Abstract
The effect of binary hydrophilic polymers on a pair of representative bio‐macromolecules in a living cell has been examined. The results showed that these bio‐macromolecules exhibited specific localization in cell‐sized droplets that were spontaneously formed through water/water microphase segregation under crowding conditions with coexisting polymers. In these experiments, a simple binary polymer system with poly(ethylene glycol) (PEG) and dextran (DEX) was used. Under the conditions of microphase segregation, DNA was entrapped within cell‐sized droplets rich in DEX. Similarly, F‐actin, linearly polymerized actin, was entrapped specifically within microdroplets rich in DEX, whereas G‐actin, a monomeric actin, was distributed evenly inside and outside these droplets. This study has been extended to a system with both F‐actin and DNA, and it was found that DNA molecules were localized separately from aligned F‐actin proteins to create microdomains inside microdroplets, reflecting the self‐emergence of a cellular morphology similar to a stage of cell division.
Keywords: DNA, liquids, microdroplets, phase separation, synthetic biology
Living cells keep a highly crowded cellular cytoplasm with 30–40 wt/wt % of cellular materials, such as DNA, RNA, and a rich variety of proteins.1 Macromolecular assemblies, such as polynucleotides and cytoskeletons, are dynamically arranged in a self‐organized manner during the cell life cycle to contribute to various biological functions, including cell motility, morphogenesis, and division, as well as the spatial arrangement and migration of organelles. It is generally considered that the systematic localization of cytoskeletal networks and the manner of polymerization is controlled by regulatory factors determined by genetic material. Various actin‐binding proteins have been found to play a role in actin network systems, through polymerization/depolymerization, resulting in the constitution of either a backbone structure or skeleton inside cells.2
Recently, it has been suggested that the crowded cellular environment plays a fundamental role in the construction of subcellular organelles and granules, as well as in the cellular morphology, that is, liquid droplets (cytoplasmic bodies) consisting of RNA and proteins,3 assembly of the bacterial cytoskeleton protein FtsZ,4 and cytoskeletal networks.5 Interestingly, it is becoming clear that these systems lack specific regulatory factors, which suggests that subcellular structures are generated spontaneously under nonspecific environmental factors in a self‐organized manner under the crowded conditions of the cytoplasm. Studies to unveil the fundamental mechanism of intracellular self‐organization under certain environments are expected to shed light on the fundamental unsolved problem of life: how can individual cells undergo appropriate differentiation in the right location under the same genetic information?
Herein, we report the characteristic behavior of DNA and/or actin in a simple binary hydrophilic polymer system, poly(ethylene glycol) (PEG)/dextran (DEX), as a simple model of a cytoplasmic solution or intracellular fluid crowded with polymeric molecules. If the two polymers were mixed at certain concentrations, cell‐sized water/water (w/w) microdroplets (referred to herein as cell‐sized aqueous/aqueous microdroplets, CAMDs), with diameters ranging from 10 to 100 μm, emerged and were sustained for more than several hours (Figure S1 and Table S1 in the Supporting Information).6 The interior and exterior of CAMDs were occupied by DEX and PEG solutions, respectively. PEG is a flexible polymer and DEX is a branched polymer with a stiff backbone, and thus, they constitute a binary aqueous two‐phase system (ATPS), which exhibits liquid–liquid phase separation (LLPS).7 Because the CAMDs apparently provide cell‐like crowded microenvironments, we have tried to examine whether microcompartmentalization has exotic effects on the behavior of fundamental bio‐macromolecules, such as DNA and actins.
