Abstract
Copy number variation (CNV) in insect genomes is a rich source of potentially-adaptive polymorphism which may help overcome the constraints of purifying selection on conserved genes and/or permit elevated transcription. Classic studies of amplified esterases and acetylcholinesterase duplication in Culex pipiens quantified evolutionary dynamics of CNV driven by insecticidal selection. A more complex and potentially medically-impactful form of CNV is found in Anopheles gambiae, with both heterogeneous duplications and homogeneous amplifications strongly linked with insecticide resistance. Metabolic gene amplification, revealed by shotgun sequencing, appears common in Aedes aegypti, but poorly understood in other mosquito species. Many methodologies have been used to detect CNV in mosquitoes, but relatively few can detect both duplications and amplifications, and contrasting methods should be combined. Genome scans for CNV have been rare to date in mosquitoes, but offer immense potential to determine the overall role of CNV as a component of resistance mechanisms.
Introduction
Duplication of genes and genomes is a major source of long-term evolutionary novelty [1], and is increasingly recognised as a key component of shorter-term adaptive responses [2]. Most duplication events are thought to be deleterious and subject to purifying selection as a result of gene expression that is either wasteful or causes imbalance in pathways [2], unless dosage is down-regulated in gene copies [3]. Genome scanning has revealed copy number variation (CNV) to be common in Drosophila melanogaster, and over half of CNVs to affect genes [4]. However, a survey of diverse fly lines found that only nine of 500 duplications surveyed were at high frequency, suggesting that unless under near-immediate positive selection duplicated genes are unlikely to spread [5]. Insecticidal selection is a source of strong pressure driving rapid phenotypic change [6], thus providing an excellent model for studying evolution on human timescales, but also serious challenges for food production and disease control programmes. Here we review how gene CNV influences insecticide resistance in mosquito disease vectors. Whilst the identification of gene CNV impacting insecticide resistance have risen markedly [7], it is currently unclear whether resistance-associated duplications are being increasingly identified because of awareness and/or methodological advances, or are genuinely becoming increasingly widespread.
Scope and definitions
We focus specifically on recent evolutionary innovation by CNV, rather than on groups of resistance genes which have arisen by duplication and are involved in insecticide resistance, which would apply to general expansions of detoxification gene families [8]. We adopt the terminology suggested by Feyereisen et al. [9] of ‘duplication’ to distinguish heterogeneous gene copies that are functionally-related but distinguishable at the protein level, from homogeneous ‘amplification’ which we define as two or more gene copies that produce identical proteins, since the functional consequences of, and selection operating on each type of CNV are likely to be different. We first focus on genes that are the target of insecticides, then move to the more complex area of genes involved in components of detoxification. A summary of methods used to identify CNV are shown in Box 1.
Box 1. Methods for detecting gene duplications (
 ) or amplifications (
) or amplifications (
 ), and the type of material required [DNA, protein, live mosquitoes - in laboratory crosses and selection regimes].
), and the type of material required [DNA, protein, live mosquitoes - in laboratory crosses and selection regimes].
Heterozygote excess at locus of interest (
 ) (i) natural populations: ideally should genotype additional markers to demonstrate signal is not demographic; scoring errors should also be ruled out. Sample sizes must be adequate to allow appropriately-powered Hardy-Weinberg tests. Useful indicator but not definitive because of possibility of selection for heterozygotes in field. [DNA] (ii) offspring from parents of known genotypes in a laboratory cross: assumes that differential mortality of homozygous offspring genotypes can be ruled out. Potentially reliable but not scalable and not tractable for field mosquito populations which are often difficult to colonise. [DNA, live mosquitoes]
) (i) natural populations: ideally should genotype additional markers to demonstrate signal is not demographic; scoring errors should also be ruled out. Sample sizes must be adequate to allow appropriately-powered Hardy-Weinberg tests. Useful indicator but not definitive because of possibility of selection for heterozygotes in field. [DNA] (ii) offspring from parents of known genotypes in a laboratory cross: assumes that differential mortality of homozygous offspring genotypes can be ruled out. Potentially reliable but not scalable and not tractable for field mosquito populations which are often difficult to colonise. [DNA, live mosquitoes]
Altered/mixed enzyme activity (
 ): biochemical detection of sensitive (wild type) and insensitive (resistant) enzymes, at a ratio deviant from 100% or 50% using inhibitor dose-response assay, which suggests an uneven mixture of alleles. Traditional technique lacking sensitivity which has been superseded by more tractable DNA-based methodologies. [protein]
): biochemical detection of sensitive (wild type) and insensitive (resistant) enzymes, at a ratio deviant from 100% or 50% using inhibitor dose-response assay, which suggests an uneven mixture of alleles. Traditional technique lacking sensitivity which has been superseded by more tractable DNA-based methodologies. [protein]
Calibrated phenotypic assays of resistance (
 ): laboratory crossing/backcrossing to produce heterozygote genotypes for which reduced mortality can be demonstrated. Requires known single-copy lines and precise calibration of expected genotype-phenotype mortality. Time-consuming and unreliable because of potential for other sources of variation in mortality. [live mosquitoes, DNA]
): laboratory crossing/backcrossing to produce heterozygote genotypes for which reduced mortality can be demonstrated. Requires known single-copy lines and precise calibration of expected genotype-phenotype mortality. Time-consuming and unreliable because of potential for other sources of variation in mortality. [live mosquitoes, DNA]
