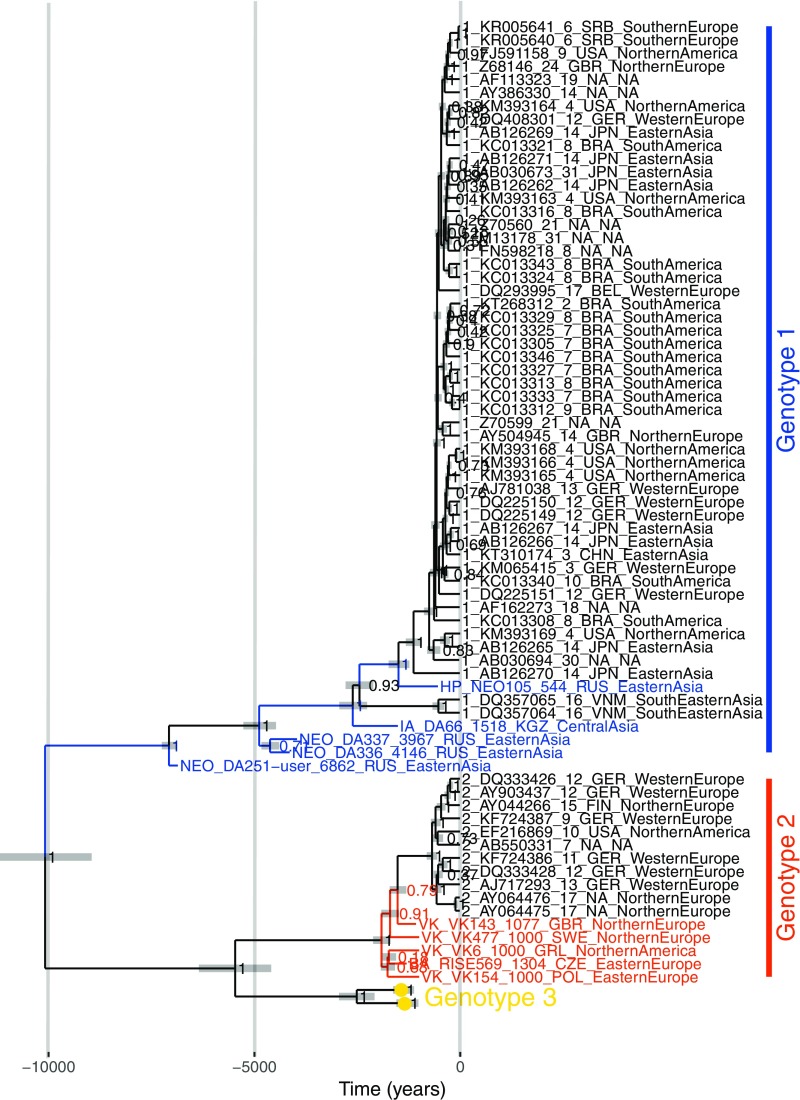
A phylogenetic tree inferred using maximum likelihood methods confirmed that the ancient sequences fall within the diversity of known B19V sequences (SI Appendix, Fig. S2C). Five ancient sequences (DA66, DA251, DA336, DA337, NEO105) fall within or basal to genotype 1, and five (RISE569, VK6, VK143, VK154, VK477) fall basal to genotype 2, consistent with the sequence similarity result presented earlier. Substitution saturation is known to affect the phylogenetic signal, and hence the inference of phylogenetic trees. However, by plotting transition and transversion frequencies against genetic distance (SI Appendix, Fig. S2D) or by testing for substitution saturation using DAMBE (SI Appendix, Table S3) (32), we did not find evidence for saturation in our sequences. To infer dated coalescent trees, sufficient temporal signal must be present in the data. A regression of root-to-tip distances from trees inferred using Neighbor Joining, maximum likelihood, and Bayesian methods all showed a temporal signal in the data (SI Appendix, Fig. S2E). Date randomization tests (33, 34) performed in BEAST2 (35) also support the presence of a temporal signal in the data (SI Appendix, Fig. S2F). Dated coalescent trees were consequently inferred using BEAST2 (Fig. 2) (35). In the following text, 95% highest posterior density (HPD) intervals are given in parentheses. We inferred a substitution rate of 1.22 × 10−5 (1.04 × 10−5–1.40 × 10−5) s/s/y and a root age of 10.1 (9.0–11.3) kya under a strict clock and a coalescent Bayesian skyline population prior (SI Appendix, Table S4) and a substitution rate of 1.67 × 10−5 (1.24 × 10−5–2.14 × 10−5) s/s/y and a root age of 8.4 (7.0–11.1) kya under a relaxed lognormal clock and a coalescent Bayesian skyline population prior, with identical topologies. The time to the most recent common ancestor of genotypes 1, 2, and 3 are 7.1 (6.9–7.3), 1.9 (1.7–2.1), and 2.5 (2.1–3.0) kya, respectively, under a strict clock and 7.3 (6.9–7.9), 1.7 (1.4–2.0), and 1.5 (0.8–2.4) kya, respectively, under a relaxed lognormal clock.
