SUMMARY
Cancer chronotherapy, treatment at specific times during circadian rhythms, endeavors to optimize anti-tumor effects and lower toxicity. However, comprehensive characterization of clock genes and their clinical relevance in cancer is lacking. We systematically characterized the alterations of clock genes across 32 cancer types by analyzing data from TCGA, CTRP, and GDSC databases. Expression alterations of clock genes are associated with key oncogenic pathways, patient survival, tumor stage and subtype in multiple cancer types. Correlations between expression of clock genes and of other genes in the genome were altered in cancerous versus normal tissues. We identified interactions between clock genes and clinically actionable genes by analyzing co-expression, protein-protein interaction and ChIP-seq data, and also found that clock gene expression is correlated to anti-cancer drug sensitivity in cancer cell lines. Our study provides a comprehensive analysis of the circadian clock across different cancer types and highlights potential clinical utility of cancer chronotherapy.
eTOC blurb
Ye et al comprehensively analyzed alterations of clock genes and circadian rhythms across multiple human cancers, and revealed strong interactions between clock genes and clinically actionable genes, which highlights the clinical utility of circadian timing in cancer chronotherapy.
INTRODUCTION
Cancer chronotherapy consists of administering treatment at an optimal time in the circadian rhythm to maximize anti-tumor effects and minimize toxicity (Dallmann et al., 2016; Levi et al., 2010). Circadian rhythms are 24-hour oscillations that coordinate a variety of biological processes in temporal precision in various organisms. Disruption of the circadian rhythm is associated with cancer development and poor prognosis. For example, patients with metastatic colorectal cancer who have normal circadian rhythms have a 5-fold higher survival rate than patients with severely disrupted circadian rhythms (Innominato et al., 2012; Mormont et al., 2000). A mathematical model based on transcription profiles of clock genes Bmal and Nr1d1 determined optimal timing and minimized the toxicity of chemotherapy in mice (Li et al., 2013b). Furthermore, early clinical trials have suggested significant clinical benefits of specific circadian timing of chemotherapy or radiotherapy by combination with vinblastine, cyclophosphamide, and methotrexate or 5-FU (Focan, 1979). Cancer chronotherapy appears to offer the potential to improve current cancer treatments and to refine the development of new anticancer drugs (Vincenzi et al., 2003). Therefore, there is an urgent need to develop systems biology approaches to further optimize and personalize cancer chronotherapy (Ballesta et al., 2017).
To develop cancer chronotherapy, the first crucial step is to understand the roles of clock genes in cancer. The mammalian circadian machinery is controlled by two main transcriptional/translational auto-regulatory feedback loops that involve 14 core clock genes. The CLOCK/ARNTL heterodimer activates the transcription of clock repressors PER1/2/3 and CRY1/2, then the PER/CRY heterodimer translocates into the nucleus to interact with CLOCK/ARNTL to inhibit their own transcription (Hurley et al., 2016; Ueda et al., 2005). In another loop, CLOCK/ARNTL is alternatively stimulated and repressed by its transcription targets, including RORs and REV-ERBs (He et al., 2016; Relógio et al., 2011). Besides core clock genes, another set of 37 clock associated genes were reported to directly or indirectly interact with these two transcriptional loops (Sahar and Sassone-Corsi, 2009). Dysregulation of clock genes promotes tumorigenesis (Lamia, 2017) through mechanisms that include the cell cycle (El-Athman et al., 2017; Wood et al., 2006), DNA damage (Sancar et al., 2010) and metabolism (Eckel-Mahan and Sassone-Corsi, 2013). For example, ARNTL, also called BMAL1, displayed a cell-autonomous tumor-suppressive role in transformation and lung tumor progression (Papagiannakopoulos et al., 2016), and BMAL1 reduced expression is associated with a decreased cell sensitivity to anticancer drugs, such as oxaliplatin and irinotecan (Dulong et al., 2015; Zeng et al., 2014). ARNTL2 are required for metastatic lung adenocarcinoma (Brady et al., 2016) and estrogen receptor-negative breast cancer (Ha et al., 2016), and is potential biomarkers for tumor aggressiveness (Mazzoccoli et al., 2012). CRY2 deficiency elevates c-MYC and enhances cancer transformation susceptibility (Huber et al., 2016). PER2 was shown to be downregulated in non-small-cell lung cancer and several human lymphoma cell lines (Hua et al., 2006; Yang and Stockwell, 2008). These studies support the important roles of clock genes in tumorigenesis.
All 14 core clock genes have at least one paralog gene that may be functionally redundant in clock regulation. Composite knockdown of paralog clock genes may cause more serious consequences than single knockdown. For example, composite PER1/2 double or PER1/2/3 triple knockdown caused complete arrhythmicity (Ramanathan et al., 2014). Similarly, knockdown of Cry1 or Cry2 displayed shorter or longer clock rhythm of locomotor activity, respectively, while Cry1/2 double knockdown caused arrhythmicity (van der Horst et al., 1999). Despite the arrhythmicity caused by double or triple knockdown, the interactive effects of these paralog genes in cancer remain to be ascertained.
On one hand, loss of circadian homeostasis increases the risk of tumorigenesis in multiple cancer types (Puram et al., 2016). Therefore, preventing disruption of the circadian rhythm has the potential to improve the efficacy of cancer therapy. On the other hand, circadian rhythms could be reprogrammed in tumors. For example, circadian metabolism in the liver is reorganized in the presence of lung adenocarcinoma (Masri et al., 2016), while hepatic circadian rhythm could be affected or reprogrammed by breast cancer (Hojo et al., 2017), suggesting that tumors may alter circadian rhythm of other cell lineages.
In this study, we systematically analyzed genomic profiling and clinical data from The Cancer Genome Atlas (TCGA) (The Cancer Genome Atlas Research Network et al., 2013) as well as drug sensitivity data from the Cancer Therapeutics Response Portal (CTRP) (Rees et al., 2015) and Genomics of Drug Sensitivity in Cancer (GDSC) (Yang et al., 2013). Our results provide a systematic analysis of clock genes across different cancer types, and highlight the significant roles of circadian clock in cancer chronotherapy.
RESULTS
The Expression of Many Clock Genes is Altered in Cancer
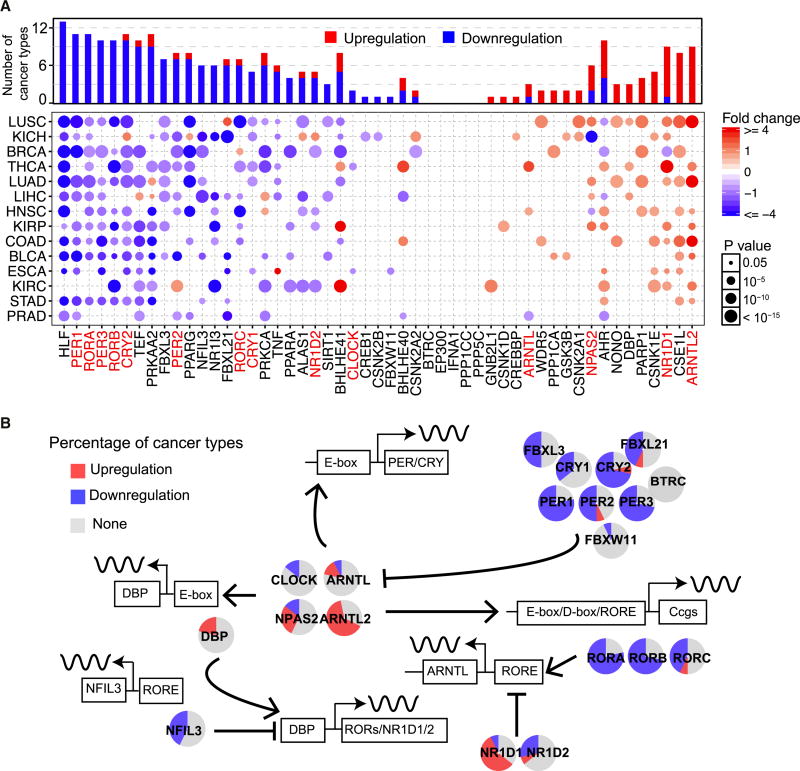
Based on literature, we defined 14 genes that control clock transcriptional/translational feedback loops as “core clock genes” (Ko and Takahashi, 2006), 37 genes that are directly or indirectly regulated by these transcriptional-translational loops as “clock-associated genes” (Relógio et al., 2014), and the union of these two gene sets as “clock genes”. We utilized TCGA data for 14 cancer types with paired RNA-seq samples (Figure S1 and Table S1) to identify genes differentially expressed between tumor and normal sample pairs for clock genes. Overall, 45 (88.2%) of the clock genes are differentially expressed in at least one cancer type. We interpret an increase in expression compared to normal samples as upregulation and a decrease in expression as downregulation. Some clock genes were upregulated in multiple cancer types. ARNTL2 and CSE1L were upregulated in nine cancer types. PER1 was downregulated in ten cancer types, PER2 was downregulated in seven cancer types, and PER3 was downregulated in nine cancer types (Figure 1A). Importantly, we identified alterations in expression of several clock genes that were not well characterized previously. For example, RORB is downregulated in five cancer types, including thyroid carcinoma (THCA; fold-change [FC] = 5.0, P = 9.1 × 10−14). RORB regulates clock rhythm and may play a role in tumor suppression. Furthermore, we observed a cancer-type-specific pattern for several genes. For example, PER2 showed overall downregulation in most cancer types, but showed upregulation in renal clear cell carcinoma (KIRC; FC = 1.9, P = 3.3 × 10−12). NPAS2, which encodes a potential tumor suppressor (Hoffman et al., 2008; Yuan et al., 2017), showed significant downregulation in kidney chromophobe (KICH) and head and neck squamous cell carcinoma (HNSC), but was upregulated in lung squamous cell carcinoma (LUSC), lung adenocarcinoma (LUAD), kidney renal papillary cell carcinoma (KIRP) and liver hepatocellular carcinoma (LIHC). This suggests that clock genes play different roles in different tumor environments (Gong et al., 2017; Han et al., 2015; Xiang et al., 2018).
Figure 1. The transcriptional dysregulation of clock genes in cancer.
