Significance
Epidemiological studies have shown a correlation between CMV infection and immune system aging, especially in elderly populations. It remains unclear whether CMV infection is a key driver of, or simply a factor associated with, aging of the immune system. We show that aging in the presence of lifelong CMV infection improves T cell immunity in old animals by broadening the immune response to a different pathogen. Animals that have aged with CMV are able to recruit novel T cells into these immune responses that are present in, but not utilized in, animals aging without CMV. These data squarely challenge the premise that CMV is solely detrimental to the aging of the adaptive immune system.
Keywords: immunity, aging, persistent viruses, CMV
Abstract
Lifelong interactions between host and the ubiquitous and persistent cytomegalovirus (CMV) have been proposed to contribute to the age-related decline in immunity. Prior work from us and others found some support for that idea, yet evidence that this led to increased vulnerability to other infections was not obtained. Moreover, evidence has accumulated that CMV infection can be beneficial to immune defense in young/adult mice and humans, dominantly via enhanced innate immunity. Here, we describe an unexpected impact of murine CMV (MCMV) upon the T cell response of old mice to Listeria monocytogenes expressing the model antigen, OVA (Lm-OVA). Single-cell sequencing of the OVA-specific CD8 T cell receptor β (TCRβ) repertoire of old mice demonstrated that old MCMV-infected mice recruited many diverse clonotypes that afforded broad and often more efficient recognition of antigenic peptide variants. This stood in contrast to old control mice, which exhibited strong narrowing and homogenization of the elicited repertoire. High-throughput sequencing of the total naïve CD8 TCRβ repertoire showed that many of these diverse OVA-specific clonotypes were present in the naïve CD8 repertoire of mice in all groups (adult, old control, and old MCMV+) yet were only recruited into the Lm-OVA response in MCMV+ old mice. These results have profound implications for our understanding of T cell immunity over a life span and suggest that our coevolution with CMV may include surprising, potentially positive impacts on adaptive heterologous immunity in late life.
Aging is associated with increased susceptibility to infectious diseases. The majority of humans age while infected with persistent microorganisms, including cytomegalovirus (CMV), which exert a disproportionally large influence on the immune system (1–4). Overall, between 40 and 95% of people within the United States are CMV seropositive before age 40 (5) and therefore live with CMV for several decades. This leads to an absolute accumulation of effector T cells specific for CMV (memory inflation) (6–8), sometimes with one or a few clonal populations occupying up to 25% of the entire CD8 T cell memory pool in elderly humans (9, 10). It has been proposed that this memory T cell inflation comes at a price to the aging immune system, although neither the exact impact nor the mechanism(s) driving this CMV effect have been precisely dissected so far (reviewed in ref. 11).
One contributing factor to impaired immunity with aging may be loss of diversity within the naïve T cell receptor (TCR) repertoire. We and others have demonstrated an age-related absolute numeric loss of naïve T cells in both mice and humans, as measured by reduction of total naïve as well as antigen-specific peptide:MHC tetramer (pMHC Tet+)-binding naïve CD8 T cells by 60–90% (7, 8, 12, 13). Moreover, a stringent assessment of human naïve T cell repertoire diversity in CMV-seronegative subjects found a twofold to fivefold reduction with aging (14). However, in light of the massive TCRαβ actual and potential diversity, which ranges between 109 and 1015, the authors concluded that this effect is minor (e.g., less than 10−8).
In aged mice, there is a marked narrowing of the elicited CD8 effector TCRβ repertoire diversity following primary infection (15–17), which may be mitigated by priming earlier in life (18). Whether this reflects lower diversity within the naïve T cell repertoire above and beyond the numeric loss of naïve T cells, or a failure of the aged immune system to recruit a diverse CD8 T cell population, remains unclear. Moreover, although quite informative, the above studies did not evaluate whether aging and persistent CMV infection in combination directly impact the naïve T cell repertoire to restrict diversity of T cell responses to new pathogens in late life.
We previously reported that old mice, infected with MCMV for life, and challenged with a novel pathogen (Listeria monocytogenes) in old age, mount primary CD8 T cell responses with modestly reduced effector function and bacterial clearance (12). However, in these, and other similar experiments (19–21), mortality from third-party infection was not increased by lifelong CMV infection.
In this work, we have utilized both single-cell PCR and high-throughput TCR sequencing platforms to conclusively evaluate whether and how lifelong CMV may affect (i) the naïve CD8 T cell repertoire with aging, and/or (ii) the elicited repertoire mobilized in response to acute infection. We find that lifelong MCMV infection has a dramatic and surprising impact on the elicited T cell repertoire. Specifically, all measured parameters of the elicited repertoire suggested that, in MCMV+ old mice, diversity was maintained at or above adult levels and was superior to that seen in MCMV-negative (MCMV−) old mice. The CD8 effector T cell populations generated in these animals following Listeria challenge often consisted of clonotypes that expressed noncanonical TCR Vβ, and/or Jβ genes relative to adult and old MCMV-negative animals. More importantly, many old MCMV+ animals exhibited broad recognition of altered peptide ligands, superior to that of MCMV-negative old mice. Within the limits of our analysis, the overall diversity of the naïve CD8 T cell pool was unchanged by either age or CMV status, and many of the diverse ovalbumin (OVA)-specific clonotypes elicited in MCMV+ old mice were present in the naïve CD8 repertoire of mice in all groups. Thus, in contrast to our expectations, lifelong MCMV infection appears to improve the TCR diversity of the elicited CD8 T cell response to a new infection in late life, by recruiting broadly diverse clonotypes with cross-reactive antigen recognition capacity that remain unutilized in MCMV-negative old animals. This unveils a paradigm whereby a lifelong, latent persistent virus modulates adaptive immunity in a manner that may benefit host defense.
Results
To evaluate how lifelong persistent infection with MCMV might influence the T cell repertoire in late life, adult mice were systemically infected with MCMV at 20 wk of age and rested until they reached 20 mo of age. [We found that infection as early as 6 or as late as 20 wk of life produced comparable magnitude of memory inflation to MCMV epitopes (a proxy measure of viral reactivation/latency) (SI Appendix, Fig. S1).] Then, adult (A, 12 wk), old mock-infected (20 mo), and MCMV-infected old mice (20 mo old, after 15 mo with MCMV) were challenged with recombinant Listeria monocytogenes (22) expressing the fragment of chicken egg albumin containing the immunodominant H-2Kb–restricted CD8 T cell epitope OVA257–264, SIINFEKL (Lm-OVA) (23). On day 7 after Lm-OVA challenge, Vβ5+ Kb-OVA257–264 tetramer+ CD8 T cells from the spleen were individually sorted for single-cell TCR Vβ CDR3 sequencing [Vβ5 = TRBV12 in International Immunogenetics Information System (IMGT) nomenclature, used hereafter] (24). The OVA-specific TRBV12 repertoires were analyzed as a dominant population elicited by this antigen in adult mice (25) (parameters of the sequencing experiments are summarized in SI Appendix, Table S1). For all analyses, “TCR clonotype” is used to describe each unique TCR Vβ amino acid sequence, whereas “TCR repertoire” is used when factoring in both the sequence and representation (copy number) of each clonotype.
Lifelong MCMV Infection Alters Gene Choice and CDR3 Length in the OVA Repertoire.
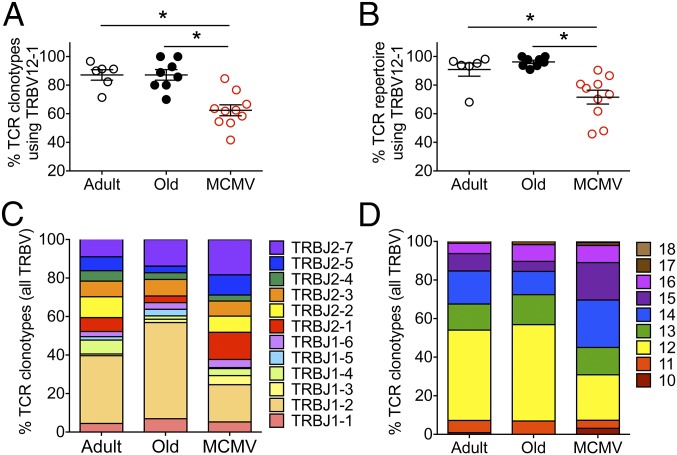
Our tetramer sorting strategy focused the analysis to Kb-OVA257–264 tetramer+ cells expressing both TRBV12-1 (Vβ5.2) and TRBV12-2 (Vβ5.1) clonotypes. The frequency of unique amino acid clonotypes (Fig. 1A) as well as the frequency of total repertoire (Fig. 1B) that expressed the favored TRBV12-1 chain was similar between old MCMV-negative and adult mice, with a preference of ∼80–90% for TRBV12-1. In contrast, old mice with lifelong MCMV infection exhibited a significant reduction in TRBV12-1 utilization, and ∼30–40% of their OVA-specific repertoire utilized the TRBV12-2 chain.
