Significance
Aminoacyl-tRNA synthetases (aaRSs) establish the rules to express the universal genetic code. During aminoacylation, each of the 20 aaRSs associates 1 of 20 amino acids with a specific trinucleotide known as anticodon. Remarkably, for alanyl-tRNAs, the synthetase makes no contact with the anticodon. Instead, it uses a “second genetic code” by picking out a single G3:U70 base pair in the tRNA acceptor stem, which is close to the amino acid attachment site, but 76 Å away from the anticodon. Here, we show that, while in the three kingdoms of life, alanyl-tRNA synthetases use G3:U70 to identify alanyl-tRNAs, surprisingly, they use three different mechanisms to achieve this. We thus suggest that, in evolution, the genetic code had a powerful and persistent preference for associating G:U with alanine.
Keywords: aminoacyl-tRNA synthetase, tRNA, evolution, specificity, second genetic code
Abstract
Throughout three domains of life, alanyl-tRNA synthetases (AlaRSs) recognize a G3:U70 base pair in the acceptor stem of tRNAAla as the major identity determinant of tRNAAla. The crystal structure of the archaeon Archaeoglobus fulgidus AlaRS in complex with tRNAAla provided the basis for G3:U70 recognition with residues (Asp and Asn) that are conserved in the three domains [Naganuma M, et al. (2014) Nature 510:507–511]. The recognition mode is unprecedented, with specific accommodation of the dyad asymmetry of the G:U wobble pair and exclusion of the dyad symmetry of a Watson–Crick pair. With this conserved mode, specificity is based more on “fit” than on direct recognition of specific atomic groups. Here, we show that, in contrast to the archaeal complex, the Escherichia coli enzyme uses direct positive (energetically favorable) minor groove recognition of the unpaired 2-amino of G3 by Asp and repulsion of a competing base pair by Asn. Strikingly, mutations that disrupted positive recognition by the E. coli enzyme had little or no effect on G:U recognition by the human enzyme. Alternatively, Homo sapiens AlaRS selects G:U without positive recognition and uses Asp instead to repel a competitor. Thus, the widely conserved Asp-plus-Asn architecture of AlaRSs can select G:U in a straightforward (bacteria) or two different unconventional (eukarya/archaea) ways. The adoption of different modes for recognition of a widely conserved G:U pair in alanine tRNAs suggests an early and insistent role for G:U in the development of the genetic code.
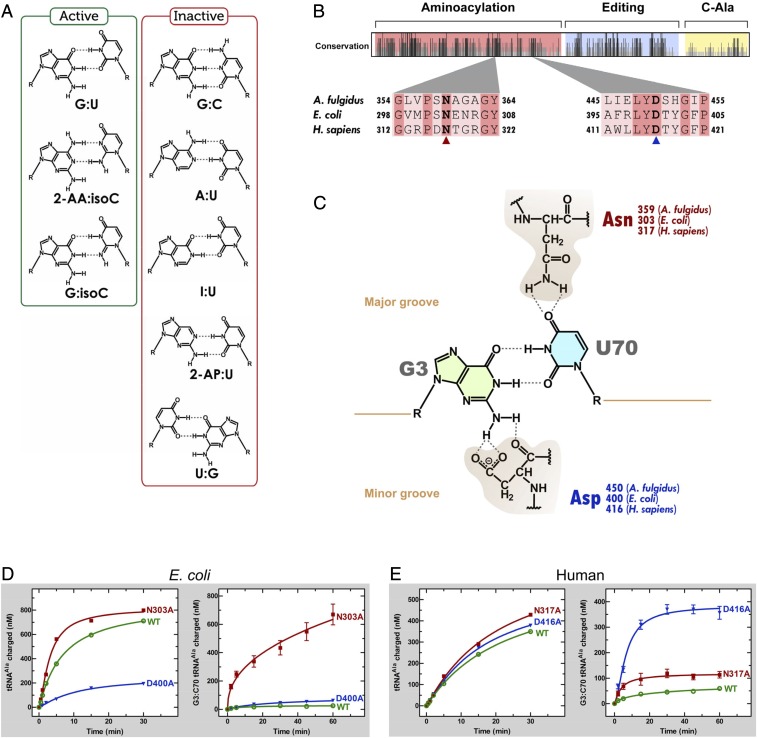
Early work showed that a single G3:U70 base pair in the acceptor stem marked tRNAAla for aminoacylation with alanine (1–4). G3:U70 is conserved through all three kingdoms of the tree of life and is so robust that its transfer into nonalanine tRNAs converts them to alanine acceptors, both in vitro and in vivo (2, 3, 5). Using synthetic RNA substrates with both natural and nonnatural substitutions at 3:70, previous investigations also showed that the unpaired exocyclic 2-amino group of G3 in the RNA minor groove was critical for aminoacylation. (With the G:C pair, the 2-amino group is H-bonded, while it is free in the G:U wobble pair.) To establish this point, nonnatural pairs that placed a free amino group in the same position were shown to be active (Fig. 1A). These included 2-amino adenosine and, separately, G paired with isocytidine (6). Ablation of the exocyclic amino group by creating an I:U pair abolished aminoacylation (7, 8). In addition, substitution of 2-aminopurine [paired with U (2-AP:U)] for G yielded an inactive substrate for aminoacylation (7, 8). The sensitivity of the system to subtle effects of the geometry of recognition was shown with the U:G substitution. Although the 2-amino group of the U:G wobble superimposes on that of G:U, its opposite orientation for projecting into the minor groove prevented aminoacylation (6). Although the role of O4 of U70 was not clear from these studies, O4 alone was clearly not sufficient to confer aminoacylation, because it is present in the inactive 2-AP:U and I:U substrates.