(A) Upregulation (red) and downregulation (blue) patterns of clock genes across different cancer types (Y-axis) compared to paired normal samples (FC > 1.5; t-test corrected p < 0.05). X-axis indicates 14 core clock genes (red) and 37 clock-associated genes (black). The color intensity indicates the fold-change, the point size indicates the significance of p. Upper bars show the frequency of cancer types, with upregulation (red) and downregulation (blue) for each clock gene. (B) Dysregulation of core clock genes in clock transcriptional loops (red, upregulation; blue, downregulation).
The mammalian circadian machinery is controlled by two main transcription auto-regulatory feedback loops. Our results provide the global view of clock transcriptional loops in multiple cancer types (Figure 1B). Among the transcriptional repressors, CRY1/2 and PER1/2/3 exhibited the most significant downregulation. Among the transcriptional activators in that ARNTL2 exhibited overall upregulation, CLOCK exhibited no alteration, whereas ARNTL and NPAS2 exhibited upregulation in several cancer types but downregulation in several other cancer types.
Effects of Clock Genes on Oncogenic Pathways
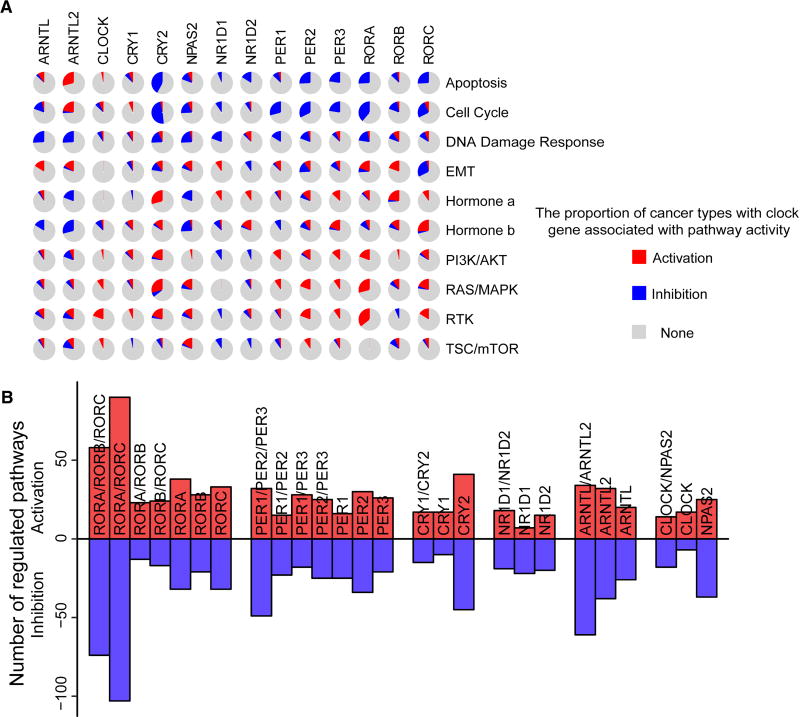
To further understand the molecular mechanisms for clock genes involved in tumorigenesis, we examined the correlation between expression of individual clock genes and activation or inhibition of ten key signaling pathways, based on a pathway score calculated from the sum of the relative protein levels of all positive regulatory components minus that of all negative regulatory components (Akbani et al., 2014) (see STAR Methods). Our results showed that clock genes are highly associated with activation or inhibition of multiple oncogenic pathways (Figures 2A and S2A). For example, CRY2, PER1, PER2, PER3, RORA and RORC are highly associated with inhibition of apoptosis and cell cycle, and activation of PI3K/AKT and RAS/MAPK signaling pathways. Despite similar repressive functions, CRY2 was associated with cancer pathway alterations in many more cancer types than CRY1 (Figure 2A). For example, CRY2 is associated with activation of the RAS/MAPK signaling pathway in nine (versus two for CRY1) cancer types and hormone a pathway in nine (versus one) cancer types, inhibition of cell cycle in fifteen (versus zero) cancer types, and DNA damage response in five (versus one) cancer types (Figures 2A and S2B). This result implies the different effects of core clock genes in the same family. Similar patterns were observed that RORA and RORB are associated with activation of epithelial-mesenchymal transition (EMT), while RORC is associated with inhibition of EMT (Figures 2A and S2B). These results suggest that clock genes are associated with alterations of multiple oncogenic pathways.
Figure 2. Functional effects of clock genes in oncogenic pathways.
(A) The proportion of cancer types with core clock genes significantly associated to activation (red) or inhibition (blue) of the ten key signaling pathways in 31 cancer types. (B) The total number of activation (red) or inhibition (blue) pathways in ten key signaling pathways across 31 cancer types, which are associated with individual or paralog clock genes. See also Figure S2.
Despite the arrhythmicity caused by double or triple knockdown, the interactive effects of these paralog genes in cancer remains unclear. Here, we compared pathway score in tumor samples with high expression of paralog clock genes in combinatoin versus tumor samples with low expression of paralog clock genes in combination (see STAR Methods). We observed that high expression levels of RORA and RORC in combination were associated with more pathways (activation of 90 and inhibition of 103 pathways) than any other combination of ROR genes (Figures 2B and S2B). Patients with low expression of RORA and RORC in combination showed significantly worse survival (Figure S2C), while low expression of each individual gene did not significantly contribute to patient survival. On the other hand, we observed that alterations of CRY1 and CRY2 in combination led to fewer alterations of signaling pathways than CRY2 alterations alone (Figure S2B), suggesting that CRY2 may play a dominant role in cancer.
Genetic and Epigenetic Alterations of Clock Genes in Cancer
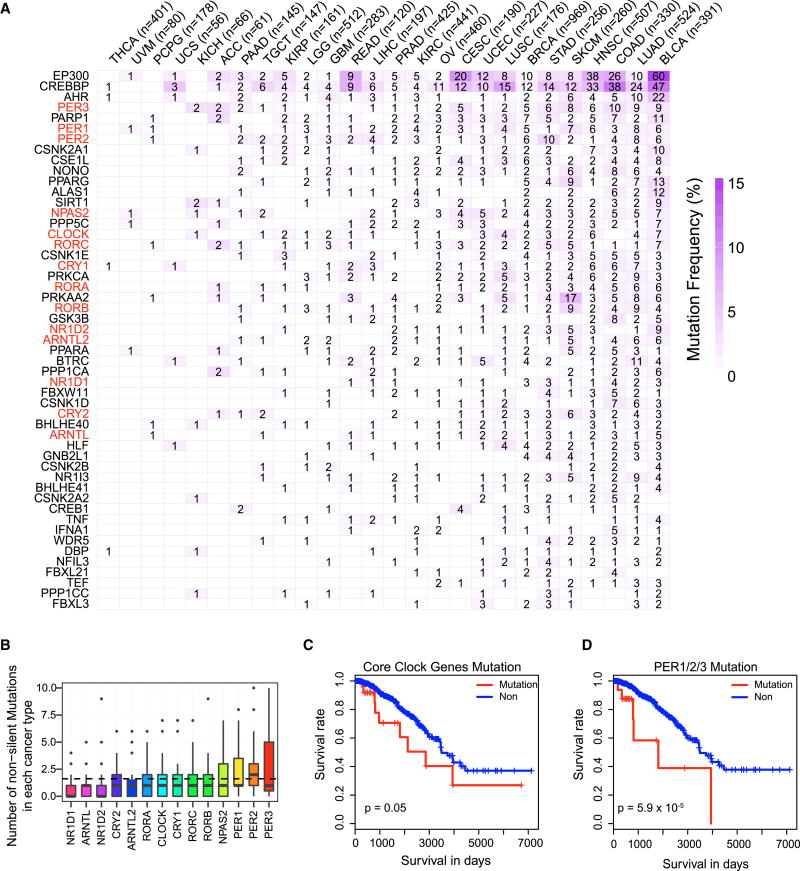
To further determine genetic alterations of clock genes in human cancer, we analyzed non-silent mutations of clock genes across cancer types. EP300 (p300) and CREBBP (CBP), which contribute to robust histone acetylation of chromatin surrounding E boxes that are regulated by the BMAL1-CLOCK complex (Lee et al., 2010), showed relatively high mutation frequencies (Figure 3A). We observed that mutation frequency is relatively low for core clock genes, with fewer than five mutations in the majority of samples (Figure 3B). However, mutations of core clock genes were associated with patients’ overall survival. For example, breast cancer patients with mutations in core clock genes had worse overall survival than those without mutations (log-rank test P = 0.05; Figure 3C). We observed the highest mutation rate for core circadian genes in PER1/2/3 (Figure 3B), which are associated with worse survival (P = 5.9 × 10−5; Figure 3D). These results suggest that genetic alterations of clock genes play functional roles in cancer.
Figure 3. Mutational landscape of clock genes in cancer.
(A) Heatmap shows the number of mutations (number in cell) and frequency of mutations (color-scale) for each clock gene in each cancer type. X-axis and Y-axis labels ordered by the sum of clock gene mutations in each cancer type and the sum of mutations in all cancer types across clock genes, respectively. Core clock genes are marked in red. (B) Box plot shows the number of mutations of core clock genes across cancer types, with outliers shown as dots. (C) Kaplan-Meier curves show overall survival between samples with (red) or without (blue) mutations in core clock genes in BRCA. (D) Kaplan-Meier curves of BRCA stratified by PER1/2/3 mutation. See also Figure S3.
DNA methylation regulates gene expression in cancer (Shen and Laird, 2013). We examined the promoter DNA methylation patterns of clock genes across different cancer types and observed an overall negative correlation between DNA methylation and gene expression (Figure S3A). We further observed multiple small alterations with a few genes showing differentially methylated patterns. For example, we observed hypermethylation in HLF, but hypomethylation in ARNTL2 (Figure S3B). In particular, ARNTL2 showed hypomethylation in three cancer types, which is consistent with upregulation of gene expression. The results suggest that promoter DNA methylation may regulate the expression of clock genes in cancer.
