An ocs element binding factor, PhOBF1, positively regulates antiviral RNA silencing mediated by salicylic acid biosynthesis and is crucial for efficient gene silencing in petunia.
Keywords: Phenylpropanoid pathways, salicylic acid biosynthesis, shikimate, tobacco mosaic virus, tobacco rattle virus, transcription factor, virus-induced gene silencing
Abstract
Virus-induced gene silencing (VIGS) is a common reverse genetics strategy for characterizing the function of genes in plants. The detailed mechanism governing RNA silencing efficiency triggered by viruses is largely unclear. Here, we reveal that a petunia (Petunia hybrida) ocs element binding factor, PhOBF1, one of the basic leucine zipper (bZIP) transcription factors, was up-regulated by Tobacco rattle virus (TRV) infection. Simultaneous silencing of PhOBF1 and a reporter gene, phytoene desaturase (PDS) or chalcone synthase (CHS), by TRV-based VIGS led to a failure of the development of leaf photobleaching or the white-corollas phenotype. PhOBF1 silencing caused down-regulation of RNA silencing-related genes, including RNA-dependent RNA polymerases (RDRs), Dicer-like RNase III enzymes (DCLs), and Argonautes (AGOs). After inoculation with the TRV-PhPDS, PhOBF1-RNAi lines exhibited a substantially impaired PDS silencing efficiency, whereas overexpression of PhOBF1 resulted in a recovery of the silencing phenotype (photobleaching) in systemic leaves. A compromised resistance to TRV and Tobacco mosaic virus was found in PhOBF1-RNAi lines, while PhOBF1-overexpressing lines displayed an enhanced resistance to their infections. Compared with wild-type plants, PhOBF1-silenced plants accumulated lower levels of free salicylic acid (SA), salicylic acid glucoside, and phenylalanine, contrarily to higher levels of those in plants overexpressing PhOBF1. Furthermore, transcripts of a number of genes associated with the shikimate and phenylpropanoid pathways were decreased or increased in PhOBF1-RNAi or PhOBF1-overexpressing lines, respectively. Taken together, the data suggest that PhOBF1 regulates TRV-induced RNA silencing efficiency through modulation of RDRs, DCLs, and AGOs mediated by the SA biosynthesis pathway.
Introduction
Virus-induced gene silencing (VIGS), related to post-transcriptional gene silencing, is an attractively fast approach for degradation of homologous RNA molecules in plants (Reid et al., 2009; Jiang et al., 2011). This RNA silencing machinery employs a number of core components, including RNA-dependent RNA polymerases (RDRs), Dicer-like RNase III enzymes (DCLs), and Argonautes (AGOs), to implement small interfering RNA (siRNA)-mediated gene knockdown in a sequence-specific manner (Schwach et al., 2005; Jaubert et al., 2011). The Arabidopsis genome contains four DCLs, six RDRs, and ten AGOs (Donaire et al., 2008; Zhang et al., 2012), respectively contributing to the siRNA biogenesis, double-stranded RNA (dsRNA) formation, and incorporation into an RNA-induced silencing complex.
DCL1 uniquely accounts for the generation of 21-nucleotide (nt) microRNAs (miRNAs), which functions as a crucial coordinator of intricate biological processes (Bartel and Bartel, 2003). The dsRNA structures can be precisely recognized and cleaved by DCL2, DCL3, and DCL4 into 22-, 24-, and 21-nt siRNAs (Donaire et al., 2008; Axtell, 2013). Depletion of DCL2, DCL3, and DCL4 enhances susceptibility to infection by RNA viruses, including Tobacco rattle virus (TRV) (Deleris et al., 2006), Cucumber mosaic virus (CMV), and Turnip crinkle virus (TCV) (Bouché et al., 2006) in Arabidopsis. It has been noted that DCL2 and DCL4 appear to impose a principle influence on virus-induced RNA silencing, whereas DCL3 acts as a minor regulator in antiviral silencing (Qu et al., 2008; Garcia-Ruiz et al., 2010). Plants use multiple RDR host pathways to generate viral secondary siRNAs by re-amplifying the cleaved single-stranded (ss) RNA complementary to the siRNA, which therefore favors the improvement of the silencing effect (Molnar et al., 2010; Alvarado and Scholthof, 2011). Moreover, RDR1, RDR2, and RDR6 are implicated in defense against various RNA viruses via production of viral secondary siRNAs (Donaire et al., 2008; Wang et al., 2010). For instance, the mutants lacking RDR1, RDR2, and RDR6 exhibit a compromised resistance to Turnip mosaic virus (TuMV) infection in Arabidopsis (Garcia-Ruiz et al., 2010). With respect to AGOs, AGO1 plays a major role in the miRNA silencing pathway (Baumberger and Baulcombe, 2005), which controls endogenous genes in terms of development, hormone regulation and stress response (Voinnet, 2009). Current evidence has uncovered the importance of AGO1 in the RNA silencing mechanism for transcriptional regulation as well as in antiviral responses (Alvarado and Scholthof, 2011). In contrast to AGO1, AGO2 seems to be a more essential factor in RNA silencing and combat for viruses. AGO2 mutation leads to an impaired resistance to invasion by CMV (Wang et al., 2011), TCV (Harvey et al., 2011), Potato virus X (PVX) (Jaubert et al., 2011), and Tomato bushy stunt virus (Scholthof et al., 2011) in Arabidopsis or Nicotiana benthamiana. To date, however, how these RNA silencing components are transcriptionally regulated remains largely unknown.
Salicylic acid (SA) is an endogenous phytohormone required for induction of systemic acquired resistance (Malamy et al., 1990). Inoculation with viral and non-viral pathogens results in increased SA accumulation (Kumar and Klessig, 2003). Exogenous application of SA inhibits virus proliferation of CMV, Tobacco mosaic virus (TMV), and TCV in Arabidopsis, tomato, tobacco and hot pepper (Shang et al., 2011). Previous studies revealed two separated pathways, the isochorismate and phenylalanine ammonia lyase pathways, for SA biosynthesis in Arabidopsis (D’Maris Amick Dempsey et al., 2011). Both pathways initiate from chorismate, which is the ultimate product of the shikimate pathway. The aromatic amino acid phenylalanine is synthesized also through the shikimate pathway (Tzin and Galili, 2010). Some of the synthesized SAs are converted into conjugated SA glucosides (SAGs) functioning as an inactive form, from which free SA can be released (Rivas-San Vicente and Plasencia, 2011). To date, the research concerning SA has mainly focused on plant defense signaling against abiotic and biotic elicitors. Several lines of evidence support that a close correlation between SA-mediated viral defense and RNA silencing pathways may exist in plants. Exposure to SA markedly promotes the activity of NtRdRP in tobacco plants (Xie et al., 2001). Silencing suppressor of CMV disrupts both RNA silencing and SA-mediated resistance to virus infection (Ji and Ding, 2001). SA-deficient transgenic tobacco plants exhibit a reduced accumulation of Plum pox virus-derived small RNAs, demonstrating that SA may serve as an enhancer of antiviral RNA silencing (Alamillo et al., 2006). The latest findings indicate that transcripts of RDR1, RDR2, DCL1, and DCL2 are up-regulated in tomato plants following Citrus exocortis viroid (CEVd), Tomato mosaic virus (ToMV) infection, and SA treatment (Campos et al., 2014).
The basic leucine zipper (bZIP) proteins constitute a large transcription factor family in eukaryotes (Riechmann et al., 2000). In Arabidopsis, the bZIP family is classified into ten subfamilies (A–I, S), of which the subfamily S is the largest group including 17 members (Jakoby et al., 2002). AtbZIP11 belongs to subfamily S and its translation is suppressed by sucrose targeting a conserved upstream open reading frame region in the 5′ untranslated regions of mRNA (Wiese et al., 2004). AtbZIP11 regulates amino acid metabolism by directly interacting with asparagine synthetase 1 (ASN1) and proline dehydrogenase 2 (ProDH2) (Hanson et al., 2008). Activation of AtbZIP11 leads to reprogramming of amino acid and sugar metabolism, leading to increased levels of phenylalanine, tryptophan, tyrosine, sucrose, fructose, and glucose, and reduced levels of trehalose, as well as promoting expression of corresponding metabolism-associated genes (Ma et al., 2011). Plant growth is inhibited in Arabidopsis plants constitutively expressing AtbZIP11 via the regulatory mechanism associating with the sucrose non-fermenting-1-related protein kinase 1 and trehalose 6-phosphate signaling systems (Ma et al., 2011). In addition, AtbZIP11 specifically activates the transcription of auxin-responsive genes via recruitment of histone acetylation machinery and contributes to the modulation of auxin-mediated responses (Weiste and Dröge-Laser, 2014). Moreover, tbz17 in tobacco, lip19 and OsOBF1 in rice, and mlip15 in maize are phylogenetically close to AtbZIP11. Transcript levels of tbz17 (Kusano et al., 1998), lip19 (Shimizu et al., 2005), and mlip15 (Kusano et al., 1995) are significantly increased under low temperature stress, while OsOBF1 shows an opposite expression pattern (Shimizu et al., 2005). In N. babacum, transcripts of tbz17 are up-regulated during leaf senescence (Yang et al., 2001). Overexpression of tbz17 results in enlarged leaf cells and increased sucrose levels compared with wild-type (WT) (Thalor et al., 2012). But the roles of bZIP11 in antiviral RNA silencing still remain enigmatic.
In previous studies, we obtained numerous up-regulated genes, including a large number of transcription factors, during flower development through transcriptomic analysis related to flower senescence in petunia (Wang et al., 2013). We have successfully used a TRV-based VIGS method to assess the function of senescence-related genes in petunia floral tissues (Chen et al., 2004; Reid et al., 2009; Jiang et al., 2011; Chang et al., 2014; Yin et al., 2015). However, when the TRV-PhCHS report system was employed to silence several candidate genes, the inoculated plants failed to show the expected white-petal phenotype of CHS silencing in the purple-petal petunia plants. Of the candidate genes, we have recently reported an ethylene-responsive element binding factor, PhERF2, which plays a critical role in TRV-induced RNA silencing and antiviral defense via transcriptional modulation of RDR2, RDR6, DCL2, and AGO2 in petunia (Sun et al., 2016). In this study, we report another regulatory gene, an ocs element binding factor of bZIP transcription factors, namely PhOBF1. Simultaneous silencing of PhOBF1 and reporter genes, PDS or CHS, led to a severe impairment of leaf photobleaching or white-flower phenotype in petunia. PhOBF1 appears to play a crucial role in the antiviral RNA silencing process through the regulation of the SA biosynthesis pathway.
Materials and methods
Plant materials and growth conditions
Petunia (Petunia×hybrida) plants were maintained in a growth chamber at 25/20 °C day/night temperatures with a 16/8 h light/dark cycle. Four-leaf-stage plantlets of cultivar ‘Primetime Blue’ were inoculated with TRV constructs to silence PhOBF1 in leaves and floral tissues. Stable transformation was obtained from ‘Mitchell Diploid’. Four-week-old transgenic plant leaves were sampled to determine the transcript levels of PhOBF1, RNA silencing-related genes and genes in the SA biosynthesis pathway, as well as concentrations of SA, SAG, and phenylalanine. To further examine the impact of PhOBF1 on silencing efficiency of the VIGS system, transgenic plants at the four-leaf stage were infiltrated with TRV-PhPDS. For multiple resistance analysis, 4-week-old WT and transformed seedlings were inoculated with TRV and TMV. For all studies of gene expression in virus assay, inoculated or uppermost unfolded leaves (the third or fourth leaf from terminal) were harvested and used for total RNA isolation. Tissue-specific expression analysis of PhOBF1 was conducted using younger leaves, stems, and roots, and flowers at anthesis (separated into sepals, petals, pistils, and stamens) of 10-week-old ‘Mitchell Diploid’ plants. Corollas at day 1, 3, 5, and 7 post-anthesis were used to study PhOBF1 expression profiles during flower senescence.
