Abstract
De novo design and construction of stimuli-responsive protein assemblies that predictably switch between discrete conformational states remains an essential but highly challenging goal in biomolecular design. We previously reported synthetic, 2D protein lattices self-assembled via disulfide bonding interactions, which endows them with a unique capacity to undergo coherent conformational changes without losing crystalline order. Here we have carried out all-atom molecular dynamics simulations to map the free-energy landscape of these lattices, validated this landscape through extensive structural characterization by electron microscopy, and established that it is predominantly governed by solvent reorganization entropy. Subsequent redesign of the protein surface with conditionally repulsive electrostatic interactions enabled us to predictably perturb the free-energy landscape, and obtain a new protein lattice whose conformational dynamics can be chemically and mechanically toggled between three different states with varying porosities and molecular densities.
Protein self-assembly is the predominant means of forming functional materials and machines in living systems1 and has been increasingly exploited in the laboratory for the bottom-up construction of synthetic materials2–5. A fundamental physical understanding of the molecular interactions which underpin protein self-assembly is critical for the rational design of targeted supramolecular architectures, as well as for engineering their structural dynamics and their response to environmental cues. To date, much research on protein or biomolecular design has centered on the optimization of energetically–or more accurately, enthalpically–favorable bonding interactions between the molecular components to achieve the thermodynamically preferred self-assembly products6,7. Such focus on enthalpy minimization has placed considerable emphasis on obtaining singular architectures that assemble predominantly via quasi-irreversible, high-affinity interactions. These approaches frequently lead to a propensity for forming aggregates or misfolded products, and even when successful, yield assemblies with no or a low degree of structural adaptiveness and stimuli-responsiveness5,8,9. Despite their critical role in every aspect of protein structure and function (e.g., folding10, recognition11, catalysis12), entropy and water-protein interactions are not explicitly considered in most design efforts, owing largely to the difficulty in their rational implementation by experiment or computation. Clearly, the design of protein assemblies with sophisticated functional and physical properties necessitates the attainment of not only structures corresponding to deep free-energy minima, but also of entire free-energy landscapes that such structures can traverse in a programmable manner.13–16 This, in turn, requires a detailed understanding and mastery of enthalpic and entropic factors that govern protein-protein and protein-solvent interactions, which we set out to achieve in this work for the first time for a synthetic protein assembly.
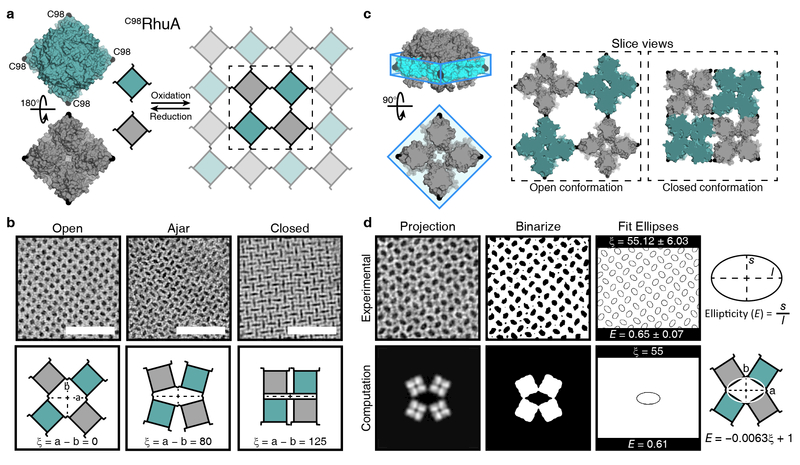
In order to streamline the protein design process and gain access to protein complexes whose structures can be modulated through chemical stimuli, our laboratory has employed strong but reversible bonding interactions (e.g., metal coordination17–19, disulfide bonds) to mediate protein self-assembly. In a recent example, a C4 symmetric tetrameric protein, L-rhamnulose-1-phosphate aldolase or RhuA, was functionalized with cysteines in its four corners at positions 98 (C98RhuA), which directed its self-assembly into 2-D crystalline lattices of μm dimensions via disulfide bond formation20 (Fig. 1a). Whereas the reversibility and redox-tunability of the disulfide bonds enabled the formation of defect-free crystals, the flexibility of these bonds allowed the C98RhuA lattices to isotropically expand and contract without losing crystallinity (Fig. 1b).
Figure 1.
Structural features of C98RhuA crystals. a, Surface representation of a C98RhuA tetramer, with positions 98 highlighted in black, and a schematic depiction of its oxidative self-assembly into porous 2D crystals. b, TEM and cartoon representations of the conformational states accessible by C98RhuA lattices and their respective ξ values. Scale bars are 50 nm. c, Description of the “interaction belt” (±10 Å of residues 98), which protrudes out from the protein. Slice views are of this belt region as viewed normal to the crystal. d, Overview of the calculation of the parameter E from experiment and its generalization to the coordinate ξ. The measured parameters and their converted values are reported for each example.
