Significance
Bacteria live in dense environments where competition for space and resources is fierce. For this reason, they often use diffusible toxins to eliminate closely related strains. Some toxins trigger systematic retaliation, raising the question of the role of provocation in bacterial warfare. We combine mathematical modeling and experiments to study the costs and benefits of provocation. In one-to-one encounters, provocation is costly as it leads to strong counterattacks. However, with three or more strains present, provocation can provide benefits via a “divide-and-conquer” effect, whereby a strain forces its opponents to wipe each other out. This effect could be harnessed as a targeted antibacterial approach; adding low levels of certain antibiotics to communities can promote warfare and cross-elimination between strains.
Keywords: provocation, competition, colicin, bacterial communities, social evolution
Abstract
Competition in animals involves a wide variety of aggressive behaviors. One of the most sophisticated strategies for a focal actor is to provoke a competitor into uncontrolled aggression toward other competitors. Like animals, bacteria rely on a broad spectrum of molecular weapons, some of which provoke potential rivals by triggering retaliation. While bacterial provocation is well documented, its potential adaptive value has received little attention. Here, we examine the costs and benefits of provocation using mathematical modeling and experiments with Escherichia coli strains encoding colicin toxins. We show that provocation is typically costly in one-to-one encounters because a provoking strain receives a strong reciprocal attack compared with nonprovoking strains. By contrast, provocation can be strongly beneficial in communities including more than two toxin-producing strains, especially when the provoker is shielded from, or resistant to, its opponents’ toxins. In these scenarios, we demonstrate that the benefit of provocation derives from a “divide-and-conquer” effect by which aggression-provoking toxin producers force their competitors into increased reciprocal aggression, leading to their cross-elimination. Furthermore, we show that this effect can be mimicked by using antibiotics that promote warfare among strains in a bacterial community, highlighting the potential of provocation as an antimicrobial approach.
Animals rely on a wide range of aggressive behaviors to deter their competitors or predators (1, 2). Aggression, however, is not without risk. Individuals that display antagonistic behaviors or are involved in power contests incur a significant energy cost and are at increased risk of injury and death (3–6). Strategic uses of provocation, by which individuals take advantage of the costs and destabilizing effects of aggression on their competitors, are therefore worth considering. Apes and other social animals are indeed known to purposely maintain or exacerbate existing conflicts between two or more of their opponents for their own benefit (7–12). These behaviors are variations on the “divide-and-conquer” strategy, famously delineated by Niccolò Machiavelli in his Art of War (Book VI), by which an individual uses provocation to pit competitors against one another to weaken them (7, 11).
Bacteria are social organisms. Most of them live in dense communities where cooperative and competitive interactions are crucial for their survival (13). Like animals, bacteria display a variety of aggressive behaviors (14). They produce a wide arsenal of molecular weapons, ranging from broad-spectrum antibiotics to strain-specific bacteriocins, comprising both free-floating toxins and membrane-bound poisoned spikes (15). Most bacteria deploy their weapons conditionally upon detection of cellular damage or physiological stress, which are often direct or indirect cues of competition (16, 17). Interestingly, some bacterial strains are known to release toxins that exacerbate toxin production in conspecifics, making systematic use of provocation (18, 19). The evolutionary rationale of such a behavior, if any, is not understood. Indeed, to date, experimental data and modeling have suggested that provocation should be costly and selected against (19).
Here we examine the costs and benefits of provocation in competitions between colicin-producing Escherichia coli strains. Colicins are a family of prototypic diffusible bacterial toxins, which target essential components of the cell, like cellular membranes, protein synthesis, or nucleic acids (20). Under normal growth conditions, the production of colicins in bacterial colonies is mostly stochastic, and it has been shown to happen in 0.1–3% of the population (SI Appendix, Fig. S1) (21–23). However, these frequencies increase dramatically when the cells are exposed to DNA damage, a common mechanism of bacterial interference and an indirect cue for increased competition in biofilms or within a host (15, 17, 21, 24, 25). Conversely, a subset of colicins kills by degrading DNA, which naturally increases toxin production in all exposed colicin-encoding competitors and singles them out as natural aggression-provoking agents (SI Appendix, Fig. S1) (18, 19, 23). Using a colony model, we examine the evolutionary significance of this form of warfare in communities.
Consistent with previous work, we find that strains using systematic provocation are generally at a disadvantage in one-to-one contests. However, within more complex communities, we show that they can provoke colicinogenic competitors into increased reciprocal aggression leading to their cross-elimination. We examine the conditions under which this form of divide-and-conquer warfare is advantageous, in which ecological scenarios it could be used in nature, and, more generally, the consequences of increased aggression in bacterial communities. Finally, we show that the addition of exogenous antibiotics can also provoke bacterial warfare. This suggests that many medical or dietary interventions are likely to modify aggression levels in human-associated microbial populations in a manner that might be harnessed to help eliminate pathogens.
Results
DNA-Degrading Toxins Increase Toxin-Mediated Aggression in Competitors.