Identification of PhOBF1
The PhOBF1 cDNA fragments were isolated by RT-PCR as previously described (Chang et al., 2014). Corresponding amino acids were deduced by the translate tool of ExPASy (http://web.expasy.org/translate/). Its conserved domain was identified using an NCBI web server (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi). Homologous proteins from petunia and other species were found through NCBI BLAST (http://blast.ncbi.nlm.nih.gov/). Multiple sequence alignment was conducted by CLUSTALW (http://www.genome.jp/tools/clustalw/). A phylogenetic tree was constructed by means of MEGA4 (version 4.0.2) software.
Abiotic stress and hormone treatments
For the cold treatment, 3-week-old seedlings of ‘Primetime blue’ were placed in small vials with distilled water and maintained at room temperature (control) and in a 4 °C room. For the salinity and dehydration treatments, the seedlings were inserted in vials containing 100 mM NaCl and no water, respectively. For the ethylene treatment, the seedlings were treated with 10 µl l−1 of ethylene gas continuously in a sealed chamber. For other hormone treatments, the plants were placed in vials containing 50 μM ABA, 50 μM gibberellic acid (GA3), 200 μM SA, or 200 μM methyl jasmonate (MeJA). Samples were collected at 0, 3, 6, 12, and 24 h post-treatment.
Construction of VIGS plasmids
The TRV-PhPDS, TRV-PhCHS, and TRV-PhPDS/CHS plasmids were generated previously using 138-bp PDS and 194-bp CHS fragments (Chen et al., 2004). To generate the TRV-PhPDS/OBF1, TRV-PhCHS/OBF1, and TRV-PhPDS/CHS/OBF1 constructs, the fragment of PhOBF1 were PCR-amplified using specific primers F1R1 (Supplementary Table S1 at JXB online) and then cloned into three respective plasmids as mentioned above. A different fragment (fragment 2) of PhOBF1 amplified by specific primers F2R2 (Supplementary Table S1) was cloned into TRV-PhPDS/CHS. Similarly, a PhbZIP44 fragment was PCR-amplified to form TRV-PhPDS/bZIP44, TRV-PhCHS/bZIP44, and TRV-PhPDS/CHS/bZIP44 constructs. Additionally, the TRV–green fluorescent protein (GFP) construct was kindly provided by Junping Gao from China Agricultural University (Beijing, China) (Tian et al., 2014).
Agro-inoculation of TRV vectors
Agrobacterium tumefaciens strain GV3101 was electrotransformed with TRV1 and TRV2 plasmids. The transformed bacteria were cultured in LB media with 40 mg l−1 kanamycin, 20 mg l−1 gentamicin, 10 mM MES, and 20 μM acetosyringone for 48 h at 28 °C. The cells were harvested via centrifugation and then resuspended in inoculation buffer containing 10 mM MgCl2, 10 mM MES, and 200 μM acetosyringone to an OD600 of 4.0, followed by incubation at room temperature for 3–5 h. Subsequently, the Agrobacterium cultures containing TRV1 and TRV2 plasmids were mixed together in a 1:1 ratio. The bacterial mixture was applied to the abaxial side of petunia leaves using a 1-ml disposal needle-free syringe (Reid et al., 2009; Jiang et al., 2011). The Agrobacterium without transformation was used as mock control.
Semi-quantitative RT-PCR and quantitative real-time PCR
TRIzol reagent (Invitrogen, Carlsbad, CA, USA) was used to isolate total RNA from petunia leaves or flowers. Purification of RNA was performed using RNase-free DNase I (Promega, Madison, WI, USA). First-strand cDNA was synthesized from total RNA (2–5 μg) with SuperScript III reverse transcriptase (Invitrogen, Carlsbad, CA, USA). PCR was carried out using Taq DNA polymerase (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s recommendations, and resulting PCR products were analysed by electrophoresis. 26S ribosomal RNA served as an internal control (Chen et al., 2004; Reid et al., 2009). Quantitative real-time PCR was performed using the SYBR Green PCR Master Mix (2X) (Applied Biosystems, Foster City, CA, USA) as previously described (Liang et al., 2014). A minimum of three biological replicates were used for determining transcript abundances of genes. Statistical significance of difference was evaluated by Duncan’s multiple range test at a P value <0.05.
Northern blot assay
Total RNA was extracted from the systemic petunia leaves infected with TRV vectors and mock control using TRIzol reagent (Invitrogen, Carlsbad, CA, USA). Small RNAs with low molecular mass were further isolated with a solution containing polyethylene glycol (PEG8000) and NaCl as previously described (Wang et al., 2004). The ethidium bromide stained 5S rRNA was served as a loading control. Blot hybridization was conducted as previously described (Donaire et al., 2008). For analysis of siRNAs derived from TRV-PhPDS, oligonucleotide probes corresponding to PDS insert (138 bp) in TRV2 vector were radiolabeled with [γ-32P]ATP using T4 polynucleotide kinase (Promega). Finally, relative siRNA accumulation was visualized through RNA blots exposed to autoradiographic film.
Generation of transgenic petunia plants
The full-length of the PhOBF1 ORF sequence was PCR-amplified using the following primers: 5′-ATCTCGAGATGG CATCTTCTAGTGGAAA-3′ (forward) with an XhoI restriction site and 5′-ATGAGCTCTCAATACTGATAGAACATAT-3′ (reverse) with a SacI restriction site. The fragment was cloned into pGSA1403 vector to form the overexpression construct (Chang et al., 2014; Yin et al., 2015). For RNAi construct, the fragment (Supplementary Fig. S1A) of PhOBF1 cDNA was amplified using the primers F1R1 (Supplementary Table S1) with addition of a sequence harboring SpeI–AscI and BamHI–SwaI restriction sites in the 5′ end of forward and reverse primers, respectively. The resulting products were ligated into pGSA1285 vector in sense and antisense orientations. The plasmids for overexpression and RNAi constructs were transformed into A. tumefaciens strain LBA4404 by electroporation. Petunia cultivar ‘Mitchell Diploid’ was used for generation of transgenic plants using the leaf-disk transformation method (Wang et al., 2013; Liang et al., 2014). Positive transgenic plants were screened on MS plates supplemented with 100 mg l−1 kanamycin (Estrada-Melo et al., 2015; Yin et al., 2015).
Measurement of SA, SAG, and phenylalanine
Petunia leaves (0.2–0.5 g) were ground to fine powder in liquid nitrogen using a mortar and pestle. Determination of free SA and SAG levels was performed using a method previously described (Lee et al., 2011) with some minor changes. Methanol–formic acid (1 ml; 95:5) was used to extract the leaf tissues. Samples were vortexed and sonicated for 20 min for a complete homogenization. The mixture was centrifuged at 15 000 g for 10 min at 4 °C. The supernatant was filtered through a 0.45 μm filter and then freeze-dried in a vacuum. The pellet was resuspended in 1 ml of 5% trichloroacetic acid. Free SA was extracted organically by supplementing 1 ml of ethylacetate–cyclopentane–isopropanol (50:50:1). The upper phase containing free SA was collected and analysed by the high-performance liquid chromatography (HPLC). The SAG-containing aqueous phase after free SA extraction was exposed to acidification treatment by hydrochloric acid to pH 1.0. Subsequent boiling of acidified solution for 30 min was implemented for the release of SA from conjugated forms. The released SA was measured as described above. For quantification of phenylalanine, the ground powder of leaves was extracted with 1 ml 60% methanol (v/v), 0.1% phosphoric acid (v/v) with 2% 3-methylsalicylic acid (w/v). Following sonication, centrifugation, and filtration, the clear solution was injected into a C18 column for HPLC separation as reported previously (Janzik et al., 2005). The HPLC assay was performed using an Agilent chromatograph (model 1100, Agilent Technologies, Santa Clara, CA, USA), equipped with a fluorescence detector. The pure SA and phenylalanine (Sigma-Aldrich, St Louis, MO, USA) were dissolved and used as reference standards.
Inoculation with TMV
Infectious sap in 0.1 M phosphate buffer (pH 7.0, 1:4 w/v) was prepared from TMV-infected leaves of N. benthamiana. Prior to inoculation, the fully expanded leaves of 4-week-old healthy petunia plantlets were dusted with 400 mesh carborundum powder. The TMV sap was used to inoculate the leaves by a mechanical method (Hull, 2009). Relative virus accumulation was checked by quantitative real-time PCR analysis of transcript levels of TMV-CP, encoding coat protein of TMV.
Results
TRV infection, abiotic stresses and hormone treatments induce PhOBF1 expression
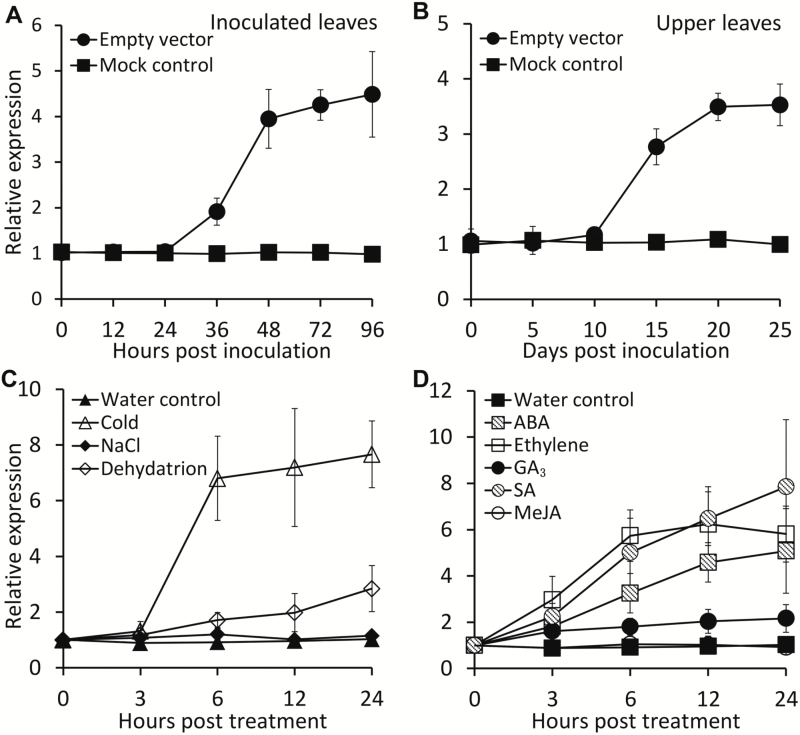
A 1029-bp cDNA corresponding to a putative ocs element binding factor 1 (OBF1) gene was isolated from a library constructed from petunia flower (Wang et al., 2013). The deduced protein, annotated as PhOBF1, was phylogenetically closed to tomato (Solanum lycopersicum) SlOBF1, potato (S. tuberosum) StOBF1, and Arabidopsis AtbZIP11. These proteins share a conserved basic leucine zipper domain with a length of 63 amino acids (Supplementary Fig. S1). PhOBF1 was constitutively expressed in leaves and roots at a higher level than in stem and floral tissues (Supplementary Fig. S2). To study the impact of TRV infection on PhOBF1 expression, the Agrobacterium bearing TRV empty vector was used to inoculate the leaves of four-leaf-stage WT petunia seedlings. Transcript abundances of PhOBF1 increased remarkably at 36 h post-inoculation (hpi) in inoculated leaves and 15 d post-inoculation (dpi) in systemically infected upper leaves, respectively (Fig. 1A, B). By contrast, the inoculation with non-transformed Agrobacterium (mock control) did not alter PhOBF1 expression levels (Fig. 1A, B). Furthermore, transcript levels of PhOBF1 increased following exposure to low temperature and drought stresses (Fig. 1C) and was also up-regulated following ABA, ethylene, GA3, and SA treatments (Fig. 1D).