Allele cloning + sequencing (
 ) detection of >2 distinct alleles. Only provides a one-way test (positive/unclear) unless many clones sequenced. Prone to false positives: must have safeguards such as detection in multiple clones obtained from entirely independent batches of cloned and sequenced amplicons. [DNA]
) detection of >2 distinct alleles. Only provides a one-way test (positive/unclear) unless many clones sequenced. Prone to false positives: must have safeguards such as detection in multiple clones obtained from entirely independent batches of cloned and sequenced amplicons. [DNA]
Intermediate patterns in genotyping assays (
 ); assay must be quantitative (i.e. not simply detect homo- or heterozygotes). Useful semi-quantitative indicator but not very reliable as a standalone without extensive assay calibration and positive controls. [DNA]
); assay must be quantitative (i.e. not simply detect homo- or heterozygotes). Useful semi-quantitative indicator but not very reliable as a standalone without extensive assay calibration and positive controls. [DNA]
Southern blotting (
 ) traditional technique typically for qualitative analysis [DNA]
) traditional technique typically for qualitative analysis [DNA]
Standard gDNA qPCR (
 ): suitable single-copy calibrator genes required and usually a single copy reference population, but will not indicate which allele in elevated copy. Quantitatively imprecise. [DNA]
): suitable single-copy calibrator genes required and usually a single copy reference population, but will not indicate which allele in elevated copy. Quantitatively imprecise. [DNA]
Droplet digital gDNA qPCR (
 ,
,
 ): excellent but expensive technique; requires calibrator genes to detect amplifications. [DNA]
): excellent but expensive technique; requires calibrator genes to detect amplifications. [DNA]
Fluorescent in situ hybridisation, FISH. (
 ) Gives an indication of the relative location of duplicates but not fine-scaled resolution when in tandem copies. Technically-challenging. [DNA; requires chromosomal preparation]
) Gives an indication of the relative location of duplicates but not fine-scaled resolution when in tandem copies. Technically-challenging. [DNA; requires chromosomal preparation]
Elevated read coverage in genome sequences (
 ,
,
 ): can be whole or targeted; qualitative and quantitative; and can identify the size of the copied genomic region, and previously unknown CNVs. Potentially very reliable if appropriate algorithm used. [DNA] See
Figure 1c.
): can be whole or targeted; qualitative and quantitative; and can identify the size of the copied genomic region, and previously unknown CNVs. Potentially very reliable if appropriate algorithm used. [DNA] See
Figure 1c.
Discordant read pair mapping in genome sequence data (
 ,
,
 ). Can identify the structure of duplications or amplifications, but does not inform on the number of copies. [DNA] See
Figure 1a,b.
). Can identify the structure of duplications or amplifications, but does not inform on the number of copies. [DNA] See
Figure 1a,b.
Long-read ‘third generation’ sequence based CNV detection (
 ,
,
 ). Potential to detect presence, copy number, sequence, and genomic location(s) of known or unknown variants. Promising, but very few applications to date in any organism. [DNA]
). Potential to detect presence, copy number, sequence, and genomic location(s) of known or unknown variants. Promising, but very few applications to date in any organism. [DNA]
Duplication and amplification: insecticide target site genes
For insecticides to be effective against a broad range of arthropod pests, their target sites must be conserved proteins, exhibiting little protein polymorphism within or between species. This has led to the popularity of neurotoxic insecticides, which are fast-acting and target sites that are highly conserved, as a result of strong purifying selection; this severely constrains the evolution of resistance by point mutation(s) [10,11]. Although after prolonged selection of a major resistance point mutation, amplifier or compensatory mutations may accrue [12,13], gene amplification may be less constrained, and a relatively rapid rate of copy number generation can provide a rich source of potentially-adaptive polymorphism [14].
Duplications of the two most important insecticide target sites - the voltage-gated sodium channel (Vgsc) and acetylcholinesterase (Ace-1) gene – have been detected in mosquitoes. Duplication of the Vgsc has been reported in the major arbovirus vector Aedes aegypti in Brazil [15] and in the lymphatic filariasis vector Culex quinquefasciatus in Tanzania [16]. In both cases, the duplication appears to involve pairing of distinct alleles, but neither appear widespread or at high frequency, and consequences for resistance are unclear at present.
In contrast, the evolutionary dynamics and fitness consequences of duplications at the Ace-1 locus have been studied intensively in mosquitoes. Many resistance-associated AChE substitutions are known in insects [9], but only three mutant codons are found in mosquitoes, G119S, F290V, and F331W. The most important is the G119S polymorphism found in Cx. pipiens pipiens, Cx. p. quinquefasicatus [17], Cx. vishnui [18], the African malaria vectors Anopheles gambiae and An. coluzzii [19] and the Latin American malaria vector An. albimanus, in which, unusually additional variants exist at the 119 codon [20]. Reported discovery of the G119S mutation in another major malaria vector, An. arabiensis from a single West African locale [21] has yet to be replicated in any location. The F290V polymorphism is found at low frequency in Mediterranean populations of Cx. pipiens [22], and the F331W polymorphism is present at very high frequency in Chinese populations of the Japanese encephalitis vector, Cx. tritaeniorhynchus [18], but no other species to date. In both Anopheles and Culex, the 119S mutation confers very strong resistance to multiple carbamate and organophosphate (OP) insecticides at both larval and adult stages, but crucially, results in a relatively inefficient enzyme, with only ≤1/3 of the activity of the wild type 119G allele [23–25]. This balance of a major fitness advantage in the presence of insecticide, but a major cost in insecticide-free environments has driven spatial and temporal variation in relative frequencies of 119G and 119S alleles in Cx. pipiens [26], and also selected for gene duplication [27].