Figure 1 shows confocal laser scanning microscopic images of CAMDs in the presence of DNA molecules. Figure 1 A reveals that fluorescence‐labeled short single‐stranded DNAs (11‐mers) are distributed evenly inside and outside of CAMDs. In contrast, long DNAs (λ‐DNA, 49 kbp) are distributed homogeneously inside DEX‐rich CAMDs (Figure 1 B). Thus, it is evident that CAMDs can entrap long DNA chains in a selective manner. Such selective entrapment of long DNA is attributable to the difference in the manner of packing of crowding polymers between PEG and DEX, that is, nanosized void space exists in the DEX‐rich phase because of its stiff backbone and branched conformation, whereas the PEG‐rich phase is fully occupied with flexible chains.8 For double‐stranded DNA with a diameter of about 2 nm, the persistence length is known to be around 170 bp, corresponding to about 50 nm. Thus, λ‐DNA behaves as a semiflexible polymer chain. It has been reported that such long DNA molecules are compressed to form a compact state and/or align in a liquid‐crystalline‐like condensate under crowded conditions with a flexible polymer, such as PEG.9 In contrast, it is expected that a semiflexible DNA chain can penetrate into the nanosized void space under crowded conditions with DEX. On the other hand, short oligomeric single‐stranded DNA has no preference for a phase rich in either PEG or DEX because it is so small.
Figure 1.
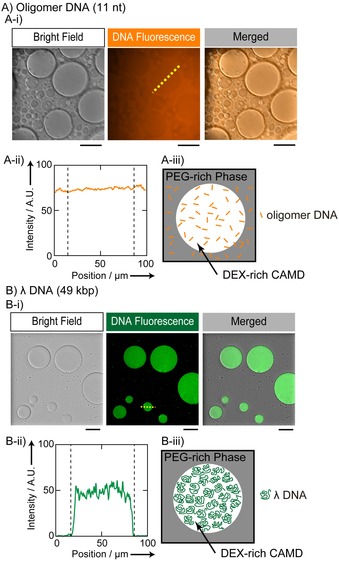
DNA localization in CAMDs in the presence of 5.0 wt % PEG and 5.0 wt % DEX observed by a confocal laser scanning microscope (CLSM). A) Single‐stranded DNAs (undecamers, 21 μm in nucleotide units) were distributed homogeneously inside and outside DEX‐rich CAMDs. Oligomer DNAs had been labeled with 3′‐TAMRA. B) Long double‐stranded λ‐DNAs (49 kbp, 38.5 μm in nt units) were observed only for DEX‐rich CAMDs, and were specifically localized inside the droplets. λ‐DNA was labeled with GelGreen. Fluorescence profiles along the dashed lines in the DNA fluorescence images (middle panels in A‐i and B‐i) are shown in A‐ii and B‐ii, and schematic representations are depicted to illustrate how both DNA molecules are located in the PEG/DEX phases (A‐iii and B‐iii). Scale bars: 50 (A‐i) and 100 μm (B‐i). (See Tables S2 and S3 for detailed information regarding the conditions.)
Figure 2 shows the distributions of actin molecules in solutions with CAMDs. Using a confocal laser scanning microscope (CLSM), we observed that an actin monomer, G‐actin, was delocalized evenly outside and inside the droplets (Figure 2 A). In contrast, a polymerized actin, F‐actin, was distributed homogeneously inside DEX‐rich CAMDs (Figure 2 B). Here, actins were induced to polymerize through the addition of potassium chloride, and their lengths were between 1 and 30 μm. Figure 2 C shows the specific localization of the bundled state of F‐actin (bundled F‐actin), which was caused by the addition of magnesium ions.10 In contrast to F‐actin in a dispersed state, as in Figure 2 B, F‐actin bundles tend to be located at the interface between the PEG‐ and DEX‐rich phases. If much higher concentrations of MgCl2 were added, F‐actins formed a cluster of large bundles that appeared to form a cytoskeletal network, and the cluster was entrapped near the periphery of DEX‐rich CAMDs (Figure S2). Interestingly, such bundled F‐actin caused deformation of the interface, which was not observed for experiments with G‐actin and was less frequently observed with F‐actin in a dispersed state. Actin plays important roles in cell motility and morphological development,11 and can transform giant liposomes, which are considered to be artificial membrane vesicles, through its interaction with the surface of lipid bilayers.12 The present observation of the deformation of the interface of CAMDs implies that actin, by nature, plays a role in the development of the morphology of interfaces of soft matter by modulating tension through its accumulation on the surface.