Disruption and Reprogramming of Circadian Rhythms in Cancer
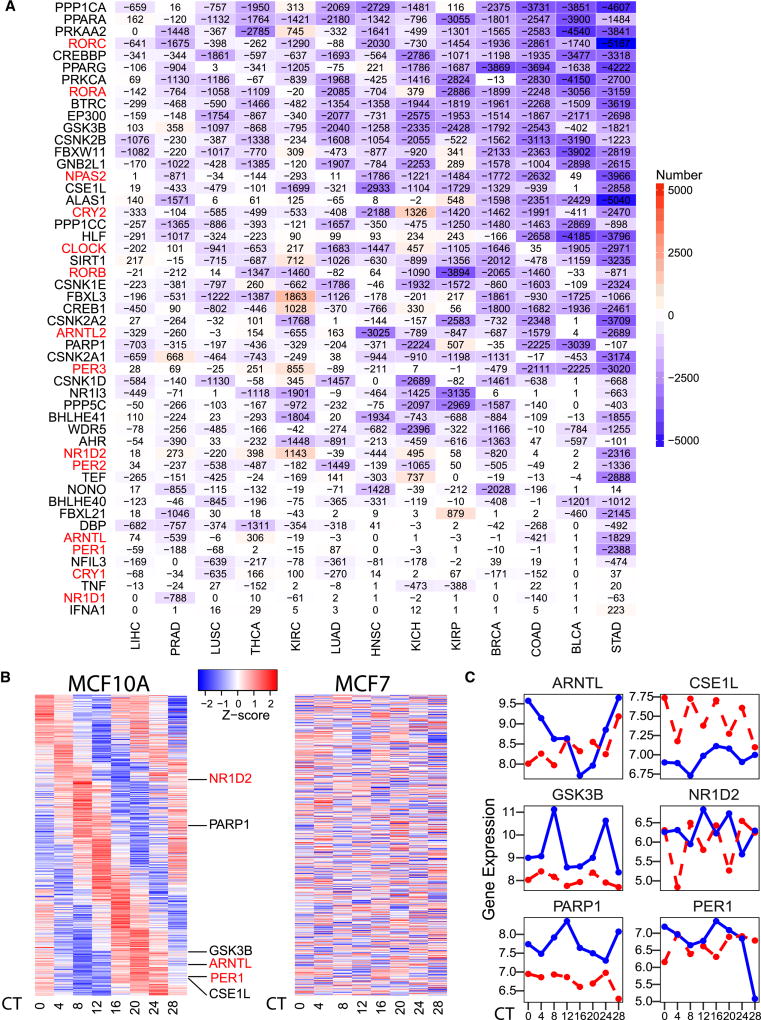
Circadian rhythms are involved in the regulation of a wide variety of physiological and behavioral rhythms by driving the expression of clock controlled genes (Guerrero-Vargas et al., 2017; Koike et al., 2012). We sought to systematically investigate the contributions of circadian rhythm disruption and impaired molecular clockworks to tumorigenesis. We calculated the correlation for gene expression between clock genes and all other genes in paired tumor and normal samples and interpret a change in correlation as associated with a disruption of the circadian rhythm. To avoid potential effects of tumor impurity, the Pearson correlation coefficient was corrected by tumor purity (Li et al., 2016). Compared to normal tissue, significantly fewer genes in tumor tissues correlated with clock genes across all cancer types, whether exhibiting negative (Figure 4A) or positive correlation (Figure S4A). Several thousand genes lost their correlation to clock genes in tumor samples. We also found that correlation among core clock genes was reduced significantly. For example, CRY2 is highly correlated with eight core clock genes in normal kidney tissue samples, but showed significantly reduced or no correlation in KIRP (Figure S4B). CRY2 is negatively correlated with ARNTL2 in normal kidney tissue samples (Pearson correlation coefficient [R] = −0.7, false discovery rate [FDR] = 0.002), but is not correlated in kidney tumor samples (R = 0.056; FDR = 0.9; Figure S4C).
Figure 4. Disruption of circadian rhythms in cancer.
(A) The number of genes which negatively correlated to clock genes (R ≤ −0.5 and FDR < 0.05, number in cell) decreased (blue) or increased (red) in tumor samples compared to paired normal samples for each clock gene in each cancer type. Pearson correlation is corrected by tumor purity in cancer samples. Core clock genes are marked in red. (B) Circadian oscillating genes as determined by JTK_CYCLE and MetaCycle::meta2d (P < 0.05) in MCF10A (left), and disrupted oscillation of these genes in MCF7 (right). Red, high expression; blue, low expression. X-axis displayed time points after serum shock. (C) Time-dependent relative expression of clock genes ARNTL, CSE1L, GSK3B, NR1D2, PARP1 and PER1 in MCF10A (blue) and MCF7 (red). See also Figure S4.
To more directly assess the effects of circadian rhythm on tumorigenesis, we reanalyzed data from a study with eight time points after serum shock in MCF7, a breast cancer cell line, and MCF10A, a non-tumorigenic breast epithelial cell line (Gutiérrez-Monreal et al., 2016). We identified 478 genes with a clear circadian oscillation pattern in MCF10A by MetaCycle::meta2d (JTK_CYCLE and meta2d adjust P < 0.05; Wu et al., 2016), while their oscillation patterns were not observed in MCF7 (Figure 4B). Detailed comparisons showed that the oscillation patterns of six clock genes in MCF10A, including PER1, ARNTL, NR1D2, PARP1, GSK3B and CSE1L, were disrupted in the MCF7 cell line (Figure 4C). These results suggested circadian rhythm of normal tissue cell line could not be maintained similarly in cancer cell line. In contrast, we also identified 552 genes with oscillation pattern in MCF7, but not in MCF10A (Figure S4D). These genes were enriched in functions including the TNF and WNT signaling pathways (Figure S4E). This suggests that disrupted and reprogrammed circadian rhythm in cancer cells may contribute to cancer development.
Clinical Relevance of Clock Genes
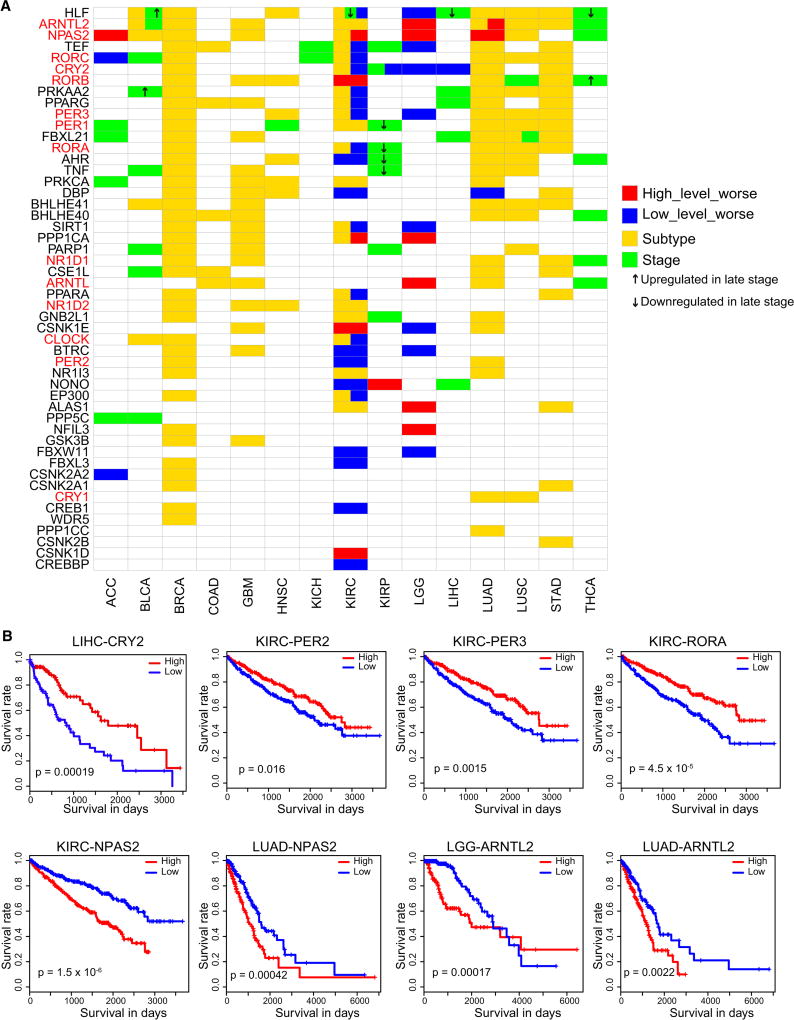
Given the global alterations of clock genes in cancer, clock genes could provide important insight for translational medicine. Here, we focused on clock genes that showed significant associations with patient survival, clinical stage or tumor subtype. We identified 31 clock genes that were associated with overall patient survival in at least one cancer type (Figure 5A). Several clock genes showed features of tumor suppressor. For example, patients with low expression level of CRY2 showed significantly worse survival in LIHC (log-rank test P = 1.9 × 10−4), KIRP (P = 4.5 × 10−4) and KIRC (P = 1.4 × 10−5; Figures 5B and S5A). In contrast, several other clock genes showed oncogenic feature, such as ARNTL2, with overexpression of ARNTL2 being associated with worse survival in LGG (P = 1.7 × 10−4) and LUAD (P = 0.0022; Figure 5B).
Figure 5. Clinical relevance of clock genes across cancer types.
(A) Clinically relevant clock genes across different cancer types. The red and blue boxes indicate high and low expression in tumors associated with worse overall survival times (log-rank test FDR < 0.05), respectively. The green box shows clock genes with significantly differential expression among tumor stages (FC > 1.5; ANOVA FDR < 0.05). The arrows represent the upregulation or downregulation of clock genes in later stages (fold change of transcriptional expression between stage III/IV and stage I/II larger than 1.5). The gold box indicates significant differential expression of clock genes among tumor subtypes (FC > 1.5; ANOVA FDR < 0.05). (B) Kaplan-Meier curves of multiple cancer types stratified by median expression levels of clock genes. See also Figure S5.
We further identified 23 clock genes that were differentially expressed among tumor stages (Figure 5A). For example, CRY2 showed significantly decreased expression in stage IV KIRP (FDR = 0.013; Figure S5B). In particular, we identified several clock genes that showed upregulation or downregulation in late cancer stages. For example, HLF (FDR = 6.2 × 10−11) and PRKAA2 (FDR = 3 × 10−3) were upregulated in late stage BLCA, while PER1 (FDR = 0.0012), CRY2 (FDR = 0.013), AHR (FDR = 0.0027), RORA (FDR = 0.0014), and TNF (FDR = 7.5 × 10−5) were downregulated in late stage KIRP. These results suggest the potential involvement of clock genes in tumor progression.
