Significance
Siderophores are small molecules used by microbes to acquire the essential metal Fe in environments ranging from infections to the open ocean. Although many bacteria use quorum sensing (QS) to regulate siderophore production, the resulting siderophore concentrations and their utility for Fe uptake have been underexplored. We report that Vibrio harveyi uses a “two-for-one,” QS- and Fe-repressed gene cluster to produce cell-associated siderophores at low cell density and accumulate soluble siderophores at high cell density. Fe-uptake experiments show that these soluble siderophores are used in Fe acquisition. We also find that QS repression helps prevent the accumulation of siderophore concentrations in excess of cellular uptake capacities, an explanation that may be generalizable to other bacteria.
Keywords: siderophores, quorum sensing, iron, Vibrio harveyi, high-resolution mass spectrometry
Abstract
The secretion of small Fe-binding molecules called siderophores is an important microbial strategy for survival in Fe-limited environments. Siderophore production is often regulated by quorum sensing (QS), a microbial counting technique that allows organisms to alter gene expression based on cell density. However, the identity and quantities of siderophores produced under QS regulation are rarely studied in the context of their roles in Fe uptake. We investigated the link between QS, siderophores, and Fe uptake in the model marine organism Vibrio harveyi where QS is thought to repress siderophore production. We find that V. harveyi uses a single QS- and Fe-repressed gene cluster to produce both cell-associated siderophores (amphiphilic enterobactins) as well as several related soluble siderophores, which we identify and quantify using liquid chromatography-coupled (LC)-MS as well as tandem high-resolution MS (LC-HR-MS/MS). Measurements of siderophore production show that soluble siderophores are present at ∼100× higher concentrations than amphi-enterobactin and that over the course of growth V. harveyi decreases amphi-enterobactin concentrations but accumulates soluble siderophores. 55Fe radio-tracer uptake experiments demonstrate that these soluble siderophores play a significant role in Fe uptake and that the QS-dictated concentrations of soluble siderophores in stationary phase are near the limit of cellular uptake capacities. We propose that cell-associated and soluble siderophores are beneficial to V. harveyi in different environmental and growth contexts and that QS allows V. harveyi to exploit “knowledge” of its population size to avoid unnecessary siderophore production.
Iron (Fe) is an essential nutrient that can limit microbial growth. Despite the abundance of Fe on the planet, the low solubility of ferric iron at circumneutral pH often leads to low bioavailability. One solution to this problem is the production of siderophores, small iron-binding molecules that solubilize Fe(III), making it bioavailable to the producing bacterium (1). Although siderophores are most commonly considered Fe-scavenging molecules, it is clear that they can also mediate microbial interactions. Many microbes take up siderophores that they do not synthesize themselves (2–5), and siderophores can be used to sequester iron from microbial competitors (5) as well as animal hosts (6). Siderophores have also been used to investigate cooperative behavior and symbiotic interactions (7–9).
The study of siderophore regulation provides a fruitful approach for understanding the multiple ways in which microbes use these molecules. One particularly intriguing finding is that in many organisms siderophore production is regulated by quorum sensing (QS), a counting technique that allows microbes to tailor gene expression to cell density (10). During the process of QS, microbes both produce and detect extracellular signaling molecules, the concentration of which is an indicator of population size. The accumulation of QS molecules often represses siderophore synthesis, leading to a decrease in their rate of production at high cell density (11–14). Conversely, increases in siderophores in response to QS have been documented in Pseudomonas aeruginosa (15).
The potential benefit of QS regulation of siderophores is widely appreciated (for example, refs. 16–19). However, we still do not fully understand why QS represses siderophore production in some organisms but stimulates it in others. In fact, arguments have been made in favor of both scenarios. Stimulation of production at high cell density might allow bacteria to ensure that siderophores are used by members of their own species (18). In contrast, repression of siderophore production at high cell density could allow bacteria to divert resources toward growth or dispersal (16). The identity and concentrations of siderophores produced under QS regulation are germane to understanding their utility in these contexts but have only been explored in a few species. Potential coregulation of siderophore production by Fe, while expected, has also been underexplored. Here, we investigate the types and quantities of siderophores produced in response to QS and Fe as well as their efficacy for Fe uptake in the model marine bacterium Vibrio harveyi BAA-1116.
QS in V. harveyi is well studied and is known to regulate numerous cellular processes, including the production of siderophores (14). Using the chrome azurol S (CAS) assay (20), a proxy for the presence of Fe-binding molecules, Lilley and Bassler (14) showed that the accumulation of QS molecules leads to a decrease in siderophore production. More recently, Zane et al. (21) demonstrated that V. harveyi produces a suite of amphiphilic enterobactin-like siderophores (amphi-enterobactins) that are associated with the cell pellet. Work by Naka et al. (22) has shown that two V. harveyi strains encode a gene cluster for biosynthesis of the much smaller, soluble siderophore anguibactin. Although the production of anguibactin in V. harveyi BAA-1116 was inferred from bioassays, it has yet to be verified by MS or other direct analytical methods. We take advantage of the detailed understanding of QS in V. harveyi as well as the recent discovery of the siderophores made by this organism to explore the relationship between QS, Fe, and siderophore production.