Fig. 1.
Specific atomic groups are important for tRNAAla identity. (A) Aminoacylation-active and -inactive tRNAAla base pair variants at the 3:70 position (cf. ref. 7). (B) Alignment of A. fulgidus, E. coli, and H. sapiens AlaRS sequences. A total of 50 sequences was aligned to give a map of relative conservation along the sequence of the AlaRS enzyme. The Asn (N) and Asp (D) residues involved in G3:U70 recognition are shown in bold. (C) Chemical representation of the G3:U70 base pair and the hydrogen bonding contacts made with the major-groove Asn and minor-groove Asp residues, according to the A. fulgidus AlaRS•tRNAAla structure. (D) Aminoacylation activities of WT and mutant E. coli AlaRS enzymes for G3:U70 (Left) and G3:C70 (Right) tRNAAla. (E) Aminoacylation activities of WT and mutant human AlaRS enzymes for G3:U70 (Left) and G3:C70 (Right) tRNAAla. In D and E, aminoacylation of G3:U70 tRNA was carried out with 5 nM of each enzyme, while that of G3:C70 tRNA was carried out with 500 nM of each enzyme. Error bars represent the SEM of triplicate experiments.
Structural insight into G3:U70 recognition by alanyl-tRNA synthetase (AlaRS) was provided by the crystal structure of the archaeal Archaeoglobus fulgidus AlaRS in complex with tRNAAla. In this structure, the tRNA-recognition domain contacts the tRNA acceptor stem from both the minor and major grooves. Asn359 and Asp450 hereby make the key H-bonding contacts with the G3:U70 wobble pair. The amide nitrogen of the Asn359 side chain contacts O4 of U70 (major-groove side), and the carboxyl side chain as well as the backbone carbonyl of Asp450 H-bonds with the 2-amino group of G3 (minor-groove side) (Fig. 1C). Both archaeal AlaRS residues, Asn359 and Asp450, that recognize the identity determinant G3:U70 base pair are conserved throughout evolution (as Asn303 and Asp400 in Escherichia coli and as Asn317 and Asp416 in Homo sapiens AlaRS). In addition, the sequences that surround these residues are highly conserved between bacteria and human (Fig. 1B).
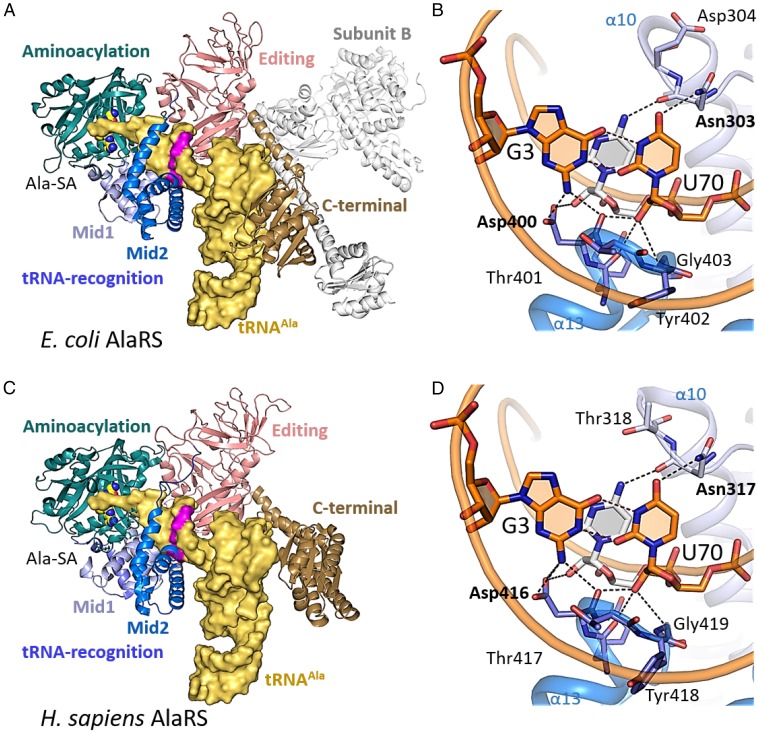
A superposition of the structures of the A. fulgidus and E. coli AlaRS catalytic fragments revealed that they share almost the same fold, except for the local difference in the orientation of the Mid2 subdomain from the tRNA-recognition domain (SI Appendix, Fig. S1). Based on sequence alignments (SI Appendix, Fig. S2) and the crystal structures of the E. coli AlaRS catalytic fragment and of the A. fulgidus AlaRS•tRNAAla complex, a homology model of E. coli AlaRS in complex with tRNAAla was constructed (Fig. 2A). The conformation of the E. coli AlaRS Mid2 subdomain was slightly adjusted for the tRNA binding. In this model, G3:U70 is recognized on the major- and minor-groove sides by the conserved amino acid residues, in the same manner as in the A. fulgidus AlaRS•tRNA complex (9) (Fig. 2B). On the minor-groove side of G3:U70, the 2-amino group of G3 hydrogen bonds with the side-chain carboxyl and main-chain carbonyl groups of Asp400. In addition, the side-chain carboxyl group of Asp400 is near the 2′-hydroxyl group of C71. The 2′-hydroxyl group of the U70 ribose hydrogen bonds with the main-chain carbonyl groups of Asp400 and Tyr399, and the amide NH group of Gly403. Asp400, Tyr399, and Gly403 are strictly conserved for all three enzymes. This observation is consistent with atomic group mutagenesis studies that demonstrated the importance of the ribose 2′-OH of U70 and other nearby atoms for aminoacylation activity (10, 11).