Tumor subtype information often provides crucial insights to help stratify patients for predicting prognosis and selecting effective treatments (Han et al., 2014). We identified 43 clock genes differentially expressed among different cancer subtypes. For example, CRY2 is differentially expressed in KIRC subtypes (FDR = 3.2 × 10−38; Figure S5C). In BRCA, triple-negative breast cancer (TNBC) does not have the receptors that are targets of current treatment leading to poor prognosis. We identified alterations of core clock genes in TBNC relative to other breast cancer subtypes: expression levels of ARNTL2 (FDR = 1.9 × 10−7) and NPAS2 (FDR = 7.9 × 10−18) are higher in TNBC (Figure S5D), while the expression levels of CRY2 (FDR = 1.1 × 10−46), PER2 (FDR = 2.4 × 10−19), NR1D1 (FDR = 4 × 10−9), RORB (FDR = 0.0034) and RORC (FDR = 3.8 × 10−20) are lower in TNBC (Figure S5E). These results suggest potential roles of clock genes as markers for particular cancer subtypes.
Interactions between Clinically Actionable Genes and Clock Genes
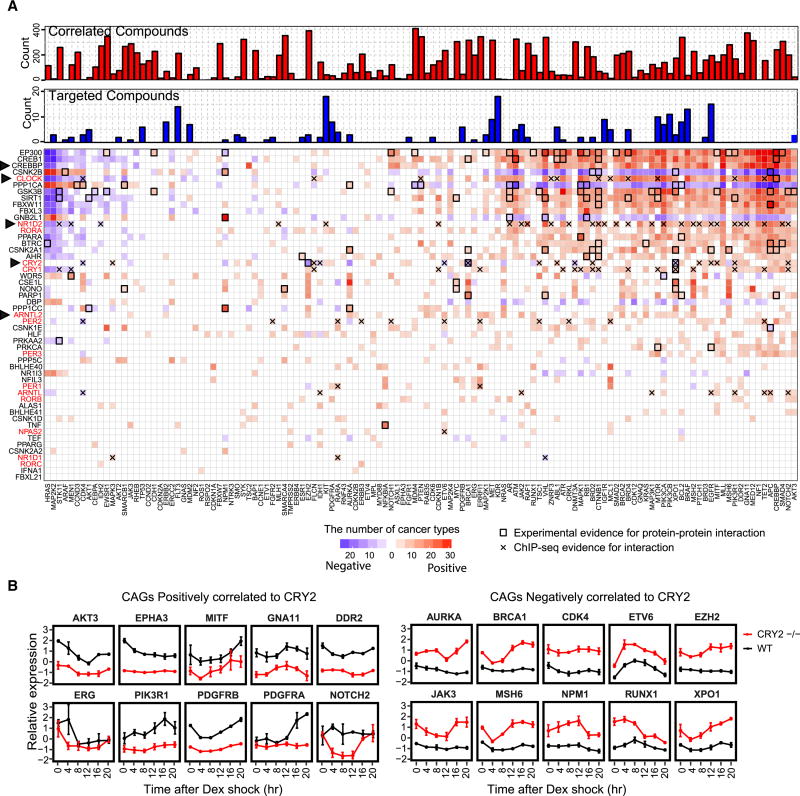
To further understand clinical implications of the clock timing, we examined correlations between transcriptional expression of clock genes and 154 clinically actionable genes (targets of FDA-approved drugs or their related marker genes), including 135 genes for targeted therapy (Van Allen et al., 2014) and 19 target genes for immunotherapy (Mak et al., 2016) across all cancer types. CREBBP, which acts as a clock transcriptional co-activator that enhances the activity of clock transcriptional activators, is also a clinically actionable gene targeted by anti-cancer drugs, such as the small molecule inhibitor C646 (Oike et al., 2014). We observed that 145 of 154 (94.2%) clinically actionable genes have significant correlations with at least one clock gene in more than five cancer types (Figures 6A and S6A). The number of clinically actionable genes correlated with clock genes ranged from 71 in LUSC to 151 in testicular germ cell tumors (TGCT; Figure S6B). The clock gene and clinically actionable gene correlation pairs ranged from 133 pairs in LUSC to 2847 pairs in thymoma (THYM; Figure S6C). For example, CRY2 shows significantly positive correlation with 31 clinically actionable genes enriched in key signaling pathways, such as PI3K/AKT, RAS and FOXO signaling pathways. CRY2 also shows significantly negative correlation with 15 clinically actionable genes (Figure 6A). Among these, CRY2 negatively correlated to CD276 and CD4, while positively correlated to TNFSF4, which are targeted genes for immunotherapy. ARNTL2 is significantly correlated to 16 of 19 (84.2%) genes targeted for immunotherapy (Figure S6A), suggesting that ARNTL2 may have an effect on cancer immunotherapy. Our results suggest that clinically actionable genes are regulated by clock genes, and highlight the significance of clock timing in the cancer treatment, including both targeted therapy and immunotherapy.
Figure 6. The expression of many clock genes and clinically actionable genes is associated with each other in cancer cells.
(A) Correlation of transcriptional expression between clock genes and clinically actionable genes. Pearson correlation coefficients |R| > 0.3; FDR < 0.05; blue: negative correlation; red: positive correlation; color scale: number of cancer types with negative or positive correlation between clock genes and clinically actionable genes. X-axis (clinically actionable genes) is ordered by the number of positively correlated clock genes minus the number of negatively correlated clock genes. Y-axis (clock genes) is ordered by the total number of correlated clinically actionable genes. If the number of cancer types is less than five, the fill color of cell is white. Triangles highlight genes discussed in the main text. Bold boxes highlight the protein-protein interactions of actionable genes and clock genes based on the experimental evidence from BioGRID and HPRD. X marks ChIP-seq evidence for interaction between clinically actionable genes and clock genes. Upper blue and red bars indicate the number of drugs that directly target (blue bars) or correlate with (red bars) actionable genes. (B) Transcript levels of clinically actionable genes (CAGs) in primary wild-type (WT) and CRY2 knock-down (CRY2 −/−) MEFs at different time points. Upper top 10 genes are positively correlated with CRY2 in cancer, while the bottom 10 genes are negatively correlated. Error bar indicates standard deviation (SD) for three biological replicates. See also Figure S6.
To further investigate the interactions between clock genes and clinically actionable genes, we reanalyzed data from a time series expression study on a CRY2 knockdown in MEF cells (Huber et al., 2016). There were decreased expression levels of clinically actionable genes that are positively correlated to CRY2, such as AKT3, EPHA3 and PIK3R1, as well as increased expression levels of clinically actionable genes that are negatively correlated to CRY2, such as JAK3, BRCA1 and CDK4, across the majority of time points in the CRY2 knockdown versus control (Figure 6B). We further used protein-protein interaction (PPI) data from the Human Protein Reference Database (Peri et al., 2004) and BioGRID (Chatr-Aryamontri et al., 2015) to identify experimentally validated protein-protein interactions between products of clock genes and clinically actionable genes (Figures 6A and S6C). For example, the clinically actionable gene EZH2 encodes a histone methyltransferase and interacts with CRY2 to regulate the expression of clock genes (Figures 6A) (Wallach et al., 2013). We further investigated the interaction between clinically actionable genes and clock genes by reanalyzing ChIP-seq data (Cho et al., 2012; Koike et al., 2012; Wu et al., 2017) (Figure 6A). Our results suggest diverse effects of clock genes on clinically actionable genes. Therefore, the significant interactions between clinically actionable genes and clock genes may affect drug responses, and the circadian timing should be necessarily considered in cancer therapy.
Potential Therapeutic Effects of Clock Genes
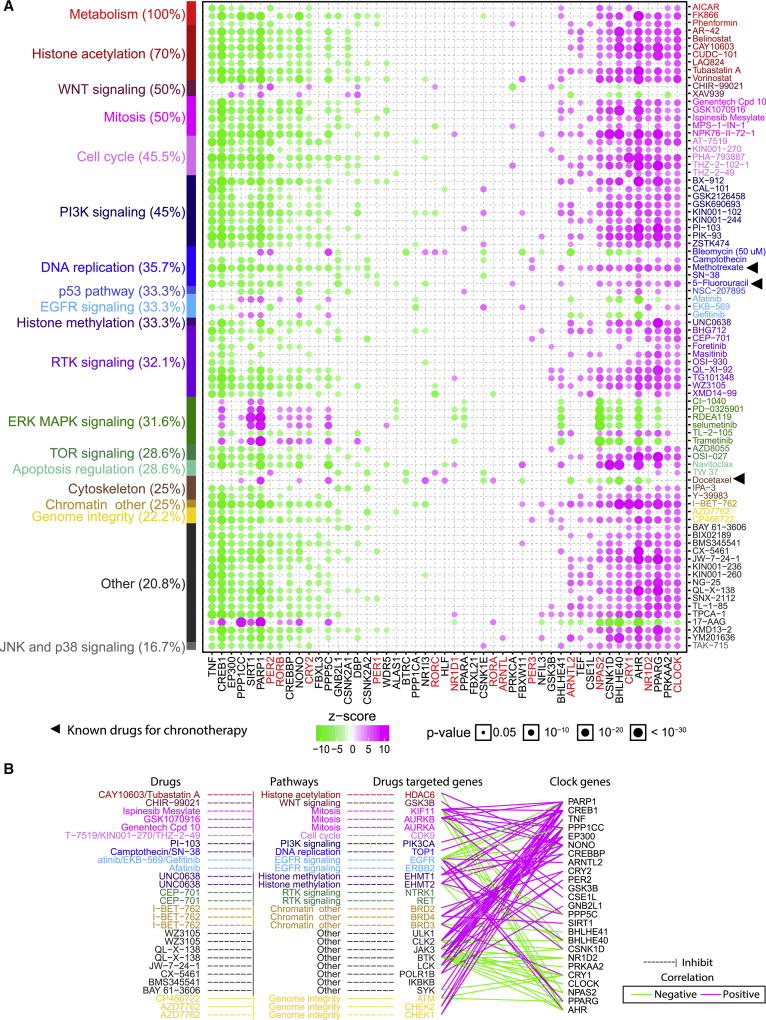
Many clinically actionable genes are targeted by anti-cancer drugs, as identified in the Cancer Therapeutics Response Portal (CTRP) and the Genomics of Drug Sensitivity in Cancer (GDSC) project. For example, EGFR, which is targeted by 15 anti-cancer drugs and correlated with 187 anti-cancer drugs (Figure 6A, upper panel), correlates with 21 clock genes, five of which showed validated protein-protein interactions (Figures 6A). Thus, following circadian timing for drug administration may largely improve the efficacy of a drug and reduce its toxicity (Levi et al., 2010). To further evaluate the potential effects of clock genes on drug response, we analyzed the correlation between drug sensitivity of 265 anti-cancer drugs and transcriptional expression of 51 clock genes across 1080 cancer cell lines from GDSC using the Z-score as previously described (Figure 7A) (Rees et al., 2015). We identified 2059 significant correlation pairs between drug sensitivity and transcriptional expression of clock genes (Figure S7B). Among these, sensitivity to 82 anti-cancer drugs significantly correlated with at least ten clock genes (Figure 7A). Previous studies showed that several drugs have utility in chronotherapy (Levi et al., 2010), such as 5-fluorouracil (Wood et al., 2006), methotrexate (Ohdo et al., 1997) and docetaxel (Tampellini et al., 1998). We confirmed these observations in our analysis, which showed that 5-flurouracil sensitivity (negative correlation) is associated with transcriptional expression of CRY2, PER2, PER1, RORB and ten other clock genes, and resistance (positive correlation) is associated with transcriptional expression of NPAS2, CLOCK, NR1D2, AHR and seven other clock genes. Furthermore, we revealed the possibility that other drugs may also be used in chronotherapy. For example, we showed that phenformin, which targets metabolism, is highly correlated with CRY2 (z-score = 4.5; P = 8 × 10−8), PER2 (z-score = −5.99; P = 2.3 × 10−9), CSNK1D (z-score = 6.04; P = 1.7 × 10−9) and 15 other clock genes. In response to anti-cancer drugs, clock genes are highly correlated with drugs which targeted important pathways, such as metabolism (100% of drugs tested), chromatin histone acetylation (70.0%), WNT signaling pathway (50.0%), cell cycle (45.5%), PI3K signaling (45.0%), DNA replication (35.7%) and p53 pathway (33.3%). Our data show two groups of clock genes with similar associations with response to drugs. One group includes CRY2 and PER2 (Figure 7A, left panel of heatmap), which are associated with sensitivity to a majority of the drugs, while another group includes CLOCK and ARNTL2 (Figure 7A, right panel of heatmap), which are associated with resistance to the majority of drugs. These two groups largely depend on high correlation among clock genes across cancer cell lines (Figure S7C).