Fig. 1.
Induction of PhOBF1 expression by TRV infection, abiotic stresses and hormone treatments in petunia leaves. (A, B) Quantitative real-time PCR analysis of PhOBF1 transcript levels in inoculated (A) and systemically infected upper (B) leaves with TRV at different time points. Four-week-old WT plants were inoculated with Agrobacterium bearing no TRV vector (mock control) or a TRV empty vector. (C, D) Quantitative real-time PCR analysis of PhOBF1 transcript levels in response to abiotic stresses (C) and hormones (D) at different time points. Three-week-old petunia seedlings were placed in vials with water at room temperature (control) or in a 4 °C room (cold), or without water at room temperature (dehydration), or with 100 mM NaCl, 50 μM ABA, 50 μM GA3, 200 μM SA, or 200 μM MeJA, or exposed to 10 μl l−1 ethylene treatment. Transcript abundances were standardized to 26S rRNA. Error bars represent standard error of the mean from three biological replicates.
Simultaneous silencing of PhOBF1 and reporter genes disrupts VIGS efficiency
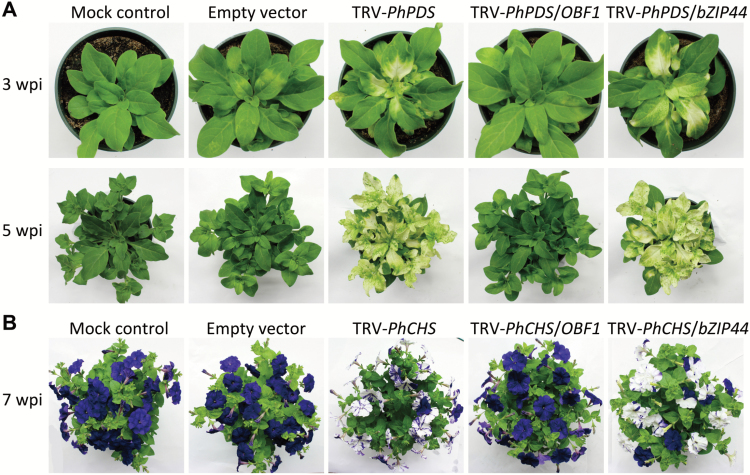
To investigate the function of PhOBF1, TRV-based VIGS with visual reporters of PDS and CHS was employed to knockdown PhOBF1 expression in the leaves and flowers of petunia, respectively. The plants inoculated with mock and empty vector displayed the normal WT phenotype with green leaves (Fig. 2A) and purple corollas (Fig. 2B). TRV-PhPDS-infected systemic leaves showed a typical PDS-silenced photobleaching phenotype. However, TRV-PhPDS/OBF1-infected systemic leaves failed to show a photobleaching phenotype at 3 and 5 weeks post-inoculation (wpi) (Fig. 2A). To examine whether the failed photobleaching phenotype is only associated with the PhOBF1 gene, a cDNA fragment from a paralog of PhOBF1, PhbZIP44 (Supplementary Fig. S1B) was used for comparison. The plants infected with TRV-PhPDS/bZIP44 showed a PDS-silenced photobleaching phenotype (Fig. 2A). To further confirm this finding, another visual reporter gene, CHS, was introduced. The CHS-silenced white-corollas phenotype, as shown in TRV-PhCHS and TRV-PhCHS/bZIP44, was not observed in the flowers infected by TRV-PhCHS/OBF1 at 7 wpi (Fig. 2B). Furthermore, the 3′ end fragment (Supplementary Fig. S1A) of PhOBF1 cDNA was inserted into a tandem construct TRV-PhPDS/CHS for VIGS assay. Both systemic leaves and flowers from TRV-PhPDS/CHS/OBF1 (F2)-infected plants did not show PDS-photobleaching and CHS-white silencing phenotypes during the entire growth periods (Supplementary Fig. S3).
Fig. 2.
Failed development of PDS-silenced leaf photobleaching and CHS-silenced white-corollas phenotypes in plants with PhOBF1-VIGS silencing. Representative phenotypes of WT plants at 3, 5 (A), and 7 (B) weeks post-inoculation (wpi) with Agrobacterium bearing no TRV vector (mock control), or a TRV empty vector, TRV-PhPDS and TRV-PhCHS, TRV-PhPDS/OBF1 and TRV-PhCHS/OBF1, TRV-PhPDS/bZIP44 and TRV-PhCHS/bZIP44 constructs, respectively.
VIGS silencing of PhOBF1 does not suppress virus proliferation
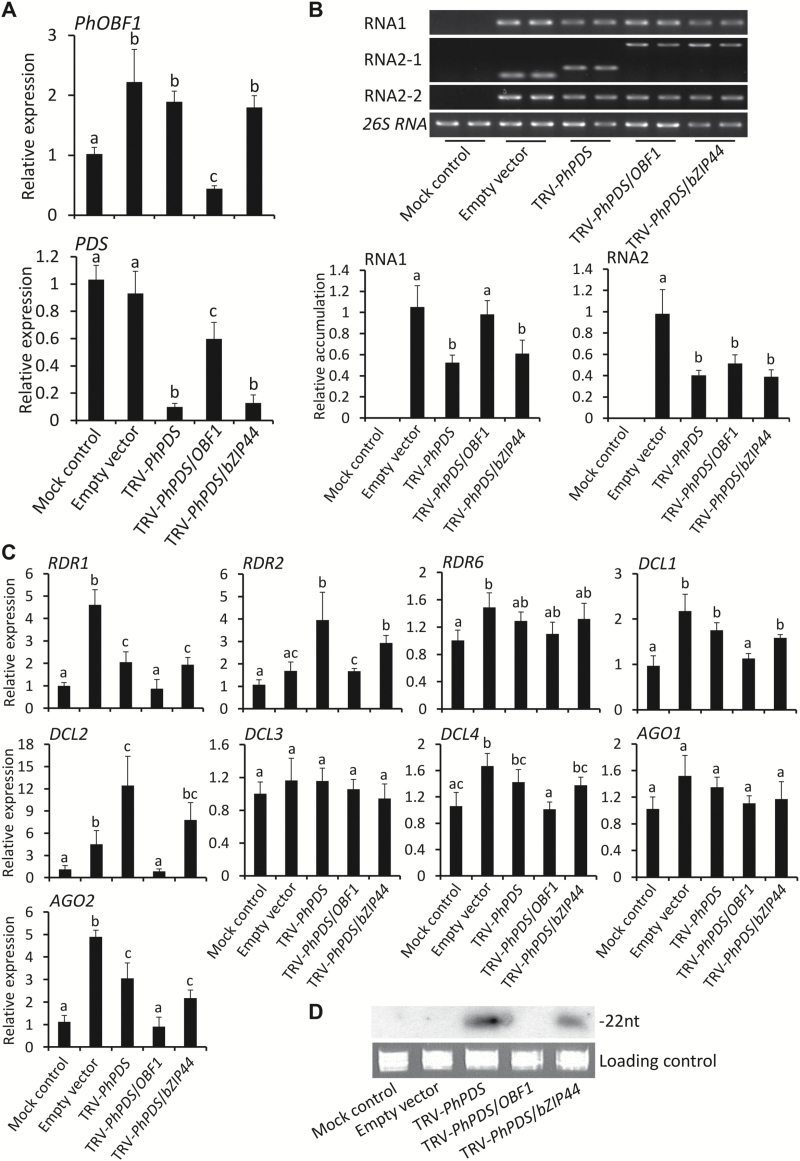
To understand why the inclusion of the PhOBF1 fragment in TRV-PhPDS and TRV-PhCHS constructs severely impaired the PDS-silenced leaf photobleaching and CHS-silenced white-corollas phenotypes, we analysed transcript abundances of PhOBF1 and PDS, and accumulation levels of TRV RNA1 and RNA2 in uppermost leaves of plants at 3 wpi. PhOBF1 transcripts were up-regulated in the leaves from plants infected with TRV constructs except TRV-PhPDS/OBF1 (Fig. 3A). The PhOBF1 expression levels in leaves infected with TRV-PhPDS/OBF1 were significantly lower than that in mock-treated leaves and other TRV construct-infected leaves (Fig. 3A), suggesting the partial knockdown of PhOBF1 expression. When examining PDS transcripts, a significantly marked reduction (greater than 90%) was found in photobleached leaves from plants infected with TRV-PhPDS and TRV-PhPDS/bZIP44 (Fig. 3A). However, only about a 40% reduction in PDS abundance was observed in leaves from TRV-PhPDS/OBF1-infected plants, compared with mock and empty vector controls (Fig. 3A).
Fig. 3.
Expression of PhOBF1, PDS, and RNA silencing-related genes and accumulation of TRV and siRNAs in the leaves of agro-infiltrated plants. (A) Quantitative real-time PCR analysis of PhOBF1 and PDS transcript levels in uppermost leaves of plants at 3 weeks post-inoculation (wpi) with Agrobacterium bearing a TRV empty vector, TRV-PhPDS, TRV-PhPDS/OBF1, and TRV-PhPDS/bZIP44 constructs. Non-transformed Agrobacterium was used as mock control. Transcript abundances were standardized to 26S rRNA. (B) Semi-quantitative RT-PCR and quantitative real-time PCR analysis of TRV RNA1 and RNA2 (RNA2-1, -2) accumulation levels in uppermost leaves of plants at 3 wpi. Primers for RNA2-1 were designed based on the region outside of multiple cloning sites (MCS) in the vector and resulted product sizes were dependent on the inserted fragments of PDS, PhOBF1/PDS, and PhOBF1/bZIP44, while RNA2-2 targets the region upstream of MCS and produced the same sizes of products. (C) Quantitative real-time PCR analysis of transcript levels of RNA silencing-related genes, including RDR1, RDR2, RDR6, DCL1, DCL2, DCL3, DCL4, AGO1, and AGO2, in uppermost leaves of plants at 3 wpi. 26S rRNA was used as internal control. (D) Northern-blot analysis of TRV-PhPDS-derived siRNA levels in uppermost leaves of plants at 3 wpi. 32P-labeled oligonucleotide probes correspond to PDS insert sequence. Ethidium bromide-stained 5S rRNA is indicated as a loading control. Error bars represent standard error of the mean from three biological replicates. Different letters indicate statistical significance as calculated by Duncan’s multiple range test at P < 0.05.
Semi-quantitative RT-PCR analysis revealed that when two primer pairs covering multiple cloning sites (MCS) (RNA2-1) and a region upstream of MCS (RNA2-2) were used (Supplementary Table S1), DNA fragments carrying PhOBF1 and PDS inserts were detected in TRV-PhPDS/OBF1-infected leaves (Fig. 3B), suggesting that the PhOBF1 silencing did not suppress the TRV replication or movement. Quantitative real-time analysis indicated that the accumulation of TRV RNA1 in empty vector- and TRV-PhPDS/OBF1-infected leaves was significantly higher than that in the leaves of plants agro-infiltrated with TRV-PhPDS and TRV-PhPDS/bZIP44 constructs (Fig. 3B).