Fig. 2.
E. coli and H. sapiens AlaRS•tRNAAla structures, modeled based on the A. fulgidus AlaRS•tRNAAla structure. (A) The overall structure of the E. coli AlaRS•tRNAAla homology model. The domains and subdomains are colored as indicated. The tRNAAla molecule is shown as a yellow surface model with the G3:U70 pair highlighted in pink. (B) The G3:U70 interaction in the E. coli AlaRS•tRNAAla homology model. The amino acid residues and the nucleotides are shown as blue and orange stick models, respectively. (C) The overall structure of the H. sapiens AlaRS•tRNAAla homology model. (D) The G3:U70 interaction in the H. sapiens AlaRS•tRNAAla homology model.
In H. sapiens AlaRS, the above-described Asp and Asn residues are conserved as Asp416 and Asn317. Using the coordinates of the H. sapiens AlaRS catalytic fragment, H. sapiens C-terminal domain, and A. fulgidus AlaRS•tRNAAla complex (Fig. 2 C and D and SI Appendix, Fig. S1), a homology model of the H. sapiens AlaRS•tRNAAla complex was created. In contrast to the archaeal and bacterial AlaRSs, which form a dimer in solution, H. sapiens AlaRS is presented as a monomer (12). To avoid steric clashes with the editing domain, the C-terminal domain (C-Ala) is reoriented relative to its archaeal and bacterial counterparts and does not form the tight contacts with the tRNA elbow region that are observed in the A. fulgidus AlaRS•tRNAAla complex, consistent with C-Ala having no effect on charging activity in the human enzyme (12). However, the interactions around the G3:U70 base pair are analogous to those in the homology model of the E. coli AlaRS•tRNAAla complex (Fig. 2D).
To understand the mechanism of G3:U70 recognition by bacterial AlaRS, we used aminoacylation kinetics with transcripts of E. coli tRNAAla and recombinant E. coli AlaRS purified from E. coli. Two mutations were introduced into E. coli AlaRS to test the role of Asp400 in minor groove recognition of the 2-amino group of G3 [Asp400:NH2(G3) interaction]. First, the hydrogen bond between the carboxyl group of Asp400 and the 2-amino group of G3 was removed by introduction of a D400A substitution, which inserts a smaller residue that should be sterically accommodated but lacks the ability to make a positive, energetically favorable interaction. The D400A mutant enzyme was virtually inactive for aminoacylation (Fig. 1D and Table 1). Next, we created D400N AlaRS. Here, the Asp-to-Asn replacement is roughly isosteric. However, while the model predicts that both carboxyl oxygens of the Asp side chain are involved in H-bonding to the 2-NH2 of G3 and the 2′-OH of C71 (Fig. 1C), the amide group of Asn cannot H-bond to G3. Like D400A AlaRS, D400N AlaRS was virtually inactive for aminoacylation (Table 1). These results mirror those in which the 2-amino of G3 was ablated (as in I:U) or shifted (as in G:C) (Fig. 1A) and are consistent with the presence of an energetically favorable contact between Asp400 and NH2(G3), which is critical for aminoacylation.
Table 1.