This coherent dynamic behavior has two unique and potentially useful physical consequences. First, owing to the rotary motion of the C98RhuA building blocks with respect to one another and the resultant retention of overall p4212 symmetry, these 2D lattices possess the lowest thermodynamically allowed Poisson’s ratio of –121 (i.e., they expand and contract by the same amount in both lateral dimensions). Materials with negative Poisson’s ratios (auxetic materials) are expected to display increased toughness and resistance to fracture and have been proposed for numerous applications22,23. Second, the C98RhuA lattices essentially are 2D molecular membranes whose porosity can be coherently changed from a pore diameter of ~4 nm in the fully open state to ~1 nm in the fully closed state (Fig. 1b). The transition from the open to closed state occurs spontaneously in aqueous suspension via a continuous population of many observable intermediate states, and is reversible upon mechanical agitation of the suspension. These unique physical properties of the 2D C98RhuA lattices have prompted the following questions: a) what are the determinants of the free-energy landscape that governs 2D C98RhuA lattice dynamics and b) can this landscape be engineered at the molecular level to modulate the collective structural and physical properties of the lattice? To address these questions, we carried out a combination of computational and experimental studies, which revealed the substantial role that entropy and water-protein interactions can play in protein structural dynamics, and allowed us to rationally modulate the lattice conformations and create mechanically and chemically switchable systems.
Results
Distinctive structural features of C98RhuA lattices
There are fundamental challenges in the computational and experimental mapping of the free energy landscape of a dynamic protein assembly. First, any protein-protein interface and the associated solvent molecules present an immense chemical complexity. This complexity, coupled with the many degrees of freedom of motion (i.e., interaction trajectories) that a dynamic interface can possess, gives rise to a multidimensional energy landscape which would be computationally intractable to model with atomistic accuracy. Second, the experimental evaluation of such a landscape or any of the trajectories it contains would be a considerable undertaking with any physical tool for studying structural dynamics. The C98RhuA lattices possess distinctive features that provide advantages in both regards:
C98RhuA lattice dynamics can be described by a simple free energy landscape with a single reaction coordinate, corresponding to the “openness” of the lattice pores. Due to the coherent dynamics of the lattice and the maintenance of its flatness during opening and closing at all times, a single parameter ξ is sufficient to define the reaction coordinate and the extent of interactions between neighboring protein units (Fig. 1b). We define ξ as the difference in the lengths of the two principal axes, a and b, of the pores, ranging from a minimum of 0 Å in the fully open state to a maximum of 125 Å in the fully closed state.
C98RhuA lattice energy landscape is further simplified by the topology and composition of RhuA surfaces. As viewed along its principal C4 symmetry axis (Fig. 1c), each C98RhuA building block is wider in its middle portion where the C98 residues are located compared to its top and bottom. This reduces the possible interaction surface between neighboring units to a small belt region (colored in cyan) as they hinge about the disulfide linkages. Additionally, the C98RhuA lateral surfaces including the belt regions are characterized by a diffuse distribution of hydrophilic residues and are expected to be non-self-interacting (Supplementary Fig. 2), thus giving rise to a landscape devoid of deep energy wells.
The reaction coordinate parameter ξ can be directly determined by transmission electron microscopy (TEM). Owing to the coherent dynamics of C98RhuA crystals, each lattice possesses a single value for pore ellipticity (E) that can be obtained accurately by TEM image processing (Fig. 1d). ξ can then be calculated through its linear relationship to E, where E = –0.0063ξ + 1. This relationship was determined by applying the same image processing procedure to calculated 2D projection maps of model structures with known ξ values (Supplementary Fig. 1). Furthermore, the fact that each TEM sample contains multiple individual C98RhuA lattices (prepared under the same condition) enables the statistical analysis of multiple “single-lattice” measurements.
Solvent entropy dominates the free-energy landscape of C98RhuA crystals
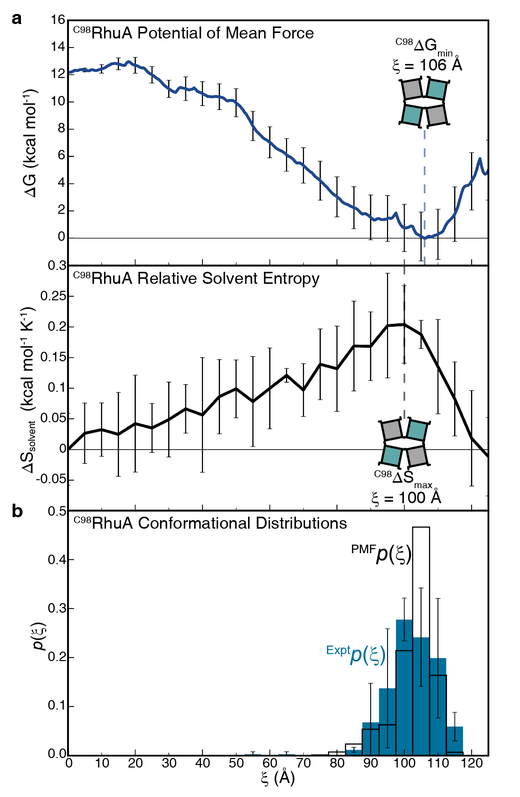
In order to evaluate the energy landscape of C98RhuA crystals, we carried out all-atom umbrella sampling (US) calculations on a set of four C98RhuA tetramers connected via intermolecular disulfide bonds (Fig. 1a). This 2×2 unit, containing 67,512 atoms, represents the minimum number of protein subunits required to explore the relevant conformational changes for C98RhuA crystals while still preserving the native lattice topology and ensuring computational tractability. As explained above, ξ was chosen as the reaction coordinate, which was divided into 26 windows from 0 to 125 Å centered at 5-Å intervals. Mild boundary conditions were employed to maintain lattice planarity imposed in the bulk crystal (Supplementary Fig. 10), and sampling was carried out at 300 K for 30 ns. Sampling was periodically tested for equilibration via the block averaging method24, and the system was judged to have equilibrated after 15 ns. Sampling statistics from the remaining 15 ns were combined and reweighted using the WHAM algorithm25 to generate a one-dimensional potential of mean force (PMF) (Fig. 2a, top).
