Flowering time is an important trait for determining the adaptability of a crop cultivar. A novel heading date regulator, OsHAPL1, interacts with DTH8-Hd1 to negatively regulate flowering time in rice.
Keywords: DTH8, Ehd1, HAP, Hd1, Hd3a, heading date, Oryza sativa, OsHAPL1, RFT1
Abstract
Heading date is an important agronomic trait related to crop yield. Many genes related to heading date have already been identified in rice (Oryza sativa), and a complicated, preliminary regulatory genetic network has also already been established, but the protein regulatory network is poorly understood. We have identified a novel heading date regulator, Heme Activator Protein like 1 (OsHAPL1), which inhibits flowering under long-day conditions. OsHAPL1 is a nuclear-localized protein that is highly expressed in leaves in a rhythmic manner. OsHAPL1 can physically interact with Days To Heading on chromosome 8 (DTH8), which physically interacts with Heading date 1 (Hd1) both in vitro and in vivo. OsHAPL1 forms a complex with DTH8 and Hd1 in Escherichia coli. OsHAPL1, DTH8, and Hd1 physically interact with the HAP complex, and also with general transcription factors in yeast (Saccharomyces cerevisiae). Further studies showed that OsHAPL1 represses the expression of the florigen genes and FLOWERING LOCUS T 1 (RFT1) and Hd3a through Early heading date 1 (Ehd1). We propose that OsHAPL1 functions as a transcriptional regulator and, together with DTH8, Hd1, the HAP complex, and general transcription factors, regulates the expression of target genes and then affects heading date by influencing the expression of Hd3a and RFT1 through Ehd1.
Introduction
Flowering (or heading in cereals) involves the transition from vegetative to reproductive growth in plants. The correct flowering time is not only important for reproductive success, but also affects crop quality and grain yield (Simpson and Dean, 2002). Multiple internal and external signals, including photoperiod, temperature, and phytohormonal levels, are involved in the flowering process (Simpson and Dean, 2002; Andrés and Coupland, 2012).
Photoperiod is the most important environmental signal determining flowering time. Previous studies have already identified many photoperiod-related genes and the regulatory genetic network of flowering in the model plants Arabidopsis and rice (Oryza sativa). In Arabidopsis (a long-day (LD) plant), the CONSTANS (CO) gene is regulated by GIGANTEA (GI, a circadian clock gene) (Park et al., 1999; Huq et al., 2000), and activates expression of FLOWERING LOCUS T (FT, a florigen gene in Arabidopsis) (Kardailsky et al., 1999). The FT protein moves to the shoot apical meristem and activates the expression of floral-determination genes to trigger flowering (Wigge et al., 2005; Corbesier et al., 2007). In rice (a short-day (SD) plant), flowering is controlled by an activation pathway in SD and a suppression pathway in LD conditions. Heading date1 (Hd1, the CO homologue in rice) is regulated by OsGI (the GI homologue in rice) (Yano et al., 2000; Hayama et al., 2003), and Hd1 promotes flowering by activating Hd3a expression under SD conditions and delays flowering by repressing Hd3a expression under LD conditions (Yano et al., 2000; Kojima et al., 2002; Komiya et al., 2008). The mechanisms are unknown. In addition, rice has a unique Early heading date 1 (Ehd1) pathway that is independent of the Hd1 pathway (Doi et al., 2004). Ehd1, which has no homologue in Arabidopsis, encodes a B-type response regulator that promotes the expression of the florigen genes and FLOWERING LOCUS T 1 (RFT1) and Hd3a (Doi et al., 2004). Many genes regulating the expression of Ehd1 have been identified. OsCOL4 (Lee et al., 2010), Grain number, plant height and heading date 7 (Ghd7) (Xue et al., 2008), Days To Heading on chromosome 8 (DTH8/Ghd8/OsHAP3H) (Xue et al., 2008; Wei et al., 2010), and OsCOL10 (Tan et al., 2016) function as repressors of Ehd1. OsMADS51 (Kim et al., 2007), Rice Indeterminate 1 (RID1)/Early heading date 2 (Ehd2) (Matsubara et al., 2008; Wu et al., 2008), Early heading date 3 (Ehd3), and Early heading date 4 (Ehd4) (Matsubara et al., 2011; Gao et al., 2013) function as positive regulators of Ehd1. Interestingly, a recent study has shown that a MYB-type protein, TaMYB72 from wheat (Triticum aestivum), a LD crop, promotes flowering under LD conditions when expressed in rice, suggesting functional conservation in flowering time control between the two diverse crops (Zhang et al., 2016).
mRNA is transcribed by RNA polymerase II, which is controlled by DNA elements in promoters and enhancers recognized by trans-acting factors and general transcription factors (GTFs; Tjian and Maniatis, 1994). Trans-acting factors regulate gene expression in a developmentally or tissue-specific cell-cycle or stimuli-dependent manner, whereas GTFs regulate all promoters as a whole multi-subunit holoenzyme (Parvin and Young, 1998). Some elements are present in most promoters, such as CCAAT boxes in animals, fungi, and plants (Bucher, 1990; Maity and de Crombrugghe, 1998; Mantovani, 1999). Previous studies have shown that CCAAT-binding proteins are NF-Y subunits composed of three parts: NF-YA (CBF-B, HAP2 in yeast), NF-YB (CBF-A, HAP3), and NF-YC (CBF-C, HAP5) (McNabb et al., 1995; Sinha et al., 1995; Mantovani, 1999). In rice, there are 11 HAP2 genes, 12 HAP3 genes, and 12 HAP5 genes (Thirumurugan et al., 2008; Li et al., 2016), each containing a conserved domain and non-conserved regions of varying length and amino acid sequence. The conserved domain is responsible for DNA binding and protein–protein interactions (Thirumurugan et al., 2008). Genes of each family member have different expression patterns, and there are various combinations of overlapping expression of HAP2, HAP3, and HAP5 (Thirumurugan et al., 2008). The biological functions of some HAP family members have recently been identified. High expression of OsHAP3E can induce a dwarf statured plant with erect leaves, whereas overexpression of OsHAP2E confers resistance to pathogens, salinity, and drought, and can increase photosynthesis rate and tiller number (Ito et al., 2011; Alam et al., 2015). OsBF-YC2 and OsNF-YC4 inhibit flowering in rice under LD conditions (Kim et al., 2016), whereas OsHAP5A, OsHAP5B, OsHAP3E, and OsHAP3D delay heading in LD conditions (Li et al., 2016). Although some HAP family members relating to heading date have been identified, the possible complexes involved in heading date are poorly understood.
Our group has been investigating the mechanisms controlling heading date by overexpressing hundreds of transcription factors in rice, and we have identified OsCOL10 and OsCOL13 as negative regulators (Sheng et al., 2016; Tan et al., 2016;). Here, we identify a new rice heading date regulator that encodes a histone-like transcription factor, Heme Activator Protein like 1 (OsHAPL1). OsHAPL1 downregulates expression of the florigen genes Hd3a and RFT1 via Ehd1, possibly by interacting with Hd1, the HAP complex, and GTFs. Our findings demonstrate a role of OsHAPL1 as a negative heading date regulator, thus providing insight into the flowering control network in rice.
Materials and methods
Plant materials and growth conditions
Transgenic plants were produced in the O. sativa japonica cv. Kita-ake background. Growth conditions included natural long days (NLD; Beijing, 40°13’N, 116°13’E), natural short days (NSD; Hainan, 18°48’N, 110°02’E), controlled long days (CLD; in growth chambers, 14 h light at 30°C/10 h darkness at 25°C), and controlled short days (CSD; in growth chambers, 10 h light at 30°C /14 h darkness at 25°C). ehd1, hd3a, and rft1 mutants in Nipponbare were created using the CRISPR-Cas9 genome editing system.
Vector construction
The ProUbi::OsHAPL1-VP64 plasmid was produced using the OsHAPL1 coding sequence (CDS) driven by the maize ubiquitin (Ubi) promoter fused to the binary vector LP042 nVP64-hyg-asRED via the Gateway cloning system. The ProUbi::OsHAPL1-Flag plasmid was produced using the OsHAPL1 CDS driven by the maize Ubi promoter fused to the binary vector pCAMBIA1390 via an In-Fusion Advantage PCR Cloning Kit (Clontech). The ProOsHAPL1::GenomeOsHAPL1-GFP plasmid was produced from the genomic DNA sequence and driven by the OsHAPL1 promoter fused to the binary vector pCAMBIA1390 using the In-Fusion Advantage PCR Cloning Kit. The oshapl1 mutant was obtained using the CRISPR-Cas9 system. The Pro35S::OsHAPL1-GFP plasmid was produced using the OsHAPL1 CDS driven by the cauliflower mosaic virus (CaMV 35S) promoter fused to the pAN580 vector. Vectors for yeast two-hybrid assays were constructed by CDS of relevant genes fused to pGBK T7 or pGAD T7 (Clontech). Vectors for pull-down assays were obtained by fusing gene CDS to the vectors pGEX-2T(GST), pET-28a(+)(His), and pMAL-c2X(MBP). Vectors for bimolecular fluorescence complementation (BiFC) were constructed by fusing the gene CDS to pSPYNE173 or pSPYCE(M) (Kerppola, 2006). All primers used in the study are listed in Supplementary Table S1, available at JXB online.
Quantitative real-time reverse transcription PCR
Total RNA was extracted using an RNA Prep Pure Kit (Zymo Research, Orange, CA, USA). A reverse transcription (RT) kit (Qiagen) was used in RT reactions. Quantitative RT-PCR (qRT-PCR) was performed using SYBR Premix Ex Taq Kit (TaKaRa; RR041A) in an ABI PRISM 7900HT (Applied Biosystems), and ubiquitin was used as the internal control. Standard errors were calculated from three biological replicates.
Subcellular localization of OsHAPL1 protein
Pro 35S ::OsHAPL1-GFP and Pro35S::OsMADS51-mCherry were co-transformed to rice protoplasts by polyethylene glycol-mediated transformation (Bart et al., 2006). Fluorescence signals were observed by confocal laser scanning microscopy (LSM 700; Carl Zeiss). Root tissues of ProOsHAPL1:: GenomeOsHAPL1-GFP transgenic plants were also analysed using the confocal laser scanning microscope with Pro35S::GFP transgenic plants as controls.
Yeast two-hybrid assays
Yeast two-hybrid assays were carried out to detect the interaction of relevant proteins using a procedure provided by Clontech. Baits consisted of proteins fused with the GAL4 DNA binding domain in pGBKT7 (Clontech). Preys were constructed by fusing proteins with the GAL4 activation domain from pGADT7. Bait and prey were co-transformed into yeast (Saccharomyces cerevisiae) AH109 cells. Positive interactions were confirmed by the ability of AH109 cells to grow on Synthetic Dropout/-Trp-Leu-His-Ade medium in a 30°C incubator.