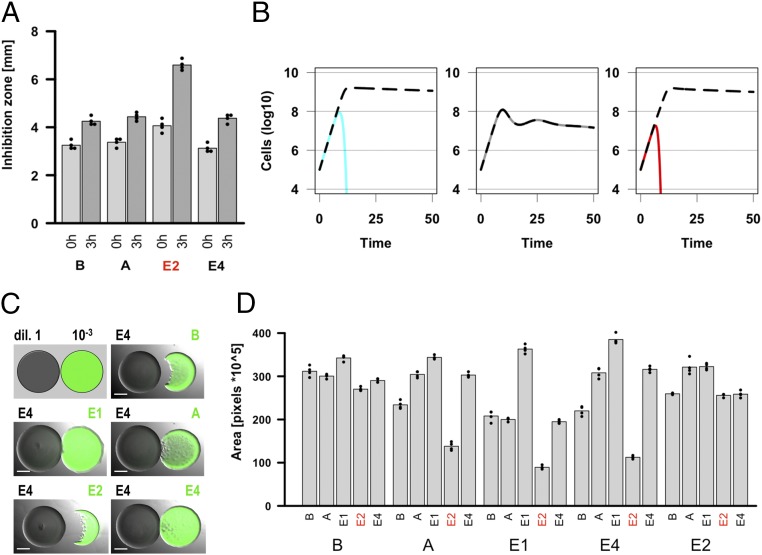
Colicinogenic E. coli strains, like many toxin-producing Proteobacteria, respond to DNA damage by increasing their investment in toxin production. In agreement with this, strains producing colicins with DNase activity (DNases) have been found to enhance toxin production in competing colicinogenic populations (18, 19). We first sought to confirm that this response happens consistently and is specific to DNases. We exposed a population of bacterial cells containing a plasmid reporting on a colicin promoter to spent medium from colicinogenic strains producing representative toxins with DNase, RNase, or pore-forming activity. An overview of the colicinogenic strains used in this study can be found in Table 1; throughout this study “B” is used to indicate our sensitive noncolicinogenic strain. We found that only the toxin with DNase activity was able to induce the production of the reporter protein (SI Appendix, Fig. S2A). To confirm that an actual colicinogenic strain would produce an excess of toxin in response to DNases, we exposed a colicinogenic strain to the same set of spent media and measured the toxicity of cell extracts from the resulting cultures. We found that, when the DNase, but not colicins of other activities, was used, the overall toxicity of the cell extract increased compared with the control, indicating that colicin had been synthesized upon DNase exposure (Fig. 1A). We confirmed that induction of DNA damage was essential to this effect by showing that a strain producing a catalytically inactive DNase toxin was unable to trigger the competitor’s response and that an exogenous DNA-damaging agent could mimic the effect of a DNase (SI Appendix, Fig. S2B). Furthermore, two additional DNase colicins gave similar results, suggesting that the ability to trigger retaliation is a general feature of DNase toxins (SI Appendix, Fig. S2B). From all of the above we conclude that DNases, in addition to their toxic activity, can function as aggression-provoking agents, exacerbating toxin production in exposed SOS-regulated colicin-encoding competitors, whereas colicins with other activities do not.
Table 1.
Overview of the colicin strains used in this study
| Strain | Abbreviation | Toxicity | Provocation | Resistance |
| BZB1011 | B | None | No | No |
| BZB1011 pColA | A | Pore-forming | No | No |
| BZB1011 pColE1 | E1 | Pore-forming | No | No |
| BZB1011 pColE2 | E2 | DNase | Yes | No |
| BZB1011 pColE4 | E4 | RNase | No | No |
| BZB1011 pColE7 | E7 | DNase | Yes | No |
| BZB1011 pColE8 | E8 | DNase | Yes | No |
| BZB1011 (btuB) | BR | None | No | Yes |
| BZB1011 (btuB) pColA | AR | Pore-forming | No | Yes |
| BZB1011 (btuB) pColE2 | E2R | DNase | Yes | Yes |
The “Abbreviation” column refers to the labeling of the strains used in the figures. The “Resistance” column indicates whether or not the strain is resistant to BtuB-binding colicins.
Fig. 1.
Aggression-provoking strains are at a disadvantage in one-to-one competitions against other toxin-producing competitors. (A) E. coli strains increase their toxin production when exposed to colicins with DNase, but not other, activities. Cultures of a colicin-E1–producing strain were exposed to spent media from four strains for 3 h. Extracts of cells were then tested for toxicity using a growth inhibition assay. (B) Strains producing aggression-provoking toxins are at a disadvantage in one-to-one competitions with other toxin-secreting strains. The graphs show the dynamics of the modeled population in pairwise encounters between strains of three types: sensitive nonproducer (cyan), nonprovoking toxin producer (black or dark gray), and aggression-provoking toxin producer (red). Against a nonprovoking toxin producer, the aggression-provoking strain (Right) decreases faster than the sensitive strain (Left). In all ODE models in this study, the maximal growth, lysis, and toxin production rates, as well as the general toxin characteristics, are identical across strains. (C and D) E. coli strains producing colicins with DNase activity are at a disadvantage when competing one-to-one against other colicin-producing strains. (C) Pairs of colicinogenic strains were spotted onto LB agar, each at a different dilution (1 vs. 10−3, respectively), and grown overnight. (D) The area occupied by the spot of the more dilute, fluorescent strain was recorded. Large and small letters on the graph correspond to the concentrated and dilute spot, respectively. For strain annotation, see Table 1; in all panels, “B” stands for BZB1011, a sensitive noncolicinogenic strain. Statistical tests can be found in SI Appendix. (Scale bars: 2,000 μm.)