Figure 2.

Actin is localized in CAMDs depending on the state of actin polymerization. All CLSM images were acquired with PEG/DEX (5.0 wt % each) and an actin concentration of 9.0 μm with 1.0 mol % Alexa Fluor 546‐labeled actin. A) G‐actins were distributed homogeneously inside and outside of DEX‐rich CAMDs. B) Polymerized F‐actins, generated by the addition of 40 mm KCl, were entrapped in DEX‐rich CAMDs. C) Bundled F‐actins, generated by the addition of 2.0 mm MgCl2, were located at the surface of DEX‐rich CAMDs, and simultaneously F‐actins existed inside the droplets. Scale bar: 50 μm. (See Tables S4, S5, and S6 for detailed information regarding each condition.)
Based on the systematic observation by microscopy, we deduced a phase diagram for the changes in the distribution and polymerization state of actins in CAMDs with different concentrations of KCl and MgCl2 (Figure 3). It has been well established that, in bulk in vitro solution, actin polymerizes into filaments in the presence of KCl (30 mm or more) or MgCl2 (a few mm or more), and F‐actins form bundles by paracrystal formation in the presence of 10 mm or more of MgCl2.10 In our experiments with the crowded solution of PEG/DEX, interestingly, the threshold concentration of KCl to induce actin polymerization decreased by about one order of magnitude (Figure 3 A), and the threshold concentration of MgCl2 to cause bundling of F‐actin decreased to about one‐third (Figure 3 B). This is attributable to a cooperative effect between the restricted localization of actin into the DEX‐rich CAMDs, as a result of its polymerization and facilitation of polymerization by accumulation due to restricted localization.
Figure 3.

Phase diagrams for the localization and morphological state of actin, depending on the concentrations of A) actin and KCl, and B) MgCl2. In B), the concentration of actin was fixed at 6.0 μm. Crosses indicate the parameter area for even distribution inside and outside CAMDs (as shown in Figure 2 A). The black and gray circles in A) show the localization of dispersed F‐actin inside CAMDs (as in Figure 2 B). The conditions indicated by closed circles caused actin localization soon after sample preparation, whereas under the conditions indicated by gray‐filled circles, several tens of minutes were required for actin localization and the reproducibility of the results was relatively low. The open diamonds show the localization of bundled F‐actin at the interface, which causes deformation of the droplets (as in Figure 2 C). The open circles indicate the clustering of bundled F‐actin inside CAMDs (as shown in Figure S2).
Based on the above‐mentioned observations of specific localization and/or morphological changes of DNA or actin, we conducted an experiment in which long DNA and F‐actin coexisted within the solution of PEG/DEX with CAMDs. Figure 4 exemplifies the results for the condition in which F‐actin is dispersed inside DEX‐rich CAMDs, as in Figure 2 B, and long double‐stranded λ‐DNA is entrapped therein, as in Figure 1 B. We found that the entrapped DNA and F‐actin assembled in a separate manner inside the droplet, that is, segregation between pair of different biopolymers was caused spontaneously. Observation by polarization microscopy, as in the lower‐right panel in Figure 4, indicates that F‐actin was assembled in parallel with some ordered (nematic) alignment. As a result, long DNAs might be located separately to form discrete domains within CAMDs in a self‐organized manner. We sometimes observed that entrapped DNAs and actins failed to be segregated in the CAMDs; this suggested that the phenomenon may occur within only a certain range of conditions and further investigation is awaited on the regime at which the microstructures emerge. Actin has also been found in the cell nucleus, and its physiological significance and role are still enigmatic, despite vigorous research.13 The nuclear actins have been argued in the context of maintenance of LLPS structures occurring in nuclei of actual cells.14 The result observed herein would help to clarify these hypotheses. Even without DEX or PEG, long DNA and F‐actin can show phase separation.15 However, in order to induce the local phase separation of DNA and F‐actin in the bulk solution, the concentration of F‐actin has to be at least about tenfold higher than that in the present result; otherwise, DNA and F‐actin should be confined to a space with a diameter comparable to that of the F‐actin length.