In vitro pull-down assays
Plasmids containing GST-, HIS-, and MBP-labelled proteins were transformed into Escherichia coli strain BL21. Transformants were grown to a concentration of OD600 = 0.6 in a 37°C concentrator, and placed in a 22°C concentrator for at least 30 min. Expression of the fusion protein was then induced by adding isopropyl β-D-1-thiogalactopyranoside to 0.5 mM, and shaking for 5–6 h in the 22°C table concentrator. E. coli cells were lysed in Tris-HCl buffer containing 25 mM Tris·HCl (pH 7.5), 150 mM NaCl, and 1 mM DTT. The sample liquid was then sonicated to destroy the cell (cycles of 3 s on, 3 s off for a total time of 2 min). If the resulting bacterial liquid was not clear, sonication was repeated. Equal amounts of the two protein solutions were mixed and centrifuged, and the supernatant was incubated with glutathione agarose beads (GE Healthcare) overnight at 4°C. After spinning (500 g, 5 min, 4°C), the beads were collected and washed five times with buffer containing 25 mM Tris·HCl (pH 7.5), 150 mM NaCl, 1 mM DTT, 1% Triton X100, and 0.1% SDS. Finally, the proteins bound to the beads were separated from the beads by boiling in 2×SDS loading buffer in a 95–100°C water bath. The proteins were separated in SDS-PAGE gels and detected by western blot analysis using anti-GST (Medical Biological Laboratories,1:2000) and anti-MBP antibodies (New England Biolabs, 1:2000).
Bimolecular fluorescence complementation assays
The interacting proteins (OsHAPL1 and DTH8, DTH8 and Hd1) referred to in this paper were separately fused with YFPC and YFPN from the expression plasmids pSPYCE(M) and pSPYNE173. The plasmids were introduced into Agrobacterium tumefaciens (strain GV3101) and infiltrated into tobacco (Nicotiana benthamiana) according to the protocol described by Walter et al. (2004). Injected leaves were visualized by laser scanning microscopy (LSM 700; Carl Zeiss) 3 days after infiltration.
Yeast one-hybrid assay
The coding region of OsHAPL1 was inserted into the pB42AD vector. The promoter region of Ehd1 were amplified and inserted into the pLacZi vector. These constructs were transformed into the yeast strain EGY48. The assay was performed as previously described (Gao et al., 2013).
Chromatin immunoprecipitation assay
ChIP assay was performed as previously described (Zhang et al., 2012). Anti-DDDDK-tag antibodies (Medical Biological Laboratories, PM020) were used for detection.
Results
Constitutive expression of the OsHAPL1 gene causes delayed flowering
Previous work has investigated the regulatory mechanism of photoperiodically determined flowering date by large-scale screening of transcriptional factors (TFs) related to flowering (Zhao et al., 2015; Tan et al., 2016). We fused different types of TFs to the VP64 activation domain, driven by the maize (UBI) promoter (Fig. 1A). These constructs were then transformed into japonica rice cultivar cv. Kita-ake. This cultivar can be transformed at high efficiency and has a very short life span due to defects in several heading date repressors; this facilitates rapid screening of flowering control genes (Xue et al., 2008; Gao et al., 2013; Koo et al., 2013; Kwon et al., 2015). In addition, using Kita-ake can help to identify regulatory networks independent of the repressors known to be defective in Kita-ake. Previous studies have shown that TFs-VP64 in this system can cause strong dominant effects, allowing the function of the TFs to be studied (Beerli et al., 1998; Hanano and Goto, 2011). More than 50 TFs affecting flowering time have previously been isolated (Sheng et al., 2016; Tan et al., 2016). From these 50 candidate TFs, we focused on a new gene, LOC_Os05g41450, with unknown function. Conserved domains analysis at the National Center for Biotechnology Information showed there was a conserved H2A superfamily domain, which is same as the transcription factor CBF/NF-Y/archaeal histone domain analysed at http://www.ebi.ac.uk/interpro/ (see Supplementary Fig. S1A, available at JXB online). Phylogenetic tree analysis showed that this protein is homologous to HAP2 subunit members (Supplementary Fig. S1B), so we named it OsHAPL1 (HAP2-LIKE 1).
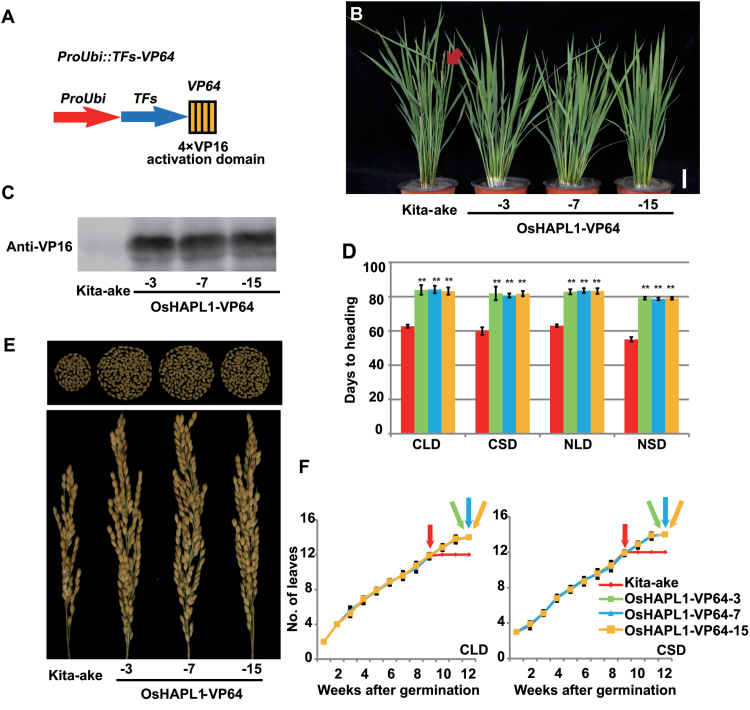
Fig. 1.
Phenotypic characterization of OsHAPL1-VP64 transgenic plants. (A) Diagram of ProUbi::TFs-VP64. (B) Heading date phenotypes of OsHAPL1-VP64 transgenic plants under LD conditions. Red arrow indicates heading. (C) Protein expression levels of OsHAPL1 in Kita-ake and three independent OsHAPL1-VP64 transgenic plants. (D) Statistical data for days to heading of Kita-ake and OsHAPL1-VP64 transgenic plants under different light conditions. See (F) for colour codes. (Student’s t-test: **P < 0.01) (E) Grains from a single panicle sampled from fully matured plants and panicle morphology of Kita-ake and OsHAPL1-VP64 transgenic lines. (F) Rate of leaf emergence of Kita-ake and transgenic plants under CLD and CSD conditions (n = 20). Arrows indicate approximate flowering time points of various lines. CLD, controlled long days; CSD, controlled short days; NLD, natural long days; NSD, natural short days.
In order to confirm the function of OsHAPL1 in flowering, we studied the phenotypes of OsHAPL1-VP64 transgenic plants. We randomly selected three independent homozygous transgenic lines to check the protein expression level; there was clear overexpression of OsHAPL1 (Fig. 1C). Transgenic plants were late heading compared to the Kita-ake wild type (WT) grown under NLD conditions (Fig. 1B). To determine whether the heading date of OsHAPL1-VP64 transgenic plants was affected by photoperiod, the plants were grown under CLD, CSD, NLD, and NSD conditions. The heading date of the transgenic plants was about 20 days later than that of Kita-ake plants under all four test conditions (Fig. 1D). OsHAPL1-VP64 transgenic plants had larger panicles and more grains than WT (Fig. 1E). Other agronomic traits, such as plant height, tiller number, panicle length, primary branches per panicle, secondary branches per panicle, grains per panicle, and 1000-grain weight were significantly increased relative to WT (Table 1). Leaf emergence rates were checked to eliminate the effect of vegetative stage on the heading date. There was no difference between these lines (Fig. 1F).
Table 1.
Phenotypic data for Kita-ake, and OsHAPL1-VP64 transgenic plants
| Genotype | Condition | Days to flowering | Plant height, cm | Tillers | Panicle length, cm | Primary branches n per panicle | Secondary branches n per panicle | Grains per panicle | 1000-grain weight, g |
|---|---|---|---|---|---|---|---|---|---|
| Kita-ake | NLD | 63.05 ± 0.76 | 70.95 ± 1.96 | 19.25 ± 2.79 | 14.23 ± 0.6 | 7.10 ± 0.72 | 11.50 ± 1.76 | 70.75 ± 6.38 | 27.07 ± 0.35 |
| OsHAPL1-VP64-3 | NLD | 82.80 ± 1.51 | 93.15 ± 2.70 | 11.05 ± 1.67 | 17.42 ± 0.82 | 14.45 ± 1.32 | 21.50 ± 4.03 | 143.85 ± 7.81 | 27.09 ± 0.57 |
| OsHAPL1-VP64-7 | NLD | 83.45 ± 1.39 | 92.70 ± 2.00 | 10.20 ± 0.95 | 17.02 ± 0.78 | 14.05 ± 1.36 | 21.25 ± 2.88 | 143.60 ± 6.64 | 26.44 ± 0.66 |
| OsHAPL1-VP64-15 | NLD | 83.22 ± 1.64 | 92.80 ± 2.58 | 10.10 ± 0.84 | 17.25 ± 0.95 | 15.02 ± 1.96 | 22.74 ± 4.26 | 140.20 ± 9.36 | 27.53 ± 1.07 |
| Kita-ake | NSD | 55.05 ± 1.31 | 59.32 ± 2.65 | 19.95 ± 1.93 | 10.58 ± 0.80 | 5.37 ± 0.60 | 5.21 ± 1.65 | 41.53 ± 5.85 | 26.31 ± 0.59 |
| OsHAPL1-VP64-3 | NSD | 79.07 ± 0.80 | 97.60 ± 1.84 | 11.53 ± 1.64 | 16.86 ± 0.90 | 13.9 ± 0.99 | 19.10 ± 1.73 | 132.40 ± 4.67 | 26.80 ± 0.54 |
| OsHAPL1-VP64-7 | NSD | 78.67 ± 0.72 | 99.40 ± 2.61 | 11.93 ± 1.10 | 17.98 ± 0.60 | 15.1.±1.52 | 27.50 ± 5.04 | 162.80 ± 10.57 | 27.08 ± 0.67 |
| OsHAPL1-VP64-15 | NSD | 79.00 ± 0.68 | 98.07 ± 2.59 | 11.43 ± 1.50 | 17.51 ± 1.06 | 14.71 ± 1.49 | 24.07 ± 6.15 | 149.07 ± 17.88 | 26.90 ± 0.65 |
Agronomic traits are based on Kita-ake and three independent homozygous OsHAPL1-VP64 transgenic plants under natural short-day (NSD) and natural long-day (NLD) conditions. Data for days to heading and other agronomic traits are presented as mean ± standard deviation (n = 22).