Strains Provoking Their Competitors into Increased Aggression Are at a Disadvantage in One-to-One Competitions.
Systematic provocation is known to be a risky strategy in animals (4, 5). Based on this, we investigated the impact of aggression-provoking toxins on the fitness of the producing strain in pairwise competitions with other strains. We built an ordinary-differential-equations (ODE)–based model of toxin-mediated competition between growing populations with finite resources. To confirm that the results were applicable to spatially structured environments, we also built a model of toxin-based competition in space (SI Appendix, Fig. S3). Both models predict that, in a competition between two strains secreting either aggression-provoking or nonprovoking toxins of equivalent toxicities and diffusion properties, the latter would always win over the former (Fig. 1B and SI Appendix, Figs. S4 and S5). In accordance with these results, we found that, in an experimental setup where two bacterial colonies compete for space (SI Appendix, Fig. S6), a DNase producer had a strong competitive disadvantage against colicinogenic competitors compared with pore-forming and RNase toxin producers (Fig. 1 C and D). By contrast, when the toxin-producing competitor could not uptake colicins, the provoking-toxin producer had no significant disadvantage compared to the nonprovoking-toxin producer (SI Appendix, Fig. S7). In addition, this effect did not seem to be dependent on colicin toxicity since our model DNase (E2) was found to have comparable, or only slightly lower, toxicity levels than its colicinogenic competitors in all of the scenarios that we tested (SI Appendix, Fig. S8). In line with this, the E2-producing strain performed much worse than the sensitive nonproducer (B) in all competitions with colicinogenic strains, confirming that growth had a negligible impact on the results (Fig. 1D and SI Appendix, Fig. S9). Similar results were obtained with a second, much weaker DNase, colicin E7 (SI Appendix, Fig. S10). We therefore conclude that toxin-producing bacteria that provoke competing strains into increased aggression are at a disadvantage in one-to-one encounters.
Strains Forcing Their Competitors into Increased Aggression Benefit When Spatially Shielded from or Resistant to Foreign Toxins.
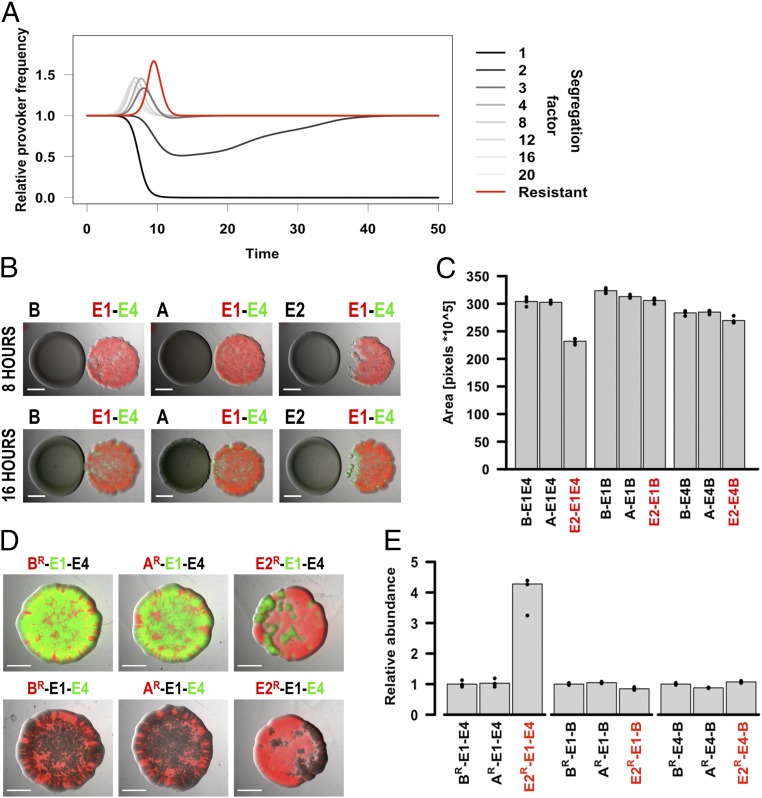
We next asked whether an aggression-provoking toxin producer could benefit from provocation in bacterial communities comprising more than two strains. Under these conditions, both our ODE and spatial models predict that aggression-provoking strains are again at a disadvantage when the toxins from all strains in the community have equal chances to reach each other (for the ODE model, see SI Appendix, Fig. S18 B and C; for the spatial model, see SI Appendix, Fig. S11). However, in cases where the probability P for the toxins of a focal strain (either an aggression-provoking or a nonprovoking strain) to reach the other competing strains is skewed in its favor (i.e., when the focal strain is shielded from its competitors’ toxins and vice versa), a provoking strain has a stronger advantage in actively growing communities than a nonprovoking strain for most values of P (Fig. 2A and SI Appendix, Figs. S12 and S13). This is because, in this case, the two competitors have a higher probability to reach each other with their toxins than the focal strain. At the same time, the increased aggression caused by provocation leads to a much quicker cross-elimination of the two adjacent competitors. We found that this effect is further enhanced when the focal strain is resistant to its competitors’ toxins; in this case, the provoker always has an advantage compared with the nonprovoker (Fig. 2A and SI Appendix, Figs. S14 and S15).
