Significance
Transthyretin (TTR) cardiac amyloidosis is characterized by the deposition of TTR amyloid fibrils in the heart. No therapy is currently available for wild-type cardiac amyloidosis. Hereditary cases are treated by liver transplantation, a crude form of gene therapy which replaces amyloidogenic mutant TTR by the more stable wild-type form, with the goal of halting further deposition and disease progression. However, wild-type TTR continues to deposit in the heart of many patients after the procedure. Until now, seeding of TTR fibril formation has not been demonstrated in vitro. We show that patient-extracted cardiac fibrils can seed both wild-type and mutant TTR fibril formation in vitro. This process can be inhibited by structure-based peptide inhibitors, thereby providing an alternative approach to therapy.
Keywords: amyloid, transthyretin, seeding, amyloidosis, cardiomyopathy
Abstract
Each of the 30 human amyloid diseases is associated with the aggregation of a particular precursor protein into amyloid fibrils. In transthyretin amyloidosis (ATTR), mutant or wild-type forms of the serum carrier protein transthyretin (TTR), synthesized and secreted by the liver, convert to amyloid fibrils deposited in the heart and other organs. The current standard of care for hereditary ATTR is liver transplantation, which replaces the mutant TTR gene with the wild-type gene. However, the procedure is often followed by cardiac deposition of wild-type TTR secreted by the new liver. Here we find that amyloid fibrils extracted from autopsied and explanted hearts of ATTR patients robustly seed wild-type TTR into amyloid fibrils in vitro. Cardiac-derived ATTR seeds can accelerate fibril formation of wild-type and monomeric TTR at acidic pH and under physiological conditions, respectively. We show that this seeding is inhibited by peptides designed to complement structures of TTR fibrils. These inhibitors cap fibril growth, suggesting an approach for halting progression of ATTR.
Amyloid diseases are characterized by the tissue deposition of proteins, synthesized as soluble precursors, as insoluble amyloid fibrils, frequently at a distance from the site of synthesis. Amyloid fibrils are robust fibrillar structures with the potential to nucleate subsequent fibril formation of the soluble parental protein. In this process, known as “amyloid seeding," fibril formation is accelerated by the presence of substoichiometric quantities of preformed fibrils. Numerous laboratories have studied and reproduced seeding of several amyloid precursors involved in both localized and systemic amyloidoses (1–4). However, despite clinical evidence suggesting that seeding of the amyloid protein transthyretin (TTR) occurs in vivo (5), the demonstration of seeding in vitro or in mouse models has not yet been achieved. Here we investigate amyloid seeding of TTR fibril formation using fibrils extracted from diseased human cardiac tissue.
TTR amyloidosis (ATTR) is caused by amyloid deposition of fibrils derived from the serum protein TTR (6–10). In hereditary ATTR, autosomal-dominant mutations destabilize the TTR tetramer, accelerating pathological aggregation and the onset of the disease (11, 12). Although more stable than most mutant forms, wild-type TTR is also found in amyloid deposits in both wild-type (13–15) and hereditary (15–17) ATTR. These deposits of TTR fibrils are found in virtually every tissue of the body and cause peripheral neuropathies and cardiomyopathies. However, the pathological mechanism is not fully understood. Studies of recombinant TTR aggregates have suggested the involvement of oligomeric intermediates in TTR cytotoxicity and subsequent pathogenicity (18).
Cardiac ATTR pathology manifests two distinct patterns (19). Type A cardiac ATTR is associated with the development of progressive infiltrative cardiomyopathy, with large, diffuse, tightly packed amyloid deposits that contain short fibrils comprised of both full-length TTR fibrils and C-terminal TTR fragments. In type B cardiac ATTR, more distinct amyloid deposits made of full-length TTR fibrils surround individual muscle cells. Although the understanding of their clinical and pathological significance is incomplete, there is a clear distinction between subtypes: type A deposits display a higher capacity to recruit wild-type TTR (16). If untreated, both subtypes of TTR deposition lead to organ failure and eventual death.
Since the liver is the main source of TTR production, beginning in 1990, patients with hereditary ATTR have been treated by liver transplantation, a crude form of gene therapy that replaces the mutant ttr gene with the wild-type gene (20). Many ATTR cases have shown prolonged life with stabilization or slowing of disease progression after transplantation, with most favorable results found for ATTR-V30M neuropathic patients at early stages (21). However, this procedure is sometimes followed by progressive cardiac deposition and death (5). Several clinical studies report that cardiac amyloid isolated from patients after liver transplantation has a predominance of wild-type over variant TTR, suggesting that wild-type TTR can be incorporated into cardiac amyloid, presumably by a process of seeding (5, 17, 22, 23).
Amyloid primary nucleation, which precedes fibril formation in the absence of seeds, is driven by specific protein segments that are the adhesive parts of amyloid proteins (24). In previous work we found that two TTR adhesive segments are the principal drivers of protein aggregation in vitro. These are the segments that form the F and H β-strands in the native TTR structure. These strands become exposed to solvent upon TTR dissociation to monomers, enabling their stacking into the steric zipper spines of amyloid fibrils (25, 26). Based on the structures of the amyloid fibrils formed by these two segments, we generated peptides designed to block fibril formation by capping the tips of TTR fibrils (25).
Here we studied amyloid seeding of ATTR fibril formation and found that amyloid fibrils extracted from ex vivo ATTR cardiac tissues can seed fibril formation of both wild-type and mutant TTR in vitro. Moreover, we find that optimized peptide inhibitors block this seeding. Our results point to the clinical importance of amyloid seeding in cardiac amyloidosis and propose inhibition of amyloid seeding as a promising therapeutic strategy for ATTR.
Results
Ex Vivo Material Extracted from ATTR-D38A Cardiac Tissue Accelerates Fibril Formation of Tetrameric TTR at Acidic pH in Vitro.
We extracted amyloid fibrils from the explanted heart of a hereditary ATTR patient carrying the ATTR-D38A mutation (SI Appendix, Table S1). Our procedures for extraction and characterization are outlined in SI Appendix, Fig. S1A. Before extraction, we confirmed the amyloid content of the explanted heart by Congo red and immunostaining of paraffin-embedded sections of the organ (SI Appendix, Fig. S1 B and C). Sections stained with Congo red displayed streaks of apple-green birefringence (SI Appendix, Fig. S1B). Tissue immunostaining revealed the presence of scattered patches of TTR deposits throughout the sample (SI Appendix, Fig. S1C). Amyloid extraction was performed on a frozen piece of the same organ by the protocol of Westermark et al. (27), using multiple cycles of homogenization with saline and water. We confirmed the presence of both short and long amyloid-like fibrils in the resultant resuspension by transmission electron microscopy (TEM) (SI Appendix, Fig. S1D). The enrichment of TTR fibrils in the extract was confirmed by anti-TTR Western blot, size-exclusion chromatography, and mass spectrometry-based protein sequencing (SI Appendix, Fig. S1 E and F and Dataset S1).
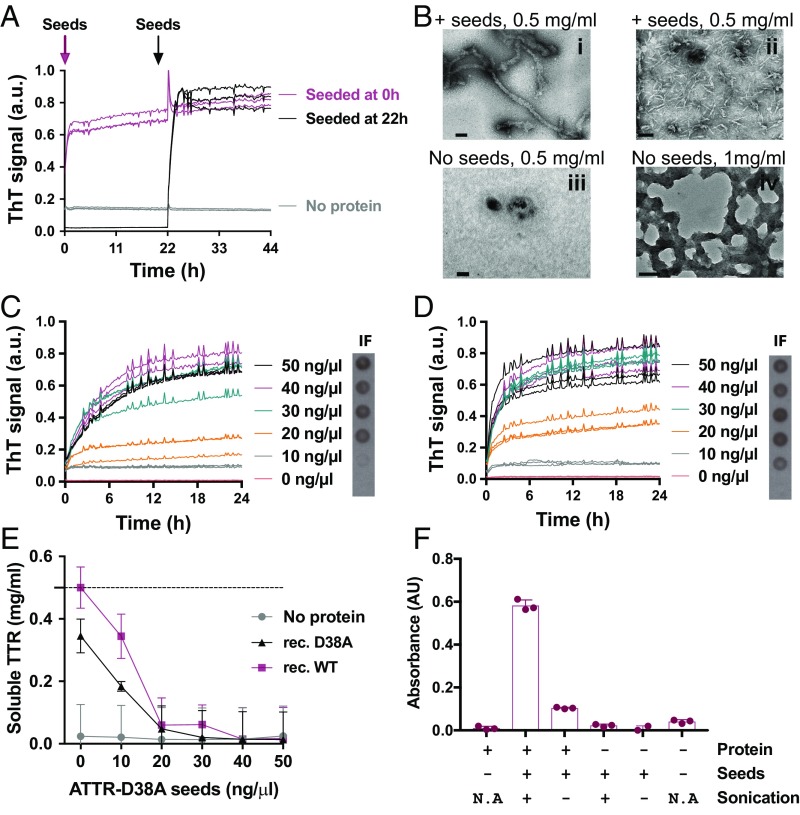
We next found that ex vivo amyloid material seeds soluble TTR into fibrils at acidic pH. We define seeds as ex vivo material obtained from ATTR patients that can accelerate recombinant TTR fibril formation. Ex vivo ATTR seeds were obtained by sonicating the resuspended fibrils. Small fibrils of various lengths were observed by TEM (SI Appendix, Fig. S1A). Total protein concentration was measured, and 30 ng/μL ex vivo seeds were added to 0.5 mg/mL native recombinant wild-type TTR (Fig. 1A). We added ATTR seeds at two different time points—at initiation or after 22 h of incubation—to evaluate the aggregation and formation of thioflavin T (ThT)-positive species of recombinant wild-type TTR before adding the seeds. The TTR tetramer must undergo rate-limiting dissociation before fibril formation (28). To render tetramer dissociation fast, we performed this assay at pH 4.3, and fibril formation was monitored by ThT fluorescence at 37 °C with constant orbital shaking. Whereas ex vivo seeds alone did not result in a substantial increase in ThT signal, the addition of seeds at either the initial or the 22-h time point resulted in a rapid increase in fluorescence (Fig. 1A). Increased fluorescence correlated with the accumulation of protein aggregates as visualized by optical microscopy (SI Appendix, Fig. S2A) and the appearance of fibrillar structures visible by TEM (Fig. 1B). In contrast, wild-type recombinant TTR does not generate aggregates at a concentration of 0.5 mg/mL after 24 h of incubation when ATTR seeds are not present (Fig. 1 B–D). Higher concentrations and longer incubation times are required for the generation of protein aggregates under these conditions (Fig. 1B). This seeding effect was dependent on TTR concentration, since the gradual addition of recombinant protein (0–1 mg/mL) to a fixed amount of seeds (30 ng/μL) results in a gradual increase in fibril formation (SI Appendix, Fig. S2B).