Ace-1 duplications in Cx. pipiens subspecies, whether involving G119S or F290V, appear to ubiquitously involve linkage of a resistant and wild type copy, probably in tandem (no recombination seems to occur between copies) to create permanent ‘heterozygotes’ which exhibit both resistance to carbamate and organophosphate insecticides, and a reduction of fitness costs [17,28]. Chromosomes with duplicated alleles can pair with other duplicants or with a single copy, leading to a range of possible genotypes. Thirteen independent duplicated genotypes have been identified to date in Cx. p. pipiens and Cx. p. quinquefasicatus [29], of which some provide similar levels of resistance to resistant (119G/G) homozygotes but with little detectable cost [27]. Studies of temporal dynamics in Cx. pipiens from southern France have demonstrated replacement of single copy resistant alleles with duplicated (Ace-1D) alleles, and spatio-temporal variation in frequencies of particular duplicates [27]. In France the duplications were first detected in the mid-1990s; being apparently very rare or absent previously [30], and in Martinique and Cuba, duplications were detected 4 and 14 years respectively post-OP spraying [31].
Owing to their major importance as disease vectors, the discovery of Ace-1 duplication in West African An. gambiae and An. coluzzii, which paired a resistant with a susceptible G119S allele, was extremely concerning because - following the model from Cx. pipiens - it suggested potential for reduced fitness costs and the spread of Ace-1 119S-based resistance [19,32]. All resistant alleles in the An. gambiae and An. coluzzii are identical, suggesting a single recent origin and introgression from one species to the other, although directionality cannot be determined [19]. In Tiassale, southern Cote d’Ivoire, where the An. coluzzi population exhibits resistance to all major adulticide classes available [33], the situation appears extreme, with all or most individuals heterozygous for G119S and resistance to bendiocarb correlating with 119S copy number [34].
Further molecular investigation revealed Ace-1 duplications to be widespread in West Africa, and encompass a full range of genotype groups differing in their balance of resistant to susceptible alleles. In contrast to the situation in Cx. pipiens, these included amplified resistant homozygotes, with up to 10 identical alleles present [11,35]. Analysis of archived G119S heterozygotes from Accra, Ghana in 2002, when 119S was very rare (<5% frequency) indicated that all were duplicated [11], a finding substantiated by wider-scale analysis of specimens from across West Africa [24]. Very strong purifying selection was detected acting on Ace-1 in Accra populations, centred on the G119S position, but with a huge selective sweep region of up to 2 megabases, which, coupled with, a rapid local increase in 119S frequency demonstrates extremely strong directional selection [11]. However, the genomic footprint detected was unexpectedly heterogeneous and asymmetrical, leading to a hypothesis that the duplicated region was at least 10-times larger than the Ace-1 gene it encompassed [11]. Genome sequence analysis has recently demonstrated that the copied region exceeds 200 kilobases and encompasses 11 additional genes, although intriguingly in some individuals a large portion of this duplicated/ amplified region is deleted, leaving only Ace-1 [24]. Importantly, acetylcholinesterase activity scales with copy number suggesting that all Ace-1 copies in An. gambiae or An. coluzzii appear to be expressed, as also observed in Culex [29]. Similarly, both resistance and, to a lesser extent fitness costs, correlate with 119S copy number [24], though the fitness costs in individuals with only amplified resistant alleles might be at least partially reduced by production of more low-activity enzyme [36]. The geographical distributions and selective drivers of each form of CNV - heterogenous duplication vs. homogeneous amplification - require further investigation as a major priority because of the current importance of the OP pirimiphos methyl for indoor residual spray-based control of malaria in Africa [37].
Duplication and amplification: genes involved in detoxification pathways
As with target sites, CNVs in metabolic genes can influence insecticide resistance by amplifying expression [38] or by providing the potential for neo-functionalisation of one copy without sacrificing the original gene [39]. Examples reported so far in mosquitoes constitute an association with increased gene expression.
The classic example of gene amplifications causing increased insecticide metabolic resistance comes from Cx. pipiens in which amplified esterase genes, Est2 and Est3, have spread across the globe, providing resistance to organophosphates [40]. Multiple alleles at this locus are linked to increased esterase levels have been reported, most of which are gene amplifications of both Est2 and Est3 or Est2 alone; copy levels range from a few- to 100-fold [40,41]. While the same genes (Est2 and Est3) are amplified in each case, other differences exist between these alleles that lead to divergent responses to selection. For example, in southern France, the amplified esterase alleles ‘A2-B2’ and ‘A4-B4’ are both present, yet have markedly different histories: A4-B4 showed a dramatic increase in frequency after its appearance in the 1980s, while A2-B2 remained at consistently low frequencies throughout the 1990s [42]. The nature of these differences and how they affect the relative success of the different alleles remains to be elucidated.