Figure 4.

Specific localization of long DNA (λ‐DNA) and F‐actin in DEX‐rich CAMDs. Fluorescence microscopic images of DNA (GelGreen), actin (Alexa Fluor 546), and those merged are shown, together with observations by polarization microscopy (four panels on the left). Images were acquired under conditions of 120 μm λ‐DNA, 10 μm actin, and 4.0 mm KCl. F‐actins were observed to be in a nematic liquid‐crystal state in the center of DEX‐rich CAMDs, and DNA molecules appeared to be segregated from the center. In other words, long DNAs are compressed by the aligned F‐actin region, as illustrated schematically (right). Scale bar: 100 μm. (See Table S8 for detailed information regarding the conditions.)
In summary, in the presence of binary polymers (PEG/DEX), 1) both semiflexible long DNAs (λ‐DNAs) and F‐actin proteins were spontaneously localized within CAMDs, in contrast to the observations with short DNAs and G‐actin proteins, which were distributed evenly both inside and outside of the droplets; 2) actin proteins exhibited various localizations in CAMDs if they had different polymerization states (monomer, fiber, and bundle), depending on the salt concentrations without proteinaceous regulatory factors; and 3) long DNAs and F‐actin, if they coexisted in the same CAMDs, exhibited microsegregation to form domains. To our surprise, despite the simplicity of the system, PEG/DEX CAMDs, or self‐emergent microcompartments under crowding, could exhibit primitive protocell‐like morphologies based on the nature of the biopolymers: first, actin proteins can deform the surfaces of CAMDs as they act on cell membranes; second, the bipolar localization of long DNAs, possibly due to the exclusion of aligned F‐actins, may also imply a primitive function of cytoskeletal proteins to generate intracellular structures. Because macromolecular crowding regulates biochemical activities,16 such confined microcompartments under macromolecular crowding are expected to change both the physicochemical properties and localization of entrapped proteins,17 and biological functions, such as enzymatic activities.18 Herein, we found that polymerization and bundling of actin in CAMDs were induced at much lower cation concentrations than that in typical aqueous medium in the absence of a crowding polymer. In the discipline of biochemistry, it is usually assumed that specific key–lock interactions determine the structure and function of living cells. However, the present results suggest that nonspecific environmental factors under confined crowding conditions may also play a decisive role in living things.
On the other hand, it should be noted that the localization of DNA, that is, entrapment of long double‐stranded DNA and almost homogeneous distribution of single‐stranded DNA, could be a specific case with the PEG/DEX CAMDs that we adopted herein. For instance, single‐stranded RNA and/or DNA can be entrapped inside microdroplets if the droplets are composed of proteins that possess some affinity to the polynucleotides.14, 19 The manner of localization of DNA and proteins would be dependent on which components mainly constitute CAMDs. Because PEG and DEX possess no particular affinity to single‐ or double‐stranded DNA, the localization observed herein could only be determined due to structural characteristics of these polymers. Moreover, it is known that the distribution of biopolymers, such as DNA, is affected by species and concentrations of coexisting salt in the bulk ATPS.7 Although we indicate concentrations in bulk solutions in Figures 2–4, at present, it remains unclear how salts such as MgCl2 and KCl would be distributed over microsystems of the CAMDs in relation to actin localization. In addition to adenosine triphosphate (ATP), it is necessary to investigate local concentrations of such small polyelectrolytes in the CAMDs; meanwhile, ATP is smaller than the oligomeric single‐stranded DNA used here, and the polymerization activity of actin was not changed, even during long‐term observations; this implies that ATP is distributed almost uniformly therein.