Figure 7. Drug effects of clock genes in chronotherapy.
(A) Correlation between drug sensitivity (AUCs) and gene expression of clock genes in at least 10 clock genes (green: negative correlation; magenta: positive correlation; size: p-value). Black triangles indicate compound verified in chronotherapy. Color bars in Y-axis indicate drugs (right) and their targeted pathways (left). (B) Correlation between clock genes and known drugs that target the genes (green: negative correlation; magenta: positive correlation), which corresponds to significant correlation between drug sensitivity and clock genes. See also Figure S7.
For drugs with known targeted genes, we further estimated the correlations between targeted genes and clock genes (Figure 7B). For example, ATM, the target of CP466722, is negatively correlated with clock genes, including PPARG (R = −0.29) and CRY1 (R = −0.28), and positively correlated with CREB1 (R = 0.28). This result suggests a potential CP466772 strategy that involves high expression levels of PPARG/CRY1 and/or low expression levels of CREB1 for optimal drug effects. Several targeted genes such as HDAC6, ERBB2, EGFR, SYR, ATM, KIT, JAK3, MTOR and JAK2 are CAGs, which are highly correlated to clock genes in cancer cell lines (Figure 7B). This further supports the concept that circadian timing could contribute to response to drug therapy.
To further investigate the potential effects of clock genes on drug sensitivity, we examined the correlation between transcriptional expression of clock genes in the Cancer Cell Line Encyclopedia (CCLE) dataset (Barretina et al., 2012) and drug sensitivity in the CTRP dataset. Among 481 compounds, we identified 329 compounds that have at least one significant correlation with clock genes, with 3577 significant correlation pairs (Figure S7D). We found 163 compounds that showed significant correlation with at least ten clock genes (Figure S7E), and 76 drugs are shared between the GDSC and CTRP databases. Among them, 53 drugs in both CRTP and GDSC have at least one pair that significantly correlates with clock genes. These results show the consistency of circadian effects on drug response.
DISCUSSION
Cancer chronotherapy has been applied to optimize clock-controlled drug metabolism, detoxification, pharmacokinetics and drug efficacy (Levi et al., 2010). However, there are limited studies in large number of patient samples showing that circadian timing can strongly improve the efficacy of a drug and reduce its toxicity. In this study, we utilized multi-dimensional omics data and clinical data across 32 cancer types from TCGA, and pharmacogenomic data from CTRP and GDSC. The schematic of the overall analysis was outlined in Figure S1. We revealed global alterations of clock genes at transcriptional, genetic and epigenetic levels. Our results suggested several clock genes may function as oncogene, such as ARNTL2, NR1D1, and NPAS2, while several other clock genes may function as tumor suppressors, such as PERs, CRYs, RORs. We revealed disruption of circadian rhythms across patient samples by showing significantly reduced number of genes correlated to clock genes. We further showed that transcriptional dysregulation of clock genes is strongly associated with patient survival, tumor stage and subtype. Then, we identified global interactions between clock genes and CAGs through co-expression, protein-protein interaction, and ChIP-seq data. This was further supported by the large-scale pharmacogenomic data from CTRP and GDSC that clock genes could largely affect anti-cancer drug sensitivity in cancer cell lines. Our study provides a comprehensive analysis of the circadian clock across different cancer types and highlights potential clinical utility of cancer chronotherapy.
Supporting the development of cancer chronotherapy, we revealed pervasive transcriptional dysregulation of clock genes across different cancer types, including changes in clock genes that are not well-characterized, such as RORB. We observed cancer-specific patterns for several genes across different cancer types, consistent with previous finding that genetic elements may play different roles in different tumor environments (Han et al., 2015). Furthermore, the expression changes in clock genes are highly associated with the activity of oncogenic pathways. We observed that paralog clock genes could play synergistic or different roles to alter oncogenic pathways, further suggesting a complex regulation of clock transcriptional loops. Despite the relatively low mutation frequency of clock genes, combinations of mutations in multiple colck genes may provide prognostic value. For example, higher frequency mutation rate of core clock genes, especially PER1/2/3, will lead to worse survival in BRCA. Our comprehensive analysis of clock genes at transcriptional, genetic and epigenetic level is the crucial step to understand the functional roles of clock genes across different cancer types.
Circadian rhythm disruption has been linked to tumorigenesis, and circadian timing is highly associated with efficacy and toxicity of cancer treatment (Fu and Lee, 2003; Kiessling and Cermakian, 2017; Levi et al., 2010; Zhang et al., 2017). Our study showed a global disruption of circadian rhythms in both patient samples and cancer cell lines. In cancer patients, number of genes correlated to clock genes is largely reduced, suggesting the disruption of circadian rhythm. Genes with circadian oscillation pattern in the cancer cell line (MCF7) are disrupted in the noncancerous cell line (MCF10A). We also showed genes that specifically established circadian rhythm in the MCF7 compare to MCF10A. Therefore, there is an urgent need to consider the disruption and/or reprogramming of circadian rhythm in cancer treatment. The present study represents a comprehensive analysis of the global alterations of circadian rhythms across a broad range of cancer types.
Furthermore, we identified clock genes that have potential clinical relevance based on associations between the expression levels of clock genes and survival times, tumor stage or subtype. Multiple clock genes are associated with patient survival and differentially expressed among tumor stages, suggesting potential alterations of clock genes during tumor progression. Furthermore, clock genes showed significant difference for distinct patient subtypes within a cancer type, and may serve as promising biomarkers. Our results highlight the possible clinical utility of clock genes in human cancer.
Finally, we showed that the majority of clinically actionable genes strongly correlate and interact with clock genes, suggesting that the circadian timing should be considered in cancer therapy. We observed the positive correlation between clinically actionable genes and several core clock genes, including PER2, CRY2 and NR1D2, which are known to be transcription suppressors in the clock transcriptional-translational loops. These core clock genes could also act as transcriptional activators. For example, NR1D2 can trans-activate the expression of Srebp-1 c (Ramakrishnan et al., 2009), and PER1/2 positively prolong the period of rhythmic gene expression by suppressing CRY activity (Akashi et al., 2014). Previous studies showed that both transcription activator and suppressor could bind to the same gene (Bugge et al., 2012; Koike et al., 2012). Furthermore, our analysis showed the expression of NR1D2 is positively correlated with CLOCK, the well-known transcriptional activator, across 24 cancer types (Figure S6D). Taken together, our results suggest the complicated regulation of transcription factors and the clock transcriptional-translational loops. Consistent with this finding, we showed that clock genes are highly correlated with sensitivity to a majority of anti-cancer drugs, as well as drug-targeted genes across cancer cell lines. A previous study demonstrated 42 anti-cancer drugs involved in chronotoxicity/chronoefficacy (Levi et al., 2010). Among these, ten drugs are also listed in GDSC. All of them are significantly correlated with the expression of at least one clock gene, while three are significantly correlated with the expression of at least ten clock genes, including 5-fluorouracil, methotrexate and docetaxel. Taken together, our results provided strong evidence that further efforts should be made to personalize or optimize cancer chronotherapy by delivering the drug at the optimal timing.
There are several limitations in our study. For example, the time points for obtaining biospecimens from cancer patients are not available. As numerous cancer patients keep circadian rhythms, the omics data may largely vary over the 24 hour period. Therefore, large-scale patient samples at different time points are necessary to further understand the critical roles of clock genes in cancer. Furthermore, drug response in cancer cell lines is not always consistent with patient survival, thus the drug response data in patient samples is necessary to further optimize cancer chronotherapy.
STAR*Methods
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact Leng Han (leng.han@uth.tmc.edu).
Quantification and Statistical Analysis
mRNA Expression, Protein Expression and Signaling Pathways
Normalized gene expression data based on expectation maximization (RSEM) (Li and Dewey, 2011) was downloaded from TCGA data portal (http://gdac.broadinstitute.org/). Genes were considered differential expression between tumor and normal paired samples if the fold-change > 1.5 and t-test FDR < 0.05. The normal sample was extracted from the same adjacent non-tumorigenic tissue in the same patient (Davis et al., 2014; The Cancer Genome Atlas Network et al., 2013).
