Abstract
Amplification of fibroblast growth factor receptor 1 (FGFR1) has been reported in many squamous cell carcinomas, and human papillomavirus (HPV)–related oropharyngeal squamous cell carcinoma has been characterized as a distinct subset with favorable prognosis. Here, we investigated the FGFR1 amplification and HPV status in tonsillar squamous cell carcinoma (TSCC) and analyzed the clinical characteristics. HPV in situ hybridization (HPV ISH) and FGFR1 fluorescence in situ hybridization (FISH) were performed using tissue microarray from 89 cases of TSCC. Fourteen of 89 (15.7%) TSCC cases had FGFR1 amplification, and HPV was detected in 59 of 89 (66.3%) cases. FGFR1 amplification status was not associated with HPV positivity (p=0.765). Outcomes were not significantly different between FGFR1 amplified and non-amplified patients. Although FGFR1 amplified patients (n=4) in the HPV ISH–negative group (n=30) had a tendency for poorer overall survival, no statistical significance was identified (p=0.150, log-rank). FGFR1 protein overexpression showed better disease-free survival (p=0.031, log-rank) in HPV-negative TSCC. This study suggests FGFR1 amplification may be important in the pathogenesis of TSCC regardless of HPV status.
Keywords: fibroblast growth factor receptor 1, fluorescence in situ hybridization, human papillomavirus, squamous cell carcinoma, tonsil
Introduction
Approximately 50,000 new cases of oral cavity and pharyngeal cancer are expected to be diagnosed, and 10,000 related deaths are estimated to occur in the United States.1 Despite enormous cancer treatment efforts, the 5-year survival rate is about 50%.2 The most important risk factors for oral cavity and pharyngeal cancer are tobacco use and alcohol consumption. Human papillomavirus (HPV) infection is an additional independent risk factor in oropharyngeal carcinoma.3 It is well known that HPV-associated head and neck squamous cell carcinoma (HNSCC) shows a good prognosis compared with non-HPV–infected patients.4
Comprehensive molecular profiling for human cancer is a major trend in the area of cancer research, and the identification of genomic alterations for specific malignancies leads to a better understanding of tumor biology and the development of target therapies.5,6 Fibroblast growth factor receptor 1 (FGFR1) is a member of four tyrosine kinase receptor family, encoded by a gene located on chromosome 8p12.1.7 Amplification of FGFR1 has been recently reported in lung,8 breast,9 and head and neck squamous cell carcinomas,10 and sensitivity to FGFR1 tyrosine kinase inhibitors in preclinical models and developed candidates of FGFR tyrosine kinase inhibitors have been reported, leading to clinical trials of these agents in FGFR1 amplified tumors.11,12
However, knowledge of the clinical and prognostic significance of FGFR1 amplification in patients with tonsillar squamous cell carcinoma (TSCC) and the relationship between HPV status and FGFR1 amplified TSCC is limited. Thus, in the present study, we investigated the prognostic significance of FGFR1 amplified TSCC and analyzed the association between HPV status and FGFR1 amplification.
Patients and Methods
Patients
This study was conducted in a cohort of patients with TSCC who underwent surgery and neoadjuvant and/or postoperative adjuvant chemoradiotherapy between January 2000 and December 2010 at Asan Medical Center, Seoul, Korea. There were 280 patients with squamous cell carcinoma diagnosed in tonsil. The exclusion criteria were patients without tissue block (n=61), patients performed only punch or needle biopsy (n=120), and patients having history of squamous cell carcinoma from larynx, tongue, esophagus, lung, and so on, previously or synchronously (n=10). The criteria used for patient selection included the availability of tumor tissue from primary TSCCs as well as data on smoking status and survival. These were summarized in Fig. 1. This study cohort was previously published by Lee et al.13 The study was approved by the Institutional Review Board of Asan Medical Center. Smoking history was measured in pack-years and classified into three categories: nonsmoker, former smoker who quit smoking more than 10 years ago, or current smoker. Alcohol consumption was defined as having no history of alcohol use, having three or less drinks per day, or having more than three drinks per day. Tumor samples were available for 89 patients. The patient population included 81 (91.0%) males and 8 (9.0%) females with a median age of 55 years. Twenty-eight patients (31.5%) were never smokers, and 53 (59.6%) were current smokers. Follow-up periods ranged from 7 to 135 months with a median follow-up of 55 months. The recurrence rate was 15.7% (14/89).
Figure 1.
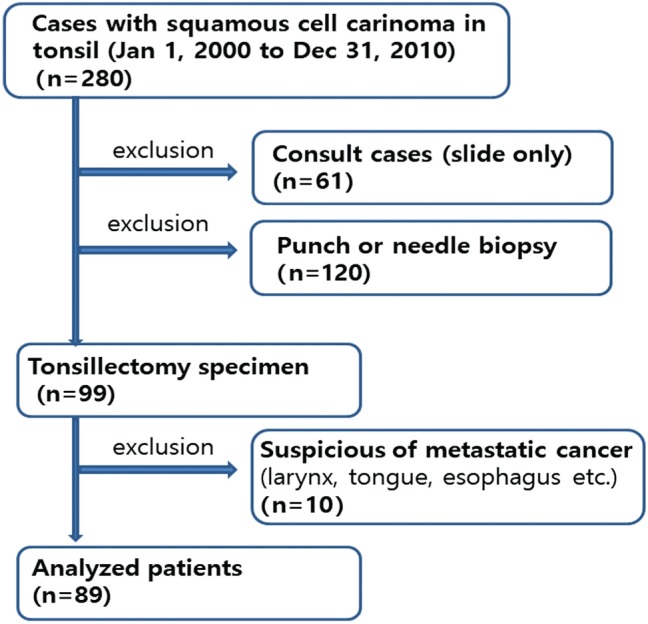
Study cohort flow diagram.
Tissue Microarray Construction and Immunohistochemistry
We used the previously published tissue microarray (TMA).13 Briefly, TMAs were constructed from 2-mm cores of representative tumor areas from paraffin-embedded blocks, in duplicate. TMAs were used in immunohistochemical staining of p16 and FGFR1, fluorescence in situ hybridization (FISH) for FGFR1 gene amplification, and HPV in situ hybridization (HPV ISH).
Immunohistochemistry (IHC) was done with antibodies for p16 (1:10, monoclonal p16INK4; Pharmingen, San Diego, California) and FGFR1 (cat. no. BS5569; 1:500, polyclonal rabbit anti-FGFR1; Bioworld, St Louis Park, Minnesota) using the Ventana NX automated IHC system (Ventana Medical Systems, Tucson, Arizona) with OptiView DAB Detection Kit (Ventana Medical Systems) according to the manufacturer’s instructions and using the reagents supplied with the kit. In brief, sections of 4 μm were mounted on silanized charged slides and allowed to dry for 10 minutes at room temperature, followed by 20 min in an incubator at 65C. After deparaffinization, heat-induced antigen retrieval was performed using pH 6.0 citrate buffer (CC1 Protocol; Ventana Medical Systems), and incubated for 32 min with primary antibodies at room temperature. The slides were counterstained with hematoxylin. As positive control, we used squamous cell carcinoma of the lung for FGFR1 and uterine cervical squamous cell carcinoma for p16. As negative control, we used adenocarcinoma of the lung for FGFR1 and normal lung tissue.
