Abstract
A growing body of studies has demonstrated that long non‐coding RNA (lncRNA) are regarded as the primary section of the ceRNA network. This is thought to be the case owing to its regulation of protein‐coding gene expression by functioning as miRNA sponges. However, functional roles and regulatory mechanisms of lncRNA‐mediated ceRNA in cervical squamous cell carcinoma (CESC), as well as their use for potential prediction of CESC prognosis, remains unknown. The aberrant expression profiles of mRNA, lncRNA, and miRNA of 306 cervical squamous cancer tissues and three adjacent cervical tissues were obtained from the TCGA database. A lncRNA‐mRNA‐miRNA ceRNA network in CESC was constructed. Meanwhile, Gene Ontology (GO) and KEGG pathway analysis were performed using Cytoscape plug‐in BinGo and DAVID database. We identified a total of 493 lncRNA, 70 miRNA, and 1921 mRNA as differentially expressed profiles. An aberrant lncRNA‐mRNA‐miRNA ceRNA network was constructed in CESC, it was composed of 50 DElncRNA, 18 DEmiRNA, and 81 DEmRNA. According to the overall survival analysis, 3 out of 50 lncRNA, 10 out of 81 mRNA, and 1 out of 18 miRNA functioned as prognostic biomarkers for patients with CESC (P value < 0.05). We extracted the sub‐network in the ceRNA network and found that two novel lncRNA were recognized as key genes. These included lncRNA MEG3 and lncRNA ADAMTS9‐AS2. The present study provides a new insight into a better understanding of the lncRNA‐related ceRNA network in CESC, and the novel recognized ceRNA network will help us to improve our understanding of lncRNA‐mediated ceRNA regulatory mechanisms in the pathogenesis of CESC.
Keywords: cervical squamous cell carcinoma, competitive endogenous RNA, long noncoding RNAs
1. INTRODUCTION
As the second most common type of cancer in females worldwide, cervical cancer carries a high risk of leading cause of mortality in females in developing countries.1 Cervical cancer has an estimated incidence of 530 000 new cases and 270 000 deaths, annually.2 Cervical squamous cell carcinoma (CESC) was responsible for 10‐15% of all females’ cancer‐related deaths worldwide that contributed to the second highest number of deaths in female cancers, exceeded only by breast cancer.3, 4 At the time of diagnosis, 80% patients have already progressed into invasive cancer stages of cervical cancer. The age at diagnosis has slowly been decreasing. Moreover, cervical cancer still carries high risks of morbidity and mortality from metastasis and recurrence.5, 6 Cervical cancer is an intricate disease, which involved in numerous reasons. There has been a great necessity and urgency in searching for novel treatment targets as well as prognosis biomarkers to improve the survival rate of cervical cancer patients.
To improve the prognosis and decrease mortality and morbidity of CESC, diagnostic biomarkers are critical for early detection and risk assessment of CESC, which can help us to select appropriate treatment. However, it has been widely accepted that CESC is a heterogeneous cancer in various aspects owing to its clinicopathological, molecular, and cellular heterogeneity. Therefore, identification of potential molecular biomarkers or therapeutic targets to conquer CESC is urgently required. Recently, accumulating evidence has suggested that, rather than being transcriptional “dark matter,” many non‐coding RNA (lncRNA) serve as master regulators that affect the expression levels of dozens or even hundreds of target genes.7, 8 Since the discovery of ceRNA regulation in human cells, multiple studies reported that lncRNA, mRNA, and other RNA act as natural miRNA sponges to suppress miRNA function. LncRNA functions as ceRNA to communicate with mRNA by competing for shared miRNA.9, 10, 11 In the study, we first acquired the aberrant expression profiles of lncRNA, mRNA, and miRNA between 360 CESC patients and 4 non‐tumor samples. After which, the aberrant lncRNA‐mRNA‐miRNA ceRNA network was constructed in CESC. A total of 3 out of 50 lncRNA, 10 out of 81 mRNA, and 1 out of 18 miRNA functioned as prognostic biomarkers for patients with CESC according to the overall survival analysis. This is the first study to investigate the cancer specific lncRNA from the TCGA and lncRNA‐mediated ceRNA network that was constructed in CESC. The present study results can be used to help clinicians to elaborate the function of lncRNA through lncRNA‐miRNA‐mRNA ceRNA network in CESC and provide novel lncRNAs as potential diagnostic biomarkers.