Figure 2.
Thermodynamic analysis of C98RhuA lattice structural dynamics. a, The calculated free energy landscape and concomitant changes in solvation entropy over a continuous range of C98RhuA lattice conformations. Dotted lines identify the free energy minimum and solvation entropy maximum, and are included to guide the eye. Free energy error was calculated for each window using the block averaging method. Solvation entropy error bars reflect the standard error of mean of 8 independent calculations (see Supplementary Methods for full details). b, Direct comparison of the equilibrium conformations of C98RhuA crystals from experimental measurement (blue) and as calculated from the PMF (colorless). Experimental error bars reflect the standard deviation determined from three statistical analyses, each comprising >100 TEM images of C98RhuA crystals.
The C98RhuA PMF reveals a smooth landscape with a monotonic 12 kcal/mol decrease in free energy from the fully open state to a close-packed state centered at ξ = 106 Å, followed by an increase upon further compaction. This landscape with a distinct minimum is consistent with the spontaneous closure of the C98RhuA lattices in aqueous suspensions, and their conversion into open forms upon gentle mechanical agitation. Importantly, the Boltzmann population distribution of C98RhuA lattice states calculated from the PMF reveals a close match to the experimental distributions determined by TEM imaging (ξmin = 102.9±7.3 Å) (Fig. 2b; see Supplementary Fig. 1 for representative binarized TEM images). This observation provides strong validation for the efficacy of both the one-dimensional PMF along ξ to describe the C98RhuA lattice dynamics and the TEM experiments to sample them.
An examination of the trajectories generated in US simulations reveals that the interfaces between C98RhuA units remain separated by a modest distance even upon full closure (ξ = 120–125 Å) (Supplementary Fig. 2). In fact, the minimum projected protein-protein distance (excluding C98) is ca. 4 Å between D39 and L64’ side chains at ξ = 125 Å, with the vast majority of all contacts >5 Å. The buried interface area between the protein units was calculated to be 361 Å2 at ξ = 125 Å, and only 115 Å2 at ξ = 106 Å. These findings led us to consider the possibility that the free energy changes observed in the PMF were not due to enthalpically favorable protein-protein interactions, but were rather dominated by entropy. Generally, the entropic contributions to the free energy can be intuitively broken up into two components, protein configurational entropy (ΔSprotein) and solvent entropy (ΔSsolvent), both of which can be critical determinants of protein-protein interactions and protein crystallization26,27. Given that the compaction of the lattice cannot be expected to increase ΔSprotein and indeed was not found to (Supplementary Fig. 11), we investigated the role of ΔSsolvent through Grid Inhomogeneous Solvation Theory (GIST) calculations28. Sampling for GIST calculations was carried out on representative protein structures selected from each US window for 20 ns each, keeping the protein coordinates maintained at their original positions with a 10 kcal/mol restraint, and saving configurations every 0.5 ps. To provide an estimation for the error, we took advantage of the inherent 4-fold symmetry of the simulated system by splitting it into four symmetrically-equivalent quadrants, each containing one protein, and applying GIST to each quadrant individually. To avoid a systematic overestimation of the entropy by the nearest-neighbor algorithm employed by GIST29, the full trajectories were split into even and odd frames (1 frame/ps) and analyzed separately, thus permitting an additional degree of averaging for error estimation. The GIST results reveal a monotonic increase in ΔSsolvent from ξ = 0 Å to 100 Å and a rapid decrease after ξ = 105 Å (Fig. 2a, bottom), a profile that is almost exactly the mirror image of the PMF (Fig. 2a, top). This striking correspondence between solvent entropy and lattice free energy, coupled with the lack of appreciable protein-protein contacts, strongly suggests that C98RhuA lattice dynamics are driven by solvent entropy.