Fig. 1.
MCMV+ old mice show divergences from canonical OVA repertoire features. (A and B) Distribution of the (A) unique TCR clonotypes (each TCR Vβ amino acid sequence) and (B) overall repertoire (factoring in both the sequence and copy number) for individual adult, old, and MCMV+ OVA-specific CD8 T cells using the TRBV12-1 gene are shown (medians shown). (C and D) Means of 6–10 mice per group show (C) J gene usage and (D) CDR3 length for all TRBV12+ OVA-specific CD8 T cells. Comparisons between groups were by Mann–Whitney U test, with Bonferroni correction for multiple pairwise comparisons (i.e., each pairwise test was assessed at the significance level of *P < 0.05/3 = 0.0167).
Similarly, there was a predominant choice of TRBJ1-2 in the adult repertoire, which became even more prevalent in the old mice (Fig. 1C). By contrast, old MCMV-infected mice showed a more balanced utilization of TRBJ segments, with lower utilization of the favored TRBJ1-2 (Fig. 1C).
CDR3 length analysis revealed a preference for CDR3 sequences of 12 aa in both adult and old MCMV-negative mice (Fig. 1D). In MCMV-infected mice, this preference was not as marked, with the emergence of many clonotypes of both shorter and longer CDR3 lengths. Of note, we found substantial variation in these repertoire features (Vβ, Jβ and CDR3) in individual MCMV+ old mice (SI Appendix, Figs. S2 and S3). Thus, while MCMV+ old mice contained clones with the prototypical TRBV12-1+, TRBJ1-2+, and 12-aa CDR3 length profile seen in adult and old MCMV− mice, their representation in the repertoire of many old MCMV+ mice was lower and TCR sequences often showed marked deviation from these dominant repertoire features.
Noncanonical Clonotypes Responding to Third-Party Infection Are Abundantly Present in Mice with Lifelong MCMV Infection.
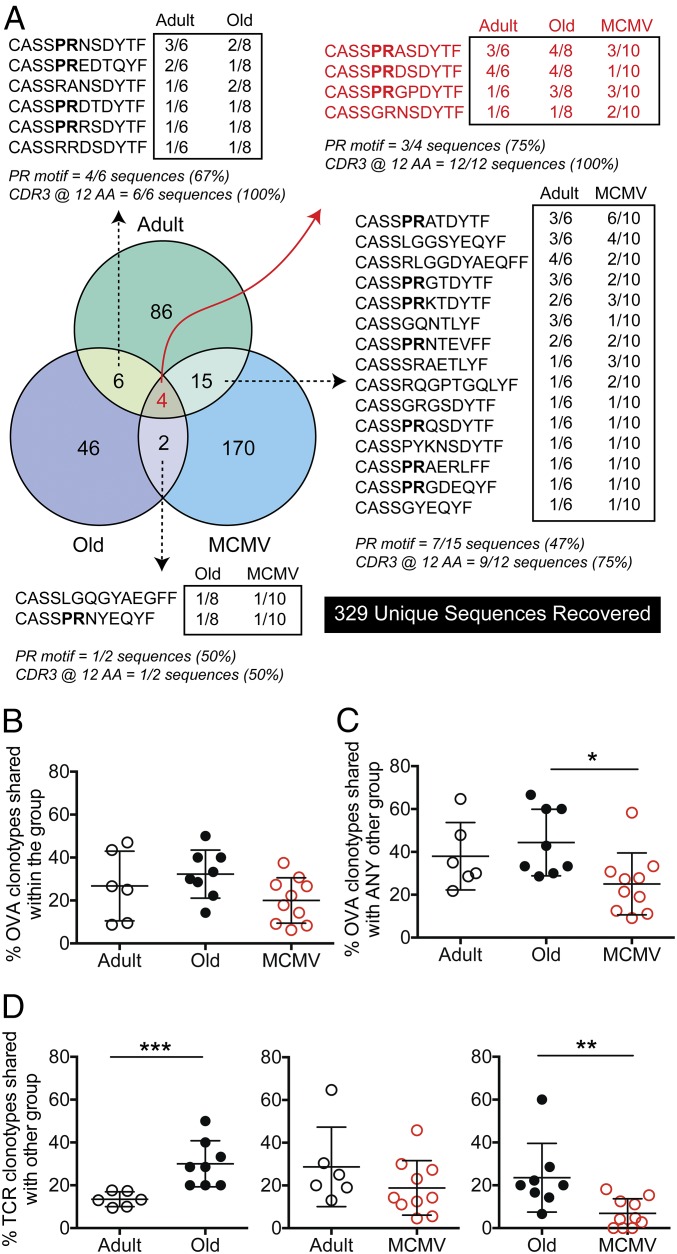
Broad TCR diversity within an antigen-specific effector population is believed to correlate with better functional protection against pathogens. We assessed clonal diversity by two measures: (i) the number of unique clonotypes within a set number of clonotypes evaluated in each sample, and (ii) the Simpson’s diversity index, which varies in value between 0 (minimum diversity, every clone identical) and 1 (maximum diversity, every clone unique). We found the expected age-related reduction in the number of unique clonotypes (Fig. 2A) in the OVA-specific repertoire (standardized to the smallest repertoire in the analysis, 21 total sequences), from an average of 11.5 ± 2.6 clonotypes (mean/SD) in adults to 6.4 ± 2.0 clonotypes in old MCMV-negative mice. Furthermore, the OVA-specific TCRβ repertoires in old MCMV-negative mice were largely dominated by one or two clonotypes, as indicated by the significant decrease in the Simpson’s diversity measures in old compared with adult mice (Fig. 2B). In contrast, old mice with lifelong MCMV infection maintained their diversity by both measures at levels at or above (14.3 ± 3.4 clonotypes) that seen in adult animals. This was surprising, as our previous studies indicated somewhat reduced functionality as measured by cytokine production and pathogen clearance within the OVA-specific repertoire in MCMV+ aged mice (12).
Fig. 2.
MCMV+ mice maintain repertoire diversity by recruitment of low-copy number unique clonotypes. (A and B) The (A) number of clonotypes and (B) Simpson’s diversity index for adult, old, and MCMV mice. (C) The clonal distribution within the overall repertoire, ranking each clone from largest to smallest in terms of additive frequency to the overall repertoire. (D and E) The frequency of unique clonotypes for adult, old, and MCMV mice utilizing P in position 5 and R in position 6 of the CDR3 sequence (D) and linear regression of the frequency of clonotypes utilizing the “PR” motif and frequency of single-copy clonotypes (MCMV P = 0.0035) (E). Individual animals are shown for A, B, D, and E, and means ± SEM in C. Comparisons between groups were by Mann–Whitney U test, with Bonferroni correction for multiple pairwise comparisons (i.e., each pairwise test was assessed at the significance level of *P < 0.05/3 = 0.0167).
We next evaluated clonal distribution within the overall repertoire, ranking the clones from largest to smallest (in terms of clonotype abundance/copy number) to estimate their additive contribution to the overall repertoire. This allowed us to ask (i) how much do the small clonotypes contribute at the tail of the curve? and (ii) how dominant are the largest clonotypes (regardless of sequence) in making up the repertoire? We found that small clonotypes, appearing only once in the response, make up about 1/5 of the repertoire in adult and only 1/10 of the repertoire in old mice, but more than 1/2 (up to 60%) of the repertoire elicited in lifelong MCMV-infected mice (SI Appendix, Fig. S4).
Direct analysis of the size of the dominant clonotypes showed that old mice mobilized few very dominant clonotypes, with the two or three largest clones making up ∼75% of their entire repertoire (Fig. 2C). In contrast, adult mice needed an average of 9 clonotypes to make up 75% of their repertoire, whereas old MCMV+ mice utilized as many as 17 different clonotypes to make up the same percentage of the repertoire, each of them present as a smaller population (Fig. 2C). These data collectively suggest that lifelong MCMV infection allows mobilization of a diverse TCRβ repertoire in late life, with recruitment of low-copy number clonotypes into a primary immune response.