2. MATERIALS AND METHODS
2.1. Patient datasets
The mRNA and miRNA expression and corresponding clinical information of PDAC patients were obtained from the TCGA data portal (https://tcga-data.nci.nih.gov/tcga/), which was imputed on IlluminaHiSeq RNA‐Seq platform, containing 306 CESC sample tissues and four adjacent non‐tumor pancreatic tissues. Both mRNA/miRNA profile data and clinical characteristics of PDAC are publicly available and available on open‐access. Therefore, approval by a local ethics committee was not needed.
2.2. Acquisition and analysis of expression profiles
The differential expression lncRNA (DElncRNA), miRNA (DEmiRNA), and mRNA (DEmRNA) between CESC and adjacent tissue were calculated using R/Bioconductor package of edgeR.12 The differentially expressed genes (DEGs) of data sets with |log2 fold change| ≥2.0 and a P‐value less than 0.05 was considered as the selection criteria for subsequent analysis.
2.3. Identification of differentially expressed lncRNA
We obtained the lncRNA expression data by repurposing the probes in the mRNA expression profiles to lncRNA based on the annotations from the GENCODE project (http://www.gencodegenes.org).13 The transformed data (antisense, lncRNA, and sense_intronic) was considered as lncRNA.
2.4. Prediction of lncRNA‐miRNA and miRNA‐mRNA interactions
First, it should be stated that, the data of lncRNA‐miRNA interactions was downloaded from a highly reliable online miRNA reference database of miRcode (http://www.mircode.org/).14 The integrated lncRNA‐miRNA pairs were predicted using miRcode, combining with the selected miRNA. Second, the prediction of targeted mRNA of miRNA was retrieved from three databases: miRTarBase,15 miRanda,16 and Targetscan.17 Finally, we established matched DElncRNA‐DEmiRNA and DEmiRNA‐DEmRNA pairs.
2.5. Reconstruction of the lncRNA‐miRNA‐mRNA ceRNA network
The DElncRNA‐DEmiRNA‐DEmRNA pairs were reconstructed by assembling all co‐expression competing triplets. The ceRNA network was visualized using Cytoscape 3.3.2.18 Simultaneously, the sub‐network of the DElncRNA‐DEmiRNA‐DEmRNA ceRNA network was estimated using Cytoscape plug‐in MCODE.
2.6. Functional enrichment analysis
To understand the underlying biological processes and pathways of aberrantly expressed mRNA in the lncRNA‐miRNA‐mRNA network, Gene Ontology (GO) Biological Process term and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis was conducted using Cytoscape plug‐in ClueGo19 and DAVID (database for annotation, visualization, and integrated discovery; https://david.ncifcrf.gov/). GO enrichment analysis was based on the threshold of P‐value < 0.05 and enrichment score >1.0. Significant enrichment results were visualized using Cytoscape software 3.3.0.
2.7. Statistical analysis
For overall survival analysis, the log‐rank test was employed to analyze the difference between CESC and normal samples among DElncRNA, DEmiRNA, and DEmRNA. A P‐value less than 0.05 was considered as statistically significance.
3. RESULTS
3.1. Differentially expressed lncRNA, miRNA, and mRNA
The expression profiles of mRNA, miRNA, and lncRNA between 304 CESC samples and four nonmalignant samples was calculated. We discovered that a total of 1921 mRNA, 493 lncRNA, and 70 miRNA were differentially expressed (|log2 fold change| ≥2.0 and FDR adjusted P less than 0.05). Of these, 712 mRNA, 128 lncRNA, and 33 miRNA were over‐expressed. A total of 1209 mRNA, 365 lncRNA, and 37 miRNA were under‐expressed. The heat map of clustering analysis of the analyzed RNA are shown in Figure 1–3.
Figure 1.
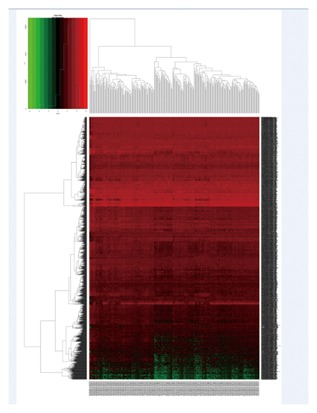
The heatmap of DEmRNA expression in CESU. A total of 1921 DEmRNA were detected. Among these DEmRNA, 712 DEmRNA were up‐regulated genes, and 1209 DEmRNA were down‐regulated genes. The color from blue to red shows the progression from low expression to high expression
Figure 3.