We used transcriptomic and metabolomic methods to test the regulation of siderophore production in V. harveyi by both Fe and QS. Surprisingly, we found that under our experimental conditions, V. harveyi has only one active siderophore gene cluster but that this cluster produces both cell-associated siderophores (amphi-enterobactins) as well as several smaller soluble amphi-enterobactin precursors and breakdown products. Over the course of growth, V. harveyi accumulates high concentrations of soluble siderophores (∼100-fold higher than measured for amphi-enterobactins) and degrades cell-associated siderophores. 55Fe radio-tracer uptake experiments show that these smaller siderophores enhance Fe uptake and that the QS-determined concentrations of siderophores found in the growth medium are likely calibrated to the maximal cellular uptake capacity. We propose that cell-associated and soluble siderophores serve different purposes during various V. harveyi life stages and that QS repression helps V. harveyi, and possibly other species, avoid unnecessary siderophore production.
Results and Discussion
Siderophore Biosynthetic Genes Are Repressed by Fe and QS.
V. harveyi produces three autoinducers: 3-hydroxybutanoyl homoserine lactone (AI-1), (S)-3-hydroxytridecan-4-one (CAI-1), and (2S,4S)-2-methyl-2,3,3,4-tetrahydroxy-tetrahydrofuran borate (AI-2) (23–25). These autoinducers are integrated additively by the same regulatory circuit (26). Using a QS mutant [ΔluxM ΔluxPQ ΔcqsS (26)] that responds only to the homoserine lactone autoinducer (hereafter HSL), we tested the effect of Fe and QS signal concentrations on the expression of anguibactin and amphi-enterobactin gene clusters. Fe limitation was achieved using a chemically defined medium in which background Fe was both low and chelated using EDTA (EDTA = 100 µM, FeT = 100 nM, unchelated Fe, Fe′ = 0.12 nM). Cells were cultured with: (i) no addition, (ii) Fe, (iii) HSL, or (iv) Fe and HSL. Fe and HSL were added at two concentrations: 100 and 500 nM. As expected, the addition of HSL did not have an effect on growth. However, Fe limitation clearly affected growth as cultures with 500 nM Fe grew faster than those with 0 or 100 nM Fe (Fig. 1A).
Fig. 1.
V. harveyi response to Fe and HSL. (A) V. harveyi growth under varying Fe and HSL concentrations. (B) Expression of amphi-enterobactin biosynthetic genes. (C) Expression of anguibactin biosynthetic genes. (D) Production of amphi-enterobactin m/z 965.3681. (E) Production of DHBA. (F) Production of DHB-Ser. Gene expression is normalized to treatments with 500 nM added Fe and HSL; rpoA was used as the housekeeping gene. Error bars represent the standard deviation for biological duplicates.
Both amphi-enterobactins and anguibactin are synthesized by nonribosomal peptide-synthetase (NRPS) enzymes. Using reverse transcription-quantitative PCR (RT-qPCR), we explored the expression of the amphi-enterobactin biosynthetic genes aebB and aebF. AebB encodes a bifunctional isochorismatase and aryl carrier protein with homology to the enterobactin gene entB (21, 27). AebF has homology to the enterobactin gene entF and encodes a four-domain NRPS (21, 28). In addition, we tested the expression of the anguibactin genes angB (a homolog of aebB and entB) that is used in the assembly of the anguibactin catechol moiety (29) and angM, which encodes peptidyl-carrier protein and condensation domains involved in the incorporation of N-hydroxyhistamine into anguibactin (29). The expression of both amphi-enterobactin genes was clearly down-regulated by 100 and 500 nM Fe (Fig. 1B). A similar result was observed with the addition of either 100 or 500 nM HSL. In contrast, angB and angM (anguibactin associated genes) did not respond to either Fe or HSL in the medium (Fig. 1C). Expression of anguibactin genes, while detected, was low [threshold cycle (CT) values of ∼30]. These results indicate that the production of amphi-enterobactins is repressed by both QS and Fe, while anguibactin synthesis responds to neither.
Detection and Quantification of Siderophores.
We next examined the production of amphi-enterobactins and anguibactin by LC-MS and LC-HR-MS/MS. In agreement with previous studies (21), we found several amphi-enterobactins with slight variations in the fatty-acid tail in cell pellet extracts (SI Appendix, Figs. S1 and S2). In addition to variants 1 and 4 reported by Zane et al. (21) (Fig. 2, see SI Appendix, Fig. S1 for a full suite of amphi-enterobactins), two amphi-enterobactins were also identified, which we suggest contain decanoyl (2, m/z = 911.3193) and tetradecanoyl (3, m/z = 967.3819) groups (Fig. 2 and SI Appendix, Fig. S1). Consistent with our RT-qPCR results, LC-MS analysis showed strong repression of amphi-enterobactins (we tracked a representative variant, m/z = 965.3681, SI Appendix, Fig. S1, hereafter amphi-enterobactin) by Fe and more modest repression by HSL (Fig. 1D). Notably, our experiments use HSL as the sole QS molecule. In wild-type cultures where all three QS molecules are produced, this response is likely stronger.
Fig. 2.

Structures of amphi-enterobactins, anguibactin, DHBA, and DHB-Ser. Variants of amphi-enterobactin fatty-acid tails are shown; 1 has been characterized by NMR (21), 4 was proposed by ref. 21 based on MS/MS data, 2 and 3 are proposed in this study based on MS/MS data (SI Appendix, Fig. S1). Several other hydroxylated and unsaturated variants also exist (SI Appendix, Fig. S1, ref. 21) but are not shown as the sites of hydroxylation and unsaturation are not known.