Fig. 1.
Amyloid seeding at pH 4.3 of wild-type TTR by ex vivo ATTR seeds extracted from the explanted heart of an ATTR-D38A patient. (A) Amyloid seeding assay of recombinant wild-type TTR when 30 ng/μL ATTR-D38A ex vivo seeds were added at 0 h and after 22 h of preincubation, as monitored by ThT fluorescence. (B) Electron micrograph of aggregates of wild-type TTR after 24 h (i and iii) or 4 d (ii and iv) of incubation. ATTR seeds were added at 0 h (i and ii). (Scale bars, 100 nm.) (C and D) Amyloid seeding assays monitored by ThT fluorescence. Increasing amounts of ATTR-D38A ex vivo seeds were added at time 0 to recombinant wild-type TTR (C) or D38A TTR (D). (Right Insets) Anti–His-tag immuno-dot blot of insoluble fractions (IF). (E) Protein concentration in the soluble fraction extracted from C and D, measured by BCA protein assay. The dashed line marks the initial protein concentration (0.5 mg/mL). (F) The 280-nm absorbance of insoluble fractions collected from an amyloid seeding assay of recombinant wild-type TTR with and without 30 ng/L sonicated or nonsonicated seeds. n = 3; error bars indicate SD. AU, absorbance units; a.u., arbitrary units; N/A, not applicable. In A, C, and D all replicates are shown.
ATTR Seeds Accelerate Fibril Formation of both Wild-Type and Mutant TTR Variants in a Dose-Dependent Manner.
Two forms of recombinant TTR were studied for this assay: wild-type TTR and the variant D38A mutant, at pH 4.3. Both forms are tetrameric in solution at pH 7.4 (SI Appendix, Fig. S2C). The addition of increasing amounts of seeds to wild-type or D38A recombinant TTR resulted in a dose-dependent increase in ThT signal after a brief lag phase of 10–20 min (Fig. 1 C and D and SI Appendix, Fig. S2 D and E). Neither recombinant TTR in the absence of ex vivo seeds nor ATTR seeds in the absence of protein caused an increase in ThT signal (Fig. 1 C and D and SI Appendix, Fig. S2 F and G). We confirmed that the resultant fibrils were derived by de novo fibril formation from recombinant protein as follows. First, we obtained the insoluble fraction of the samples by centrifugation and resuspension in guanidinium hydrochloride. These fractions were spotted onto a nitrocellulose membrane for immuno-dot blot analysis with an antibody that recognizes the histidine tag of recombinant protein. We found seed-dependent enhancement of recombinant TTR content in the insoluble fraction (Fig. 1 C and D, Right Insets), consistent with an increase in absorbance at 280 nm (SI Appendix, Fig. S2H). As expected, the increase in insoluble material was accompanied by a gradual decrease of soluble TTR, as detected by BCA protein assay (Fig. 1E) and anti–His-tag Western blot (SI Appendix, Fig. S2I). We could not detect any remaining soluble protein after 24 h incubation with seeds at 30 ng/μL or higher (Fig. 1E). We observed that the soluble fraction depletes faster than the ThT-positive species appear (Fig. 1E vs. Fig. 1 C and D, respectively). We reason that, at low seed concentrations, the sample may contain both highly ordered and less well-ordered assemblies. Taken together, these results suggest that ex vivo extracts obtained from ATTR-D38A cardiac tissue can accelerate fibril formation of recombinant wild-type and D38A TTR at acidic pH where tetramer dissociation is fast.
Amyloid Seeding Is Caused by the TTR Fibrillar Content of ex Vivo Extracts and Not by Other Coextracted Insoluble Material.
We first observed that sonication greatly potentiates TTR amyloid seeding, suggesting the involvement of fibril tips in templating TTR fibril formation (Fig. 1F). In addition, immunodepletion of ex vivo extracts of TTR reduced their capacity to seed recombinant wild-type TTR (SI Appendix, Fig. S3 A and B). TTR was sequentially removed from the ex vivo extract by several cycles of binding to an anti-TTR surface. The seeding capacity diminishes as TTR is gradually removed from the extract. As an additional control, we obtained cardiac tissue explanted after ATTR-unrelated heart failure (SI Appendix, Table S1). We confirmed the absence of amyloid TTR deposits by Congo red and immunostaining of tissue sections (SI Appendix, Fig. S3 C and D). After performing the saline-water extraction described above, we evaluated TTR content in the extract by anti-TTR dot blot (SI Appendix, Fig. S3E) and confirmed the absence of amyloid fibrils by TEM (SI Appendix, Fig. S3F). We then performed an amyloid seeding assay and found that non-ATTR ex vivo extract does not accelerate native recombinant TTR fibril formation (SI Appendix, Fig. S3G). Together these findings suggest that amyloid seeding of recombinant TTR is caused by fibrillar TTR-derived material found in ATTR cardiac tissue and not by other insoluble material extracted from cardiac specimens, such as collagen (SI Appendix, Fig. S3F).
Under Physiological Conditions, ATTR-D38A Cardiac Seeds Also Accelerate Fibril Formation of TTR When Monomerized by Mutagenesis.
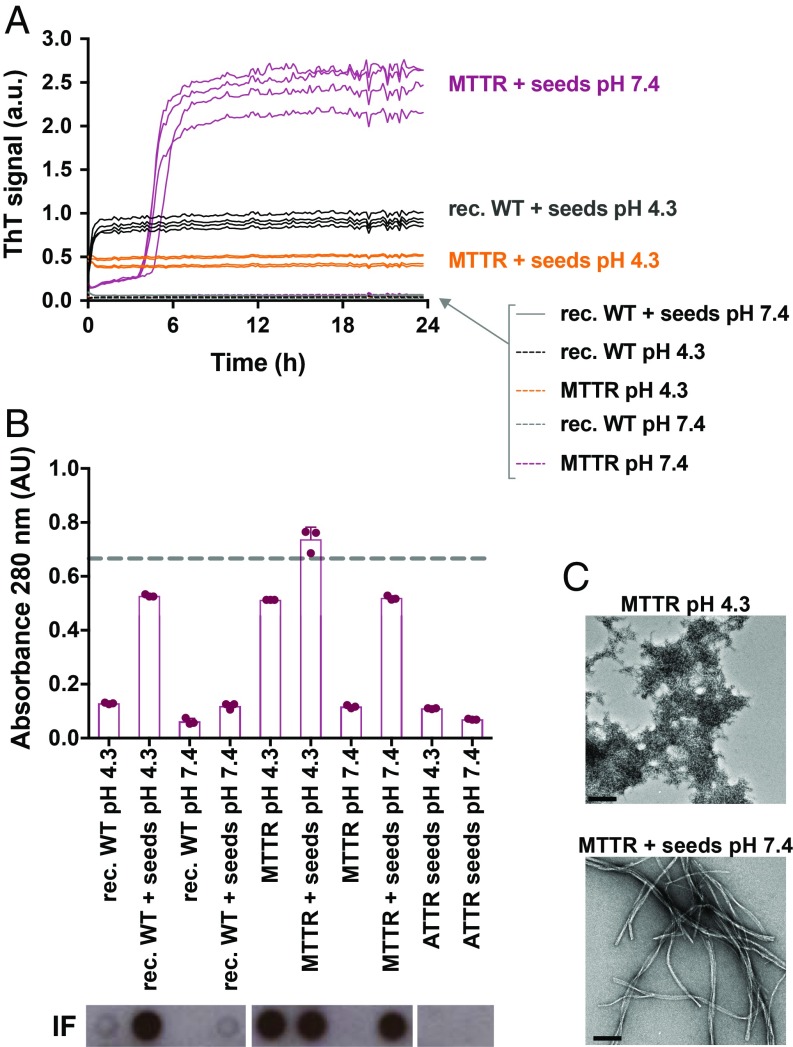
In seeking to evaluate amyloid seeding of ATTR fibrils under physiological conditions, we chose to work with a monomeric variant of TTR engineered by Kelly and coworkers (29), herein referred to as “MTTR” (25). This variant carries the double mutation F87M/L110M, is soluble after purification, and displays faster aggregation than wild-type TTR or familial mutants because it bypasses the time requirement for tetramer dissociation (29). Kelly and coworkers (30) found that, when subjected to low pH, MTTR unfolds into an amyloidogenic variant that nucleates into aggregates in a process that is not affected by the presence of seeds. In that study, seeds were prepared by aggregation of MTTR at low pH in vitro. We wondered whether seeds obtained from ATTR patients differ from those originating in vitro. We evaluated the seeding capacity of ATTR-D38A fibrils with recombinant tetrameric and monomeric TTR at physiological and acidic pH. We followed amyloid fibril formation by ThT fluorescence, protein quantification, anti–His-tag dot blot of insoluble fractions, and TEM, as described above (Fig. 2). Consistent with our results in Fig. 1, wild-type TTR aggregation was accelerated by ATTR seeds at pH 4.3 (Fig. 2). We also found that the addition of ATTR seeds to MTTR at pH 7.4 resulted in the formation of ThT-positive species (Fig. 2A). However, the addition of these seeds to recombinant wild-type TTR did not show a significant effect when tested at physiological pH (Fig. 2A), suggesting that seeded fibril formation requires the dissociation of tetramers into monomers. This hypothesis is supported by the observation that ATTR-D38A seeds are capable of converting MTTR into ThT-positive species under physiological conditions with a lag phase of 4–5 h (Fig. 2A). Under these conditions, TEM analysis revealed that 24 h of incubation of MTTR with ATTR seeds resulted in elongated amyloid-like fibrils (Fig. 2C). For completeness we note that MTTR aggregated under acidic conditions in the absence of ATTR seeds, but these aggregates were ThT-negative and appear as amorphous, nonfibrillar structures when analyzed by TEM (Fig. 2 A and C). The addition of seeds to MTTR at pH 4.3 resulted in the formation of ThT-positive species, but no lag phase was observed, similar to recombinant wild-type TTR in the presence of seeds at pH 4.3 (Fig. 2 A and C). We did not find significant differences in the seeding capacity of seeds that were preincubated at pH 4.3 vs. pH 7.5 when added to MTTR. Taken together, our results suggest that ATTR-D38A seeds accelerate monomeric TTR fibril formation under physiological conditions.
Fig. 2.
Amyloid seeding of monomeric TTR by ex vivo ATTR-D38A seeds under physiological conditions. (A) Amyloid seeding assay followed by ThT fluorescence. ATTR-D38A ex vivo seeds (30 ng/μL) were added to recombinant wild-type (rec, WT) TTR or MTTR at pH 4.3 or 7.4, as labeled. All replicates are shown; n = 4. a.u., arbitrary units. (B) Absorbance at 280 nm of insoluble fractions collected from A. The dashed line marks the initial protein concentration (0.5 mg/mL). Error bars indicate SD. n = 3. AU, absorbance units. (Bottom Insets) Anti–His-tag immuno-dot blot of insoluble fractions. (C) Electron micrographs of MTTR aggregates obtained at pH 4.3 and pH 7.4. (Scale bars, 200 nm.)