Fig. 2.
Aggression-provoking strains benefit from provocation, provided they are shielded from or resistant to their competitors’ toxins. (A) We modeled competitions of three toxin-producing strains, including a focal strain segregated from its competitors or resistant to their toxins. Segregation means that the probability for the focal strain to reach or be reached by its competitors is lower by a factor u (segregation factor) compared with the probability of its two competitors to reach each other. The graph shows the ratio between the frequency of a focal provoker relative to a focal nonprovoker against the same competitors as a function of time for different values of u. For u > 2, an aggression-provoking strain benefits compared with a nonprovoker (gray lines). A resistant provoker has an even stronger advantage (red line). We note that the differences between provoker and nonprovoker appear for a limited time frame, after the toxins reach relevant concentrations and before the community stops growing because of nutrient exhaustion (SI Appendix, Fig. S12). (B and C) When segregated and opposed to mixed colonies of two colicin-producing strains, an E. coli strain producing a DNase colicin (E2) has a strong advantage compared with a strain producing a non-DNase colicin (A). (B) An undiluted focal strain was spotted onto LB agar next to a mixed spot of two other colicin producers, diluted 1,000-fold, and grown overnight. (C) The area occupied by the mixed spot was measured. For each focal strain (B, A, or E2), in the absence of interaction, the area occupied by the mixed colicin producers (E1E4) is expected to be no lower than the lowest of the values for single competitors (E1B or E4B). The disproportionate decrease in the case of E2 is evidence for provocation. (D and E) In communities of three toxin producers, a colicin-resistant E. coli strain producing a DNase colicin (E2) has an overwhelming advantage compared with a resistant strain producing a non-DNase colicin (A). (D) A mixture of three strains, a focal resistant strain labeled with red fluorescent protein (RFP) and each of its competitors alternatively labeled with GFP or unlabeled, was spotted onto LB agar and grown overnight. (E) The fluorescence intensity of each strain was recorded. The graph shows the ratios between the focal strain (BR, AR, and E2R) and both of its competitors (E1 and E4), normalized to the focal nonproducer (BR). For strain annotation, see Table 1; in all panels “B” stands for BZB1011, a sensitive noncolicinogenic strain. Statistical tests can be found in the SI Appendix. (Scale bars: 2,000 μm.)
We sought to test whether a DNase-producing strain would benefit in experimental setups reproducing the conditions of the two modeled scenarios, the biased toxin exposure and the resistant provoker. To create conditions corresponding to the first scenario, we spotted a focal DNase or pore-forming colicin producer next to a spot containing an equal ratio of two competitors (an RNase producer and a second pore-forming toxin producer). In this setting, the competitors are on average closer to each other than to the focal strain, which agrees with the mathematical model shown in Fig. 2A. We could observe that the focal provoking strain had a notable advantage compared with the focal nonprovoking strain (Fig. 2 B and C), even though their toxicities were similar (SI Appendix, Fig. S8). Interestingly, in some cases, the competitive balance between the two mixed strains shifted exactly at the interface of the competition with the aggression-provoking strain. This indicates that the local increase in aggression levels also has an impact on the relative fitness of the competing strains (Fig. 2B, 16 h).
To investigate the second scenario, we mixed equal amounts of colicin-resistant colicinogenic strains (btuB mutants, Table 1) with two other sensitive competitors in a single colony. We observed that the resistant DNase producer had an overwhelming advantage only when both its competitors were colicinogenic strains (SI Appendix, Fig. S16), in which case it was able to completely outgrow them (Fig. 2 D and E). This occurred because, in the presence of the resistant DNase producer, the competitor strains were provoked into increasing their toxin production, reciprocally killing each other without harming the resistant focal strain. By contrast, neither the resistant nonproducer nor the resistant pore-forming toxin producer seemed to have a significant growth advantage over their competitors despite their resistance to colicins (SI Appendix, Fig. S17).
We conclude that strains producing aggression-provoking toxins can have a distinct selective advantage over their toxin-producing competitors when they are either spatially protected from or resistant to their competitors’ toxins. Under such conditions, these strains can exacerbate the aggression levels of nearby competitors and induce them to kill each other, which then allows them to occupy the cleared space. Such scenarios are analogous to conflicts among social animals or humans in which divide-and-conquer strategies are used.
Chemical Manipulation of Bacterial Aggression Levels Can Decrease Productivity and Change Competition Outcomes in Communities.