Water structure reorganization during C98RhuA lattice motions
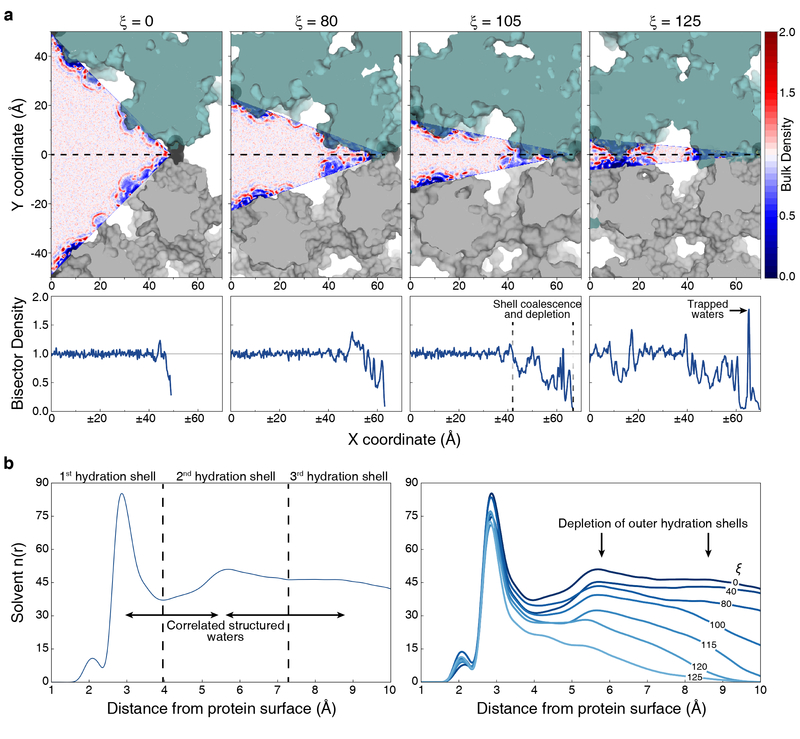
As the microscopic detail afforded by molecular dynamics simulations allows solvent reorganization to be rigorously interrogated, we sought to rationalize the entropic changes accompanying C98RhuA lattice dynamics in the context of molecular hydration theory. We initially characterized the density profiles as a function of ξ, which is conveniently projected into a 2D plot due to the low dimensionality of the system (Fig. 3a, top; Supplementary Fig. 3). Consistent with previous computational30,31 and experimental32,33 studies, the anomalous structure of water within the hydration shells of biomolecules are clearly visible as density variations which line the perimeter of the pore and propagate ~10 Å away from the protein surface. Strong protein-water interactions give rise to the high density primary hydration layer, which in turn induces additional structure in the form of concentric density peaks. As neighboring RhuA subunits draw closer with increasing ξ, their hydration shells coalesce, leading to an ejection of these semi-structured water molecules into the bulk solvent (Fig. 3a, bottom). Inspection of the number distribution of water molecules as a function of distance from the protein surface reveals that it is primarily the secondary and tertiary hydration shells which are released into the bulk (Fig. 3b, right). Upon further narrowing of the pore, isolated pockets of high density manifest at the acute hinge regions (Fig. 3a, bottom), which reflects a trapping of water molecules by the protein surfaces. This trapping effect may contribute to the sharp decrease in solvent entropy34 for ξ >105 Å.
Figure 3.
Consequences of lattice compaction on solvent structure within the pore. a, Two-dimensional plots of the normalized water density within the pore. 1D plots of the density along the pore bisector are shown below to facilitate quantitative interpretation of the data. Density scales are relative to the bulk density of neat water. Labels here and in b identify hydration effects of interest. b, Number distributions of solvent molecules proximal to the protein surface. Concentric hydration shells around the protein are identified as peaks in the distribution for ξ = 0 (left).
The computational tractability afforded by the protein structure provides an opportunity to examine the structure of water under confinement for a non-idealized system. We calculated two additional order parameters for water within the pore (see Supplementary Methods): qtetra, a measure of local tetrahedral structure, and coordination number (CN), the number of neighboring solvent molecules within 3.5 Å of a given water. We find that qtetra decreases on average as ξ increases, although at very high values of ξ there is also an increase in the fraction of ice-like (tetrahedral) water molecules, with qtetra values close to 1 (Supplementary Fig. 4). Similarly, we find that the average CN decreases with increasing ξ (Supplementary Fig. 4), which is indicative of the isolation of water molecules from other solvent molecules by the proteins. When these parameters are averaged over increasingly thick slabs within the pores, water molecules become progressively more bulk-like, further emphasizing that most of the perturbation to the water structure occurs within the interaction belt and thus directly reflects the effect of nearby protein surfaces.
Rational perturbation of the free energy landscape of C98RhuA lattices
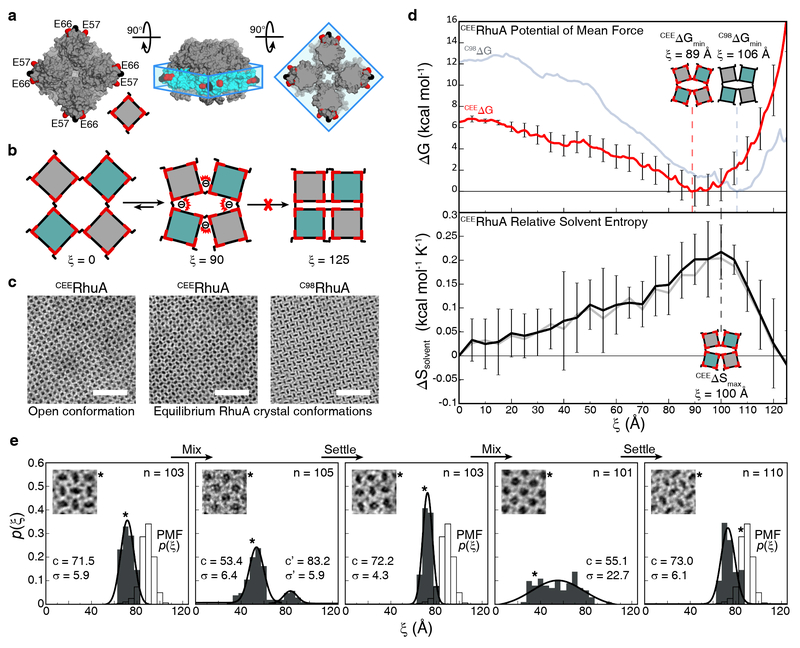
We next set out to rationally modulate the free energy landscape of C98RhuA lattices through site-specific modification of protein interaction surfaces. Given the non-trivial nature of engineering entropic factors, we envisioned that electrostatically repulsive (i.e., enthalpically unfavorable) interactions placed near the C98-C98 hinges could hinder the closure of the lattices independently of solvent entropy, thus yielding an altered equilibrium state compared to C98RhuA. To achieve this effect, we chose to replace the residues P57 and A66 with negatively charged glutamates; these positions lie at comparable heights within the “interaction belt” and are immediately adjacent to the C98-C98 hinges to maximize the range over which their electrostatic repulsion can influence the lattice dynamics (Fig. 4a).