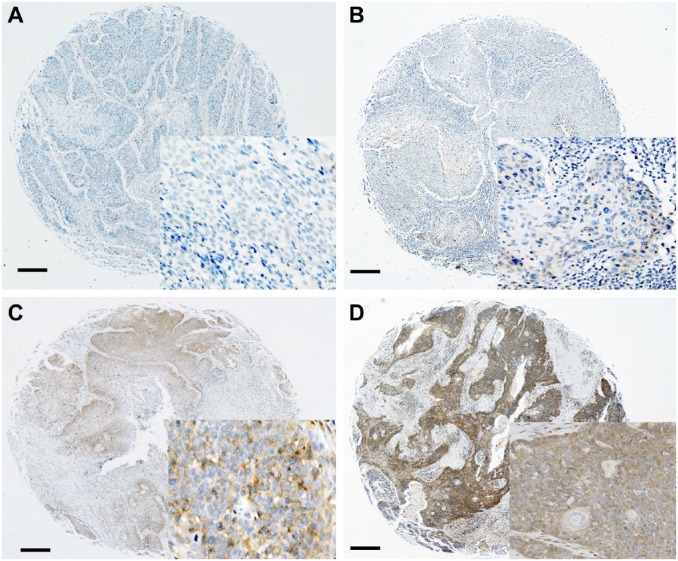
p16INK4 expression was regarded as positive if the nuclei and/or cytoplasm were strongly and diffusely stained in ≥70% of the tumor cells.14 FGFR1 expression was semiquantitatively scored according to cytoplasmic and membranous stain, as described previously15: 0, no expression; 1, ≤10% positive; 2, 10–50% positive; and 3, ≥50% positive. Representative IHC images were shown in Fig. 2. For statistical analysis, the FGFR1 scores were divided into two groups: positive (2 and 3) versus negative (0 and 1).
Figure 2.
Representative images of FGFR1 expression from negative (score 0, A), weakly positive (score 1, B), and intermediately positive (score 2, C) to strongly positive (score 3, D) are shown by immunohistochemistry. Scale bar represents 50 µm. Abbreviation: FGFR1, fibroblast growth factor receptor 1.
FGFR1 Fluorescence In Situ Hybridization
We performed the FISH assay using a commercially available FGFR1 probe (cat. no. LPS018, Aquarius, FGFR1 Breakapart/Amplification probe; Cytocell, Cambridge, United Kingdom). The FGFR1 probe mix consists of a green 267-kb probe telomeric to the FGFR1 gene (8p12) spanning the LETM2 gene and D8S135 marker, and a red probe that is 272 kb centromeric to the FGFR1 gene (8p12) spanning a 108-kb region, including the D8S389 and D8S2317 markers. An accompanying 8-centromere probe (D8Z2, 8p11.1-q11.1) in blue acted as a control for chromosome 8.
FISH analyses were interpreted by two experienced evaluators (J.S.S. and M.L.) who were blinded to the clinical data. At least 40 nuclei per patient were evaluated. As there are currently no standard criteria for the definition of FGFR1 FISH positivity, we defined it as follows: For the FGFR1 gene amplified group, more than 2.2 copies of the gene was considered to be positive. The copy number control ratio and FGFR1 non-amplified group had fewer than 2.2 copies.16
HPV In Situ Hybridization
The Ventana INFORM HPV III Family 16 Probe (B) was used in conjunction with the ISH iView Blue Plus Detection Kit (Ventana Medical Systems). The INFORM HPV III Family 16 Probe (B) detects the following high-risk HPV genotypes: 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, and 66. By light microscopy, any blue nuclear dots in the tumor cells were regarded as positive staining. All cases were classified in a binary manner as either positive or negative.
DNA Extraction
HPV ISH–negative, p16-positive cases were retested by HPV DNA chip. We obtained the blocks for HPV genotyping with the same samples previously used for TMA. DNA was extracted from formalin-fixed paraffin-embedded tissue using a LaboPass Tissue Mini DNA Purification Kit (Cosmo Genetech, Seoul, Korea). Paraffin-embedded tumor tissues were cut into 20-µm-thick sections, using disposable microtome blades, and three consequent sections were collected using microcentrifuge tubes. Then, two extractions were mixed with 1.2 mL of xylene, and excess xylene was removed by two 1.2-mL 100% ethanol washes. Dried tissue samples were incubated with lysis buffer and proteinase K at 56C for 30 min. Subsequently, the mixture was applied to the spin column and centrifuged into a collection tube according to the manufacturer’s protocol. The purified DNA was used directly for polymerase chain reaction (PCR).
HPV Genotyping
A commercially available HPV DNA chip (Goodgene, Seoul, Korea) was used for HPV genotyping. The HPV DNA chip contained 40 type-specific probes, including 21 types of high-risk HPV (16, 18, 26, 31, 33, 35, 39, 45, 51, 52, 53, 56, 58, 59, 66, 67, 68, 69, 70, 73, and 82) and 19 types of low-risk HPV (6, 11, 30, 32, 34, 40, 41, 42, 43, 44, 54, 55, 61, 62, 72, 81, 83, 84, and 90). Briefly, DNA amplification was performed in a 2720 Thermal Cycler (Applied Biosystems, Foster City, CA) by PCR with primer sets, which target L1 and L2 regions of HPV DNA. The amount of DNA was 10 µl. As a control gene, the human β-globin gene was also amplified. The PCR products from all samples were detected by electrophoresis using 2% agarose gels, and the HPV DNA product size was 185 bp. Hybridized HPV DNA was visualized using a DNA chip scanner (GeneScan; Goodgene). To avoid contamination that may yield a false-positive result, all PCR-related work was performed in specialized zones within a PCR laboratory.13
Statistical Analysis
Statistical analyses were performed using SPSS version 18.0 (Statistical Package for Social Science, SPSS Inc., Chicago, IL). Comparisons of clinicopathologic variables between the FGFR1 amplified and non-amplified groups were made, using chi-square or Fisher’s exact test for nominal variables. The overall survival (OS) and disease-free survival (DFS) were compared using the Kaplan–Meier method, and the survival differences were calculated by the log-rank test. All p values less than .05 were considered to be statistically significant.
Results
Frequency of FGFR1 Gene Amplification and Association With Clinical Characteristics
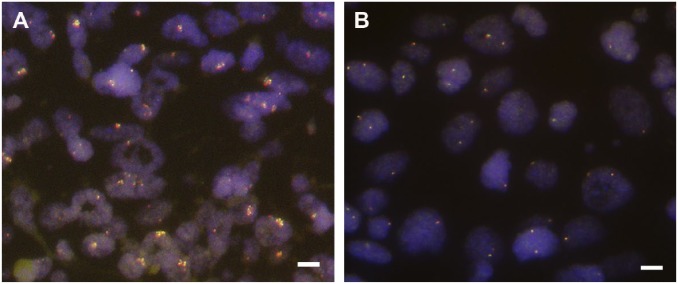
Fourteen of 89 (15.7%) TSCC cases showed FGFR1 gene amplification by FISH, and the average amplification ratio was 4.78 (Fig. 3). The clinicopathologic characteristics were compared between the 14 FGFR1 gene amplified cases and the 75 non-amplified FGFR1 gene cases, summarized in Table 1. The mean age of the FGFR1 gene amplified and non-amplified patients was 52.8 and 55.6 years old, respectively. However, these ages were not significantly different (p=0.489). The FGFR1 gene amplified group was not associated with any clinicopathologic variables, including pT stage (p=0.609), pN stage (p=0.790), pTNM stage (p=0.795), and alcohol history (p=0.297). With respect to smoking habits, five of 14 (17.9%) FGFR1 gene amplified cases were never smokers, and 9 of 14 (48.8%) FGFR1 gene amplified cases were smokers, but no statistical significance was observed between the two groups (p=0.155).