We were unable to detect anguibactin (5) in V. harveyi supernatants prepared under several conditions, although it was readily detected in supernatant extracts from the known anguibactin producer Vibrio anguillarum 775. The absence of both an anguibactin gene expression response and anguibactin production implies that this gene cluster is not active under our experimental conditions (it remains unknown whether this gene cluster is active in other contexts). In contrast, the V. harveyi aeb gene cluster is expressed and the biosynthesis of amphi-enterobactin clearly responds to exogenous Fe and QS molecules.
To assess the presence of other siderophores, searches for catechol-containing compounds (using UV-vis absorbance at 310 nm, Materials and Methods) in V. harveyi supernatants were conducted, revealing peaks that corresponded to two smaller siderophores: 2,3-dihydroxybenzoic acid (6, DHBA) and 2,3-dihydroxybenzoyl serine (7, DHB-Ser, Fig. 2). The identity of these molecules was confirmed using a synthetic standard (DHBA) and LC-HR-MS/MS (DHB-Ser, SI Appendix, Fig. S3). LC-MS analysis showed repression of DHBA and DHB-Ser synthesis by the addition of either Fe or HSL (Fig. 1 E and F). These siderophores were present at much higher concentrations than amphi-enterobactin, but their response to Fe and HSL was remarkably similar to that of the larger siderophore (Fig. 1D), suggesting these products are all derived from the same gene cluster.
Two possible sources of DHBA and DHB-Ser can be envisioned: premature release from the biosynthetic pathway and degradation of amphi-enterobactins. As is common for catechol containing siderophores, DHBA is synthesized from chorismate (21) and then incorporated into the larger siderophore. In V. harveyi, a fraction of this DHBA could be released into the medium rather than being used in the assembly of amphi-enterobactins. In view of the much larger concentrations of DHB-Ser as opposed to amphi-enterobactin (compare Fig. 1D to Fig. 1F), some direct production of DHB-Ser by premature release from the NRPS is likely occurring as well, consistent with the complex, multistep biosynthesis of the siderophore (30). Release of several intermediates has been observed by in vitro mutation studies of the NRPS for enterobactin (28, 31). The accumulation of such high quantities of DHB-Ser and DHBA implies an inefficient amphi-enterobactin biosynthetic process, which may be a beneficial feature of the V. harveyi gene cluster. Indeed, the propensity for the release of catechol monomers by other triscatechol producers has recently been reviewed (32).
Untargeted LC-HR-MS/MS Search for Biosynthetic Intermediates.
The Fe–enterobactin complex is exceedingly stable and Fe uptake requires the activity of esterases (33) that hydrolyze the siderophore. Suspecting that hydrolysis of amphi-enterobactins might also contribute to the soluble siderophore pool in V. harveyi cultures, we used LC-HR-MS/MS to search for potential breakdown products of amphi-enterobactins in culture supernatants. Rather than look for all possible biosynthetic intermediates, we limited the search using our workflow for untargeted identification of iron-binding molecules (34). Specifically, searches for masses corresponding to the free ligand ([M+H]+) as well as the 56Fe and 54Fe complexes ([M-3H++56/54Fe3+]+ or [M-2H++56/54Fe2+]+) revealed several potential iron chelates. The corresponding apo-siderophore precursor ions were further restricted to those with a product ion corresponding to m/z = 224.0553 ([M+H]+), a characteristic fragment of enterobactins and amphi-enterobactins (21) (SI Appendix, Figs. S1 and S4). In doing so, we identified several related structures (Fig. 3 and SI Appendix, Fig. S4 and Table S1), many corresponding to linearized amphi-enterobactin fragments. Collectively, our data imply that, in addition to producing amphi-enterobactins, V. harveyi secretes significantly larger quantities of DHBA and DHB-Ser, as well as biosynthetic intermediates, a process that can be attributed to both premature release and degradation of the full-length siderophore.
Fig. 3.
Amphi-enterobactin breakdown product (m/z = 527.2597) found in V. harveyi supernatant. (A) Fe-isotope pattern showing 54/56Fe complexes as well as 13C isotopes. (B) MS/MS spectra and (C) proposed structure. See SI Appendix, Fig. S4 and Tables S1 and S2 for further breakdown products.
QS Repression of Amphi-enterobactin, DHBA, and DHB-Ser During Growth.