Consistent with previous data, in many of the CDR3 sequences we found a conserved “PR” amino acid motif with a proline (P) in position 5 and arginine (R) in position 6 (counting from the conserved cysteine), where the R is likely to make a salt bridge to the exposed E residue of the OVA peptide (25, 26). Of interest, the PR motif in the OVA repertoire cannot be fully germline encoded (SI Appendix, Table S2); although the arginine in the motif is often fully or partially encoded by the D gene (red), the proline in the PR motif requires N additions (black). Furthermore, although many instances of PR motif utilization involved the use of TRBV12-1 and TRBJ1-2, favored by many clonotypes in adult and old mice, the PR motif was also created by recombination with other TRBJ genes, suggesting postrecombinatorial, likely antigen-driven, selection.
We found the frequency of clonotypes expressing the PR motif to be comparable between adult and old MCMV-negative mice (Fig. 2D), even though the old animals had many fewer different clonotypes making up their response, with higher variability in the usage of the PR motif (Fig. 2D). This further supports the idea that this motif remains favored with aging. Of interest, the frequency of PR motif utilization was notably reduced in most MCMV+ old mice (Fig. 2D; twofold lower and significantly different from adult mice, P < 0.05), and there was a significant relationship by linear regression between the loss of the PR motif and the emergence of single-copy clonotypes (P = 0.0035, R2 = 0.676 for MCMV+ old mice; Fig. 2E).
TRBV Sequences in the OVA-Specific Repertoire Vary Significantly with both Age and CMV Infection.
There is strong evidence that the primary effectors stimulated after infection are clonally less diverse in aged mice (13, 15, 17) (Fig. 2), and it was speculated that this phenomenon could be further pronounced by CMV infection (27, 28). We compared the “sharing” of specific clonotypes between animals within an experimental group (i.e., across all of the adult mice), as well as the sharing of clonotypes across different experimental groups (i.e., adult vs. old; Fig. 3). Of the 24 individual animal repertoires evaluated, a total of 329 unique CDR3 sequences were recovered. Fig. 3A depicts the level of CDR3 amino acid sequence and PR motif sharing found across all experimental groups, as well as within each group. For example, the center of the Venn diagram indicates (in red) that only four CDR3 sequences were shared between adult, old MCMV−, and old MCMV+ mice. Three out of four of these sequences contained the conserved PR amino acid motif, and all of these shared sequences were of the preferred CDR3 length of 12 aa (red text). However, aging with and without MCMV seemed to differentially influence the CDR3 OVA-specific repertoire recovered. Only two CDR3 sequences were shared between old MCMV− and old MCMV+ mice, and each of these sequences only appeared in a single animal within each experimental group (Fig. 3A). In contrast, sharing between adult and old mice, and adult and MCMV+ old mice was more extensive, with more sequences being shared between groups, and with multiple animals within each experimental group harboring the shared clonotypes. When we compared interindividual sharing of clonotypes within each of the three experimental groups (i.e., how many adult mice shared clonotypes with other adult mice), we found a comparable level of sharing of clonotypes within the groups (Fig. 3B). In contrast, when we assessed how frequently clonotypes were shared with other experimental groups, we found that clonotypes in MCMV+ old mice were shared less often. Specifically, although adult and old mice without MCMV shared ∼40% of their clonotypes with animals in other groups, old mice with MCMV showed a reduced frequency of clonotype sharing (Fig. 3C). We also examined cross-group sharing by asking “if an adult has a given TCRβ clonotype, is that clonotype also found in any old mice?” (Fig. 3D, Left), and then repeating for all other group combinations. We found that 28.6 ± 10.8% of clonotypes found in an old mouse would also be found in an adult mouse (Fig. 3D, Left), whereas only 13.4 ± 3.5% of clonotypes from an adult mouse would also be found in an old mouse. This independently confirms that the elicited adult repertoire narrows as the mouse ages. However, this effect was not seen if the animal aged with MCMV infection (Fig. 3D, Center), where no difference was seen in sharing of clonotypes between the adult and MCMV+ old groups. Finally, only 6.9 ± 6.8 clonotypes in MCMV+ old mice were found in uninfected old mice, whereas 23.6 ± 16.0 clonotypes in old mice were found in MCMV+ old mice (Fig. 3D, Right). Thus, the elicited repertoire within the MCMV+ old mice included many clonotypes seen in adult animals, but these clonotypes were lost in the old mice.
Fig. 3.
Minimal overlap between the OVA repertoires in adult, old, and MCMV+ mice. The degree of amino acid CDR3 sequence sharing between experimental groups (adult, old, MCMV) and within each group is shown. Unique clonotypes only appearing in one animal (regardless of group) were omitted for simplicity (see SI Appendix, Fig. S4 for a full list of CDR3 sequences). (A) Venn diagram showing shared clonotypes across experimental groups. Boxed numbers represent how many animals within that group contained that clonotype (i.e., sharing within the group). (B and C) The frequency of clonotypes within each mouse that are shared with an animal in either (B) the same or (C) any other experimental group. Comparisons between groups were by Kruskal–Wallis test, with Dunn correction for multiple pairwise comparisons. (D) The frequency of clonotypes within each mouse that are shared with an animal in the other experimental group shown. Comparisons between groups were by Mann–Whitney U test.
Lifelong MCMV Affords a Broader Spectrum of Functional TCR Antigen Recognition.
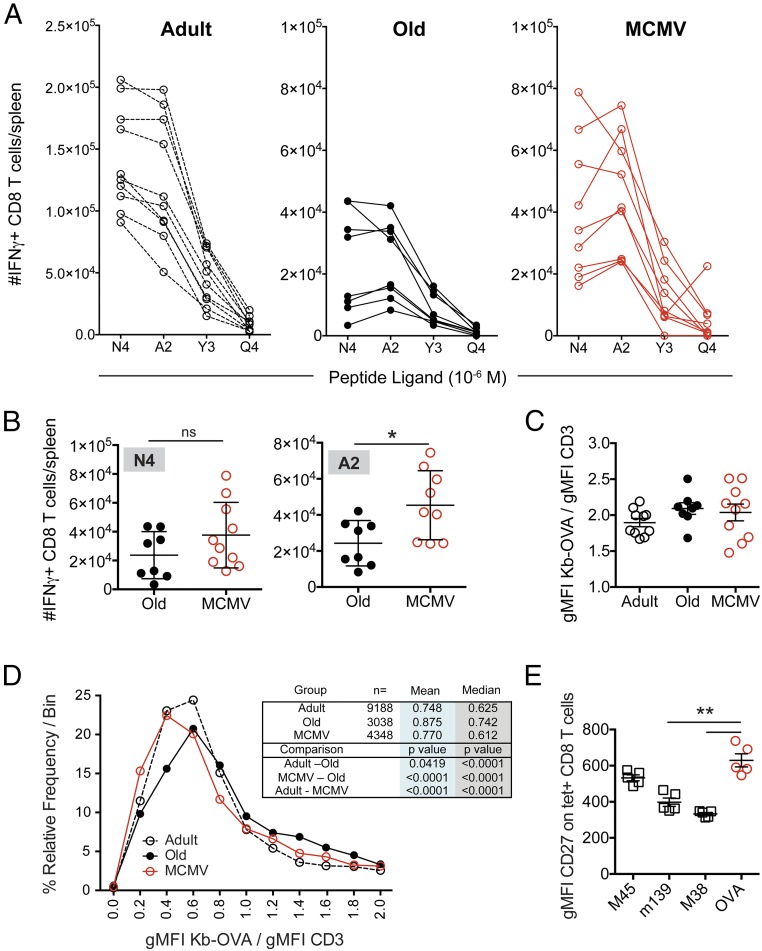
Is recruitment of noncanonical low-copy clonotypes into the repertoire biologically meaningful? To address this, we measured IFN-γ production by effector T cells following stimulation with OVA257–264-derived altered peptide ligands (APLs) that show a ∼20-fold range in their ability to stimulate TCR transgenic OT-I CD8 T cells specific for the native SIINFEKL epitope (29). Fig. 4A show the absolute number of splenocytes responding to the native ligand (N4), and to the APL [decreasing sensitivity from A2 > Y3 > Q4 (29)], for individual animals in each group. Although the overall sizes of the IFN-γ–producing populations were comparable between the old vs. MCMV+ groups when restimulated with the native N4 epitope (Fig. 4B), there was marked variation in the magnitude of the APL responses within the MCMV+ animals (Fig. 4 A and B), and some (albeit not all) MCMV+ animals exhibited greater responses to the A2 APL relative to the N4 epitope. Of interest, many MCMV+ animals responded significantly better to the “A2” ligand than old MCMV-negative mice (Fig. 4B). We assessed the intensity of Kb-OVA257–264 tetramer staining when normalized to CD3 levels (as a proxy measure of avidity for the OVA-specific CD8 T cells) and failed to detect any differences between the experimental groups (Fig. 4C). Therefore, there were no gross differences in the avidity of CD8 T cells elicited from adult, old, or MCMV-infected old mice.
Fig. 4.