Figure 3.
FGFR1 gene copy number assessment by FISH. (A) The cells show FGFR1 gene amplification. (B) The cells show normal disomic signals. Scale bar represents 10 µm. Abbreviations: FGFR1, fibroblast growth factor receptor 1; FISH, fluorescence in situ hybridization.
Table 1.
Patients Characteristics According to FGFR1 Amplification Status and Immunohistochemistry.
| Characteristics | FGFR1 FISH |
p Value | FGFR1 Protein
Expression |
p Value | ||
|---|---|---|---|---|---|---|
| Amplified (%) | Nonamplified (%) | High (%) | Low (%) | |||
| Number of patients | 14 (15.7%) | 75 (84.3%) | 35 (39.3%) | 53 (59.6%) | ||
| Age, years | ||||||
| Median (range) | 52.8 (±12.02) | 55.6 (±10.01) | 0.489a | 52.3 (±9.82) | 57.0 (±10.41) | 0.227a |
| Sex | ||||||
| Male | 13 (16.0%) | 68 (84.0%) | 0.793 | 32 (40.0%) | 48 (60.0%) | 0.890 |
| Female | 1 (12.5%) | 7 (87.5%) | 3 (37.5%) | 5 (62.5%) | ||
| Size (cm) | ||||||
| ≤4 | 13 (16.7%) | 65 (83%) | 0.518b | 34 (44.2%) | 43 (55.8%) | 0.044b |
| >4 | 1 (9.1%) | 10 (90.9%) | 1 (9.1%) | 10 (90.9%) | ||
| Differentiation | ||||||
| WD | 3 (18.8%) | 13 (81.3%) | 0.353 | 7 (43.8%) | 9 (56.3%) | 0.695 |
| MD | 8 (12.7%) | 55 (87.3%) | 23 (37.1%) | 39 (629%) | ||
| PD | 3 (30.0%) | 7 (70.0%) | 5 (50.0%) | 5 (50.0%) | ||
| Keratinization | ||||||
| Keratinizing | 4 (12.9%) | 27 (87.1%) | 0.763 | 16 (53.3%) | 14 (46.7%) | 0.071 |
| Non-keratinizing | 10 (17.2%) | 48 (82.8%) | 19 (32.8%) | 39 (60.2%) | ||
| LN metastasis | ||||||
| Present | 12 (15.2%) | 67 (84.7%) | 0.654b | 32 (41.0%) | 46 (59.0%) | 0.734b |
| Absent | 2 (20.0%) | 8 (80.0%) | 3 (30.0%) | 7 (70.0%) | ||
| pT stage | ||||||
| T1 | 4 (11.4%) | 31 (88.6%) | 0.609 | 19 (54.3%) | 16 (45.7%) | 0.080 |
| T2 | 8 (21.6%) | 29 (78.4%) | 13 (36.2%) | 23 (63.9%) | ||
| T3 | 1 (9.1%) | 10 (90.9%) | 2 (18.2%) | 9 (81.8%) | ||
| T4 | 1 (9.1%) | 5 (83.3%) | 1 (16.7%) | 5 (83.3%) | ||
| pN stage | ||||||
| N0 | 2 (20.0%) | 8 (80.0%) | 0.790 | 3 (30.0%) | 7 (70.0%) | 0.279 |
| N1 | 2 (12.5%) | 14 (87.5%) | 10 (62.5%) | 6 (37.5%) | ||
| N2a | 1 (11.1%) | 8 (88.9%) | 4 (44.4%) | 5 (55.6%) | ||
| N2b | 9 (19.6%) | 37 (80.4%) | 15 (33.3%) | 30 (66.7%) | ||
| N2c | 0 (0.0%) | 6 (100.0%) | 3 (50.0%) | 3 (50.0%) | ||
| N3 | 0 (0.0%) | 2 (100.0%) | 0 (0.0%) | 2 (100.0%) | ||
| pTNM stage | ||||||
| I | 1 (20.0%) | 4 (80.0%) | 0.795 | 3 (60.0%) | 2 (40.0%) | 0.092 |
| II | 0 (0.0%) | 4 (100.0%) | 0 (0.0%) | 4 (100.0%) | ||
| III | 3 (20.0%) | 12 (80.0%) | 9 (60.0%) | 6 (40.0%) | ||
| IV | 10 (15.4%) | 55 (84.6%) | 23 (35.9%) | 41 (64.1%) | ||
| Smoking history | ||||||
| Never smoker | 5 (17.9%) | 23 (82.1%) | 0.155 | 12 (42.9%) | 16 (57.1%) | 0.920 |
| Former smoker | 3 (37.5%) | 5 (62.5%) | 3 (37.5%) | 5 (62.5%) | ||
| Current smoker | 6 (11.3%) | 47 (88.7%) | 20 (38.5%) | 32 (61.5%) | ||
| Alcohol history | ||||||
| Never/social | 6 (11.3%) | 47 (88.7%) | 0.297 | 21 (39.6%) | 32 (60.4%) | 0.985 |
| Yes | 8 (22.2%) | 28 (77.8%) | 14 (40.0%) | 21 (60.0%) | ||
| HPV ISH | ||||||
| Positive | 10 (16.9%) | 49 (83.1%) | 0.765b | 24 (41.4%) | 34 (58.6%) | 0.819b |
| Negative | 4 (13.3%) | 26 (86.7%) | 11 (36.7%) | 19 (63.3%) | ||
| p16 IHC | ||||||
| Positive | 9 (12.2%) | 65 (87.8%) | 0.055b | 31 (42.5%) | 42 (57.5%) | 0.386b |
| Negative | 5 (33.3%) | 10 (66.7%) | 4 (26.7%) | 11 (73.3%) | ||
| FGFR1 IHC | ||||||
| Positive | 6 (17.4%) | 29 (82.6%) | 0.797 | |||
| Negative | 8 (15.1%) | 45 (84.9%) | ||||
Abbreviations: FGFR1, fibroblast growth factor receptor 1; FISH, fluorescence in situ hybridization; HPV, human papillomavirus; ISH, in situ hybridization; IHC, immunohistochemistry; WD, well differentiated; MD, moderatedly differentiated; PD, poorly differentiated; LN, lymph node.
t-test.
Fisher’s exact test.
FGFR1 Protein Expression and Association With Clinicopathologic Variables
The results are summarized in Table 1. Thirty-seven (41.6%), 16 (18.0%), 9 (10.1%), and 26 (29.2%) of TSCC patients divided to the following scores of 0, 1, 2, and 3, respectively, by FGFR1 IHC. Thirty-five of 89 (39.3%) TSCC patients categorized as the high FGFR1 protein expression group. No correlation was found between FGFR1 gene amplification and FGFR1 protein expression (r = 0.027, p=0.797). The tumor size was inversely correlated with FGFR1 high expression (p=0.044), although the pT stage (p=0.080) was not correlated with FGFR1 expression. On the analysis between clinical parameter and FGFR1 protein expression, 24 (27.2%) patients with FGFR1 protein high expression showed positive HPV ISH results, whereas 11 (12.4%) patients with FGFR1 protein high expression showed negative HPV ISH results. However, HPV status and FGFR1 protein expression were not correlated in this study (p=0.669). Other clinicopathologic variables, including sex (p=0.890), tumor differentiation (p=0.695), keratinization (p=0.071), lymph node metastasis (p=0.734), pN stage (p=0.279), smoking history (p=0.920), and alcohol history (p=0.985), were not correlated with FGFR1 protein expression.