VIGS silencing of PhOBF1 inhibits expression of RNA silencing-related genes and accumulation of siRNAs
To further understand failed detection of PDS and CHS silencing phenotypes in TRV-PhPDS/OBF1- and TRV-PhCHS/OBF1-infected plants, we examined transcript abundances of genes associated with the RNA silencing pathway. RDRs, DCLs and AGOs are indispensable components participating in dsRNA generation, cleavage and siRNA binding in the RNA silencing pathway (Burch-Smith et al., 2006; Gould and Kramer, 2007; Jaubert et al., 2011; Tian et al., 2014). We measured the transcript abundances of RDRs (1, 2, 6), DCLs (1–4) and AGOs (1, 2) in uppermost leaves of plants at 3 wpi. The majority of the selected genes, except RDR6, DCL3, and AGO1, were significantly up-regulated in the leaves from empty vector-, TRV-PhPDS- or TRV-PhPDS/bZIP44-infected plants but not in TRV-PhPDS/OBF1-infected plants (Fig. 3C).
To determine TRV-PhPDS-derived siRNA abundance, northern blot analysis was performed using a DNA fragment of the PDS reporter as probe. No signals were detectable in the leaves from TRV-PhPDS/OBF1-infected plants. By comparison, one exclusive band with a length of 22 nt was detected in the leaves from plants infected with TRV-PhPDS and TRV-PhPDS/bZIP44 (Fig. 3D).
PhOBF1 silencing by RNAi impairs VIGS efficiency
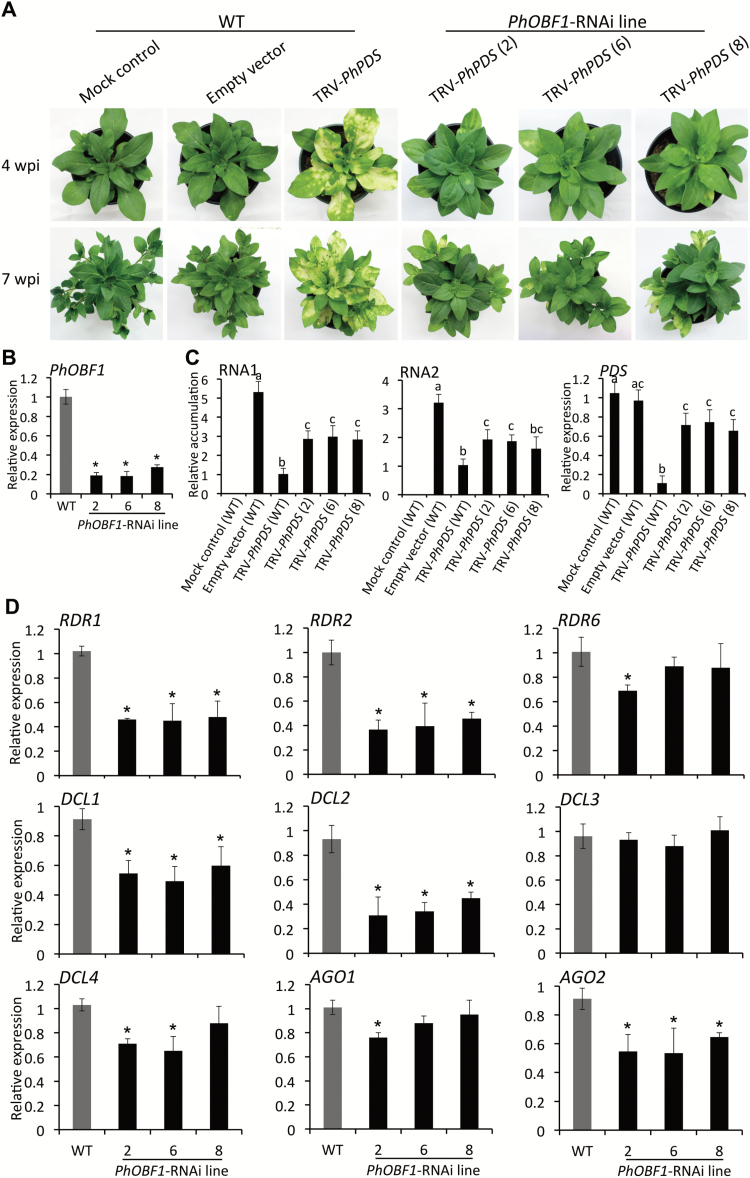
To better dissect PhOBF1’s function, transgenic petunia plants with down-regulated PhOBF1 expression by RNAi were generated (lines 2, 6 and 8) and infected with TRV constructs. A striking impairment of the photobleaching phenotype was detected in the leaves of the transgenic plants infected with TRV-PhPDS reporter constructs (Fig. 4A), confirming the failed photobleaching phenotype in the VIGS assay. Transcript levels of PhOBF1 in PhOBF1-RNAi lines were reduced by 70–80%, compared with WT (Fig. 4B). Accumulation levels of TRV RNA1 and RNA2 were higher in PhOBF1-silenced lines than that in WT plants infected with TRV-PhPDS (Fig. 4C). Following infection with TRV-PhPDS, approximately 90% reduction in PDS transcripts was found in WT leaves, whereas only 20–30% reduction in PDS transcripts was detected in PhOBF1-RNAi lines (Fig. 4C). Silencing of PhOBF1 led to significant down-regulation of RDR1, RDR2, DCL1, DCL2, and AGO2 in PhOBF1-RNAi, compared with the control lines (Fig. 4D). Transcript levels of RDR6, DCL4, and AGO1 were slightly reduced in certain PhOBF1-RNAi lines (Fig. 4D). PhOBF1 silencing did not affect transcript abundances of DCL3 (Fig. 4D).
Fig. 4.
Impairment of leaf photobleaching phenotype of PDS silencing in PhERF2-RNAi lines inoculated with Agrobacterium bearing TRV-PhPDS. (A) Representative phenotypes of WT and PhOBF1-RNAi lines (2, 6 and 8) inoculated with non-transformed Agrobacterium (mock control), or Agrobacterium bearing a TRV empty vector and TRV-PhPDS construct. Photographs were taken at 4 and 7 weeks post-inoculation (wpi). (B) Quantitative real-time PCR analysis of transcript abundances of PhOBF1 in uppermost leaves of 4-week-old WT and PhOBF1-RNAi lines (2, 6 and 8). Expression levels were normalized to 26S rRNA. (C) Quantitative real-time PCR analysis of TRV (RNA1 and RNA2) accumulation and PDS transcript levels in uppermost leaves of WT and PhOBF1-RNAi lines (2, 6 and 8) at 4 wpi. Accumulation and transcript levels were standardized to 26S rRNA. (D) Quantitative real-time PCR analysis of transcript abundances of RNA silencing-related genes, including RDR1, RDR2, RDR6, DCL1, DCL2, DCL3, DCL4, AGO1, and AGO2, in uppermost leaves of 4-week-old WT and PhOBF1-RNAi lines (2, 6 and 8). 26S rRNA was used as internal standard. Error bars represent standard error of the mean from three biological replicates. Significance of difference was calculated using Duncan’s multiple range test (P<0.05) and shown as asterisks or different letters.
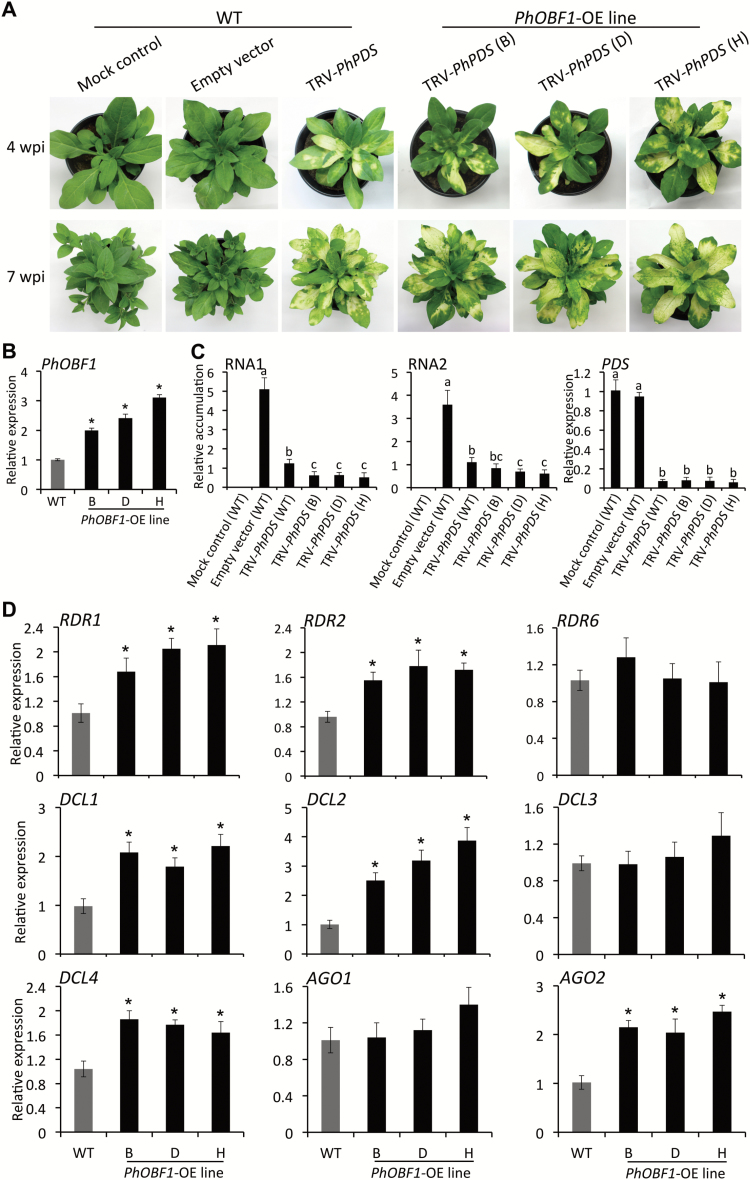
PhOBF1 overexpression restores silencing phenotype
To further study the function of PhOBF1 in the RNA silencing process, we generated PhOBF1-overexpressing (OE) lines (B, D and H) in petunia. A prominent PDS-silenced leaf photobleaching phenotype emerged at 4 and 7 wpi in systemic leaves of WT and OE lines infected with TRV-PhPDS (Fig. 5A). A two- to three-fold increase in PhOBF1 transcript levels was detected in three independent lines, compared with WT (Fig. 5B). TRV RNA1 and RNA2 accumulation was significantly decreased in PhOBF1-OE lines compared with WT (Fig. 5C). A dramatic and significant decrease in PDS transcript levels was found in the uppermost photobleached leaves of control and PhOBF1-OE lines infected with TRV-PhPDS at 4 wpi (Fig. 5C). Moreover, overexpression of PhOBF1 significantly increased the transcript levels of RDR1, RDR2, DCL1, DCL2, DCL4, and AGO2 in all transgenic lines (Fig. 5D). Overexpression of PhOBF1 did not affect transcript abundances of RDR6, DCL3, and AGO1.
Fig. 5.
