Abstract
A variety of approaches have been developed for the derivation of hepatocyte-like cells from pluripotent stem cells. Currently, most of these strategies employ step-wise differentiation approaches with recombinant growth-factors or small-molecule analogs to recapitulate developmental signaling pathways. Here, we tested the efficacy of a small-molecule based differentiation protocol for the generation of hepatocyte-like cells from human pluripotent stem cells. Quantitative gene-expression, immunohistochemical, and western blot analyses for SOX17, FOXA2, CXCR4, HNF4A, AFP, indicated the stage-specific differentiation into definitive endoderm, hepatoblast and hepatocyte-like derivatives. Furthermore, hepatocyte-like cells displayed morphological and functional features characteristic of primary hepatocytes, as indicated by the production of ALB (albumin) and α-1-antitrypsin (A1AT), as well as glycogen storage capacity by periodic acid-Schiff staining. Together, these data support that the small-molecule based hepatic differentiation protocol is a simple, reproducible, and inexpensive method to efficiently drive the differentiation of human pluripotent stem cells towards a hepatocyte-like phenotype, for downstream pharmacogenomic and regenerative medicine applications.
Keywords: Human pluripotent stem cell differentiation, Hepatocyte, Hepatocyte-like cells, Small molecules
1. Introduction
Purified primary human hepatocytes (PHHs) are invaluable basic research reagents for drug metabolism, toxicity, liver function, patho-physiology and antiviral response in vitro studies. However, a major roadblock toward the use of donor-derived PHHs as a model system, is their limited accessibility, batch-to-batch variability and their inability to readily proliferate ex vivo (Wu et al., 2012; Lohmann et al., 1999; Blight et al., 2002; Hannoun et al., 2016). A potential alternative approach has been the establishment of tumor-derived and immortalized human hepatocyte cell lines such as the hepatoma cell line Huh-7 and the HepaRG cell line (Lohmann et al., 1999; Blight et al., 2002). However, these cell lines are limited by the fact that they are derived from already existing tumors or are immortalized ex vivo (Lohmann et al., 1999; Blight et al., 2002; Breiman et al., 2005; Wu et al., 2012; Sa-Ngiamsuntorn et al., 2016).
To this end, human pluripotent stem cells (hPSCs) have revolutionized the in vitro generation of unlimited quantities of hepatocyte-like cells (HLCs) for pharmacogenomic and regenerative medicine applications (Goldring et al., 2017; Palakkan et al., 2017; Sakurai et al., 2017; Rashid et al., 2010). Early protocols for the generation of HLCs from hPSCs relied on the use of embryoid body formation (Imamura et al., 2004; Baharvand et al., 2006; Basma et al., 2009). These methods are based on the spontaneous formation of cell aggregates (embryoid bodies) from pluripotent cells which represent all three germ layers (Siller et al., 2016). The drawback to these early methods is the reliance on spontaneous differentiation of the EBs to produce hepatocytes which leads to low differentiation efficiency and the heterogeneous nature of these spontaneously differentiating cultures producing many undesired cell types in addition to the hepatocytes. Currently, the most efficient method to produce HLCs is a monolayer culture of hPSCs exposed in a step-wise manner to specific cocktails of recombinant growth factors and cytokines, including Activin A,Wnt3a, hepatocyte growth factor (HGF), oncostatin M (OSM), fibroblast growth factor 4 (FGF4), vascular endothelial growth factor (VEGF), epidermal growth factor (EGF) and bone morphogenic protein 4 (BMP4) (Agarwal et al., 2008; Si-Tayeb et al., 2010; Sullivan et al., 2010; Chen et al., 2012; Wu et al., 2012; Hannan et al., 2013; Carpenter et al., 2014; Sun et al., 2015; Lang et al., 2016; Carpenter et al., 2016; Hannoun et al., 2016; Sa-Ngiamsuntorn et al., 2016; Sakurai et al., 2017). These approaches were designed to mimic the embryonic stages of hepatocyte development (Supporting Information Fig. S1).
Compared to recombinant growth factors, small molecules offer a simple, highly efficient, and cost-effective alternative for generation of HLCs from hPSCs (Touboul et al., 2010; Borowiak et al., 2009; Sullivan et al., 2010; Shan et al., 2013; Siller et al., 2015, 2016; Tasnim et al., 2015; Mathapati et al., 2016). Specifically, effective modulation of Wnt signaling (Dravid et al., 2005; Engert et al., 2013; Nakanishi et al., 2009) and glycogen synthase kinase 3 (GSK-3) inhibition (Bone et al., 2011; Tahamtani et al., 2013; Sullivan et al., 2010; Shan et al., 2013; Siller et al., 2015, 2016; Tasnim et al., 2015; Mathapati et al., 2016) using small molecules efficiently produced DE populations. Accordingly, here we tested the efficacy of a previously published (Siller et al., 2015; Mathapati et al., 2016) small-molecule based differentiation protocol for deriving HLCs with a mature phenotype from hPSCs. We show that GSK-3 inhibition with CHIR99021 efficiently directs hPSCs toward the DE lineage, from which hepatoblasts and hepatocyte-like cells are subsequently derived in a stage-specific manner, via a combination of small-molecule treatments with DMSO, dexamethasone and the HGF receptor agonist N-hexanoic-Tyr, Ile-(6) aminohexanoic amide (dihexa) (Siller et al., 2016; Mathapati et al., 2016; McCoy et al., 2013).