Seeded TTR Fibrils Exhibit Molecular Properties in Common with Amyloid Fibrils.
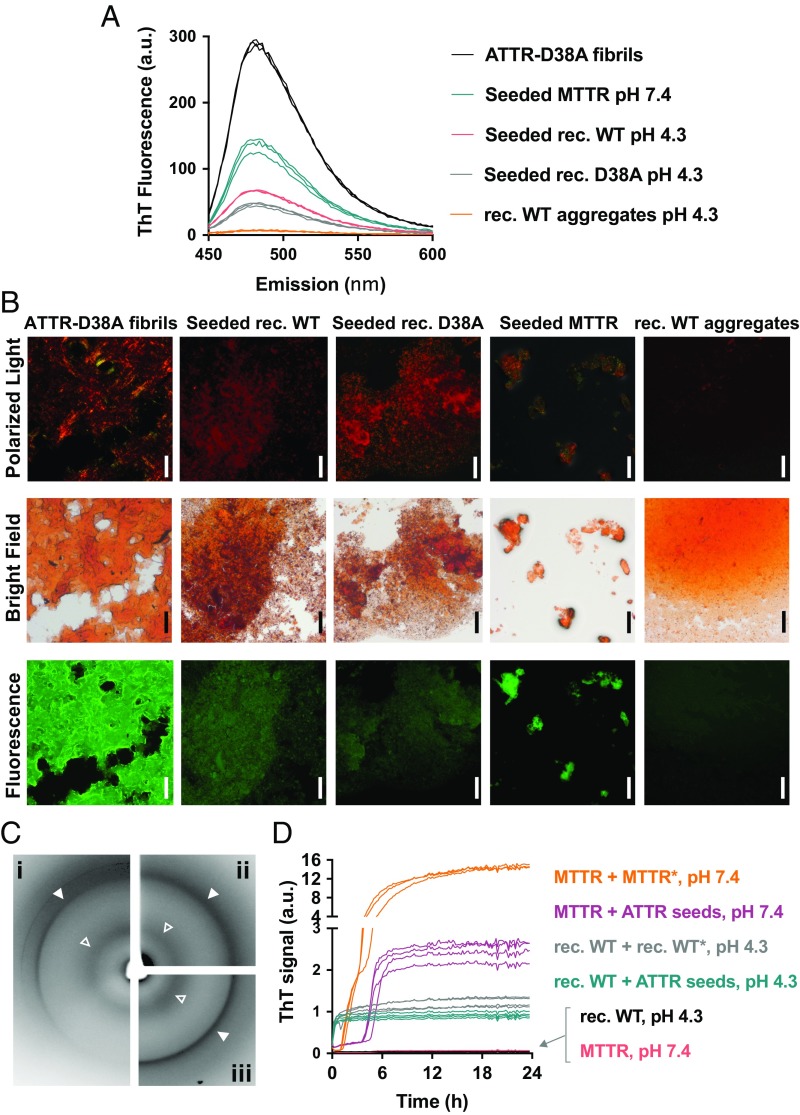
The analysis of the ThT fluorescence spectrum of individual fibril samples shows that the affinity of ThT is greatest for ATTR fibrils, followed in order by MTTR seeded fibrils, wild-type TTR seeded fibrils, and D38A seeded fibrils (Fig. 3A). In contrast, protein aggregates obtained from unseeded wild-type TTR after 4 d of incubation at pH 4.3 did not present any significant affinity for ThT (Fig. 3A). We also found that ATTR fibrils and seeded fibrils exhibit birefringence when stained with Congo red, but wild-type TTR aggregates do not (Fig. 3B). Moreover, seeded TTR fibrils and ATTR fibrils both display a cross-β diffraction pattern when analyzed by X-rays (Fig. 3C). The diffraction patterns of both fibrils exhibit the two characteristic strong reflections of amyloid: the reflection at 4.7 Å that results from the stacking of β-strands along the fibril and the reflection at near 11 Å that results from the mating of adjacent β-sheets (Fig. 3C and SI Appendix, Fig. S4A and Table S2). In addition, we found that, similar to ex vivo fibrils, seeded fibrils obtained by the addition of ATTR-D38A seeds to wild-type TTR or MTTR also accelerate fibril formation of both wild-type TTR and MTTR (Fig. 3D), suggesting that the conformation of the ex vivo fibrils is encoded in the products of our fibril formation assay.
Fig. 3.
Characterization of fibrils obtained from wild-type TTR and MTTR by amyloid seeding. (A) ThT spectra of 0.25 mg/mL TTR fibrils and aggregates. Samples were obtained from recombinant wild-type, D38A, or MTTR variants when incubated with ATTR-D38A ex vivo seeds for 24 h. MTTR seeded fibrils were obtained at pH 7.4; D38A and wild-type TTR-seeded fibrils were obtained at pH 4.3. Protein aggregates were obtained from 1 mg/mL recombinant wild-type TTR incubated for 4 d at pH 4.3 in the absence of seeds. a.u., arbitrary units. (B) Congo red staining of TTR fibrils and aggregates analyzed in A and visualized by optical microscopy under fluorescence [540-nm filter, colored in green for better contrast (Bottom), bright-field (Middle), and polarized light (Top)]. (Scale bars, 100 μm.) (C) X-ray diffraction of ex vivo seeds (i) and fibrils obtained from wild-type TTR (ii) and MTTR (iii) when incubated with ATTR-D38A for 24 h at pH 4.3 and 7.4, respectively. Open and closed arrowheads point to 4.7-Å and ∼11-Å reflections, respectively. (D) Amyloid seeding assay of 0.5 mg/mL TTR variants by the addition of several fibril samples. MTTR*, fibrils obtained after incubation of 0.5 mg/mL MTTR with ATTR-D38A seeds for 24 h at pH 7.4; rec. WT*, fibrils obtained after incubation of 0.5 mg/mL recombinant wild-type TTR with ATTR-D38A seeds for 24 h at pH 4.3. Both MTTR and recombinant wild-type fibrils were sonicated before this assay and were used as seeds, as labeled. All replicates are shown; n = 3. a.u., arbitrary units.
ATTR ex Vivo Fibrils Carrying Various Genotypes also Seed Wild-Type TTR Fibril Formation.
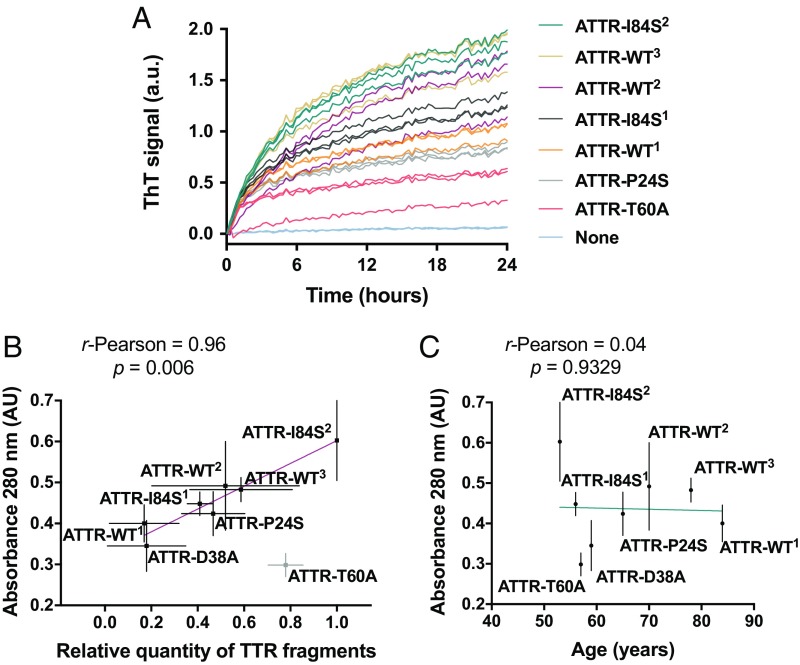
We extracted ex vivo fibrils from the cardiac tissues of seven additional ATTR patients and found that they also were capable of seeding fibril formation of wild-type TTR under partially denaturing conditions (Fig. 4). Among these patients, there were three cases of wild-type amyloidosis and four cases of hereditary amyloidosis other than ATTR-D38A (SI Appendix, Table S1). All patients presented advanced amyloidosis symptomatology or were deceased as a consequence of ATTR. The tissue specimens came from either explanted or autopsied hearts. We confirmed the presence of amyloid-like fibrils by TEM after the extraction (SI Appendix, Fig. S5A). We also confirmed TTR content in the resultant fibril resuspension by Western blot and mass spectrometry sequencing (SI Appendix, Fig. S5 B and C and Dataset S1). The addition of these ex vivo seeds to wild-type TTR under acidic conditions resulted in the formation of ThT-positive species, as observed for ATTR-D38A ex vivo seeds (Fig. 4A).
Fig. 4.
Amyloid seeding by ATTR material extracted from the hearts of eight ATTR patients. (A) Amyloid seeding assay at pH 4.3 of wild-type recombinant TTR in the presence of ATTR seeds extracted from the hearts of ATTR patients with various genetic backgrounds, monitored by ThT fluorescence. Superscripts denote particular ATTR patients. All replicates are shown; n = 3. a.u., arbitrary units. (B) Comparison of amyloid seeding capacity (vertical axis) and relative quantity of truncated TTR of ex vivo seeds. Truncated TTR content was quantified by ImageJ from two independent Western blots. (C) Comparison of amyloid seeding capacity (vertical axis) and ATTR patient age. In both B and C, amyloid seeding capacity was measured as follows: 30 ng/μL seeds were incubated with 0.5 mg/mL recombinant wild-type TTR at pH 4.3, and 280-nm absorbance of the insoluble fraction collected after seeding for 24 h was measured. Linear regression and Pearson r were obtained by OriginLab.
We next analyzed whether amyloid seeding capacity correlates with any cardiac pathology subtype, gender, or age, three factors previously associated with ATTR severity (31–33). The presence of fragmented TTR, present in type A cardiac amyloidosis, was evaluated by Western blot using an antibody that specifically recognizes TTR C-terminal fragments (SI Appendix, Fig. S5B). With the exception of ATTR-T60A, we observed an apparent correlation between seeding capacity and fragmented TTR content, with a Pearson correlation coefficient of 0.96 (Fig. 4B). We did not observe correlation with ATTR patient age or gender in our assays (Fig. 4C); detecting such a correlation, if one exists, may require a larger cohort.