Kinetics of aminoacylation by E. coli AlaRS mutants
| E. coli AlaRS | kcat, s−1 | Km, μM | kcat/Km, μM−1⋅s−1 | Rel kcat/Km |
| WT | 0.36 ± 0.02 | 0.12 ± 0.03 | 3.0 ± 0.9 | 1 |
| N303A | 0.82 ± 0.05 | 0.14 ± 0.04 | 5.7 ± 1.9 | 1.9 |
| N303D | 0.12 ± 0.01 | 3.4 ± 0.4 | 0.034 ± 0.006 | 0.012 |
| D400A | 0.17 ± 0.01 | 1.4 ± 0.2 | 0.13 ± 0.03 | 0.042 |
| D400N | 0.14 ± 0.01 | 1.2 ± 0.1 | 0.12 ± 0.02 | 0.040 |
| N303A/D400A | 0.30 ± 0.02 | 0.67 ± 0.12 | 0.44 ± 0.11 | 0.15 |
Assays were carried out at 25 °C and pH 7.5, with tRNAAla concentrations ranging from 62.5 nM to 24.7 μM. Enzyme concentrations were 10 nM (WT), 12.5 nM (N303A), 1 μM (N303D), 80 nM (D400A), 60 nM (D400N), and 20 nM (N303A/D400A). The data represent the mean of triplicate experiments ± SEM.
For the recognition of O4 of U70 by Asn303 of E. coli AlaRS, two additional mutants of E. coli AlaRS were cloned and purified. The N303D substitution was selected as an approximately isosteric substitution to disrupt the interaction with O4 of U70. Consistent with the universal conservation of Asn303 and with an electrostatic repulsion between the introduced carboxyl of D303 and O4 of U70, N303D AlaRS was almost completely inactive for aminoacylation (Table 1). Next, we made an N303A replacement that substituted Asn with the smaller methyl group side chain of Ala. Remarkably, N303A AlaRS was twofold higher in activity than WT AlaRS (Fig. 1D and Table 1). Most of this change came from a change in kcat. Interestingly, the apparent dissociation constant Km was the same [0.12 µM (WT) versus 0.14 µM (N303A)] when the H bond from Asn303 to O4 of U70 was removed. Thus, and surprisingly, the highly conserved H bond of Asn303 to U70 is a somewhat negative contribution to recognition of tRNAAla.
We then considered the possibility that the effects of N303A were dependent on, or coupled to, the presence of Asp400. To investigate this possibility, we asked whether the N303A substitution could rescue in part the deleterious effect of the D400A replacement. Indeed, the N303A substitution into D400A AlaRS (to give N303A/D400A AlaRS) promoted the activity over that of D400A AlaRS alone (Table 1 and SI Appendix, Fig. S3A). Remarkably, this enhancement amounted to an activity for N303A/D400A AlaRS that was only sixfold to sevenfold less than that of WT AlaRS. Thus, the effects of N303A are not dependent on the presence of Asp400. The result gave further motivation to understand the rationale for the presence of the highly conserved Asn303 H bond with O4 of U70. In particular, we considered the possibility that conservation of Asn303 throughout evolution was not solely for recognition of O4 of U70.
The most prominent possibility is that the universally conserved Asn303 is for repelling the major competitor 3:70 base pair in nonalanine-tRNAs of E. coli. Competitor tRNAs are a major challenge for AARSs (13, 14). The dissociation constant for many noncognate tRNA:AARS complexes is within 10- to 100-fold of that for the cognate complex (15–18). The Kd values for synthetase-tRNA complexes are high—1 µM at pH 7.5 is typical. This high Kd facilitates efficient turnover during protein synthesis but leaves less “room” for specificity. To achieve accuracy of aminoacylation, discrimination by kcat has a major role. In E. coli, the major competitors for AlaRS are those tRNAs that have a G3:C70 base pair. For example, 19 isoacceptors for 10 different amino acids in E. coli have G3:C70. Because simple substitution of G3:C70 with G:U confers aminoacylation with alanine, the other parts of the tRNA in these noncognate substrates are of lesser significance.
To investigate the role of Asn303 in rejecting a G3:C70-containing competitor, we made a transcript of E. coli G3:C70 tRNAAla and investigated its activity with the different recombinant AlaRSs that we had created. As expected (2), G3:C70 tRNAAla was completely inactive for aminoacylation by WT AlaRS. In contrast, the N303A single-point substitution gave an enzyme that was active on G3:C70 tRNAAla (Fig. 1D). Importantly, while D400A AlaRS, like WT AlaRS, does not acylate G3:C70 tRNAAla, the double-mutant N303A/D400A AlaRS, like N303A AlaRS, was active on G3:C70 tRNAAla (SI Appendix, Fig. S3B).