2. Materials and methods
2.1. Small molecules, chemicals and antibodies
Human pluripotent stem cells (SC101A) was from Systems Biosciences (Palo Alto, CA), Dihexa (N-hexanoic-Tyr-Ile-(6) aminohex-anoic amide) (DC9760) was from DC Chemicals (Shanghai, China). Stemolecule CHIR99021 (04–0004) was from Stemgent (Lexington, MA). Selective p160ROCK Inhibitor Y-27632 (1254) was from Tocris (Minneapolis, MN). L-15 Medium (L1518), 2-Mercaptoethanol (M6250), Tryptose Phosphate Broth (T8159), Hydrocortisone-21- hemisuccinate (H4881), Sodium-L-Ascorbate (A4034), and Dexamethasone (D4902) were from Sigma (St. Louis, MO). Essential 8 medium (A1517001), RPMI/B27 medium (61870036), B27 supplement (17504044), GlutaMax supplement (35050061), Insulin- Transferrin-Selenium supplement (41400045), Knock Out DMEM (10829018), KnockOut Serum Replacement (10828010), Fetal Bovine Serum (16000036), MEM Non-Essential Amino Acids Solution (11140050), DMSO (D12345), Geltrex (A1569601), 0.5M EDTA (15575020), Dulbecco's phosphate-buffered saline, calcium and magnesium free DPBS−−(14190144), ProLong Gold Antifade Mountant with DAPI (P36931), Trizol Reagent (15596018) were from Thermo Fisher Scientific (Rockford, IL). QuantiFast SYBR Green PCR Kit (204056) and QuantiNova Reverse Transcription Kit (205411) were from Qiagen (Germantown, MD). Human Serum Albumin ELISA Kit (1190) was from Alpha Diagnostic International (San Antonio, TX). Human alpha 1 Antitrypsin ELISA Kit (ab108799) was from Abcam (Cambridge, MA). Periodic Acid-Schif (PAS) Kit (395B-1KT) was from Sigma (St. Louis, MO). Antibodies against OCT4 (ab19857), SOX2 (ab92494), SOX17 (ab84990) were from Abcam (Cambridge, MA). Antibodies against NANOG (sc-293121), SSEA-1 (sc-21702), TRA-160 (sc-21705), TRA-181 (sc-21706), FOXA2 (sc-271103), CXCR4 (sc- 53534), HNF4A (sc-374229), A1AT (sc-30121) were from Santa Cruz Biotechnology, Inc (Santa Cruz, CA). Antibody against AFP (A8452) and ALBUMIN (A6684) was from Sigma (St. Louis, MO). Secondary FITC-conjugated anti-rabbit (A21206), anti-mouse (A11059), TRITC-conjugated anti-rabbit (R37117) and anti-mouse (A11005) antibodies were from Thermo Fisher Scientific (Rockford, IL).
2.2. Human pluripotent stem cell culture
Human induced pluripotent stem cells (SC101A) were obtained from Systems Biosciences (Palo Alto, CA). For the purpose of our experiments, hPSCs were adapted into feeder-free conditions by plating on Geltrex (Thermo Fisher Scientific, Rockford, IL) coated plates, and fed daily with Essential 8 Medium (Thermo Fisher Scientific, Rockford, IL). At 60–70% confluence, cells were passaged using 0.5mM EDTA (Thermo Fisher Scientific, Rockford, IL). The E8 medium was supplemented with 10 μM of p160ROCK inhibitor Y- 27632 (ROCKi, Tocris) during the first 24 h after passaging. Pluripotency was monitored by qRT-PCR and immunostaining the cells for the pluripotency markers Nanog, OCT4, SOX2, Tra-161, Tra- 181 and SSEA4 and qRT-PCR for OCT4, c-Myc, REX-1, Nanog, SOX2 and KLF4.
2.3. In vitro differentiation of iPSCs into hepatic cells
hPSCs were differentiated to HLCs using a slightly modified version of the previously described protocol (Siller et al., 2016; Mathapati et al., 2016). There were minor adjustments in timing and concentrations. Briefly, hPSCs were seeded on Geltrex-coated 6-well plates or cover slips in E8 medium with ROCKi at a final concentration of 10 μM. hPSCs were cultured until 80% confluence was achieved (3 days).
2.3.1. Definitive endoderm formation
For Definitive Endoderm (DE) differentiation, hPSCs were washed with 5 mL DPBS−/− and cultured in 2 mL RPMI/B-27 medium (Thermo Fisher Scientific) with insulin and 10uM CHIR99021 (Stemgent) for 72 h at 37 °C, 5% CO2 with daily media changes (Mathapati et al., 2015; Siller et al., 2015; Siller et al., 2016). After 72 h, CHIR99021 was removed and DE cells were cultured in RPMI/B27 +Insulin for 24 h.
2.3.2. Hepatoblast specification
For hepatoblast specification, DE cells were cultured in Knockout DMEM (Thermo Fisher Scientific) containing 20% Knockout Serum Replacement (Thermo Fisher Scientific), 1% DMSO (Thermo Fisher Scientific), 5mM GlutaMAX (Thermo Fisher Scientific), 1 ×MEM non-essential amino acids (Thermo Fisher Scientific) and 100 μM 2- mercaptoethanol (Sigma). DE cells were incubated with hepatoblast specification media at 37 °C, 5% CO2 for 6 days with media changes every 48 h (Mathapati et al., 2015; Siller et al., 2015; Siller et al., 2016).
2.3.3. Hepatocyte-like cell differentiation and maturation
For hepatocyte-like cell differentiation, hepatoblasts were cultured in maturation medium consisting of L-15 Leibovitz medium (Thermo Fisher Scientific), 100 nM Dihexa (DC Chemicals), 6% Insulin- Transferrin-Selenium supplement (Thermo Fisher Scientific), 10% Fetal bovine serum (Thermo Fisher Scientific), 2mM GlutaMAX (Thermo Fisher Scientific), 10% Tryptose Phosphate Broth (Thermo Fisher Scientific), 10 μM hydrocortisone-21-hemisuccinate (Sigma), 100 nM Dexamethasone (Sigma) and 50 μg/mL Sodium-L-Ascorbate (Sigma). Hepatoblasts were incubated in hepatic maturation media at 37 °C, 5% CO2 for 12 days with media changes every 48 h (Mathapati et al., 2015; Siller et al., 2015; Siller et al., 2016). All small molecules were resuspended in DMSO.
2.4. Immunocytochemistry
Differentiated and undifferentiated cells were grown on glass cover slips, washed with PBS, fixed with 2% paraformaldehyde for 10min at room temperature, washed with PBS and quenched with 100 mmol/l glycine (pH 7) for 5 mins. They were permeabilized with 0.1% Triton-X- 100 for 5min, blocked with 1% BSA in PBS for 20min at room temperature, and sequentially incubated with respective primary antibodies diluted in blocking solution (1% BSA in PBS) at 4 °C overnight and then with appropriate fluorophore labeled secondary antibodies diluted in blocking solution (1% BSA in PBS) for 1 h at 37 °C in the dark. PBS was used for washes between steps. Controls were incubated with secondary antibodies only. Coverslips were mounted onto glass microscope slides with Prolong Gold Antifade with DAPI (Thermo Fisher Scientific). Confocal images of immunostained cells were obtained using 60 × oil objective on a Leica SP5 Scanning Confocal Microscope (Frankfurt, Germany). Digitized confocal images were processed with Image J image processing and analysis software. For quantification, differentiated cells stained with respective antibodies were manually counted using confocal microscopy. To minimize bias, the images were randomized. Cells staining positive and negative for respective antibodies were counted in 4 fields of view, and percentages were then determined. A minimum of 300 cells were counted in each field of view. The data is presented as the average of the 4 fields of view + /− the standard deviations (Asumda and Chase, 2012; Siller et al., 2016).