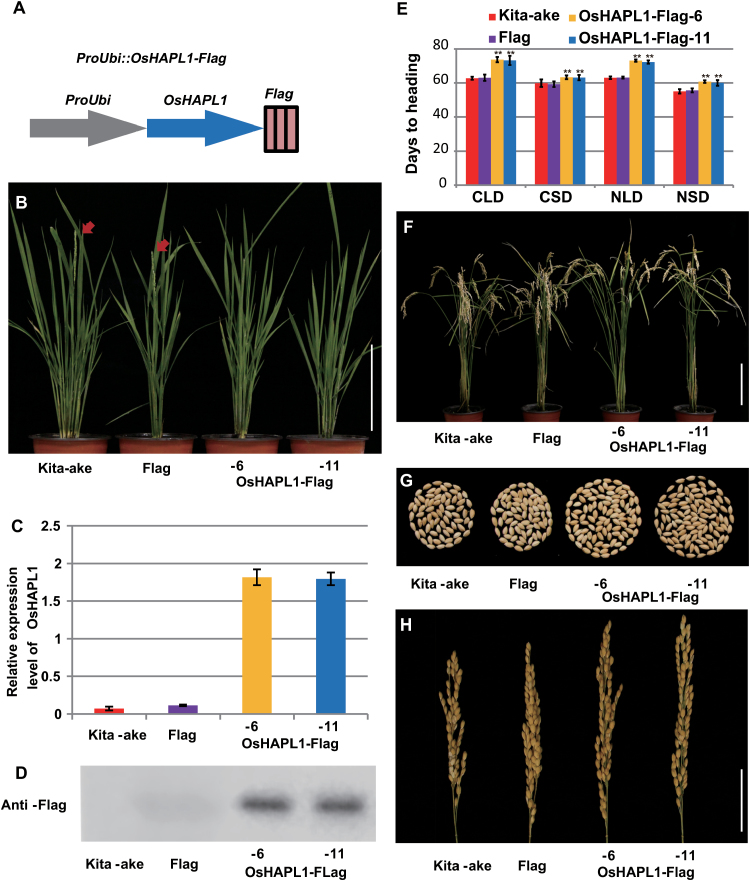
To further confirm the function of OsHAPL1 for flowering, the constitutive expression vector ProUbi::OsHAPL1-Flag was constructed and transformed into Kita-ake WT (Fig. 2A). We randomly selected two independent homozygous transgenic lines to determine the mRNA and protein expression levels; there was obvious mRNA and protein overexpression of OsHAPL1 (Fig. 2C, D). The transgenic plants were late flowering relative to Kita-ake and Proubi::Flag (control) transgenic plants under NLD conditions (Fig. 2B). Similarly, to check whether the heading dates of OsHAPL1-Flag transgenic plants were affected by photoperiod, heading date was obtained under different conditions. The transgenic plants had later heading dates than Kita-ake and Proubi::Flag transgenic plants in all four environments (Fig. 2E). Transgenic plants were taller and produced larger panicles with more grains (Fig. 2F, G, H). Other agronomic traits were similar to those of OsHAPL1-VP64 transgenic plants (Table 2). These data show that OsHAPL1 functions as a flowering repressor.
Fig. 2.
Phenotypic characterization of OsHAPL1-Flag transgenic plants. (A) Diagram of ProUbi::OsHAPL1-Flag. (B) Heading date phenotypes of OsHAPL1-Flag transgenic plants under LD conditions. Red arrows indicate heading. (C, D) mRNA (C) and protein (D) expression levels of OsHAPL1 in Kita-ake, Flag, and two independent OsHAPL1-Flag transgenic plants. (E) Statistical data for wild-type Kita-ake, Flag, and OsHAPL1-Flag transgenic plants under different light conditions. (Student’s t-test: **P < 0.01). (F) Mature stage of Kita-ake, Flag, and OsHAPL1-Flag transgenic plants under NLD conditions. (G) Grains from a single panicle sampled the fully matured plants of Kita-ake, Flag, and OsHAPL1-Flag transgenic plants. (H) Panicle morphology of Kita-ake and OsHAPL1-Flag transgenic plants. CLD, controlled long days; CSD, controlled short days; NLD, natural long days; NSD, natural short days.
Table 2.
Phenotypic data for Kita-ake, Flag, and OsHAPL1-Flag transgenic plants
| Genotype | Condition | Days to flowering | Plant height, cm | Tillers | Panicle length, cm | Primary branches n per panicle | Secondary branches per panicle | Grains per panicle | 1000-grain weight, g |
|---|---|---|---|---|---|---|---|---|---|
| Kita-ake | NLD | 63.05 ± 0.76 | 70.95 ± 1.96 | 19.25 ± 2.79 | 14.23 ± 0.6 | 7.10 ± 0.72 | 11.50 ± 1.76 | 70.75 ± 6.38 | 27.07 ± 0.35 |
| Flag | NLD | 63.17 ± 0.58 | 71.08 ± 2.15 | 18.83 ± 3.16 | 14.25 ± 0.64 | 7.08 ± 0.67 | 11.64 ± 1.44 | 69.33 ± 5.26 | 26.94 ± 0.36 |
| OsHAPL1-Flag-6 | NLD | 73.15 ± 0.67 | 76.10 ± 3.40 | 14.65 ± 2.78 | 16.07 ± 0.42 | 10.15 ± 1.6 | 15.70 ± 3.47 | 99.20 ± 11.06 | 26.94 ± 0.72 |
| OsHAPL1-Flag-11 | NLD | 72.30 ± 0.98 | 74.20 ± 2.98 | 15.42 ± 2.99 | 15.94 ± 0.46 | 11.52 ± 2.25 | 16.35 ± 4.66 | 95.50 ± 8.53 | 27.04 ± 1.43 |
| Kita-ake | NSD | 55.05 ± 1.31 | 59.32 ± 2.65 | 19.95 ± 1.93 | 10.58 ± 0.80 | 5.37 ± 0.60 | 5.21 ± 1.65 | 41.53 ± 5.85 | 26.31 ± 0.59 |
| Flag | NSD | 55.64 ± 1.21 | 59.27 ± 2.37 | 20.27 ± 1.85 | 10.59 ± 0.97 | 5.18 ± 0.40 | 5.09 ± 1.58 | 41.36 ± 5.43 | 26.09 ± 0.86 |
| OsHAPL1-Flag-6 | NSD | 61.40 ± 0.70 | 77.10 ± 2.42 | 12.70 ± 2.00 | 12.84 ± 0.81 | 6.63 ± 0.92 | 9.50 ± 1.60 | 59.50 ± 8.52 | 26.31 ± 0.59 |
| OsHAPL1-Flag-11 | NSD | 60.01 ± 0.88 | 74.50 ± 3.44 | 12.20 ± 4.10 | 12.25 ± 0.65 | 6.13 ± 0.64 | 8.75 ± 1.91 | 56.00 ± 8.00 | 26.81 ± 0.56 |
Agronomic traits are based on Kita-ake, Flag, and two independent homozygous OsHAPL1-Flag transgenic plants under natural short-day (NSD) and natural long-day (NLD) conditions. Data for days to heading and other agronomic traits are presented as mean ± standard deviation (n = 25).
Downregulation of the OsHAPL1 gene causes early flowering in long-day conditions
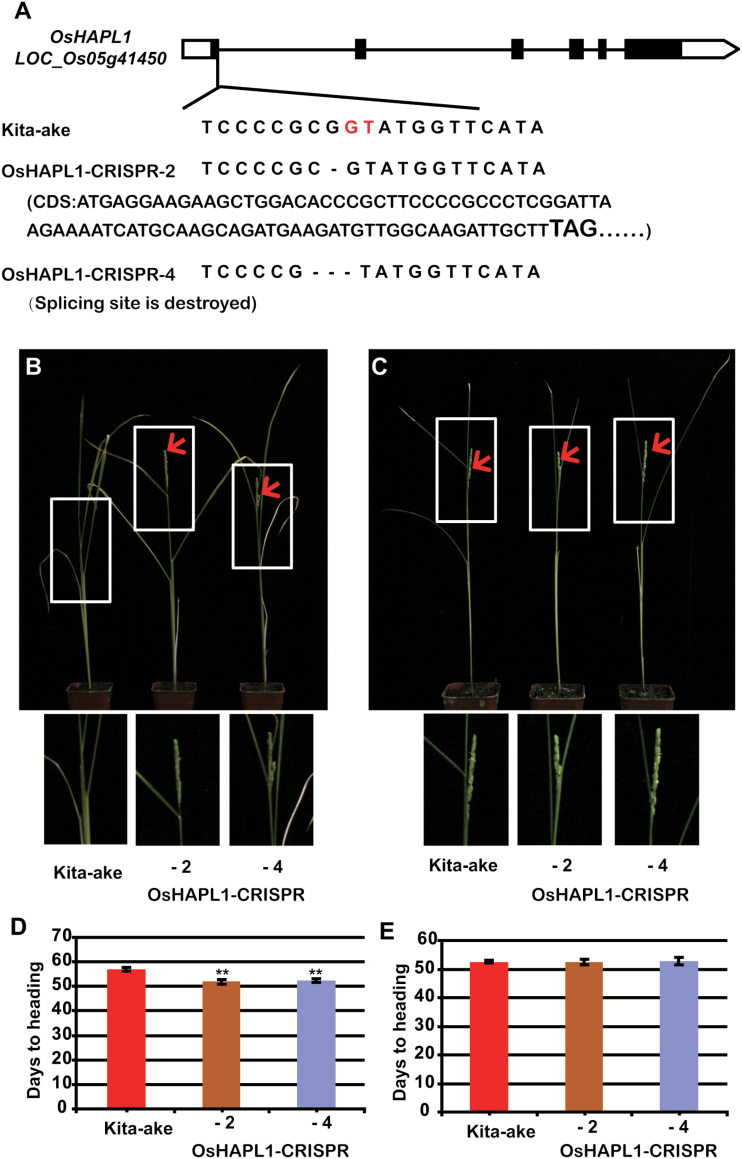
To further confirm the function of OsHAPL1, an oshapl1 mutant was constructed using the CRISPR-Cas9 method (Miao et al., 2013). Two homozygous oshapl1 mutant lines were obtained (Fig. 3A). These plants flowered earlier than WT under CLD conditions (Fig. 3B, D) but not under CSD conditions (Fig. 3C, E). We also created oshapl1 mutants using the same method in Nipponbare, a day length-sensitive cultivar, and observed a similar earlier flowering phenotype under NLD conditions (see Supplementary Fig. S2, available at JXB online). Thus, OsHAPL1 has a role in controlling flowering time under LD.
Fig. 3.
The phenotype of oshapl1 mutants. (A) Mutant site in oshapl1 induced by the CRISPR-Cas9 genome editing system. The one-base deletion in line 2 resulted in an early stop codon (bold letters), and the three-base deletion in line 4 disrupted the slicing site. The splicing site is highlighted in red. (B) Phenotype of oshapl1 mutants under CLD conditions. A close-up of the top part of the corresponding plants is shown below the panel. Red arrows indicate heading. (C) Phenotype of oshapl1 mutants under CSD conditions. A close-up of the top part of the corresponding plants is shown below the panel. Red arrows indicate heading. (D) Statistical data for heading date of Kita-ake and oshapl1 mutants under CLD. (E) Statistical data for heading date of Kita-ake and oshapl1 mutants under CSD. CLD, controlled long days; CSD, controlled short days; NLD, natural long days; NSD, natural short days.