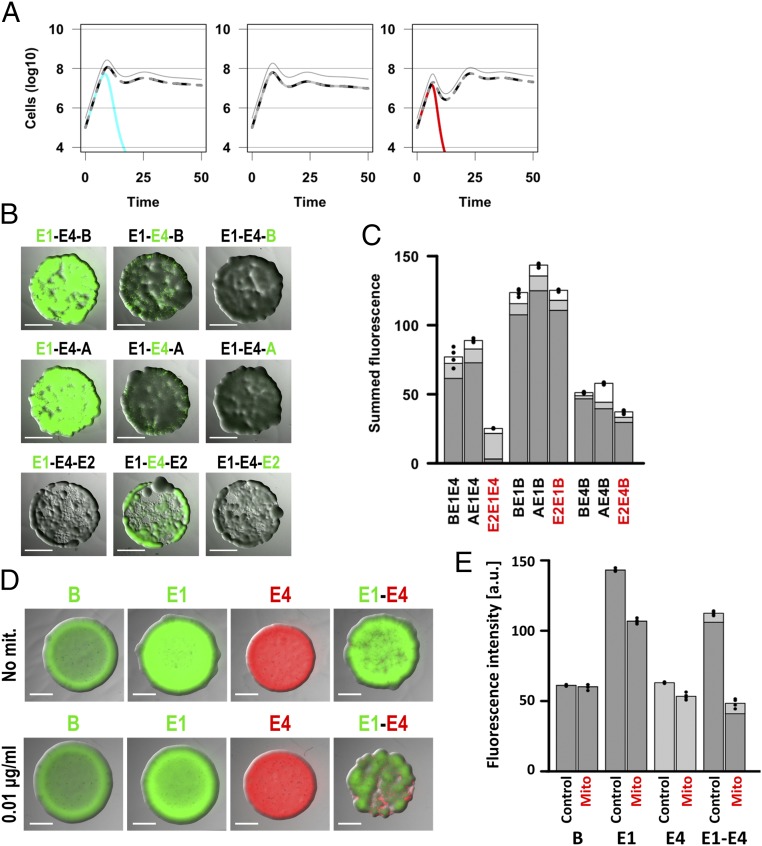
Provocation, whether beneficial to the provoking strain or not, has a significant impact on the overall dynamics of complex bacterial communities. Both our mathematical models predict that, provided that one strain does not dominate entirely, the productivity of three-strain communities is much worse if an aggression-provoking toxin producer and at least a second toxin producer are included in the community (Fig. 3A and SI Appendix, Figs. S18 and S19). We tested this prediction in our experimental system by spotting mixtures of three different colicin-producing and nonproducing strains, each marked in turn with GFP. We found that, as predicted, productivity was worst when the mixture included a DNase producer and two other colicinogenic strains (Fig. 3 B and C). In addition, we could observe important shifts in competition outcomes when an aggression-provoking strain was present in the mixture. In Fig. 3B, the colicin-E4–producing strain dominates in the presence of a DNase while the colicin-E1–producing strain dominates in its absence. This shows that DNA-damaging toxins strongly affect and often cripple the growth of entire bacterial communities because of their aggression-provoking properties.
Fig. 3.
Increase in aggression has a strong impact on the productivity and composition of bacterial communities. (A–C) The productivity of three-member communities is hampered when an aggression-provoking strain is present. (A) We modeled the competition of three-strain communities and report the dynamics of each strain (nonproducer in cyan, nonprovoking producers in black and dark or light gray, aggression-provoking producer in red) and of the whole population (thin gray line). While an aggression-provoking strain is frequent in the community (Right), the overall community productivity is heavily impacted (time points 1–15). (B) Equal amounts of three strains, each in turn labeled with GFP, were mixed and spotted onto LB agar. (C) After overnight incubation, the fluorescence intensity for each strain was measured, and intensities were summed up for each community (strains B, A, or E2 in white; first and second competitors in dark and light gray, respectively). (D and E) DNA-damaging chemicals have a similar effect on bacterial communities as aggression-provoking toxins. (D) Equal amounts of colicin-E1– and colicin-E4–producing strains (GFP- and RFP-labeled, respectively) were mixed and spotted, together with controls, onto LB agar with or without added mitomycin C at a final concentration of 0.01 μg ml−1. After overnight incubation, the intensity of the fluorescence was measured. (E) The summed RFP and GFP fluorescence values are plotted. For strain annotation see Table 1; in all panels “B” stands for BZB1011, a sensitive non colicinogenic strain. Statistical tests can be found in the SI Appendix. (Scale bars: 2000 μm.)
We reasoned that such effects should not be specific to DNA-damaging colicins but should also arise with the use of any DNA-damaging agent eliciting a sustained LexA-mediated response (SOS response). To test this, we grew two-member mixed communities of colicin-producing strains on subinhibitory concentrations of mitomycin C, well known for its capacity to trigger an SOS response and colicin production (26) (Fig. 3 D and E). We used 0.01 μg/mL of mitomycin C, while we found that the minimal inhibitory concentration of this chemical for our background strain was 2 μg/mL (SI Appendix, Fig. S20). The overall productivity of the community decreased much more in the presence of mitomycin C in mixtures including two reciprocally sensitive toxin producers than in the relevant pure strain controls (Fig. 3 D and E). This was also found to be true for two other pairs of colicinogenic strains although the strength of the effect differed (SI Appendix, Fig. S21). In addition, a significant difference in the relative abundance of each of the two strains was observed in two of the three pairs (Fig. 3D and SI Appendix, Fig. S21A), indicating again that not only the overall productivity, but also the competition outcome in the mixed community was altered when the aggression levels increased.