Figure 4.
Design and analysis of the designed construct CEERhuA. a, Surface representation of a CEERhuA tetramer, with residues 98 and 57/66 colored in black and red, respectively. The installed residues are coplanar with C98 and lie within the “interaction belt”. b, Cartoon of CEERhuA lattice dynamics and the anticipated effects of the design. c, TEM images of RhuA lattices, showing the open and equilibrium states for CEERhuA alongside that for a fully closed C98RhuA crystal. Scale bars are 50 nm. d, Relative to C98RhuA, the thermodynamic analysis of CEERhuA dynamics reveals a shifted free energy minimum towards the open state, but retention of a nearly identical solvent entropy profile, demonstrating the purely enthalpic consequences of the design. The free energy and solvent entropy profiles for CEERhuA are shown as red and black lines; those for C98RhuA are depicted as faint blue and grey lines. Dotted lines identify the free energy minimum and solvation entropy maximum, and are included to guide the eye. Free energy error was calculated for each window using the block averaging method. Solvation entropy error bars reflect the standard error of mean of 8 independent calculations (see Supplementary Methods for full details). e, Experimental distributions of CEERhuA conformations over multiple cycles of sedimentation and resuspension. Equilibrium distributions are significantly more open than C98RhuA, and slightly more open than predicted by the PMF. Gaussian fits to each distribution are labeled with their center (c) and standard deviation (σ). n is the number of crystals analyzed. The lattice conformation of each inset is marked with an asterisk.
We first evaluated the consequences of the designed mutations in silico. The US calculations for the E57/E66/C98-RhuA mutant (CEERhuA) were carried out in an identical fashion to those for C98RhuA, with the exception that stronger force constants were required at ξ >110 Å (Supplementary Table 1), an early indication that the mutations disfavored the closed conformations. The PMF of the CEERhuA lattice is shown in Fig. 4b overlaid with that of C98RhuA to highlight their differences. The driving force for CEERhuA lattice closure (–6 kcal/mol) is halved compared to the C98RhuA lattice due to repulsive electrostatic interactions, which give rise to a shallower landscape at low ξ values and a steep increase at ξ >100 Å due the proximity of E57 and E66 sidechains upon lattice closure. GIST calculations (Fig. 4d, bottom) revealed that the solvent entropy profile for the CEERhuA lattice is nearly identical to that of C98RhuA (faint underlay), indicating that the observed changes in the CEERhuA energy landscape are entirely enthalpic in origin.
An important outcome of E57 and E66 mutations is that the PMF energy minimum is shifted from ξmin = 106 Å for C98RhuA to ξmin = 89 Å for CEERhuA (Fig. 4d, top), which predicts a considerably less compact lattice conformation at equilibrium for the latter. To experimentally probe this prediction, we prepared the CEERhuA mutant and optimized the solution conditions for its self-assembly into μm-sized, 2D lattices in high yields (Supplementary Fig. 5). To attain the equilibrium state for these lattices, the crystal suspensions were left unperturbed for three days during which they sedimented to the bottom of the container, as was also observed for the C98RhuA crystals. In accordance with the predictions, TEM analysis of equilibrated CEERhuA lattices revealed a significantly more relaxed conformation (termed “ajar”) compared to C98RhuA, with a tight distribution of states centered at ξmin = 72.1±5.2 Å (Fig. 4e, “settled” states). This value is smaller than that predicted by the PMF, but corresponds to deviation of only ~2 kcal/mol, within the error of experimental measurements and the precision of US calculations. The reduced accuracy of the predicted minimum may be exacerbated by the shallowness of the CEERhuA landscape. A statistical analysis of the distribution of CEERhuA lattice conformations was carried out over multiple rounds of vigorous mixing followed by sedimentation (Fig. 4e), establishing that the opening-closing dynamics of this variant were also reversible and mechanically actuated.