OsHAPLl is a nuclear-localized protein and highly expressed in leaves
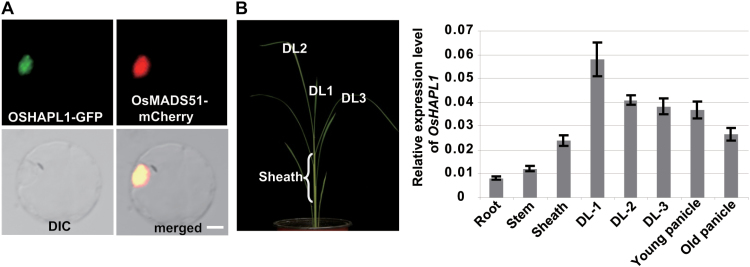
To investigate the subcellular localization of OsHAPLl, the OsHAPLl-GFP fusion protein was introduced into rice protoplasts. GFP fluorescence signals were detected in the nucleus, and the signals merged with known nuclear-localized signals (Fig. 4A). GFP signals were also detected in the nucleus of root cells of ProOsHAPL1::GenomicOsHAPL1-GFP transgenic rice (see Supplementary Fig. S3, available at JXB online).
Fig. 4.
Properties of OsHAPL1 protein and expression pattern of OsHAPL1. (A) Subcellular localization of OsHAPL1 in the rice protoplast. Scale bar = 5 μm. (B) Expression pattern of OsHAPL1 in different tissues. DL1 (uppermost leaf), DL2 (second uppermost leaf), DL3 (third uppermost leaf), and leaf sheath were sampled as shown in the left picture.
Real-time qRT-PCR assays were performed with mRNA from various tissues to analyse the expression patterns of OsHAPL1. OsHAPL1 was generally expressed in all tissues, but more highly expressed in the uppermost and second uppermost leaves (DL1, DL2) (Fig. 4B). High expression of OsHAPL1 in leaves is consistent with functions related to heading date.
Expression of OsHAPL1 is photoperiod responsive
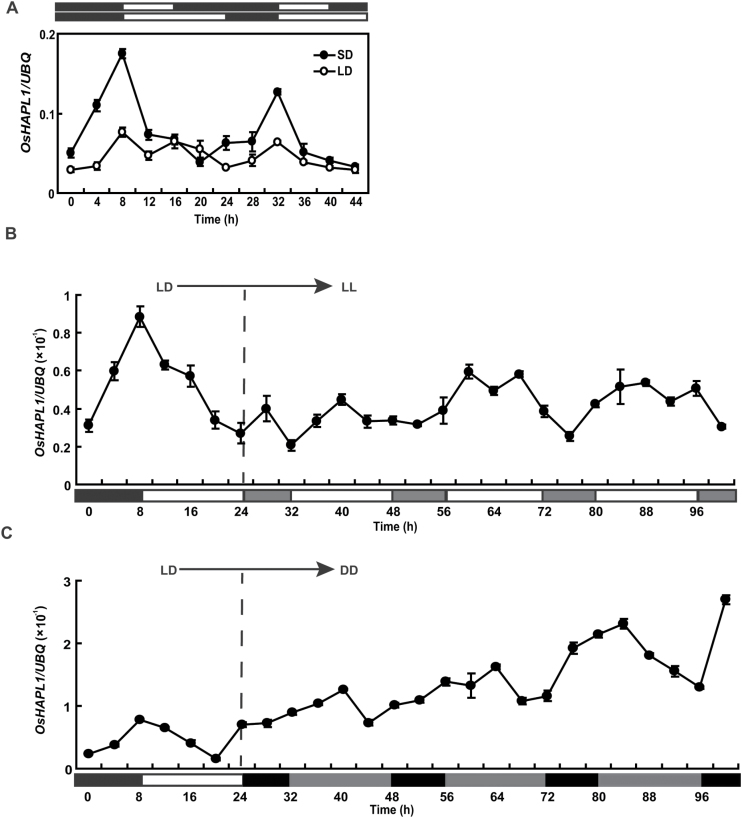
OsHAPL1 exhibited the same diurnal rhythmic expression patterns under SD and LD conditions. It accumulated from the beginning of darkness, reached a peak at the start of the light period, and then rapidly decreased (Fig. 5A). To ascertain if OsHAPL1 transcript levels were controlled by the circadian clock, plants grown in LD conditions were transferred to continuous light or continuous darkness, and then checked for expression of OsHAPL1. When subjected to continuous darkness or light conditions, the rhythmic expression patterns disappeared (Fig. 5B, C). Weakening of the rhythm in continuous darkness and continuous light suggests that OsHAPL1 is not controlled by the circadian clock. These results indicate instead that OsHAPL1 is expressed rhythmically and affected by photoperiod.
Fig. 5.
Expression analysis of OsHAPL1. (A) Rhythmic expression of OsHAPL1. (B, C) Expression analysis of OsHAPL1 under continuous light (LL) (B) and continuous dark (DD) (C) conditions. The plants were grown in growth chambers under LD conditions (14 h light /10 h darkness) for 30 days and then transferred to LL or DD conditions. White bars indicate light; black bars indicate darkness. The rice Ubiquitin gene was used as the internal control. Values represent means ± standard deviation (SD) from three independent biological replicates.
OsHAPL1 interacts with DTH8-Ghd8-OsHAP3H
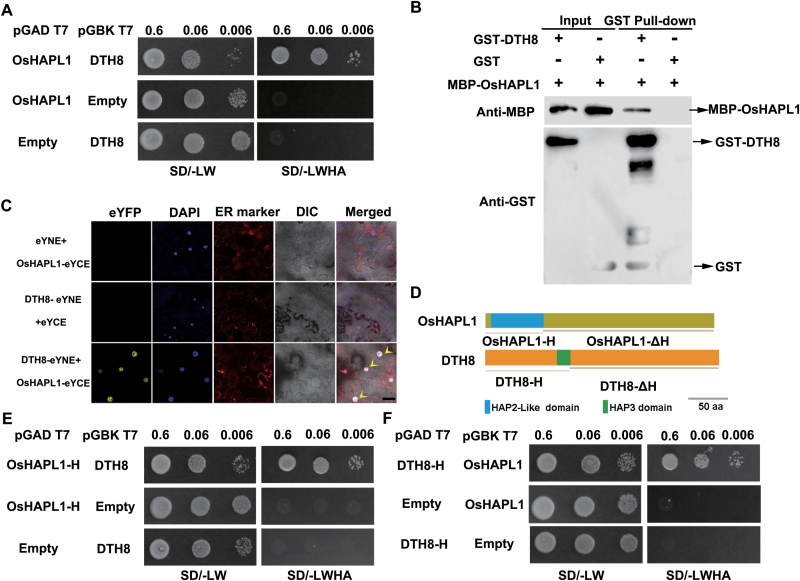
To investigate the potential functional role of OsHAPLl on heading date, we looked for proteins that interact with OsHAPLl using yeast two-hybrid screening of the yeast strain AH109. Using pGBKT7-OsHAPLl (a fusion protein consisting of the GAL4 DNA binding domain and OsHAPLl) as bait, we identified an amino acid sequence corresponding to residues 3–125 of DTH8-Ghd8-OsHAP3H (LOC-Os08g07740.1) (Wei et al., 2010; Yan et al., 2011) from a rice GAL4 activation fusion library. Previous studies have shown that DTH8 is a CCAAT-box-binding TF encoding a putative HAP3 subunit, and has a role in determining heading date in rice. Overexpression of DTH8 delays flowering (Wei et al., 2010); this phenotype is consistent with overexpression of OsHAPL1.
To confirm the results from library screening, the interaction between the full-length amino acid sequences of OsHAPLl and DTH8 was tested on selective media plates (Fig. 6A). An in vitro pull-down assay was performed to further confirm the interaction of OsHAPLl and DTH8. GST-tagged DTH8 could pull down MBP-tagged OsHAPLl, whereas GST alone could not pull down MBP-tagged OsHAPLl (Fig. 6B). To investigate whether OsHAPLl interacts with DTH8 in planta, a BiFC assay was performed in N. benthamiana. Fluorescence signals were observed in the epidermal cells of tobacco leaves transformed transiently with pSPYCE(M)-OsHAPLl and pSPYNE173-DTH8 (Fig. 6C), whereas no signals were present in the negative control (transformed with pSPYCE(M)-OsHAPLl and pSPYNE173, pSPYCE(M) and pSPYNE(173)-DTH8) (Fig. 6C). We also used qRT-PCR to investigate the expression of DTH8 and found that DTH8 has a similar diurnal expression pattern to OsHAPL1 (see Supplementary Fig. S4, available at JXB online). Together, these results suggest a possible interaction in planta between OsHAPLl and DTH8.
Fig. 6.
Interactions between OsHAPL1 and DTH8. (A) OsHAPL1 interacted with DTH8 in yeast two-hybrid assays. Yeast was grown at 30°C for 4 days. (B) OsHAPL1 interacted with DTH8 in GST pull-down assays. Arrow indicates the target protein. (C) OsHAPL1 interacted with DTH8 in tobacco BiFC assays. Scale bar = 20 μm. (D) Protein structural diagram of OsHAPL1 and DTH8. aa, amino acids. (E) OsHAPL1 interacted with DTH8 in yeast two-hybrid assays owing to its HAP2-like domain. Yeast was grown at 30°C for 4 days. (F) DTH8 interacted with OsHAPL1 in yeast two-hybrid assays owing to its HAP3 domain. Yeast was grown at 30°C for 4 days. DAPI, 4'-6-diamidino-2-phenylindole; DIC, differential interference contrast microscope; ER, endoplasmic reticulum; eYCE, pSPYCE(M); eYFP, enhanced yellow fluorescent protein; eYNE, pSPYNE173; SD/-LW, Synthetic Dropout/-Trp-Leu; SD/-LWHA, Synthetic Dropout/-Trp-Leu-His-Ade.
To identify the OsHAPLl domain involved in the interaction with DTH8, we performed yeast two-hybrid assays using either OsHAPLl with the HAP domain only (OsHAPLl-H) or OsHAPLl with the HAP domain deleted (OsHAPLl-∆H) and DTH8 (Fig. 6D). OsHAPLl-H interacted with the DTH8 protein, whereas OsHAPLl-∆H did not (Fig. 6E). DTH8 also interacted with OsHAPLl by means of its HAP domain (Fig. 6F). These experiments indicate that the HAP domain is essential for the interaction between OsHAPLl and DTH8.
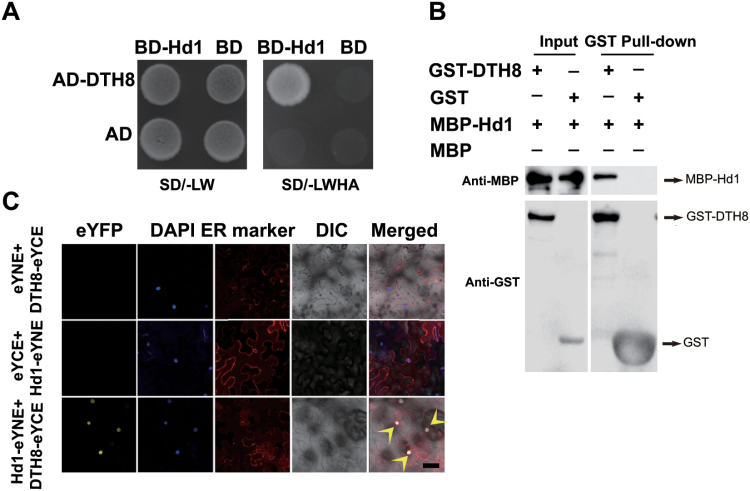
OsHAPL1, Hd1, and DTH8 form a complex in E. coli
Hd1 is an important protein involved in determining the heading date in rice (Yano et al., 2000). Hd1 can delay flowering in LD conditions, and this function is consistent with those of OsHAPL1 and DTH8, so we speculated whether Hd1 can interact with OsHAPL1 and DTH8. A yeast two-hybrid assay showed that Hd1 cannot interact with OsHAPL1 (data not shown), where it can interact with DTH8 (Fig. 7A). We further confirmed the interaction between Hd1 and DTH8 by in vitro pull-down and BiFC assays (Fig. 7B, C).