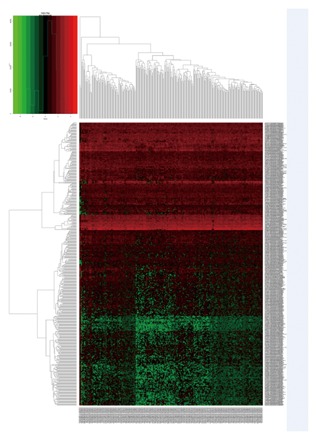
The heatmap of DElncRNA expression in CESU. A total of 493 DElncRNA were detected. Among these DElncRNA, 128 DEmRNA were up‐regulated genes and 365 DElncRNAs were down‐regulated genes. The color from blue to red shows the progression from low expression to high expression
Figure 2.
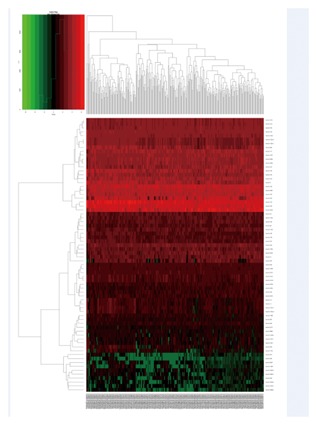
The heatmap of DEmiRNA expression in CESU. A total of 70 DEmiRNA were detected. Among these DEmiRNA, 33 DEmRNA were up‐regulated genes and 37 DEmiRNA were down‐regulated genes. The color from blue to red shows the progression from low expression to high expression
3.2. Construction and analysis of lncRNA‐miRNA‐mRNA ceRNA network
The lncRNA‐miRNA‐mRNA ceRNA network was established based on the relationship between DElncRNA, DEmiRNA, DEmRNA. In the ceRNA network, we identified a total of 50 lncRNA nodes, 18 miRNA nodes, 81 mRNA nodes, and 2484 edges as differentially expressed profiles. The network is presented in Figure 4. We noticed that DElncRNA ADAMTS9‐AS2 interacted with as many as 13 DEmiRNA, including: hsa‐mir‐145, hsa‐mir‐182, hsa‐mir‐31, hsa‐mir‐96, hsa‐mir‐106a, hsa‐mir‐140, hsa‐mir‐141, hsa‐mir‐143, hsa‐mir‐183, hsa‐mir‐200a, hsa‐mir‐204, hsa‐mir‐205, and hsa‐mir‐32. Therefore, lncRNA ADAMTS9‐AS2 may greatly contribute to the pathogenesis of CESC.
Figure 4.
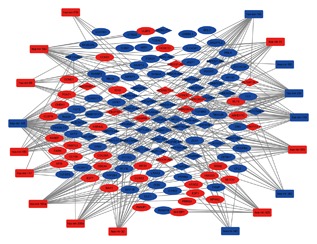
The dysregulated lncRNA‐mRNA‐miRNA ceRNA network. Diamond denotes lncRNA, square represents mRNA, and round rectangle represents miRNA. All shapes in red and green stand for up‐regulation and down‐regulation, respectively
3.3. Prognostic overall survival assessment of lncRNA, miRNA, and mRNA
A total of 3 out of 50 DElncRNAs including AC097717.1, C20orf203, and EMX2OS were significantly associated with overall survival (P < 0.05) and exhibited positive effects. A total of 10 out of 81 mRNA significantly correlated with CESC overall survival (P < 0.05). Four DElncRNA of high expression (BCL2, E2F1, HMGB3, and RASSF2), and 6 DElncRNA of low level expression (ERG, FASN, PTPRM, TBXA2R, ULBP2, and ZCCHC24) were significantly related to the progression of CESC overall survival. Only 1 DElncmiRNA (hsa‐mir‐14) was significantly associated with CESC overall survival (P < 0.05) (See Figure 5).
Figure 5.