OVA-specific cells in MCMV+ mice show altered avidity for cognate antigen. (A) On day 7 after Lm-OVA infection, splenocytes were stimulated with SIINFEKL (N4) peptide or three different altered peptide ligands (A2, Y3, and Q4) at 10−6 M. The number of cells able to produce IFN-γ in response to each was assessed by FCM. Data show the individual responses for adult (open circles, n = 10) and old animals (black circles, n = 8), or MCMV+ mice (red circles, n = 9). (B) Comparison of the number of IFN-γ+ cells responding to N4 (Left) or A2 (Right) peptides in old vs. MCMV+ old mice. Significance determined by Mann–Whitney U test. (C) The gMFI of Kb-OVA257–264 tetramer binding normalized to total CD3 staining. Data depict mean ± SEM. (D) The distribution of binned Kb-OVA/CD3 gMFI of individual cells (as a measurement of TCR avidity) for adult (open, dashed line), old (black), and MCMV+ old (red) mice. (E) Splenocytes from MCMV+ old mice were stained with tetramers against three MCMV epitopes (M45, noninflating; M38 and m139, inflating), as well as the Kb-OVA257–264 tetramer. The gMFI of CD27 on each tetramer+ population was assessed. *P < 0.05, **P < 0.01. For all analyses, data were pooled from two independent experiments with n = 4–5 mice/experiment.
However, given that we were trying to assess the influence on avidity of the single-copy noncanonical clonotypes in the MCMV+ animals, this measurement was likely too blunt to detect such changes. We deconvoluted our FCM data files (FCS Extract software: research.stowers.org/mcm/efg/ScientificSoftware/Utility/FCSExtract/index.htm) so that we could measure the tetramer/CD3 binding ratio on individual cells within each OVA tetramer+ population. We then evaluated the distribution of these binding intensities across the population, to measure the representation of low-avidity cells in the MCMV+ old mice relative to the two other groups (Fig. 4D). We found that the TCR repertoire of MCMV+ old mice exhibits a significant negative shift in their relative distribution, with more clonotypes of lower avidity (increased tetramer/CD3 ratio). In contrast, the TCR repertoires of old mice contained a greater representation of higher-avidity clonotypes [consistent with our prior results (16)]. Collectively, these data suggest that the increased clonotypic diversity in the MCMV+ old mice confers variable, but overall increased, TCR cross-recognition of antigenic variants, potentially useful for control of escape variants and/or heterologous immunity, although the biological significance of this observation will need to be tested directly.
The OVA Repertoire Is Unlikely to Be Cross-Reactive with MCMV Epitopes.
One mechanism that might underlie the recruitment of noncanonical clonotypes from MCMV+ old mice would be if they were cross-reactive with MCMV antigen(s). This is a very difficult possibility to rule out, as the ∼230-kb MCMV genome is predicted to encode between 170–750 ORFs, many of unknown expression and function (30, 31). It has been reported that inflationary, persistently stimulated MCMV-specific T cell populations down-regulate CD27 expression as a consequence of repeated antigenic stimulation (32). To assess the potential that clonotypes within the OVA-specific repertoire might be cross-reactive with MCMV antigens, we assessed CD27 expression on three different MCMV tetramer+ populations, as well as the newly stimulated OVA-specific T cells. We utilized tetramers for Db-M45985–993, an acute-phase, “noninflating” MCMV response, as well as tetramers specific for two “inflationary” epitopes, Kb-m139419–426 and Kb-M38316–323 T cells (33). If the OVA response was cross-reactive with MCMV-derived antigens, the expression of CD27 on the Kb-OVA257–264 tetramer+ cells (as surrogate markers for antigenic experience) should resemble the expression seen on the inflating MCMV tetramer populations (M38 and m139). We found the opposite—that the Kb-OVA257–264 tetramer+ CD8 T cells in MCMV-infected animals exhibited high CD27 expression, similar to the noninflating M45 epitope (Fig. 4E), and significantly different from the inflating m139 and M38 epitopes. Moreover, most of the noncanonical clonotypes found in the anti-OVA CD8 response are present at very low or single copy number and thus could not have been expanded in response to repeated antigen exposure, as might be expected if they were cross-reactive with MCMV. Although this does not formally exclude cross-reactive recognition of MCMV antigens, these data strongly argue against cross-reactivity to MCMV as an explanation for the presence of these novel OVA-specific clonotypes in the MCMV+ old mice.
Neither Age nor CMV Status Impacts Naive CD8 T Cell Repertoire Diversity.
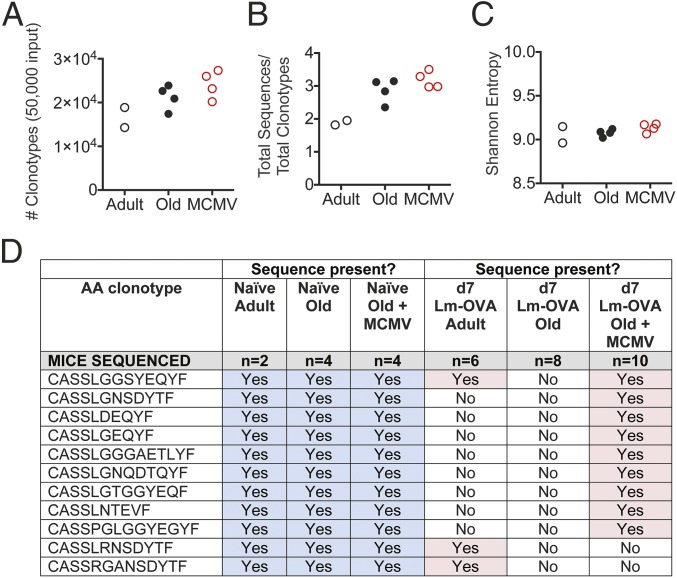
Changes in the elicited OVA-specific TCR repertoire in the MCMV+ old mice could be due to changes in the naïve, preimmune repertoire; alternatively, they could be due to differential recruitment of the existing repertoire as a consequence of MCMV infection. To distinguish between these possibilities, we sequenced the naïve CD8 T cell pool in adult, old MCMV−, and old MCMV+ animals. A total of 50,000 splenic naïve CD8 T cells (CD8+ CD44− CD62L+)/mouse was FACS sorted for high-throughput TCRβ repertoire analysis using a well-validated multiplex primer set (34) and unique molecular tags for each sample. We analyzed a total of 886,074 naïve CD8 TCRβ sequences from adult (118,585 sequences from two mice), old (368,148 sequences, four mice), and MCMV+ old mice (342,677 sequences, four mice). For each input sample of 50,000 sorted naïve CD8 T cells, between 14,278 and 25,993 unique clonotypes were recovered (DNA sequence) (Fig. 5A). The coverage was similar among all samples—the average number of sequences recovered per amino acid clonotype was between 2 and 3 (Fig. 5B). This allowed us to compare both the number of sequences and their distribution, by calculating the Shannon entropy for each sample. We found no difference in the entropy values, suggesting that neither age nor CMV status greatly influence the diversity of the naïve repertoire as measured by entropy (Fig. 5C).
Fig. 5.
Neither age nor MCMV infection reduces naïve CD8 TCRβ repertoire diversity. A total of 50,000 splenic naïve CD8 T cells (CD8+CD44−CD62L+) was FACS sorted from adult, old, and MCMV+ old mice for high-throughput TCRβ repertoire analysis by Illumina MiSeq. (A and B) Total number of unique nucleic acid clonotypes (A) and number of sequences recovered per nucleic acid clonotype for each sample (B) are shown. (C) Calculated Shannon entropy values for each sample. (D) Comparison of whether noncanonical sequences were recovered in the elicited OVA-specific repertoire on day 7 after Lm-OVA infection (Right, pink) vs. their presence in the total naïve CD8 TCRβ repertoire (Left, blue).
These data indicate that the naïve CD8 T cell repertoire is of similar diversity in adult, old, and MCMV+ old mice, yet the elicited OVA-specific repertoire is highly restricted in old mice (Figs. 1–3). We interrogated the naïve CD8 repertoire dataset for all OVA-specific sequences that we had recovered in the elicited OVA-specific response (SI Appendix, Fig. S4). Even with a limited sample of 50,000 total naïve CD8 T cells (not restricted to the TRBV12 family), we could detect many of the OVA-specific TRBV12 naïve precursors (Fig. 5D). We found within the naïve repertoire of all three groups many of the “unique,” single-copy clonotypes that were mobilized only in the MCMV+ old mice infected with Lm-OVA (Fig. 5D). Thus, the narrow TCR repertoire mounted in old MCMV-negative mice in response to infection is not due to the absence of available clonotypes, but rather due to a failure of the aged immune system to effectively recruit and/or maintain these T cells in the effector response.