Association of FGFR1 Gene Amplification With HPV Status and p16
We used partly previously published data13 of HPV ISH, p16, and HPV genotyping. Fifty-nine of 89 (66.3%) TSCC patients showed HPV ISH positivity, and 74 of 89 (83.1%) TSCCs showed p16 immunopositivity (Supplementary Figure 1). A strong positive correlation between p16 expression and HPV status (p<0.001) was identified (Supplementary Table 1). However, no survival benefit according to HPV and p16 status was observed (Supplementary Figure 3.).
Thirty-six of 89 (40.4%) TSCC patients showed immunopositivity for FGFR1 protein, but no correlation was found between FGFR1 gene amplification and FGFR1 protein expression (p=0.115).
HPV genotyping using a DNA chip was performed. No HPV DNA was detected by genotyping in all 17 cases. To overcome the restricted HPV genotyping (p16+/HPV ISH−, n=17), we analyzed HPV genotyping with validation set (n=83) and the results described in Supplementary Material 2, Supplementary Figure 2, and Supplementary Tables 1 and 2.
Ten of 14 (71.4%) patients of the FGFR1 gene amplified group showed HPV ISH positivity, and 9 (64.3%) of the FGFR1 gene amplified patients showed immunopositivity for p16. However, no correlations were observed between HPV ISH and FGFR1 gene amplification (p=0.765) or p16 expression and FGFR1 gene amplification (p=0.055).
Survival Outcomes According to FGFR1 Amplification and FGFR1 Protein Expression
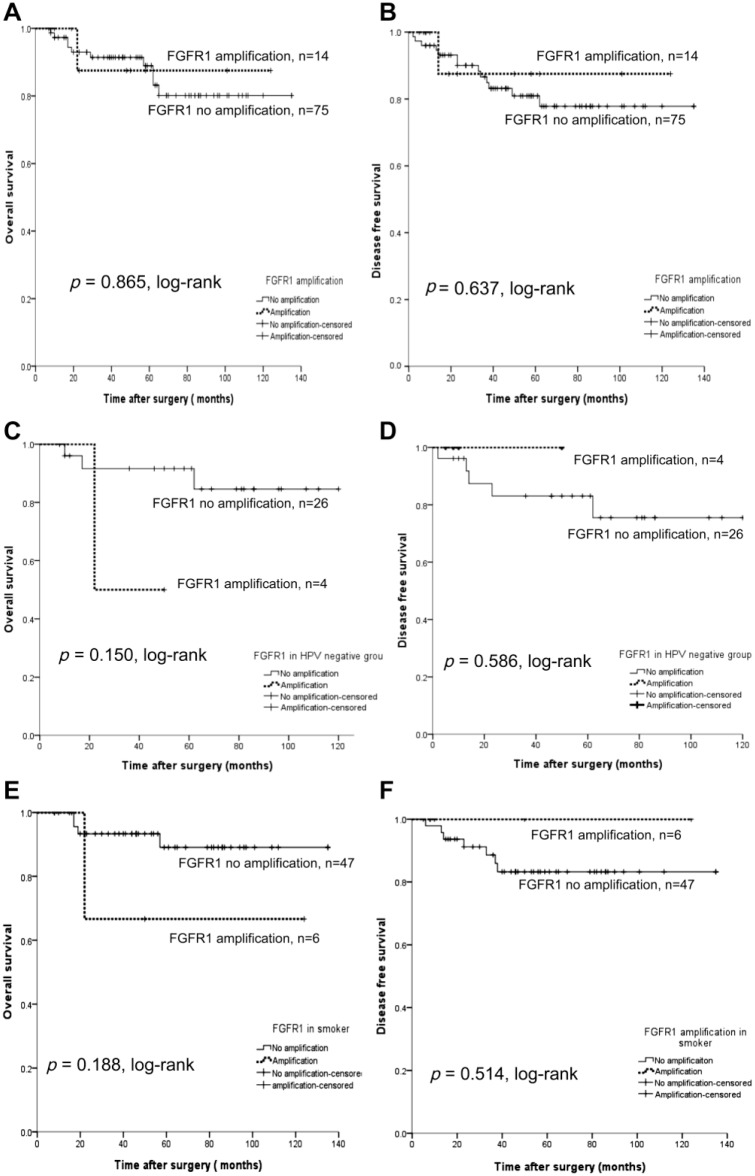
The OS and DFS rates (DFS) between the amplified FGFR1 group and non-amplified group did not show statistical significance (p=0.865, log-rank, and p=0.637, log-rank, respectively, Fig. 4). When restricting the analysis to the HPV-negative group only (n=30), a survival benefit of the amplified versus the non-amplified FGFR1 group was not observed (p=0.150, log-rank, and p=0.586, log-rank, respectively). In addition, no significant survival benefit was observed among smokers (n=53), whether the patients had the amplified FGFR1 gene or not.
Figure 4.
OS and DFS for FGFR1 gene amplification determined by FISH. (A, B) FGFR1 amplification is not associated with prognosis. (C, D) When restricting analysis to the HPV-negative group only (n=30), there is still no association with OS (C) or DFS (D). (E, F) When restricting the analysis to smokers (n=53), there is no association with OS (E) or DFS (F). Abbreviations: OS, overall survival; DFS, disease-free survival; FGFR1, fibroblast growth factor receptor 1; FISH, fluorescence in situ hybridization; HPV, human papillomavirus.
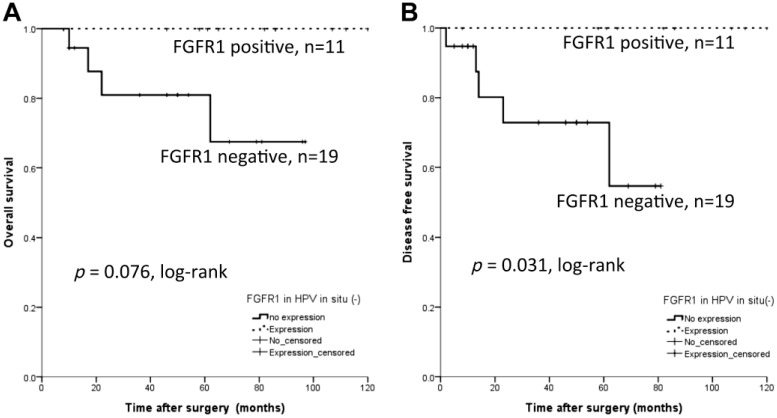
For FGFR1 protein expression, the OS (p=0.082, log-rank) and DFS (p=0.458, log-rank) were not statistically different whether FGFR1 was expressed or not. However, when restricting the analysis to the HPV-negative group only (n=30, Fig. 5), the FGFR1 protein positive expression group (n=11) had better DFS (p=0.031, log-rank), while OS was not different from the FGFR1 protein expression negative group (p=0.076, log-rank).
Figure 5.