Amplification of esterases has also been implicated in larval resistance to the organochloride temephos in the Asian tiger mosquito Ae. albopictus, with the genes CCEae3a and CCEae6a being amplified around 10-fold in pooled cDNA from a resistant compared to a susceptible strain [43]. As with Cx. pipiens, amplifications are not geographically restricted, being found in populations from both Greece and Florida [44].
In Cx. quinquefasciatus, metabolic resistance to permethrin is mediated by overexpression of the cytochrome P450 Cyp9M10 both in laboratory strains [45] and in the wild [46]. Overexpression is primarily caused by cis-acting regulatory changes, but a duplication of the region comprising both the gene and its regulatory region further increases expression and resistance levels [47,48]. Interestingly, the expression differences between the duplicated and unduplicated forms are greater than can be explained by the duplication alone [47], indicating that further cis-regulatory differences must exist.
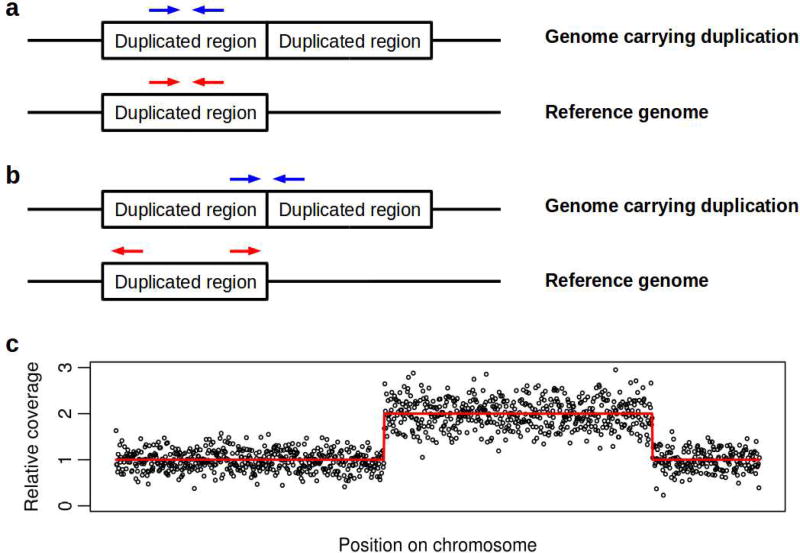
The above examples of metabolic gene amplification were detected by studies focused on individual genes to which attention was drawn by their high levels of expression [43,48,49]. Increasingly, next generation sequencing combined with computational methods for detecting CNVs from sequencing data allow systematic population-scale studies searching across genomes for amplifications and possible links to insecticide resistance. The many computational tools available for detecting CNVs from sequencing data have been reviewed in detail [50], but the majority rely on one of two basic principles: (1) detecting changes in the amount of DNA coming from the amplified region, and (2) detecting the alignment of reads on or around the CNV breakpoint. A gene duplication in a region will increase the number of sequencing reads aligning to that region on the (unduplicated) reference genome, thus increasing the sequencing coverage in that region. Detecting this anomalous coverage indicates the presence of a CNV and gives an idea of the number of copies involved in a duplication event. Identifying reads that map across breakpoints, or pairs of reads mapping either side of a breakpoint (Figure 1), provides accurate information on the start and end points of the CNV and can provide base-pair level resolution of the breakpoint position.
Figure 1.
Detecting a tandem gene duplication (or amplification) using discordant read alignment (a,b), or an amplification from read coverage irrespective of relative genomic positions. a) Example of concordant read alignment. Paired end sequencing of a DNA fragment produces a pair of reads (blue arrows) which map facing towards each-other when aligned to the reference genome (red arrows). b) Example of discordant read alignment. Paired end sequencing of a DNA fragment that overlaps the duplication break point produces a pair of reads which map facing away from each other at either end of the duplication when aligned to the reference genome. c) Simulated data showing patterns used to detect CNVs using sequencing coverage. Each point represents a genomic window (usually in the order of a hundred or more base pairs) over which coverage was calculated and normalised such that 1 represents the unduplicated state. The red line indicates the true underlying copy number state. The CNV is visible as a marked transient increase in normalised coverage (here from 1 to 2).
In the world’s most important arbovirus vector, Aedes aegypti, a targeted deep-sequencing analysis of 763 candidate genes was used to compare sequencing coverage between laboratory strains, which were resistant or susceptible to insecticides [51]. Forty-one genes showed evidence of more amplification in resistant than susceptible mosquitoes. Among amplified genes, P450s were significantly over-represented, suggesting a possible association between these gene amplifications and metabolic resistance. Furthermore, some of the amplifications were shown to be correlated with gene expression [51,52]. However, this targeted sequencing was neither able to describe the structure of the duplications in detail, nor identify independent duplication events that might be tracked across populations. In contrast, using untargeted whole-genome sequencing increases the chances that the breakpoints of the duplication are included in the sequenced regions, thus permitting precise and accurate identification of the extent of a duplication and matching of duplication types between individuals. Such research is currently under-way using data resources from the Anopheles gambiae 1000 genomes (Ag1000G) project [13] (https://www.malariagen.net/projects/ag1000g).