Leaf photobleaching phenotype of PDS silencing in PhOBF1-overexpressing lines inoculated with Agrobacterium bearing TRV-PhPDS. (A) Representative phenotypes of WT and PhOBF1-overexpressing (OE) lines (B, D and H) inoculated with Agrobacterium bearing no TRV vector (mock control), or a TRV empty vector and TRV-PhPDS construct. Photographs were taken at 4 and 7 weeks post-inoculation (wpi). (B) Quantitative real-time PCR analysis of expression levels of PhOBF1 in the leaves of WT and PhOBF1-OE lines (B, D and H). Samples were harvested from uppermost leaves of 4-week-old plants. 26S rRNA was used as normalization control. (C) Quantitative real-time PCR analysis of TRV (RNA1 and RNA2) accumulation and PDS transcript abundances in uppermost leaves of WT and PhOBF1-OE lines (B, D and H) 4 wpi. 26S rRNA was used as internal control. (D) Quantitative real-time PCR analysis of transcript abundances of RNA silencing-related genes, including RDR1, RDR2, RDR6, DCL1, DCL2, DCL3, DCL4, AGO1, and AGO2, in uppermost leaves of 4-week-old WT and PhOBF1-OE lines (B, D and H). Transcript levels were standardized to 26S rRNA. Error bars represent standard error of the mean from three biological replicates. Asterisks or different letters indicate statistical significance using Duncan’s multiple range test at P<0.05.
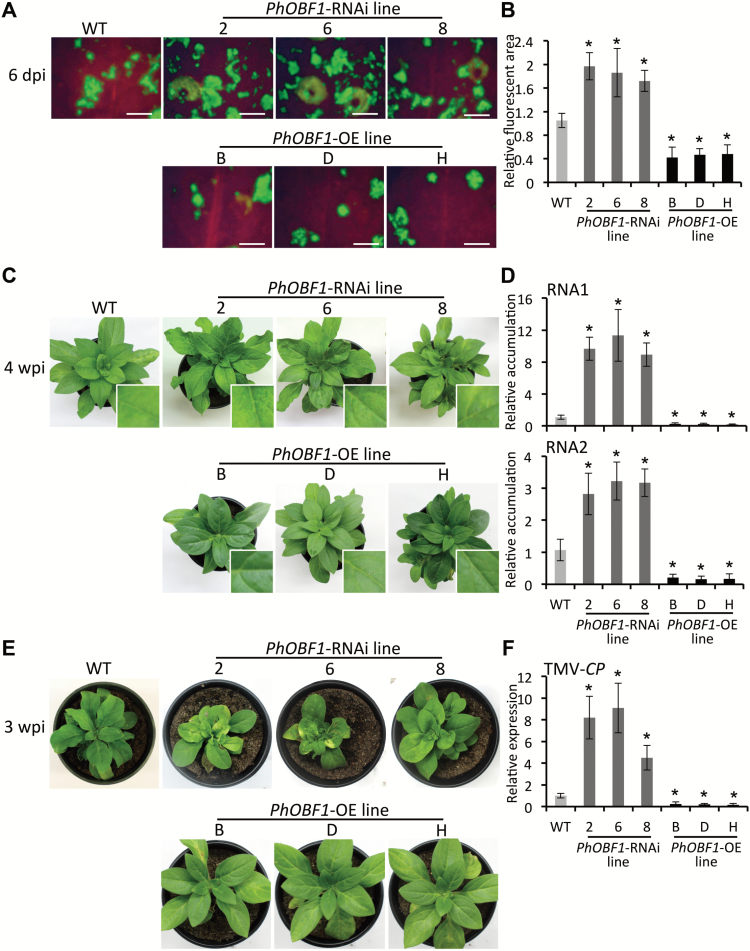
PhOBF1 affects plant defense against TRV and TMV
To examine the role of PhOBF1 in plant defense against viral infections, a TRV vector expressing green fluorescent protein (TRV-GFP) (Tian et al., 2014) was used to inoculate transgenic and WT leaves to visualize the accumulation of TRV under UV irradiation. At 6 dpi, the fluorescent area was visibly larger in PhOBF1-RNAi but smaller in PhOBF1-OE lines than in the control line (Fig. 6A, B). Furthermore, TRV empty vector was used for inoculation to observe virus symptom development in the transgenic and WT plants. Compared with WT, PhOBF1-RNAi lines exhibited a severer leaf mottling, chlorosis and curling symptoms at 4 wpi with TRV, whereas the virus symptoms were considerably reduced in PhOBF1-OE lines (Fig. 6C). Consistent with that, higher accumulation levels of TRV RNA1 and RNA2 were found in PhOBF1-RNAi lines. On the other hand, lower accumulation levels were detected in the plants overexpressing PhOBF1 than in WT plants (Fig. 6D and Supplementary Table S2). Furthermore, the WT plants inoculated with TMV developed a symptom shown as slightly mottled and curled leaves, while leaf yellowing, necrosis, and dwarfing were observed in PhOBF1-RNAi lines. A much milder virus symptom was observed in PhOBF1-OE lines than in WT plants (Fig. 6E). At 3 wpi, PhOBF1-RNAi lines accumulated more transcripts of TMV-CP, encoding coat protein of TMV, but fewer TMV-CP transcripts were detected in PhOBF1-OE lines than in the control line (Fig. 6F and Supplementary Table S2).
Fig. 6.
Contribution of PhOBF1 to petunia plant defense against TRV and TMV. GFP fluorescent foci (A) and relative fluorescent area (B) in inoculated leaves of WT, PhOBF1-RNAi (2, 6 and 8), and PhOBF1-overexpressing (OE) (B, D and H) lines at 6 days post-inoculation (dpi) with Agrobacterium bearing a TRV-GFP construct. Photographs were taken under UV light. Scale bars (A): 5 mm. Images of fluorescent area from three plants were analysed using Photoshop software. (C) Disease symptoms of WT, PhOBF1-RNAi (2, 6 and 8) and PhOBF1-OE (B, D and H) lines at 4 weeks post-inoculation (wpi) with Agrobacterium bearing a TRV empty vector. The insets are magnified views of viral symptoms in uppermost leaves infected with TRV. (D) Quantitative real-time PCR analysis of TRV (RNA1 and RNA2) accumulation levels in systemically infected (uppermost) leaves of WT, PhOBF1-RNAi (2, 6 and 8), and PhOBF1-OE (B, D and H) lines at 4 wpi. Transcript abundances were normalized to 26S rRNA. (E) Disease symptoms of WT, PhOBF1-RNAi (2, 6 and 8), and PhOBF1-OE (B, D and H) lines at 3 wpi with TMV. (F) Quantitative real-time PCR analysis of transcript levels of TMV-CP, encoding TMV coat protein, in systemically infected (uppermost) leaves of WT, PhOBF1-RNAi (2, 6 and 8), and PhOBF1-OE (B, D and H) lines at 3 wpi. 26S rRNA was used as an internal control. Error bars represent standard error of the mean from three biological replicates. Asterisks denote statistical significance as determined by Duncan’s multiple range test at P<0.05.
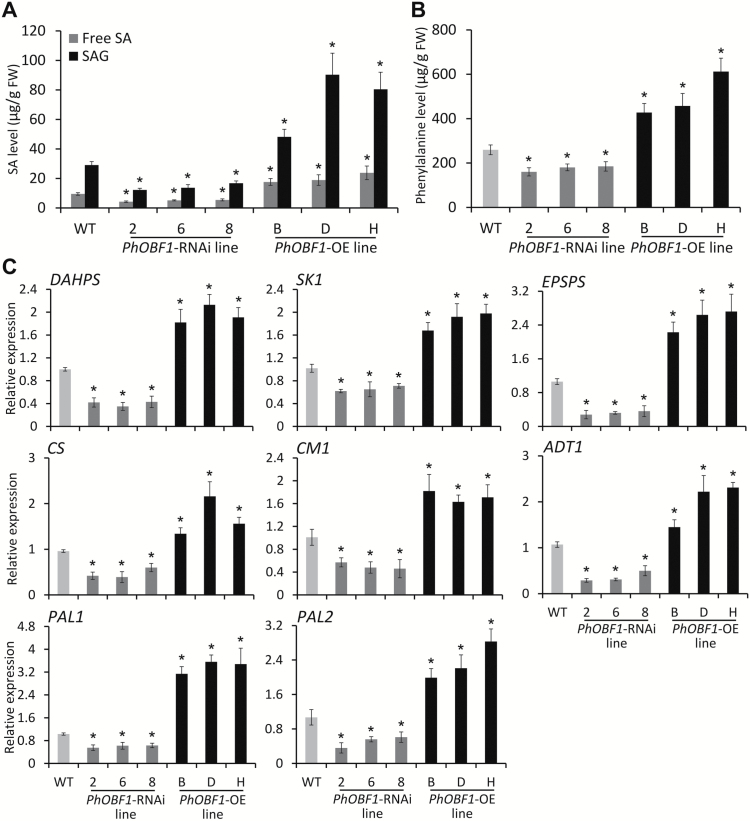
PhOBF1 modulates the biosynthesis of SA
Since the latest evidence suggests that transcripts of RDR1, RDR2, DCL1, and DCL2 are up-regulated in tomato plants following CEVd, ToMV infection, and SA treatment (Campos et al., 2014), we decided to examine the expression profile of RNA silencing-related genes in petunia plants treated with exogenous SA, and to measure the free SA, SAG, and phenylalanine contents in WT and transgenic plants. SA treatment resulted in a dramatic induction of RDR1, RDR2, DCL1, DCL2, DCL4, and AGO2 transcripts in petunia (Supplementary Fig. S4). Importantly, concentrations of free SA and conjugated SA were significantly reduced in PhOBF1-RNAi lines, but increased in PhOBF1-OE lines, compared with WT (Fig. 7A). Transgenic lines also showed similar alteration in concentrations of phenylalanine (Fig. 7B). Furthermore, we determined transcript abundances of the core genes in the shikimate and phenylpropanoid pathways that are associated with these biochemical changes. We found that transcript abundances of genes such as 3-deoxy-d-arabino-heptulosonate-7-phosphate synthase (DAHPS), shikimate kinase (SK1), 5-enolpyruvylshikimate 3-phosphate synthase (EPSPS), chorismate synthase (CS), chorismate mutase 1 (CM1), arogenate dehydratase 1 (ADT1), PAL1, and PAL2 were significantly down-regulated in PhOBF1-RNAi lines, but up-regulated in PhOBF1-OE lines (Fig. 7C). These results suggested that PhOBF1 may play important roles in the regulation of the RNA silencing pathway through biosynthesis of the hormone SA.
Fig. 7.
Involvement of PhOBF1 in the biosynthesis of salicylic acid. (A, B) Free SA, SA glucoside (SAG), and phenylalanine levels in the leaves of WT, PhOBF1-RNAi (2, 6 and 8), and PhOBF1-overexpressing (OE) (B, D and H) lines. Samples were harvested from all the leaves of 4-week-old WT and transgenic seedlings. (C) Quantitative real-time PCR analysis of transcript abundances of genes, including DAHPS, SK1, EPSPS, CS, CM1, ADT1, PAL1, and PAL2, in the shikimate and phenylpropanoid pathways in the leaves of WT, PhOBF1-RNAi (2, 6 and 8), and PhOBF1-OE (B, D and H) lines. Samples were harvested from the uppermost leaves of 4-week-old WT and transgenic seedlings. Expression levels were normalized to 26S rRNA. Error bars represent standard error of the mean from three biological replicates. Asterisks indicate statistical significance using Duncan’s multiple range test at P<0.05.