Actual cells contain highly complicated macromolecular systems, and this motivated us to simplify these systems to realize an artificial cell model.20 A PEG/DEX LLPS that can provide microdroplets, such as CAMDs, has recently gained attention because of its active21 and morphological22 behavior and usefulness in cell biotechnology.23 The present results of coexisting DNA/actin suggest that CAMDs are compartments that develop interactions among bio‐macromolecules, and thus, are useful for artificial cell studies. In other words, herein we showed that microdroplets caused by simple depletion effects served a simple real‐world model of a cellular system. These systems are generated under markedly different physicochemical interactions from that of coacervates, that is, aggregation products among macromolecules.24 Recently, it was found that nuclei could express LLPS with multiphase structures.14 Actually, biological macromolecules could interact both attractively and exclusively inside such microcompartments in a complicated manner. CAMDs can provide cell models in which attraction to crowd molecules is avoided, and it is expected that the result of the present study may help to open up new horizons on the significant role of phase separation in the formation of organelles.5, 6, 25
Experimental Section
Polymers: We used a PEG/DEX ATPS. PEG 6000 was purchased from Wako Pure Chemical Industries (Osaka, Japan), with an average molecular weight (MW) of 7300–9300 Da. DEX was also purchased from Wako Pure Chemical Industries, with an average MW of 180 000 to 210 000 Da. These two polymers were dissolved in nuclease‐free water (Milli‐Q, 18.2 MΩ cm) to prepare 20 wt % stock solutions. For use as a tracer for PEG‐rich domains, methoxyl PEG fluorescein (mPEG‐Fluorescein) was purchased from Nanocs Inc. (New York, NY), with an average MW of 10 kDa, an excitation wavelength (λ ex) of 490 nm, and an emission wavelength (λ em) of 520 nm. The mPEG‐Fluorescein was dissolved in nuclease‐free water to give a 10 wt % stock solution.
DNA: Single‐stranded oligomer DNA was purchased from Hokkaido System Science (Sapporo, Japan). The 3′‐labeled single‐stranded oligomer DNA had the following properties: 11‐mer (ATG CTG ATC GC), MW=3953.84 Da, 3′‐carboxytetramethylrhodamine (3′‐TAMRA; λ ex=542 nm, λ em=568 nm). It was dissolved in nuclease‐free water at 100 μm. λ‐DNA with a MW of 31.5 MDa (48 502 bp) was purchased from Nippon Gene Co., Ltd. (Tokyo, Japan). It was dissolved at a concentration of 0.25 g L−1 (nucleotide concentration of 770 μm). To label λ‐DNA, GelGreen, as a fluorescent dye (λ ex=500 nm, λ em=530 nm), was purchased from Biotium Inc. (Fremont, CA). It was dissolved in nuclease‐free water as a 0.5 mm stock solution.
Actin: Monomeric actin (globular actin, G‐actin) was prepared from acetone powder obtained from chicken breast muscle.26 To obtain filamentous actin (F‐actin), G‐actin was polymerized in G‐buffer (1 mm NaHCO3, pH 8.0, 0.2 mm CaCl2, 0.2 mm ATP) containing salt (potassium chloride or magnesium chloride). Whole actin (1.0 mol %) was labeled with Alexa Fluor 546 C5 maleimide (λ ex=537 nm, λ em=556 nm; Thermo Fisher Scientific, Inc., Waltham, MA), against actin monomer, as reported previously.27 Experiments with actin were performed in G‐buffer with or without salt.
Other materials: The antioxidant 2‐mercaptoethanol (2ME) was purchased from Wako Pure Chemical Industries. The buffer solution tris(hydroxymethyl)aminomethane⋅HCl (Tris⋅HCl 1 m, pH 7.5) was purchased from Nippon Gene, and diluted with nuclease‐free water to obtain 200 mm stock solutions. Potassium chloride and magnesium chloride were purchased from Wako Pure Chemical Industries and dissolved in nuclease‐free water at 1.0 m as stock solutions.