2.5. Western blot analysis
Total cell lysates were collected and prepared as previously described (). Protein content of lysates was determined and stored at − 20 °C. Total cell proteins (25 μg) were resolved on 8–12% sodium dodecylsulphate polyacrylamide gel and transferred to a polyvinylidene difluoride (PVDF) membrane. Blocking was done with 5% non-fat skimmed milk in Tris-Buffered saline (TBS, pH 7.4) containing 0.1% Tween-20 (TBST). The membranes were probed with respective primary antibodies overnight at 4 °C. Blotting membranes were washed three times with TBST and incubated with secondary antibodies for 2 h at room temperature. Immunoreactive bands were visualized using chemiluminescence reagent (Yoneda et al., 2016) followed by autoradiography. β-actin was used as the loading control.
2.6. RNA isolation, cDNA synthesis and RT-PCR analysis
Total RNA was extracted from hPSCs using Trizol Reagent (Thermo Fisher Scientific) according to the manufacturer's protocol. For qPCR, 2 μg of total RNA was reverse transcribed with the QuantiNova Reverse Transcription Kit (Qiagen) according to the manufacturer's protocol. qPCR was performed using the QuantiFast SYBR Green PCR Kit (Qiagen) according to the manufacturer's instructions. The cycling profile for real-time PCR (40 cycles) was as follows: 30 s at 95 °C for enzyme activation, 5 s at 95 °C for initial denaturation, 5 s at 65 °C for annealing/extension and a 5 s melt curve step at 65–95 °C. Gene analysis was performed with the Bio-Rad CFX Manager software (Bio-Rad). Gene expression is normalized relative to unstimulated cells and fold variation is GAPDH normalized. The primer sequences used are shown in Additional file 1.
2.7. Albumin production
Albumin production was assessed on matured HLCs using the human Serum Albumin ELISA Kit (Alpha Diagnostics) according to the manufacturer's instructions. Maturation media (L15 medium containing dexamethasone and Dihexa) was collected at 24 h intervals during the maturation and flash frozen at − 80 °C for further analysis. All data was normalized to total protein in the well. Data is presented at the mean of two experiments + /− the standard deviation.
2.8. A1AT production
Human alpha 1 Antitrypsin production was assessed on matured HLCs using the human alpha 1 antitrysin ELISA Kit (Abcam) according to the manufacturer's instructions. Maturation media (L15 medium containing dexamethasone and Dihexa) was collected at 24 h intervals during the maturation and flash frozen at − 80 °C for further analysis. All data was normalized to total protein in the well. Data is presented at the mean of two experiments + /− the standard deviation.
2.9. Periodic acid-Schiff staining
The Periodic Acid-Schiff Staining Kit (Sigma) was using to assess differentiated HLCs for glycogen storage according to the manufacturer's instructions.
2.10. Imaging
Phase contrast imaging was performed on the IncuCyte S3 Live-Cell Analysis System (Essen Bioscience, London, UK).
2.11. Statistics
All differentiation experiments were carried out in triplicate (n = 3). Data are presented as mean ± SEM. Statistical significance was determined using two tailed student's t-test with p < 0.05 determined to be significant.
3. Results
3.1. Characterization of human induced pluripotent stem cells
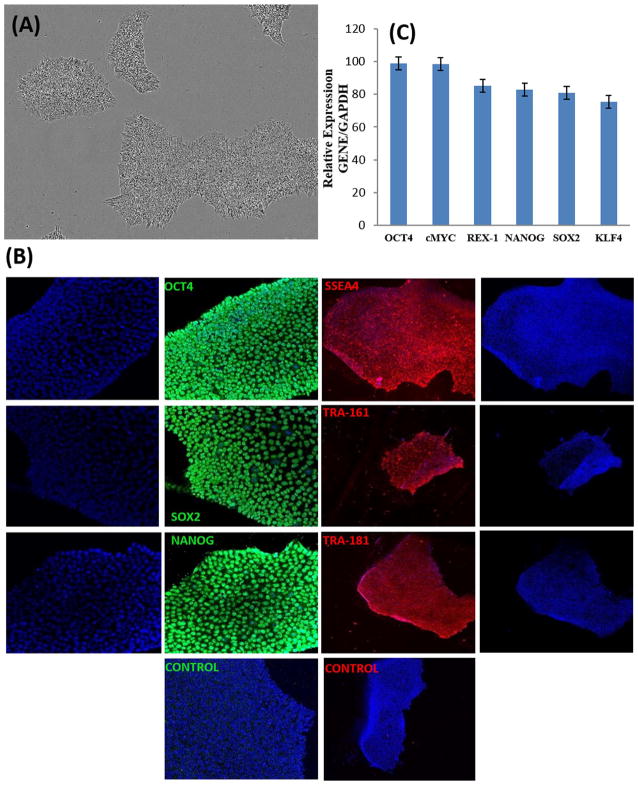
Phase contrast micrograph of hPSC colonies displaying typical morphology is shown in Fig. 1A. Expression of embryonic stem cell and pluripotency markers OCT4, SOX2, NANOG, SSEA4, TRA-1–61 and TRA-1–81 by hPSCs was detected by immunostaining (Fig. 1B). qRT-PCR analysis of hPSCs is demonstrating the expression of OCT4, SOX2, cMYC, REX4, NANOG and KLF4 is shown in Fig. 1C.
Fig. 1. Characterization of human iPSCs.
(A) Phase Contrast Micrograph of human iPSC colonies (20 ×). (B) Immunostaining of human iPSC clones for pluripotency markers OCT4, SOX2, NANOG, SSEA-4, Tra-1–60, and Tra-1–81. Nuclei were stained with DAPI in all. (C) qRT-PCR analysis for relative expression of OCT-4, cMYC, REX-4, NANOG, SOX-2 and KLF4. Columns show the combined mean ΔΔCq values for each marker. Data represent relative expression of transcripts normalized relative to GAPDH and expressed at the Mean ± SEM. Representative data from three independent experiments are shown.