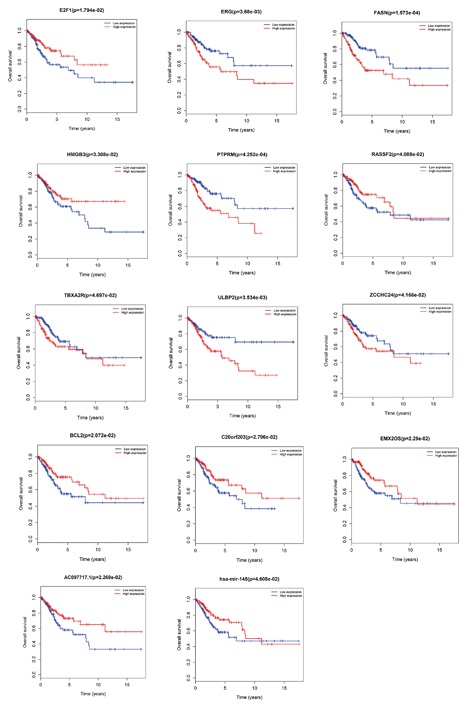
Kaplan‐Meier survival curves of 10 DElncRNA, 3 DElncRNA, and DEmiRNA for the overall survival in CESC
3.4. Functional prediction of lncRNA‐miRNA‐mRNA ceRNA network
In order to predict the function of aberrantly expressed genes in the ceRNA network, the intersection mRNA was analyzed by Cytoscape plug‐in ClueGo and DAVID. Therefore, 81 DEmRNA were conducted to analyze their functions. The results revealed that enrichment of 140 GO categories occurred in the biological process (see Figure 6 and Table 1). The top 5 GO terms were a response to mechanical stimulus, regulation of transcription involved in the G1/S transition of mitotic cell cycle, intrinsic apoptotic signaling pathways in response to endoplasmic reticulum stress, fibroblast proliferation, and muscle cell proliferation. KEGG analysis, focusing on the biological pathways, showed that 12 pathways were significantly enriched, particularly apoptosis, miRNAs in cancer, cell cycle, p53 signaling pathway, and prostate cancer pathways (Table 2). The mentioned pathways are exhibited in Figure 7.
Figure 6.
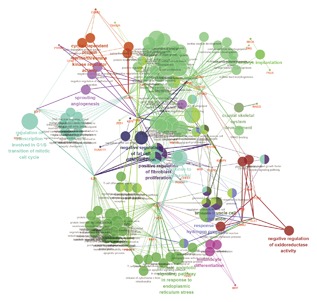
GO terms show as an interaction network using Cytoscape plug‐in ClueGO
Table 1.
The top five gene ontology analysis of differentially expressed mRNA (DEmRNA) associated with CESC
| GO id | GO terms | Ontology source | Gene count | Genes | P value |
|---|---|---|---|---|---|
| GO:0009612 | Response to mechanical stimulus | GO_BiologicalProcess‐GOA_23.02.2017_10h01 | 8 | BAK1, BCL2, DLC1, FOXO1, LRRK2, MYB, PDGFD, TXNIP | 210.0E‐9 |
| GO:0033002 | Muscle cell Proliferation | GO_BiologicalProcess‐GOA_23.02.2017_10h01 | 9 | CCNB1, FGF2, IGFBP5, MYB, PDGFD, TGFBR2, TGFBR3, THBS1, ZFPM2 | 71.0E‐9 |
| GO:0048144 | Fibroblast proliferation | GO_BiologicalProcess‐GOA_23.02.2017_10h01 | 4 | [BAK1, BCL2, E2F1, PMAIP1] | 28.0E‐6 |
| GO:0000083 | Regulation of transcription involved in G1/S transition of Mitotic cell cycle | GO_BiologicalProcess‐GOA_23.02.2017_10h01 | 5 | CCNB1, E2F1, MYB, PDGFD, PDGFRA | 4.3E‐6 |
| GO:0070059 | Intrinsic apoptotic signaling pathway in response to endoplasmic reticulum stress | GO_BiologicalProcess‐GOA_23.02.2017_10h01 | 5 | CCNB1, E2F1, MYB, PDGFD, PDGFRA | 4.3E‐6 |
Table 2.
The Top Five KEGG Analysis of Differentially Expressed mRNA (DEmRNA) Associated with CESC
| GO id | KEGG terms | Ontology source | Gene count | Genes | P value |
|---|---|---|---|---|---|
| GO:0004215 | Apoptosis | KEGG_01.03.2017 | 3 | BAK1, BCL2, PMAIP1 | 1.3E‐3 |
| GO:0005206 | MicroRNAs in cancer | KEGG_01.03.2017 | 13 | BAK1, BCL2, CCNE1, CCNE2, CDC25A, E2F1, KIF23, PDGFRA, RECK, THBS1, ZEB1, ZEB2, ZFPM2 | 44.0E‐9 |
| GO:0004110 | Cell cycle | KEGG_01.03.2017 | 6 | CCNB1, CCNE1, CCNE2, CDC25A, CHEK1, E2F1 | 150.0E‐6 |
| GO:0004115 | p53 signaling pathway | KEGG_01.03.2017 | 7 | CCNB1, CCNE1, CCNE2, CHEK1, PMAIP1, RRM2, THBS1 | 290.0E‐9 |
| GO:0005215 | Prostate cancer | KEGG_01.03.2017 | 8 | AKT3, BCL2, CCNE1, CCNE2, E2F1, FOXO1, PDGFD, PDGFRA | 83.0E‐9 |
Figure 7.