Amyloid Seeding Can Be Halted by Peptide Inhibitors of TTR Aggregation.
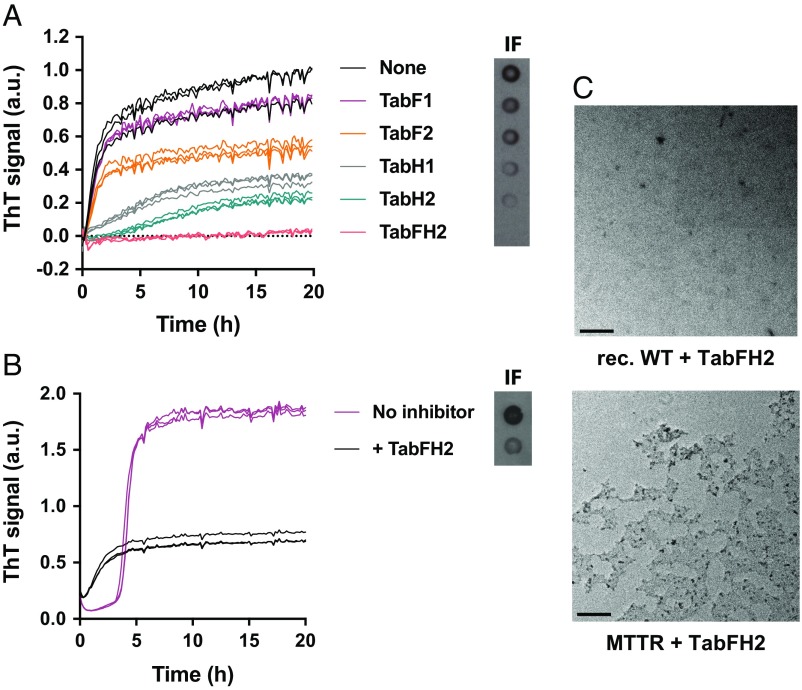
In our previous study, we identified β-strands F and H of native TTR as driving TTR amyloid aggregation in vitro (25). From the atomic structures of these strands in their amyloid forms, we designed two 16-residue peptides intended to cap fibrils of the F and H strands. Together these two peptides were able to reduce fibril formation of wild-type recombinant TTR. In the present work, we optimized these peptides by reducing their amyloidogenicity without compromising their inhibiting activity, as detailed in Materials and Methods and SI Appendix, Fig. S6. All evaluated sequences including both original and optimized peptides are listed in SI Appendix, Table S3. We then evaluated the effectiveness of the best-scored inhibitors in our seeding assays (SI Appendix, Fig. S6B and Table S3) and found that two optimized peptides arrested TTR fibril formation more efficiently than our earlier inhibitors (Fig. 5A and SI Appendix, Fig. S6B). Both the original and optimized peptide inhibitors are named as “Tab” (transthyretin aggregation blockers) followed by the letters F and/or H referring to the target strand(s). The name ends with a number that depends on whether the peptide was obtained in the original design (number 1) (25) or was optimized in the present work (number 2). For instance, TabF1 corresponds to the peptide inhibitor that targets strand F as described by Saelices et al. (25), and TabFH2 refers to a mixture of the two optimized peptide inhibitors that target strands F (TabF2, RRRRHVAHPFV-N-me-Glu-FTE) and H (TabH2, RRRRSYVTNPTSY-N-me-Ala-VT) designed and characterized in this study (SI Appendix, Table S3). We evaluated these inhibitors in our amyloid seeding assay containing 0.5 mg/mL recombinant wild-type TTR (∼35 μM) and 30 ng/μL ATTR-D38A seeds in the presence or the absence of 180 μM peptide inhibitors. We found that targeting strand H is more effective than targeting strand F (Fig. 5A). Nonetheless, the combination of peptide blockers for both strands F and H, TabFH2, resulted in full inhibition of seeding from ex vivo ATTR-D38A fibrils (Fig. 5A). The inhibiting activity of TabFH2 was found to be dose dependent (SI Appendix, Fig. S7). In contrast, the addition of 180 μM diflunisal or tafamidis did not inhibit amyloid seeding under the same conditions (SI Appendix, Fig. S8A). Preincubation of TTR with the stabilizing compounds for 22 h did not affect this outcome (SI Appendix, Fig. S8B). Others have shown that the presence of these compounds at similar concentrations fully inhibits aggregation of recombinant TTR at acidic pH in the absence of seeds (34, 35). Finally, TabFH2 was found to be efficient at halting amyloid seeding of MTTR under physiological conditions (Fig. 5B). Inhibition of amyloid seeding was confirmed by TEM (Fig. 5C).
Fig. 5.
Inhibition of amyloid seeding by designed peptide inhibitors. (A) ThT assay to evaluate inhibition of seeding of ex vivo ATTR-D38A at pH 4.3 by 180 μM designed inhibitors of TTR aggregation when added to 0.5 mg/mL recombinant wild-type TTR. All replicates are shown; n = 3. (Right Inset) Anti-TTR dot blot of insoluble fractions. (B) Inhibition of amyloid seeding of 0.5 mg/mL MTTR by 180 μM TabFH2 at physiological pH, monitored by ThT fluorescence. All replicates are shown; n = 3. (C) Electron micrographs of samples containing TabFH2 obtained in A and B confirming inhibition of amyloid seeding.
Discussion
Seeding in Amyloid Disease.
The importance of seeding for conversion of soluble proteins to pathogenic amyloid fibrils was first recognized by Ranlov and coworkers (36–39) in the late 1960s, followed by Robert Kisilevsky and coworkers (4, 40–42) beginning in the late 1970s. They described the so-called “amyloid enhancing factor” in a mouse model of AA amyloidosis, which they defined as a transferable activity that reduces the induction time of splenic deposition of the amyloid protein. Later, the pioneering study of Jarrett and Lansbury recognized the role of seeding in the progression and transmission of other amyloid diseases (43) by demonstrating that introduction of a seed—a preformed amyloid fibril—into a solution of the same dissolved but supersaturated protein nucleates the formation of identical amyloid fibrils (44–46). In vitro experiments have shown that tissues extracted from the autopsied brains of Alzheimer’s patients accelerate the aggregation of dissolved β-amyloid (47, 48). In a dramatic demonstration of the power of amyloid seeding, serial in vitro seeding has been developed into a method of amplifying infectious prion fibrils (49, 50).
In vivo amyloid seeding has been proposed to be the mechanism of spreading from cell to cell of amyloid fibrils associated with neurodegenerative diseases (51–54). For example, the injection of fibrils of amyloid-β, tau, or α-synuclein into the brains of animals has been shown to induce the formation of inclusions at the injection sites, from which they spread to distant regions of the brain, presumably by a mechanism of seeding (2, 55, 56). Amyloid seeding may also account for the iatrogenic transmission of Creutzfeldt–Jakob disease (CJD) via cornea transplant, contaminated neurosurgical instruments, blood products from CJD patients, or contaminated growth hormone (57).
In short, amyloid seeding appears to be a potent mechanism of amplification and spread of disease-related fibrils. However, despite clinical evidence of amyloid seeding of TTR in vivo, demonstration of TTR amyloid seeding has remained elusive in the laboratory. Here we have investigated the potential of ex vivo fibrils extracted from ATTR patients to seed recombinant TTR in vitro and its inhibition by structure-based peptide inhibitors.
Potent Power of Seeds Extracted from ATTR Patients.
The first of our two principal findings is that insoluble material containing amyloid fibrils of TTR, extracted from cardiac tissues of ATTR patients (SI Appendix, Figs. S1 and S5), seeds both wild-type and familial variants of TTR into amyloid fibrils (Fig. 1). Table 1 summarizes the relevant experiments and controls. At pH 4.3, where acid destabilizes the native tetrameric structure of TTR, forming monomers and thereby exposing the amyloid-driving segments of β-strands F and H, the ATTR-extracted seeds cause immediate aggregation into TTR amyloid fibrils. Under the same conditions in the absence of these seeds, we observe the gradual formation of amorphous aggregates of TTR, as others have observed (30, 58). The lineal increase in aggregation of TTR, lacking the commonly observed lag phase in amyloid aggregation, has been explained by others as a “downhill polymerization” in which monomers gradually add to growing aggregates (30). Similar lineal formation of aggregates has been observed for mutant MTTR either seeded by MTTR recombinant aggregates or in the absence of seeds (30). In contrast, our ATTR ex vivo seeds produce an immediate rapid formation of MTTR amyloid at acidic pH. Apparently, the patient-extracted ATTR seeds possess a conformation differing from recombinant TTR fibrils that endows ATTR fibrils with potent seeding potential, at least at low pH.
Table 1.
Summary of relevant experiments and controls
| pH | Seeded material | Seeds | Result | Data |
| 4.3 | Wild-type native TTR | Extracted ATTR | Immediate TTR amyloid fibrils; no lag | Fig. 1 A and B |
| No seeds | Gradual TTR amorphous aggregation | Figs. 1B, 3 A and B, and 5 A and B | ||
| MTTR | Extracted ATTR | Immediate TTR amyloid fibrils; no lag | Fig. 2A | |
| No seeds | Gradual TTR amorphous aggregation | Hurshman et al. (30) | ||
| MTTR fibrils | Gradual TTR amorphous aggregation | Hurshman et al. (30) | ||
| 7.4 | Wild-type native TTR | Extracted ATTR | No aggregates | Fig. 2 |
| No seeds | No aggregates | Fig. 2 | ||
| MTTR | Extracted ATTR | Lag, then immediate formation of amyloid fibrils upon seeding | Figs. 2 and 3 | |
| No seeds | Slow formation of amorphous aggregates | Hurshman et al. (30) |
It is important to know if this potent seeding potential of patient-extracted seeds operates under physiological conditions that more accurately represent in vivo environments. Consequently, we repeated seeding experiments at pH 7.4 (Fig. 2 and Table 1). At this higher pH, the seeds extracted from ATTR patients again nucleate MTTR into amyloid fibrils. The kinetic profile in this experiment (Fig. 2A) shows a typical amyloid lag phase shortened by seeding. Others have shown that ThT binds to MTTR aggregates at acidic pH with an affinity that is only 0.6% higher than physiological pH—or baseline (29). We find that ThT binds to MTTR when seeded both at acidic pH, with 15% binding compared with baseline, and at physiological pH, with 91% binding compared with baseline. Taken together, our experiments at low and physiological pH suggest that, once ATTR forms amyloid fibrils, these have the capacity to convert wild-type or variant TTR into additional amyloid fibrils upon TTR dissociation.