Survival analysis for FGFR1 protein expression in patients with HPV in situ hybridization negativity. FGFR1 is a poor prognostic factor for recurrence (p=0.031, log-rank, B), while no prognostic significance is observed in overall survival (p=0.076, log-rank, A). Abbreviations: FGFR1, fibroblast growth factor receptor 1; HPV, human papillomavirus.
Discussion
We reported FGFR1 amplification and protein expression in patients with TSCC and analyzed the association between FGFR1 amplification or protein expression and HPV status. We reviewed the published studies on PubMed for FGFR1 amplification and FGFR1 protein expression in HNSCC and summarized in Table 2. In brief, the frequency of FGFR1 amplification ranged from 0.8% to 20%, and FGFR1 amplified patients were free of HPV infection. Association with smoking in amplified patients was controversial. No OS differences between the FGFR1 amplified and non-amplified groups were noted, although previous studies revealed that high protein expression was a poor prognostic factor.
Table 2.
Summary of Published Series for FGFR1 Amplification and FGFR1 Protein Expression in Head and Neck Squamous Cell Carcinoma.
| Study | Location | Sample No. | Methods | Probe/Ab | Cutoff | Frequency | A/w HPV Status | A/w Smoking | Comments |
|---|---|---|---|---|---|---|---|---|---|
| Ozretic et al.17 | OP | 231 | FISH | ND | FGFR1/CEN8 ≥2 or cell displaying >6 signals | Amplification, 5.6% (13/231) | 8% (1/13) shows HPV 16 DNA positivity | 92% (12/13) are smoker | 26.4% (61/234) are c-myc expression 2.6% (6/231) are both c-myc positivity and FGFR1 amplification |
| Koole et al.18-molecular diagnostic theory | OC, OP | 951 | IHC | Abcam, 1:2000, ab10646 | High expression, ≥10% | High expression, 64% | 67% in HPV-positive 85% in HPV-negative |
NE | High FGFR1 expression shows poor prognosis (p=0.018) |
| Koole et al.19-CCR | OC, OP | 452 | FISH | Cytocell Aquarius | FGFR1/CEN8 >2 | Amplification, 3% (3/94) | All amplified cases show HPV negativity | NE | |
| IHC | Abcam, 1:2000, ab10646 | High expression, ≥ 10% | High expression, 74.5% (330/443) | 82% (36/45) in HPV-positive 75% (294/398) in HPV- negative |
NE | FGFR1 protein expression is a poor prognostic factor in HPV-negative HNSCC (p=0.001) | |||
| Seiwert et al.20 | OC, OP, L, HP | 120 | Parallel sequencing | Illumina HiSeq 2000/2500 sequencer | ND | Amplification, 0.8% (1/120) | An amplified case shows HPV negativity | NE | 17.6% of HPV-positive tumors harbor somatic mutations in FGFR2/3 |
| Feldman et al.21 | OC, OP, NP, HP, L, UP | 735 | Multiplatform profilinga | Illumina MiSeq platform for NGS | ND | No mutation (0/337) | 0% (0/37) in HPV -positive 0% (0/47) in HPV -negative |
NE | |
| TCGA Network22 | OC, OP, L | 279 | Comprehensive multiplatform genetic profiling | ND | ND | Amplification, 10% (28/279) | All amplified cases show HPV negativity | NE | No differences of overall survival between amplified group and non-amplified group (p=0.924) |
| Schröck et al.23 | Sinonasal cancer | 138 (57 of SqCC) | FISH | Metasystems | ≥20% (100 cells) High: cell displaying >9, low: 2–9 signals |
Amplification, 20% (9/45) | All amplified cases show p16 immunonegativity | 3% (1/32) is smoker, but N-S (p=0.30) | No differences of overall survival between amplified group and non-amplified group (p>0.05) |
| Young et al.24 | Tongue | 123 | FISH | Abnova | FGFR1/CEN8 >2% or >50% of cell displaying >5 signals | Amplification, 9.3% (10/107) | NE | All amplified patients (10) are smoker (p=0.03) | No prognostic significance of FGFR1 amplification for overall survival (p=0.90) |
| Göke et al.10 | OC, OP, HP, L, UP | 555 | FISH | Metasystems | ≥20% High: cell displaying >9 Low: 2–9 signals |
Amplification, 15% (68/452) | All amplified cases show HPV negativity (p=0.003) | 96.5% (55/57) are smoker (p=0.04) | No prognostic significance of FGFR1 amplified group for overall survival (p=0.71) |
| Freier et al.15 | OC | 92 (FISH)/178 (IHC) | FISH | BAC clone RP11-350N15 | ≥10% Cell displaying >6 signals |
Amplification, 17.4% (16/92) | NE | NE | No differences of overall survival between amplified group and non-amplified group (p>0.05) |
| IHC | Abcam, 1:100 | Divided by intensity | High expression, 11.8% (21/178) | NE | NE | High expression is related to early stage. (p=0.002) | |||
| Hase et al25 | OC | 61 | IHC | Santa Cruz, 1:100 | Any FGFR1 expression cells | High expression 55.7% (34/61) | NE | NE |
Abbreviations: FGFR1, fibroblast growth factor receptor 1; No., number; HPV, human papillomavirus; OP, oropharynx; FISH, fluorescence in situ hybridization; ND, not described; OC, oral cavity; IHC, immunohistochemistry; NE, not evaluated; HNSCC, head and neck squamous cell carcinoma; L, larynx; HP, hypopharynx; NP, nasopharynx; UP, unknown primary; NGS, next-generation sequencing; SqCC, squamous cell carcinoma; N-S, statistically not significant. A/w, association with.
Multiple-platform includes gene sequencing (exome sequencing, RNA-Seq, methylation), gene copy number alteration, and protein expression.
We performed in silico analysis for TCGA Head and Neck Squamous Cell Carcinoma (TCGA-HNSCC) and compared TCGA data with our data (Supplementary Material 1). Our study revealed a relatively high frequency of FGFR1 amplification (15.7%) compared with TCGA data, which showed FGFR1 amplification in 10.0% of HNSCCs. Freier et al reported the FGFR1 amplification frequency to be 17.4% in oral squamous cell carcinoma,15 but Ozretic et al reported 5.6% in oropharyngeal squamous cell carcinoma.17 These differences could be explained by the following factors. First, the definition of amplified FGFR1 was not fully standardized. The cutoff values ranged from 2.0 to 9.0 in the published English literature.16,26,27 Second, the specimen populations of the previous studies were heterogeneous in that they reported on various squamous cell carcinomas of the head and neck, including the oral cavity, larynx, salivary gland, oropharynx, and even sinonasal cavity. However, our study was the most homogeneous given that we focused only on TSCC. Third, different detection methods were used among the studies; the applied methods included FISH/silver in situ hybridization (SISH), next-generation sequencing, mRNA expression, and so on.
For the relationship between FGFR1 amplification and HPV status, published TCGA data22 revealed that there were no HPV-positive patients among those with FGFR1 amplification, but our data showed that 10 (71.4%) patients with FGFR1 amplification had positive HPV ISH results. As shown in Table 2, FGFR1 amplified patients had a tendency to lack HPV infection; however, not all FGFR1 amplified patients showed HPV negativity. Ozretic et al.17 reported that 8% (1/13) of FGFR1 amplified patients had HPV-16 DNA positivity. A reason for these results may be racial differences and varying specimen purity. The incidence of HPV in the Korean population ranges from 37% to 67%,4,13 whereas Western data have shown a range of 37–80%.28,29 In general, oropharyngeal squamous cell carcinoma showed a higher frequency of HPV than other HNSCCs (79% vs. 21%, respectively).21 The detection method used cannot explain the differences in HPV status among the FGFR1 amplified patients. The sensitivity of HPV ISH was known to be 88%, whereas that of DNA quantitative PCR was 97%.30 In other words, HPV ISH had higher false negativity. Therefore, our results support the notions that FGFR1 amplification was not associated with HPV status, and that FGFR1 amplification occurred regardless of HPV status.