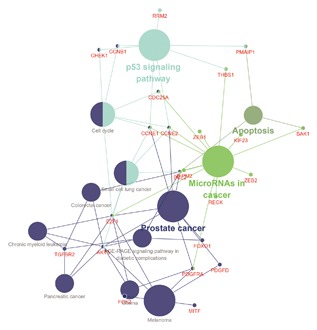
KEGG pathways displayed as an interaction network Cytoscape plug‐in ClueGO
3.5. Key lncRNA‐miRNA‐mRNA in sub‐network
We analyzed the hub gene in the ceRNA network using Cytoscape plug‐in MCODE. A total of 10 nodes could be selected hub nodes, including lncRNA MEG3, lncRNA ADAMTS9‐AS2, hsa‐mir‐141, hsa‐mir‐96, RECK, ZFPM2, OSBPL3, RPLR, AP1S2, and TPM2. The sub‐network is shown in Figure 8.
Figure 8.
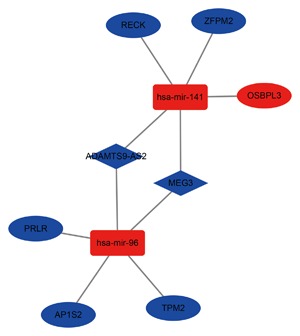
The sub‐network of DElncRNA‐mediated ceRNA network. Diamond denotes lncRNA, ellipse represents mRNA, and round rectangle represents miRNA. All shapes in red and green stand for up‐regulation and down‐regulation, respectively
4. DISCUSSION
Cervical cancer in females contributes to the second highest number of deaths in the world, exceeded only by breast cancer. Cervical cancer carries a high risks of morbidity and mortality. It goes without saying that recurrence and metastasis are the most significant challenge to overcome in the treatment of cervical cancer.5, 6 Therefore, looking for available biomarker markers of cervical cancer is essential for improving its prognosis in patients. At present, an increasing amount of evidence indicates that lncRNA plays an important molecular role in apoptosis, proliferation, progression, metastasis, invasion, and relapse and relapse of tumor.20, 21, 22, 23, 24 LncRNAs acts as the potential biomarkers for diagnosis and prognosis in various cancers.25, 26, 27 Numerous studies have indicated that the dysregulated lncRNA expression profiles are associated with the development of various cancers, includes CESC, as well as respective patients survival rate. This hence uncovers the potential for lncRNA to be utilized as a potential prognostic cancer biomarker.28, 29, 30, 31, 32, 33 Recent studies have reported that lncRNA can regulate miRNA abundance by binding and sequestering them, making lncRNA the titular name of miRNA sponges.34, 35 Meanwhile, lncRNA also regulates the expression of target mRNA. Based on the theory Salmena proposed; a competing endogenous RNA (ceNRA) hypothesis that lncRNA functions as ceRNA to react with mRNA by competing for shared miRNA.36 Experimental evidence indicates that lncRNA, that harbors similar sequences to its targeted miRNA, can sequester miRNA away from mRNA. LncRNA CASC2 up‐regulates PTEN as a ceRNA of miR‐21 and plays an important role in cervical cancer sensitivity to DDP, and may serve as a potential target for cancer diagnosis and treatment.37 Gao et al38 demonstrated that the tumor‐promoting role of lncRNA PVT1, is that it acts as competing endogenous RNA (ceRNA), or a molecular sponge in negatively modulating miR‐424. This tumor‐promoting role of lncRNA PVT1 might provide a novel therapeutic target for cervical cancer. However, a comprehensive analysis of CESC‐related lncRNA and miRNA in a whole genome‐wide, especially based on high through detection with large‐scale sample size, has always been lacking. Therefore, it is crucial to explore the functional roles and regulatory mechanisms of the lncRNA‐miRNA‐mRNA ceRNA network in the development of CESC. In this presented study, a total of 494 lncRNAs, 70 miRNA, and 1921 mRNA differentially expressed profiles were identified from the TCGA database. An aberrant lncRNA‐mRNA‐miRNA ceRNA network was constructed in CESC. The lncRNA‐miRNA‐mRNA ceRNA network was composed of 50 DElncRNAs, 18 DEmiRNA, and 81 DEmRNA. We extracted the sub‐network in the ceRNA network and found that four nodes were recognized as key genes, including: lncRNA MEG3, lncRNA ADAMTS9‐AS2, hsa‐mir‐141, and hsa‐mir‐96. As for lncRNA MEG3, several studies have suggested its involvement in various cancers via acting on cell apoptosis, including cervical cancer. The lncRNA ADAMTS9‐AS2, hsa‐mir‐141, and hsa‐mir‐96 can also be regarded as potential biomarkers for CESC. Hsa‐mir‐141 and hsa‐mir‐96, lncRNA ADAMTS9‐AS2 originally served as prognostic predictors, providing reasonable implications in future clinical practice regarding targeted treatment of CESC. Three out of 50 lncRNAs, 10 out of 81 mRNAs, and 1 out of 18 miRNAs functioned as prognostic biomarkers for patients with CESC according to the overall survival analysis (P value < 0.05). GO analysis and KEGG pathway analysis have been used to evaluate the biological functions enriched among differentially expressed coding genes. The results of DEmRNA related GO analysis revealed that enrichment of 140 GO categories in the biological process was significant with P‐value < 0.05. These significant GO terms involved a response to mechanical stimulus, regulation of transcription involved in the G1/S transition of mitotic cell cycle, intrinsic apoptotic signaling pathway in response to endoplasmic reticulum stress, fibroblast proliferation, and muscle cell proliferation. The pathway analysis further demonstrated that 12 pathways were enriched, and primarily involved: apoptosis, miRNAs in cancer, cell cycle, p53 signaling pathway, and prostate cancer pathways.