Fig. 2.
Maximum clade credibility tree inferred in BEAST2. A strict clock and coalescent Bayesian skyline population prior were used. The substitution rate is inferred as 1.22 × 10−5 s/s/y (95% HPD interval, 1.04 × 10−5–1.40 × 10−5 s/s/y), and the root age as 10.1 kya (95% HPD interval, 9.0–11.3 kya). The x axis denotes time into the past, in years. Taxon names: genotype/historical period, accession number/sample identifier, sample age, country abbreviation of sequence origin and region of sequence origin, as determined by the Standard country or area codes for statistical use (42). Horizontal gray bars indicate 95% HPD intervals of node ages.
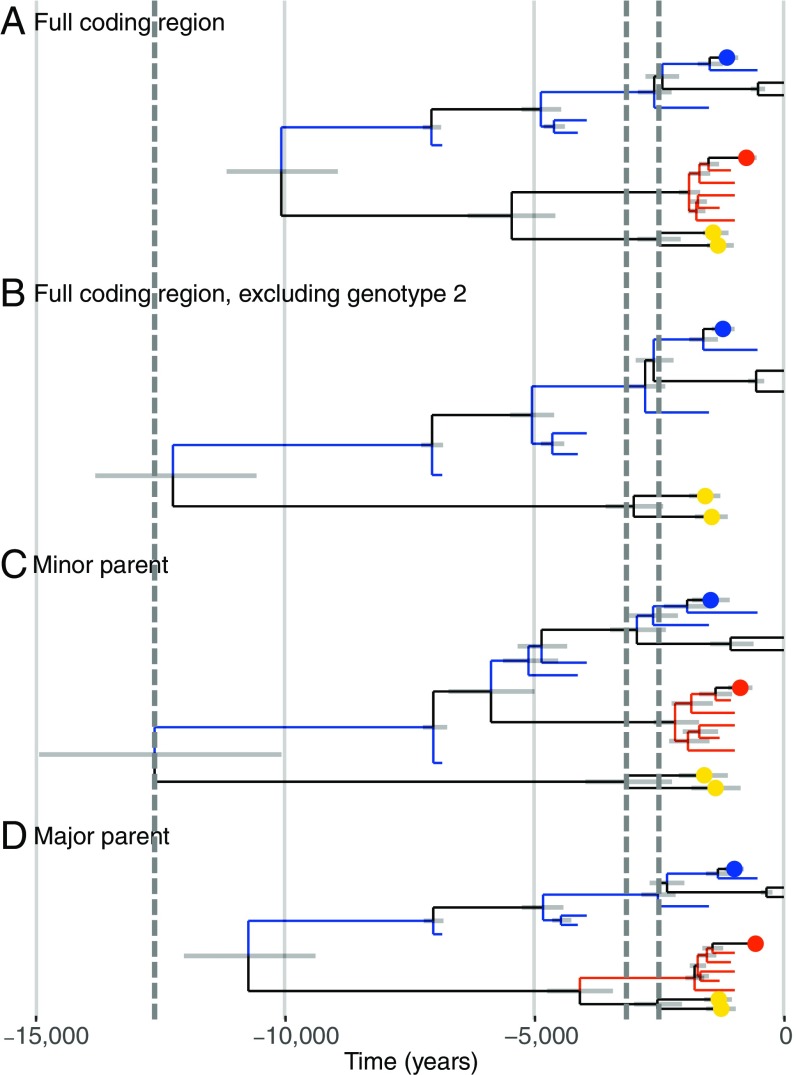
Recombination is known to affect the accuracy of phylogenetic analyses. Hence, to better understand the effect of recombination on the dated B19V tree, we inferred trees under a strict clock and coalescent Bayesian skyline population prior for the three genotypes individually, dating the most recent common ancestor of genotypes 1, 2, and 3 to 6.9 (6.9–6.9) kya, 1.6 (1.4–1.8) kya, and 0.6 (0.1, 6.1) kya, respectively. Further, we inferred trees under a strict clock and coalescent Bayesian skyline population prior for the full genome, but excluding all genotype 2 sequences (Fig. 3B), and separately for the region of the minor (Fig. 3C) and major (Fig. 3D) parent. The maximum root age was inferred to be 12.6 (10.4–15.2) kya when only the minor parent was considered (Fig. 3C); however, the 95% HPD interval of the root ages overlapped in all cases. The 95% HPD interval on the split of genotype 2 from genotype 1 in Fig. 3C suggests that the recombination event that formed genotype 2 occurred between 5.0 and 6.8 kya.