Thus, Asn303 appears to be essential for repelling G3:C70-containing competitor tRNAs through its interaction on the major-groove side. This repulsion compensates for Asn303 being the nonoptimal amino acid for charging tRNAAla. At the same time, H bonding by Asp400 to the 2-NH2 of G3 on the minor groove side appears optimal for “positive” recognition of the G:U pair. Thus, one contact—Asp400—is a strong recruiter of the G3:U70 pair, while the other contact—Asn303—protects the Asp400 interaction by repelling competitor tRNAs. These characteristics are expected for a straightforward system of protein–nucleic acid recognition (19–22).
Next, we turned to the human system, where the same G3:U70 base pair in the acceptor stem of human tRNAAla is essential for aminoacylation with alanine in vitro and in vivo. In these assays, we kept fixed the E. coli tRNAAla transcript, which is efficiently aminoacylated in a G3:U70-dependent manner by WT human AlaRS. To our surprise, except for the N317D substitution that introduces an electrostatically repulsive interaction with O4 of U70 (see above), all of the other mutations that disrupted aminoacylation of tRNAAla by the bacterial AlaRS resulted in little change in the efficiency of aminoacylation of H. sapiens AlaRS (Fig. 1E, Table 2, and SI Appendix, Fig. S3C). Examples included the N317A, D416A, D416N, and N317A/D416A substitutions. Thus, neither Asn317 nor Asp416 plays the role of “positive recognition.” The human enzyme hereby shows a striking similarity to A. fulgidus AlaRS, in which neither the N359A nor D450A mutation disrupts aminoacylation of the WT tRNAAla (9).
Table 2.
Kinetics of aminoacylation by human AlaRS mutants
| Human AlaRS | kcat, s−1 | Km, μM | kcat/Km, μM−1⋅s−1 | Rel kcat/Km |
| WT | 0.23 ± 0.01 | 0.26 ± 0.04 | 0.89 ± 0.18 | 1 |
| N317A | 0.20 ± 0.01 | 0.29 ± 0.05 | 0.69 ± 0.15 | 0.78 |
| N317D | 0.024 ± 0.001 | 0.41 ± 0.11 | 0.058 ± 0.018 | 0.065 |
| D416A | 0.32 ± 0.02 | 0.35 ± 0.08 | 0.92 ± 0.26 | 1.0 |
| D416N | 0.20 ± 0.01 | 0.47 ± 0.07 | 0.43 ± 0.08 | 0.48 |
| N317A/D416A | 0.21 ± 0.01 | 0.23 ± 0.06 | 0.90 ± 0.31 | 1.0 |
Assays were carried out at 25 °C and pH 7.5, with tRNAAla concentrations ranging from 62.5 nM to 24.7 μM. Enzyme concentrations were 20 nM (WT), 43 nM (N317A), 250 nM (N317D), 50 nM (D416A), 50 nM (D416N), and 25 nM (N317A/D416A). The data represent the mean of triplicate experiments ± SEM.
These results raised the question of whether the human enzyme used Asn317 and Asp416 to reject noncognate tRNAs, so that the specificity for G3:U70 was because it fits the best into the shape-specific site for the 3:70 base pair and not because of any determinants for positive recognition as seen in the bacterial system. To address this question, we used G3:C70 tRNAAla as substrate for H. sapiens AlaRS. Strikingly, while the WT enzyme was totally inactive on the noncognate substrates, all substitutions—N317A, N317D, D416A, D416N, and N317A/D416A—reduced the specificity of aminoacylation by now allowing charging of G3:C70 tRNAAla (Fig. 1E). Among the mutants that effectively aminoacylated G3:U70 tRNAAla (all except N317D), the most dramatic loss of specificity occurred with the N317A/D416A mutant enzyme (SI Appendix, Fig. S3D). Interestingly, this observation contrasts to that from the archaeal system, where only substitutions of Asp450 cause reduced specificity on G3:C70 tRNAAla, while substitutions in Asn359 have no effect (9).