Among these key lncRNA, several studies have reported that lncRNA MEG3 played crucial roles in the development of various cancers, such as: non‐small cell lung cancer, cervical cancer, colorectal cancer, esophageal cancer.39, 40, 41, 42, 43 The lncRNA MEG3 had effects to suppress cervical cancer by regulation of PI3K/AKT/Bcl‐2/Bax/P21 and PI3K/AKT/MMP‐2/9 signaling pathways. However, only two studies demonstrated that lncRNA ADAMTS9‐AS2 was associated with development of gliomas,44 and colorectal cancer.45 However, the contribution of lncRNA ADAMTS9‐AS2 to the development of CESC is still not certain from the current available study. Further studies should be performed to address these issues. According to the lncRNA‐miRNA‐mRNA sub‐network, we speculated that downregulated‐lncRNA ADAMTS9‐AS2 might have a role in altering the upregulated‐hsa‐mir‐141 and upregulated‐hsa‐mir‐96. Recent studies demonstrated that miR‐141 down‐regulated TM4SF1 expression to inhibit invasion and migration of prostate cancer cells.46 Wang Y suggested that hsa‐miR‐96 may affect the growth of bladder cancer cells by up‐regulating IRS1 and MAP4K1 levels, functioning as a promising diagnostic marker in human bladder urothelial carcinomas.47 Based on these findings, lncRNA ADAMTS9‐AS2 may be involve the invasion and migration of CESC. These results also indicated that lncRNA ADAMTS9‐AS2 and MEG3 is a critical lncRNA in the development of CESC.
In summary, we first reconstructed the lncRNA‐miRNA‐mRNA ceRNA network, and analyzed the lncRNA related ceRNA in the development of CESC. Our results demonstrated that lncRNA plays an important role in the development of CESC. Two novel lncRNA ADAMTS9‐AS2 and MEG3 may be chosen as key lncRNA. Further studies are needed to explore the biological mechanisms of these two lncRNAs in CESC.
AUTHORS’ CONTRIBUTION
Authors JKS, AZY, JGZ, and JGL wrote the main manuscript text. XHY, ZC, and ELJ prepared Figures 1–8. XHY and ELJ contributed on data analysis and all authors reviewed the manuscript.
CONFLICTS OF INTEREST
The authors declare no competing financial interests.
Song J, Ye A, Jiang E, et al. Reconstruction and analysis of the aberrant lncRNA‐miRNA‐mRNA network based on competitive endogenous RNA in CESC. J Cell Biochem. 2018;119:6665–6673. 10.1002/jcb.26850
Jukun Song, Aizhu Ye, Zhu Chen, and Enli Jiang contributed equally to this article.
Contributor Information
Yu Zhou, Email: 2694492281@qq.com.
Jianguo Liu, Email: liujg_001@163.com.