Fig. 7.
Interaction between DTH8 and Hd1. (A) DTH8 interacted with Hd1 in yeast two-hybrid assays. Yeast was grown at 30°C for 4 days. (B) DTH8 interacted with Hd1 in GST pull-down assays. Arrows indicate target proteins. (C) OsHAPL1 interacted with DTH8 in tobacco BiFC assays. Scale bar = 20 μm. DAPI, 4'-6-diamidino-2-phenylindole; DIC, differential interference contrast microscope; ER, endoplasmic reticulum; eYCE, pSPYCE(M); eYFP, enhanced yellow fluorescent protein; eYNE, pSPYNE173; SD/-LW, Synthetic Dropout/-Trp-Leu; SD/-LWHA, Synthetic Dropout/-Trp-Leu-His-Ade.
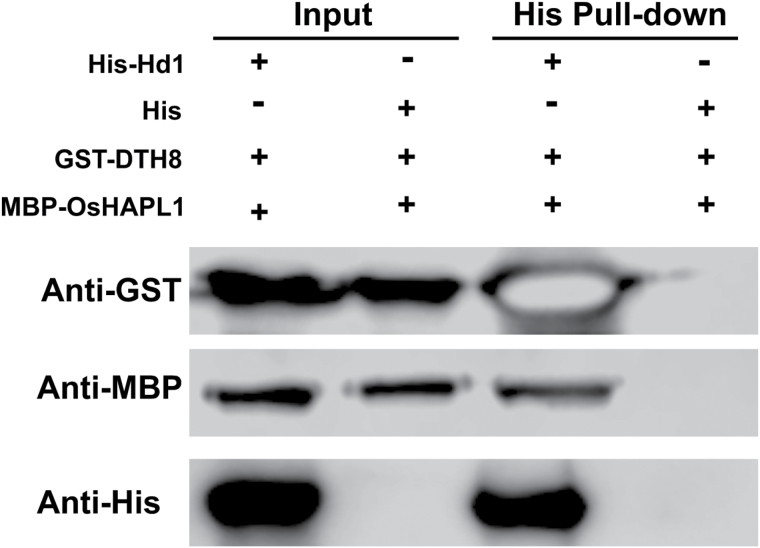
The interaction among OsHAPL1, DTH8, and Hd1 implies that these three proteins may form a complex, and that DTH8 may function as a bridge. To confirm this, histidine pull-down of these three proteins was performed. DTH8 and OsHAPL1 were pulled down by Hd1 (Fig. 8), suggesting that OsHAPL1, DTH8, and Hd1 can form a complex in E. coli.
Fig. 8.
OsHAPL1, DTH8, and Hd1 form a complex in His pull-down assays. In vitro binding assay with the indicated recombinant proteins showed direct interaction between OsHAPL1, DTH8, and Hd1.
OsHAPL1, DTH8, and Hd1 interact with HAP family proteins in yeast
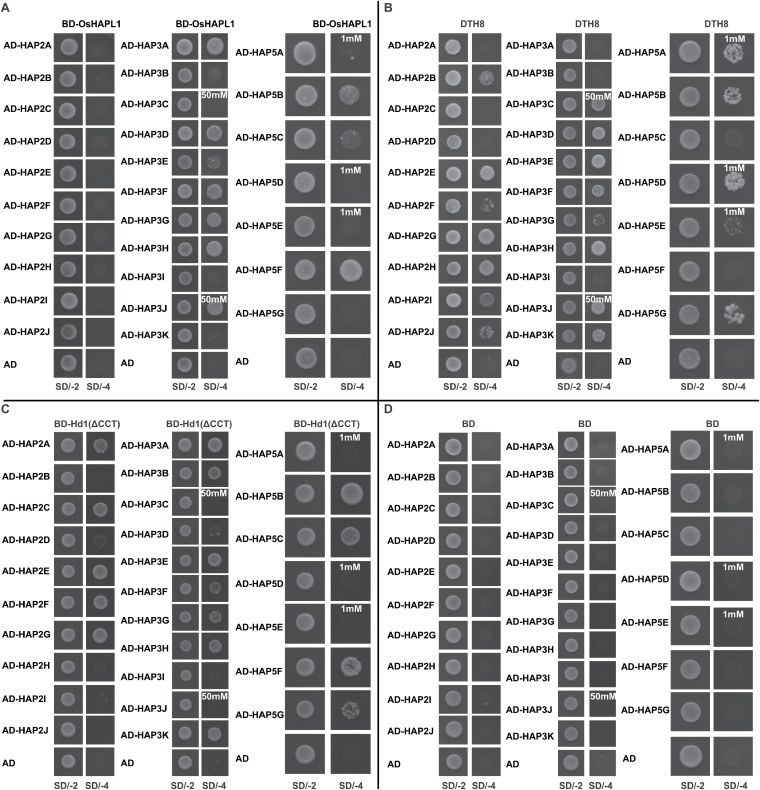
Previous studies have shown that various HAP complexes control different growth and developmental processes by tissue-specific expression and complex formation of the three subunits (Thirumurugan et al., 2008; Laloum et al., 2013). To preliminarily identify the possible HAP members involved in the heading date related to OsHAPL1, DTH8, and Hd1, we performed yeast two-hybrid assays. OsHAPL1 interacted with HAP3 family members HAP3A, HAP3D, HAP3F, HAP3G, HAP3H, and HAP3J, and with HAP5 members HAP5B and HAP5F (Fig. 9A). DTH8 interacted with HAP2 family members HAP2B, HAP2E, HAP2G, HAP2H, HAP2I, and HAP2J; with HAP3 members HAP3C, HAP3D, HAP3E, HAP3F, HAP3H, HAP3J, and HAP3K; and with HAP5 members HAP5A, HAP5B, HAP5D, and HAP5G (Fig. 9B). Because Hd1 can self-activatie, Hd1(ΔCCT) fused with the GAL4 DNA binding domain. Yeast data showed that Hd1(ΔCCT) can interact with HAP2 family members HAP2A, HAP2C, HAP2E, HAP2F, and HAP2G; with HAP3 members HAP3A, HAP3B, HAP3E, HAP3F, HAP3G, HAP3H, and HAP3K; and with HAP5 members HAP5B, HAP5C, HAP5F, and HAP5G (Fig. 9C). Figure 9D shows the negative control. These data suggest many HAP family members can interact with OsHAPL1, DTH8, and Hd1, and suggest that these HAP members may be involved in the heading date.
Fig. 9.
Interactions between OsHAPL1, DTH8, Hd1, and HAP subunit members. (A) Interactions between OsHAPL1 and HAP in yeast two-hybrid assays. (B) Interactions between DTH8 and HAP in yeast two-hybrid assays. (C) Interactions between Hd1 and HAP in yeast-two-hybrid assays. (D) Negative control. AD, pGAD T7; BD, pGBK T7; SD/-2, Synthetic Dropout/-Trp-Leu; SD/-4, Synthetic Dropout/-Trp-Leu-His-Ade.
OsHAPL1, DTH8, and Hd1 interact with components of the transcription start complex in yeast
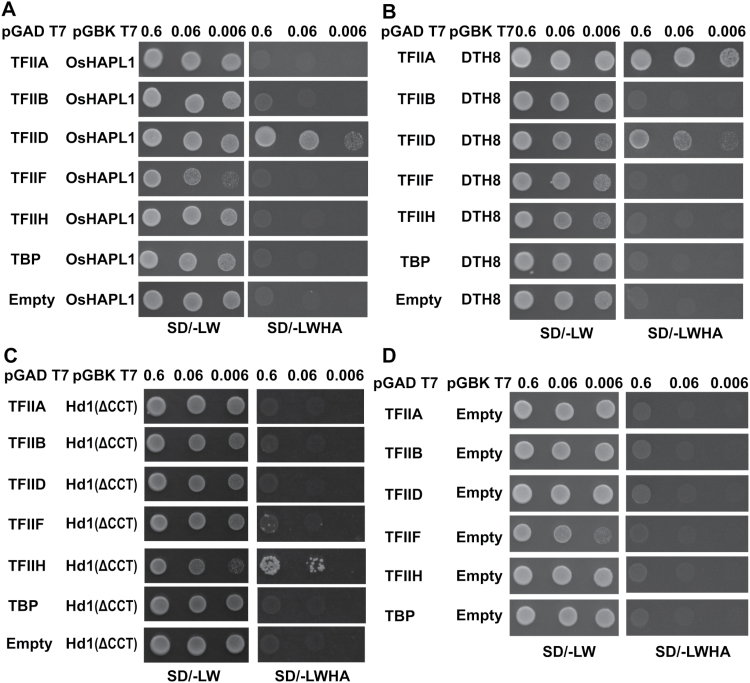
Because a previous study indicated that the interaction of HAP3 and HAP5 with TATA-Binding Protein (TBP) depends on short domains adjacent to their histone fold motifs (Bellorini et al., 1997), we speculated that HAP proteins could be involved in transcriptional start complex formation. To verify this possibility, we performed a yeast two-hybrid assay to determine whether OsHAPL1 can interact with GTFs. Results showed that OsHAPL1 could interact with TFIID but not with TFIIA, TFIIB, TFIIF, TFIIH or TBP (Fig. 10A). Because OsHAPL1, DTH8, and Hd1 can form a complex, we speculated that DTH8 and Hd1 could also interact with GTFs. DTH8 interacted with TFIIA and TFIID, but could not interact with TFIIB, TFIIF, TFIIH, or TBP (Fig. 10B). Hd1 interacted with TFIIH, but could not interact with TFIIA, TFIIB, TFIID, TFIIF, or TBP (Fig. 10C). Figure 10D shows the negative control. These preliminary data suggest that OsHAPL1, DTH8, and Hd1 serve as co-regulators of transcription.
Fig. 10.
Interactions between OsHAPL1, DTH8, Hd1, and GTFs. (A) Interactions between OsHAPL1 and GTFs in yeast two-hybrid assays. (B) Interactions between DTH8 and GTFs in yeast two-hybrid assays. (C) Interactions between Hd1 and GTFs in yeast two-hybrid assays. (D) Negative control. SD/-LW, Synthetic Dropout/-Trp-Leu; SD/-LWHA, Synthetic Dropout/-Trp-Leu-His-Ade.