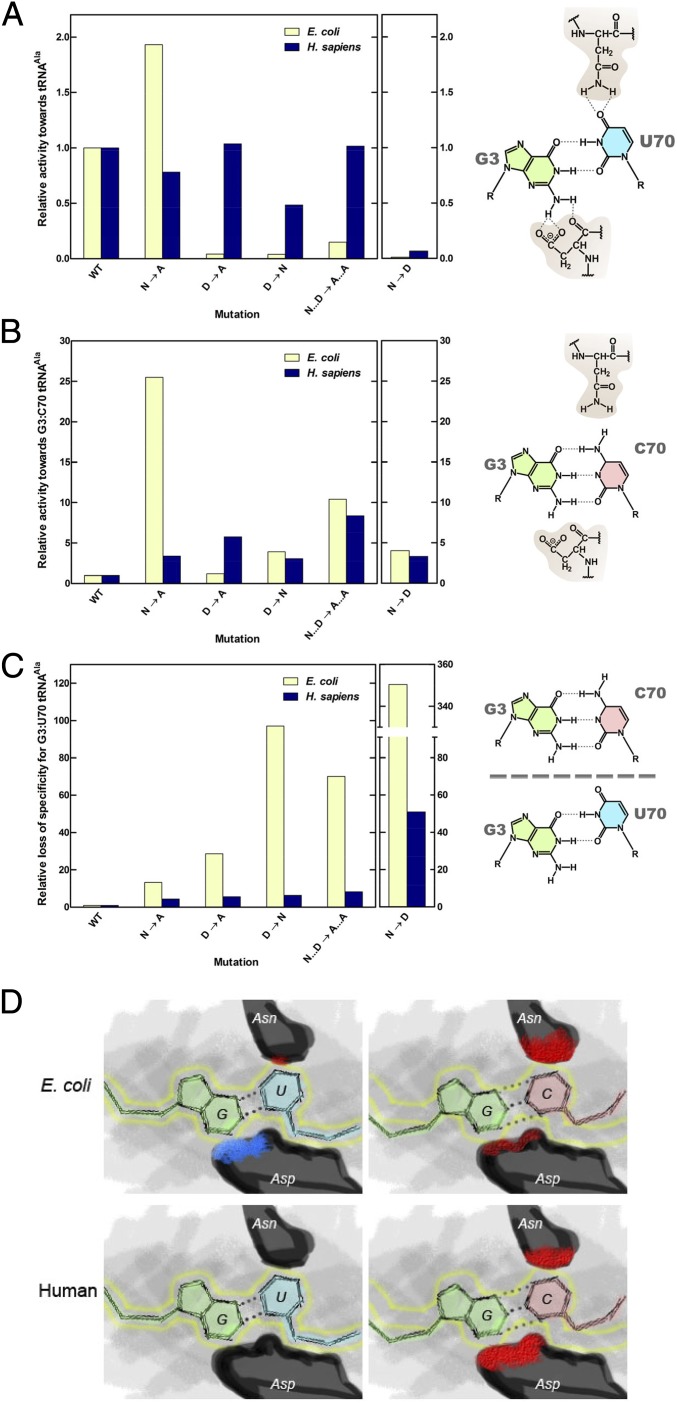
To measure the loss of specificity for G3:U70 for each of the mutations in the E. coli and H. sapiens enzymes, we compared the relative activities of the mutant versus the WT enzymes (Fig. 3 A–C). To compare the relative loss of discriminatory power of the mutant AlaRSs for G3:U70 tRNAAla, the relative activity for each mutant for G3:C70 tRNAAla (Fig. 3B) was divided by the relative activity for G3:U70 tRNAAla (Fig. 3A) (WT AlaRS is normalized to 1). Strikingly, the effect of the mutations on the loss of discriminatory power in every instance is more profound with the bacterial enzyme (Fig. 3C). This result is explained by the greater negative effect of the mutations on aminoacylation of WT G3:U70 tRNAAla by the bacterial AlaRS. Thus, the “direct” system of positive recognition of specific nucleotide determinants in bacteria is more vulnerable to specificity-changing mutations. The human enzyme instead suffers less loss of specificity when N317 or D416 is altered, mainly because some substitutions have little effect on the activity toward the natural G3:U70 tRNAAla. These observations raise the possibility that the human enzyme evolved its “shape-related” specificity mechanism to withstand what would otherwise be the inactivating effects of mutations of the highly conserved N317 and D416.
Fig. 3.
E. coli and human AlaRS enzymes display different methods of ensuring G3:U70 specificity. (A) Relative aminoacylation activities of E. coli and human AlaRS enzymes toward G3:U70 tRNAAla. (B) Relative aminoacylation activities toward G3:C70 tRNAAla. (C) Relative loss of discriminatory power of the mutant AlaRSs for G3:U70 tRNAAla. The loss of specificity is measured by the relative activity for G3:C70 tRNAAla divided by the relative activity for G3:U70 tRNAAla. WT AlaRS is normalized to 1. (D) Representation of the different modes of tRNA specificity by E. coli and human AlaRSs. Blue patches signify positive interactions between enzyme and tRNA, and red patches signify negative interactions. The sizes of the patches are roughly proportional to the strength of the positive or negative interactions. The position of the G:U wobble pair is outlined in yellow.
The G3:U70 recognition modes are distinct in E. coli, H. sapiens (Fig. 3D), and A. fulgidus AlaRSs (9). Particularly significant is the lack of any positive functional interaction in the eukaryal and archaeal systems, such as that provided by the favorable contact between the carboxyl of D416 and the 2-NH2 of G3 in E. coli. The congruency of the conserved Asn-plus-Asp structure in ensuring G3:U70 recognition in all three domains of life, as exemplified in this work, support the early origin of the G:U base pair as an identity element for alanine. The conservation of this G:U pair as an identity element but not with a specific mechanism of its recognition suggests that the G:U pair was fixed early during genetic code evolution and raises the possibility it predated recognition by the protein enzyme. Thus, the genetic code ostensibly had a powerful and persistent preference for associating G:U with alanine.