REFERENCES
- 1. Joshi PK, Esko T, Mattsson H, et al. Directional dominance on stature and cognition in diverse human populations. Nature. 2015;523:459–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lahue BJ, Baginska E, Li SS, Parisi M. Health technology assessment on cervical cancer screening, 2000–2014. Int J Technol Assess Health Care. 2015;31:171–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Liu S, Zhang P, Chen Z, Liu M, Li X, Tang H. MicroRNA‐7 downregulates XIAP expression to suppress cell growth and promote apoptosis in cervical cancer cells. FEBS Lett 2013;587:2247–2253. [DOI] [PubMed] [Google Scholar]
- 4. Ojesina AI, Lichtenstein L, Freeman SS, et al. Landscape of genomic alterations in cervical carcinomas. Nature. 2014;506:371–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kulkarni PR, Rani H, Vimalambike MG, Ravishankar S. Opportunistic screening for cervical cancer in a tertiary hospital in Karnataka. India. Asian Pac J Cancer Prev. 2013;14:5101–5105. [DOI] [PubMed] [Google Scholar]
- 6. Wang IT, Chou SC, Lin YC. Zoledronic acid induces apoptosis and autophagy in cervical cancer cells. Tumour Biol. 2014;35:11913–11920. [DOI] [PubMed] [Google Scholar]
- 7. Caley DP, Pink RC, Trujillano D, Carter DR. Long noncoding RNAs, chromatin, and development. Sci World J. 2010;10:90–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yang L, Duff MO, Graveley BR, Carmichael GG, Chen LL. Genomewide characterization of non‐polyadenylated RNAs. Genome Biol. 2011;12:R16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146:353–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ergun S, Oztuzcu S. Oncocers: ceRNA‐mediated cross‐talk by sponging miRNAs in oncogenic pathways. Tumour Biol. 2015;36:3129–3136. [DOI] [PubMed] [Google Scholar]
- 11. Qi X, Zhang DH, Wu N, Xiao JH, Wang X, Ma W. CeRNA in cancer: possible functions and clinical implications. J Med Genet. 2015;52:710–718. [DOI] [PubMed] [Google Scholar]
- 12. Robinson MD, McCarthy DJ, Smyth GK. EdgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Derrien T, Johnson R, Bussotti G, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775–1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jeggari A, Marks DS, Larsson E. MiRcode: a map of putative microRNA target sites in the long non‐coding transcriptome. Bioinformatics 2012;28:2062–2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hsu SD, Tseng YT, Shrestha S, et al. MiRTarBase update 2014: an information resource for experimentally validated miRNA‐target interactions. Nucleic Acids Res. 2014;42:D78–D85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. Human MicroRNA targets. PLoS Biol 2004;2:e363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Park K, Kim KB. MiRTar Hunter: a prediction system for identifying human microRNA target sites. Mol Cells. 2013;35:195–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Maere S, Heymans K, Kuiper M. BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics. 2005;21:3448–3449. [DOI] [PubMed] [Google Scholar]
- 20. Jin L, Fu H, Quan J, et al. Overexpression of long non‐coding RNA differentiation antagonizing non‐protein coding RNA inhibits the proliferation, migration and invasion and promotes apoptosis of renal cell carcinoma. Mol Med Rep. 2017;16:4463–4468. [DOI] [PubMed] [Google Scholar]
- 21. Li D, Li H, Yang Y, Kang L. Long noncoding RNA urothelial carcinoma associated 1 promotes the proliferation and metastasis of human lung tumor cells by regulating microRNA‐144. Oncol Res. 2017. 10.3727/096504017X15009792179602 [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 22. Li S, Yang J, Xia Y, Fan Q, Yang KP. LncRNA NEAT1 promotes proliferation and invasion via targeting miR‐181a‐5p in non‐small cell lung cancer. Oncol Res. 2017;26:289–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhou M, Xu W, Yue X, et al. Relapse‐related long non‐coding RNA signature to improve prognosis prediction of lung adenocarcinoma. Oncotarget. 2016;7:29720–29738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lu S, Zhou J, Sun Y, et al. The noncoding RNA HOXD‐AS1 is a critical regulator of the metastasis and apoptosis phenotype in human hepatocellular carcinoma. Mol Cancer. 2017;16:125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hu Y, Chen HY, Yu CY, et al. A long non‐coding RNA signature to improve prognosis prediction of colorectal cancer. Oncotarget. 2014;5:2230–2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tong YS, Wang XW, Zhou XL, et al. Identification of the long non‐coding RNA POU3F3 in plasma as a novel biomarker for diagnosis of esophageal squamous cell carcinoma. Mol Cancer. 