Microscopy: Images were obtained with a CLSM or an epifluorescent microscope. The CLSM was a Nikon A1 (Nikon Corp., Tokyo, Japan) instrument equipped with a 10× objective. The acquired images were analyzed by using a NIS Elements Viewer (Nikon). For phase contrast and polarized epifluorescent microscopic images, we used a BX60 microscope with a 40× objective (NA=0.75, Olympus, Tokyo, Japan). The BX60 microscope was equipped with a charge‐coupled device (CCD) camera (WAT‐910HX; Watec Co., Ltd., Yamagata, Japan). The obtained images were analyzed by using ImageJ software (Rasband, W.S., ImageJ, US National Institutes of Health, Bethesda, Maryland, USA, http://imagej.nih.gov/ij/, 1997–2016).
Procedures: We first prepared stock solutions as described above, and then mixed them in each microtube, as shown in the Supporting Information (experimental solution details), prior to microscopic observations.
Conflict of interest
The authors declare no conflict of interest.
Supporting information
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supplementary
Acknowledgements
This work was supported, in part, by JSPS KAKENHI grants no. JP15H02121 and JP16K07293, and by MEXT KAKENHI grants no. JP25103012 and JP24104004. H.S. received support as Research Fellow of the Japan Society for the Promotion of Science.
N. Nakatani, H. Sakuta, M. Hayashi, S. Tanaka, K. Takiguchi, K. Tsumoto, K. Yoshikawa, ChemBioChem 2018, 19, 1370.
Contributor Information
Dr. Kingo Takiguchi, Email: j46037a@cc.nagoya-u.ac.jp.
Dr. Kanta Tsumoto, Email: tsumoto@chem.mie-u.ac.jp.
References
- 1.
- 1a. Zimmerman S. B., Minton A. P., Annu. Rev. Biophys. Biomol. Struct. 1993, 22, 27–65; [DOI] [PubMed] [Google Scholar]
- 1b. Ellis R. J., Curr. Opin. Struct. Biol. 2001, 11, 114–119. [DOI] [PubMed] [Google Scholar]
- 2. Pollard T. D., Cooper J. A., Science 2009, 326, 1208–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.
- 3a. W. M. Aumiller, Jr. , Keating C. D., Adv. Colloid Interface Sci. 2017, 239, 75–87; [DOI] [PubMed] [Google Scholar]
- 3b. Nielsen F. C., Hansen H. T., Christiansen J., BioEssays 2016, 38, 674–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Monterroso B., Zorrilla S., Sobrinos-Sanguino M., Keating C. D., Rivas G., Sci. Rep. 2016, 6, 35140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Courchaine E. M., Lu A., Neugebauer K. M., EMBO J. 2016, 35, 1603–1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hyman A. A., Weber C. A., Jülicher F., Annu. Rev. Cell Dev. Biol. 2014, 30, 39–58. [DOI] [PubMed] [Google Scholar]
- 7. Albertsson P.-Å., Partition of Cell Particles and Macromolecules , 2nd ed., Wiley, New York, 1971. [Google Scholar]
- 8. Biswas N., Ichikawa M., Datta A., Sato Y. T., Yanagisawa M., Yoshikawa K., Chem. Phys. Lett. 2012, 539–540, 157–162. [Google Scholar]
- 9.
- 9a. Lerman L. S., Proc. Natl. Acad. Sci. USA 1971, 68, 1886–1890; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9b. Bloomfield V. A., Curr. Opin. Struct. Biol. 1996, 6, 334–341; [DOI] [PubMed] [Google Scholar]
- 9c. Vasilevskaya V. V., Khokhlov A. R., Matsuzawa Y., Yoshikawa K., J. Chem. Phys. 1995, 102, 6595–6602; [Google Scholar]
- 9d. Kojima M., Kubo K., Yoshikawa K., J. Chem. Phys. 2006, 124, 024902. [DOI] [PubMed] [Google Scholar]
- 10.