Kim et al.26 and Seo et al.31 et al revealed that high copy FGFR1 amplification was associated with heavy smoking in squamous cell carcinoma of the lung. In contrast, Heist et al.16 suggested that FGFR1 amplification was not associated with smoking. To this end, the association of smoking with FGFR1 amplification in HNSCC is still controversial. Ozretic et al.,17 Göke et al.,10 and Young et al.24 reported that high FGFR1 amplification was associated with smoking in HNSCC, but Schröck et al.23 reported that there was no such association. Our study showed that there was no association between FGFR1 amplification and smoking.
Gene amplification and protein expression show different correlations according to the target. Amplification of Her2/neu (human epidermal growth factor receptor 2) gene amplification in breast cancer and its protein c-erb2 expression are well correlated; thus, IHC for c-erb2 is used as a screening method for the selection of patients who have to be treated with trastuzumab, which is only used on patients with Her2/neu gene amplification by FISH. However, as seen in our report and others, FGFR1 amplification and FGFR1 protein expression in HNSCC are not correlated with each other.15 FGFR1 protein expression was more frequent (12–75%) than gene amplification.15,18 Our study revealed FGFR1 protein in 39.3% and FGFR1 gene amplification in 15.7% of cases and there was no correlation between protein expression and gene amplification (p=0.797).
The prognosis of FGFR1 amplification is controversial not only in HNSCC but also in squamous cell carcinoma of the lung. Three previous studies26,31,32 reported that amplified FGFR1 was a poor prognostic factor in non–small cell carcinoma, but Jiang et al reported by meta-analysis of four studies33 that FGFR1 amplification had no influence on OS. In addition, all published studies of FGFR1 amplification in HNSCC showed no prognostic significance for OS (Table 2). However, Koole et al.18,19 reported that high expression of FGFR1 protein was related with poor prognosis.
Our study had several limitations: First, the numbers of the cases were relatively small; second, selection bias was present. The submitted cohort contained patients only with an adequate amount of specimen from tonsillectomy for TMA excluding inoperable cases with higher stage, which may affect the analysis of relationship between FGFR1 amplified squamous cell carcinoma and survival rate. Third, incomplete test for HPV genotyping was performed. Although we tried to additionally test for the other samples except HPV ISH−/p16+ cases at the time of writing, we could not buy the same genotyping chip kit that we previously tested due to the company failure. Moreover, half of the samples were too old to extract DNA and the quality of the DNA was very poor. To overcome this issue, we tried to validate the results of HPV genotyping based on the medical record. These were described in Supplementary Material 2 and Supplementary Tables 1 and 2. Last, we could not determine the sensitivity and specificity of anti-FGFR1 antibody because of discordance between FGFR1 amplification and FGFR1 protein expression (p=0.797) and just fully relied on the manufacturer’s instruction.
The identification of genetic alterations in multiple FGFR family members in human cancers highlights pan-FGFR inhibition as a promising therapeutic approach in a variety of malignancies. Several small-molecule FGFR inhibitors with differing selectivity and potency profiles, such as brivanib, dovitinib, AZD4547, BGJ398, and erdafitinib, are in various stages of clinical development. Dovitinib,34 AZD4547,18 and BJG39835,36 showed good responses in HNSCC at the preclinical level.
In conclusion, our study showed that FGFR1 amplification was frequently observed in patients with TSCC, not associated with HPV status and not associated with prognosis. Our study suggests that FGFR1 amplification may be important in pathogenesis regardless of HPV status. Further study is required that FGFR1 amplification in TSCC correlates with response to targeted anti-FGFRs drugs.
Supplemental Material
Supplemental material, JHC761652_Supplemental_Material for Association Between Fibroblast Growth Factor Receptor 1 Gene Amplification and Human Papillomavirus Prevalence in Tonsillar Squamous Cell Carcinoma With Clinicopathologic Analysis by Soonchan Park, Miji Lee, Kyung-Ja Cho, Sung Bae Kim, Jong-Lyel Roh, Seung-Ho Choi, Soon Yuhl Nam, Sang Yoon Kim and Joon Seon Song in Journal of Histochemistry & Cytochemistry
Acknowledgments
The authors thank Hee Jin Lee, MD, PhD, from the Department of Pathology, University of Ulsan College of Medicine, Asan Medical Center, Seoul, South Korea, for her critical review and technical assistance in the statistical analysis.
Footnotes
Competing Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Author Contributions: SP, ML, K-JC, and JSS designed and carried out the histological and biochemical experiments and data analysis, and drafted the manuscript. SBK, J-LR, S-HC, SYN, and SYK provided tonsillar squamous cell carcinoma of human samples and clinical data and contributed to the data analysis and revision of the manuscript. All authors have read and approved the final manuscript.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by a grant from Kyung Hee University #KHU-20150821 (S.P.).
Contributor Information
Soonchan Park, Department of Radiology, Kyung Hee University Hospital at Gangdong, College of Medicine, Kyung Hee University, Seoul, Republic of Korea.
Miji Lee, Department of Pathology, Veterans Health Service Medical Center, Seoul, Republic of Korea.
Kyung-Ja Cho, Departments of Pathology, University of Ulsan College of Medicine, Asan Medical Center, Seoul, Republic of Korea.
Sung Bae Kim, Medical Oncology, University of Ulsan College of Medicine, Asan Medical Center, Seoul, Republic of Korea.
Jong-Lyel Roh, Head and Neck Surgery, University of Ulsan College of Medicine, Asan Medical Center, Seoul, Republic of Korea.
Seung-Ho Choi, Head and Neck Surgery, University of Ulsan College of Medicine, Asan Medical Center, Seoul, Republic of Korea.
Soon Yuhl Nam, Head and Neck Surgery, University of Ulsan College of Medicine, Asan Medical Center, Seoul, Republic of Korea.
Sang Yoon Kim, Head and Neck Surgery, University of Ulsan College of Medicine, Asan Medical Center, Seoul, Republic of Korea.
Joon Seon Song, Departments of Pathology, University of Ulsan College of Medicine, Asan Medical Center, Seoul, Republic of Korea.