Fig. 3.
Trees inferred using BEAST2 from different regions of the B19V genome. Trees were inferred using a strict clock and coalescent Bayesian skyline population prior. Positions of ancient genotype 1 and ancient genotype 2 sequences are shown as blue or red tips, respectively. Collapsed modern genotype 1, 2, and 3 sequences are shown as blue, red, and yellow dots, respectively. 95% HPD intervals of node ages are shown by gray horizontal bars. The x axis denotes time into the past, in years. Vertical gray dashed lines indicate, from left to right, the maximum height of the root and the maximum and minimum height of the genotype 3 clade. (A) Tree from Fig. 2 using the full coding region. (B) Tree using the full coding region, excluding genotype 2 sequences. (C) Tree using the minor parent (positions 1,877–3,352 of the alignment). (D) Tree using the major parent (positions 1–1,876 and 3,353–4,354 of the alignment).
Discussion
We show that it is possible to recover and sequence single-stranded ancient viral DNA molecules from human skeletons that are up to ∼6.9 thousand years old. Although Toppinen et al. (20) found that human B19V DNA can persist in human bone for decades, the fact that ssDNA is able to persist for ∼6.9 kya may seem surprising, given that depurination and deamination occur about four times faster in ssDNA than in double-stranded DNA (36). However, single-stranded B19V DNA may be protected by the viral capsid, by self-annealing of ssDNA during preservation, or through the binding of ssDNA to the inside of the viral capsid, as observed in canine Parvovirus (37). This finding also provides hope for the retrieval of RNA viruses from archaeological samples, notwithstanding the traditional view of RNA as too unstable for long-term survival (36).
By computational screening of shotgun data from 1,578 ancient individuals, we found 20 samples with reads matching 30–99.7% of the B19V coding region, eight of which had a majority of reads matching genotype 1 and 12 matching genotype 2. No sample had a majority of reads matching genotype 3. The phylogenetic positioning of the ancient genotype 1 sequences may suggest a ladder-like evolution, in which older virus strains go extinct as they are replaced by novel variants, as observed in modern genotype 1 sequences (18).
The ancient sequences allow us to revise the current timelines for B19V evolution. Specifically, we show that B19V has been associated with humans for thousands of years, given our finding of ancient sequences as old as ∼6.9 kya that are still most similar to genotype 1. This rules out the suggestion that genotype 1 only arose after World War II (18, 20). However, endogenous parvovirus-derived genomes found in a large range of mammals suggests that the virus may be considerably older than 12.6 kya (38). Furthermore, we find a substitution rate of 1.22 × 10−5 s/s/y, approximately an order of magnitude lower than rates inferred solely from modern B19V sequences (18–20). Our rate estimate is in line with rates estimated for other single- and double-stranded DNA viruses (21). The fact that we find lower rates when sampling over the course of a longer period, as opposed to sampling over the course of a shorter period, is consistent with the phenomenon of time dependency of substitution rates (39, 40). This phenomenon has been attributed to unfixed changes at the tips of the tree, different selection pressures over time, or method errors (41). Inferred B19V substitution rates may decrease as older ancient sequences are discovered and incorporated into future analyses.