Selective, metal-mediated switching of CEERhuA lattices
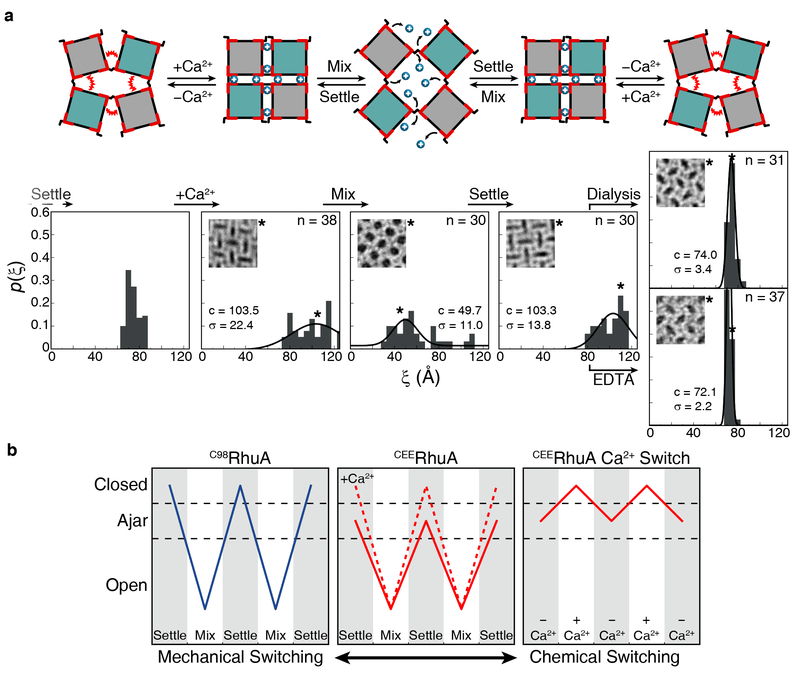
An inspection of the CEERhuA conformations from US simulations indicates that E57 and E66 carboxylate groups could come within sufficient proximity (<5 Å) upon lattice compaction to enable metal coordination across protein-protein interfaces (Supplementary Fig. 6). Thus, we reasoned that the interfacial E57 and E66 dicarboxylate motifs may engender a chemically switchable system whereby the full closure of the lattice may be induced by metal binding (Fig. 5a). Indeed, we found that the addition of Ca2+ to CEERhuA lattices in the equilibrium “ajar” state led to a shift in the population of lattices to higher ξ values in a concentration-dependent manner (Supplementary Fig. 8), reaching a distribution that is similar to the closed form of C98RhuA lattices at 20 mM Ca2+ (Fig. 5a). In contrast, similar amounts of Mg2+ or up to 150 mM monovalent cations (Na+ or K+) did not cause any noticeable deviation from the “ajar” state (Supplementary Fig. 7), but the addition of 20 mM Ca2+ to CEERhuA lattices pre-treated with 20 mM Mg2+ induced their closure (Supplementary Fig. 8). These results indicate that observed effect of Ca2+ on the lattice conformational state is selective and not due to a non-specific electrostatic screening effect. Like C98RhuA lattices, the Ca2+-incubated CEERhuA lattices could be reverted into a fully open state upon mechanical mixing (Fig. 5a). Importantly, these lattices could be switched to the “ajar” state upon dialysis or treatment with ethylenediaminetetraacetate (EDTA) to remove Ca2+, and this process could be reversed upon re-incubation with Ca2+.
Figure 5.
Chemical and mechanical switching behavior of CEERhuA crystals. a, Cartoon depicting all possible switching modes of CEERhuA lattices, with each cartoon state directly corresponding to the experimental distribution(s) below it. Addition of 20 mM Ca2+ to the equilibrium “ajar” population of CEERhuA crystals induces a shift towards more closed conformations, from which C98RhuA-like mechanical switching is possible. The ajar conformation is fully recoverable upon removal of Ca2+ via dialysis or EDTA, thus providing three distinct switching modes. Gaussian fits to each distribution are labeled with their center (c) and standard deviation (σ). n is the number of crystals analyzed. The lattice conformation of each inset is marked with an asterisk. b, Summary of switching modes for RhuA crystals. In contrast with C98RhuA, CEERhuA has two mechanical modes dictated by the presence of Ca2+, as well as a purely chemical mode via the addition/removal of Ca2+.
The conformational properties of C98RhuA and CEERhuA lattices are summarized in Fig. 5b. Whereas the C98RhuA system could only be mechanically switched between open and closed states, CEERhuA lattices can adapt two different mechanical switching modes (open-closed or open-ajar) or a chemical switching mode (ajar-closed), depending on the presence or the absence of Ca2+ in the system. Thus, CEERhuA lattices provide a unique 2D membrane system (or molecular display) whose porosity (or display density) can be chemically and mechanically toggled between three different states in a fully coherent fashion.