3.2. Induction of homogeneous definitive endoderm population
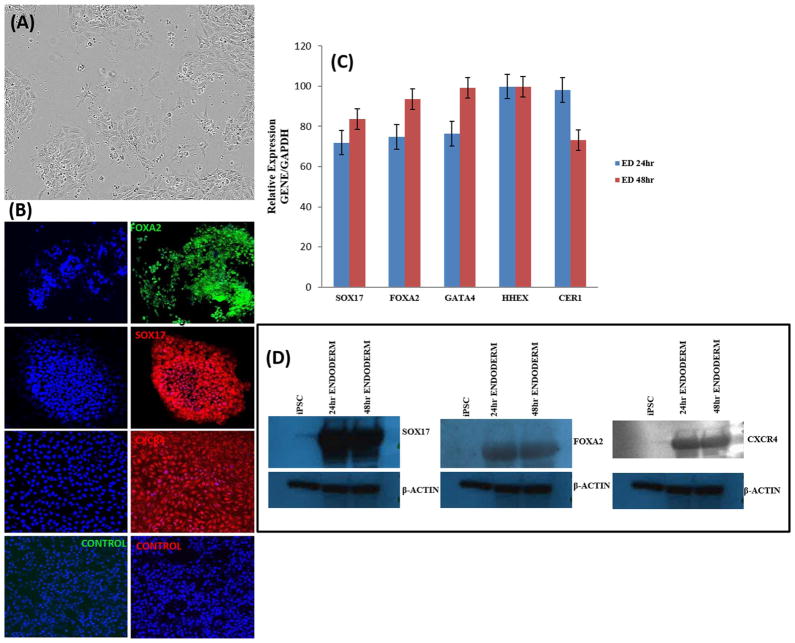
We induced DE differentiation as a starting point for the generation of functional hepatocytes from hPSCs. hPSCs at 80% confluence were treated with 10 μM of the GSK3β inhibitor CHIR99021 (Stemgent) for 72 hrs. We compared the effect of CHIR99021 at 3uM and 10uM in inducing endoderm formation by assessing the expression of SOX17, FOXA2, GATA4, HHEX and CER1 transcripts (Supporting Information Fig. S4). There was no significant difference in gene expression between the two concentrations of CHIR99021. We therefore settled on a 10uM concentration of CHIR99021 for all experiments in the current study. We observed significant morphological changes over the 72 h period (Fig. 1A; Supporting Information Fig. S2). After 24 h of differentiation, the typical flat hPSC colonies changed to a continuous monolayer with a compact, domed, bright 3D morphology (Fig. 1A; Supporting Information Fig. S2) (D'Amour et al., 2005; Siller et al. 2016). There was a decrease in the high nuclear to cytoplasmic ratio in hPSCs within 24 h of treatment, indicative of epithelial-to-mesenchymal transition in response to directed differentiation (Fig. 1A; Supporting Information Fig. S2). Significant cellular migration and proliferation was evident by 48 h after differentiation (Supporting Information Fig. S2). At 48 h, we observed the prototypical DE morphology— single-cell thick epithelial sheets with a cobblestone/ petal appearance (Supporting Information Fig. S2) (D'Amour et al., 2005; Siller et al. 2016). Successful induction of DE differentiation was indicated by the expression of the DE markers sex determining region Y-box 17 (SOX17), chemokine (C-X-C motif) receptor type 4 (CXCR4), cerberus (CER1), hematopoietically expressed homeobox (HHEX) and FOXA2 which encodes the hepatocyte nuclear factor (HNF3β) at 24h and 48 h (Fig. 2A; Fig. 2B; Fig. 2C). We assessed the efficiency of DE formation by analyzing FOXA2, SOX17 and CXCR4 expression by immunostaining (Fig. 2B) and western blot analysis (Fig. 2C). The overall proportion of FOXA2, SOX17 and CXCR4 positive cells was 97 ± 2% 96 ± 1% and 96 ± 1% respectively (Supporting Information Fig. S3). Taken together, our data confirms that treatment of hPSCs with the GSK-3 inhibitor CHIR99021 definitively generates a large population of DE cells.
Fig. 2. Characterization of Definitive Endoderm differentiation.
(A) Representative phase contrast micrograph of definitive endoderm cells (20 ×). (B) Expression of FOXA2, SOX17 and CXCR4 after treatment with CHIR99021, imaged with confocal microscopy. Nuclei were stained with DAPI in all. (C) Relative gene expression after 24 h and 48 h of differentiation as measured by qRT-PCR. Data represent relative expression of transcripts normalized relative to GAPDH and undifferentiated controls. Data are represented as Mean ± SEM for three biologically independent experiments (n = 3). (D) Western blot assessment of FOXA2, SOX17 and CXCR4 expression at 24 h and 48 h of Definitive Endoderm differentiation. Representative data from three independent experiments are shown.
3.3. Differentiation of definitive endoderm into hepatoblasts
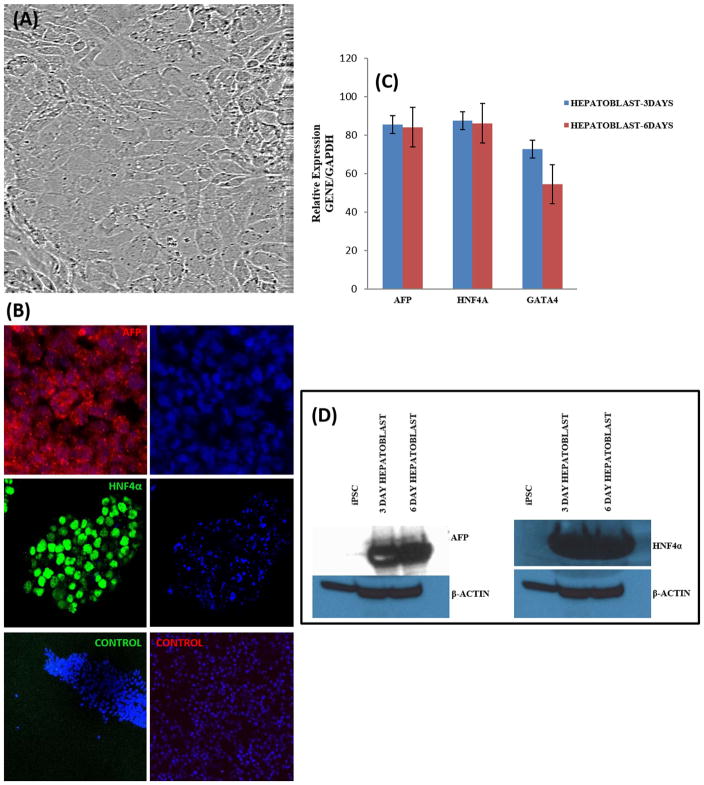
Hepatic specification was achieved by treatment of DE cells for 7 days with 1% DMSO in Knockout DMEM (Thermo Fisher Scientific) containing 20% Knockout Serum Replacement (Thermo Fisher Scientific). Over 6 days of differentiation, cells adopted morphological features characteristic of hepatoblasts (Fig. 3A; Supporting Information Fig. S2C). We observed rapid proliferation, cytoplasmic enlargement of individual cells to form a cuboidal shape with well-defined boundaries (Fig. 3A; Supporting Information Fig. S2). We also observed a mesenchymal-like morphology with cytoplasmic granularity after 48 h of induction with hepatic specification media (Supporting Information Fig. S2C). We analyzed hepatoblasts at 3 days and 6 days for expression of AFP, HNF4A and GATA4 by qRT-PCR (Fig. 3B). Hepatoblast expression of AFP and HNF4A proteins was confirmed by immunostaining (Fig. 3C) and Western blot analysis (Fig. 3D). We assessed the proportion of AFP (97 ± 1%) and HNF4A (97 ± 0.5%) positive cells by immunostaining (Supporting Information Fig. S3). The expression of AFP and HNF4A confirmed successful transition from DE to early hepatic progenitor fate (hepatoblasts).