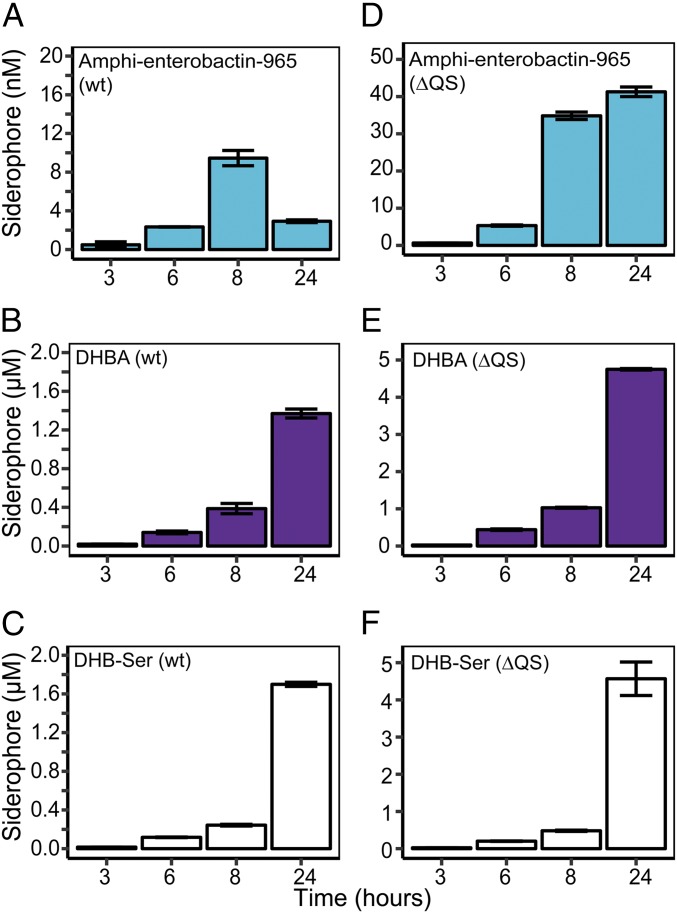
To gain further insight into the production and use of amphi-enterobactin, DHBA, and DHB-Ser, we quantified these molecules during growth of wild-type (WT) cultures as well as a mutant strain that does not respond to QS (ΔQS) (14). Consistent with QS repression of siderophore production (14), concentrations of siderophores were much higher in the supernatants of the ΔQS mutant than in WT cultures (compare Fig. 4 A–C and Fig. 4 D–F). Time-resolved experiments using the ΔQS strain showed a steady increase in all three siderophores (Fig. 4 D–F). However, in WT cultures, amphi-enterobactin concentrations increased during exponential growth but decreased in stationary phase (Fig. 4A), whereas DHBA and DHB-Ser steadily accumulated (Fig. 4 B and C). As was the case for short-term experiments (Fig. 1 D–F), DHBA and DHB-Ser were both produced at much higher concentrations (micromolar) than amphi-enterobactin (nanomolar). The decrease in amphi-enterobactin in stationary phase provides direct evidence for amphi-enterobactin breakdown by V. harveyi (Fig. 4A). Here, repression by QS reduces the production rate of amphi-enterobactins below the breakdown rate. As a result, amphi-enterobactins are degraded in late growth stages, whereas soluble siderophores accumulate.
Fig. 4.
Siderophore production over time. (A) Amphi-enterobactin m/z 965.3681, (B) DHBA, and (C) DHB-Ser production in WT cultures. (D) Amphi-enterobactin m/z 965.3681, (E) DHBA, and (F) DHB-Ser production in mutants that do not respond to QS [ΔQS, luxO D47E (14)]. Error bars represent the standard deviation for biological duplicates.
Small Soluble Siderophores Aid in Fe Uptake.
To test the utility of DHBA and DHB-Ser in Fe acquisition, we compared 55Fe-uptake rates in the presence and absence of 1 μM synthetic DHBA, or spent medium containing V.-harveyi–produced DHBA, in both WT cultures and cultures deficient in the production of amphi-enterobactins (ΔaebF). Spent medium was added to a final DHBA concentration of 1 μM, comparable to the concentrations produced by WT cultures (1–2 μM, Fig. 4B). As expected, in the absence of DHBA, Fe uptake from 55Fe-EDTA was slow in both WT and ΔaebF cultures (Fig. 5A). In contrast, the addition of either spent medium or synthetic DHBA significantly enhanced the rate of Fe uptake in both strains (P < 0.005, one-way ANOVA). There was no significant difference in Fe uptake between the spent medium and DHBA treatments (P = 0.92, Fig. 5A). These results are also consistent with studies of Escherichia coli, which produces enterobactin, accumulates enterobactin fragments in the growth medium, and exhibits enhanced Fe uptake in the presence of DHB-Ser and DHBA (35, 36). In our experiments, amphi-enterobactins are found almost exclusively in the cell pellet. Our results from spent medium additions therefore solely reflect the capacity for V. harveyi to utilize the added soluble siderophores and synthetic DHBA. The enhanced rates of Fe uptake in treatments with both DHBA and spent medium suggest that DHBA, and possibly DHB-Ser, as well as other siderophore fragments form biologically available Fe complexes that are used by V. harveyi (Fig. 5A). Notably, Fe-uptake rates were nearly identical between spent medium and DHBA treatments, implying that DHBA may be responsible for most of the enhancement in Fe uptake.
Fig. 5.
Radio-tracer (55Fe) uptake experiments. (A) Fe-uptake rates from Fe-EDTA, Fe-EDTA + 1 µM DHBA, and Fe-EDTA + spent medium (containing 1 µM DHBA and breakdown products) in WT cultures (green) and ΔaebF mutants (white). Different letters indicate statistically significant differences (P < 0.05). (B) Fe uptake in response to varying levels of DHBA. Gray line: theoretical linear response; red line: observed response. Error bars represent the standard deviation for biological duplicates.
There are two potential routes for Fe uptake from DHBA and DHB-Ser: direct transport of Fe-siderophore complexes and exchange of Fe between weaker siderophores and amphi-enterobactins at the cell surface. Our data suggest that both processes occur. BLAST searches of the V. harveyi genome revealed hits to CirA (VIBHAR_01404 e-value of 7e-105) and Fiu (VIBHAR_06299, e-value of 1e-30), which transport DHB-Ser and other catechols (35, 37). In addition, comparisons of Fe-uptake rates in WT and ΔaebF V. harveyi hint at Fe exchange. These experiments show that the presence of amphi-enterobactins aids in Fe uptake from soluble siderophores: Rates of Fe uptake from DHBA and spent medium were slower in mutants than in WT cultures (Fig. 5A). In contrast, mutant and WT strains showed no difference in Fe-uptake rate from Fe-EDTA buffered medium, suggesting that the mutant is only deficient in Fe uptake from soluble siderophores. Our experiments imply that amphi-enterobactins may be involved in some of the uptake of Fe from weaker siderophores (Fig. 5A), possibly through a ligand-exchange mechanism whereby Fe is transferred from weaker siderophores to amphi-enterobactins at the cell surface.