Discussion
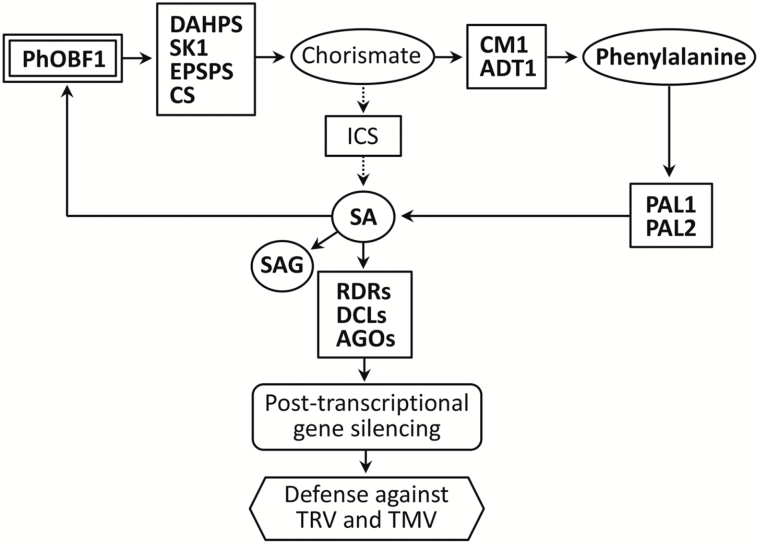
In this study, our data suggest that the ocs element binding factor PhOBF1 positively affects TRV-induced RNA silencing efficiency via the SA biosynthesis pathway (Fig. 8). OBF proteins bind to a class of DNA promoter sequences known as ocs elements or ACGT elements, which are essential for expression of bacterial and viral pathogen genes (Katagiri and Chua, 1992), and genes involved in the plant defense response (Ellis et al., 1993; Chen and Singh, 1999). OBFs belong to a specific group of the bZIP transcription factor family in plants. A total of 75 members of the bZIP family have been identified in Arabidopsis (Jakoby et al., 2002). The bZIP transcription factors induce the expression of genes whose promoter regions commonly share a core ACGT element, such as the G-box motif (CACGTG) (Hanson et al., 2008). They participate in a variety of plant-specific processes, including pathogen resistance, stress response, light signaling, seed maturation and germination, and flower development (Jakoby et al., 2002; Nijhawan et al., 2008).
Fig. 8.
A proposed model for PhOBF1 function in antiviral RNA silencing. PhOBF1 positively regulates the genes in the shikimate and phenylpropanoid pathways, as well as phenylalanine and SA accumulation. Ultimately, PhOBF1 contributes to extensive resistance to TRV and TMV through SA-mediated post-transcriptional gene silencing. Solid arrows indicate well-described relationships, and dotted arrows indicate undescribed steps in this work. The SA, SAG, phenylalanine, and related genes analysed in PhOBF1-RNAi and -OE lines are shown in bold.
We have been investigating the role of regulatory genes, differentially expressed during petunia flower development, in the control of the corolla senescence process (Wang et al., 2013). We employed the TRV-based VIGS method to silence these candidate genes, including bZIPs, to functionally study flower senescence-associated genes (Chang et al., 2014; Yin et al., 2015). When inoculated with TRV construct carrying PhOBF1 and reporter gene (PDS or CHS) fragments, the purple-flower petunia cultivar plants were incapable of developing the PDS- or CHS-silenced white leaves or flowers throughout the entire growth period (Fig. 2A, B), suggesting a dramatic reduction in VIGS efficiency. PhOBF1 transcripts were partially down-regulated in the leaves from TRV-PhPDS/OBF1-infected plants, in which a reduction by approximately 40% in PDS levels occurred, compared with up to 90% reduction in PDS levels in photobleached leaves in the control (Fig. 3A). These data led us to hypothesize that PhOBF1 probably functions as an important transcriptional regulator of antiviral RNA silencing.
RNA silencing triggered by virus dsRNA requires a few key components such as RDRs, DCLs and AGOs in plants (Costa et al., 2013; Kasai et al., 2013). Our data showed that the abundances of some RNA silencing-related genes, including RDR1, RDR2, DCL1, DCL2, DCL4, and AGO2, were decreased in the PhOBF1-silenced plants (Figs 3C and 4D) or increased in the PhOBF1-overexpressing transgenic plants (Fig. 5D). It is quite likely that PhOBF1 contributes to the RNA silencing response by modulating the transcription of these genes.
RNA silencing serves as one type of antiviral mechanism, and exerts virus-derived siRNAs (vsiRNAs) as guides to target endogenous mRNA and simultaneously viral RNA (Ma et al., 2015). Interference with the RNA silencing mechanism causes an impairment of resistance to diverse viruses. Many studies have been performed to examine the impact of key genes in the silencing complex on virus resistance. For example, mutation in the DCL genes results in more susceptibility to TCV and CMV (Bouché et al., 2006), TRV (Donaire et al., 2008), TMV (Lewsey and Carr, 2009), TuMV (Garcia-Ruiz et al., 2010), and PVX (Andika et al., 2015). A novel argonaute protein, AGO18, has been identified as conferring a broad-spectrum virus resistance in rice (Wu et al., 2015). Given PhOBF1’s importance in RNA silencing, it is expected that PhOBF1 is highly likely to be involved in plant resistance against a wide range of viral pathogens. Our findings of the compromised and enhanced resistance to TRV and TMV infections in PhOBF1-silenced and -overexpressing transgenic lines of petunia, respectively (Fig. 6), support this notion, and further prove the hypothesis that PhOBF1 plays a pivotal role in the antiviral RNA silencing process. Moreover, we observed quite strong effects on VIGS silencing of reporter genes (PDS or CHS) and antiviral responses given the moderate down-regulation or overexpression of PhOBF1. The altered transcripts of multiple RNA silencing-related genes by PhOBF1 probably contribute to these strong effects. DCLs and AGOs play different roles in antiviral RNA silencing, and the mutant lines ago1-27 and dcl2/dcl3/dcl4 infected with TRV-PhPDS show impaired leaf photobleaching and increased TRV accumulation (Ma et al., 2015). But it appears that those single or triple mutants have a relatively mild impact on gene silencing and antiviral defense compared with the PhOBF1 silencing or overexpression. As a powerful transcription factor, PhOBF1 may greatly affect the antiviral RNA silencing process by regulating other critical factors, such as SA.
Many pieces of evidence have revealed a close association between antiviral RNA silencing and the plant endogenous hormone SA. Both ToMV and CEVd infections promote the accumulation of SA (Bellés et al., 1999), as well as transcript levels of a number of RDR and DCL genes (Campos et al., 2014). Exogenous SA treatment reduces the infection caused by Alfalfa mosaic virus (AlMV) (Van Huijsduijnen et al., 1986), TMV (Chivasa et al., 1997; Murphy and Carr, 2002; Shang et al., 2011), CMV and TCV (Shang et al., 2011), and PVX (Falcioni et al., 2014). Degradation of SA in tobacco results in a reduced accumulation of virus-derived siRNAs and enhanced virus multiplication (Alamillo et al., 2006). Moreover, exogenous SA treatment not only increases the levels of RDR1 in Arabidopsis or tobacco (Yang et al., 2004; Quilis et al., 2008), but also RDR2, DCL1, and DCL2 transcript abundances in tomato (Campos et al., 2014). Our observations of significantly increased expression levels of RDR1, RDR2, DCL1, DCL2, DCL4, and AGO2 in petunia following treatment with SA (Supplementary Fig. S4) are consistent with these findings. In addition, we found that the impaired and recovered PDS-silenced leaf photobleaching phenotypes in PhOBF1-silenced and -overexpressing petunias were accompanied by reduced and increased endogenous SA levels, respectively (Fig. 7A). It seems possible that SA serves as an intermediate signal linking PhOBF1 with the antiviral RNA silencing pathway. Future studies will determine if exogenous SA treatment can recover the impaired PDS-silenced leaf photobleaching or CHS-silenced white-corollas phenotype in PhOBF1-silenced petunia plants inoculated with TRV constructs carrying reporter genes (PDS or CHS).
SA biosynthesis involves the shikimate and phenylpropanoid pathways, which are controlled by some critical genes, such as DAHPS, SK1, EPSPS, CS, CM1, ADT1, PAL1, and PAL2. Apart from SA content, we found that PhOBF1 silencing and overexpression led to down-regulation and up-regulation of these genes, respectively (Fig. 7C). Interestingly, silencing of R2R3-MYB-like transcription factors, ODO1 (Verdonk et al., 2005), EOBI and EOBII (Spitzer-Rimon et al., 2012), also results in reduction of a set of genes transcripts in the shikimate and phenylpropanoid pathways. ODO1, transcriptionally regulated by EOBI and EOBII (Spitzer-Rimon et al., 2012), is involved in floral fragrance production of petunia, and its suppression causes a decreased level of methyl salicylate (Verdonk et al., 2005). It appears likely that one of these three MYB-like genes might be a crucial target of PhOBF1 for the SA-mediated RNA silencing mechanism. However, we found that the petunia plants inoculated with TRV-PhCHS/ODO1 in the VIGS assay showed the CHS-silenced white-corollas phenotype (data not shown), suggesting that PhOBF1 is unlikely to regulate the SA-mediated antiviral RNA silencing system by activating downstream ODO1 expression. The involvement of PhOBF1 in floral scent biosynthesis requires further examination in the future.
It should be mentioned that TRV 16 kDa (16K) protein, encoded by genomic RNA1, functions as a suppressor of RNA silencing. However, unlike the strong silencing suppressors CMV 2b (Goto et al., 2007) and Tombusvirus P19 (Lakatos et al., 2004), TRV 16K acts as a weak suppressor that transiently blocks local RNA silencing and enhances viral RNA accumulation early (Martínez-Priego et al., 2008). In this study, we mainly focused on the silencing phenotypes in systemic leaves or flowers infected with various TRV constructs. It seems likely that the silencing of endogenous genes and antiviral defenses in inoculated leaves are more complex. In the early infection process, 16K suppressor may impose effects on PhOBF1-involved antiviral RNA silencing, and it remains unclear whether these effects can lead to the changes of final responses in systemically infected tissues. Furthermore, a possible relationship between PhOBF1 and TRV 16K suppressor should be explored in future work.
We are interested in the role of PhOBF1 in the regulation of flower senescence. Ethylene treatment increased the transcripts of PhOBF1 (Fig. 1D). Its transcript levels were also up-regulated at various developmental stages of petunia corollas (Supplementary Fig. S5). Our analysis of flower longevity revealed that silencing of PhOBF1 accelerated flower senescence and its overexpression extended flower longevity (Supplementary Table S3). Furthermore, both ABA and GA3 treatments increased the expression levels of PhOBF1 (Fig. 1D). The crosstalk between ethylene and ABA (Chang et al., 2014) or GA3 (Yin et al., 2015) during flower senescence has been discussed in petunia. More studies are required to clarify the role of PhOBF1 in the regulation of petunia flower senescence. Additionally, the role of PhOBF1 in plant growth and development should be further investigated, because we found that down-regulation of PhOBF1 resulted in increased plant height and reduced stem diameter, while overexpression of PhOBF1 caused slightly smaller and thicker leaves (data not shown).
It is worth mentioning that PhERF2, an ERF transcription factor we previously reported, plays a quite similar role in antiviral RNA silencing to PhOBF1. PhERF2 silencing also substantially impaired the PDS-silenced leaf photobleaching phenotype and PhERF2 overexpression restored it (Sun et al., 2016). The same involvements of PhOBF1 and PhERF2 in resistance to TRV were found. Thus, we hypothesize that a close association between PhOBF1 and PhERF2 probably occurs. In Arabidopsis, OBF4 interacts with an ethylene-responsive element binding factor AtEBP and may collaboratively contribute to the plant defense response (Büttner and Singh, 1997). However, we did not find altered PhERF2 expression in PhOBF1-silenced and -overexpressing lines, or altered PhOBF1 expression in PhERF2 transgenic lines (data not shown). PhOBF1 and PhERF2 affected transcript levels of different RNA silencing-related genes, and for example, PhOBF1 rather than PhERF2 affected expression levels of RDR1, DCL1, and DCL4. Moreover, PhOBF1, not PhERF2, plays a role in the regulation of petunia flower senescence. One explanation is that they perhaps regulate TRV-induced RNA silencing through two distinct pathways. ERF proteins specifically bind to the GCC box, which commonly presents in the promoter of downstream defensive genes of the ethylene signaling pathway (Ohme-Takagi and Shinshi, 1995). It seems likely that PhOBF1 and PhERF2 may be a positive regulator in the SA biosynthesis pathway and ethylene signaling pathway, respectively.