2015;14:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Xu MD, Qi P, Weng WW, et al. Long non‐coding RNA LSINCT5 predicts negative prognosis and exhibits oncogenic activity in gastric cancer. Medicine (Baltimore). 2014;93:e303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Fu XL, Liu DJ, Yan TT, et al. Analysis of long non‐coding RNA expression profiles in pancreatic ductal adenocarcinoma. Sci Rep. 2016;6:33535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Han D, Gao X, Wang M, et al. Long noncoding RNA H19 indicates a poor prognosis of colorectal cancer and promotes tumor growth by recruiting and binding to eIF4A3. Oncotarget. 2016;7:22159–22173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Huang C, Yu W, Wang Q, et al. Increased expression of the lncRNA PVT1 is associated with poor prognosis in pancreatic cancer patients. Minerva Med. 2015;106:143–149. [PubMed] [Google Scholar]
- 31. Li C, Zhou L, He J, Fang XQ, Zhu SW, Xiong MM. Increased long noncoding RNA SNHG20 predicts poor prognosis in colorectal cancer. BMC Cancer. 2016;16:655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Liu T, Zhang X, Yang YM, Du LT, Wang CX. Increased expression of the long noncoding RNA CRNDE‐h indicates a poor prognosis in colorectal cancer, and is positively correlated with IRX5 mRNA expression. Onco Targets Ther. 2016;9:1437–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yong S, Yabin Y, Bing Z, et al. Reciprocal regulation of DGCR5 and miR‐320a affects the cellular malignant phenotype and 5‐FU response in pancreatic ductal adenocarcinoma. Oncotarget. 2017;8:90868–90878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tan JY, Sirey T, Honti F, et al. Extensive microRNA‐mediated crosstalk between lncRNAs and mRNAs in mouse embryonic stem cells. Genome Res. 2015;25:655–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505:344–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Militello G, Weirick T, John D, Doring C, Dimmeler S, Uchida S. Screening and validation of lncRNAs and circRNAs as miRNA sponges. Brief Bioinform. 2016;18:780–788. [DOI] [PubMed] [Google Scholar]
- 37. Feng Y, Zou W, Hu C, et al. Modulation of CASC2/miR‐21/PTEN pathway sensitizes cervical cancer to cisplatin. Arch Biochem Biophys. 2017;623–624:20–30. [DOI] [PubMed] [Google Scholar]
- 38. Gao YL, Zhao ZS, Zhang MY, Han LJ, Dong YJ, Xu B. Long non‐coding RNA PVT1 facilitates cervical cancer progression via negative regulating of miR‐424. Oncol Res. 2017;25:1391–1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Dong Z, Zhang A, Liu S, et al. Aberrant methylation‐mediated silencing of lncRNA MEG3 functions as a ceRNA in esophageal cancer. Mol Cancer Res. 2017;15:800–810. [DOI] [PubMed] [Google Scholar]
- 40. Li L, Shang J, Zhang Y, et al. MEG3 is a prognostic factor for CRC and promotes chemosensitivity by enhancing oxaliplatin‐induced cell apoptosis. Oncol Rep. 2017;38:1383–1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wang X, Wang Z, Wang J, Wang Y, Liu L, Xu X. LncRNA MEG3 has anti‐activity effects of cervical cancer. Biomed Pharmacother. 2017;94:636–643. [DOI] [PubMed] [Google Scholar]
- 42. Zhang J, Yao T, Lin Z, Gao Y. Aberrant methylation of MEG3 functions as a potential plasma‐based biomarker for cervical cancer. Sci Rep. 2017;7:6271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zhang Z, Liu T, Wang K, et al. Down‐regulation of long non‐coding RNA MEG3 indicates an unfavorable prognosis in non‐small cell lung cancer: evidence from the GEO database. Gene. 2017;630:49–58. [DOI] [PubMed] [Google Scholar]
- 44. Yao J, Zhou B, Zhang J, et al. A new tumor suppressor LncRNA ADAMTS9‐AS2 is regulated by DNMT1 and inhibits migration of glioma cells. Tumour Biol. 2014;35:7935–7944. [DOI] [PubMed] [Google Scholar]
- 45. Li Q, Dai Y, Wang F, Hou S. Differentially expressed long non‐coding RNAs and the prognostic potential in colorectal cancer. Neoplasma. 2016;63:977–983. [DOI] [PubMed] [Google Scholar]
- 46. Xu L, Li Q, Xu D, et al. Hsa‐miR‐141 downregulates TM4SF1 to inhibit pancreatic cancer cell invasion and migration. Int J Oncol. 2014;44:459–466. [DOI] [PubMed] [Google Scholar]
- 47. Chang X, Yu C, Li J, Yu S, Chen J. Hsa‐miR‐96 and hsa‐miR‐217 Expression Down‐Regulates with Increasing Dysplasia in Pancreatic Intraepithelial Neoplasias and Intraductal Papillary Mucinous Neoplasms. Int J Med Sci. 2017;14:412–418. [DOI] [PMC free article] [PubMed] [Google Scholar]