Differences between ATTR fibrils and in vitro-produced TTR aggregates may explain the lack of seeding of the latter (30, 59). Visual inspection of species by electron microscopy shows clear morphological differences (SI Appendix, Fig. S1D vs. Fig. 1B). Whereas ATTR ex vivo fibrils appear as fibrillar structures, TTR aggregates obtained in the absence of seeds appear as amorphous aggregates. In addition, we found that ATTR ex vivo fibrils exhibit higher Congo red and thioflavin T reactivity than TTR aggregates (Fig. 3 A and B), suggesting a different supramolecular organization. Of special note is that the daughter fibrils formed by ATTR seeding can themselves seed additional wild-type or variant TTR into amyloid fibrils (Fig. 3D). That is, the fibril conformation that endows the fibrils extracted from ATTR tissue with potent seeding power is transmitted by a process of serial templating to a new generation of fibrils, conferring affinity for Congo red and ThT as well as seeding capacity (Fig. 3D). We reason that ATTR fibrils form in an environment that differs from in vitro conditions and that they mature after months or years of environment exposure. The reason why other laboratories did not observe acceleration of TTR deposition in vivo after the injection of ex vivo ATTR fibrils into transgenic mouse models remains unclear (59, 60), but we note that these studies included only a limited number of animals and conditions.
Implications of Seeding for ATTR.
The powerful seeding effect of disease-related TTR fibrils is consistent with key observations about ATTR pathogenesis. Of particular importance, our findings provide a potential explanation for why the treatment of liver transplantation for ATTR is not always effective and sometimes results in cardiac deposition of wild-type TTR (5, 17, 22, 23, 61, 62). Because TTR is largely biosynthesized in the liver, transplantation with a liver that secretes wild-type TTR should replace most circulating mutant TTR with wild-type TTR. However, if mutant amyloid seeds remain in other tissues, our experiments suggest that the seeds are able to convert the wild-type TTR provided by the new liver to amyloid fibrils, perpetuating disease. There are also cases of deposition of TTR amyloid bearing the donor mutation in recipients of so-called “domino” liver transplantations (63). In this procedure, explanted livers from patients with familial ATTR are implanted in patients with fatal hepatic disorders. Some previous reports propose age as a significant contributor to secondary cardiac deposition in domino liver transplants, resulting from a compromised chaperoning function of the aged implanted liver (64, 65). Although explanted livers do not contain congophilic fibrillar structures, the presence of undetectable seeds in the explanted livers has also been proposed to be a cause of disease transmission in domino liver transplant recipients (66–68).
Our finding that ATTR amyloid material seeds fibril formation of wild-type TTR (Fig. 4) illuminates other puzzling observations about ATTR pathogenesis. Although found to be more stable than mutant variants in vitro, wild-type TTR deposits in both hereditary and wild-type ATTR (12). Previous studies have found a correlation of more rapid ATTR pathogenesis with less stable mutant TTR variants (11). However, if the stability of TTR is the principal factor in the formation of amyloid fibrils, it is puzzling that TTR amyloid deposits in hereditary ATTR patients contain 30–70% of the more stable wild-type protein (17, 61, 69, 70). However, the presence of wild-type TTR is expected as a result of seeding of the wild type by disease-related seeds, as shown in Figs. 1A and 4A.
In ATTR patients, TTR seeding may be affected by a variety of factors such as age, gender, tissue type, pathology, or genotype. Westermark and coworkers (19) discovered two distinct pathological patterns of amyloid deposition in ATTR patients (9). In type A amyloidosis, TTR depositions contain, in addition to full-length TTR, a 79-residue truncated C-terminal fragment of TTR; in type B patients, TTR depositions contain only full-length TTR. The type A phenotype has been found to carry a higher risk of progressive cardiac deposition after liver transplantation (16). In our analyses of ATTR cardiac seeds, six of eight samples contained traceable amounts of fragmented TTR (SI Appendix, Fig. S5B). We observed that, with the exception of one ATTR case, amyloid seeding capacity correlated with the presence of C-terminal fragments in the extract (Fig. 4B). In our assays, neither patient gender nor age correlated with seeding capacity (Fig. 4C). However, our limited number of specimens does not permit definite conclusions. We are also aware that this study does not include the evaluation of amyloid seeding in tissue samples other than the heart. It is possible that the tissue type may influence protein fragmentation and/or amyloid seeding. In any case, we suggest that amyloid seeding may become pathologically relevant with advancing disease after amyloid nuclei are formed from dissociated TTR.
Inhibition of TTR Fibril Formation.
Our second principal finding is that the capping of TTR amyloid fibrils by designed peptides inhibits TTR amyloid seeding (Fig. 5). This may be of special importance for cardiac ATTR patients, who are often diagnosed when manifesting advanced TTR deposition and have limited treatment options. The general approach of fibril capping for amyloid inhibition has previously been shown to be effective for blocking fibrils of other proteins: constructs of tau in vitro and p53 in vitro and in mice (71, 72). In our previous work on inhibition of TTR, we used the steric zipper structures of β-strands F and H to design the TabFH1 peptide blockers (25). However, a limitation of the TabFH1 blockers is that they themselves form fibrils at high concentration. The TabFH2 inhibitors designed in the present study do not share this limitation and show improved efficacy.
In summary, here we document the capacity of disease-related seeds of TTR amyloid to convert native tetrameric wild-type TTR to amyloid in vitro. This finding helps to explain why disease symptoms persist in some patients even after the main source of disease-prone TTR has been removed. Because seeding tetrameric TTR into fibrils in vitro requires an acidic environment not necessarily present in vivo, further scrutiny of the relevance of these results to human pathology is warranted. Our results also suggest that the inhibition of the elongation of TTR fibrils by designed inhibitors may be a plausible treatment for ATTR cases. We envision combinatory treatments for ATTR, targeting protein expression, protein stability, deposit clearance, and amyloid seeding (35, 73–78).
Materials and Methods
Antibodies.
Antibodies used were rabbit anti-human TTR polyclonal antibody (Western blots and dot blots, 1:10,000; immunohistochemistry, 1:2,000; ELISA, 1:4,000; DAKO, Agilent Technologies), anti–His-tag HRP-conjugated HisProbe (dot blots, 1:10,000; Thermo Fisher Scientific), and HRP-conjugated goat anti-rabbit IgG antibody (Western blots and dot blots, 1:5,000; immunohistochemistry, 1:2,000; DAKO, Agilent Technologies). Anti-truncated TTR was obtained as a gift from Gunilla Westermark (Uppsala University, Uppsala, Sweden) (79) (labeled as “1898”; 1:5,000). Ig fraction from nonimmunized rabbits was used as negative control (immunohistochemistry, 1:2,000; DAKO, Agilent Technologies).
Patients and Tissue Material.
Tissues from eight ATTR patients carrying wild-type TTR (n = 3) or TTR mutations (n = 5) were included in the study (see SI Appendix, Table S1 for patient and tissue material characteristics). Specimens from either explanted hearts or autopsy were obtained from several laboratories. The University of California, Los Angeles Office of the Human Research Protection Program granted exemption from Internal Review Board review because all specimens were anonymized.
Congo Red Staining of Cardiac Tissue.
Congo red staining was performed to verify the presence of amyloid deposition in tissues. Cardiac sections10 μm thick were deparaffinized by heating, xylene baths, and gradual hydration. The samples were stained using the Amyloid Stain, Congo Red Kit (Sigma Aldrich) as described by the manufacturer. Briefly, slides were incubated in a freshly made alkaline sodium chloride solution for 30 min and then were transferred to and incubated in an alkaline Congo red solution for another 30 min. Slides were dehydrated, mounted with Permount (Thermo Fisher Scientific) and then were analyzed under bright-field and polarized light. Amyloid was identified by its characteristic apple-green birefringence.
Immunohistochemistry of Cardiac Tissue.
Immunostaining of 4-μm tissue sections was performed using polyclonal rabbit anti-human TTR antibodies (DAKO, Agilent Technologies) or Ig fraction from nonimmunized rabbits as negative control (DAKO, Agilent Technologies) diluted 1:2,000 in blocking buffer. HRP-conjugated goat anti-rabbit IgG antibody (DAKO, Agilent Technologies) was used diluted 1:2,000 in blocking buffer. Reactivity visualization was made with the EnVision+ System/HRP, Rb (DAB+) (DAKO, Agilent Technologies) as described by the manufacturer.
Extraction of Amyloid Material from Human Tissue.
Amyloid fibrils were extracted from fresh-frozen human tissue as described earlier (27). Briefly, 1–5 g of amyloid-positive cardiac tissue was thawed and minced with a modular motorized homogenizer in 10 mL of 0.15 M NaCl. The sample was centrifuged at 15,000 rpm for 30 min (rotor F18S-12x50y, Thermo Fisher Scientific), and the resulting pellet was homogenized in 0.15 M NaCl and centrifuged again. This process was repeated seven times. The pellet was further homogenized in distilled water and centrifuged three times; the final pellet was lyophilized. Amyloid fibril load was confirmed by TEM and X-ray fiber diffraction. TTR content was analyzed by Western blot and liquid sample-electrospray ionization-MS/MS. Non-ATTR cardiac control tissue was subjected to the same extraction procedure.
TEM.
TTR aggregation was confirmed by TEM as described by Saelices et al. (25). Briefly, a 2- to 5-μL sample was spotted onto freshly glow-discharged carbon-coated grids (Ted Pella), incubated for 4 min, and then rinsed three times with distilled water. Grids were finally negatively stained with 2% uranyl acetate for 2 min. An FEI T12 Quick CryoEM electron microscope at an accelerating voltage of 120 kV was used to examine the specimens. Images were recorded digitally by a Gatan 2K×2K CCD camera.
Size-Exclusion Chromatography.
A 20-μL sample containing 5 μg of sonicated ATTR-D38A ex vivo seeds was run over a Superose 6 Increase 3.2/300 column (GE Healthcare), which separates molecules with masses between ∼5,000 and ∼5,000,000 Da, using PBS (pH 7.4) as elution buffer. Gel filtration standard mixture was purchased from Bio-Rad.
X-Ray Fibril Diffraction.
X-ray fibril diffraction patterns were collected as described previously (80). Briefly, seeded fibrils and ex vivo extracted fibrils were concentrated by centrifugation, washed, and oriented while drying between two glass capillaries. The glass capillaries were mounted on a brass pin for diffraction at room temperature using X-rays produced by a Rigaku FR-E+ rotating anode generator equipped with a Rigaku R-AXIS HTC imaging plate detector. Diffraction patterns were collected at a distance of 180 mm and analyzed using the Adxv software package (81). X-ray reflection peaks were obtained by radial intensity averaging.
Sequencing of Proteins Contained in Patient-Derived Samples.