Discussion
In mammals, aging is associated with a dysfunction in T cell responses to numerous pathogens. Defects within these aged primary T cell responses against infection have been attributed to various combinations of the following: reduced and altered populations of naïve T cell precursors to recruit into the response (13); impaired antigen presentation function by dendritic cells (35, 36); increased apoptosis of old T cell effectors (37); and reduced effector T cell expansion and maturation (15, 37–39).
In contrast to the controlled environment of laboratory animals, the human immune system is constantly assaulted by infectious exposure. Many microorganisms, and all herpesviruses, are known to manipulate their host immune systems. Herpesviruses deploy multiple molecular mechanisms in place to avoid recognition and elimination by both innate and adaptive immunity (reviewed in ref. 3). Microbial pathogens (40, 41) including herpesviruses (42, 43) have been also shown to benefit the host by providing protection against heterologous infection, and improving vaccine efficacy. In some cases, the mechanism was shown to involve persistent activation of the innate immune system (42). Such effects were either not studied in aging (40, 44) or were found to not affect immune responses in older adults (43).
An age-associated contraction in TCR repertoire diversity was inferred following the discovery and characterization of T cell clonal expansions in mice (45, 46) and humans (47, 48), and the latter were subsequently associated with CMV (9, 28). Observational data from aging human cohorts has revealed associations between peripheral CD4/CD8 T cell ratios, CMV antibody titers, and all-cause mortality (reviewed in ref. 11). However, the above associations remain to be causally and mechanistically dissected.
In this report, we demonstrate an unexpected effect of a persistent herpesvirus infection on adaptive T cell immunity in older animals. Specifically, the OVA-specific Vβ5+ (TRBV12) CD8 T cell repertoire mobilized in response to Listeria infection was remarkably broad in the presence, but highly restricted in the absence, of a lifelong CMV infection. The response consisted of many single-copy clonotypes that did not carry one or more of the conventional signature elements of this response (TRBV12.1, TRBJ1.2, 12-aa CDR3 length, and a conserved PR motif in the CDR3; Figs. 1–3). This broad repertoire in MCMV+ old mice provided a heterologous cytokine response against variants of the original antigen, a benefit not afforded to old animals in the absence of CMV infection. Of interest, this diverse repertoire recruitment was not based on differences in the available preimmune naïve T cell repertoire. High-throughput sequencing of naïve CD8 T cells revealed that all animal groups had at their disposal many of the same CDR3 sequences that were elicited in CMV lifelong-infected old mice after Listeria challenge, but these precursors were not mobilized and/or maintained in the response in either adult or old mice without CMV infection.
In young mice, the recruitment of naïve T cell precursors into an immune response is believed to be virtually complete, with the T cell effector response reflecting the naïve precursor population (49–51). In youth (the only model studied), differences in clonotype recruitment are typically not seen at early time points in the response (i.e., day 3), when both high- and low-avidity T cells become activated and enter into primary expansion (29). However, differences become evident at the peak of the response (day 7–9), where low-avidity T cells have already entered into the contraction stage and are therefore present in fewer numbers (29). Thus, a protective effector T cell population is generally held to be composed of large expansions of high-avidity clonotypes, with smaller populations of clonotypes with reduced avidity, but which are critical for defense against pathogen escape variants or infection with heterologous pathogens (52–54). To our knowledge, assessments of aged antigen-specific repertoires have not been performed at early time points. Thus, one possibility is that old mice (without MCMV) may also show a diverse TCRβ repertoire at early time points after Lm-OVA challenge, but that recruitment is not sustained throughout the clonotype expansion phase between days 3 and 7. Indeed, prior findings of restricted TCR repertoire in effector CD8 T cells in old mice following HSV (16) and influenza infections (15, 17) are consistent with a failure of the aged organism to effectively recruit all available precursors, rather than an absolute loss of them. Similarly, we did not find differences in kinetics, but rather only in magnitude, of the response against Lm-OVA without (12) MCMV. We speculate that old mice with persistent MCMV infection can better recruit and/or maintain lower avidity effectors by an unknown mechanism. Experiments are in progress to identify that putative mechanism(s) and test whether it may operate in adult mice too.
Our previous study found some functional defects in the OVA-specific effector CD8 T cells in MCMV+ old mice, which correlated to a modest delay in pathogen clearance early, but not later, in the response; this did not result in increased morbidity and mortality (12), a finding echoed in other studies (19–21). The repertoire sequence data presented here suggest that these functional defects are completely disconnected from standard measures of repertoire diversity. Indeed, old mice aged with persistent MCMV infection show many TCRβ repertoire diversity features that more closely resemble young adult animals than to their age-matched, MCMV-free controls (Fig. 2 A and B).
Perhaps of most importance, the data presented here suggest that the ability to mount a diverse effector response can be maintained into late life, and that lifelong persistent MCMV infection can positively influence this capacity. This may reflect changes in antigen presentation capacity (impaired in aged mice; ref. 35) or some other unknown mechanism that lifelong MCMV infection mediates. Mechanistically, MCMV could impact the recruitment of diverse TCR clonotypes into immune responses by infecting or otherwise altering dendritic cells as the key mediators of T cell priming. Myeloid progenitors and dendritic cells (DCs) are known reservoirs of CMV (55), and over a lifetime, the frequency of DCs that harbor virus would likely increase. We attempted to assess phenotypic changes in the expression levels of CD27, CD40, CD80, CD86, or MHC-II on the surface of CD8α+ DCs in adult, old, or MCMV+ old mice either before, or 24 h after Lm-OVA infection. The CD8α+ DC subset is the primary population used to prime CD8 T cells following infection with Listeria (56, 57). Unfortunately, on a bulk population level, no differences were seen either at baseline, or 24 h postinfection in the degree to which the CD8α DCs up-regulated these costimulatory molecules. However, until better tools are available to determine the frequency of DCs that harbor virus in these animals and to allow us to examine responses of the infected DCs only, these negative results will remain difficult to interpret. Future studies to assess how lifelong CMV infection influences DC maturation and priming capacity are underway.
Overall, our data significantly challenge the hypothesis that CMV has a simple negative impact upon the immune response with aging, as MCMV+ mice were able to recruit clonotypes of greater diversity and with sequences never seen in the elicited response of MCMV-negative old animals. We propose that the recruitment of such diverse clonotypes into the immune response is likely to be beneficial in a broader context of immune defense (outside of the pathogen-free laboratory environment), where exposure to pathogen variants is much more likely (52–54).
Materials and Methods
Mice and Lifelong MCMV Infection.
Eighteen-week-old adult C57BL/6 (B6, H-2b) male mice were purchased from The Jackson Laboratory. At 20 wk of age, adult mice were infected with 105 pfu of MCMV intraperitoneally (Smith strain, originally obtained from M. Jarvis and J. Nelson, Oregon Health and Science University, Portland, OR, passage 3 on M210B4 cells), and then followed to 20 mo of age. Twenty-month-old male C57BL/6 mice were purchased from the National Institute of Aging Aged Rodent Colony (Charles River Laboratories). All mice were maintained under specific pathogen-free conditions in the animal facility at the University of Arizona, and experiments were conducted under guidelines set by the University of Arizona Institutional Animal Care and Use Committee.
L. monocytogenes Infections.
Mice were systemically infected by i.v. injection in the lateral tail vein with 1–8 × 105 cfu of recombinant L. monocytogenes expressing the OVA protein [Lm-OVA (22)] in a volume of 100 μL of sterile PBS.
Single-Cell Sorting for OVA-Specific CD8 TCRβ Sequencing.
Seven days following Lm-OVA challenge, splenocytes were collected and passed through a 40-μm mesh screen to prepare single-cell suspensions, and then negatively enriched for CD8+ T cells using magnetic beads (Miltenyi Biotec). Highly enriched CD8+ T cells were stained with fluorochrome-conjugated anti-CD8α (53-6.7), anti-CD4 (GK1.5), anti-CD44 (IM7), anti-Vβ5.1/5.2 (MR9-4), and Kb-OVA257–264 tetramers (NIH Tetramer Core Facility) for 45 min on ice, and then washed twice. Cells were resuspended in sorting buffer and CD8+CD4−CD44+Kb-OVA+Vβ5+ lymphocytes were sorted as single cells into 96-well plates using a FACSAria cell sorter (BD Biosciences). Wells without sorted cells were included on every plate to control for contamination.
Single-Cell cDNA Synthesis and RT-PCR.