Fig. 3. Characterization of Hepatoblast differentiation.
(A) Representative phase contrast micrograph of hepatoblast cells (20 ×). (B) Expression of AFP and HNF4A at 3 and 6 days, imaged with confocal microscopy. Nuclei were stained with DAPI in all. (C) Relative gene expression after 3 and 6 days of differentiation as measured by qRT-PCR. Data represent relative expression of transcripts normalized relative to GAPDH and undifferentiated controls. Data are represented as Mean ± SEM for three biologically independent experiments (n = 3). (D) Western blot assessment of AFP and HNF4A expression at 3 and 6 days of hepatoblast differentiation. Representative data from three independent experiments are shown.
3.4. Maturation of hepatoblasts into hepatocyte-Like cells
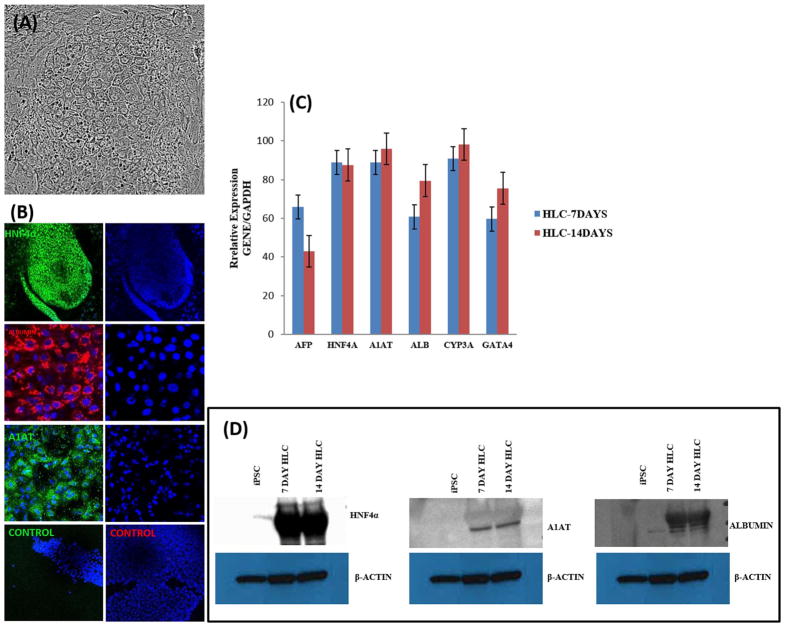
N-hexanoic-Tyr, Ile-(6) aminohexanoic amide (dihexa) is a small molecule mimetic that was originally developed to treat neurodegenerative diseases (McCoy et al., 2013; Benoist et al., 2014; Uribe et al., 2015). It is a potent and stable HGF receptor agonist that is proven to be involved in hepatocyte maturation (McCoy et al., 2013; Siller et la, 2015; Mathapati et al., 2016; Hannoun et al., 2016). To promote terminal differentiation, hepatoblasts were treated for 14 days with 100 nM Dihexa, 6% Insulin-Transferrin-Selenium supplement, 10% Fetal bovine serum, 10% Tryptose Phosphate Broth, 10 μM hydrocortisone- 21-hemisuccinate, 100 nM Dexamethasone and 50 μg/mL Sodium-L-Ascorbate (Siller et la, 2015; Mathapati et al., 2016). Within the first 3 days of maturation, cells adopted a large angular polygonal morphology, distinct round nuclei with one or two prominent nucleoli and binucleate cells with bright junctions characteristic of mature hepatocytes (Fig. 4A; Supporting Information Fig. S2). Gene expression for liver-specific genes such as HNF4A, albumin, (ALB), α1-antitrypsin (A1AT) and CYP3A4 was analyzed by qRT-PCR (Fig. 4B). We further assessed the expression of the hepatic transcripts CYP71a, K18, and K19 (Supporting Information Fig. S5). The respective proteins for albumin (ALB), HNF4A and α1-antitrypsin (A1AT) were assessed by immunostaining (Fig. 4C) and Western blot analysis (Fig. 4D). We assessed the proportion of ALB (97 ± 1%), A1AT (98 ± 0.5%) and HNF4A (98 ± 0.3%) positive cells by immunostaining (Supporting Information Fig. S3).
Fig. 4.
Characterization of Hepatocyte-like cell differentiation. (A) Representative phase contrast micrograph of HLC cells (20 ×). (B) Expression of HNF4A, A1AT and ALBUMIN at 7 and 14 days, imaged with confocal microscopy. (C) Relative gene expression after 7 and 14 days of differentiation as measured by qRT-PCR. Data represent relative expression of transcripts normalized relative to GAPDH and undifferentiated controls. Data are represented as Mean ± SEM for three biologically independent experiments (n = 3). (D) Western blot assessment of HNF4A, A1AT and ALBUMIN expression at 7 and 14 days of HLC maturation. Representative data from three independent experiments are shown.
3.5. Functional assessment of hepatocyte-like cells
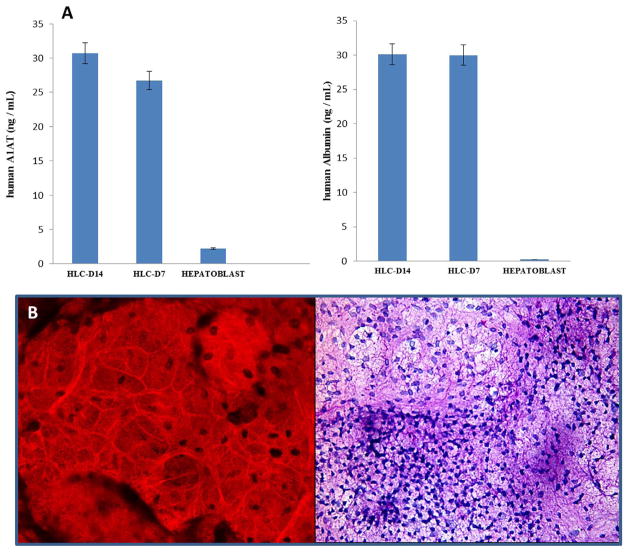
To determine the functionality of these small-molecule derived HLCs, we assessed the secretion of albumin and α1-antitrypsin by ELISA. We detected significant levels of both proteins in the culture medium (Fig. 5A; B). We next tested the ability of these small-molecule derived HLCs to store glycogen by staining with periodic acid Schiff (PAS) and counter-staining with H& E. Extensive cytoplasmic staining (pink to purple) was observed (Fig. 5C) which is consistent with glycogen storage.