It has been shown previously that genotype 2 was formed by a recombination between genotypes 1 and 3 (31). Using the position of the ancient sequences as shown in Fig. 3C, we were able to date this recombination event to between 5.0 and 6.8 kya. Recombination leads to the underestimation of root ages in dated coalescent trees, which is what we observed when comparing trees inferred from the full genomes (Fig. 3A) with trees inferred from only genotypes 1 and 3 (Fig. 3B) or from the minor parent (Fig. 3C). Interestingly, we did not find the same when inferring a tree using the major parent (Fig. 3D), where the root age is underestimated, similar to the full tree in Fig. 3A. However, in all cases, the 95% HPD intervals of the root ages overlap.
On the basis of our data, it is possible to hypothesize a geographical separation between the ancient genotype 1 sequences, found in Central Asia, and ancient genotype 2 sequences, found in Europe and Greenland. However, this may simply be a result of sampling bias due to the small number of positive samples. Although no ancient genotype 3 sequences have been found, there appears to be no feature inherent to the biology of genotype 3 that inhibits long-term preservation of its DNA (16), as the genotype has been found in remains of World War II soldiers by Toppinen et al. (20).
This small number of ancient samples of B19V has allowed us to revise previous best estimates of rate of evolution and the timespan of the association of B19V with humans, both by an order of magnitude. Similar gains in our understanding of the evolutionary processes of other pathogens, and for improving phylogenetic inference methods when no ancient sequences exist, surely await.
Methods
Screening of Ancient Samples.
Next-generation sequencing datasets were screened with Burrows–Wheeler Aligner and BLASTn. Details are provided in SI Appendix, Supplementary Information Methods.
Phylogenetic Analyses.
Phylogenetic analyses were performed in PhyML and BEAST2. Details are provided in SI Appendix, Supplementary Information Methods.
Data Availability.
All B19V DNA consensus sequences that support the findings of this study are available under European Nucleotide Archive accession number PRJEB26712 before publication. The alignments and XML files used to perform the analysis presented in this paper are available at https://github.com/acorg/parvo-2018.
Supplementary Material
Acknowledgments
We thank Stuart Rankin and the staff of the University of Cambridge High Performance Computing service and the National High-throughput Sequencing Centre (Copenhagen). S.S. and A.M. thank Lena Ideström, Curator/Collection Manager, Gotland Museum, Sweden, for all her help with the Viking Age osteological material in the museum’s collection. E.W. thanks St. Johns College, Cambridge, for providing an excellent environment for scientific discussions. This work was supported by The Danish National Research Foundation, The Danish National Advanced Technology Foundation (The Genome Denmark platform, Grant 019-2011-2), The Lundbeck Foundation, The Villum Kann Rasmussen Foundation, KU2016, European Union FP7 Programme ANTIGONE (Grant agreement 278976), the European Union Horizon 2020 Research and Innovation Programme, COMPARE (Grant agreement 643476), the Zoological Institute of the Russian Academy of Sciences (state assignment AAAA-A17-117022810195-3), the Social Sciences and Humanities Research Council of Canada (Major Collaborative Research Initiative Grants 410-2000-1000, 412-2005-1004, and 412-2011-1001), and the National Science Foundation: Comparative Island Ecodynamics (RF40C41.00.01).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Sequences used in this study have been made available on the European Nucleotide Archive (accession no. PRJEB26712). Alignments and XML files are available on GitHub at https://github.com/acorg/parvo-2018.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1804921115/-/DCSupplemental.