Literature Cited
- 1. Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin. 2017;67(1):7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 2. Marur S, Forastiere AA. Head and neck cancer: changing epidemiology, diagnosis, and treatment. Mayo Clin Proc. 2008;83(4):489–501. doi: 10.4065/83.4.489. [DOI] [PubMed] [Google Scholar]
- 3. D’Souza G, Kreimer AR, Viscidi R, Pawlita M, Fakhry C, Koch WM, Westra WH, Gillison ML. Case-control study of human papillomavirus and oropharyngeal cancer. N Engl J Med. 2007;356(19):1944–56. doi: 10.1056/NEJMoa065497. [DOI] [PubMed] [Google Scholar]
- 4. Song JS, Kim MS, Park JW, Lee YS, Kang CS. Expression of human papillomavirus-related proteins and its clinical implication in tonsillar squamous cell carcinoma. Korean J Pathol. 2012;46(2):177–86. doi: 10.4132/KoreanJPathol.2012.46.2.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Weir B, Zhao X, Meyerson M. Somatic alterations in the human cancer genome. Cancer Cell. 2004;6(5):433–8. doi: 10.1016/j.ccr.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 6. Beroukhim R, Mermel CH, Porter D, Wei G, Raychaudhuri S, Donovan J, Barretina J, Boehm JS, Dobson J, Urashima M, Mc Henry KT, Pinchback RM, Ligon AH, Cho YJ, Haery L, Greulich H, Reich M, Winckler W, Lawrence MS, Weir BA, Tanaka KE, Chiang DY, Bass AJ, Loo A, Hoffman C, Prensner J, Liefeld T, Gao Q, Yecies D, Signoretti S, Maher E, Kaye FJ, Sasaki H, Tepper JE, Fletcher JA, Tabernero J, Baselga J, Tsao MS, Demichelis F, Rubin MA, Janne PA, Daly MJ, Nucera C, Levine RL, Ebert BL, Gabriel S, Rustgi AK, Antonescu CR, Ladanyi M, Letai A, Garraway LA, Loda M, Beer DG, True LD, Okamoto A, Pomeroy SL, Singer S, Golub TR, Lander ES, Getz G, Sellers WR, Meyerson M. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463(7283):899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Eswarakumar VP, Lax I, Schlessinger J. Cellular signaling by fibroblast growth factor receptors. Cytokine Growth Factor Rev. 2005;16(2):139–49. doi: 10.1016/j.cytogfr.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 8. Zhang J, Zhang L, Su X, Li M, Xie L, Malchers F, Fan S, Yin X, Xu Y, Liu K, Dong Z, Zhu G, Qian Z, Tang L, Schottle J, Zhan P, Ji Q, Kilgour E, Smith PD, Brooks AN, Thomas RK, Gavine PR. Translating the therapeutic potential of AZD4547 in FGFR1-amplified non-small cell lung cancer through the use of patient-derived tumor xenograft models. Clin Cancer Res. 2012;18(24):6658–67. doi: 10.1158/1078-0432.CCR-12-2694. [DOI] [PubMed] [Google Scholar]
- 9. Lee HJ, Seo AN, Park SY, Kim JY, Park JY, Yu JH, Ahn JH, Gong G. Low prognostic implication of fibroblast growth factor family activation in triple-negative breast cancer subsets. Ann Surg Oncol. 2014;21(5):1561–8. doi: 10.1245/s10434-013-3456-x. [DOI] [PubMed] [Google Scholar]
- 10. Göke F, Bode M, Franzen A, Kirsten R, Goltz D, Göke A, Sharma R, Boehm D, Vogel W, Wagner P, Lengerke C, Kristiansen G, Kirfel J, Van Bremen T, Bootz F, Heasley LE, Schröck A, Perner S. Fibroblast growth factor receptor 1 amplification is a common event in squamous cell carcinoma of the head and neck. Mod Pathol. 2013;26(10):1298–306. doi: 10.1038/modpathol.2013.58. [DOI] [PubMed] [Google Scholar]
- 11. Andre F, Bachelot T, Campone M, Dalenc F, Perez-Garcia JM, Hurvitz SA, Turner N, Rugo H, Smith JW, Deudon S, Shi M, Zhang Y, Kay A, Porta DG, Yovine A, Baselga J. Targeting FGFR with dovitinib (TKI258): preclinical and clinical data in breast cancer. Clin Cancer Res. 2013;19(13):3693–702. doi: 10.1158/1078-0432.CCR-13-0190. [DOI] [PubMed] [Google Scholar]
- 12. Soria JC, DeBraud F, Bahleda R, Adamo B, Andre F, Dienstmann R, Delmonte A, Cereda R, Isaacson J, Litten J, Allen A, Dubois F, Saba C, Robert R, D’Incalci M, Zucchetti M, Camboni MG, Tabernero J. Phase I/IIa study evaluating the safety, efficacy, pharmacokinetics, and pharmacodynamics of lucitanib in advanced solid tumors. Ann Oncol. 2014;25(11):2244–51. doi: 10.1093/annonc/mdu390. [DOI] [PubMed] [Google Scholar]
- 13. Lee M, Kim SB, Lee SW, Roh JL, Choi SH, Nam SY, Kim SY, Cho KJ. Human papillomavirus prevalence and cell cycle related protein expression in tonsillar squamous cell carcinomas of korean patients with clinicopathologic analysis. Korean J Pathol. 2013;47(2):148–57. doi: 10.4132/KoreanJPathol.2013.47.2.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Begum S, Gillison ML, Ansari-Lari MA, Shah K, Westra WH. Detection of human papillomavirus in cervical lymph nodes: a highly effective strategy for localizing site of tumor origin. Clin Cancer Res. 2003;9(17):6469–75. [PubMed] [Google Scholar]
- 15. Freier K, Schwaenen C, Sticht C, Flechtenmacher C, Muhling J, Hofele C, Radlwimmer B, Lichter P, Joos S. Recurrent FGFR1 amplification and high FGFR1 protein expression in oral squamous cell carcinoma (OSCC). Oral Oncol. 2007;43(1):60–6. doi: 10.1016/j.oraloncology.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 16. Heist RS, Mino-Kenudson M, Sequist LV, Tammireddy S, Morrissey L, Christiani DC, Engelman JA, Iafrate AJ. FGFR1 amplification in squamous cell carcinoma of the lung. J Thorac Oncol. 2012;7(12):1775–80. doi: 10.1097/JTO.0b013e31826aed28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ozretic L, Wagner S, Huebbers CU, Gattenlohner S, Klussmann JP, Beutner D, Zander T, Buettner R, Quaas A. FGFR1 amplification and co-overexpression of c-MYC in oropharyngeal squamous cell carcinoma. Oral Oncol. 2016;54:e7–9. doi: 10.1016/j.oraloncology.2015.12.006. [DOI] [PubMed] [Google Scholar]
- 18. Koole K, Brunen D, van Kempen PM, Noorlag R, de Bree R, Lieftink C, van Es RJ, Bernards R, Willems SM. FGFR1 is a potential prognostic biomarker and therapeutic target in head and neck squamous cell carcinoma. Clin Cancer Res. 2016;22(15):3884–93. doi: 10.1158/1078-0432.CCR-15-1874. [DOI] [PubMed] [Google Scholar]