Our RT-PCR protocol was adapted from those of Kedzierska and Hamrouni (58, 59). Single CD8+CD44+Kb-OVA+Vβ5+ cells were sorted directly into 96-well PCR plates containing 5 μL of cDNA reaction mix made of the following: 0.25 μL of Sensiscript reverse transcriptase (Qiagen), 1× cDNA buffer (Qiagen), 0.5 mM 2′-deoxynucleoside 5′-triphosphate (Qiagen), 100 μg/mL tRNA (Invitrogen), 50 ng of oligo-dT12–18 (Invitrogen), 20 U of RNase Out (Invitrogen), and 0.1% Triton X-100 (Sigma-Aldrich). cDNA synthesis was performed immediately after sorting by incubating plates at 37 °C for 90 min, followed by 5 min at 95 °C. Plates were immediately stored at −80 °C. Vβ5 (TRBV12 in IMGT nomenclature) transcripts were amplified by nested PCR, and the entire 5-μL cDNA reaction was used for the first PCR in a final 25-μL volume containing 1.25 U of DreamTaq polymerase (Fisher Scientific) in the manufacturer’s 1× Buffer with 200 μM each 2′-deoxynucleoside 5′-triphosphate (Fisher Scientific), and 100 nM external degenerate sense TRBV12 primer (5′-GGGGTTGTCCAGTCTCC-3′) and external antisense TRBC primer (5′-CCAGAAGGTAGCAGAGACCC-3′). The PCR cycling program began with 5 min at 95 °C, followed by 40 cycles of 20 s at 95 °C, 20 s at 56 °C, and 45 s at 72 °C, ending with 5 min at 72 °C. A 4-μL aliquot of the first PCR product was used for the second PCR with the internal degenerate TRBV12 sense primer (5′-CCAGCAGATTCTCAGTCC-3′) and the internal antisense TRBC primer (5′-GGGTAGCCTTTTGTTTGTTTG-3′). The second PCR program was the same as the first, with 35 rounds of amplification. PCR products were resolved on a 1.8% agarose gel, purified with the MinElute 96 UF PCR purification kit (Qiagen), and sequenced with 12 pmol of the internal degenerate TRBV12 sense primer, using an Applied Biosystems 3730XL DNA Analyzer at the University of Arizona Genomics Core.
OVA-Specific TCRβ Clonotype Analysis.
The OVA257–264–specific CD8+ TCRβ clonotypes were characterized by sequentially aligning each TCRβ sequence with the best match of either the TRBV12-1 or TRBV12-2 gene and then the best-match Jβ gene, using the IMGT reference alleles for the Mus musculus TRB genes (24). The CDR3 sequence was then identified between, and inclusive of, the conserved cysteine in the TRBV region and the conserved phenylalanine in the TRBJ region.
Lm-OVA–Specific TCRβ Repertoire Diversity Analysis.
The diversities of the CD8+ TCRβ repertoires specific for the OVA257–264 epitope in each mouse were evaluated using two different measures of diversity: the number of different TRB amino acid sequence clonotypes and the Simpson’s diversity index (60). The Simpson’s diversity index accounts for both the variety of amino acid sequence clonotypes and their clone sizes. This relative diversity index ranges in value from 0 (minimal diversity) to 1 (maximal diversity). To account for the differences in the TRBV repertoire samples sizes obtained between mice, the diversity of a TCRβ repertoire was estimated as the median value of 10,000 random draws of subsamples of 21 TCRβ sequences from the total TCRβ repertoires obtained for each mouse (60, 61). The diversity analysis was performed using MATLAB (The MathWorks).
Flow Cytometry and Intracellular Cytokine Staining.
On day 7 after Lm-OVA challenge, splenocytes were collected and passed through a 40-μm mesh screen to prepare single-cell suspensions. Cells were stained for 1 h at 4 °C with fluorochrome-conjugated antibodies against CD3 (clone 1B11), CD4 (GK1.5), CD8α (53-6.7), Vβ5.1/5.2 (MR9-4), CD27 (LG.3A10), KLRG1 (2F1), and the Kb-OVA257–264 tetramers, or MCMV Db-M45985–993, Kb-m139419–426, and Kb-M38316–323 tetramers (NIH Tetramer Core Facility).
For avidity measurements by tetramer staining, the geometric mean fluorescent intensity (gMFI) of Kb-OVA was normalized to the gMFI of CD3. To deconvolute flow cytometry (FCM) data, the gated Kb-OVA+ CD8+ T cell subset was exported in FlowJo, and then extracted to evaluate expression of each marker on individual events with FCS Extractor software (research.stowers.org/mcm/efg/ScientificSoftware/Utility/FCSExtract/index.htm). These single event values were pooled across all animals within each experimental group (adult, old, MCMV). These values were binned to assess the relative distribution of staining ratios for each experimental mouse group. P values in the table were calculated by ANOVA with Bonferonni adjustment (means) or by Kruskal–Wallis test (medians).
To measure responsiveness to APLs, splenocytes were incubated for 6 h at 37 °C in a total volume of 100 μL of RPMI 1640 plus 5% FCS containing 0.1 μg/well brefeldin A (eBioscience), and 10−6 M of the following OVA257–264-derived APLs: native “N4” (SIINFEKL), APL “A2” (SAINFEKL), APL “Y3” (SIYNFEKL), APL “Q4” (SIIQFEKL), or no peptide. Cells were washed, stained overnight at 4 °C with fluorochrome-conjugated antibodies specific for the surface markers CD4, and CD8α, washed, fixed, and permeabilized, and then stained for intracellular IFN-γ (XMG1.2) using the BD Fix/Perm buffer kit according to the manufacturer’s directions.
Data acquisition was performed on a custom-made, four-laser BD Fortessa flow cytometer (Becton Dickinson), and was analyzed using FlowJo software (Tree Star). A minimum of 10,000 CD8+ events within the lymphocyte gate was collected for all files.
Naïve CD8 T Cell Sorting for TCRβ Repertoire Analysis.
Splenocytes were collected and passed through a 40-μm mesh screen to prepare single-cell suspensions, and then negatively enriched for CD8+ T cells using magnetic beads (Miltenyi Biotec). Highly enriched CD8+ T cells were stained with fluorochrome-conjugated anti-CD8α (53-6.7), anti-CD4 (GK1.5), anti-CD62L (MEL-14), and anti-CD44 (IM7) for 45 min on ice, and then washed twice. Cells were resuspended in sorting buffer and 50,000 CD8+CD4−CD44−CD62L+ lymphocytes were sorted into TRI Reagent (Molecular Research Center) using a FACSAria cell sorter (BD Biosciences). At least three replicate samples (50,000 each) were collected from each animal and stored at −80 °C.
Naïve CD8 TCRβ Library Construction, Molecular Barcoding, and Sequencing.
RNA was chloroform extracted and alcohol coprecipitated with 40 μg of RNase-free glycogen. The RNA was reverse transcribed (Superscript III/Platinum Taq) at 50 °C for 30 min, followed by one cycle of PCR in the presence of a multiplex set of mouse TRBV and TRBC primers (62). The 5′ ends of the TRBV primers were appended with molecular tags (9 × N-nucleotides) and Illumina MiSeq adaptors, and the TRBC primer was appended with an additional molecular tag (4 × N-nucleotides). The single-cycle PCR conditions contained a single melting step of 94 °C of 30 s, followed by a single annealing stage of 51 °C for 45 s and an extension cycle of 72 °C for 3 min. The double-stranded DNA product was purified by Ampure XP beads and further amplified with oligos containing Illumina MiSeq adaptors and standard Illumina barcodes derived from the Illumina N501-N504, N701-N704 series. The following PCR conditions were utilized: 95 °C (30 s), 58 °C (45 s), and 72 °C (45 s); for 35 cycles. The double-stranded DNA product was purified by Agilent Ampure XP beads, yielding a mouse TCR β-chain (mTCRβ) library whose size is between 320 and 540 bp. The mTCRβ library was further purified by Pippin Prep (Sage Science) and analyzed for the presence of residual primer dimer artifacts utilizing Advanced Analytical Technologies High Sensitivity NGS Fragment Analyzer. Each of the 10 mTCRβ libraries was quantified by qPCR and sequenced on an Illumina MiSeq instrument with 2 × 300-bp paired-end v3 chemistry in the University of Arizona Genome Sequencing Core.
To informatically control for amplification bias, PCR error, and sequencing error, an analysis was carried out with MT-Toolbox to deconvolute the molecular tags. Using this program, TCR sequences with identical molecular tags were deemed to have derived from the same input mRNA template molecule (63). The variable (V) gene and joining (J) gene usage for each TCR was determined utilizing IgBLAST (64) emulated within the VDJserver infrastructure (https://vdjserver.org/, according to the germline reference gene sequences annotated in IMGT [www.imgt.org/IMGTrepertoire (24)].