Fig. 5. Functional assays of hepatocyte-like cells.
(A) Serum protein excretion of A1AT and ALBUMIN in HLCs and Hepatoblasts. A1AT and ALB were normalized to total protein content in each well. (B) Glycogen storage in HLCs as indicated by PAS staining. Data represent the Mean ± SEM for three biologically independent experiments. Representative data from three independent experiments are shown.
4. Discussion
We presented data here that demonstrate efficient HLC generation from hPSCs in vitro via a small-molecule based differentiation protocol. The morphology, molecular and functional profile of the HLCs we generated is consistent with hepatocytes. We demonstrate that hepatocyte- like cells derived through this protocol express markers of definitive endoderm (SOX17, FOXA2, CXCR4), hepatoblast (HNF4A, AFP) and hepatocyte (HNF4A, AFP, ALB (albumin)), A1AT (α − 1-antitrypsin (A1AT)). The HLCs we produced with this protocol retained critical hepatic functions such as ALB, A1AT secretion and glycogen storage. This data is further proof of the efficacy of small molecules as a viable alternative to growth factors. The differentiated cells obtained with the current protocol are duly named “hepatocyte-like” because we did not test their expression of the complete repertoire of hepatic genes and metabolic proteins. Other labs have demonstrated that HLCs produced via this small-molecule driven approach maintain robust Cytochrome P450, CYP3A4, CYP1A2 activity, secrete fibronectin, and maintain uptake of indocyanine (Siller et al., 2015; Mathapati et al., 2016). But the continued expression of AFP in the HLCs produced in this current study indicates that they are not completely synonymous with primary adult hepatocytes.
PHHs maintain a regenerative capacity in their native tissue environment which is lost ex vivo. A stem cell based in vitro model system that maintains the full repertoire of hepatocellular functions, and responses is invaluable for the field from the standpoint of mechanistic work. It is now possible to successfully recapitulate liver disorders in a controlled in vitro environment with hPSCs (Siller et al., 2013). A critical first step in harnessing hPSCs to model liver disease is the development of a simple and efficient differentiation system. The small-molecule based protocol we used in this study was first reported by Siller et al. (2015). It was slightly modified and improved by Mathapati et al. (2016). This protocol is devoid of growth factors is chemically defined. The protocol proved to be rather straightforward in our hands. We discovered that it is flexible with regards to specific concentrations of the key small-molecules required to drive the differentiation process. Its simplicity lies in the fact that a single chemical entity (GSK-3 inhibitor CHIR99021) is required to generate a considerably large pure population of DE cells with a hepatic predisposition (Dravid et al., 2005; Bone et al., 2011; Tahamtani et al., 2013). Presumably, the inhibition of GSK-3 by CHIR99021 mimics the action of Wnt signaling and induces the endogenous NODAL signaling pathway (Bone et al., 2011). These actions enhance the differentiation of pluripotent cells to a state akin to the primitive streak and then to definitive endoderm (Bone et al., 2011). The second step in this small-molecule based protocol relies on sequential treatment with DMSO. DMSO is a well-established driver of hepatic specification (Hay et al., 2008; Sullivan et al., 2010; Siller et al., 2015; Mathapati et al., 2016). Both the first and second steps of this small-molecule based protocol present a great opportunity for cost saving in HLC production.
In conclusion, we demonstrate here that sequential culture of hPSCs as a monolayer in chemically defined medium supplemented with only small-molecules is sufficient to efficiently generate HLCs with adult-liver characteristics and functionality. Our data is further evidence that the small-molecule based approach is a simple, inexpensive and reproducible platform for the derivation of hepatocytes from hPSCs.
Supplementary Material
Acknowledgments
The authors greatly acknowledge Maria De Medina, Yaima de la Fuente, and Sandy Dostaler at the University of Miami Schiff Center for Liver Disease for their invaluable assistance; Maria Boulina for her invaluable assistance with confocal microscopy at the University of Miami School of Medicine Diabetes Research Institute Microscopy Core Lab.
Abbreviations
- A1AT
α1-antitrypsin (A1AT)
- ALB
albumin
- BMP-2
bone morphogenetic protein 2
- BSA
bovine serum albumin
- DAPI
4',6-diamidino-2-phenylindole
- BMP-4
bone morphogenic protein 4
- DE
definitive endoderm
- DMEM-HG
Dubelcco's modified Eagle's medium—high glucose
- DMSO
dimethyl sulfoxide
- EDTA
ethylenediaminetetraacetic acid
- EGF
epidermal growth factor
- FBS
fetal bovine serum
- FGF
fibroblast growth factor
- FITC
fluorescein isothiocyanate
- GSK-3
glycogen synthase kinase 3
- iPSC
induced pluripotent stem cell
- PBS
phosphate buffered saline
- TRITC
tetramethylrhodamine isothiocyanate
- HCV
hepatitis C virus
- HBV
hepatitis B virus
- HGF
hepatocyte growth factor
- OSM
oncostatin M
- hPSC
human pluripotent stem cells
- HLCs
hepatocyte-like cells
- ROCK
rho-associated protein kinase
- PHH
primary human hepatocytes
- RPMI
Roswell Park Memorial Institute Medium
- VEGF
vascular endothelial growth factor
Appendix A. Supplementary material
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.diff.2018.03.002.