Finally, we tested whether the concentrations of DHBA produced by WT cultures (1–2 µM) were tuned to the cellular Fe-uptake capacity. We measured Fe-uptake rates for a range of DHBA concentrations (1–10 μM). Fe-uptake rates increased with increasing concentrations of DHBA but saturated at concentrations greater than a few micromolar (Fig. 5B).
Ecological Utility of Cell-Associated and Soluble Siderophores.
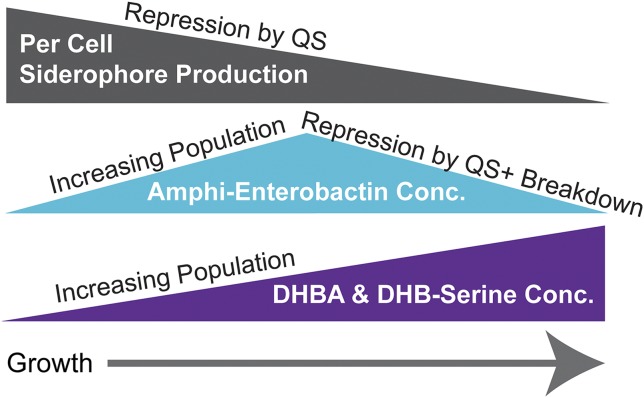
In the wild, V. harveyi has a complex lifestyle, existing as a free-living and a pathogenic microbe (38, 39). During growth, V. harveyi decreases concentrations of amphi-enterobactins but accumulates soluble siderophores. This pattern of siderophore production––which is due to the breakdown of amphi-enterobactins during uptake, decreased synthesis of amphi-enterobactins due to QS, and what appears to be a diversity-oriented biosynthetic gene cluster (Figs. 6 and 7)––may provide beneficial versatility for different V. harveyi life stages.
Fig. 6.
Amphi-enterobactin (aeb) gene cluster. Proposed sources of fragments and breakdown products due to direct biosynthesis/premature release (step a) or degradation (step b) are shown. Genes are color coded: red, long-chain fatty-acid CoA ligase; blue, DHBA biosynthesis; green, NRPS; brown, P-pant transferase; gray, hypothetical protein; after ref. 21.
Fig. 7.
Siderophore production during V. harveyi growth. Over the course of growth per cell siderophore production decreases, concentrations of amphi-enterobactins initially increase due to growth but ultimately decrease due to repression by QS as well as breakdown. Despite repression by QS, DHBA and DHB-Ser accumulate due to increasing population size.
It is of particular interest that V. harveyi adds a hydrophobic tail to enterobactin, a siderophore that is primarily produced by terrestrial species (40, 41). The increased solubility of charged molecules in high ionic strength seawater may necessitate the presence of a fatty-acid tail to anchor siderophores at the cell surface. The production of amphiphilic siderophores is in fact common among marine bacteria and has been proposed to reduce the loss of siderophores to diffusion in the dilute marine environment (42). During planktonic growth, V. harveyi must rely chiefly on amphi-enterobactins for Fe uptake as the bulk of DHBA and DHB-Ser will be lost to diffusion. The small size of amphi-enterobactins allows for tighter packing on the cell surface (compared with membrane-bound transporters) and could create an Fe “sink” at the cell surface. During growth in spatially confined environments (such as on particles, in biofilms, or during infections) where soluble siderophores can accumulate, V. harveyi may rely on DHBA and DHB-Ser for Fe uptake. Here, in addition to direct uptake from DHBA and DHB-Ser, Fe uptake could involve a “bucket brigade” mechanism (43–45) whereby Fe is shuttled by these soluble siderophores to amphi-enterobactins at the cell surface (Fig. 5A).
Competition with other species is another reason for the production of both soluble and cell-associated siderophores. This has not been explored in V. harveyi, but competition assays using E. coli mutants that are deficient in enterobactin biosynthesis suggest that enterobactin is privatized at low cell density but shared at high cell density, and that this helps to protect against siderophore piracy (46). Our study supports the proposal of this previous work that enterobactin could be produced at low cell density while more soluble enterobactin derivatives or breakdown products (33, 35, 40) accumulate at high cell density. Measurements of amphi-enterobactin, DHBA, and DHB-Ser show exactly this type of transition in V. harveyi: Amphi-enterobactin concentrations increase during exponential growth but decrease during stationary phase, while the concentrations of DHBA and DHB-Ser continuously increase (Figs. 4 A–C and 7). In some contexts, amphi-enterobactins may therefore function as siderophores available only to V. harveyi whereas the weaker, more soluble DHBA and DHB-Ser are accessed by multiple bacteria.
Repression by QS and Fe Optimizes Siderophore Production.