Protein expression based on reverse phase protein array (RPPA) from The Cancer Proteome Atlas (http://tcpaportal.org/tcpa/download.html) (Li et al., 2013a). RPPA data were median-centered and normalized by standard deviation across all tumor samples for each component to access normalized protein expression data. The pathway score of ten key signaling pathways based on protein expression was calculated for each tumor sample as the sum of the relative protein levels of all positive regulatory components minus the equivalent sum for the negative regulatory components (Akbani et al., 2014). Antibodies targeting different phosphorylated forms for the same protein with Pearson correlation coefficient larger than 0.85 were averaged. Based on median expression of each clock gene in each cancer type, tumor samples are classified as high expression and low expression groups. For interactive effects of clock paralog genes, tumor samples were divided into two groups with low expression or high expression of all paralog genes. Then we used a Student t test to assess statistical differences in pathway score between these two groups with p value < 0.05. Under statistical significance, the expression of individual or paralog clock genes positively correlated with the pathway score indicates their association with activation of the pathway, while the negative correlation indicates the association with inhibition of the pathway.
Disruption of Circadian Rhythm in Cancer
To evaluate the disruption of clock genes in tumor, we used the number of genes correlated with clock genes in tumor minus the number of genes correlated with clock genes in normal samples. Correlation between the transcriptional expression of clock genes and other genes was calculated based on Pearson’s correlation, and considered significant if |R| > 0.5 and FDR < 0.05. The correlation in tumor samples is corrected by tumor purity (http://cistrome.org/TIMER/download.html) (Li et al., 2016).
Mutation and DNA Methylation Analysis
We obtained the mutation data (MAF files) and DNA methylation 450K data from the TCGA data portal (http://gdac.broadinstitute.org/). We filtered out the samples with more than 1000 mutations in the exome to eliminate the potential bias induced by ultra-mutation samples (Yuan et al., 2016). The number and frequency of non-silent somatic mutations across all samples calculated per gene in each cancer type. To assess the effect of promoter DNA methylation on clock gene expression, we kept the methylation probes involved in −1,500 bp to +500 bp around TSS (Amabile et al., 2015), and generated a one-to-one gene-methylation probe mapped by retaining the maximal Beta value change between normal samples and tumor samples. We defined the Beta value of DNA methylation as a change greater than 0.2 between normal and tumor pairs, and FDR < 0.05 in the promoter as dysregulated promoter DNA methylation (Bibikova et al., 2011). We performed Spearman’s correlation analysis for the promoter DNA methylation and clock gene expression and retained the minimum Spearman correlation coefficient.
Interactions and Correlations between Clock Genes and Clinically Actionable Genes
To obtain a list of clinically actionable genes (CAGs), we first obtained 135 genes from a previous study (http://archive.broadinstitute.org/cancer/cga/target) (Van Allen et al., 2014). We also obtained 19 targeted genes for immunotherapy from a previous study (Mak et al., 2016). For correlation between transcriptional expression of clock genes and clinically actionable genes in tumor samples, we used Pearson’s correlation and considered |R| > 0.3 and FDR < 0.05 as significant correlation.
Protein-protein interaction (PPI) data downloaded from the Human Protein Reference Database (HPRD; http://www.hprd.org/) and BioGRID (https://thebiogrid.org/). ChIP-seq data for clock genes were downloaded from Gene Expression Omnibus (GEO, https://www.ncbi.nlm.nih.gov/geo/; GSE39860, GSE69100 and GSE34019) (Cho et al., 2012; Koike et al., 2012b; Wu et al., 2017). The ChIP-seq data was analyzed by HOMER (http://homer.salk.edu/homer/) (Heinz et al., 2010).
Identification of Clinically Relevant Clock Genes
Clinical information for cancer patients, including patients’ overall survival, disease stages, and tumor subtypes, were obtained from TCGA data portal (http://gdac.broadinstitute.org/). A log-rank test was performed to assess the association between transcript expression of clock genes and overall survival. Transcript expression levels were dichotomized by the median level, and FDR < 0.05 was considered as statistical significance. We used Kruskal-Wallis nonparametric analysis of variance (ANOVA) to detect clock genes with differential expression among different tumor subtypes and disease stages, and considered FDR < 0.05 and fold-change of maximal differential expression > 1.5 to be a significant difference.
Time Series Expression Analysis for Cancerous and Non-cancerous Cell Lines
We downloaded time series cDNA microarray data of MCF7 and MCF10A from GSE76370 (Gutiérrez-Monreal et al., 2016). Oscillation patterns of genes were predicted by MetaCycle::meta2d (JTK_CYCLE and meta2d adjust P < 0.05; Wu et al., 2016). We also downloaded time series CRY2 knockdown RNA-seq data for the MEF cell line from GEO (GSE89018) (Huber et al., 2016).
Analysis of Drug Sensitivity and Gene Expression
Normalized gene expression profiles for ~1,000 cancer cell lines and drug response measurements as area under the curves (AUCs) were downloaded from GDSC (http://www.cancerrxgene.org/downloads). Gene-centric RMA-normalized mRNA expression data measured on the Affymetrix GeneChip Human Genome U133 Plus 2.0 Array were downloaded from the CCLE (https://portals.broadinstitute.org/ccle/data). Drug response (AUCs) for 481 small molecules across cancer cell lines was downloaded from CTRP (https://ocg.cancer.gov/programs/ctd2/data-portal). To evaluate the correlation between small-molecule sensitivity and basal gene expression levels, we performed an analysis as previously described (Rees et al., 2015). Briefly, the Pearson correlation coefficients of transcript levels and AUCs were normalized using Fisher’s Z transformation. A Bonferroni-corrected, two-tailed distribution with family-wise error rate less than 0.025 in each tail for z-scored Pearson correlation coefficients of annotated drug-target pairs (242 pairs in GDSC; 662 pairs in CTRP) were compared to the same number of correlation pairs generated by randomly sampling correlations (Figure S7A). We determined the threshold as |z| > 3.89 and |z| > 4.13 for drug sensitivity from GDSC and CTRP, respectively.
Supplementary Material
Key Resource Table
Highlight.
Transcription dysregulation and clinical relevance of clock genes in cancer.
Disruption and reprogramming of circadian rhythms in cancer.
Strong interactions between clock genes and clinically actionable genes.
Potential therapeutic effects of clock genes in cancer chronotherapy.
Acknowledgments
This work was supported by Cancer Prevention & Research Institute of Texas (RR150085 to L.H.); National Institute of Health (AG045828 to Z.C., and GM114424 to S.-H.Y.); The Welch Foundation (AU-1731 to Z.C.); National Institutes of Health (R00DK094981 and R01 CA218025 to C. Lin); China Scholarship Council (201606160058 to C. Liu); National Natural Science Foundation of China (31771458 to A. Guo). J.S.T. is an Investigator in the Howard Hughes Medical Institute. We gratefully acknowledge contributions from the TCGA Research Network. We thank LeeAnn Chastain for editorial assistance.
DECLARATION OF INTERESTS
G.B.M. has sponsored research support from AstraZeneca and is on the Scientific Advisory Board for AstraZeneca, ImmunoMet, Nuevolution, and Precision Medicine. J.S.T is a cofounder and Scientific Advisory Board member of Reset Therapeutics, Inc., a biotech company working on circadian rhythms and metabolism.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
L.Han conceived and supervised the project. Y. Ye, S. Yoo, and L.Han designed and performed the research. Y. Xiang, F.M.O., Y. Kim, P.K. Park, Q. Hu, C. Lin, C. Liu, L. Diao, Y. Lou, A. Guo, B. Zhou and L. Wang performed data analysis. Y. Ye, Z.C., J.S.T., G.B.M., S. Yoo, and L. Han interpreted results. Y. Ye, G.B.M., and L.Han wrote the manuscript with input from all other authors.