References
- Agarwal S, Holton KL, Lanza R. Efficient differentiation of functional hepatocytes from human embryonic stem cells. Stem Cells. 2008;26:1117–1127. doi: 10.1634/stemcells.2007-1102. [DOI] [PubMed] [Google Scholar]
- Asumda FZ, Chase PB. Nuclear cardiac troponin and tropomyosin are expressed early in cardiac differentiation of rat mesenchymal stem cells. Differentiation. 2012;83:106–115. doi: 10.1016/j.diff.2011.10.002. [DOI] [PubMed] [Google Scholar]
- Baharvand H, Hashemi SM, Kazemi, Ashtiani S, Farrokhi A. Differentiation of human embryonic stem cells into hepatocytes in 2D and 3D culture systems in vitro. Int J Dev Biololgy. 2006;50:645–652. doi: 10.1387/ijdb.052072hb. [DOI] [PubMed] [Google Scholar]
- Basma H, Soto-Gutiérrez A, Yannam GR, Liu L, Ito R, Yamamoto T, Ellis E, Carson SD, Sato S, Chen Y, Muirhead D, Navarro-Alvarez N, Wong RJ, Roy-Chowdhury J, Platt JL, Mercer DF, Miller JD, Strom SC, Kobayashi N, Fox IJ. Differentiation and transplantation of human embryonic stem cell-derived hepatocytes. Gastroenterology. 2009;136:990–999. doi: 10.1053/j.gastro.2008.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benoist CC, Kawas LH, Zhu M, Tyson KA, Stillmaker L, Appleyard SM, Wright JW, Wayman GA, Harding JW. The pro-cognitive and synaptogenic effects of angiotensin IV-derived peptides are dependent on activation of the hepatocyte growth factor/c-Met system. J Pharmacol Exp Ther. 2014;351:390–402. doi: 10.1124/jpet.114.218735. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Blight KJ, McKeating JA, Rice CM. Highly permissive cell lines for subgenomic and genomic hepatitis C virus RNA replication. J Virol. 2002;76:13001–13014. doi: 10.1128/JVI.76.24.13001-13014.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bone HK, Nelson AS, Goldring CE, Tosh D, Welham MJ. A novel chemically directed route for the generation of definitive endoderm from human embryonic stem cells based on inhibition of GSK-3. J Cell Sci. 2011;124:1992–2000. doi: 10.1242/jcs.081679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borowiak M, Maehr R, Chen S, Chen AE, Tang W, Fox JL, Schreiber SL, Melton DA. Small molecules efficiently direct endodermal differentiation of mouse and human embryonic stem cells. Cell Stem Cell. 2009;4:348–358. doi: 10.1016/j.stem.2009.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breiman A, Grandvaux N, Lin R, Ottone C, Akira S, Yoneyama M, Fujita T, Hiscott J, Meurs EF. Inhibition of RIG-I-dependent signaling to the interferon pathway during hepatitis C virus expression and restoration of signaling by IKKepsilon. J Virol. 2005;79:3969–3978. doi: 10.1128/JVI.79.7.3969-3978.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpentier A, Tesfaye A, Chu V, Nimgaonkar I, Zhang F, Lee SB, Thorgeirsson SS, Feinstone SM, Liang TJ. Engrafted human cell-derived hepatocytes establish an infectious HCV murine model. J Clin Investig. 2014;124:4953–4964. doi: 10.1172/JCI75456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpentier A, Nimgaonkar I, Chu V, Xia Y, Hu Z, Liang TJ. Hepatic differentiation of human pluripotent stem cells in miniaturized format suitable for high-throughput screen. Stem Cell Res. 2016;16:640–650. doi: 10.1016/j.scr.2016.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YF, Tseng CY, Wang HW, Kuo HC, Yang VW, Lee OK. Rapid generation of mature hepatocyte-like cells from human induced pluripotent stem cells by an efficient three step protocol. Hepatology. 2012;55:1193–1203. doi: 10.1002/hep.24790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Amour KA, Agulnick AD, Eliazer S, Kelly OG, Kroon E, Baetge EE. Efficient differentiation of human embryonic stem cells to definitive endoderm. Nat Biotechnol. 2005;23:1543–1541. doi: 10.1038/nbt1163. [DOI] [PubMed] [Google Scholar]
- Dravid G, Ye Z, Hammond H, Chen G, Pyle A, Donovan P, Yu X, Cheng L. Defining the role of Wnt/beta-catenin signaling in the survival, proliferation, and self-renewal of human embryonic stem cells. Stem Cells. 2005;23:1489–1501. doi: 10.1634/stemcells.2005-0034. [DOI] [PubMed] [Google Scholar]
- Engert S, Burtscher I, Liao WP, Dulev S, Schotta G, Lickert H. Wnt/b-catenin signaling regulates Sox17 expression and is essential for organizer and endoderm formation in the mouse. Development. 2013;140:3128–3138. doi: 10.1242/dev.088765. [DOI] [PubMed] [Google Scholar]
- Goldring C, Antoine DJ, Bonner F, Crozier J, Denning C, Fontana RJ, Hanley NA, Hay DC, Ingelman-Sundberg M, Juhila S, Kitteringham N, Silva-Lima B, Norris A, Pridgeon C, Ross JA, Young RS, Tagle D, Tornesi B, van de Water B, Weaver RJ, Zhang F, Park BK. Stem cell-derived models to improve mechanistic understanding and prediction of human drug-induced livery injury. Hepatology. 2017;2:710–721. doi: 10.1002/hep.28886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannan NR, Segeritz CP, Touboul T, Vallier L. Production of hepatocyte-like cells from human pluripotent stem cells. Nat Protoc. 2013;8:430–437. doi: 10.1038/nprot.2012.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannoun Z, Steichen C, Dianat N, Weber A, Dubart-Kupperschmitt A. The potential of induced pluripotent stem cell derived hepatocytes. J Hepatol. 2016;1:182–199. doi: 10.1016/j.jhep.2016.02.025. [DOI] [PubMed] [Google Scholar]
- Hay DC, Fletcher J, Payne C, Terrace JD, Gallagher RC, Snoeys J, Black JR, Wojtacha D, Samuel K, Hannoun Z, Pryde A, Filippi C, Currie IS, Forbes SJ, Ross JA, Newsome PN, Iredale JP. Highly efficient differentiation of hESCs to functional hepatic endoderm requires activin A and Wnt3a signaling. Proc Natl Acad Sci USA. 2008;105:12301–12306. doi: 10.1073/pnas.0806522105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imamura T, Cui L, Teng R, Johkura K, Okouchi Y, Asanuma K, Ogiwara N, Sasaki K. Embryonic stem cell-derived embryoid bodies in three-dimensional culture system form hepatocyte-like cells in vitro and in vivo. Tissue Eng. 2004;10:1716–1724. doi: 10.1089/ten.2004.10.1716. [DOI] [PubMed] [Google Scholar]