- 10a. Kasai M., Asakura S., Oosawa F., Biochim. Biophys. Acta 1962, 57, 13–21; [DOI] [PubMed] [Google Scholar]
- 10b. Hanson J., Proc. R. Soc. London Ser. B 1973, 183, 39–58. [DOI] [PubMed] [Google Scholar]
- 11. Blanchoin L., Boujemaa-Paterski R., Sykes C., Plastino J., Physiol. Rev. 2014, 94, 235–263. [DOI] [PubMed] [Google Scholar]
- 12. Honda M., Takiguchi K., Ishikawa S., Hotani H., J. Mol. Biol. 1999, 287, 293–300. [DOI] [PubMed] [Google Scholar]
- 13. Misu S., Takebayashi M., Miyamoto K., Front. Genet. 2017, 8, 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Feric M., Vaidya N., Harmon T. S., Mitrea D. M., Zhu L., Richardson T. M., Kriwacki R. W., Pappu R. V., Brangwynne C. P., Cell 2016, 165, 1686–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Negishi M., Sakaue T., Takiguchi K., Yoshikawa K., Phys. Rev. E 2010, 81, 051921. [DOI] [PubMed] [Google Scholar]
- 16. Nakano S.-i., Miyoshi D., Sugimoto N., Chem. Rev. 2014, 114, 2733–2758. [DOI] [PubMed] [Google Scholar]
- 17.
- 17a. Hase M., Yoshikawa K., J. Chem. Phys. 2006, 124, 104903; [DOI] [PubMed] [Google Scholar]
- 17b. Kato A., Shindo E., Sakaue T., Tsuji A., Yoshikawa K., Biophys. J. 2009, 97, 1678–1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kato A., Yanagisawa M., Sato Y. T., Fujiwara K., Yoshikawa K., Sci. Rep. 2012, 2, 283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Nott T. J., Craggs T. D., Baldwin A. J., Nat. Chem. 2016, 8, 569–575. [DOI] [PubMed] [Google Scholar]
- 20.
- 20a. Chiarabelli C., Stano P., Luisi P. L., Curr. Opin. Biotechnol. 2009, 20, 492–497; [DOI] [PubMed] [Google Scholar]
- 20b. Suzuki K., Miyazaki M., Takagi J., Itabashi T., Ishiwata S. i., Proc. Natl. Acad. Sci. USA 2017, 114, 2922–2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ban T., Fukuyama T., Makino S., Nawa E., Nagatsu Y., Langmuir 2016, 32, 2574–2581. [DOI] [PubMed] [Google Scholar]
- 22. Yuan H., Ma Q., Song Y., Tang M. Y. H., Chan Y. K., Shum H. C., Macromol. Chem. Phys. 2017, 218, 1600422. [Google Scholar]
- 23. Han C., Takayama S., Park J., Sci. Rep. 2015, 5, 11891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.
- 24a. Black K. A., Priftis D., Perry S. L., Yip J., Byun W. Y., Tirrell M., ACS Macro Lett. 2014, 3, 1088–1091; [DOI] [PubMed] [Google Scholar]
- 24b. Lindhoud S., Claessens M. M. A. E., Soft Matter 2016, 12, 408–413. [DOI] [PubMed] [Google Scholar]
- 25.
- 25a. Weber S. C., Brangwynne C. P., Cell 2012, 149, 1188–1191; [DOI] [PubMed] [Google Scholar]
- 25b. Mitrea D. M., Kriwacki R. W., Cell Commun. Signaling 2016, 14, 1; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25c. Brangwynne C. P., Tompa P., Pappu R. V., Nat. Phys. 2015, 11, 899–904. [Google Scholar]
- 26. Spudich J. A., Watt S., J. Biol. Chem. 1971, 246, 4866–4871. [PubMed] [Google Scholar]
- 27. Fujiwara I., Takahashi S., Tadakuma H., Funatsu T., Ishiwata S. i., Nat. Cell Biol. 2002, 4, 666–673. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supplementary