References
- 1.Gallinella G. Parvovirus B19 achievements and challenges. ISRN Virol. 2013;2013:1–33. [Google Scholar]
- 2.Young NS, Brown KE. Parvovirus B19. N Engl J Med. 2004;350:586–597. doi: 10.1056/NEJMra030840. [DOI] [PubMed] [Google Scholar]
- 3.Ozawa K, Kurtzman G, Young N. Replication of the B19 parvovirus in human bone marrow cell cultures. Science. 1986;233:883–886. doi: 10.1126/science.3738514. [DOI] [PubMed] [Google Scholar]
- 4.Söderlund M, et al. Persistence of parvovirus B19 DNA in synovial membranes of young patients with and without chronic arthropathy. Lancet. 1997;349:1063–1065. doi: 10.1016/S0140-6736(96)09110-6. [DOI] [PubMed] [Google Scholar]
- 5.Pyöriä L, et al. Extinct type of human parvovirus B19 persists in tonsillar B cells. Nat Commun. 2017;8:14930. doi: 10.1038/ncomms14930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schenk T, Enders M, Pollak S, Hahn R, Huzly D. High prevalence of human parvovirus B19 DNA in myocardial autopsy samples from subjects without myocarditis or dilative cardiomyopathy. J Clin Microbiol. 2009;47:106–110. doi: 10.1128/JCM.01672-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tanawattanacharoen S, Falk RJ, Jennette JC, Kopp JB. Parvovirus B19 DNA in kidney tissue of patients with focal segmental glomerulosclerosis. Am J Kidney Dis. 2000;35:1166–1174. doi: 10.1016/s0272-6386(00)70055-2. [DOI] [PubMed] [Google Scholar]
- 8.Gray A, et al. Persistence of parvovirus B19 DNA in testis of patients with testicular germ cell tumours. J Gen Virol. 1998;79:573–579. doi: 10.1099/0022-1317-79-3-573. [DOI] [PubMed] [Google Scholar]
- 9.Adamson LA, Fowler LJ, Ewald AS, Clare-Salzler MJ, Hobbs JA. Infection and persistence of erythrovirus B19 in benign and cancerous thyroid tissues. J Med Virol. 2014;86:1614–1620. doi: 10.1002/jmv.23852. [DOI] [PubMed] [Google Scholar]
- 10.Norja P, et al. Bioportfolio: Lifelong persistence of variant and prototypic erythrovirus DNA genomes in human tissue. Proc Natl Acad Sci USA. 2006;103:7450–7453. doi: 10.1073/pnas.0602259103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blümel J, et al. Parvovirus B19–Revised. Transfus Med Hemother. 2010;37:339–350. doi: 10.1159/000322190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brown KE, Simmonds P. Parvoviruses and blood transfusion. Transfusion. 2007;47:1745–1750. doi: 10.1111/j.1537-2995.2007.01459.x. [DOI] [PubMed] [Google Scholar]
- 13.Servant A, et al. Genetic diversity within human erythroviruses: Identification of three genotypes. J Virol. 2002;76:9124–9134. doi: 10.1128/JVI.76.18.9124-9134.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Parsyan A, Szmaragd C, Allain J-P, Candotti D. Identification and genetic diversity of two human parvovirus B19 genotype 3 subtypes. J Gen Virol. 2007;88:428–431. doi: 10.1099/vir.0.82496-0. [DOI] [PubMed] [Google Scholar]
- 15.Toan NL, et al. Phylogenetic analysis of human parvovirus B19, indicating two subgroups of genotype 1 in Vietnamese patients. J Gen Virol. 2006;87:2941–2949. doi: 10.1099/vir.0.82037-0. [DOI] [PubMed] [Google Scholar]
- 16.Ekman A, et al. Biological and immunological relations among human parvovirus B19 genotypes 1 to 3. J Virol. 2007;81:6927–6935. doi: 10.1128/JVI.02713-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hübschen JM, et al. Phylogenetic analysis of human parvovirus b19 sequences from eleven different countries confirms the predominance of genotype 1 and suggests the spread of genotype 3b. J Clin Microbiol. 2009;47:3735–3738. doi: 10.1128/JCM.01201-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Norja P, Eis-Hübinger AM, Söderlund-Venermo M, Hedman K, Simmonds P. Rapid sequence change and geographical spread of human parvovirus B19: Comparison of B19 virus evolution in acute and persistent infections. J Virol. 2008;82:6427–6433. doi: 10.1128/JVI.00471-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shackelton LA, Holmes EC. Phylogenetic evidence for the rapid evolution of human B19 erythrovirus. J Virol. 2006;80:3666–3669. doi: 10.1128/JVI.80.7.3666-3669.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Toppinen M, et al. Bones hold the key to DNA virus history and epidemiology. Sci Rep. 2015;5:17226. doi: 10.1038/srep17226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Duffy S, Shackelton LA, Holmes EC. Rates of evolutionary change in viruses: Patterns and determinants. Nat Rev Genet. 2008;9:267–276. doi: 10.1038/nrg2323. [DOI] [PubMed] [Google Scholar]