V. harveyi represses siderophore production when Fe levels are high. This result has been seen in many organisms and is a straightforward mechanism to avoid unnecessary secretion of siderophores. In V. harveyi, QS also represses siderophore production at high cell density. This may allow V. harveyi to devote resources to other processes such as growth and dispersal, as has been suggested in the case of QS repression of polymer secretion at high cell density in Vibrio cholerae (16). In addition, once cell density becomes sufficiently high, large per-cell siderophore production rates are simply no longer necessary to achieve maximal Fe uptake. It is the concentration of siderophores in the medium, not the per-cell production rate, that matters for Fe uptake. And, this process has limits: cellular Fe transport is not infinitely fast and cells do not have infinite surface area for siderophore transporters (47). The repression of siderophore production by QS may therefore allow V. harveyi to leverage “knowledge” of its population size to achieve a useful, but not profligate, siderophore concentration. This is seen in our experiments where increases in the concentrations of DHBA above ∼2 µM (the concentration produced in stationary phase, Fig. 4), lead to increased Fe-uptake rates, but with diminishing returns (Fig. 5B). Indeed, the QS-dictated ∼2 µM DHBA concentration seems well tuned to the maximal uptake capacity of the cells.
QS regulates siderophores in species other than V. harveyi. For example, QS represses ornibactin in Burkholderia cepacia (11, 12) and vulnibactin in Vibrio vulnificus (13). In contrast, some Pseudomonas aeruginosa QS signals enhance the production of pyoverdine (15). In light of our proposal that increased siderophore production at high cell density is not beneficial for Fe uptake, we speculate that the direction of QS regulation may offer some broad insight into siderophore function. For example, in P. aeruginosa, siderophores are part of a strategy to accumulate high concentrations of Fe which trigger the production of biofilms and enable persistent infections (48). Siderophores repressed by QS may be primarily devoted to Fe acquisition for growth, while those stimulated by QS may have more complex roles, acting for example as signals or virulence factors or aiding in the establishment of chronic infections. This is certainly the case for pyoverdine (48, 49).
Conclusion
We have shown that V. harveyi has an Fe-and QS-repressed “two-for-one” siderophore gene cluster, that releases both cell-associated and soluble products. The most abundant of these soluble siderophores, DHBA and DHB-Ser, are found at concentrations nearly 100× higher than measured for amphi-enterobactin and are responsible for a large fraction of Fe uptake by V. harveyi. During planktonic growth, cell-associated siderophores may serve as transporters, allowing efficient Fe uptake from dilute marine environments. In contrast, during infections or growth on particles, where DHBA and DHB-Ser accumulate to high concentrations, these soluble siderophores become important for Fe uptake. Our results show that soluble and cell-associated siderophores may also work in concert. Finally, we find that QS repression of siderophore production allows V. harveyi to synthesize “just the right amount” of siderophores for Fe uptake. Our study is one of few to quantitatively investigate siderophore production in response to QS. Based on our findings, we make some more general predictions about the link between QS and siderophore production: namely that siderophores which are repressed by QS (as is the case in V. harveyi) are most likely used in Fe uptake for growth, whereas those stimulated by QS may serve other purposes acting for example as signals or virulence factors. Quantitative information about siderophore production and QS in other organisms will allow for the expansion, verification, and modification of these ideas.
Materials and Methods
Materials and methods including culturing conditions, BLAST searches, RNA extraction, RT-qPCR, siderophore quantification, LC-HR-MS/MS experiments, Fe-isotope studies, and 55Fe-uptake experiments are detailed in SI Appendix.
Supplementary Material
Acknowledgments
We are grateful to A. Butler and C. S. Harwood for insightful reviews. We thank the Bassler Lab (especially J. S. Valastyan) for the kind gift of WT and QS mutant V. harveyi strains; M. G. Haygood and H. Naka for providing the ΔaebF mutant; T. Srikumar and S. Kyin (Princeton Proteomics and Mass Spectrometry Core facility) as well as Y. Wu (M.R.S. laboratory) for help with LC-HR-MS/MS; J. R. Reinfelder for guidance with radio-isotope experiments; and E. P. Greenberg for helpful comments on the manuscript. This research was supported by the Princeton Environmental Institute and National Science Foundation Grant OCE 1657639 (to F.M.M.M.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1805791115/-/DCSupplemental.
References
- 1.Hider RC, Kong X. Chemistry and biology of siderophores. Nat Prod Rep. 2010;27:637–657. doi: 10.1039/b906679a. [DOI] [PubMed] [Google Scholar]
- 2.Granger J, Price NM. The importance of siderophores in iron nutrition of heterotrophic marine bacteria. Limnol Oceanogr. 1999;44:541–555. [Google Scholar]