OsHAPL1 delays flowering by repressing Ehd1 as well as the downstream florigen genes Hd3a and RFT1
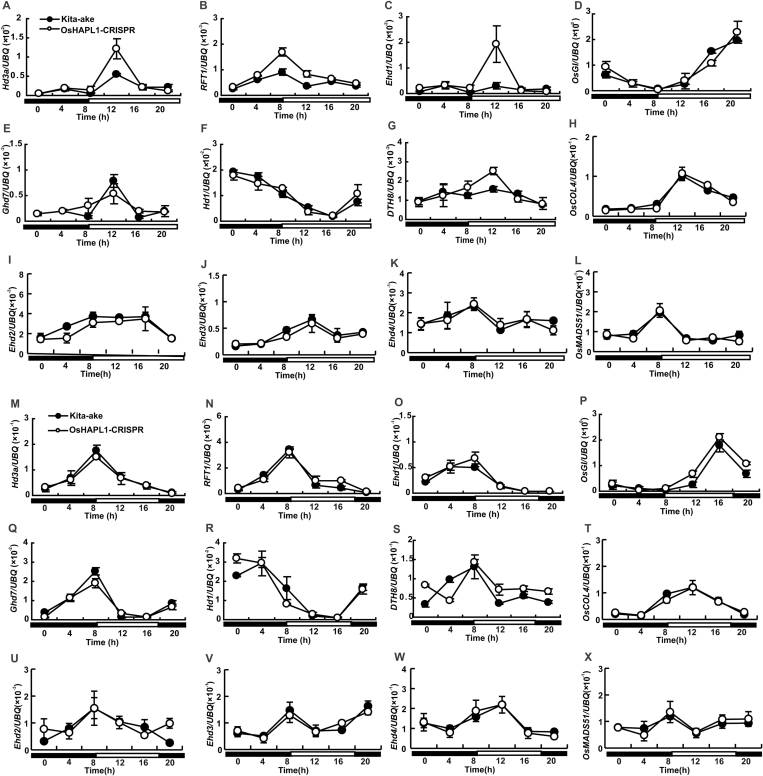
To study downstream genes regulated by OsHAPL1, we analysed the expression of key genes related to the heading date in 30-day-old WT and oshapl1 mutant plants grown in LD and SD conditions using qRT-PCR. The expression levels of Ehd1 were upregulated in oshapl1 mutant plants relative to WT in LD conditions. The florigen genes Hd3a and RFT1 in oshapl1 mutant plants were also upregulated in LD conditions (Fig. 11). The expression of these three genes was not affected in oshapl1 mutant plants in SD conditions. The expression of OsGI, Ghd7, Hd1, DTH8, Ehd2/3/4, OsMADS51, and OsCOL4 was not significantly different between oshapl1 mutant plants and WT in either environment (Fig. 11). These data suggest that OsHAPL1 can affect heading date via Ehd1, Hd3a, and RFT1. To further confirm that OsHAPL1 functions upstream of Ehd1, Hd3a, and RFT1, we performed expression analysis for OsHAPL1 in ehd1, hd3a, and rft1 mutants. The expression of OsHAPL1 was not changed in these mutants (see Supplementary Fig. S5, available at JXB online). These results suggest that OsHAPL1 regulates heading date by controlling the expression of Ehd1, Hd3a, and RFT1 in LD conditions.
Fig. 11.
The rhythmic expression patterns of Hd3a, RFT1, Ehd1, OsGI, Ghd7, Hd1, DTH8, OsCOL4, Ehd2,3,4, and OsMADS51 in the wild-type Kita-ake and oshapl1 mutants under LD conditions (A–L) and SD conditions (M–X). The plants were grown in growth chambers under LD conditions (14 h light /10 h darkness) and SD conditions (10 h light /14 h darkness) for 30 days. White bars indicate light; black bars indicate darkness. The rice Ubiquitin gene was used as the internal control. Values represent means ± standard deviation from three independent biological replicates.
Discussion
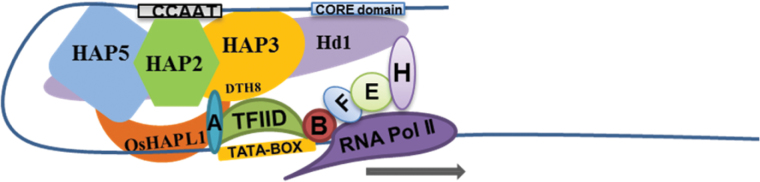
We report a functional analysis of OsHAPL1, which is a novel repressor of heading date. Our studies showed that OsHAPL1 can form a complex with DTH8 and Hd1 in E. coli. In addition, OsHAPL1, DTH8, and Hd1 also interact with some members of the HAP subfamily groups and with GTFs. The data strongly indicate that the OsHAPL1-DTH8-Hd1 complex acts as a transcriptional regulator of heading date by interacting with the HAP complex and GTFs (Fig. 12). Furthermore, qPCR analysis of genes related to heading date showed that the expression of Hd3a, RFT1, and Ehd1 was downregulated by OsHAPL1. Thus, our studies suggest that the OsHAPL1-DTH8-Hd1 complex regulates the transcription of target genes to control heading date in rice.
Fig. 12.
Proposed model for the OsHAPL1-DTH8-Hd1 complex that affects heading date in rice. The complex may function as a transcriptional regulator, together with the HAP complex and GTFs, to control expression of downstream genes, and then to affect heading date by influencing the expression of Hd3a, RFT1, and Ehd1.
OsHAPL1 is a HAP2-like transcription regulator
The HAP complex consists of three subunits, namely HAP2 (also called NF-YA or CBF-B), HAP3 (NF-YB or CBF-A), and HAP5 (NF-YC or CBF-C) (Thirumurugan et al., 2008). Each subunit contains a conserved domain responsible for DNA binding and protein interaction as well as conserved regions (Thirumurugan et al., 2008). The HAP complex binds to CCAAT sequences, which are among the most general trans-acting elements in many genes in fungi, animal, and plants (Maity and de Crombrugghe, 1998; Mantovani, 1999). In yeast, each subunit of HAP2, HAP3, and HAP5 has only one gene-encoded protein, and together they form a heterotrimeric CCAAT-box-binding complex, which can recruit a fourth polypeptide, ScHAP4, that associates with the HAP complex and activates transcription via its acidic domain, but it cannot bind to DNA (Forsburg and Guarente, 1989). In animals, homologues of the yeast HAP complex are called NF-Y, CP1, or CBP (Vuorio et al., 1990; Becker et al., 1991; Li et al., 1992; Sinha et al., 1995); there is no HAP4 homologue and the HAP complex possibly interacts with other TFs to regulate transcription of target genes (Bellorini et al., 1997). In plants each subunit consists of multiple proteins. In rice, there are 11 HAP2 genes, 12 HAP3 genes, and 12 HAP5 genes (Thirumurugan et al., 2008; Li et al., 2016). OsHAPL1, a HAP2-like TF, identified by its effect on heading date, associates with certain members of the HAP3 and HAP5 family proteins. We speculated that OsHAPL1 is a protein homologue of ScHAP4, but blast analysis of the amino acid sequences showed only 9% homology (see Supplementary Fig. S6, available at JXB online), indicating they are not homologues. It seems that OsHAPL1 may function like other HAP family members. In addition, OsHAPL1 also interacts with the GTF TFIID, suggesting that it functions as a transcription regulator in the heading pathway in rice. The CCT domain of CO in Arabidopsis is similar to HAP2 in yeast, and CO may replace AtHAP2 in the HAP complex to form a trimeric CO-AtHAP3-AtHAP5 complex to regulate flowering-related gene expression (Wenkel et al., 2006). When AtHAP3 and AtHAP5 were overexpressed, flowering was delayed. Overexpression of AtHAP2 or AtHAP3 could impair the formation of a CO-AtHAP3-AtHAP5 complex, leading to delayed flowering (Wenkel et al., 2006). Here, OsHAPL1 functioned in the same way as a HAP2-like transcriptional regulator, interacting with some HAP3 and HAP5 proteins to form complexes. DTH8-OsHAP3H and Hd1 can also interact with some HAP2, 3, and 5 proteins. The OsHAPL1-DTH8-Hd1 complex could interact with the HAP complex to regulate the expression of flowering genes, and hence control heading date. More evidence is needed to confirm whether the mechanism is similar to that in Arabidopsis.
The OsHAPL1-DTH8-Hd1 complex may function as a transcription regulator through direct interaction with general transcription factors
We found that OsHAPL1, DTH8, and Hd1 form a complex in E. coli. OsHAPL1 interacts with the GTF TFIID, DTH8 interacts with TFIIA and TFIID, and Hd1 interacts with TFIIH. GTFs and RNA polymerase II play important roles in transcription in eukaryotes. Six GTFs and RNA polymerase II assemble into a stable pre-initiation complex responsible for RNA synthesis (Zawel and Reinberg, 1993). TFIIA plays an important role in transcriptional activation and facilitation of TFIID to its promoter (Buratowski et al., 1989; Maldonado et al., 1990; Ma et al., 1993; Lieberman and Berk, 1994). TFIID binding to the TATA motif helps to bind TFIIB (Buratowski et al., 1989; Maldonado et al., 1990); The TFIID-TFIIB complex then recruits TFIIF and RNA polymerase II to the promoter to form a TFIID-TFIIB-TFIIF-PolII complex intermediate (Flores et al., 1990). TFIIE binds to the promoter by interacting with GTFs, and recruits THIIH to the complex (Flores et al., 1992). The TFIIA-TFIID-TFIIB-TFIIF-PolII-TFIIE-THIIH complex then initiates transcription (Zawel and Reinberg, 1995; Mermelstein et al., 1996). A previous study showed that assembly of the transcription initiation complex requires many regulatory proteins that directly interact with the GTFs (Zawel and Reinberg, 1995). The transcription initiation process also needs many regulatory factors such as activators, repressors, co-activators, and co-repressors (Herschbach and Johnson, 1993). For example, Dr1 in yeast represses transcription of RNA polymerase II and RNA polymerase III (Inostroza et al., 1992; White et al., 1994). Dr1 represses RNA polymerase II transcription because it interacts with TBP and prevents TFIIA and TFIIB entering the pre-initiation complex (Inostroza et al., 1992; Kim et al., 1995), thus preventing formation of an active complex, and initiation of transcription by RNA polymerase II (Inostroza et al., 1992). Dr1-associated protein 1 (DrAp1) forms a heterotetramer with Dr1, but cannot repress transcription. DrAp1 functions as a co-repressor that enhances the repression activity of Dr1 (White et al., 1994; Kim et al., 1995; Mermelstein et al., 1996). dDr1/dDrAp1 in Drosophila have a different function, repressing the transcription of TATA-type promoters and activating transcription of a TATA-less promoter (Willy et al., 2000). In rice, Dr1 or DrAp1 separately interact with the TBP-DNA complex, forming heterodimers that strongly interact with the TBP-DNA complex to form larger complexes (Song et al., 2002). OsDrAp1 has strong repression activity and OsDr1 has weak repression activity (Song et al., 2002). OsHAPL1 encodes a histone-like TF, and its homologues in maize and Brachypodium are Dr1-associated co-repressors also named OsDrAp2 (Thirumurugan et al., 2008). Taken together, the interaction between OsHAPL1, DTH8, Hd1, and GTFs suggests that OsHAPL1-DTH8-Hd1 may regulate transcription pre-initiation by interacting with some GTFs.