Naïve CD8 TCRβ Repertoire Diversity Analysis.
Shannon’s entropy calculations were performed according to Diz et al. (65). Briefly, Shannon entropy is utilized as a readout of population diversity of the TCRβ clonotypes within each sample. The number of unique species of TCRβ clonotypes and the frequency of each of those unique clonotypes are utilized to calculate Shannon’s entropy. We define S as the total number of unique TCRβ clonotypes in the sample, and Pi as the proportion of the sample corresponding to the ith TCRβ clonotype, leading to the definition of Shannon entropy H as follows:
Entropy is highest when the number of unique TCRβ clonotypes is high and when the proportion of each TCRβ clonotype does not dominate the frequency landscape.
Supplementary Material
Acknowledgments
We thank past and present members of the J.N.-Ž. laboratory for help and stimulating discussion. Expert cell-sorting assistance was provided by Paula Campbell at the University of Arizona Cancer Center/Arizona Research Laboratories–Division of Biotechnology Cytometry Core Facility (supported by CCSG-CA 023074). This work was supported by US Public Health Service Grants U54 AI081680 (Pacific Northwest Research Center of Excellence in Biodefense and Emerging Diseases), National Institute of Allergy and Infectious Diseases Grant HHSN27220110017C, and National Institute on Aging Grant R01 AG048021 from the National Institutes of Health. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the BioProject database (accession no. PRJNA436418).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1719451115/-/DCSupplemental.
References
- 1.Nikolich-Žugich J, van Lier RAW. Cytomegalovirus (CMV) research in immune senescence comes of age: Overview of the 6th International Workshop on CMV and Immunosenescence. Geroscience. 2017;39:245–249. doi: 10.1007/s11357-017-9984-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Souquette A, Frere J, Smithey M, Sauce D, Thomas PG. A constant companion: Immune recognition and response to cytomegalovirus with aging and implications for immune fitness. Geroscience. 2017;39:293–303. doi: 10.1007/s11357-017-9982-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jackson SE, et al. CMV immune evasion and manipulation of the immune system with aging. Geroscience. 2017;39:273–291. doi: 10.1007/s11357-017-9986-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nikolich-Žugich J, Goodrum F, Knox K, Smithey MJ. Known unknowns: How might the persistent herpesvirome shape immunity and aging? Curr Opin Immunol. 2017;48:23–30. doi: 10.1016/j.coi.2017.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bate SL, Dollard SC, Cannon MJ. Cytomegalovirus seroprevalence in the United States: The National Health and Nutrition Examination Surveys, 1988–2004. Clin Infect Dis. 2010;50:1439–1447. doi: 10.1086/652438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Holtappels R, et al. Control of murine cytomegalovirus in the lungs: Relative but not absolute immunodominance of the immediate-early 1 nonapeptide during the antiviral cytolytic T-lymphocyte response in pulmonary infiltrates. J Virol. 1998;72:7201–7212. doi: 10.1128/jvi.72.9.7201-7212.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wertheimer AM, et al. Aging and cytomegalovirus infection differentially and jointly affect distinct circulating T cell subsets in humans. J Immunol. 2014;192:2143–2155. doi: 10.4049/jimmunol.1301721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vescovini R, et al. Naïve and memory CD8 T cell pool homeostasis in advanced aging: Impact of age and of antigen-specific responses to cytomegalovirus. Age (Dordr) 2014;36:625–640. doi: 10.1007/s11357-013-9594-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Khan N, et al. Cytomegalovirus seropositivity drives the CD8 T cell repertoire toward greater clonality in healthy elderly individuals. J Immunol. 2002;169:1984–1992. doi: 10.4049/jimmunol.169.4.1984. [DOI] [PubMed] [Google Scholar]
- 10.Sylwester AW, et al. Broadly targeted human cytomegalovirus-specific CD4+ and CD8+ T cells dominate the memory compartments of exposed subjects. J Exp Med. 2005;202:673–685. doi: 10.1084/jem.20050882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pawelec G. Immunosenenescence: Role of cytomegalovirus. Exp Gerontol. 2014;54:1–5. doi: 10.1016/j.exger.2013.11.010. [DOI] [PubMed] [Google Scholar]
- 12.Smithey MJ, Li G, Venturi V, Davenport MP, Nikolich-Žugich J. Lifelong persistent viral infection alters the naive T cell pool, impairing CD8 T cell immunity in late life. J Immunol. 2012;189:5356–5366. doi: 10.4049/jimmunol.1201867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rudd BD, et al. Nonrandom attrition of the naive CD8+ T-cell pool with aging governed by T-cell receptor:pMHC interactions. Proc Natl Acad Sci USA. 2011;108:13694–13699. doi: 10.1073/pnas.1107594108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qi Q, et al. Diversity and clonal selection in the human T-cell repertoire. Proc Natl Acad Sci USA. 2014;111:13139–13144. doi: 10.1073/pnas.1409155111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yager EJ, et al. Age-associated decline in T cell repertoire diversity leads to holes in the repertoire and impaired immunity to influenza virus. J Exp Med. 2008;205:711–723. doi: 10.1084/jem.20071140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rudd BD, Venturi V, Davenport MP, Nikolich-Žugich J. Evolution of the antigen-specific CD8+ TCR repertoire across the life span: Evidence for clonal homogenization of the old TCR repertoire. J Immunol. 2011;186:2056–2064. doi: 10.4049/jimmunol.1003013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Quinn KM, et al. Heightened self-reactivity associated with selective survival, but not expansion, of naïve virus-specific CD8+ T cells in aged mice. Proc Natl Acad Sci USA. 2016;113:1333–1338. doi: 10.1073/pnas.1525167113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Valkenburg SA, et al. Early priming minimizes the age-related immune compromise of CD8+ T cell diversity and function. PLoS Pathog. 2012;8:e1002544. doi: 10.1371/journal.ppat.1002544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cicin-Sain L, et al. Cytomegalovirus infection impairs immune responses and accentuates T-cell pool changes observed in mice with aging. PLoS Pathog. 2012;8:e1002849. doi: 10.1371/journal.ppat.1002849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marandu TF, et al. Immune protection against virus challenge in aging mice is not affected by latent herpesviral infections. J Virol. 2015;89:11715–11717. doi: 10.1128/JVI.01989-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mekker A, et al. Immune senescence: Relative contributions of age and cytomegalovirus infection. PLoS Pathog. 2012;8:e1002850. doi: 10.1371/journal.ppat.1002850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pope C, et al. Organ-specific regulation of the CD8 T cell response to Listeria monocytogenes infection. J Immunol. 2001;166:3402–3409. doi: 10.4049/jimmunol.166.5.3402. [DOI] [PubMed] [Google Scholar]
- 23.Carbone FR, Bevan MJ. Induction of ovalbumin-specific cytotoxic T cells by in vivo peptide immunization. J Exp Med. 1989;169:603–612. doi: 10.1084/jem.169.3.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lefranc MP, et al. IMGT, the international ImMunoGeneTics database. Nucleic Acids Res. 1999;27:209–212. doi: 10.1093/nar/27.1.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kelly JM, et al. Identification of conserved T cell receptor CDR3 residues contacting known exposed peptide side chains from a major histocompatibility complex class I-bound determinant. Eur J Immunol. 1993;23:3318–3326. doi: 10.1002/eji.1830231239. [DOI] [PubMed] [Google Scholar]
- 26.Fremont DH, Stura EA, Matsumura M, Peterson PA, Wilson IA. Crystal structure of an H-2Kb-ovalbumin peptide complex reveals the interplay of primary and secondary anchor positions in the major histocompatibility complex binding groove. Proc Natl Acad Sci USA. 1995;92:2479–2483. doi: 10.1073/pnas.92.7.2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nikolich-Žugich J. Ageing and life-long maintenance of T-cell subsets in the face of latent persistent infections. Nat Rev Immunol. 2008;8:512–522. doi: 10.1038/nri2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hadrup SR, et al. Longitudinal studies of clonally expanded CD8 T cells reveal a repertoire shrinkage predicting mortality and an increased number of dysfunctional cytomegalovirus-specific T cells in the very elderly. J Immunol. 2006;176:2645–2653. doi: 10.4049/jimmunol.176.4.2645. [DOI] [PubMed] [Google Scholar]