Conclusion: copy number variation is an increasing problem for mosquito control
Studies of Cx. pipiens subspecies provided a template for understanding CNV-mediated resistance at one of the most important insecticide target sites and major-effect metabolic resistance loci. In both cases the superb long-term datasets combined with classical laboratory and population genetic analysis techniques yielded results which clearly indicate that duplications appear to rise in frequency as a result of insecticide application. With the widespread removal of OP-use from most of the range where Cx. pipiens are a major vector, however, the applied importance has greatly diminished. A far greater current threat comes from the recently-increasing CNVs in the An. gambiae species pair, the consequences of which are not yet fully understood for Ace-1 (but appear of major concern), and are all-but unknown for metabolic genes. An. gambiae Ace-1 CNV evolution is very different from that of Culex, and appears to perfectly track Ace-1 resistant allele increases from a point of extreme rarity about 15 years ago [11]. The causal element which generated an increase in gene duplication/ amplification from this time-point is unknown. A notable feature of major methods for detecting CNV (Box 1) is that many are capable of detecting heterogeneous duplications, typically at single known candidate genes of interest. Recent studies from An. gambiae [11,24] and Ae. aegypti [51] have shown how extending the region scanned beyond the mutation, gene or immediate genomic locale can bring insights into selection, CNV size and identification of novel CNVs, including amplifications. Population genomic whole genome sequencing projects, especially those targeting sampling areas with anti-vector interventions via time-series should soon generate a vastly improved understanding of both the extent and importance of CNVs as part of the mosquito insecticide resistance armoury.
CNVs are a major source of polymorphism in genomes and arise frequently
Copies may be heterogeneous (duplications) or homogeneous (amplifications)
AChE gene CNV in An. gambiae is of primary applied importance, but is complex
Correspondence between CNV appearance and dynamics and insecticide application
Whole genome analyses are beginning to revolutionise understanding of CNVs
Acknowledgments
We are grateful to Craig Wilding and Chris Jones for comments on an earlier version of the manuscript. DW’s work on mosquito CNV is supported by Wellcome Trust (094960/Z/10/Z; 110236/Z/15/Z) and NIAID (1R011AI082734-01; 5R01AI116811-02). LSD’s work is supported by Wellcome Trust (109917/Z/15/Z). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIAID.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
Papers of particular interest, published within the last two years, have been highlighted as:
* of special interest
** of outstanding interest
- 1.Otto SP, Yong P. The evolution of gene duplicates. Adv Genet. 2002;46:451–483. doi: 10.1016/s0065-2660(02)46017-8. [DOI] [PubMed] [Google Scholar]
- 2.Kondrashov FA. Gene duplication as a mechanism of genomic adaptation to a changing environment. Proc Biol Sci. 2012;279:5048–5057. doi: 10.1098/rspb.2012.1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Qian W, Liao BY, Chang AY, Zhang J. Maintenance of duplicate genes and their functional redundancy by reduced expression. Trends Genet. 2010;26:425–430. doi: 10.1016/j.tig.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Emerson JJ, Cardoso-Moreira M, Borevitz JO, Long M. Natural selection shapes genome-wide patterns of copy-number polymorphism in Drosophila melanogaster. Science. 2008;320:1629–1631. doi: 10.1126/science.1158078. [DOI] [PubMed] [Google Scholar]
- 5.Cardoso-Moreira M, Emerson JJ, Clark AG, Long M. Drosophila duplication hotspots are associated with late-replicating regions of the genome. PLoS Genet. 2011;7:e1002340. doi: 10.1371/journal.pgen.1002340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Denholm I, Devine GJ, Williamson MS. Evolutionary genetics. Insecticide resistance on the move. Science. 2002;297:2222–2223. doi: 10.1126/science.1077266. [DOI] [PubMed] [Google Scholar]
- 7.Bass C, Field LM. Gene amplification and insecticide resistance. Pest Manag Sci. 2011;67:886–890. doi: 10.1002/ps.2189. [DOI] [PubMed] [Google Scholar]
- 8.Ranson H, Claudianos C, Ortelli F, Abgrall C, Hemingway J, Sharakhova MV, Unger MF, Collins FH, Feyereisen R. Evolution of supergene families associated with insecticide resistance. Science. 2002;298:179–181. doi: 10.1126/science.1076781. [DOI] [PubMed] [Google Scholar]