- 3.D’Onofrio A, et al. Siderophores from neighboring organisms promote the growth of uncultured bacteria. Chem Biol. 2010;17:254–264. doi: 10.1016/j.chembiol.2010.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cordero OX, Ventouras L-A, DeLong EF, Polz MF. Public good dynamics drive evolution of iron acquisition strategies in natural bacterioplankton populations. Proc Natl Acad Sci USA. 2012;109:20059–20064. doi: 10.1073/pnas.1213344109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Traxler MF, Seyedsayamdost MR, Clardy J, Kolter R. Interspecies modulation of bacterial development through iron competition and siderophore piracy. Mol Microbiol. 2012;86:628–644. doi: 10.1111/mmi.12008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ratledge C, Dover LG. Iron metabolism in pathogenic bacteria. Annu Rev Microbiol. 2000;54:881–941. doi: 10.1146/annurev.micro.54.1.881. [DOI] [PubMed] [Google Scholar]
- 7.Griffin AS, West SA, Buckling A. Cooperation and competition in pathogenic bacteria. Nature. 2004;430:1024–1027. doi: 10.1038/nature02744. [DOI] [PubMed] [Google Scholar]
- 8.Nadell CD, Bassler BL, Levin SA. Observing bacteria through the lens of social evolution. J Biol. 2008;7:27. doi: 10.1186/jbiol87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schuster M, Sexton DJ, Diggle SP, Greenberg EP. Acyl-homoserine lactone quorum sensing: From evolution to application. Annu Rev Microbiol. 2013;67:43–63. doi: 10.1146/annurev-micro-092412-155635. [DOI] [PubMed] [Google Scholar]
- 10.Waters CM, Bassler BL. Quorum sensing: Cell-to-cell communication in bacteria. Annu Rev Cell Dev Biol. 2005;21:319–346. doi: 10.1146/annurev.cellbio.21.012704.131001. [DOI] [PubMed] [Google Scholar]
- 11.Lewenza S, Conway B, Greenberg EP, Sokol PA. Quorum sensing in Burkholderia cepacia: Identification of the LuxRI homologs CepRI. J Bacteriol. 1999;181:748–756. doi: 10.1128/jb.181.3.748-756.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lewenza S, Sokol PA. Regulation of ornibactin biosynthesis and N-acyl-L-homoserine lactone production by CepR in Burkholderia cepacia. J Bacteriol. 2001;183:2212–2218. doi: 10.1128/JB.183.7.2212-2218.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wen Y, Kim IH, Son J-S, Lee B-H, Kim K-S. Iron and quorum sensing coordinately regulate the expression of vulnibactin biosynthesis in Vibrio vulnificus. J Biol Chem. 2012;287:26727–26739. doi: 10.1074/jbc.M112.374165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lilley BN, Bassler BL. Regulation of quorum sensing in Vibrio harveyi by LuxO and sigma-54. Mol Microbiol. 2000;36:940–954. doi: 10.1046/j.1365-2958.2000.01913.x. [DOI] [PubMed] [Google Scholar]
- 15.Stintzi A, Evans K, Meyer JM, Poole K. Quorum-sensing and siderophore biosynthesis in Pseudomonas aeruginosa: LasR/lasI mutants exhibit reduced pyoverdine biosynthesis. FEMS Microbiol Lett. 1998;166:341–345. doi: 10.1111/j.1574-6968.1998.tb13910.x. [DOI] [PubMed] [Google Scholar]
- 16.Nadell CD, Xavier JB, Levin SA, Foster KR. The evolution of quorum sensing in bacterial biofilms. PLoS Biol. 2008;6:e14–e19. doi: 10.1371/journal.pbio.0060014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xavier JB, Kim W, Foster KR. A molecular mechanism that stabilizes cooperative secretions in Pseudomonas aeruginosa. Mol Microbiol. 2011;79:166–179. doi: 10.1111/j.1365-2958.2010.07436.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Darch SE, West SA, Winzer K, Diggle SP. Density-dependent fitness benefits in quorum-sensing bacterial populations. Proc Natl Acad Sci USA. 2012;109:8259–8263. doi: 10.1073/pnas.1118131109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heilmann S, Krishna S, Kerr B. Why do bacteria regulate public goods by quorum sensing?-How the shapes of cost and benefit functions determine the form of optimal regulation. Front Microbiol. 2015;6:767. doi: 10.3389/fmicb.2015.00767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schwyn B, Neilands JB. Universal chemical assay for the detection and determination of siderophores. Anal Biochem. 1987;160:47–56. doi: 10.1016/0003-2697(87)90612-9. [DOI] [PubMed] [Google Scholar]
- 21.Zane HK, et al. Biosynthesis of amphi-enterobactin siderophores by Vibrio harveyi BAA-1116: Identification of a bifunctional nonribosomal peptide synthetase condensation domain. J Am Chem Soc. 2014;136:5615–5618. doi: 10.1021/ja5019942. [DOI] [PubMed] [Google Scholar]
- 22.Naka H, Actis LA, Crosa JH. The anguibactin biosynthesis and transport genes are encoded in the chromosome of Vibrio harveyi: A possible evolutionary origin for the pJM1 plasmid-encoded system of Vibrio anguillarum? Microbiology Open. 2013;2:182–194. doi: 10.1002/mbo3.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Higgins DA, et al. The major Vibrio cholerae autoinducer and its role in virulence factor production. Nature. 2007;450:883–886. doi: 10.1038/nature06284. [DOI] [PubMed] [Google Scholar]
- 24.Chen X, et al. Structural identification of a bacterial quorum-sensing signal containing boron. Nature. 2002;415:545–549. doi: 10.1038/415545a. [DOI] [PubMed] [Google Scholar]
- 25.Cao JG, Meighen EA. Purification and structural identification of an autoinducer for the luminescence system of Vibrio harveyi. J Biol Chem. 1989;264:21670–21676. [PubMed] [Google Scholar]
- 26.Long T, et al. Quantifying the integration of quorum-sensing signals with single-cell resolution. PLoS Biol. 2009;7:e68. doi: 10.1371/journal.pbio.1000068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gehring AM, Bradley KA, Walsh CT. Enterobactin biosynthesis in Escherichia coli: Isochorismate lyase (EntB) is a bifunctional enzyme that is phosphopantetheinylated by EntD and then acylated by EntE using ATP and 2,3-dihydroxybenzoate. Biochemistry. 1997;36:8495–8503. doi: 10.1021/bi970453p. [DOI] [PubMed] [Google Scholar]