Ex vivo seeds were analyzed by mass spectrometry, as described elsewhere (82). Before a trypsin digest, a chloroform/methanol precipitation was performed to precipitate the protein in the original ex vivo seeds. Four volumes of methanol, one volume of chloroform, and three volumes of water were sequentially added to one volume of each protein sample with vortexing after each addition. The samples were centrifuged at 16,000 × g for 5 min, and the aqueous supernatant was removed. The protein remained at the interface between the upper and lower phases. Four volumes of methanol were again added, and the samples were vortexed. The samples were centrifuged at 16,000 × g for 5 min, and the supernatant was removed without disturbing the pellet. The protein pellets in each sample were solubilized in an in-solution digest buffer consisting of 0.5% sodium deoxycholate, 12 mM sodium lauroyl sarcosinate, and 50 mM triethylammonium bicarbonate (TEAB). Samples were heated for 5 min at 95 °C, and the protein concentrations were measured using the Pierce BCA Protein Assay Kit (Thermo Fisher Scientific). Protein disulfides were then reduced with 5 mM Tris(2-carboxyethyl)phosphine (TCEP) in 50 mM TEAB for 30 min at room temperature and alkylated with 10 mM iodoacetamide in 50 mM TEAB for 30 min at room temperature in the dark. Protein solutions were diluted fivefold with 50 mM TEAB. Lyophilized porcine trypsin (Promega) was solubilized in 50 mM TEAB and was used in a mass ratio of 1:100 (trypsin:protein) for a double enzymatic digestion of 4 h and then 16 h at 37 °C. Sodium deoxycholate was precipitated from the peptide solutions with trifluoroacetic acid at a final concentration of 0.5%. The samples were then centrifuged at 16,000 × g for 5 min, and the supernatant was transferred into new tubes before lyophilization in a centrifugal evaporator. The samples were reconstituted in 100 μL of a buffer solution (2% acetonitrile, 0.5% acetic acid) and desalted with C18 stage tips as previously described elsewhere (83). Eluted samples were lyophilized and solubilized with 10 μL of 2% acetonitrile, 0.1% formic acid and then were placed into injection vials for MS analysis.
The samples were analyzed with an Eksigent 2D nanoLC system attached to a Q-Exactive Plus (Thermo Fisher Scientific). Peptides were injected into a laser-pulled nanobore 20 cm × 1.8 μm C18 column (AcuTech Scientific) in buffer A (2% acetonitrile, 0.15% formic acid) and were eluted using a 3-h linear gradient from 3 to 80% buffer B (98% acetonitrile, 0.15% formic acid). The Q-Exactive Plus was operated in Full MS/dd-MS2 mode with a resolution of 70,000 and an auto gain control target of 3e6 for the parent scan. The top 20 ions above +1 charge were subjected to higher collision dissociation (HCD) set to a value of 30 with a resolution of 17,500 and an auto gain control target of 1e5 as well as a dynamic exclusion of 15 s. Tandem mass spectrometry data were matched to a protein fasta file (UniProt-Human 20161005; 21,047 sequences; 11,462,872 residues) using Mascot software (Matrix Sciences). Identified proteins ordered by exponentially modified protein abundance index (emPAI) are listed in Dataset S1. emPAI offers approximate, label-free, relative quantitation of the proteins in the mixture based on protein coverage by the peptide matches in the database search result. All ex vivo extracts were enriched in TTR but also contained a mixture of other proteins, with the consistent presence of serum amyloid-P component, myosin, collagen, and a TRAJ56 fragment.
Western Blot of TTR from ex Vivo Material.
Equal amounts of total protein were loaded and separated by electrophoresis under denaturing conditions and immunoblotted with polyclonal anti-human TTR (1:10,000; DAKO, Agilent Technologies) or anti-truncated TTR obtained from Gunilla Westermark (1:5,000). HRP-conjugated goat anti-rabbit secondary antibody was diluted to 1:5,000, and SuperSignal West Pico Chemiluminescent Substrate (Thermo Fisher Scientific) was used to detect TTR as described by the manufacturer.
Recombinant TTR Preparation.
Recombinant protein preparations were obtained as described previously (25). Briefly, Escherichia coli-expressed proteins were purified by affinity in a HisTrap column (GE Healthcare Life Science). Peak fractions were combined and further purified by size exclusion on a Superdex S75 prep grade column (GE Healthcare Life Science). Final samples were stored in 10 mM sodium phosphate (pH 7.5), 100 mM KCl, and 1 mM EDTA at −20 °C.
Nonseeded TTR Aggregation Assay.
TTR aggregation assays are described elsewhere (25). Briefly, 1 mg/mL TTR sample in 10 mM sodium acetate (pH 4.3), 100 mM KCl, and 1 mM EDTA was incubated at 37 °C for a maximum of 4 d. Protein aggregation was followed by absorbance at 400 nm, anti-TTR immuno-dot blot of the insoluble fraction, and/or TEM.
Amyloid Seeding Assays.
Fibril extracts were used to seed the formation of new aggregates from recombinant TTR. To further purify the extracts, we treated them with 1% sodium dodecyl sulfate, and the soluble fraction was discarded after centrifugation at 13,000 rpm for 5 min (rotor FA-24x2, Eppendorf). This process was repeated two times, and soluble fractions were discarded. The sample was washed with 10 mM sodium acetate (pH 7.5), 100 mM KCl, and 1 mM EDTA three times by centrifugation and then was sonicated in cycles of 5 s on/5 s off for a total of 10 min at the minimum intensity (18%). The protein content was measured using the Pierce BCA Protein Assay Kit (Thermo Fisher Scientific). Seeds were added to 0.5 mg/mL recombinant TTR in a final volume of 200 μL of 5 μM ThT, 10 mM sodium acetate (pH 4.3 or pH 7.4), 100 mM KCl, and 1 mM EDTA. Unless labeled otherwise, cardiac seeds were analyzed at a concentration of 30 ng/μL. ThT fluorescence emission was measured at 482 nm with absorption at 440 nm in a Varioskan Flash reader (Thermo Fisher Scientific) or a FLUOstar Omega (BMG LabTech) microplate reader. Plates were incubated at 37 °C for 20–72 h as labeled with cycles of 9 min shaking (700 rpm double orbital) and 1 min rest throughout the incubation. Measurements were taken every 10 min (bottom read) with automatic gain if read in a Varioskan Flash reader or with a manual gain of 1,000 if read in a FLUOstar Omega microplate reader. We normalized all our assays to a reference positive control included in every run. Figures show ThT signal in arbitrary units (a.u.). Fibril formation was confirmed by TEM. The insoluble fraction was extracted by two cycles of centrifugation and final resuspension with 6 M guanidinium hydrochloride. Soluble fractions were saved from the first centrifugation cycle, and protein concentration was measured by the Pierce BCA protein assay kit (Thermo Fisher Scientific). Absorbance at 280 nm of insoluble fractions was measured in absorbance units (AU) in a BioPhotometer UV/Vis spectrophotometer (Eppendorf) or in a Nanodrop 1000 spectrophotometer (Thermo Fisher Scientific). Insoluble and soluble fractions were further analyzed by immunoblots.
Ex Vivo Extract Immunodepletion of TTR Fibrils.
TTR fibrils were depleted from ex vivo extract by immunoreaction to anti-TTR microplate strips from the Prealbumin ELISA kit (Abcam) as follows. A 100-μL sample containing 1.2 μg/μL ATTR-D38A was added to an anti-TTR–precoated well and incubated at room temperature for 30 min. The sample was transferred to a new well and incubated again at room temperature for 30 min. This procedure was repeated eight times. After every incubation, 5-μL aliquots were transferred to test tubes and saved. A 5-μL aliquot was also collected from the original sample, before incubation. The amyloid seeding assay was performed as described above. Collected samples were added to 0.5 mg/mL recombinant wild-type TTR in a final volume of 200 μL, and ThT fluorescence was read every 10 min for 24 h in a FLUOstar Omega microplate with a manual gain of 1,000. Optic micrographs of resulting aggregates were taken after incubation using a Celigo S Imaging system under bright-field and UV channels.
Immuno-Dot Blot Analysis.
TTR aggregation was visualized by dot-blot analysis as described previously (25). The insoluble fraction of the samples was obtained by cycles of centrifugation as described above and dotted onto nitrocellulose membranes (0.2 μm; Bio-Rad). TTR was observed by using HisProbe (Thermo Scientific Fisher) or polyclonal rabbit anti-human TTR (DAKO, Agilent Technologies) antibodies at a concentration of 1:10,000.
Design of Aggregation Blockers.
The first generation of TTR aggregation blockers was designed to bind strands F and H as described previously (25). They shared a 12-residue-long wild-type sequence with the addition of an N-methyl group and a four-residue arginine tag. The use of the nonnatural N-methyl groups to protect aggregation blockers from proteolysis has been reported elsewhere (81). The addition of a poly-arginine tag confers higher solubility that is presumed to hinder self-aggregation. These peptides were further optimized in silico to decrease potential self-association. Two in silico optimization strategies were performed. (i) Each residue of each peptide was subsequently substituted by arginine, and (ii) the overall composition of the peptides was maintained, but the order of residues was changed. The overall propensity of every sequence to form steric zippers was calculated by the algorithm ZipperDB, developed previously (84). ZipperDB calculates the propensity of each six-residue segment of any given sequence to form steric zippers. The predicted full-atom energies of interactions across the zipper interface formed by possible self-association for each peptide inhibitor are shown in SI Appendix, Table S3. We found that arginine substitutions did not result in a significant energy difference (SI Appendix, Table S3A). Sequence swapping resulted in peptide sequences with the highest energy scores, which were less prone to aggregate. The C-terminal residue of F15 was eliminated to obtain a higher energy score, resulting in TabF2. The optimized inhibitors were then selected and tested in vitro.
The best-scoring peptides were evaluated in vitro for their efficacy at inhibiting amyloid seeding (SI Appendix, Fig. S6 and Table S3B). Amyloid seeding assays were performed as described above. Peptide (180 μM) was added to 0.5 mg/mL recombinant wild-type TTR and 30 ng/μL ATTR-D38A seeds. Amyloid seeding was measured after 24 h of incubation at 37 °C by ThT fluorescence. The ThT signal obtained in the absence of peptide was considered 100%. The best two inhibitors, TabF2 (RRRRHVAHPFV-N-me-Glu-FTE) and TabH2 (RRRRSYVTNPTSY-N-me-Ala-VT), were selected for further analysis and used as a mixture, referred to herein as “TabFH2.”
Statistical Analysis.
Statistical analysis of absorbance, TTR aggregation, and ThT signal was performed with Prism 7 for Mac (GraphPad Software) using an unpaired t test. All samples were included in the analysis. All quantitative experiments are presented as independent replicates or as means ± SD of at least three independent experiments.
Data and Materials Availability.
The authors declare that all data generated or analyzed during this study that support the findings are available within this published article and its SI Appendix files.