- 22.Nielsen R, et al. Tracing the peopling of the world through genomics. Nature. 2017;541:302–310. doi: 10.1038/nature21347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rasmussen S, et al. Early divergent strains of Yersinia pestis in Eurasia 5,000 years ago. Cell. 2015;163:571–582. doi: 10.1016/j.cell.2015.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vågene ÅJ, et al. Salmonella enterica genomes from victims of a major sixteenth-century epidemic in Mexico. Nat Ecol Evol. 2018;2:520–528. doi: 10.1038/s41559-017-0446-6. [DOI] [PubMed] [Google Scholar]
- 25.Mühlemann B, et al. Ancient hepatitis B viruses from the Bronze age to the Medieval period. Nature. 2018;557:418–423. doi: 10.1038/s41586-018-0097-z. [DOI] [PubMed] [Google Scholar]
- 26.Duggan AT, et al. 17th century Variola virus reveals the recent history of Smallpox. Curr Biol. 2016;26:3407–3412. doi: 10.1016/j.cub.2016.10.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heegaard ED, Brown KE. Human parvovirus B19. Clin Microbiol Rev. 2002;15:485–505. doi: 10.1128/CMR.15.3.485-505.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Orlando L, Gilbert MTP, Willerslev E. Reconstructing ancient genomes and epigenomes. Nat Rev Genet. 2015;16:395–408. doi: 10.1038/nrg3935. [DOI] [PubMed] [Google Scholar]
- 29.Willerslev E, Cooper A. Ancient DNA. Proc Biol Sci. 2005;272:3–16. doi: 10.1098/rspb.2004.2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martin DP, Murrell B, Golden M, Khoosal A, Muhire B. RDP4: Detection and analysis of recombination patterns in virus genomes. Virus Evol. 2015;1:vev003. doi: 10.1093/ve/vev003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shen H, Zhang W, Wang H, Shao S. Identification of recombination in the NS1 and VPs genes of parvovirus B19. J Med Virol. 2016;88:1457–1461. doi: 10.1002/jmv.24471. [DOI] [PubMed] [Google Scholar]
- 32.Xia X. Assessing substitution saturation with DAMBE. In: Lemey P, Salemi M, Vandamme A-M, editors. The Phylogenetic Handbook: A Practical Approach to Phylogenetic Analysis and Hypothesis Testing. 2nd Ed. Cambridge Univ Press; Cambridge, UK: 2009. pp. 615–630. [Google Scholar]
- 33.Ramsden C, Holmes EC, Charleston MA. Hantavirus evolution in relation to its rodent and insectivore hosts: No evidence for codivergence. Mol Biol Evol. 2009;26:143–153. doi: 10.1093/molbev/msn234. [DOI] [PubMed] [Google Scholar]
- 34.Duchêne S, Duchêne D, Holmes EC, Ho SYW. The performance of the date-randomization test in phylogenetic analyses of time-structured virus data. Mol Biol Evol. 2015;32:1895–1906. doi: 10.1093/molbev/msv056. [DOI] [PubMed] [Google Scholar]
- 35.Bouckaert R, et al. BEAST 2: A software platform for Bayesian evolutionary analysis. PLoS Comput Biol. 2014;10:e1003537. doi: 10.1371/journal.pcbi.1003537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362:709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 37.Chapman MS, Rossmann MG. Single-stranded DNA-protein interactions in canine parvovirus. Structure. 1995;3:151–162. doi: 10.1016/s0969-2126(01)00146-0. [DOI] [PubMed] [Google Scholar]
- 38.Katzourakis A, Gifford RJ. Endogenous viral elements in animal genomes. PLoS Genet. 2010;6:e1001191. doi: 10.1371/journal.pgen.1001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Aiewsakun P, Katzourakis A. Time-dependent rate phenomenon in viruses. J Virol. 2016;90:7184–7195. doi: 10.1128/JVI.00593-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Duchêne S, Holmes EC, Ho SYW. Analyses of evolutionary dynamics in viruses are hindered by a time-dependent bias in rate estimates. Proc Biol Sci. 2014;281:20140732. doi: 10.1098/rspb.2014.0732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ho SYW, et al. Time-dependent rates of molecular evolution. Mol Ecol. 2011;20:3087–3101. doi: 10.1111/j.1365-294X.2011.05178.x. [DOI] [PubMed] [Google Scholar]
- 42.United Nations Statistics Division 1998 Standard country or area codes for statistical use (M49): Geographic regions. Available at https://unstats.un.org/unsd/methodology/m49/. Accessed March 12, 2018.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All B19V DNA consensus sequences that support the findings of this study are available under European Nucleotide Archive accession number PRJEB26712 before publication. The alignments and XML files used to perform the analysis presented in this paper are available at https://github.com/acorg/parvo-2018.