OsHAPL1-DTH8-Hd1 functions as a repressor of heading in rice under long-day conditions
Heading in rice is regulated by a complicated genetic network involving many genes. Rice is a SD plant that flowers rapidly under SD conditions, but is late flowering under LD conditions (Takimoto and Ikeda, 1961). We found that knockout of OsHAPL1 promoted flowering under LD conditions, and overexpression of OsHAPL1 delayed flowering. We also found that OsHAPL1, DTH8, and Hd1 form a complex in E. coli. Previous studies have shown that Hd1 promotes flowering by regulating the expression of Hd3a (a florigen in rice) in SD conditions, whereas it delays flowering by downregulating expression of Hd3a under LD conditions through unknown mechanisms (Yano et al., 2000; Kojima et al., 2002; Tamaki et al., 2007; Komiya et al., 2008). DTH8 suppresses flowering in rice by downregulating expression of Ehd1 and Hd3a in LD conditions, but has no influence on flowering in SD conditions (Wei et al., 2010). A previous study of DTH8 also showed a reduced grain number per panicle and yield per plant in the loss-of-function line CSSL61 (Wei et al., 2010). We characterized the yield features by measuring agronomic traits, such as tiller number, grain number per panicle, 1000-grain weight, and yield per plant. We found that grain number per panicle and grain yield per plant were significantly reduced in oshapl1 (Supplementary Table S2, available at JXB online). This is a consistent yield feature between oshapl1 and dth8. Altogether, OsHAPL1, DTH8, and Hd1 function in the same pathway and consistently behave as repressors of heading in LD conditions while having no influence on heading date in SD conditions. The Ehd1 pathway is a unique, Hd1-independent flowering pathway. Ehd1 encodes a B-type response regulator that is highly conserved in cultivated rice, but has no homologue in Arabidopsis (Doi et al., 2004; Takahashi et al., 2009). It positively regulates expression of Hd3a and the florigen RFT1 (Doi et al., 2004; Komiya et al., 2008; Komiya et al., 2009). Many Ehd1-repressors have already identified, including DTH8. Here, we found another Ehd1-repressor, OsHAPL1, which forms a complex with DTH8 and Hd1, and negatively regulates expression of Ehd1, and then inhibits flowering by downregulating expression of RFT1 and Hd3a. Recent studies have shown that Hd1 interacts with Ghd7 to form a complex that can bind to the promoter of Ehd1 and repress its expression (Nemoto et al., 2016). In our study, Hd1 interacted with DTH8 and OsHAPL1, so we speculate that Hd1, DTH8, OsHAPL1, and Ghd7 may interact with the same complex. Our preliminary yeast data showed that DTH8 can interact with Ghd7, whereas OsHAPL1 cannot (data not shown). Additionally, our ChIP-qPCR and yeast one-hybrid assays showed that OsHAPL1 cannot bind to the promoter of Ehd1. OsHAPL1 may regulate other target genes to control heading date via Ehd1. Nevertheless, our results have demonstrated the possibility of using OsHAPL1 to increase the adaptability of rice varieties. Combinational manipulation of this gene, together with other trait genes such as GS3 for grain size, Gn1a for grain number, and AFD1 for plant height (Ren et al., 2016; Shen et al., 2016), may provide a useful practice for crop improvement.
Supplementary data
Supplementary data are available at JXB online.
Fig. S1. Protein structure and phylogenetic tree of OsHAPL1.
Fig. S2. The phenotype of oshapl1 mutants in Nipponbare background.
Fig. S3. Subcellular localization of OsHAPL1.
Fig. S4. Rhythmic expression analysis of DTH8.
Fig. S5. Expression analysis of OsHAPL1 between Nipponbare and ehd1, hd3a, and rft1 mutants, under CLD and CSD conditions.
Fig. S6. Blast analysis of the amino acid sequences of OsHAPL1 and ScHAP4.
Table S1. Primers used in this study.
Table S2. Comparison of major agricultural traits between Kia-ake and oshapl1 under NLD conditions.
Supplementary Material
Acknowledgements
This work was supported by the National Natural Science Foundation (31401466), the National Key Research and Development Program (2016YFD0100301), and the National Transgenic Major Program (2016ZX08001006-007 and 2016ZX08009-003). We thank Dr Robert Alexander McIntosh (The University of Sydney) for critical reading of the manuscript.
References
- Alam MM, Tanaka T, Nakamura H, et al. 2015. Overexpression of a rice heme activator protein gene (OsHAP2E) confers resistance to pathogens, salinity and drought, and increases photosynthesis and tiller number. Plant Biotechnology Journal 13, 85–96. [DOI] [PubMed] [Google Scholar]
- Andrés F, Coupland G. 2012. The genetic basis of flowering responses to seasonal cues. Nature Reviews. Genetics 13, 627–639. [DOI] [PubMed] [Google Scholar]
- Bart R, Chern M, Park CJ, Bartley L, Ronald PC. 2006. A novel system for gene silencing using siRNAs in rice leaf and stem-derived protoplasts. Plant Methods 2, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker DM, Fikes JD, Guarente L. 1991. A cDNA encoding a human CCAAT-binding protein cloned by functional complementation in yeast. Proceedings of the National Academy of Sciences of the United States of America 88, 1968–1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beerli RR, Segal DJ, Dreier B, Barbas CF., 3rd 1998. Toward controlling gene expression at will: specific regulation of the erbB-2/HER-2 promoter by using polydactyl zinc finger proteins constructed from modular building blocks. Proceedings of the National Academy of Sciences of the United States of America 95, 14628–14633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellorini M, Lee DK, Dantonel JC, Zemzoumi K, Roeder RG, Tora L, Mantovani R. 1997. CCAAT binding NF-Y-TBP interactions: NF-YB and NF-YC require short domains adjacent to their histone fold motifs for association with TBP basic residues. Nucleic Acids Research 25, 2174–2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucher P. 1990. Weight matrix descriptions of four eukaryotic RNA polymerase II promoter elements derived from 502 unrelated promoter sequences. Journal of Molecular Biology 212, 563–578. [DOI] [PubMed] [Google Scholar]
- Buratowski S, Hahn S, Guarente L, Sharp PA. 1989. Five intermediate complexes in transcription initiation by RNA polymerase II. Cell 56, 549–561. [DOI] [PubMed] [Google Scholar]
- Corbesier L, Vincent C, Jang S, et al. 2007. FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 316, 1030–1033. [DOI] [PubMed] [Google Scholar]
- Doi K, Izawa T, Fuse T, Yamanouchi U, Kubo T, Shimatani Z, Yano M, Yoshimura A. 2004. Ehd1, a B-type response regulator in rice, confers short-day promotion of flowering and controls FT-like gene expression independently of Hd1. Genes & Development 18, 926–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores O, Ha I, Reinberg D. 1990. Factors involved in specific transcription by mammalian RNA polymerase II. Purification and subunit composition of transcription factor IIF. The Journal of Biological Chemistry 265, 5629–5634. [PubMed] [Google Scholar]
- Flores O, Lu H, Reinberg D. 1992. Factors involved in specific transcription by mammalian RNA polymerase II. Identification and characterization of factor IIH. The Journal of Biological Chemistry 267, 2786–2793. [PubMed] [Google Scholar]
- Forsburg SL, Guarente L. 1989. Identification and characterization of HAP4: a third component of the CCAAT-bound HAP2/HAP3 heteromer. Genes & Development 3, 1166–1178. [DOI] [PubMed] [Google Scholar]
- Gao H, Zheng XM, Fei G, et al. 2013. Ehd4 encodes a novel and Oryza-genus-specific regulator of photoperiodic flowering in rice. PLoS Genetics 9, e1003281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanano S, Goto K. 2011. Arabidopsis TERMINAL FLOWER1 is involved in the regulation of flowering time and inflorescence development through transcriptional repression. The Plant Cell 23, 3172–3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayama R, Yokoi S, Tamaki S, Yano M, Shimamoto K. 2003. Adaptation of photoperiodic control pathways produces short-day flowering in rice. Nature 422, 719–722. [DOI] [PubMed] [Google Scholar]
- Herschbach BM, Johnson AD. 1993. Transcriptional repression in eukaryotes. Annual Review of Cell Biology 9, 479–509. [DOI] [PubMed] [Google Scholar]
- Huq E, Tepperman JM, Quail PH. 2000. GIGANTEA is a nuclear protein involved in phytochrome signaling in Arabidopsis. Proceedings of the National Academy of Sciences of the United States of America 97, 9789–9794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inostroza JA, Mermelstein FH, Ha I, Lane WS, Reinberg D. 1992. Dr1, a TATA-binding protein-associated phosphoprotein and inhibitor of class II gene transcription. Cell 70, 477–489. [DOI] [PubMed] [Google Scholar]
- Ito Y, Thirumurugan T, Serizawa A, Hiratsu K, Ohme-Takagi M, Kurata N. 2011. Aberrant vegetative and reproductive development by overexpression and lethality by silencing of OsHAP3E in rice. Plant Science 181, 105–110. [DOI] [PubMed] [Google Scholar]
- Kardailsky I, Shukla VK, Ahn JH, Dagenais N, Christensen SK, Nguyen JT, Chory J, Harrison MJ, Weigel D. 1999. Activation tagging of the floral inducer FT. Science 286, 1962–1965. [DOI] [PubMed] [Google Scholar]
- Kerppola TK. 2006. Design and implementation of bimolecular fluorescence complementation (BiFC) assays for the visualization of protein interactions in living cells. Nature Protocols 1, 1278–1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SK, Park HY, Jang YH, Lee KC, Chung YS, Lee JH, Kim JK. 2016. OsNF-YC2 and OsNF-YC4 proteins inhibit flowering under long-day conditions in rice. Planta 243, 563–576. [DOI] [PubMed] [Google Scholar]
- Kim SL, Lee S, Kim HJ, Nam HG, An G. 2007. OsMADS51 is a short-day flowering promoter that functions upstream of Ehd1, OsMADS14, and Hd3a. Plant Physiology 145, 1484–1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TK, Zhao Y, Ge H, Bernstein R, Roeder RG. 1995. TATA-binding protein residues implicated in a functional interplay between negative cofactor NC2 (Dr1) and general factors TFIIA and TFIIB. The Journal of Biological Chemistry 270, 10976–10981. [DOI] [PubMed] [Google Scholar]
- Kojima S, Takahashi Y, Kobayashi Y, Monna L, Sasaki T, Araki T, Yano M. 2002. Hd3a, a rice ortholog of the Arabidopsis FT gene, promotes transition to flowering downstream of Hd1 under short-day conditions. Plant & Cell Physiology 43, 1096–1105. [DOI] [PubMed] [Google Scholar]
- Komiya R, Ikegami A, Tamaki S, Yokoi S, Shimamoto K. 2008. Hd3a and RFT1 are essential for flowering in rice. Development 135, 767–774. [DOI] [PubMed] [Google Scholar]