We speculate that the different recognition modes of the bacterial, archaeal, and human AlaRSs may be a consequence of the effects of domain–domain interactions, rather than of local interactions. E. coli AlaRS can be divided into an N-terminal catalytic fragment and another half (the editing domain and the C-terminal domain), which are both soluble (13–25). In contrast, A. fulgidus AlaRS cannot be similarly divided, probably because of the hydrophobic interface between the catalytic fragment and the editing domain (26). Consequently, the interaction between the catalytic fragment and the editing domain of E. coli AlaRS is probably more flexible than that of A. fulgidus AlaRS and possibly than that of H. sapiens AlaRS. The flexibility of this interface could provide the accommodation of a direct system of recognition for the bacterial enzyme, while its rigidity could enforce a mechanism based on steric fit.
In examining more than 2,500 deposited sequences of AlaRSs from the three kingdoms of life, we found that the common Asn-plus-Asp structure for recognition of G3:U70 is highly conserved (>95%) in all three domains of life (SI Appendix, Table S1). Interestingly, for both the bacterial and human enzyme, replacement of the residue that is proximal to O4 of U70—N303A (E. coli) or N317A (human)—yields an enzyme with robust activity on each respective native tRNAAla substrate (Fig. 3A). However, in both instances, specificity is lost (Fig. 3C). These observations are consistent with the idea that repulsion of the competitor tRNAs was of paramount importance in the selection and retention of the conserved Asn. Similarly, unlike the bacterial enzyme, in the human system the effect of substitution of Asp416 with either Ala or Asn is minimal, and yet those substitutions still result in a decline in specificity and selective pressure to retain the conserved Asp.
Methods Summary
E. coli and H. sapiens AlaRS enzymes were overexpressed in E. coli and purified as previously described (25). Concentrations of purified AlaRS enzymes were measured by active-site titration assays (27). E. coli tRNAAla and G3:C70 tRNAAla were in vitro transcribed using T7 polymerase. Aminoacylation with WT E. coli tRNAAla was performed by incubating purified enzyme (5 nM) in reaction buffer (50 mM Hepes, pH 7.5, 20 mM KCl, 5 mM MgCl2, 1 mM DTT), with 4 mM ATP, 20 μM l-[3H]Ala, and 2 μM tRNAAla at 25 °C. For reactions with G3:C70 tRNAAla, 500 nM enzyme and 10 μM G3:C70 tRNAAla was used, at 37 °C. Incorporation of l-[3H]Ala was measured by scintillation counting in a MicroBeta plate reader (27).
Supplementary Material
Acknowledgments
This work was supported by NIH Grants GM15539 and GM23562 and by a fellowship from the National Foundation for Cancer Research.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1807109115/-/DCSupplemental.
References
- 1.Rich A, RajBhandary UL. Transfer RNA: Molecular structure, sequence, and properties. Annu Rev Biochem. 1976;45:805–860. doi: 10.1146/annurev.bi.45.070176.004105. [DOI] [PubMed] [Google Scholar]
- 2.Hou YM, Schimmel P. A simple structural feature is a major determinant of the identity of a transfer RNA. Nature. 1988;333:140–145. doi: 10.1038/333140a0. [DOI] [PubMed] [Google Scholar]
- 3.McClain WH, Foss K. Changing the identity of a tRNA by introducing a G-U wobble pair near the 3′ acceptor end. Science. 1988;240:793–796. doi: 10.1126/science.2452483. [DOI] [PubMed] [Google Scholar]
- 4.Giegé R. Toward a more complete view of tRNA biology. Nat Struct Mol Biol. 2008;15:1007–1014. doi: 10.1038/nsmb.1498. [DOI] [PubMed] [Google Scholar]
- 5.Hou YM, Schimmel P. Evidence that a major determinant for the identity of a transfer RNA is conserved in evolution. Biochemistry. 1989;28:6800–6804. doi: 10.1021/bi00443a003. [DOI] [PubMed] [Google Scholar]
- 6.Park SJ, Hou YM, Schimmel P. A single base pair affects binding and catalytic parameters in the molecular recognition of a transfer RNA. Biochemistry. 1989;28:2740–2746. doi: 10.1021/bi00432a056. [DOI] [PubMed] [Google Scholar]
- 7.Musier-Forsyth K, et al. Specificity for aminoacylation of an RNA helix: An unpaired, exocyclic amino group in the minor groove. Science. 1991;253:784–786. doi: 10.1126/science.1876835. [DOI] [PubMed] [Google Scholar]