Our data demonstrate that PhOBF1 plays an important role in antiviral RNA silencing. The positive regulation of PhOBF1 in the TRV-induced gene silencing process via SA may provide a feasible approach to improve the efficiency of the TRV-based VIGS system, through a transient overexpression of PhOBF1 or its homologs, or an appropriate application of exogenous SA. Further confirmation should be sought for whether PhOBF1 also plays a regulatory role in other virus-based VIGS systems besides TRV. It is noteworthy that abiotic stresses, such as salt, drought, oxidation and low temperature, induce the expression of a number of RNA silencing-related genes in tomato (Bai et al., 2012). We also found that low temperature and dehydration treatments increased the transcript levels of PhOBF1 (Fig. 1C). The existing evidence supports that low temperature and low humidity enhance TRV-induced gene silencing efficiency in tomato (Fu et al., 2006). The effects of these abiotic stresses on the RNA silencing response still require more study in the future.
Supplementary data
Supplementary data are available at JXB online.
Fig. S1. Petunia PhOBF1 cDNA and deduced amino acid sequence analysis.
Fig. S2. Expression of PhOBF1 in various tissues of petunia plant.
Fig. S3. Failed development of leaf photobleaching and white-corollas phenotypes in petunia plants inoculated with a PhPDS/CHS/OBF1 tandem TRV construct.
Fig. S4. Induction of RNA silencing-related genes in petunia leaves treated with salicylic acid.
Fig. S5. Induction of PhOBF1 expression during petunia flower senescence.
Table S1. Primers used for semi-quantitative RT-PCR and quantitative real-time PCR.
Table S2. The numerical data for relative accumulation or expression levels of TRV RNAs and TMV-CP in WT, PhOBF1-RNAi and PhOBF1-overexpressing lines infected with TRV empty vector and TMV, respectively.
Table S3. The longevity of attached flowers from WT, PhOBF1-RNAi and PhOBF1-overexpressing lines.
Supplementary Material
Acknowledgements
We thank Bryce Falk and Raja Sekhar Nandety for experimental assistance in northern blot analysis. We are grateful to Lee Ann Richmond for operation of the laboratory instruments. We thank Weixing Shan for kindly providing the TMV strains. We also thank the anonymous reviewers for their invaluable suggestions for further improvement of the manuscript. This study was partially funded by United States Department of Agriculture (USDA) CRIS project 5306-21000-019-00D.
References
- Alamillo JM, Saénz P, García JA. 2006. Salicylic acid-mediated and RNA-silencing defense mechanisms cooperate in the restriction of systemic spread of plum pox virus in tobacco. The Plant Journal 48, 217–227. [DOI] [PubMed] [Google Scholar]
- Alvarado VY, Scholthof HB. 2011. AGO2: A new argonaute compromising plant virus accumulation. Frontiers in Plant Science 2, 112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andika IB, Maruyama K, Sun L, Kondo H, Tamada T, Suzuki N. 2015. Differential contributions of plant Dicer-like proteins to antiviral defences against Potato virus X in leaves and roots. The Plant Journal 81, 781–793. [DOI] [PubMed] [Google Scholar]
- Axtell MJ. 2013. Classification and comparison of small RNAs from plants. Annual Review of Plant Biology 64, 137–159. [DOI] [PubMed] [Google Scholar]
- Bai M, Yang GS, Chen WT, Mao ZC, Kang HX, Chen GH, Yang YH, Xie BY. 2012. Genome-wide identification of Dicer-like, Argonaute and RNA-dependent RNA polymerase gene families and their expression analyses in response to viral infection and abiotic stresses in Solanum lycopersicum. Gene 501, 52–62. [DOI] [PubMed] [Google Scholar]
- Bartel B, Bartel DP. 2003. MicroRNAs: at the root of plant development?Plant Physiology 132, 709–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumberger N, Baulcombe D. 2005. Arabidopsis Argonaute1 is an RNA Slicer that selectively recruits microRNAs and short interfering RNAs. Proceedings of the National Academy of Sciences, USA 102, 11928–11933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellés JM, Garro R, Fayos J, Navarro P, Primo J, Conejero V. 1999. Gentisic acid as a pathogen-inducible signal, additional to salicylic acid for activation of plant defenses in tomato. Molecular Plant-Microbe Interactions 12, 227–235. [Google Scholar]
- Bouché N, Lauressergues D, Gasciolli V, Vaucheret H. 2006. An antagonistic function for Arabidopsis DCL2 in development and a new function for DCL4 in generating viral siRNAs. The EMBO Journal 25, 3347–3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burch-Smith TM, Schiff M, Liu Y, Dinesh-Kumar SP. 2006. Efficient virus-induced gene silencing in Arabidopsis. Plant Physiology 142, 21–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Büttner M, Singh KB. 1997. Arabidopsis thaliana ethylene-responsive element binding protein (AtEBP), an ethylene-inducible, GCC box DNA-binding protein interacts with an ocs element binding protein. Proceedings of the National Academy of Sciences, USA 94, 5961–5966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos L, Granell P, Tárraga S, López-Gresa P, Conejero V, Bellés JM, Rodrigo I, Lisón P. 2014. Salicylic acid and gentisic acid induce RNA silencing-related genes and plant resistance to RNA pathogens. Plant Physiology and Biochemistry 77, 35–43. [DOI] [PubMed] [Google Scholar]
- Chang X, Donnelly L, Sun D, Rao J, Reid MS, Jiang CZ. 2014. A Petunia homeodomain-leucine zipper protein, PhHD-Zip, plays an important role in flower senescence. PLoS One 9, e88320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JC, Jiang CZ, Gookin TE, Hunter DA, Clark DG, Reid MS. 2004. Chalcone synthase as a reporter in virus-induced gene silencing studies of flower senescence. Plant Molecular Biology 55, 521–530. [DOI] [PubMed] [Google Scholar]
- Chen W, Singh KB. 1999. The auxin, hydrogen peroxide and salicylic acid induced expression of the Arabidopsis GST6 promoter is mediated in part by an ocs element. The Plant Journal 19, 667–677. [DOI] [PubMed] [Google Scholar]
- Chivasa S, Murphy AM, Naylor M, Carr JP. 1997. Salicylic acid interferes with tobacco mosaic virus replication via a novel salicylhydroxamic acid-sensitive mechanism. The Plant Cell 9, 547–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa AT, Bravo JP, Makiyama RK, Nunes AV, Maia IG. 2013. Viral counter defense X antiviral immunity in plants: mechanisms for survival. In: Romanowski C, ed. Current issues in molecular virology – Viral genetics and biotechnological applications. Rijeka, Croatia: Intech, 251–285. [Google Scholar]
- Deleris A, Gallego-Bartolome J, Bao J, Kasschau KD, Carrington JC, Voinnet O. 2006. Hierarchical action and inhibition of plant Dicer-like proteins in antiviral defense. Science 313, 68–71. [DOI] [PubMed] [Google Scholar]
- D’Maris Amick Dempsey AC, Vlot MCW, Daniel FK. 2011. Salicylic acid biosynthesis and metabolism. The Arabidopsis Book 9, e0156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaire L, Barajas D, Martínez-García B, Martínez-Priego L, Pagán I, Llave C. 2008. Structural and genetic requirements for the biogenesis of tobacco rattle virus-derived small interfering RNAs. Journal of Virology 82, 5167–5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis JG, Tokuhisa JG, Llewellyn DJ, Bouchez D, Singh K, Dennis ES, Peacock WJ. 1993. Does the ocs-element occur as a functional component of the promoters of plant genes?The Plant Journal 4, 433–443. [DOI] [PubMed] [Google Scholar]
- Estrada-Melo AC, Ma C, Reid MS, Jiang CZ. 2015. Overexpression of an ABA biosynthesis gene using a stress-inducible promoter enhances drought resistance in petunia. Horticulture Research 2, 15013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falcioni T, Ferrio JP, Del Cueto AI, Giné J, Achón MÁ, Medina V. 2014. Effect of salicylic acid treatment on tomato plant physiology and tolerance to Potato virus X infection. European Journal of Plant Pathology 138, 331–345. [Google Scholar]
- Fu DQ, Zhu BZ, Zhu HL, Zhang HX, Xie YH, Jiang WB, Zhao XD, Luo KB. 2006. Enhancement of virus-induced gene silencing in tomato by low temperature and low humidity. Molecules and Cells 21, 153–160. [PubMed] [Google Scholar]
- Garcia-Ruiz H, Takeda A, Chapman EJ, Sullivan CM, Fahlgren N, Brempelis KJ, Carrington JC. 2010. Arabidopsis RNA-dependent RNA polymerases and dicer-like proteins in antiviral defense and small interfering RNA biogenesis during Turnip Mosaic Virus infection. The Plant Cell 22, 481–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto K, Kobori T, Kosaka Y, Natsuaki T, Masuta C. 2007. Characterization of silencing suppressor 2b of cucumber mosaic virus based on examination of its small RNA-binding abilities. Plant & Cell Physiology 48, 1050–1060. [DOI] [PubMed] [Google Scholar]
- Gould B, Kramer EM. 2007. Virus-induced gene silencing as a tool for functional analyses in the emerging model plant Aquilegia (columbine, Ranunculaceae). Plant Methods 3, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson J, Hanssen M, Wiese A, Hendriks MM, Smeekens S. 2008. The sucrose regulated transcription factor bZIP11 affects amino acid metabolism by regulating the expression of ASPARAGINE SYNTHETASE1 and PROLINE DEHYDROGENASE2. The Plant Journal 53, 935–949. [DOI] [PubMed] [Google Scholar]
- Harvey JJ, Lewsey MG, Patel K, Westwood J, Heimstädt S, Carr JP, Baulcombe DC. 2011. An antiviral defense role of AGO2 in plants. PLoS One 6, e14639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hull R. 2009. Mechanical inoculation of plant viruses. Current Protocols in Microbiology 16B.6, doi:10.1002/9780471729259.mc16b06s13. [DOI] [PubMed] [Google Scholar]
- Jakoby M, Weisshaar B, Dröge-Laser W, Vicente-Carbajosa J, Tiedemann J, Kroj T, Parcy F; bZIP Research Group 2002. bZIP transcription factors in Arabidopsis. Trends in Plant Science 7, 106–111. [DOI] [PubMed] [Google Scholar]
- Janzik I, Preiskowski S, Kneifel H. 2005. Ozone has dramatic effects on the regulation of the prechorismate pathway in tobacco (Nicotiana tabacum L. cv. Bel W3). Planta 223, 20–27. [DOI] [PubMed] [Google Scholar]
- Jaubert M, Bhattacharjee S, Mello AF, Perry KL, Moffett P. 2011. ARGONAUTE2 mediates RNA-silencing antiviral defenses against Potato virus X in Arabidopsis. Plant Physiology 156, 1556–1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji LH, Ding SW. 2001. The suppressor of transgene RNA silencing encoded by Cucumber mosaic virus interferes with salicylic acid-mediated virus resistance. Molecular Plant-Microbe Interactions 14, 715–724. [DOI] [PubMed] [Google Scholar]
- Jiang CZ, Chen JC, Reid MS. 2011. Virus-induced gene silencing in ornamental plants. In: Kodama H, Komamine A, eds. RNAi and plant gene function analysis. New York: Humana Press, 81–96. [DOI] [PubMed] [Google Scholar]