- 29.Zehn D, Lee SY, Bevan MJ. Complete but curtailed T-cell response to very low-affinity antigen. Nature. 2009;458:211–214. doi: 10.1038/nature07657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rawlinson WD, Farrell HE, Barrell BG. Analysis of the complete DNA sequence of murine cytomegalovirus. J Virol. 1996;70:8833–8849. doi: 10.1128/jvi.70.12.8833-8849.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Juranic Lisnic V, et al. Dual analysis of the murine cytomegalovirus and host cell transcriptomes reveal new aspects of the virus-host cell interface. PLoS Pathog. 2013;9:e1003611. doi: 10.1371/journal.ppat.1003611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Welten SPM, et al. CD27-CD70 costimulation controls T cell immunity during acute and persistent cytomegalovirus infection. J Virol. 2013;87:6851–6865. doi: 10.1128/JVI.03305-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Munks MW, et al. Four distinct patterns of memory CD8 T cell responses to chronic murine cytomegalovirus infection. J Immunol. 2006;177:450–458. doi: 10.4049/jimmunol.177.1.450. [DOI] [PubMed] [Google Scholar]
- 34.Bunztman A, Vincent BG, Krovi H, Steele S, Frelinger JA. The LCMV gp33-specific memory T cell repertoire narrows with age. Immun Ageing. 2012;9:17. doi: 10.1186/1742-4933-9-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li G, Smithey MJ, Rudd BD, Nikolich-Žugich J. Age-associated alterations in CD8α+ dendritic cells impair CD8 T-cell expansion in response to an intracellular bacterium. Aging Cell. 2012;11:968–977. doi: 10.1111/j.1474-9726.2012.00867.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gigley JP, Khan IA. Plasmacytoid DC from aged mice down-regulate CD8 T cell responses by inhibiting cDC maturation after Encephalitozoon cuniculi infection. PLoS One. 2011;6:e20838. doi: 10.1371/journal.pone.0020838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smithey MJ, Renkema KR, Rudd BD, Nikolich-Žugich J. Increased apoptosis, curtailed expansion and incomplete differentiation of CD8+ T cells combine to decrease clearance of L. monocytogenes in old mice. Eur J Immunol. 2011;41:1352–1364. doi: 10.1002/eji.201041141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brien JD, Uhrlaub JL, Hirsch A, Wiley CA, Nikolich-Zugich J. Key role of T cell defects in age-related vulnerability to West Nile virus. J Exp Med. 2009;206:2735–2745. doi: 10.1084/jem.20090222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jiang J, et al. Limited expansion of virus-specific CD8 T cells in the aged environment. Mech Ageing Dev. 2009;130:713–721. doi: 10.1016/j.mad.2009.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reese TA, et al. Sequential infection with common pathogens promotes human-like immune gene expression and altered vaccine response. Cell Host Microbe. 2016;19:713–719. doi: 10.1016/j.chom.2016.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Oh JZ, et al. TLR5-mediated sensing of gut microbiota is necessary for antibody responses to seasonal influenza vaccination. Immunity. 2014;41:478–492. doi: 10.1016/j.immuni.2014.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Barton ES, et al. Herpesvirus latency confers symbiotic protection from bacterial infection. Nature. 2007;447:326–329. doi: 10.1038/nature05762. [DOI] [PubMed] [Google Scholar]
- 43.Furman D, et al. Cytomegalovirus infection enhances the immune response to influenza. Sci Transl Med. 2015;7:281ra43. doi: 10.1126/scitranslmed.aaa2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Barton ES, Rajkarnikar S, Langston PK, Price MJ, Grayson JM. Gammaherpesvirus latency differentially impacts the generation of primary versus secondary memory CD8+ T cells during subsequent infection. J Virol. 2014;88:12740–12751. doi: 10.1128/JVI.02106-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Callahan JE, Kappler JW, Marrack P. Unexpected expansions of CD8-bearing cells in old mice. J Immunol. 1993;151:6657–6669. [PubMed] [Google Scholar]
- 46.LeMaoult J, et al. Age-related dysregulation in CD8 T cell homeostasis: Kinetics of a diversity loss. J Immunol. 2000;165:2367–2373. doi: 10.4049/jimmunol.165.5.2367. [DOI] [PubMed] [Google Scholar]
- 47.Hingorani R, et al. Clonal predominance of T cell receptors within the CD8+ CD45RO+ subset in normal human subjects. J Immunol. 1993;151:5762–5769. [PubMed] [Google Scholar]
- 48.Posnett DN, Sinha R, Kabak S, Russo C. Clonal populations of T cells in normal elderly humans: The T cell equivalent to “benign monoclonal gammapathy.”. J Exp Med. 1994;179:609–618. doi: 10.1084/jem.179.2.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moon JJ, et al. Naive CD4+ T cell frequency varies for different epitopes and predicts repertoire diversity and response magnitude. Immunity. 2007;27:203–213. doi: 10.1016/j.immuni.2007.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Obar JJ, Khanna KM, Lefrançois L. Endogenous naive CD8+ T cell precursor frequency regulates primary and memory responses to infection. Immunity. 2008;28:859–869. doi: 10.1016/j.immuni.2008.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.van Heijst JWJ, et al. Recruitment of antigen-specific CD8+ T cells in response to infection is markedly efficient. Science. 2009;325:1265–1269. doi: 10.1126/science.1175455. [DOI] [PubMed] [Google Scholar]
- 52.Messaoudi I, Guevara Patiño JA, Dyall R, LeMaoult J, Nikolich-Žugich J. Direct link between mhc polymorphism, T cell avidity, and diversity in immune defense. Science. 2002;298:1797–1800. doi: 10.1126/science.1076064. [DOI] [PubMed] [Google Scholar]
- 53.Price DA, et al. T cell receptor recognition motifs govern immune escape patterns in acute SIV infection. Immunity. 2004;21:793–803. doi: 10.1016/j.immuni.2004.10.010. [DOI] [PubMed] [Google Scholar]
- 54.Cornberg M, et al. Narrowed TCR repertoire and viral escape as a consequence of heterologous immunity. J Clin Invest. 2006;116:1443–1456. doi: 10.1172/JCI27804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hahn G, Jores R, Mocarski ES. Cytomegalovirus remains latent in a common precursor of dendritic and myeloid cells. Proc Natl Acad Sci USA. 1998;95:3937–3942. doi: 10.1073/pnas.95.7.3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Belz GT, et al. Cutting edge: Conventional CD8 alpha+ dendritic cells are generally involved in priming CTL immunity to viruses. J Immunol. 2004;172:1996–2000. doi: 10.4049/jimmunol.172.4.1996. [DOI] [PubMed] [Google Scholar]
- 57.Campisi L, et al. Splenic CD8α+ dendritic cells undergo rapid programming by cytosolic bacteria and inflammation to induce protective CD8+ T-cell memory. Eur J Immunol. 2011;41:1594–1605. doi: 10.1002/eji.201041036. [DOI] [PubMed] [Google Scholar]
- 58.Kedzierska K, Turner SJ, Doherty PC. Conserved T cell receptor usage in primary and recall responses to an immunodominant influenza virus nucleoprotein epitope. Proc Natl Acad Sci USA. 2004;101:4942–4947. doi: 10.1073/pnas.0401279101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hamrouni A, Aublin A, Guillaume P, Maryanski JL. T cell receptor gene rearrangement lineage analysis reveals clues for the origin of highly restricted antigen-specific repertoires. J Exp Med. 2003;197:601–614. doi: 10.1084/jem.20021945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Venturi V, Kedzierska K, Turner SJ, Doherty PC, Davenport MP. Methods for comparing the diversity of samples of the T cell receptor repertoire. J Immunol Methods. 2007;321:182–195. doi: 10.1016/j.jim.2007.01.019. [DOI] [PubMed] [Google Scholar]
- 61.Venturi V, et al. Method for assessing the similarity between subsets of the T cell receptor repertoire. J Immunol Methods. 2008;329:67–80. doi: 10.1016/j.jim.2007.09.016. [DOI] [PubMed] [Google Scholar]
- 62.Baker FJ, Lee M, Chien Y-H, Davis MM. Restricted islet-cell reactive T cell repertoire of early pancreatic islet infiltrates in NOD mice. Proc Natl Acad Sci USA. 2002;99:9374–9379. doi: 10.1073/pnas.142284899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yourstone SM, Lundberg DS, Dangl JL, Jones CD. MT-Toolbox: Improved amplicon sequencing using molecule tags. BMC Bioinformatics. 2014;15:284. doi: 10.1186/1471-2105-15-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ye J, Ma N, Madden TL, Ostell JM. IgBLAST: An immunoglobulin variable domain sequence analysis tool. Nucleic Acids Res. 2013;41:W34–W40. doi: 10.1093/nar/gkt382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Diz R, et al. Autoreactive effector/memory CD4+ and CD8+ T cells infiltrating grafted and endogenous islets in diabetic NOD mice exhibit similar T cell receptor usage. PLoS One. 2012;7:e52054. doi: 10.1371/journal.pone.0052054. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.