- 9*.Feyereisen R, Dermauw W, Van Leeuwen T. Genotype to phenotype, the molecular and physiological dimensions of resistance in arthropods. Pestic Biochem Physiol. 2015;121:61–77. doi: 10.1016/j.pestbp.2015.01.004. Excellent review of resistance mechanisms including some coverage of CNV. [DOI] [PubMed] [Google Scholar]
- 10.Remnant EJ, Good RT, Schmidt JM, Lumb C, Robin C, Daborn PJ, Batterham P. Gene duplication in the major insecticide target site Rdl, in Drosophila melanogaster. Proc Natl Acad Sci U S A. 2013;110:14705–14710. doi: 10.1073/pnas.1311341110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11**.Weetman D, Mitchell SN, Wilding CS, Birks DP, Yawson AE, Essandoh J, Mawejje HD, Djogbenou LS, Steen K, Rippon EJ, et al. Contemporary evolution of resistance at the major insecticide target site gene Ace-1 by mutation and copy number variation in the malaria mosquito Anopheles gambiae. Mol Ecol. 2015;24:2656–2672. doi: 10.1111/mec.13197. Paper quantifying the strength of selection on the Ace-1 G119S mutation in Anopheles and highlighting the magnitude of, the CNV region. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kasai S, Sun H, Scott JG. Diversity of knockdown resistance alleles in a single house fly population facilitates adaptation to pyrethroid insecticides. Insect Mol Biol. 2017;26:13–24. doi: 10.1111/imb.12267. [DOI] [PubMed] [Google Scholar]
- 13*.The Anopheles gambiae Genomes C. Genetic diversity of the African malaria vector Anopheles gambiae. Nature. 2017;552:96. doi: 10.1038/nature24995. Landmark paper describing the extraordinary genomic diversity of Anopheles gambiae from across sub-Saharan Africa. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schrider DR, Houle D, Lynch M, Hahn MW. Rates and genomic consequences of spontaneous mutational events in Drosophila melanogaster. Genetics. 2013;194:937–954. doi: 10.1534/genetics.113.151670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martins AJ, Brito LP, Linss JG, Rivas GB, Machado R, Bruno RV, Lima JB, Valle D, Peixoto AA. Evidence for gene duplication in the voltage-gated sodium channel gene of Aedes aegypti. Evol Med Public Health. 2013;2013:148–160. doi: 10.1093/emph/eot012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martins WFS, Subramaniam K, Steen K, Mawejje H, Liloglou T, Donnelly MJ, Wilding CS. Detection and quantitation of copy number variation in the voltage-gated sodium channel gene of the mosquito Culex quinquefasciatus. Sci Rep. 2017;7:5821. doi: 10.1038/s41598-017-06080-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Labbe P, Berthomieu A, Berticat C, Alout H, Raymond M, Lenormand T, Weill M. Independent duplications of the acetylcholinesterase gene conferring insecticide resistance in the mosquito Culex pipiens. Mol Biol Evol. 2007;24:1056–1067. doi: 10.1093/molbev/msm025. [DOI] [PubMed] [Google Scholar]
- 18.Alout H, Berthomieu A, Cui F, Tan Y, Berticat C, Qiao C, Weill M. Different amino-acid substitutions confer insecticide resistance through acetylcholinesterase 1 insensitivity in Culex vishnui and Culex tritaeniorhynchus (Diptera: Culicidae) from China. J Med Entomol. 2007;44:463–469. doi: 10.1603/0022-2585(2007)44[463:dascir]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 19.Djogbenou L, Chandre F, Berthomieu A, Dabire R, Koffi A, Alout H, Weill M. Evidence of introgression of the ace-1(R) mutation and of the ace-1 duplication in West African Anopheles gambiae s. s. PLoS One. 2008;3:e2172. doi: 10.1371/journal.pone.0002172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liebman KA, Pinto J, Valle J, Palomino M, Vizcaino L, Brogdon W, Lenhart A. Novel mutations on the ace-1 gene of the malaria vector Anopheles albimanus provide evidence for balancing selection in an area of high insecticide resistance in Peru. Malar J. 2015;14:74. doi: 10.1186/s12936-015-0599-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dabire RK, Namountougou M, Diabate A, Soma DD, Bado J, Toe HK, Bass C, Combary P. Distribution and frequency of kdr mutations within Anopheles gambiae s.l. populations and first report of the ace.1 G119S mutation in Anopheles arabiensis from Burkina Faso (West Africa) PLoS One. 2014;9:e101484. doi: 10.1371/journal.pone.0101484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alout H, Berthomieu A, Hadjivassilis A, Weill M. A new amino-acid substitution in acetylcholinesterase 1 confers insecticide resistance to Culex pipiens mosquitoes from Cyprus. Insect Biochem Mol Biol. 2007;37:41–47. doi: 10.1016/j.ibmb.2006.10.001. [DOI] [PubMed] [Google Scholar]
- 23.Alout H, Djogbenou L, Berticat C, Chandre F, Weill M. Comparison of Anopheles gambiae and Culex pipiens acetycholinesterase 1 biochemical properties. Comp Biochem Physiol B Biochem Mol Biol. 2008;150:271–277. doi: 10.1016/j.cbpb.2008.03.008. [DOI] [PubMed] [Google Scholar]