Discussion
When sensing physiological stress or direct competition cues like DNA damage, bacteria often react by increasing their investment in defense mechanisms. Here we study strains producing DNA-damaging toxins that exacerbate the aggressive responses of competitors before killing them. We use this model system to elucidate the adaptive determinants, positive or negative, of bacterial provocation. We find that strains that release aggression-provoking toxins are at a clear disadvantage when they compete pairwise with other toxin-producing competitors because they enhance their competitors’ toxin production without being able to adjust their own toxin synthesis accordingly (Fig. 1). On the other hand, when growing in communities comprising more than two colicinogenic strains—and if they are shielded from or resistant to their competitors’ toxins—provokers can overcome their competitors by inducing them to eradicate each other in a way reminiscent of divide-and-conquer schemes (Fig. 2). Finally, because they induce an overall increase in aggression, provoking toxin producers have a negative impact on the productivity of the communities in which they grow (Fig. 3).
Our experimental system and mathematical models converge in demonstrating that provocation significantly affects the competition of provokers (Figs. 1 and 2) and the composition of bacterial communities (Fig. 3). The only requirement for this to happen is that two or more toxin producers, including a provoker, interact. This condition is likely to be realized often in nature. Existing literature and our bioinformatic analyses show that several types of toxin producers do coexist in the microbiota of murine and human populations (27–29) (Dataset S1). Moreover, through analyzing metagenomic data from the FijiCOMP project, we found that in more than 50% of individual human microbiota where colicin genes were identified, two or more colicin genes were present (30) (Dataset S2). This agrees with models of the evolution of toxin-producing metacommunities, which predict the coexistence of multiple toxin producers with their resistant counterparts (SI Appendix, Fig. S22) (31, 32). Provocation is therefore expected to have a significant impact in natural communities.
From an evolutionary perspective, we have shown that provocation is a double-edged sword: it is costly in some scenarios and advantageous in others. Whether provocation evolved as a competitive strategy remains an open question. For provocation to have an adaptive value in nature, the scenarios in which it is beneficial must outweigh the ones in which it is detrimental. Our results show that provocation is beneficial to the provoker if two conditions are met: (i) in addition to the focal strain, at least two other toxin-producing strains need to be present and (ii) the focal strain must be either resistant to foreign toxins or asymmetrically exposed to toxins compared with its opponents. Based on the above, the first condition is likely to be met in natural environments. The occurrence of the second condition, however, is more challenging to assess due to our lack of knowledge of the small-scale ecology and evolution of bacterial communities. Asymmetric toxin exposure should occur in biofilms or colonies, which are known for their strong spatial structure, provided that segregation patterns similar to the ones in Fig. 2C arise (33–35). Toxin resistance, on the other hand, is frequently observed in the laboratory and in natural communities. We, and others, readily see the emergence of cells resistant to colicins both in noncolicinogenic and colicinogenic laboratory populations, often driven by insertion sequences in the toxin receptor (20) (SI Appendix, Fig. S23). Resistant strains have also been observed in the gut; in mice-associated E. coli populations, phenotypic resistance to one or more colicins has been shown to be widespread (27). Consistent with this, about 0.7% of sequenced E. coli strains (National Center for Biotechnology Information database) lack a functional btuB gene, making them potentially resistant to all BtuB-binding colicins (Dataset S3), and in about 7% of E. coli genomes, the btuB gene is very divergent (SI Appendix, Fig. S24 and Dataset S3). We tested one of these divergent alleles and found that it imparted a degree of resistance to at least one colicin (SI Appendix, Fig. S25). This suggests that partial colicin resistance can be associated also with functional receptors in nature, which would lower the cost of resistance and promote the evolution of provocation. Finally, assuming the same rate of resistance emergence, we find that provoking strains would benefit more than nonprovoking strains since they have a competitive advantage in communities comprising more than two colicinogenic strains and do not pay additional costs in other environments (Fig. 2A and SI Appendix, Fig. S26). In sum, conditions where provocation is beneficial seem likely to occur in natural systems, although we cannot comment on their frequency. Nonetheless, phases of coexistence of multiple toxin-producing strains and their resistant counterparts, like the ones predicted by published models, could provide favorable selection conditions for resistant provoker strains and warrant the maintenance of the provocation trait in the population (SI Appendix, Fig. S22) (31, 32).
In natural communities, bacteria commonly keep each other in check through competitive interactions (17). Our results indicate that aggression levels, and in particular the rate of production of natural antimicrobials, can influence the productivity of communities (Fig. 3 A–C). Moreover, we show that aggression levels can be manipulated through chemical agents that interfere with the regulation of bacterial defense mechanisms (Fig. 3 D and E). We suggest that exacerbating aggression in nonclonal bacterial communities, especially biofilms found in industrial settings or in specific chronic infections, could help to lower microbial load and undermine the evolution of resistance (36–38). This could be done using either chemical agents like DNA-damaging antibiotics at subinhibitory concentrations or strains engineered to secrete aggression-provoking toxins that would function akin to a Trojan horse. At any rate, we hope that this research will draw the attention of microbiologists and clinicians to the possibility of manipulating microbial aggressive behaviors and its potential as an antimicrobial intervention.