- 19. Koole K, Clausen MJ, van Es RJ, van Kempen PM, Melchers LJ, Koole R, Langendijk JA, van Diest PJ, Roodenburg JL, Schuuring E, Willems SM. FGFR family members protein expression as prognostic markers in oral cavity and oropharyngeal squamous cell carcinoma. Mol Diagn Ther. 2016;20(4):363–74. doi: 10.1007/s40291-016-0204-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Seiwert TY, Zuo Z, Keck MK, Khattri A, Pedamallu CS, Stricker T, Brown C, Pugh TJ, Stojanov P, Cho J, Lawrence MS, Getz G, Bragelmann J, DeBoer R, Weichselbaum RR, Langerman A, Portugal L, Blair E, Stenson K, Lingen MW, Cohen EE, Vokes EE, White KP, Hammerman PS. Integrative and comparative genomic analysis of HPV-positive and HPV-negative head and neck squamous cell carcinomas. Clin Cancer Res. 2015;21(3):632–41. doi: 10.1158/1078-0432.CCR-13-3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Feldman R, Gatalica Z, Knezetic J, Reddy S, Nathan CA, Javadi N, Teknos T. Molecular profiling of head and neck squamous cell carcinoma. Head Neck. 2016;38(Suppl 1):E1625–38. doi: 10.1002/hed.24290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. The Cancer Genome Atlas Network. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature. 2015;517(7536):576–82. doi: 10.1038/nature14129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Schröck A, Goke F, Wagner P, Bode M, Franzen A, Huss S, Agaimy A, Ihrler S, Kirsten R, Kristiansen G, Bootz F, Lengerke C, Perner S. Fibroblast growth factor receptor-1 as a potential therapeutic target in sinonasal cancer. Head Neck. 2014;36(9):1253–7. doi: 10.1002/hed.23443. [DOI] [PubMed] [Google Scholar]
- 24. Young RJ, Lim AM, Angel C, Collins M, Deb S, Corry J, Wiesenfeld D, Kleid S, Sigston E, Lyons B, Russell PA, Wright G, McArthur GA, Fox SB, Rischin D, Solomon B. Frequency of fibroblast growth factor receptor 1 gene amplification in oral tongue squamous cell carcinomas and associations with clinical features and patient outcome. Oral Oncol. 2013;49(6):576–81. doi: 10.1016/j.oraloncology.2013.01.006. [DOI] [PubMed] [Google Scholar]
- 25. Hase T, Kawashiri S, Tanaka A, Nozaki S, Noguchi N, Kato K, Nakaya H, Nakagawa K. Correlation of basic fibroblast growth factor expression with the invasion and the prognosis of oral squamous cell carcinoma. J Oral Pathol Med. 2006;35(3):136–9. doi: 10.1111/j.1600-0714.2006.00397.x. [DOI] [PubMed] [Google Scholar]
- 26. Kim HR, Kim DJ, Kang DR, Lee JG, Lim SM, Lee CY, Rha SY, Bae MK, Lee YJ, Kim SH, Ha SJ, Soo RA, Chung KY, Kim JH, Lee JH, Shim HS, Cho BC. Fibroblast growth factor receptor 1 gene amplification is associated with poor survival and cigarette smoking dosage in patients with resected squamous cell lung cancer. J Clin Oncol. 2013;31(6):731–7. doi: 10.1200/JCO.2012.43.8622. [DOI] [PubMed] [Google Scholar]
- 27. Schildhaus HU, Heukamp LC, Merkelbach-Bruse S, Riesner K, Schmitz K, Binot E, Paggen E, Albus K, Schulte W, Ko YD, Schlesinger A, Ansen S, Engel-Riedel W, Brockmann M, Serke M, Gerigk U, Huss S, Goke F, Perner S, Hekmat K, Frank KF, Reiser M, Schnell R, Bos M, Mattonet C, Sos M, Stoelben E, Wolf J, Zander T, Buettner R. Definition of a fluorescence in-situ hybridization score identifies high- and low-level FGFR1 amplification types in squamous cell lung cancer. Mod Pathol. 2012;25(11):1473–80. doi: 10.1038/modpathol.2012.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ragin CC, Taioli E. Survival of squamous cell carcinoma of the head and neck in relation to human papillomavirus infection: review and meta-analysis. Int J Cancer. 2007;121(8):1813–20. doi: 10.1002/ijc.22851. [DOI] [PubMed] [Google Scholar]
- 29. Adelstein DJ, Ridge JA, Gillison ML, Chaturvedi AK, D’Souza G, Gravitt PE, Westra W, Psyrri A, Kast WM, Koutsky LA, Giuliano A, Krosnick S, Trotti A, Schuller DE, Forastiere A, Ullmann CD. Head and neck squamous cell cancer and the human papillomavirus: summary of a National Cancer Institute State of the Science Meeting, November 9-10, 2008, Washington, D.C: Head Neck; 2009;31(11):1393–422. doi: 10.1002/hed.21269. [DOI] [PubMed] [Google Scholar]
- 30. Schache AG, Liloglou T, Risk JM, Filia A, Jones TM, Sheard J, Woolgar JA, Helliwell TR, Triantafyllou A, Robinson M, Sloan P, Harvey-Woodworth C, Sisson D, Shaw RJ. Evaluation of human papilloma virus diagnostic testing in oropharyngeal squamous cell carcinoma: sensitivity, specificity, and prognostic discrimination. Clin Cancer Res. 2011;17(19):6262–71. doi: 10.1158/1078-0432.CCR-11-0388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Seo AN, Jin Y, Lee HJ, Sun PL, Kim H, Jheon S, Kim K, Lee CT, Chung JH. FGFR1 amplification is associated with poor prognosis and smoking in non-small-cell lung cancer. Virchows Arch. 2014;465(5):547–58. doi: 10.1007/s00428-014-1634-2. [DOI] [PubMed] [Google Scholar]
- 32. Xie FJ, Lu HY, Zheng QQ, Qin J, Gao Y, Zhang YP, Hu X, Mao WM. The clinical pathological characteristics and prognosis of FGFR1 gene amplification in non-small-cell lung cancer: a meta-analysis. Onco Targets Ther. 2016;9:171–81. doi: 10.2147/OTT.S91848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Jiang T, Gao G, Fan G, Li M, Zhou C. FGFR1 amplification in lung squamous cell carcinoma: a systematic review with meta-analysis. Lung Cancer. 2015;87(1):1–7. doi: 10.1016/j.lungcan.2014.11.009. [DOI] [PubMed] [Google Scholar]
- 34. Sweeny L, Zimmermann TM, Liu Z, Rosenthal EL. Evaluation of tyrosine receptor kinases in the interactions of head and neck squamous cell carcinoma cells and fibroblasts. Oral Oncol. 2012;48(12):1242–9. doi: 10.1016/j.oraloncology.2012.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. von Massenhausen A, Deng M, Billig H, Queisser A, Vogel W, Kristiansen G, Schröck A, Bootz F, Göke F, Franzen A, Heasley L, Kirfel J, Brägelmann J, Perner S. Evaluation of FGFR3 as a therapeutic target in head and neck squamous cell carcinoma. Target Oncol. 2016;11(5):631–42. doi: 10.1007/s11523-016-0431-z. [DOI] [PubMed] [Google Scholar]
- 36. Goke F, Franzen A, Hinz TK, Marek LA, Yoon P, Sharma R, Bode M, von Maessenhausen A, Lankat-Buttgereit B, Göke A, Golletz C, Kirsten R, Boehm D, Vogel W, Kleczko EK, Eagles JR, Hirsch FR, Van Bremen T, Bootz F, Schroeck A, Kim J, Tan AC, Jimeno A, Heasley LE, Perner S. FGFR1 expression levels predict BGJ398 sensitivity of FGFR1-dependent head and neck squamous cell cancers. Clin Cancer Res. 2015;21(19):4356–64. doi: 10.1158/1078-0432.CCR-14-3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, JHC761652_Supplemental_Material for Association Between Fibroblast Growth Factor Receptor 1 Gene Amplification and Human Papillomavirus Prevalence in Tonsillar Squamous Cell Carcinoma With Clinicopathologic Analysis by Soonchan Park, Miji Lee, Kyung-Ja Cho, Sung Bae Kim, Jong-Lyel Roh, Seung-Ho Choi, Soon Yuhl Nam, Sang Yoon Kim and Joon Seon Song in Journal of Histochemistry & Cytochemistry