- Komiya R, Yokoi S, Shimamoto K. 2009. A gene network for long-day flowering activates RFT1 encoding a mobile flowering signal in rice. Development 136, 3443–3450. [DOI] [PubMed] [Google Scholar]
- Koo BH, Yoo SC, Park JW, et al. 2013. Natural variation in OsPRR37 regulates heading date and contributes to rice cultivation at a wide range of latitudes. Molecular Plant 6, 1877–1888. [DOI] [PubMed] [Google Scholar]
- Kwon CT, Koo BH, Kim D, Yoo SC, Paek NC. 2015. Casein kinases I and 2α phosphorylate Oryza sativa pseudo-response regulator 37 (OsPRR37) in photoperiodic flowering in rice. Molecules and Cells 38, 81–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laloum T, De Mita S, Gamas P, Baudin M, Niebel A. 2013. CCAAT-box binding transcription factors in plants: Y so many?Trends in Plant Science 18, 157–166. [DOI] [PubMed] [Google Scholar]
- Lee YS, Jeong DH, Lee DY, et al. 2010. OsCOL4 is a constitutive flowering repressor upstream of Ehd1 and downstream of OsphyB. The Plant Journal 63, 18–30. [DOI] [PubMed] [Google Scholar]
- Lieberman PM, Berk AJ. 1994. A mechanism for TAFs in transcriptional activation: activation domain enhancement of TFIID-TFIIA–promoter DNA complex formation. Genes & Development 8, 995–1006. [DOI] [PubMed] [Google Scholar]
- Li Q, Yan W, Chen H, Tan C, Han Z, Yao W, Li G, Yuan M, Xing Y. 2016. Duplication of OsHAP family genes and their association with heading date in rice. Journal of Experimental Botany 67, 1759–1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XY, Mantovani R, Hooft van Huijsduijnen R, Andre I, Benoist C, Mathis D. 1992. Evolutionary variation of the CCAAT-binding transcription factor NF-Y. Nucleic Acids Research 20, 1087–1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma D, Watanabe H, Mermelstein F, et al. 1993. Isolation of a cDNA encoding the largest subunit of TFIIA reveals functions important for activated transcription. Genes & Development 7, 2246–2257. [DOI] [PubMed] [Google Scholar]
- Maity SN, de Crombrugghe B. 1998. Role of the CCAAT-binding protein CBF/NF-Y in transcription. Trends in Biochemical Sciences 23, 174–178. [DOI] [PubMed] [Google Scholar]
- Maldonado E, Ha I, Cortes P, Weis L, Reinberg D. 1990. Factors involved in specific transcription by mammalian RNA polymerase II: role of transcription factors IIA, IID, and IIB during formation of a transcription-competent complex. Molecular and Cellular Biology 10, 6335–6347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantovani R. 1999. The molecular biology of the CCAAT-binding factor NF-Y. Gene 239, 15–27. [DOI] [PubMed] [Google Scholar]
- Matsubara K, Yamanouchi U, Nonoue Y, Sugimoto K, Wang ZX, Minobe Y, Yano M. 2011. Ehd3, encoding a plant homeodomain finger-containing protein, is a critical promoter of rice flowering. The Plant Journal 66, 603–612. [DOI] [PubMed] [Google Scholar]
- Matsubara K, Yamanouchi U, Wang ZX, Minobe Y, Izawa T, Yano M. 2008. Ehd2, a rice ortholog of the maize INDETERMINATE1 gene, promotes flowering by up-regulating Ehd1. Plant Physiology 148, 1425–1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNabb DS, Xing Y, Guarente L. 1995. Cloning of yeast HAP5: a novel subunit of a heterotrimeric complex required for CCAAT binding. Genes & Development 9, 47–58. [DOI] [PubMed] [Google Scholar]
- Mermelstein F, Yeung K, Cao J, et al. 1996. Requirement of a corepressor for Dr1-mediated repression of transcription. Genes & Development 10, 1033–1048. [DOI] [PubMed] [Google Scholar]
- Miao J, Guo DS, Zhang JZ, Huang QP, Qin GJ, Zhang X, Wan JM, Gu HY, Qu LJ. 2013. Targeted mutagenesis in rice using CRISPR-Cas system. Cell Research 23, 1233–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemoto Y, Nonoue Y, Yano M, Izawa T. 2016. Hd1,a CONSTANS ortholog in rice, functions as an Ehd1 repressor through interaction with monocot-specific CCT-domain protein Ghd7. The Plant Journal 86, 221–233. [DOI] [PubMed] [Google Scholar]
- Park DH, Somers DE, Kim YS, et al. 1999. Control of circadian rhythms and photoperiodic flowering by the Arabidopsis GIGANTEA gene. Science 285, 1579–1582. [DOI] [PubMed] [Google Scholar]
- Parvin JD, Young RA. 1998. Regulatory targets in the RNA polymerase II holoenzyme. Current Opinion in Genetics & Development 8, 565–570. [DOI] [PubMed] [Google Scholar]
- Ren D, Rao Y, Wu L, et al. 2016. The pleiotropic ABNORMAL FLOWER AND DWARF1 affects plant height, floral development and grain yield in rice. Journal of Integrative Plant Biology 58, 529–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L, Wang C, Fu Y, Wang J, Liu Q, Zhang X, Yan C, Qian Q, Wang K. 2016. QTL editing confers opposing yield performance in different rice varieties. Journal of Integrative Plant Biology. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- Sheng P, Wu F, Tan J, et al. 2016. A CONSTANS-like transcriptional activator, OsCOL13, functions as a negative regulator of flowering downstream of OsphyB and upstream of Ehd1 in rice. Plant Molecular Biology 92, 209–222. [DOI] [PubMed] [Google Scholar]
- Simpson GG, Dean C. 2002. Arabidopsis, the Rosetta stone of flowering time?Science 296, 285–289. [DOI] [PubMed] [Google Scholar]
- Sinha S, Maity SN, Lu J, de Crombrugghe B. 1995. Recombinant rat CBF-C, the third subunit of CBF/NFY, allows formation of a protein-DNA complex with CBF-A and CBF-B and with yeast HAP2 and HAP3. Proceedings of the National Academy of Sciences of the United States of America 92, 1624–1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song W, Solimeo H, Rupert RA, Yadav NS, Zhu Q. 2002. Functional dissection of a Rice Dr1/DrAp1 transcriptional repression complex. The Plant Cell 14, 181–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y, Teshima KM, Yokoi S, Innan H, Shimamoto K. 2009. Variations in Hd1 proteins, Hd3a promoters, and Ehd1 expression levels contribute to diversity of flowering time in cultivated rice. Proceedings of the National Academy of Sciences of the United States of America 106, 4555–4560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takimoto A, Ikeda K. 1961. Effect of twilight on photoperiodic induction in some short day plants. Plant and Cell Physiology 2, 213–229. [Google Scholar]
- Tamaki S, Matsuo S, Wong HL, Yokoi S, Shimamoto K. 2007. Hd3a protein is a mobile flowering signal in rice. Science 316, 1033–1036. [DOI] [PubMed] [Google Scholar]
- Tan J, Jin M, Wang J, et al. 2016. OsCOL10, a CONSTANS-like gene, functions as a flowering time repressor downstream of Ghd7 in rice. Plant & Cell Physiology 57, 798–812. [DOI] [PubMed] [Google Scholar]
- Thirumurugan T, Ito Y, Kubo T, Serizawa A, Kurata N. 2008. Identification, characterization and interaction of HAP family genes in rice. Molecular Genetics and Genomics: MGG 279, 279–289. [DOI] [PubMed] [Google Scholar]
- Tjian R, Maniatis T. 1994. Transcriptional activation: a complex puzzle with few easy pieces. Cell 77, 5–8. [DOI] [PubMed] [Google Scholar]
- Vuorio T, Maity SN, de Crombrugghe B. 1990. Purification and molecular cloning of the “A” chain of a rat heteromeric CCAAT-binding protein. Sequence identity with the yeast HAP3 transcription factor. The Journal of Biological Chemistry 265, 22480–22486. [PubMed] [Google Scholar]
- Walter M, Chaban C, Schütze K, et al. 2004. Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. The Plant Journal 40, 428–438. [DOI] [PubMed] [Google Scholar]
- Wei X, Xu J, Guo H, et al. 2010. DTH8 suppresses flowering in rice, influencing plant height and yield potential simultaneously. Plant Physiology 153, 1747–1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenkel S, Turck F, Singer K, Gissot L, Le Gourrierec J, Samach A, Coupland G. 2006. CONSTANS and the CCAAT box binding complex share a functionally important domain and interact to regulate flowering of Arabidopsis. The Plant Cell 18, 2971–2984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White RJ, Khoo BC, Inostroza JA, Reinberg D, Jackson SP. 1994. Differential regulation of RNA polymerases I, II, and III by the TBP-binding repressor Dr1. Science 266, 448–450. [DOI] [PubMed] [Google Scholar]
- Wigge PA, Kim MC, Jaeger KE, Busch W, Schmid M, Lohmann JU, Weigel D. 2005. Integration of spatial and temporal information during floral induction in Arabidopsis. Science 309, 1056–1059. [DOI] [PubMed] [Google Scholar]
- Willy PJ, Kobayashi R, Kadonaga JT. 2000. A basal transcription factor that activates or represses transcription. Science 290, 982–985. [DOI] [PubMed] [Google Scholar]
- Wu CY, You CJ, Li CS, Long T, Chen GX, Byrne ME, Zhang QF. 2008. RID1, encoding a Cys2/His2-type zinc finger transcription factor, acts as a master switch from vegetative to floral development in rice. Proceedings of the National Academy of Sciences of the United States of America 105, 12915–12920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue W, Xing Y, Weng X, et al. 2008. Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nature Genetics 40, 761–767. [DOI] [PubMed] [Google Scholar]
- Yan WH, Wang P, Chen HX, et al. 2011. A major QTL, Ghd8, plays pleiotropic roles in regulating grain productivity, plant height, and heading date in rice. Molecular Plant 4, 319–330. [DOI] [PubMed] [Google Scholar]
- Yano M, Katayose Y, Ashikari M, et al. 2000. Hd1, a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the Arabidopsis flowering time gene CONSTANS. The Plant Cell 12, 2473–2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zawel L, Reinberg D. 1993. Initiation of transcription by RNA polymerase II: a multi-step process. Progress in Nucleic Acid Research and Molecular Biology 44, 67–108. [DOI] [PubMed] [Google Scholar]
- Zawel L, Reinberg D. 1995. Common themes in assembly and function of eukaryotic transcription complexes. Annual Review of Biochemistry 64, 533–561. [DOI] [PubMed] [Google Scholar]
- Zhang L, Cheng Z, Qin R, et al. 2012. Identification and characterization of an epi-allele of FIE1 reveals a regulatory linkage between two epigenetic marks in rice. The Plant Cell 24, 4407–4421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Liu G, Jia J, et al. 2016. The wheat MYB-related transcription factor TaMYB72 promotes flowering in rice. Journal of Integrative Plant Biology 58, 701–704. [DOI] [PubMed] [Google Scholar]
- Zhao T, Liu J, Li HY, et al. 2015. Using hybrid transcription factors to study gene function in rice. Science China. Life Sciences 58, 1160–1162. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.