Materials and Methods
A detailed description of the methods used for modeling, bioinformatics analyses, genetics, microbiological assays, and data analysis can be found in SI Appendix. Unless otherwise indicated, E. coli strains were grown overnight in 5 mL lysogeny broth (LB) broth in 50-mL polypropylene tubes at 37 °C with agitation (220 rpm). For competition experiments, cells were harvested from overnight cultures, washed twice with LB broth, and normalized in LB to an optical density at 600 nm (OD600) of 1.2. Competitions and mitomycin C experiments were carried out on 0.8% wt/vol LB agar medium, while 1.5% wt/vol LB agar medium was used for determination of colicin toxicity and genetics. Unless otherwise stated, LB agar plates were incubated statically at 37 °C for 16 h. Imaging was performed with a Leica M205FA fluorescent stereomicroscope (Leica Microsystems) using a 10× (N.A. 0.5) dry objective and the associated LAS X software. Images were processed using FIJI/ImageJ software (https://fiji.sc/).
Supplementary Material
Acknowledgments
We thank B. Kerr for the colicin A reporter plasmid and the wild-type colicinogenic strains and N. Craig for pGRG25; S. Mostowy for use of his Leica M205FA microscope; A. Willis and V. Torraca for their help with the imaging; R. C. D. Furniss for his insightful comments on the manuscript; and S. Mitri for her help with the ODE model. We express our gratitude to Ph. Engel and his group for hosting D.G. in difficult times. This study was funded by Medical Research Council Career Development Award MR/M009505/1 (to D.A.I.M.) and Swiss National Science Foundation Postdoc Mobility Fellowships P2LAP3_155109 and P300PA_167703 (to D.G.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1801028115/-/DCSupplemental.
References
- 1.Lorenz K. 1974. On Aggression (Harcourt Brace Jovanovich, New York), p xiv, 306 p.
- 2.Hammerstein P. What theoretical biology has to say on aggression in humans and animals. In: Kortüm HH, Heinze JR, editors. Aggression in Humans and Other Primates: Biology, Psychology, Sociology. de Gruyter; Berlin: 2013. pp. 23–39. [Google Scholar]
- 3.Maynard Smith J, Price GR. The logic of animal conflict. Nature. 1973;246:15–18. [Google Scholar]
- 4.Dugatkin LA. Tendency to inspect predators predicts mortality risk in the guppy, Poecilia reticulata. Behav Ecol. 1992;3:124–128. [Google Scholar]
- 5.Sloan Wilson D, Clark AB, Coleman K, Dearstyne T. Shyness and boldness in humans and other animals. Trends Ecol Evol. 1994;9:442–446. doi: 10.1016/0169-5347(94)90134-1. [DOI] [PubMed] [Google Scholar]
- 6.Georgiev AV, Klimczuk ACE, Traficonte DM, Maestripieri D. When violence pays: A cost-benefit analysis of aggressive behavior in animals and humans. Evol Psychol. 2013;11:678–699. doi: 10.1177/147470491301100313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Almeida RM, Cabral JC, Narvaes R. Behavioural, hormonal and neurobiological mechanisms of aggressive behaviour in human and nonhuman primates. Physiol Behav. 2015;143:121–135. doi: 10.1016/j.physbeh.2015.02.053. [DOI] [PubMed] [Google Scholar]
- 8.Ramirez JM, Andreu JM. Aggression’s typologies. Rev Int Psychol Soc. 2003;3:125–141. [Google Scholar]
- 9.Posner EA, Spier KE, Vermeule A. Divide and conquer. Journal of Legal Analysis. 2010;2:417–471. [Google Scholar]
- 10.Case CR, Maner JK. Divide and conquer: When and why leaders undermine the cohesive fabric of their group. J Pers Soc Psychol. 2014;107:1033–1050. doi: 10.1037/a0038201. [DOI] [PubMed] [Google Scholar]
- 11.de Waal F. Chimpanzee Politics. Power and Sex Among Apes. Jonathan Cape; London: 1982. [Google Scholar]
- 12.Massen JJ, Szipl G, Spreafico M, Bugnyar T. Ravens intervene in others’ bonding attempts. Curr Biol. 2014;24:2733–2736. doi: 10.1016/j.cub.2014.09.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nadell CD, Xavier JB, Foster KR. The sociobiology of biofilms. FEMS Microbiol Rev. 2009;33:206–224. doi: 10.1111/j.1574-6976.2008.00150.x. [DOI] [PubMed] [Google Scholar]
- 14.Hibbing ME, Fuqua C, Parsek MR, Peterson SB. Bacterial competition: Surviving and thriving in the microbial jungle. Nat Rev Microbiol. 2010;8:15–25. doi: 10.1038/nrmicro2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jamet A, Nassif X. New players in the toxin field: Polymorphic toxin systems in bacteria. MBio. 2015;6:e00285-15. doi: 10.1128/mBio.00285-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ghazaryan L, Tonoyan L, Ashhab AA, Soares MI, Gillor O. The role of stress in colicin regulation. Arch Microbiol. 2014;196:753–764. doi: 10.1007/s00203-014-1017-8. [DOI] [PubMed] [Google Scholar]