- Kasai M, Matsumura H, Yoshida K, Terauchi R, Taneda A, Kanazawa A. 2013. Deep sequencing uncovers commonality in small RNA profiles between transgene-induced and naturally occurring RNA silencing of chalcone synthase-A gene in petunia. BMC Genomics 14, 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katagiri F, Chua NH. 1992. Plant transcription factors: present knowledge and future challenges. Trends in Genetics 8, 22–27. [DOI] [PubMed] [Google Scholar]
- Kumar D, Klessig DF. 2003. High-affinity salicylic acid-binding protein 2 is required for plant innate immunity and has salicylic acid-stimulated lipase activity. Proceedings of the National Academy of Sciences, USA 100, 16101–16106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusano T, Berberich T, Harada M, Suzuki N, Sugawara K. 1995. A maize DNA-binding factor with a bZIP motif is induced by low temperature. Molecular & General Genetics 248, 507–517. [DOI] [PubMed] [Google Scholar]
- Kusano T, Sugawara K, Harada M, Berberich T. 1998. Molecular cloning and partial characterization of a tobacco cDNA encoding a small bZIP protein. Biochimica et Biophysica Acta 1395, 171–175. [DOI] [PubMed] [Google Scholar]
- Lakatos L, Szittya G, Silhavy D, Burgyán J. 2004. Molecular mechanism of RNA silencing suppression mediated by p19 protein of tombusviruses. The EMBO Journal 23, 876–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DH, Choi HW, Hwang BK. 2011. The pepper E3 ubiquitin ligase RING1 gene, CaRING1, is required for cell death and the salicylic acid-dependent defense response. Plant Physiology 156, 2011–2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewsey MG, Carr JP. 2009. Effects of DICER-like proteins 2, 3 and 4 on cucumber mosaic virus and tobacco mosaic virus infections in salicylic acid-treated plants. The Journal of General Virology 90, 3010–3014. [DOI] [PubMed] [Google Scholar]
- Liang YC, Reid MS, Jiang CZ. 2014. Controlling plant architecture by manipulation of gibberellic acid signalling in petunia. Horticulture Research 1, 14061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J, Hanssen M, Lundgren K, et al. 2011. The sucrose-regulated Arabidopsis transcription factor bZIP11 reprograms metabolism and regulates trehalose metabolism. The New Phytologist 191, 733–745. [DOI] [PubMed] [Google Scholar]
- Ma X, Nicole MC, Meteignier LV, Hong N, Wang G, Moffett P. 2015. Different roles for RNA silencing and RNA processing components in virus recovery and virus-induced gene silencing in plants. Journal of Experimental Botany 66, 919–932. [DOI] [PubMed] [Google Scholar]
- Malamy J, Carr JP, Klessig DF, Raskin I. 1990. Salicylic acid: a likely endogenous signal in the resistance response of tobacco to viral infection. Science 250, 1002–1004. [DOI] [PubMed] [Google Scholar]
- Martínez-Priego L, Donaire L, Barajas D, Llave C. 2008. Silencing suppressor activity of the Tobacco rattle virus-encoded 16-kDa protein and interference with endogenous small RNA-guided regulatory pathways. Virology 376, 346–356. [DOI] [PubMed] [Google Scholar]
- Molnar A, Melnyk CW, Bassett A, Hardcastle TJ, Dunn R, Baulcombe DC. 2010. Small silencing RNAs in plants are mobile and direct epigenetic modification in recipient cells. Science 328, 872–875. [DOI] [PubMed] [Google Scholar]
- Murphy AM, Carr JP. 2002. Salicylic acid has cell-specific effects on tobacco mosaic virus replication and cell-to-cell movement. Plant Physiology 128, 552–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nijhawan A, Jain M, Tyagi AK, Khurana JP. 2008. Genomic survey and gene expression analysis of the basic leucine zipper transcription factor family in rice. Plant Physiology 146, 333–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohme-Takagi M, Shinshi H. 1995. Ethylene-inducible DNA binding proteins that interact with an ethylene-responsive element. The Plant Cell 7, 173–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu F, Ye X, Morris TJ. 2008. Arabidopsis DRB4, AGO1, AGO7, and RDR6 participate in a DCL4-initiated antiviral RNA silencing pathway negatively regulated by DCL1. Proceedings of the National Academy of Sciences, USA 105, 14732–14737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quilis J, Peñas G, Messeguer J, Brugidou C, San Segundo B. 2008. The Arabidopsis AtNPR1 inversely modulates defense responses against fungal, bacterial, or viral pathogens while conferring hypersensitivity to abiotic stresses in transgenic rice. Molecular Plant-Microbe Interactions 21, 1215–1231. [DOI] [PubMed] [Google Scholar]
- Reid MS, Chen JC, Jiang CZ. 2009. Virus-induced gene silencing for functional characterization of genes in petunia. In: Gerats T, Strommer J, eds. Petunia. Berlin Heidelberg: Springer, 381–394. [Google Scholar]
- Riechmann JL, Heard J, Martin G, et al. 2000. Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science 290, 2105–2110. [DOI] [PubMed] [Google Scholar]
- Rivas-San Vicente M, Plasencia J. 2011. Salicylic acid beyond defence: its role in plant growth and development. Journal of Experimental Botany 62, 3321–3338. [DOI] [PubMed] [Google Scholar]
- Scholthof HB, Alvarado VY, Vega-Arreguin JC, Ciomperlik J, Odokonyero D, Brosseau C, Jaubert M, Zamora A, Moffett P. 2011. Identification of an ARGONAUTE for antiviral RNA silencing in Nicotiana benthamiana. Plant Physiology 156, 1548–1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwach F, Vaistij FE, Jones L, Baulcombe DC. 2005. An RNA-dependent RNA polymerase prevents meristem invasion by Potato virus X and is required for the activity but not the production of a systemic silencing signal. Plant Physiology 138, 1842–1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang J, Xi DH, Xu F, et al. 2011. A broad-spectrum, efficient and nontransgenic approach to control plant viruses by application of salicylic acid and jasmonic acid. Planta 233, 299–308. [DOI] [PubMed] [Google Scholar]
- Shimizu H, Sato K, Berberich T, Miyazaki A, Ozaki R, Imai R, Kusano T. 2005. LIP19, a basic region leucine zipper protein, is a Fos-like molecular switch in the cold signaling of rice plants. Plant & Cell Physiology 46, 1623–1634. [DOI] [PubMed] [Google Scholar]
- Spitzer-Rimon B, Farhi M, Albo B, et al. 2012. The R2R3-MYB-like regulatory factor EOBI, acting downstream of EOBII, regulates scent production by activating ODO1 and structural scent-related genes in petunia. The Plant Cell 24, 5089–5105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun D, Nandety RS, Zhang Y, Reid MS, Niu L, Jiang CZ. 2016. A petunia ethylene-responsive element binding factor, PhERF2, plays an important role in antiviral RNA silencing. Journal of Experimental Botany 67, 3353–3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thalor SK, Berberich T, Lee SS, Yang SH, Zhu X, Imai R, Takahashi Y, Kusano T. 2012. Deregulation of sucrose-controlled translation of a bZIP-type transcription factor results in sucrose accumulation in leaves. PLoS One 7, e33111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian J, Pei H, Zhang S, Chen J, Chen W, Yang R, Meng Y, You J, Gao J, Ma N. 2014. TRV-GFP: a modified Tobacco rattle virus vector for efficient and visualizable analysis of gene function. Journal of Experimental Botany 65, 311–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzin V, Galili G. 2010. The Biosynthetic pathways for shikimate and aromatic amino acids in Arabidopsis thaliana. The Arabidopsis Book 8, e0132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Huijsduijnen RH, Alblas S, De Rijk R, Bol J. 1986. Induction by salicylic acid of pathogenesis-related proteins and resistance to Alfalfa mosaic virus infection in various plant species. Journal of General Virology 67, 2135–2143. [Google Scholar]
- Verdonk JC, Haring MA, van Tunen AJ, Schuurink RC. 2005. ODORANT1 regulates fragrance biosynthesis in petunia flowers. The Plant Cell 17, 1612–1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voinnet O. 2009. Origin, biogenesis, and activity of plant microRNAs. Cell 136, 669–687. [DOI] [PubMed] [Google Scholar]
- Wang H, Stier G, Lin J, Liu G, Zhang Z, Chang Y, Reid MS, Jiang CZ. 2013. Transcriptome changes associated with delayed flower senescence on transgenic petunia by inducing expression of etr1-1, a mutant ethylene receptor. PLoS One 8, e65800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JF, Zhou H, Chen YQ, Luo QJ, Qu LH. 2004. Identification of 20 microRNAs from Oryza sativa. Nucleic Acids Research 32, 1688–1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XB, Jovel J, Udomporn P, Wang Y, Wu Q, Li WX, Gasciolli V, Vaucheret H, Ding SW. 2011. The 21-nucleotide, but not 22-nucleotide, viral secondary small interfering RNAs direct potent antiviral defense by two cooperative argonautes in Arabidopsis thaliana. The Plant Cell 23, 1625–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XB, Wu Q, Ito T, Cillo F, Li WX, Chen X, Yu JL, Ding SW. 2010. RNAi-mediated viral immunity requires amplification of virus-derived siRNAs in Arabidopsis thaliana. Proceedings of the National Academy of Sciences, USA 107, 484–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiste C, Dröge-Laser W. 2014. The Arabidopsis transcription factor bZIP11 activates auxin-mediated transcription by recruiting the histone acetylation machinery. Nature Communications 5, 3883. [DOI] [PubMed] [Google Scholar]
- Wiese A, Elzinga N, Wobbes B, Smeekens S. 2004. A conserved upstream open reading frame mediates sucrose-induced repression of translation. The Plant Cell 16, 1717–1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Yang Z, Wang Y, Zheng L, Ye R, Ji Y, Zhao S, Ji S, Liu R, Xu L. 2015. Viral-inducible Argonaute18 confers broad-spectrum virus resistance in rice by sequestering a host microRNA. eLife 4, e05733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Fan B, Chen C, Chen Z. 2001. An important role of an inducible RNA-dependent RNA polymerase in plant antiviral defense. Proceedings of the National Academy of Sciences, USA 98, 6516–6521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang SJ, Carter SA, Cole AB, Cheng NH, Nelson RS. 2004. A natural variant of a host RNA-dependent RNA polymerase is associated with increased susceptibility to viruses by Nicotiana benthamiana. Proceedings of the National Academy of Sciences, USA 101, 6297–6302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang SH, Berberich T, Sano H, Kusano T. 2001. Specific association of transcripts of tbzF and tbz17, tobacco genes encoding basic region leucine zipper-type transcriptional activators, with guard cells of senescing leaves and/or flowers. Plant Physiology 127, 23–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin J, Chang X, Kasuga T, Bui M, Reid MS, Jiang CZ. 2015. A basic helix-loop-helix transcription factor, PhFBH4, regulates flower senescence by modulating ethylene biosynthesis pathway in petunia. Horticulture Research 2, 15059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Zhang X, Singh J, Li D, Qu F. 2012. Temperature-dependent survival of Turnip crinkle virus-infected arabidopsis plants relies on an RNA silencing-based defense that requires dcl2, AGO2, and HEN1. Journal of Virology 86, 6847–6854. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.