Supplementary Material
Acknowledgments
We thank Dr. Michael Sawaya for X-ray fibril diffraction studies, Dr. Jeffery Kelly for a gift of tafamidis and discussions, Dr. Gunilla Westermark for the gift of anti-fragmented TTR antibody 1898 and discussions, Drs. Joel Buxbaum and Duilio Cascio for helpful discussions, Dr. James LeBlanc and Dr. Daniel Anderson for technical support, Dr. Sarah Dry for assistance in collecting and storing specimens, Steve Yu for his help on electron microscopy, and the patients who generously donated tissues. This work was supported by the Amyloidosis Foundation, the NIH, the Department of Energy, and The Howard Hughes Medical Institute.
Footnotes
Conflict of interest statement: The authors and the University of California, Los Angeles have filed an international patent application (no. PCT/US17/40103) for the transthyretin inhibitors. D.S.E. is an advisor to and equity holder in ADRx, Inc. L.S. is a consultant for ADRx, Inc. D.S.E. and Gunilla Westermark are coauthors on a 2017 paper.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1805131115/-/DCSupplemental.
References
- 1.Holmes BB, et al. Proteopathic tau seeding predicts tauopathy in vivo. Proc Natl Acad Sci USA. 2014;111:E4376–E4385. doi: 10.1073/pnas.1411649111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Meyer-Luehmann M, et al. Exogenous induction of cerebral beta-amyloidogenesis is governed by agent and host. Science. 2006;313:1781–1784. doi: 10.1126/science.1131864. [DOI] [PubMed] [Google Scholar]
- 3.Marin-Argany M, et al. Cell damage in light chain amyloidosis: Fibril internalization, toxicity and cell-mediated seeding. J Biol Chem. 2016;291:19813–19825. doi: 10.1074/jbc.M116.736736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kisilevsky R, Boudreau L. Kinetics of amyloid deposition. I. The effects of amyloid-enhancing factor and splenectomy. Lab Invest. 1983;48:53–59. [PubMed] [Google Scholar]
- 5.Pomfret EA, et al. Effect of orthotopic liver transplantation on the progression of familial amyloidotic polyneuropathy. Transplantation. 1998;65:918–925. doi: 10.1097/00007890-199804150-00010. [DOI] [PubMed] [Google Scholar]
- 6.Costa PP, Figueira AS, Bravo FR. Amyloid fibril protein related to prealbumin in familial amyloidotic polyneuropathy. Proc Natl Acad Sci USA. 1978;75:4499–4503. doi: 10.1073/pnas.75.9.4499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sletten K, Westermark P, Natvig JB. Senile cardiac amyloid is related to prealbumin. Scand J Immunol. 1980;12:503–506. doi: 10.1111/j.1365-3083.1980.tb00098.x. [DOI] [PubMed] [Google Scholar]
- 8.Benson MD. Partial amino acid sequence homology between an heredofamilial amyloid protein and human plasma prealbumin. J Clin Invest. 1981;67:1035–1041. doi: 10.1172/JCI110114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pras M, Prelli F, Franklin EC, Frangione B. Primary structure of an amyloid prealbumin variant in familial polyneuropathy of Jewish origin. Proc Natl Acad Sci USA. 1983;80:539–542. doi: 10.1073/pnas.80.2.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Merlini G, Bellotti V. Molecular mechanisms of amyloidosis. N Engl J Med. 2003;349:583–596. doi: 10.1056/NEJMra023144. [DOI] [PubMed] [Google Scholar]
- 11.Hurshman Babbes AR, Powers ET, Kelly JW. Quantification of the thermodynamically linked quaternary and tertiary structural stabilities of transthyretin and its disease-associated variants: The relationship between stability and amyloidosis. Biochemistry. 2008;47:6969–6984. doi: 10.1021/bi800636q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Benson MD. Pathogenesis of transthyretin amyloidosis. Amyloid. 2012;19:14–15. doi: 10.3109/13506129.2012.668501. [DOI] [PubMed] [Google Scholar]
- 13.Gorevic PD, Prelli FC, Wright J, Pras M, Frangione B. Systemic senile amyloidosis. Identification of a new prealbumin (transthyretin) variant in cardiac tissue: Immunologic and biochemical similarity to one form of familial amyloidotic polyneuropathy. J Clin Invest. 1989;83:836–843. doi: 10.1172/JCI113966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Westermark P, Bergström J, Solomon A, Murphy C, Sletten K. Transthyretin-derived senile systemic amyloidosis: Clinicopathologic and structural considerations. Amyloid. 2003;10:48–54. [PubMed] [Google Scholar]
- 15.Su Y, et al. Transthyretin-derived amyloid deposition in the heart of an elderly Japanese population. Amyloid. 2011;18:180–181. doi: 10.3109/13506129.2011.574354067. [DOI] [PubMed] [Google Scholar]
- 16.Ihse E, Suhr OB, Hellman U, Westermark P. Variation in amount of wild-type transthyretin in different fibril and tissue types in ATTR amyloidosis. J Mol Med (Berl) 2011;89:171–180. doi: 10.1007/s00109-010-0695-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yazaki M, et al. Cardiac amyloid in patients with familial amyloid polyneuropathy consists of abundant wild-type transthyretin. Biochem Biophys Res Commun. 2000;274:702–706. doi: 10.1006/bbrc.2000.3203. [DOI] [PubMed] [Google Scholar]
- 18.Reixach N, Deechongkit S, Jiang X, Kelly JW, Buxbaum JN. Tissue damage in the amyloidoses: Transthyretin monomers and nonnative oligomers are the major cytotoxic species in tissue culture. Proc Natl Acad Sci USA. 2004;101:2817–2822. doi: 10.1073/pnas.0400062101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bergström J, et al. Amyloid deposits in transthyretin-derived amyloidosis: Cleaved transthyretin is associated with distinct amyloid morphology. J Pathol. 2005;206:224–232. doi: 10.1002/path.1759. [DOI] [PubMed] [Google Scholar]
- 20.Benson MD. In: Amyloid and Related Disorders. Picken MM, Dogan A, editors. Humana; Herrera, GA: 2012. pp. 53–67. [Google Scholar]
- 21.Benson MD. Liver transplantation and transthyretin amyloidosis. Muscle Nerve. 2013;47:157–162. doi: 10.1002/mus.23521. [DOI] [PubMed] [Google Scholar]
- 22.Stangou AJ, et al. Progressive cardiac amyloidosis following liver transplantation for familial amyloid polyneuropathy: Implications for amyloid fibrillogenesis. Transplantation. 1998;66:229–233. doi: 10.1097/00007890-199807270-00016. [DOI] [PubMed] [Google Scholar]
- 23.Stangou AJ, Hawkins PN. Liver transplantation in transthyretin-related familial amyloid polyneuropathy. Curr Opin Neurol. 2004;17:615–620. doi: 10.1097/00019052-200410000-00012. [DOI] [PubMed] [Google Scholar]
- 24.Eisenberg D, Jucker M. The amyloid state of proteins in human diseases. Cell. 2012;148:1188–1203. doi: 10.1016/j.cell.2012.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saelices L, et al. Uncovering the mechanism of aggregation of human transthyretin. J Biol Chem. 2015;290:28932–28943. doi: 10.1074/jbc.M115.659912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nelson R, et al. Structure of the cross-beta spine of amyloid-like fibrils. Nature. 2005;435:773–778. doi: 10.1038/nature03680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Westermark P, Sletten K, Johansson B, Cornwell GG., 3rd Fibril in senile systemic amyloidosis is derived from normal transthyretin. Proc Natl Acad Sci USA. 1990;87:2843–2845. doi: 10.1073/pnas.87.7.2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Foss TR, Wiseman RL, Kelly JW. The pathway by which the tetrameric protein transthyretin dissociates. Biochemistry. 2005;44:15525–15533. doi: 10.1021/bi051608t. [DOI] [PubMed] [Google Scholar]
- 29.Jiang X, et al. An engineered transthyretin monomer that is nonamyloidogenic, unless it is partially denatured. Biochemistry. 2001;40:11442–11452. doi: 10.1021/bi011194d. [DOI] [PubMed] [Google Scholar]
- 30.Hurshman AR, White JT, Powers ET, Kelly JW. Transthyretin aggregation under partially denaturing conditions is a downhill polymerization. Biochemistry. 2004;43:7365–7381. doi: 10.1021/bi049621l. [DOI] [PubMed] [Google Scholar]
- 31.Galant NJ, Westermark P, Higaki JN, Chakrabartty A. Transthyretin amyloidosis: An under-recognized neuropathy and cardiomyopathy. Clin Sci (Lond) 2017;131:395–409. doi: 10.1042/CS20160413. [DOI] [PubMed] [Google Scholar]
- 32.Damy T, et al. Clinical, ECG and echocardiographic clues to the diagnosis of TTR-related cardiomyopathy. Open Heart. 2016;3:e000289. doi: 10.1136/openhrt-2015-000289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rapezzi C, et al. Disease profile and differential diagnosis of hereditary transthyretin-related amyloidosis with exclusively cardiac phenotype: An Italian perspective. Eur Heart J. 2013;34:520–528. doi: 10.1093/eurheartj/ehs123. [DOI] [PubMed] [Google Scholar]
- 34.Adamski-Werner SL, Palaninathan SK, Sacchettini JC, Kelly JW. Diflunisal analogues stabilize the native state of transthyretin. Potent inhibition of amyloidogenesis. J Med Chem. 2004;47:355–374. doi: 10.1021/jm030347n. [DOI] [PubMed] [Google Scholar]
- 35.Bulawa CE, et al. Tafamidis, a potent and selective transthyretin kinetic stabilizer that inhibits the amyloid cascade. Proc Natl Acad Sci USA. 2012;109:9629–9634. doi: 10.1073/pnas.1121005109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Werdelin O, Ranlov P. Amyloidosis in mice produced by transplantation of spleen cells from casein-treated mice. Acta Pathol Microbiol Scand. 1966;68:1–18. doi: 10.1111/apm.1966.68.1.1. [DOI] [PubMed] [Google Scholar]
- 37.Ranlov P. The adoptive transfer of experimental mouse amyloidosis by intravenous injections of spleen cell extracts from casein-treated syngeneic donor mice. Acta Pathol Microbiol Scand. 1967;70:321–335. [PubMed] [Google Scholar]
- 38.Hardt F, Ranlov P. Transfer amyloidosis. Local and systemic amyloidosis in recipients of syngeneic spleen grafts from non-amyloidotic, casein-sensitized donor mice. Acta Pathol Microbiol Scand. 1968;73:549–558. [PubMed] [Google Scholar]