References
- Akashi M, Okamoto A, Tsuchiya Y, Todo T, Nishida E, Node K. A Positive Role for PERIOD in Mammalian Circadian Gene Expression. Cell Rep. 2014;7:1056–1064. doi: 10.1016/j.celrep.2014.03.072. [DOI] [PubMed] [Google Scholar]
- Akbani R, Ng PKS, Werner HMJ, Shahmoradgoli M, Zhang F, Ju Z, Liu W, Yang J-Y, Yoshihara K, Li J, et al. A pan-cancer proteomic perspective on The Cancer Genome Atlas. Nat. Commun. 2014;5:3887. doi: 10.1038/ncomms4887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Allen EM, Wagle N, Stojanov P, Perrin DL, Cibulskis K, Marlow S, Jane-Valbuena J, Friedrich DC, Kryukov G, Carter SL, et al. Whole-exome sequencing and clinical interpretation of formalin-fixed, paraffin-embedded tumor samples to guide precision cancer medicine. Nat. Med. 2014;20:682–688. doi: 10.1038/nm.3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amabile G, Di Ruscio A, Müller F, Welner RS, Yang H, Ebralidze AK, Zhang H, Levantini E, Qi L, Martinelli G, et al. Dissecting the role of aberrant DNA methylation in human leukaemia. Nat. Commun. 2015;6:7091. doi: 10.1038/ncomms8091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballesta A, Innominato PF, Dallmann R, Rand DA, Lévi FA. Systems Chronotherapeutics. Pharmacol. Rev. 2017;69:161–199. doi: 10.1124/pr.116.013441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin Aa, Kim S, Wilson CJ, Lehár J, Kryukov GV, Sonkin D, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity Supp. Nature. 2012;483:603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibikova M, Barnes B, Tsan C, Ho V, Klotzle B, Le JM, Delano D, Zhang L, Schroth GP, Gunderson KL, et al. High density DNA methylation array with single CpG site resolution. Genomics. 2011;98:288–295. doi: 10.1016/j.ygeno.2011.07.007. [DOI] [PubMed] [Google Scholar]
- Brady JJ, Chuang CH, Greenside PG, Rogers ZN, Murray CW, Caswell DR, Hartmann U, Connolly AJ, Sweet-Cordero EA, Kundaje A, et al. An Arntl2-Driven Secretome Enables Lung Adenocarcinoma Metastatic Self-Sufficiency. Cancer Cell. 2016;29:697–710. doi: 10.1016/j.ccell.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugge A, Feng D, Everett LJ, Briggs ER, Mullican SE, Wang F, Jager J, Lazar MA. Rev-Erbα and Rev-Erbβ Protect the Circadian Clock and Metabolic Function. 2012:657–667. doi: 10.1101/gad.186858.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatr-Aryamontri A, Breitkreutz BJ, Oughtred R, Boucher L, Heinicke S, Chen D, Stark C, Breitkreutz A, Kolas N, O’Donnell L, et al. The BioGRID interaction database: 2015 update. Nucleic Acids Res. 2015;43:D470–D478. doi: 10.1093/nar/gku1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H, Zhao X, Hatori M, Yu RT, Barish GD, Lam MT, Chong L-W, DiTacchio L, Atkins AR, Glass CK, et al. Regulation of circadian behaviour and metabolism by REV-ERB-α and REV-ERB-β. Nature. 2012;485:123–127. doi: 10.1038/nature11048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dallmann R, Okyar A, Lévi F. Dosing-Time Makes the Poison: Circadian Regulation and Pharmacotherapy. Trends Mol. Med. 2016;22:430–445. doi: 10.1016/j.molmed.2016.03.004. [DOI] [PubMed] [Google Scholar]
- Davis CF, Ricketts CJ, Wang M, Yang L, Cherniack AD, Shen H, Buhay C, Kang H, Kim SC, Fahey CC, et al. The Somatic Genomic Landscape of Chromophobe Renal Cell Carcinoma. Cancer Cell. 2014;26:319–330. doi: 10.1016/j.ccr.2014.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulong S, Ballesta A, Okyar A, Levi F. Identification of Circadian Determinants of Cancer Chronotherapy through In Vitro Chronopharmacology and Mathematical Modeling. Mol. Cancer Ther. 2015;14:2154–2164. doi: 10.1158/1535-7163.MCT-15-0129. [DOI] [PubMed] [Google Scholar]
- Eckel-Mahan K, Sassone-Corsi P. Metabolism and the Circadian Clock Converge. Am. Physiol. Soc. 2013;93:107–135. doi: 10.1152/physrev.00016.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Athman R, Genov NN, Mazuch J, Zhang K, Yu Y, Fuhr L, Abreu M, Li Y, Wallach Thomas, et al. The Ink4a/Arf locus operates as a regulator of the circadian clock modulating RAS activity. 2017 doi: 10.1371/journal.pbio.2002940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Focan C. Sequential chemotherapy and circadian rhythm in human solid tumours. A randomised trial. Cancer Chemother. Pharmacol. 1979;3:197–202. doi: 10.1007/BF00262422. [DOI] [PubMed] [Google Scholar]
- Fu L, Lee CC. The circadian clock: pacemaker and tumour suppressor. Nat. Rev. Cancer. 2003;3:350–361. doi: 10.1038/nrc1072. [DOI] [PubMed] [Google Scholar]
- Fu L, Pelicano H, Liu J, Huang P, Lee CC. The circadian gene Period2 plays an important role in tumor suppression and DNA damage response in vivo. Cell. 2002;111:41–50. doi: 10.1016/s0092-8674(02)00961-3. [DOI] [PubMed] [Google Scholar]
- Gong J, Li Y, Liu C, Xiang Y, Li C, Ye Y, Zhang Z, Hawke DH, Park PK, Diao L, et al. A Pan-cancer Analysis of the Expression and Clinical Relevance of Small Nucleolar RNAs in Human Cancer. Cell Rep. 2017;21:1–14. doi: 10.1016/j.celrep.2017.10.070. [DOI] [PubMed] [Google Scholar]
- Guerrero-Vargas NN, Navarro-Espíndola R, Guzmán-Ruíz MA, Basualdo M del C, Espitia-Bautista E, López-Bago A, Lascurain R, Córdoba-Manilla C, Buijs RM, Escobar C. Circadian disruption promotes tumor growth by anabolic host metabolism; experimental evidence in a rat model. BMC Cancer. 2017;17:625. doi: 10.1186/s12885-017-3636-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutiérrez-Monreal MA, Treviño V, Moreno-Cuevas JE, Scott S-P. Identification of circadian-related gene expression profiles in entrained breast cancer cell lines. Chronobiol. Int. 2016;33:392–405. doi: 10.3109/07420528.2016.1152976. [DOI] [PubMed] [Google Scholar]
- Ha N, Long J, Cai Q, Shu XO, Hunter KW. The Circadian Rhythm Gene Arntl2 Is a Metastasis Susceptibility Gene for Estrogen Receptor-Negative Breast Cancer. 2016:1–20. doi: 10.1371/journal.pgen.1006267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han L, Yuan Y, Zheng S, Yang Y, Li J, Edgerton ME, Diao L, Xu Y, Verhaak RGW, Liang H. The Pan-Cancer analysis of pseudogene expression reveals biologically and clinically relevant tumour subtypes. Nat. Commun. 2014;5:3963. doi: 10.1038/ncomms4963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han L, Diao L, Yu S, Xu X, Li J, Zhang R, Yang Y, Werner HMJ, Eterovic AK, Yuan Y, et al. The Genomic Landscape and Clinical Relevance of A-to-I RNA Editing in Human Cancers. Cancer Cell. 2015;28:515–528. doi: 10.1016/j.ccell.2015.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He B, Nohara K, Park N, Park YS, Guillory B, Zhao Z, Garcia JM, Koike N, Lee CC, Takahashi JS, et al. The Small Molecule Nobiletin Targets the Molecular Oscillator to Enhance Circadian Rhythms and Protect against Metabolic Syndrome. Cell Metab. 2016;23:610–621. doi: 10.1016/j.cmet.2016.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. Simple Combinations of Lineage-Determining Transcription Factors Prime cis-Regulatory Elements Required for Macrophage and B Cell Identities. Mol. Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman AE, Zheng T, Ba Y, Zhu Y. The circadian gene NPAS2, a putative tumor suppressor, is involved in DNA damage response. Mol. Cancer Res. 2008;6:1461–1468. doi: 10.1158/1541-7786.MCR-07-2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hojo H, Enya S, Arai M, Suzuki Y, Nojiri T, Kangawa K, Koyama S, Kawaoka S. Remote reprogramming of hepatic circadian transcriptome by breast cancer. Oncotarget. 2017;8:34128–34140. doi: 10.18632/oncotarget.16699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Horst GT, Muijtjens M, Kobayashi K, Takano R, Kanno S, Takao M, de Wit J, Verkerk A, Eker aP, van Leenen D, et al. Mammalian Cry1 and Cry2 are essential for maintenance of circadian rhythms. Nature. 1999;398:627–630. doi: 10.1038/19323. [DOI] [PubMed] [Google Scholar]
- Hua H, Wang Y, Wan C, Liu Y, Zhu B, Yang C, Wang X, Wang Z, Cornelissen-Guillaume G, Halberg F. Circadian gene mPer2 overexpression induces cancer cell apoptosis. Cancer Sci. 2006;97:589–596. doi: 10.1111/j.1349-7006.2006.00225.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber AL, Papp SJ, Chan AB, Henriksson E, Jordan SD, Kriebs A, Nguyen M, Wallace M, Li Z, Metallo CM, et al. CRY2 and FBXL3 Cooperatively Degrade c-MYC. Mol. Cell. 2016;64:774–789. doi: 10.1016/j.molcel.2016.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley JM, Loros JJ, Dunlap JC. Circadian Oscillators: Around the Transcription–Translation Feedback Loop and on to Output. Trends Biochem. Sci. 2016;41:834–846. doi: 10.1016/j.tibs.2016.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innominato PF, Giacchetti S, Bjarnason GA, Focan C, Garufi C, Coudert B, Iacobelli S, Tampellini M, Durando X, Mormont MC, et al. Prediction of overall survival through circadian rest-activity monitoring during chemotherapy for metastatic colorectal cancer. Int. J. Cancer. 2012;131:2684–2692. doi: 10.1002/ijc.27574. [DOI] [PubMed] [Google Scholar]
- Kiessling S, Cermakian N. The tumor circadian clock : a new target for cancer therapy ? Futur. Oncol. 2017;13:2607–2610. doi: 10.2217/fon-2017-0456. [DOI] [PubMed] [Google Scholar]
- Ko CH, Takahashi JS. Molecular components of the mammalian circadian clock. Hum. Mol. Genet. 2006;15:271–277. doi: 10.1093/hmg/ddl207. [DOI] [PubMed] [Google Scholar]
- Koike N, Yoo S-H, Huang H-C, Kumar V, Lee C, Kim T-K, Takahashi JS. Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science. 2012;338:349–354. doi: 10.1126/science.1226339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamia KA. Ticking time bombs: connections between circadian clocks and cancer. F1000Research. 2017;6:1910. doi: 10.12688/f1000research.11770.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Lee J, Kwon I, Nakajima Y, Ohmiya Y, Son GH, Lee KH, Kim K. Coactivation of the CLOCK-BMAL1 complex by CBP mediates resetting of the circadian clock. J. Cell Sci. 2010;123:3547–3557. doi: 10.1242/jcs.070300. [DOI] [PubMed] [Google Scholar]