- 17.Cornforth DM, Foster KR. Competition sensing: The social side of bacterial stress responses. Nat Rev Microbiol. 2013;11:285–293. doi: 10.1038/nrmicro2977. [DOI] [PubMed] [Google Scholar]
- 18.Majeed H, Gillor O, Kerr B, Riley MA. Competitive interactions in Escherichia coli populations: The role of bacteriocins. ISME J. 2011;5:71–81. doi: 10.1038/ismej.2010.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Majeed H, Lampert A, Ghazaryan L, Gillor O. The weak shall inherit: Bacteriocin-mediated interactions in bacterial populations. PLoS One. 2013;8:e63837. doi: 10.1371/journal.pone.0063837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cascales E, et al. Colicin biology. Microbiol Mol Biol Rev. 2007;71:158–229. doi: 10.1128/MMBR.00036-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gillor O, Vriezen JA, Riley MA. The role of SOS boxes in enteric bacteriocin regulation. Microbiology. 2008;154:1783–1792. doi: 10.1099/mic.0.2007/016139-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ghazaryan L, Soares MI, Gillor O. Auto-regulation of DNA degrading bacteriocins: Molecular and ecological aspects. Antonie Van Leeuwenhoek. 2014;105:823–834. doi: 10.1007/s10482-014-0136-1. [DOI] [PubMed] [Google Scholar]
- 23.Mavridou DAI, Gonzalez D, Kim W, West SA, Foster KR. Bacteria use collective behavior to generate diverse combat strategies. Curr Biol. 2018;28:345–355.e4. doi: 10.1016/j.cub.2017.12.030. [DOI] [PubMed] [Google Scholar]
- 24.Boles BR, Singh PK. Endogenous oxidative stress produces diversity and adaptability in biofilm communities. Proc Natl Acad Sci USA. 2008;105:12503–12508. doi: 10.1073/pnas.0801499105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jones RM, Mercante JW, Neish AS. Reactive oxygen production induced by the gut microbiota: Pharmacotherapeutic implications. Curr Med Chem. 2012;19:1519–1529. doi: 10.2174/092986712799828283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hardy KG, Meynell GG. Colicin factors and mitomycin-C. J Gen Microbiol. 1972;73:547–549. doi: 10.1099/00221287-73-3-547. [DOI] [PubMed] [Google Scholar]
- 27.Gordon DM, Riley MA, Pinou T. Temporal changes in the frequency of colicinogeny in Escherichia coli from house mice. Microbiology. 1998;144:2233–2240. doi: 10.1099/00221287-144-8-2233. [DOI] [PubMed] [Google Scholar]
- 28.Gordon DM, Riley MA. A theoretical and empirical investigation of the invasion dynamics of colicinogeny. Microbiology. 1999;145:655–661. doi: 10.1099/13500872-145-3-655. [DOI] [PubMed] [Google Scholar]
- 29.Riley MA, Gordon DM. The ecological role of bacteriocins in bacterial competition. Trends Microbiol. 1999;7:129–133. doi: 10.1016/s0966-842x(99)01459-6. [DOI] [PubMed] [Google Scholar]
- 30.Brito IL, et al. Mobile genes in the human microbiome are structured from global to individual scales. Nature. 2016;535:435–439. doi: 10.1038/nature18927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Biernaskie JM, Gardner A, West SA. Multicoloured greenbeards, bacteriocin diversity and the rock-paper-scissors game. J Evol Biol. 2013;26:2081–2094. doi: 10.1111/jeb.12222. [DOI] [PubMed] [Google Scholar]
- 32.Kerr B, Riley MA, Feldman MW, Bohannan BJ. Local dispersal promotes biodiversity in a real-life game of rock-paper-scissors. Nature. 2002;418:171–174. doi: 10.1038/nature00823. [DOI] [PubMed] [Google Scholar]
- 33.Nadell CD, Drescher K, Foster KR. Spatial structure, cooperation and competition in biofilms. Nat Rev Microbiol. 2016;14:589–600. doi: 10.1038/nrmicro.2016.84. [DOI] [PubMed] [Google Scholar]
- 34.Stacy A, McNally L, Darch SE, Brown SP, Whiteley M. The biogeography of polymicrobial infection. Nat Rev Microbiol. 2016;14:93–105. doi: 10.1038/nrmicro.2015.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mark Welch JL, Hasegawa Y, McNulty NP, Gordon JI, Borisy GG. Spatial organization of a model 15-member human gut microbiota established in gnotobiotic mice. Proc Natl Acad Sci USA. 2017;114:E9105–E9114. doi: 10.1073/pnas.1711596114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ganesh K, et al. Chronic wound biofilm model. Adv Wound Care (New Rochelle) 2015;4:382–388. doi: 10.1089/wound.2014.0587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wolcott R, Costerton JW, Raoult D, Cutler SJ. The polymicrobial nature of biofilm infection. Clin Microbiol Infect. 2013;19:107–112. doi: 10.1111/j.1469-0691.2012.04001.x. [DOI] [PubMed] [Google Scholar]
- 38.Xu D, Jia R, Li Y, Gu T. Advances in the treatment of problematic industrial biofilms. World J Microbiol Biotechnol. 2017;33:97. doi: 10.1007/s11274-016-2203-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.