- 28.Shaw-Reid CA, et al. Assembly line enzymology by multimodular nonribosomal peptide synthetases: The thioesterase domain of E. coli EntF catalyzes both elongation and cyclolactonization. Chem Biol. 1999;6:385–400. doi: 10.1016/S1074-5521(99)80050-7. [DOI] [PubMed] [Google Scholar]
- 29.Crosa JH, Walsh CT. Genetics and assembly line enzymology of siderophore biosynthesis in bacteria. Microbiol Mol Biol Rev. 2002;66:223–249. doi: 10.1128/MMBR.66.2.223-249.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Challis GL, Hopwood DA. Synergy and contingency as driving forces for the evolution of multiple secondary metabolite production by Streptomyces species. Proc Natl Acad Sci USA. 2003;100:14555–14561. doi: 10.1073/pnas.1934677100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gehring AM, Mori I, Walsh CT. Reconstitution and characterization of the Escherichia coli enterobactin synthetase from EntB, EntE, and EntF. Biochemistry. 1998;37:2648–2659. doi: 10.1021/bi9726584. [DOI] [PubMed] [Google Scholar]
- 32.Reitz ZL, Sandy M, Butler A. Biosynthetic considerations of triscatechol siderophores framed on serine and threonine macrolactone scaffolds. Metallomics. 2017;9:824–839. doi: 10.1039/c7mt00111h. [DOI] [PubMed] [Google Scholar]
- 33.Lin H, Fischbach MA, Liu DR, Walsh CT. In vitro characterization of salmochelin and enterobactin trilactone hydrolases IroD, IroE, and Fes. J Am Chem Soc. 2005;127:11075–11084. doi: 10.1021/ja0522027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baars O, Morel FMM, Perlman DH. ChelomEx: Isotope-assisted discovery of metal chelates in complex media using high-resolution LC-MS. Anal Chem. 2014;86:11298–11305. doi: 10.1021/ac503000e. [DOI] [PubMed] [Google Scholar]
- 35.Hantke K. Dihydroxybenzoylserine–A siderophore for E. coli. FEMS Microbiol Lett. 1990;55:5–8. doi: 10.1016/0378-1097(90)90158-m. [DOI] [PubMed] [Google Scholar]
- 36.Hancock RE, Hantke K, Braun V. Iron transport in Escherichia coli K-12. 2,3-dihydroxybenzoate-promoted iron uptake. Arch Microbiol. 1977;114:231–239. doi: 10.1007/BF00446867. [DOI] [PubMed] [Google Scholar]
- 37.Nikaido H, Rosenberg EY. Cir and Fiu proteins in the outer membrane of Escherichia coli catalyze transport of monomeric catechols: Study with β-lactam antibiotics containing catechol and analogous groups. J Bacteriol. 1990;172:1361–1367. doi: 10.1128/jb.172.3.1361-1367.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Austin B, Zhang X-H. Vibrio harveyi: A significant pathogen of marine vertebrates and invertebrates. Lett Appl Microbiol. 2006;43:119–124. doi: 10.1111/j.1472-765X.2006.01989.x. [DOI] [PubMed] [Google Scholar]
- 39.Farmer J, Janda J, Brenner F, Cameron D, Birkhead K. Bergey’s Manual of Systematic Bacteriology. Vol 2 Springer; New York: 2005. Genus I. Vibrio Pacini 1854, 411AL. [Google Scholar]
- 40.O’Brien IG, Gibson F. The structure of enterochelin and related 2,3-dihydroxy-N-benzoylserine conjugates from Escherichia coli. Biochim Biophys Acta. 1970;215:393–402. doi: 10.1016/0304-4165(70)90038-3. [DOI] [PubMed] [Google Scholar]
- 41.Pollack JR, Neilands JB. Enterobactin, an iron transport compound from Salmonella typhimurium. Biochem Biophys Res Commun. 1970;38:989–992. doi: 10.1016/0006-291x(70)90819-3. [DOI] [PubMed] [Google Scholar]
- 42.Sandy M, Butler A. Microbial iron acquisition: Marine and terrestrial siderophores. Chem Rev. 2009;109:4580–4595. doi: 10.1021/cr9002787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Martinez JS, et al. Structure and membrane affinity of a suite of amphiphilic siderophores produced by a marine bacterium. Proc Natl Acad Sci USA. 2003;100:3754–3759. doi: 10.1073/pnas.0637444100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gobin J, Horwitz MA. Exochelins of Mycobacterium tuberculosis remove iron from human iron-binding proteins and donate iron to mycobactins in the M. tuberculosis cell wall. J Exp Med. 1996;183:1527–1532. doi: 10.1084/jem.183.4.1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fukushima T, et al. Gram-positive siderophore-shuttle with iron-exchange from Fe-siderophore to apo-siderophore by Bacillus cereus YxeB. Proc Natl Acad Sci USA. 2013;110:13821–13826. doi: 10.1073/pnas.1304235110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Scholz RL, Greenberg EP. Sociality in Escherichia coli: Enterochelin is a private good at low cell density and can be shared at high cell density. J Bacteriol. 2015;197:2122–2128. doi: 10.1128/JB.02596-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hudson RJM, Morel FMM. Trace metal transport by marine microorganisms: Implications of metal coordination kinetics. Deep Sea Res Part I Oceanogr Res Pap. 1993;40:129–150. [Google Scholar]
- 48.Banin E, Vasil ML, Greenberg EP. Iron and Pseudomonas aeruginosa biofilm formation. Proc Natl Acad Sci USA. 2005;102:11076–11081. doi: 10.1073/pnas.0504266102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lamont IL, Beare PA, Ochsner U, Vasil AI, Vasil ML. Siderophore-mediated signaling regulates virulence factor production in Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 2002;99:7072–7077. doi: 10.1073/pnas.092016999. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.