Discussion
An essential feature of natural protein assemblies is their ability to adapt their structures and functions with high fidelity and coherence in specific response to environmental cues. Perhaps one of the best-known examples is the cooperative O2 binding behavior of hemoglobin that is differentially regulated by pH or CO2 concentration, as well as by specific metabolites like 2,3-biphosphoglyceric acid35, although essentially every cellular protein is subject to some type of regulation by external stimuli. Accordingly, a major goal in the de novo design of proteins is to construct dynamic systems whose structural adaptability can be predicted and programmed at a molecular level. Toward this end, we have reported here the first example of a synthetic protein assembly whose free energy landscape was fully delineated and whose mechano- and chemo-responsive structural switching behavior was predictably engineered. In this regard, a key design element was the use of strong but reversible and flexible bonding interactions to assemble the 2D RhuA lattices. The small footprint of the disulfide bonds on the protein surfaces not only endowed the RhuA lattices with an immense range of conformational flexibility, but it also allowed other types of interactions (electrostatic/metal coordination) to be “dialed in” to predictably alter its free-energy landscape and control its structural dynamics. In an alternative strategy, one can envision the en-masse design of more extensive, non-covalent interfaces between protein subunits that could accommodate multiple discrete conformations which interchange upon application of stimuli of interest, as found in natural functional complexes. This remains an exciting but challenging goal for the future that will require considerable advances in the understanding and control of non-covalent interactions, as well as of protein-solvent interactions and entropy, as our study indicates. In addition to these general protein design considerations, the dynamic two-dimensionality of RhuA lattices also has specific functional implications. Owing to their high surface-to-volume ratios and uniform porosities, 2D nanomaterials are particularly well-suited for applications in separation/filtration36, templating37, sensing38, catalysis39, coatings40, among many others41. The unique ability to control and tune the porosity or molecular density of RhuA lattices could offer important advantages in such applications and the fabrication of adaptive molecular devices.
Supplementary Material
Acknowledgements
We thank M. Gilson, T. Kurtzman, and S. Ramsey for helpful discussions regarding GIST, R. Subramanian for assistance with generation of projection maps from computational models, and T. Baker for use of EM facilities. This work was primarily supported by the US Department of Energy (DOE) (Division of Materials Sciences, Office of Basic Energy Sciences, Award DE-SC0003844 to F.A.T.). F.P. was supported by the National Science Foundation through grant CHE-1453204 (computation). R.A. was supported in part by the UCSD NIH Molecular Biophysics Training Grant (T32-GM08326). All computer simulations were performed on the Extreme Science and Engineering Discovery Environment (XSEDE), which is supported by the National Science Foundation through grant ACI-1053575.
Footnotes
Data Availability
The principal data supporting the findings of this work are available within the figures and the Supplementary Information. Additional data that support the findings of this study are available from the corresponding authors on request.
Author contributions
R.A. co-initiated the project, designed and performed all of the experiments, simulations, and data analysis, and co-wrote the paper. Y.S. performed TEM data collection and analysis. F.P. guided simulation design and computational data analysis, and co-wrote the paper. F.A.T. initiated the project, guided experiment design and data analysis, and co-wrote the paper.
Additional information
Supplementary information is available in the online version of the paper. Reprints and permissions information is available online at www.nature.com/reprints. Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations. Correspondence and requests for materials should be addressed to F.A.T and F. P.
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Marsh JA & Teichmann SA Structure, Dynamics, Assembly, and Evolution of Protein Complexes. Annu. Rev. Biochem 84, 551–575 (2015). [DOI] [PubMed] [Google Scholar]
- 2.Salgado EN, Radford RJ & Tezcan FA Metal-directed protein self-assembly. Acc. Chem. Res 43, 661–672 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Song WJ & Tezcan FA A designed supramolecular protein assembly with in vivo enzymatic activity. Science 346, 1525–1528 (2014). [DOI] [PubMed] [Google Scholar]
- 4.Yeates TO Geometric Principles for Designing Highly Symmetric Self-Assembling Protein Nanomaterials. Annu. Rev. Biophys 46, 23–42 (2017). [DOI] [PubMed] [Google Scholar]
- 5.Bale JB et al. Accurate design of megadalton-scale two-component icosahedral protein complexes. Science 353, 389–394 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Koga N et al. Principles for designing ideal protein structures. Nature 491, 222 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Norn CH & André I Computational design of protein self-assembly. Curr. Opin. Struct. Biol 39, 39–45 (2016). [DOI] [PubMed] [Google Scholar]
- 8.Ringler P & Schulz GE Self-Assembly of Proteins into Designed Networks. Science 302, 106–109 (2003). [DOI] [PubMed] [Google Scholar]
- 9.Sinclair JC, Davies KM, Venien-Bryan C & Noble MEM Generation of protein lattices by fusing proteins with matching rotational symmetry. Nat. Nanotechnol 6, 558–562 (2011). [DOI] [PubMed] [Google Scholar]