- Levi F, Okyar A, Dulong S, Innominato PF, Clairambault J, Lévi F. Circadian timing in cancer treatments. Annu. Rev. Pharmacol. Toxicol. 2010;50:377–421. doi: 10.1146/annurev.pharmtox.48.113006.094626. [DOI] [PubMed] [Google Scholar]
- Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Severson E, Pignon J-C, Zhao H, Li T, Novak J, Peng J, Shen H, Aster JC, Rodig S, et al. Comprehensive analyses of tumor immunity: implications for cancer immunotherapy. Genome Biol. 2016:1–16. doi: 10.1186/s13059-016-1028-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Lu Y, Akbani R, Ju Z, Roebuck PL, Liu W, Yang J-Y, Broom BM, Verhaak RGW, Kane DW, et al. TCPA: a resource for cancer functional proteomics data. Nat. Methods. 2013a;10:1046–1047. doi: 10.1038/nmeth.2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XM, Mohammad-Djafari A, Dumitru M, Dulong S, Filipski E, Siffroi-Fernandez S, Mteyrek A, Scaglione F, Guettier C, Delaunay F, et al. A circadian clock transcription model for the personalization of cancer chronotherapy. Cancer Res. 2013b;73:7176–7188. doi: 10.1158/0008-5472.CAN-13-1528. [DOI] [PubMed] [Google Scholar]
- Mak MP, Tong P, Diao L, Cardnell RJ, Gibbons DL, William WN, Skoulidis F, Parra ER, Rodriguez-Canales J, Wistuba II, et al. A Patient-Derived, Pan- Cancer EMT Signature Identifies Global Molecular Alterations and Immune Target Enrichment Following Epithelial-to-Mesenchymal Transition. 2016 doi: 10.1158/1078-0432.CCR-15-0876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masri S, Papagiannakopoulos T, Kinouchi K, Liu Y, Cervantes M, Baldi P, Jacks T, Sassone-Corsi P. Lung Adenocarcinoma Distally Rewires Hepatic Circadian Homeostasis. Cell. 2016;165:896–909. doi: 10.1016/j.cell.2016.04.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzoccoli G, Pazienza V, Panza A, Valvano MR, Benegiamo G, Vinciguerra M, Andriulli A, Piepoli A. ARNTL2 and SERPINE1: Potential biomarkers for tumor aggressiveness in colorectal cancer. J. Cancer Res. Clin. Oncol. 2012;138:501–511. doi: 10.1007/s00432-011-1126-6. [DOI] [PubMed] [Google Scholar]
- Mormont M, Waterhouse J, Bleuzen P, Mormont M, Waterhouse J, Bleuzen P, Giacchetti S, Jami A, Misset J, Touitou Y, et al. Marked 24-h Rest / Activity Rhythms Are Associated with Better Quality of Life , Better Response , and Longer Survival in Patients with Metastatic Colorectal Cancer and Good Performance Status. Clin. Cancer Res. 2000;6:3038–3045. [PubMed] [Google Scholar]
- Ohdo S, K I, E Y, S H, S N, N O. Chronotoxicity of methotrexate in mice and its relation to circadian rhythm of DNA synthesis and pharmacokinetics. 1997;75:283–290. doi: 10.1254/jjp.75.283. [DOI] [PubMed] [Google Scholar]
- Oike T, Komachi M, Ogiwara H, Amornwichet N, Saitoh Y, Torikai K, Kubo N, Nakano T, Kohno T. C646, a selective small molecule inhibitor of histone acetyltransferase p300, radiosensitizes lung cancer cells by enhancing mitotic catastrophe. Radiother. Oncol. 2014;111:222–227. doi: 10.1016/j.radonc.2014.03.015. [DOI] [PubMed] [Google Scholar]
- Papagiannakopoulos T, Bauer MR, Davidson SM, Heimann M, Subbaraj L, Bhutkar A, Bartlebaugh J, Vander Heiden MG, Jacks T. Circadian Rhythm Disruption Promotes Lung Tumorigenesis. Cell Metab. 2016;24:324–331. doi: 10.1016/j.cmet.2016.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peri S, Navarro JD, Kristiansen TZ, Amanchy R, Surendranath V, Muthusamy B, Gandhi TKB, Chandrika KN, Deshpande N, Suresh S, et al. Human protein reference database as a discovery resource for proteomics. Nucleic Acids Res. 2004;32:D497–D501. doi: 10.1093/nar/gkh070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puram RV, Kowalczyk MS, De Boer CG, Schneider RK, Miller PG, McConkey M, Tothova Z, Tejero H, Heckl D, Järås M, et al. Core Circadian Clock Genes Regulate Leukemia Stem Cells in AML. Cell. 2016;165:303–316. doi: 10.1016/j.cell.2016.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakrishnan SN, Lau P, Crowther LM, Cleasby ME, Millard S, Leong GM, Cooney GJ, Muscat GEO. Rev-erb beta regulates the Srebp-1c promoter and mRNA expression in skeletal muscle cells. Biochem. Biophys. Res. Commun. 2009;388:654–659. doi: 10.1016/j.bbrc.2009.08.045. [DOI] [PubMed] [Google Scholar]
- Ramanathan C, Xu H, Khan SK, Shen Y, Gitis PJ, Welsh DK, Hogenesch JB, Liu AC. Cell Type-Specific Functions of Period Genes Revealed by Novel Adipocyte and Hepatocyte Circadian Clock Models. PLoS Genet. 2014;10 doi: 10.1371/journal.pgen.1004244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rees MG, Seashore-Ludlow B, Cheah JH, Adams DJ, Price EV, Gill S, Javaid S, Coletti ME, Jones VL, Bodycombe NE, et al. Correlating chemical sensitivity and basal gene expression reveals mechanism of action. Nat. Chem. Biol. 2015;12:1–10. doi: 10.1038/nchembio.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Relógio A, Westermark PO, Wallach T, Schellenberg K, Kramer A, Herzel H. Tuning the mammalian circadian clock: Robust synergy of two loops. PLoS Comput. Biol. 2011;7:1–18. doi: 10.1371/journal.pcbi.1002309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Relógio A, Thomas P, Medina-Pérez P, Reischl S, Bervoets S, Gloc E, Riemer P, Mang-Fatehi S, Maier B, Schäfer R, et al. Ras-Mediated Deregulation of the Circadian Clock in Cancer. PLoS Genet. 2014;10 doi: 10.1371/journal.pgen.1004338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahar S, Sassone-Corsi P. Metabolism and cancer: the circadian clock connection. Nat. Rev. Cancer. 2009;9:886–896. doi: 10.1038/nrc2747. [DOI] [PubMed] [Google Scholar]
- Sancar A, Lindsey-Boltz LA, Kang T-H, Reardon JT, Lee JH, Ozturk N. Circadian clock control of the cellular response to DNA damage. FEBS Lett. 2010;584:2618–2625. doi: 10.1016/j.febslet.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H, Laird PW. Interplay between the cancer genome and epigenome. Cell. 2013;153:38–55. doi: 10.1016/j.cell.2013.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tampellini M, Filipski E, Liu XH, Lemaigre G, Li XM, Vrignaud P, François E, Bissery MC, Lévi F. Docetaxel chronopharmacology in mice. Cancer Res. 1998;58:3896–3904. [PubMed] [Google Scholar]
- The Cancer Genome Atlas Research Network. Weinstein JN, Collisson EA, Mills GB, Shaw KRM, Stuart JM, Ozenberger BA, Ellrott K, Shmulevich I, Sander C. The Cancer Genome Atlas Pan-Cancer analysis project. Nat. Genet. 2013;45:1113–1120. doi: 10.1038/ng.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda HR, Hayashi S, Chen W, Sano M, Machida M, Shigeyoshi Y, Iino M, Hashimoto S. System-level identification of transcriptional circuits underlying mammalian circadian clocks. Nat. Genet. 2005;37:187–192. doi: 10.1038/ng1504. [DOI] [PubMed] [Google Scholar]
- Vincenzi B, Santini D, La Cesa A, Tonini G, Levi F, Mormont MC. Cancer chronotherapy: Principles, applications, and perspectives. Cancer. 2003;98:881–883. doi: 10.1002/cncr.11600. [DOI] [PubMed] [Google Scholar]
- Wallach T, Schellenberg K, Maier B, Kalathur RKR, Porras P, Wanker EE, Futschik ME, Kramer A. Dynamic Circadian Protein-Protein Interaction Networks Predict Temporal Organization of Cellular Functions. PLoS Genet. 2013;9 doi: 10.1371/journal.pgen.1003398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood PA, Du-Quiton J, You S, Hrushesky WJM. Circadian clock coordinates cancer cell cycle progression, thymidylate synthase, and 5-fluorouracil therapeutic index. Mol. Cancer Ther. 2006;5:2023–2033. doi: 10.1158/1535-7163.MCT-06-0177. [DOI] [PubMed] [Google Scholar]
- Wu G, Anafi RC, Hughes ME, Kornacker K, Hogenesch JB. MetaCycle: An integrated R package to evaluate periodicity in large scale data. Bioinformatics. 2016;32:3351–3353. doi: 10.1093/bioinformatics/btw405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Tang D, Liu N, Xiong W, Huang H, Li Y, Ma Z, Zhao H, Chen P, Qi X, et al. Reciprocal Regulation between the Circadian Clock and Hypoxia Signaling at the Genome Level in Mammals. Cell Metab. 2017;25:73–85. doi: 10.1016/j.cmet.2016.09.009. [DOI] [PubMed] [Google Scholar]
- Xiang Y, Ye Y, Lou Y, Yang Y, Cai C, Zhang Z, Mills T, Chen N, Kim Y, Ozguc FM, et al. Comprehensive characterization of alternative polyadenylation in human cancer. J. Natl. Cancer Inst. 2018;110:1–22. doi: 10.1093/jnci/djx223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang WS, Stockwell BR. Inhibition of casein kinase 1-epsilon induces cancer-cell-selective, PERIOD2-dependent growth arrest. Genome Biol. 2008;9:R92. doi: 10.1186/gb-2008-9-6-r92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Soares J, Greninger P, Edelman EJ, Lightfoot H, Forbes S, Bindal N, Beare D, Smith JA, Thompson IR, et al. Genomics of Drug Sensitivity in Cancer (GDSC): A resource for therapeutic biomarker discovery in cancer cells. Nucleic Acids Res. 2013;41:955–961. doi: 10.1093/nar/gks1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan P, Li J, Zhou F, Huang Q, Zhang J, Guo X, Lyu Z, Zhang H, Xing J. NPAS2 promotes cell survival of hepatocellular carcinoma by transactivating CDC25A. Cell Death Dis. 2017;8:e2704. doi: 10.1038/cddis.2017.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y, Liu L, Chen H, Wang Y, Xu Y, Mao H, Li J, Mills GB, Shu Y, Li L, et al. Comprehensive Characterization of Molecular Differences in Cancer between Male and Female Patients. Cancer Cell. 2016;29:711–722. doi: 10.1016/j.ccell.2016.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng ZL, Luo HY, Yang J, Wu WJ, Chen DL, Huang P, Xu RH. Overexpression of the circadian clock gene bmal1 increases sensitivity to oxaliplatin in colorectal cancer. Clin. Cancer Res. 2014;20:1042–1052. doi: 10.1158/1078-0432.CCR-13-0171. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Giacchetti S, Parouchev A, Hadadi E, Li X, Dallmann R, Xandri-Monje H, Portier L, Adam R, Lévi F, et al. Dosing time dependent in vitro pharmacodynamics of Everolimus despite a defective circadian clock. Cell Cycle. 2017;4101 doi: 10.1080/15384101.2017.1387695. 00-00. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.