- Lang J, Vera D, Cheng Y, Tang H. Modeling dengue virus-hepatic cell interactions using human pluripotent stem cell-derived hepatocyte-like cells. Stem Cell Rep. 2016;7:341–354. doi: 10.1016/j.stemcr.2016.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohmann V, Korner F, Koch J, Herian U, Theilmann L, Bartenschlager R. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science. 1999;285:110–113. doi: 10.1126/science.285.5424.110. [DOI] [PubMed] [Google Scholar]
- Mathapati S, Siller R, Impellizzeri AA, Lycke M, Vegheim K, Almaas R, Sullivan GJ. Small-molecule-directed hepatocyte-like cell differentiation of pluripotent stem cells. Curr Protoc Stem Cell Biol. 2016;38:1G.6–1 G.6.18. doi: 10.1002/cpsc.13. [DOI] [PubMed] [Google Scholar]
- McCoy AT, Benoist CC, Wright JW, Kawas LH, Bule-Ghogare JM, Zhu M, Appleyard SM, Wayman GA, Harding JW. Evaluation of metabolically stabilized angiotensin IV analogs as procognitive/antidementia agents. J Pharmacol Exp Ther. 2013;344:141–154. doi: 10.1124/jpet.112.199497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi M, Kurisaki A, Hayashi Y, Warashina M, Ishiura S, Kusuda-Furue M, Asashima M. Directed induction of anterior and posterior primitive streak by Wnt from embryonic stem cells cultured in a chemically defined serum-free medium. FASEB J. 2009;23:114–122. doi: 10.1096/fj.08-111203. [DOI] [PubMed] [Google Scholar]
- Palakkan AA, Nanda J, Ross JA. Pluripotent stem cells to hepatocytes, the journey so far. Biomed Rep. 2017;4:367–373. doi: 10.3892/br.2017.867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashid ST, Corbineau S, Hannan N, Marciniak SJ, Miranda E, Alexander G, Huang-Doran I, Griffin J, Ahrlund-Richter L, Skepper J, Semple R, Weber A, Lomas DA, Vallier L. Model. Inherit. Metab. Disord. liver Using Human. Induc. pluripotent stem. Cells. 2010;9:3127–3126. doi: 10.1172/JCI43122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sa-Ngiamsuntorn K, Wongkajornsilp A, Phanthong P, Borwornpinyo S, Kitiyanant N, Chantratita W, Hongeng S. A robust model of natural hepatitis C infection using hepatocyte-like cells derived from induced pluripotent stem cells as long along-term host. Virol J. 2016;13:59. doi: 10.1186/s12985-016-0519-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurai F, Mitani S, Yamamoto T, Takayama K, Tachibana M, Watashi K, Wakita T, Iijima S, Tanaka Y, Mizuguchi H. Human induced-pluripotent stem cell-derived hepatocyte-like cells as an in vitro model of human hepatitis B virus infection. Sci Rep. 2017;7:45698. doi: 10.1038/srep45698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan J, Schwartz RE, Ross NT, Logan DJ, Thomas D, Duncan SA, North TE, Goessling W, Carpenter AE, Bhatia SN. Identification of small molecules for human hepatocyte expansion and iPS differentiation. Nat Chem Biol. 2013;9:514–520. doi: 10.1038/nchembio.1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Si-Tayeb K, Noto FK, Nagaoka M, Li J, Battle MA, Duris C, North PE, Dalton S, Duncan SA. Highly efficient generation of human hepatocyte-like cells from induced pluripotent stem cells. Hepatology. 2010;51:297–305. doi: 10.1002/hep.23354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siller R, Greenhough S, Park IH, Sullivan GJ. Modelling human disease with pluripotent stem cells. Curr Gene Ther. 2013;13:99–110. doi: 10.2174/1566523211313020004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siller R, Greenhough S, Naumovska E, Sullivan GJ. Small-molecule-driven hepatocyte differentiation of human pluripotent stem cells. Stem Cell Rep. 2015;4:939–952. doi: 10.1016/j.stemcr.2015.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siller R, Naumovska E, Mathapati S, Lycke M, Greenhough S, Sullivan GJ. Development of a rapid screen for the endoderm differentiation potential of human pluripotent stem cell lines. Sci Rep. 2016;6:37178. doi: 10.1038/srep37178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan GJ, Hay DC, Park IH, Fletcher J, Hannoun Z, Payne CM, Dalgetty D, Black JR, Ross JA, Samuel K, Wang G, Daley GQ, Lee JH, Church GM, Forbes SJ, Iredale JP, Wilmut I. Generation of functional human hepatic endoderm from human induced pluripotent stem cells. Hepatology. 2010;51:329–335. doi: 10.1002/hep.23335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun C, Hu JJ, Pan Q, Cao Y, Fan JG, Li GM. Hepatic differentiation of rat induced pluripotent stem cells in vitro. World J Gastroenterol. 2015;21:11118–111126. doi: 10.3748/wjg.v21.i39.11118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasnim F, Phan D, Toh YC, Yu H. Cost-effective differentiation of hepatocyte-like cells from human pluripotent stem cells using small molecules. Biomaterials. 2015;70:115–125. doi: 10.1016/j.biomaterials.2015.08.002. [DOI] [PubMed] [Google Scholar]
- Tahamtani Y, Azarnia M, Farrokhi A, Sharifi-Zarchi A, Aghdami N, Baharvand H. Treatment of human embryonic stem cells with different combinations of priming and inducing factors toward definitive endoderm. Stem Cells Dev. 2013;22:1419–1432. doi: 10.1089/scd.2012.0453. [DOI] [PubMed] [Google Scholar]
- Touboul T, Hannan NRF, Corbineau S, Martinez A, Martinet C, Branchereau S, Mainot S, Strick-Marchand H, Pedersen R, Di Santo J, Weber A, Vallier L. Generation of functional hepatocytes from human embryonic stem cells under chemically defined conditions that recapitulate liver development. Hepatology. 2010;51:1754–1765. doi: 10.1002/hep.23506. [DOI] [PubMed] [Google Scholar]
- Uribe PM, Kawas LH, Harding JW, Coffin AB. Hepatocyte growth factor mimetic protects lateral line hair cells from aminoglycoside exposure. Front Cell Neurosci. 2015;9:3. doi: 10.3389/fncel.2015.00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Robotham JM, Lee E, Dalton S, Kneteman NM, Gilbert DM, Tang H. Productive hepatitis C virus infection of stem cell-derived hepatocytes reveals a critical transition to viral permissiveness during differentiation. PLoS Pathog. 2012;8:e1002617. doi: 10.1371/journal.ppat.1002617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneda M, Hyun J, Jakubski S, Saito S, Nakajima A, Schiff ER, Thomas E. Hepatitis B Virus and DNA Stimulation Trigger a Rapid Innate Immune Response through NF-κB. J Immunol. 2016;197:630–643. doi: 10.4049/jimmunol.1502677. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.