- 10.Levy Y & Onuchic JN Water Mediation in Protein Folding and Molecular Recognition. Annu. Rev. Biophys. Biomol. Struct 35, 389–415 (2006). [DOI] [PubMed] [Google Scholar]
- 11.Young T, Abel R, Kim B, Berne BJ & Friesner RA Motifs for molecular recognition exploiting hydrophobic enclosure in protein–ligand binding. Proc. Natl. Acad. Sci 104, 808–813 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hedstrom L Serine Protease Mechanism and Specificity. Chem. Rev 102, 4501–4524 (2002). [DOI] [PubMed] [Google Scholar]
- 13.Davey JA, Damry AM, Goto NK & Chica RA Rational design of proteins that exchange on functional timescales. Nat. Chem. Biol 13, 1280 (2017). [DOI] [PubMed] [Google Scholar]
- 14.Joh NH et al. De novo design of a transmembrane Zn2+-transporting four-helix bundle. Science 346, 1520–1524 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cerasoli E, Sharpe BK & Woolfson DN ZiCo: A Peptide Designed to Switch Folded State upon Binding Zinc. J. Am. Chem. Soc 127, 15008–15009 (2005). [DOI] [PubMed] [Google Scholar]
- 16.Ambroggio XI & Kuhlman B Computational design of a single amino acid sequence that can switch between two distinct protein folds. J. Am. Chem. Soc 128, 1154–1161 (2006). [DOI] [PubMed] [Google Scholar]
- 17.Brodin JD et al. Metal-directed, chemically tunable assembly of one-, two- and three-dimensional crystalline protein arrays. Nat. Chem 4, 375–382 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sontz PA, Bailey JB, Ahn S & Tezcan FA A metal organic framework with spherical protein nodes: rational chemical design of 3D protein crystals. J. Am. Chem. Soc 137, 11598–11601 (2015). [DOI] [PubMed] [Google Scholar]
- 19.Bailey JB, Subramanian RH, Churchfield LA & Tezcan FA Metal-Directed Design of Supramolecular Protein Assemblies. Methods Enzymol. 580, 223–250 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Suzuki Y et al. Self-assembly of coherently dynamic, auxetic, two-dimensional protein crystals. Nature 533, 369–373 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grima JN & Evans KE Auxetic behavior from rotating squares. J. Mater. Sci. Lett 19, 1563–1565 (2000). [Google Scholar]
- 22.Yang W, Li Z-M, Shi W, Xie B-H & Yang M-B Review on auxetic materials. J. Mater. Sci 39, 3269–3279 (2004). [Google Scholar]
- 23.Evans KE & Alderson A Auxetic Materials: Functional Materials and Structures from Lateral Thinking! Adv. Mater 12, 617–628 (2000). [Google Scholar]
- 24.Zhu F & Hummer G Convergence and error estimation in free energy calculations using the weighted histogram analysis method. J. Comput. Chem 33, 453–465 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kumar S, Rosenberg JM, Bouzida D, Swendsen RH & Kollman PA The Weighted Histogram Analysis Method for Free-Energy Calculations on Biomolecules. I. The Method. J. Comput. Chem 13, 1011–1021 (1992). [Google Scholar]
- 26.De Yoreo JJ & Vekilov PG Principles of Crystal Nucleation and Growth. Rev. Mineral Geochem 54, 57–93 (2003). [Google Scholar]
- 27.Derewenda ZS & Vekilov PG Entropy and surface engineering in protein crystallization. Acta Crystallogr. D 62, 116–124 (2006). [DOI] [PubMed] [Google Scholar]
- 28.Nguyen CN, Young TK & Gilson MK Grid inhomogeneous solvation theory: Hydration structure and thermodynamics of the miniature receptor cucurbit[7]uril. J. Chem. Phys 137, 044101 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huggins DJ Comparing distance metrics for rotation using the k-nearest neighbors algorithm for entropy estimation. J. Comput. Chem 35, 377–385 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Giovambattista N, Rossky PJ & Debenedetti PG Effect of Temperature on the Structure and Phase Behavior of Water Confined by Hydrophobic, Hydrophilic, and Heterogeneous Surfaces. J. Phys. Chem. B 113, 13723–13734 (2009). [DOI] [PubMed] [Google Scholar]
- 31.Tarek M & Tobias DJ The Dynamics of Protein Hydration Water: A Quantitative Comparison of Molecular Dynamics Simulations and Neutron-scattering Experiments. Biophys. J 79, 3244–3257 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ebbinghaus S et al. An extended dynamical hydration shell around proteins. Proc. Natl. Acad. Sci 104, 20749–20752 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heyden M et al. Dissecting the THz spectrum of liquid water from first principles via correlations in time and space. Proc. Natl. Acad. Sci 107, 12068–12073 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dunitz JD The Entropic Cost of Bound Water in Crystals and Biomolecules. Science 264, 670–670 (1994). [DOI] [PubMed] [Google Scholar]
- 35.Yuan Y, Tam MF, Simplaceanu V & Ho C New Look at Hemoglobin Allostery. Chem. Rev 115, 1702–1724 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Joshi RK et al. Precise and Ultrafast Molecular Sieving Through Graphene Oxide Membranes. Science 343, 752–754 (2014). [DOI] [PubMed] [Google Scholar]
- 37.Tahara K, Nakatani K, Iritani K, De Feyter S & Tobe Y Periodic Functionalization of Surface-Confined Pores in a Two-Dimensional Porous Network Using a Tailored Molecular Building Block. ACS Nano 10, 2113–2120 (2016). [DOI] [PubMed] [Google Scholar]
- 38.Huber C et al. Heterotetramers Formed by an S-Layer–Streptavidin Fusion Protein and Core-Streptavidin as a Nanoarrayed Template for Biochip Development. Small 2, 142–150 (2006). [DOI] [PubMed] [Google Scholar]
- 39.Li Y et al. MoS2 Nanoparticles Grown on Graphene: An Advanced Catalyst for the Hydrogen Evolution Reaction. J. Am. Chem. Soc 133, 7296–7299 (2011). [DOI] [PubMed] [Google Scholar]
- 40.Mader C, Küpcü S, Sleytr UB & Sára M S-layer-coated liposomes as a versatile system for entrapping and binding target molecules. Biochim. Biophys. Acta. - Biomembranes 1463, 142–150 (2000). [DOI] [PubMed] [Google Scholar]
- 41.Butler SZ et al. Progress, Challenges, and Opportunities in Two-Dimensional Materials Beyond Graphene. ACS Nano 7, 2898–2926 (2013). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.