- 39.Wanstrup J, Ranlov P. Transfer amyloidosis: Ultrastructure of the transferred subcellular fractions. Acta Pathol Microbiol Scand. 1968;74:303–304. doi: 10.1111/j.1699-0463.1968.tb03482.x. [DOI] [PubMed] [Google Scholar]
- 40.Kisilevsky R, Axelrad M. The pathogenesis of amyloid deposition: A new hypothesis. Med Hypotheses. 1976;2:233–237. doi: 10.1016/s0306-9877(76)80002-3. [DOI] [PubMed] [Google Scholar]
- 41.Kisilevsky R, Axelrad M, Corbett W, Brunet S, Scott F. The role of inflammatory cells in the pathogenesis of amyloidosis. Lab Invest. 1977;37:544–553. [PubMed] [Google Scholar]
- 42.Axelrad MA, Kisilevsky R, Willmer J, Chen SJ, Skinner M. Further characterization of amyloid-enhancing factor. Lab Invest. 1982;47:139–146. [PubMed] [Google Scholar]
- 43.Jarrett JT, Lansbury PTJ., Jr Seeding “one-dimensional crystallization” of amyloid: A pathogenic mechanism in Alzheimer’s disease and scrapie? Cell. 1993;73:1055–1058. doi: 10.1016/0092-8674(93)90635-4. [DOI] [PubMed] [Google Scholar]
- 44.Bessen RA, et al. Non-genetic propagation of strain-specific properties of scrapie prion protein. Nature. 1995;375:698–700. doi: 10.1038/375698a0. [DOI] [PubMed] [Google Scholar]
- 45.Petkova AT, et al. Self-propagating, molecular-level polymorphism in Alzheimer’s beta-amyloid fibrils. Science. 2005;307:262–265. doi: 10.1126/science.1105850. [DOI] [PubMed] [Google Scholar]
- 46.Colby DW, et al. Prion detection by an amyloid seeding assay. Proc Natl Acad Sci USA. 2007;104:20914–20919. doi: 10.1073/pnas.0710152105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Paravastu AK, Qahwash I, Leapman RD, Meredith SC, Tycko R. Seeded growth of beta-amyloid fibrils from Alzheimer’s brain-derived fibrils produces a distinct fibril structure. Proc Natl Acad Sci USA. 2009;106:7443–7448. doi: 10.1073/pnas.0812033106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lu JX, et al. Molecular structure of β-amyloid fibrils in Alzheimer’s disease brain tissue. Cell. 2013;154:1257–1268. doi: 10.1016/j.cell.2013.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Saborio GP, Permanne B, Soto C. Sensitive detection of pathological prion protein by cyclic amplification of protein misfolding. Nature. 2001;411:810–813. doi: 10.1038/35081095. [DOI] [PubMed] [Google Scholar]
- 50.Castilla J, Saá P, Hetz C, Soto C. In vitro generation of infectious scrapie prions. Cell. 2005;121:195–206. doi: 10.1016/j.cell.2005.02.011. [DOI] [PubMed] [Google Scholar]
- 51.Prusiner SB. Cell biology. A unifying role for prions in neurodegenerative diseases. Science. 2012;336:1511–1513. doi: 10.1126/science.1222951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Goedert M. Neurodegeneration. Alzheimer’s and Parkinson’s diseases: The prion concept in relation to assembled Aβ, tau, and α-synuclein. Science. 2015;349:1255555. doi: 10.1126/science.1255555. [DOI] [PubMed] [Google Scholar]
- 53.Goedert M, Eisenberg DS, Crowther RA. Propagation of tau aggregates and neurodegeneration. Annu Rev Neurosci. 2017;40:189–210. doi: 10.1146/annurev-neuro-072116-031153. [DOI] [PubMed] [Google Scholar]
- 54.Goedert M, Jakes R, Spillantini MG. The synucleinopathies: Twenty years on on. J Parkinsons Dis. 2017;7(Suppl 1):S53–S71. doi: 10.3233/JPD-179005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Clavaguera F, et al. Transmission and spreading of tauopathy in transgenic mouse brain. Nat Cell Biol. 2009;11:909–913. doi: 10.1038/ncb1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Desplats P, et al. Inclusion formation and neuronal cell death through neuron-to-neuron transmission of alpha-synuclein. Proc Natl Acad Sci USA. 2009;106:13010–13015. doi: 10.1073/pnas.0903691106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Brown P, et al. Iatrogenic Creutzfeldt-Jakob disease, final assessment. Emerg Infect Dis. 2012;18:901–907. doi: 10.3201/eid1806.120116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Colon W, Kelly JW. Partial denaturation of transthyretin is sufficient for amyloid fibril formation in vitro. Biochemistry. 1992;31:8654–8660. doi: 10.1021/bi00151a036. [DOI] [PubMed] [Google Scholar]
- 59.Tagoe CE, French D, Gallo G, Buxbaum JN. Amyloidogenesis is neither accelerated nor enhanced by injections of preformed fibrils in mice transgenic for wild-type human transthyretin: The question of infectivity. Amyloid. 2004;11:21–26. doi: 10.1080/13506120410001674982. [DOI] [PubMed] [Google Scholar]
- 60.Wei L, et al. Deposition of transthyretin amyloid is not accelerated by the same amyloid in vivo. Amyloid. 2004;11:113–120. doi: 10.1080/13506120410001726344. [DOI] [PubMed] [Google Scholar]
- 61.Liepnieks JJ, Benson MD. Progression of cardiac amyloid deposition in hereditary transthyretin amyloidosis patients after liver transplantation. Amyloid. 2007;14:277–282. doi: 10.1080/13506120701614032. [DOI] [PubMed] [Google Scholar]
- 62.Carvalho A, Rocha A, Lobato L. Liver transplantation in transthyretin amyloidosis: Issues and challenges. Liver Transpl. 2015;21:282–292. doi: 10.1002/lt.24058. [DOI] [PubMed] [Google Scholar]
- 63.Lladó L, et al. Risk of transmission of systemic transthyretin amyloidosis after domino liver transplantation. Liver Transpl. 2010;16:1386–1392. doi: 10.1002/lt.22174. [DOI] [PubMed] [Google Scholar]
- 64.Buxbaum JN, et al. Why are some amyloidoses systemic? Does hepatic “chaperoning at a distance” prevent cardiac deposition in a transgenic model of human senile systemic (transthyretin) amyloidosis? FASEB J. 2012;26:2283–2293. doi: 10.1096/fj.11-189571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Misumi Y, et al. Recipient aging accelerates acquired transthyretin amyloidosis after domino liver transplantation. Liver Transpl. 2016;22:656–664. doi: 10.1002/lt.24371. [DOI] [PubMed] [Google Scholar]
- 66.Stangou AJ, Heaton ND, Hawkins PN. Transmission of systemic transthyretin amyloidosis by means of domino liver transplantation. N Engl J Med. 2005;352:2356. doi: 10.1056/NEJM200506023522219. [DOI] [PubMed] [Google Scholar]
- 67.Westermark GT, Westermark P. Prion-like aggregates: Infectious agents in human disease. Trends Mol Med. 2010;16:501–507. doi: 10.1016/j.molmed.2010.08.004. [DOI] [PubMed] [Google Scholar]
- 68.Westermark GT, Fändrich M, Lundmark K, Westermark P. Noncerebral amyloidoses: Aspects on seeding, cross-seeding, and transmission. Cold Spring Harb Perspect Med. 2017;8:a024323. doi: 10.1101/cshperspect.a024323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Obayashi K, et al. Amyloid turnover after liver transplantation in FAP. Amyloid. 2011;18:187–189. doi: 10.3109/13506129.2011.574354070. [DOI] [PubMed] [Google Scholar]
- 70.Westermark P, Sletten K, Olofsson BO. Prealbumin variants in the amyloid fibrils of Swedish familial amyloidotic polyneuropathy. Clin Exp Immunol. 1987;69:695–701. [PMC free article] [PubMed] [Google Scholar]
- 71.Sievers SA, et al. Structure-based design of non-natural amino-acid inhibitors of amyloid fibril formation. Nature. 2011;475:96–100. doi: 10.1038/nature10154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Soragni A, et al. A designed inhibitor of p53 aggregation rescues p53 tumor suppression in ovarian carcinomas. Cancer Cell. 2016;29:90–103. doi: 10.1016/j.ccell.2015.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ackermann EJ, et al. Suppressing transthyretin production in mice, monkeys and humans using 2nd-generation antisense oligonucleotides. Amyloid. 2016;23:148–157. doi: 10.1080/13506129.2016.1191458. [DOI] [PubMed] [Google Scholar]
- 74.Ackermann EJ, et al. Clinical development of an antisense therapy for the treatment of transthyretin-associated polyneuropathy. Amyloid. 2012;19:43–44. doi: 10.3109/13506129.2012.673140. [DOI] [PubMed] [Google Scholar]
- 75.Coelho T, et al. Safety and efficacy of RNAi therapy for transthyretin amyloidosis. N Engl J Med. 2013;369:819–829. doi: 10.1056/NEJMoa1208760. [DOI] [PubMed] [Google Scholar]
- 76.Sekijima Y, Tojo K, Morita H, Koyama J, Ikeda S. Safety and efficacy of long-term diflunisal administration in hereditary transthyretin (ATTR) amyloidosis. Amyloid. 2015;22:79–83. doi: 10.3109/13506129.2014.997872. [DOI] [PubMed] [Google Scholar]
- 77.Higaki JN, et al. Novel conformation-specific monoclonal antibodies against amyloidogenic forms of transthyretin. Amyloid. 2016;23:86–97. doi: 10.3109/13506129.2016.1148025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hosoi A, et al. Novel antibody for the treatment of transthyretin amyloidosis. J Biol Chem. 2016;291:25096–25105. doi: 10.1074/jbc.M116.738138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ihse E, et al. Amyloid fibrils containing fragmented ATTR may be the standard fibril composition in ATTR amyloidosis. Amyloid. 2013;20:142–150. doi: 10.3109/13506129.2013.797890. [DOI] [PubMed] [Google Scholar]
- 80.Rodriguez JA, et al. Structure of the toxic core of α-synuclein from invisible crystals. Nature. 2015;525:486–490. doi: 10.1038/nature15368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Arvai A. 2015 Adxv–A Program to Display X-ray Diffraction Images. Available at http://www.scripps.edu/∼arvai/adxv.html. Accessed June 15, 2018.
- 82.Shaw BF, et al. Detergent-insoluble aggregates associated with amyotrophic lateral sclerosis in transgenic mice contain primarily full-length, unmodified superoxide dismutase-1. J Biol Chem. 2008;283:8340–8350. doi: 10.1074/jbc.M707751200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Rappsilber J, Mann M, Ishihama Y. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat Protoc. 2007;2:1896–1906. doi: 10.1038/nprot.2007.261. [DOI] [PubMed] [Google Scholar]
- 84.Thompson MJ, et al. The 3D profile method for identifying fibril-forming segments of proteins. Proc Natl Acad Sci USA. 2006;103:4074–4078. doi: 10.1073/pnas.0511295103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.