- 24**.Assogba BS, Milesi P, Djogbenou LS, Berthomieu A, Makoundou P, Baba-Moussa LS, Fiston-Lavier AS, Belkhir K, Labbe P, Weill M. The ace-1 locus Is amplified in all resistant Anopheles gambiae mosquitoes: fitness consequences of homogeneous and heterogeneous duplications. PLoS Biol. 2016;14:e2000618. doi: 10.1371/journal.pbio.2000618. Excellent paper confirming the ubiquity of duplication (or amplification) of Ace-1 resistant alleles, quantifying resistance and costs and establishing hypotheses for CNV occurrence under different granularities of selection. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Assogba BS, Djogbenou LS, Milesi P, Berthomieu A, Perez J, Ayala D, Chandre F, Makoutode M, Labbe P, Weill M. An ace-1 gene duplication resorbs the fitness cost associated with resistance in Anopheles gambiae, the main malaria mosquito. Sci Rep. 2015;5:14529. doi: 10.1038/srep14529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lenormand T, Bourguet D, Guillemaud T, Raymond M. Tracking the evolution of insecticide resistance in the mosquito Culex pipiens. Nature. 1999;400:861–864. doi: 10.1038/23685. [DOI] [PubMed] [Google Scholar]
- 27.Labbe P, Berticat C, Berthomieu A, Unal S, Bernard C, Weill M, Lenormand T. Forty years of erratic insecticide resistance evolution in the mosquito Culex pipiens. PLoS Genet. 2007;3:e205. doi: 10.1371/journal.pgen.0030205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alout H, Labbe P, Berthomieu A, Pasteur N, Weill M. Multiple duplications of the rare ace-1 mutation F290V in Culex pipiens natural populations. Insect Biochem Mol Biol. 2009;39:884–891. doi: 10.1016/j.ibmb.2009.10.005. [DOI] [PubMed] [Google Scholar]
- 29.Labbe P, Milesi P, Yebakima A, Pasteur N, Weill M, Lenormand T. Gene-dosage effects on fitness in recent adaptive duplications: ace-1 in the mosquito Culex pipiens. Evolution. 2014;68:2092–2101. doi: 10.1111/evo.12372. [DOI] [PubMed] [Google Scholar]
- 30.Lenormand T, Guillemaud T, Bourguet D, Raymond M. Appearance and sweep of a gene duplication: adaptive response and potential for new functions in the mosquito Culex pipiens. Evolution. 1998;52:1705–1712. doi: 10.1111/j.1558-5646.1998.tb02250.x. [DOI] [PubMed] [Google Scholar]
- 31.Bourguet D, Raymond M, Bisset J, Pasteur N, Arpagaus M. Duplication of the Ace.1 locus in Culex pipiens mosquitoes from the Caribbean. Biochem Genet. 1996;34:351–362. doi: 10.1007/BF00554410. [DOI] [PubMed] [Google Scholar]
- 32.Djogbenou L, Labbe P, Chandre F, Pasteur N, Weill M. Ace-1 duplication in Anopheles gambiae: a challenge for malaria control. Malar J. 2009;8:70. doi: 10.1186/1475-2875-8-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Edi CV, Koudou BG, Jones CM, Weetman D, Ranson H. Multiple-insecticide resistance in Anopheles gambiae mosquitoes, Southern Cote d'Ivoire. Emerg Infect Dis. 2012;18:1508–1511. doi: 10.3201/eid1809.120262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Edi CV, Djogbenou L, Jenkins AM, Regna K, Muskavitch MA, Poupardin R, Jones CM, Essandoh J, Ketoh GK, Paine MJ, et al. CYP6 P450 enzymes and ACE-1 duplication produce extreme and multiple insecticide resistance in the malaria mosquito Anopheles gambiae. PLoS Genet. 2014;10:e1004236. doi: 10.1371/journal.pgen.1004236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Djogbenou LS, Assogba B, Essandoh J, Constant EA, Makoutode M, Akogbeto M, Donnelly MJ, Weetman D. Estimation of allele-specific Ace-1 duplication in insecticide-resistant Anopheles mosquitoes from West Africa. Malar J. 2015;14:507. doi: 10.1186/s12936-015-1026-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kwon DH, Choi JY, Je YH, Lee SH. The overexpression of acetylcholinesterase compensates for the reduced catalytic activity caused by resistance-conferring mutations in Tetranychus urticae. Insect Biochem Mol Biol. 2012;42:212–219. doi: 10.1016/j.ibmb.2011.12.003. [DOI] [PubMed] [Google Scholar]
- 37.Coleman S, Dadzie SK, Seyoum A, Yihdego Y, Mumba P, Dengela D, Ricks P, George K, Fornadel C, Szumlas D, et al. A reduction in malaria transmission intensity in Northern Ghana after 7 years of indoor residual spraying. Malar J. 2017;16:324. doi: 10.1186/s12936-017-1971-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schmidt JM, Good RT, Appleton B, Sherrard J, Raymant GC, Bogwitz MR, Martin J, Daborn PJ, Goddard ME, Batterham P, et al. Copy number variation and transposable elements feature in recent, ongoing adaptation at the Cyp6g1 locus. PLoS Genet. 2010;6:e1000998. doi: 10.1371/journal.pgen.1000998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39**.Zimmer CTGW, Singh KS, Randall E, Lueke B, Gutbrod O, Kohler M, Nauen R, Davies TGE, Bass C. Neofunctionalization of duplicated P450 genes drives the evolution of insecticide resistance in the brown planthopper. Current Biology. doi: 10.1016/j.cub.2017.11.060. in press. To our knowledge the only paper to date demonstrating a link between recent neofunctionalisation of a CNV and insecticide resistance, with comprehensive functional analyses. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Raymond M, Berticat C, Weill M, Pasteur N, Chevillon C. Insecticide resistance in the mosquito culex pipiens: what have we learned about adaptation? Genetica. 2001;112–113:287–296. [PubMed] [Google Scholar]