- 8.Musier-Forsyth K, Scaringe S, Usman N, Schimmel P. Enzymatic aminoacylation of single-stranded RNA with an RNA cofactor. Proc Natl Acad Sci USA. 1991;88:209–213. doi: 10.1073/pnas.88.1.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Naganuma M, et al. The selective tRNA aminoacylation mechanism based on a single G•U pair. Nature. 2014;510:507–511. doi: 10.1038/nature13440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beuning PJ, Gulotta M, Musier-Forsyth K. Atomic group “mutagenesis” reveals major groove fine interactions of a tRNA synthetase with an RNA helix. J Am Chem Soc. 1997;119:8397–8402. [Google Scholar]
- 11.Musier-Forsyth K, Schimmel P. Functional contacts of a transfer RNA synthetase with 2′-hydroxyl groups in the RNA minor groove. Nature. 1992;357:513–515. doi: 10.1038/357513a0. [DOI] [PubMed] [Google Scholar]
- 12.Sun L, Song Y, Blocquel D, Yang XL, Schimmel P. Two crystal structures reveal design for repurposing the C-Ala domain of human AlaRS. Proc Natl Acad Sci USA. 2016;113:14300–14305. doi: 10.1073/pnas.1617316113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ibba M, Söll D. Quality control mechanisms during translation. Science. 1999;286:1893–1897. doi: 10.1126/science.286.5446.1893. [DOI] [PubMed] [Google Scholar]
- 14.Ling J, et al. Resampling and editing of mischarged tRNA prior to translation elongation. Mol Cell. 2009;33:654–660. doi: 10.1016/j.molcel.2009.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krauss G, Riesner D, Maass G. Mechanism of discrimination between cognate and non-cognate tRNAs by phenylalanyl-tRNA synthetase from yeast. Eur J Biochem. 1976;68:81–93. doi: 10.1111/j.1432-1033.1976.tb10766.x. [DOI] [PubMed] [Google Scholar]
- 16.Carter CW., Jr Cognition, mechanism, and evolutionary relationships in aminoacyl-tRNA synthetases. Annu Rev Biochem. 1993;62:715–748. doi: 10.1146/annurev.bi.62.070193.003435. [DOI] [PubMed] [Google Scholar]
- 17.Ryckelynck M, Giegé R, Frugier M. Yeast tRNA(Asp) charging accuracy is threatened by the N-terminal extension of aspartyl-tRNA synthetase. J Biol Chem. 2003;278:9683–9690. doi: 10.1074/jbc.M211035200. [DOI] [PubMed] [Google Scholar]
- 18.Fender A, Sissler M, Florentz C, Giegé R. Functional idiosyncrasies of tRNA isoacceptors in cognate and noncognate aminoacylation systems. Biochimie. 2004;86:21–29. doi: 10.1016/j.biochi.2003.11.011. [DOI] [PubMed] [Google Scholar]
- 19.Pabo CO, Sauer RT. Protein-DNA recognition. Annu Rev Biochem. 1984;53:293–321. doi: 10.1146/annurev.bi.53.070184.001453. [DOI] [PubMed] [Google Scholar]
- 20.Ollis DL, While SW. Structural basis of protein-nucleic acid interactions. Chem Rev. 1987;87:981–995. [Google Scholar]
- 21.Patel DJ. Protein-nucleic-acid interaction. A molecular handshake. Nature. 1994;367:688–690. doi: 10.1038/367688a0. [DOI] [PubMed] [Google Scholar]
- 22.Patel DJ. Adaptive recognition in RNA complexes with peptides and protein modules. Curr Opin Struct Biol. 1999;9:74–87. doi: 10.1016/s0959-440x(99)80010-4. [DOI] [PubMed] [Google Scholar]
- 23.Guo M, et al. Paradox of mistranslation of serine for alanine caused by AlaRS recognition dilemma. Nature. 2009;462:808–812. doi: 10.1038/nature08612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beebe K, Mock M, Merriman E, Schimmel P. Distinct domains of tRNA synthetase recognize the same base pair. Nature. 2008;451:90–93. doi: 10.1038/nature06454. [DOI] [PubMed] [Google Scholar]
- 25.Guo M, et al. The C-Ala domain brings together editing and aminoacylation functions on one tRNA. Science. 2009;325:744–747. doi: 10.1126/science.1174343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Naganuma M, Sekine S, Fukunaga R, Yokoyama S. Unique protein architecture of alanyl-tRNA synthetase for aminoacylation, editing, and dimerization. Proc Natl Acad Sci USA. 2009;106:8489–8494. doi: 10.1073/pnas.0901572106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Beebe K, Waas W, Druzina Z, Guo M, Schimmel P. A universal plate format for increased throughput of assays that monitor multiple aminoacyl transfer RNA synthetase activities. Anal Biochem. 2007;368:111–121. doi: 10.1016/j.ab.2007.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.