- 41.Rooker S, Guillemaud T, Berge J, Pasteur N, Raymond M. Coamplification of esterase A and B genes as a single unit in Culex pipiens mosquitoes. Heredity (Edinb) 1996;77(Pt 5):555–561. [PubMed] [Google Scholar]
- 42.Guillemaud T, Lenormand T, Bourguet D, Chevillon C, Pasteur N, Raymond M. Evolution of resistance in Culex pipiens: allele replacement and changing environment. Evolution. 1998;52:443–453. doi: 10.1111/j.1558-5646.1998.tb01644.x. [DOI] [PubMed] [Google Scholar]
- 43.Grigoraki L, Lagnel J, Kioulos I, Kampouraki A, Morou E, Labbe P, Weill M, Vontas J. Transcriptome profiling and genetic study reveal amplified carboxylesterase genes implicated in temephos resistance, in the asian tiger mosquito Aedes albopictus. PLoS Negl Trop Dis. 2015;9:e0003771. doi: 10.1371/journal.pntd.0003771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44*.Grigoraki L, Pipini D, Labbe P, Chaskopoulou A, Weill M, Vontas J. Carboxylesterase gene amplifications associated with insecticide resistance in Aedes albopictus: geographical distribution and evolutionary origin. PLoS Negl Trop Dis. 2017;11:e0005533. doi: 10.1371/journal.pntd.0005533. Paper demonstrating the widespread geographical distribution of the temephos-linked carboxylesterase duplication, likely linked with the extraordinary recent invasive spread of Ae. albopictus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hardstone MC, Komagata O, Kasai S, Tomita T, Scott JG. Use of isogenic strains indicates CYP9M10 is linked to permethrin resistance in Culex pipiens quinquefasciatus. Insect Mol Biol. 2010;19:717–726. doi: 10.1111/j.1365-2583.2010.01030.x. [DOI] [PubMed] [Google Scholar]
- 46.Wilding CS, Smith I, Lynd A, Yawson AE, Weetman D, Paine MJ, Donnelly MJ. A cis-regulatory sequence driving metabolic insecticide resistance in mosquitoes: functional characterisation and signatures of selection. Insect Biochem Mol Biol. 2012;42:699–707. doi: 10.1016/j.ibmb.2012.06.003. [DOI] [PubMed] [Google Scholar]
- 47.Itokawa K, Komagata O, Kasai S, Masada M, Tomita T. Cis-acting mutation and duplication: History of molecular evolution in a P450 haplotype responsible for insecticide resistance in Culex quinquefasciatus. Insect Biochem Mol Biol. 2011;41:503–512. doi: 10.1016/j.ibmb.2011.04.002. [DOI] [PubMed] [Google Scholar]
- 48.Itokawa K, Komagata O, Kasai S, Okamura Y, Masada M, Tomita T. Genomic structures of Cyp9m10 in pyrethroid resistant and susceptible strains of Culex quinquefasciatus. Insect Biochem Mol Biol. 2010;40:631–640. doi: 10.1016/j.ibmb.2010.06.001. [DOI] [PubMed] [Google Scholar]
- 49.Mouches C, Pasteur N, Berge JB, Hyrien O, Raymond M, de Saint Vincent BR, de Silvestri M, Georghiou GP. Amplification of an esterase gene is responsible for insecticide resistance in a California Culex mosquito. Science. 1986;233:778–780. doi: 10.1126/science.3755546. [DOI] [PubMed] [Google Scholar]
- 50.Zhao M, Wang Q, Wang Q, Jia P, Zhao Z. Computational tools for copy number variation (CNV) detection using next-generation sequencing data: features and perspectives. BMC Bioinformatics. 2013;14(Suppl 11):S1. doi: 10.1186/1471-2105-14-S11-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51**.Faucon F, Dusfour I, Gaude T, Navratil V, Boyer F, Chandre F, Sirisopa P, Thanispong K, Juntarajumnong W, Poupardin R, et al. Identifying genomic changes associated with insecticide resistance in the dengue mosquito Aedes aegypti by deep targeted sequencing. Genome Res. 2015;25:1347–1359. doi: 10.1101/gr.189225.115. Groundbreaking work in mosquitoes applying targeted genome sequencing to discover the extent of CNVs in metabolic genes in Ae. aegypti. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52*.Faucon F, Gaude T, Dusfour I, Navratil V, Corbel V, Juntarajumnong W, Girod R, Poupardin R, Boyer F, Reynaud S, et al. In the hunt for genomic markers of metabolic resistance to pyrethroids in the mosquito Aedes aegypti: an integrated next-generation sequencing approach. PLoS Negl Trop Dis. 2017;11:e0005526. doi: 10.1371/journal.pntd.0005526. Follow-up work to ref 51, extending the CNV discovery phase and developing DNA-based markers for CNV-driven gene expression variation. [DOI] [PMC free article] [PubMed] [Google Scholar]