Abstract
Proteins in the fibrous amyloid state are a major hallmark of neurodegenerative disease. Understanding the multiple conformations, or polymorphs, of amyloid proteins at the molecular level is a challenge of amyloid research. Here, we detail the wide range of polymorphs formed by a segment of human TAR DNA-binding protein 43 (TDP-43) as a model for the polymorphic capabilities of pathological amyloid aggregation. Using X-ray diffraction, microelectron diffraction (MicroED) and single-particle cryo-EM, we show that the 247DLIIKGISVHI257 segment from the second RNA-recognition motif (RRM2) forms an array of amyloid polymorphs. These associations include seven distinct interfaces displaying five different symmetry classes of steric zippers. Additionally, we find that this segment can adopt three different backbone conformations that contribute to its polymorphic capabilities. The polymorphic nature of this segment illustrates at the molecular level how amyloid proteins can form diverse fibril structures.
The formation of elongated, unbranched fibrillar protein aggregates, termed amyloid, has been linked to neurodegenerative diseases including Alzheimer’s diseases, Parkinson’s disease, prion diseases and amyotrophic lateral sclerosis (ALS), among others1–4. For decades, scientists have sought molecular structures for amyloid aggregates and have attempted to understand the range of polymorphic fibrillar structures formed by proteins in the amyloid state5–7. These questions have been approached in recent years using diffraction methods8–10, solid-state NMR11–15, and cryo-EM16–18. These studies have shown that amyloid fibrils consist of packed β-sheets that run parallel or roughly parallel to the fibril axes. Each β-sheet adheres to its neighboring sheets through the side chains that project roughly perpendicular to the fibril axis, toward the neighboring sheet. Because the side chains of mating sheets can interdigitate like the teeth of a zipper, this interaction has been called a steric zipper8.
Steric zippers can form between identical or nonidentical β-sheets. A common property of β-sheets in the amyloid state is the ability to pack in a variety of ways to form several different fibril structures. Such polymorphic amyloid assemblies are well known. Research on amyloid-β has demonstrated how conformational variations can lead to distinct assemblies of amyloid aggregates5,6. Different polymorphs of the same amyloid protein can exhibit different biophysical or biomedical properties. In the case of prion protein, distinct strains can confer different toxic effects as well as transmission capabilities on the basis of the fibrillar structure adopted by the protein19. Recently, tau has also been shown to form over a dozen different self-propagating amyloid strains20. Because polymorphs can determine transmission and severity of disease, understanding polymorph structure has important medical implications.
Structural studies of amyloid at the atomic level have offered evidence for two basic models to explain the frequent observation that a single protein produces amyloid fibrils of varying morphologies or polymorphs. In the first model, termed segmental polymorphism, the segment forming the zipper spine differs between amyloid fibrils21,22. Segmental polymorphism has been observed in a number of proteins, including islet amyloid polypeptide (IAPP), in which various six- and eleven-residue segments of IAPP are each capable of forming zippers independently23,24. In a second model, termed packing polymorphism, a single zipper-forming segment packs differently in different amyloid fibrils21,22. Outside of amyloid research, this type of packing is commonly referred to as modal polymorphism. We have observed this polymorphism by the amyloid-β segment, 16KLVFFA21, in which three distinct zipper interfaces were revealed by three crystal forms25. Both types of polymorphism presume homotypic zipper interfaces, i.e., two mated sheets both composed of the same segment. However, analogous polymorphisms are also likely to exist for heterotypic zippers, in which the two mated sheets are derived from sequentially distant segments21,22, as in solid-state NMR structures of α-synuclein and amyloid-β11–13. The abundance of examples of segmental and packing variations observed between distinct fibrils raises the question of whether these same variations play a role within a single fibril, a phenomenon termed positional polymorphism. If so, to what extent does positional polymorphism expand the repertoire of amyloid polymorphs accessible to a given sequence?
Aggregates of TDP-43 have been described in patients with ALS and frontotemporal lobar degeneration (FTLD)26,27 and more recently in patients with Alzheimer’s, Parkinson’s and Huntington’s diseases28–31. Distinct from other amyloid and amyloid-like proteins, where loss of function is not deadly, TDP-43 is essential for cellular homeostasis, and knockdown of TDP-43 in mice results in embryonic lethality32–34. In addition, changes in TDP-43 expression levels lead to motor deficits and loss of body fat in mouse models32,35. This finding has led to the hypothesis that TDP-43 aggregation can confer toxicity through a loss-of-function mechanism. In this case, TDP-43 is redistributed from the nucleus to the cytoplasm, as seen in neuronal cell culture experiments36,37. While the effects of the nuclear clearance of TDP-43 are still being investigated, it has been shown that loss of TDP-43 function results in the inclusion of cryptic exons in transcripts, improper regulation of the Microprocessor complex and dysregulation of Rho family GTPases35,38,39. Thus, understanding the mechanism by which TDP-43 aggregates has become an essential focus of research in ALS and other neurodegenerative diseases. By understanding how the protein aggregates, aggregation inhibitors can be developed, and protein function can be restored, as demonstrated with p53 in ovarian carcinomas40.
Here, we apply three structural methods, X-ray diffraction, MicroED, and single-particle cryo-EM, to determine three different assemblies of a segment in RRM2 of TDP-43. We find that the extensive positional polymorphism of this segment offers insight into the diverse structures this peptide can form. We propose such positional polymorphism is not limited to TDP-43, but a common phenomenon among amyloid proteins in general.
Results
TDP-43 RRM2 segments exhibit amyloid properties
Several studies have pointed to the C-terminal low-complexity domain (LCD) of TDP-43 as responsible for pathological amyloid aggregation. Specifically, segment 286–331 has been shown to form amyloid fibrils and confer neurotoxicity on primary cortical neurons41. Residues 311–360 have been proposed to be the amyloid core through NMR and CD studies42,43. Finally, it has been shown that residues 341–367 can drive pathological aggregation of TDP-43 in neuronal cell lines44, residues 342–366 transition from a random coil to a β-sheet45, and that deletion of residues 318–343 delays aggregation of full-length TDP-43 (ref. 42). These findings offer strong support for the role of the C terminus in TDP-43 aggregation.
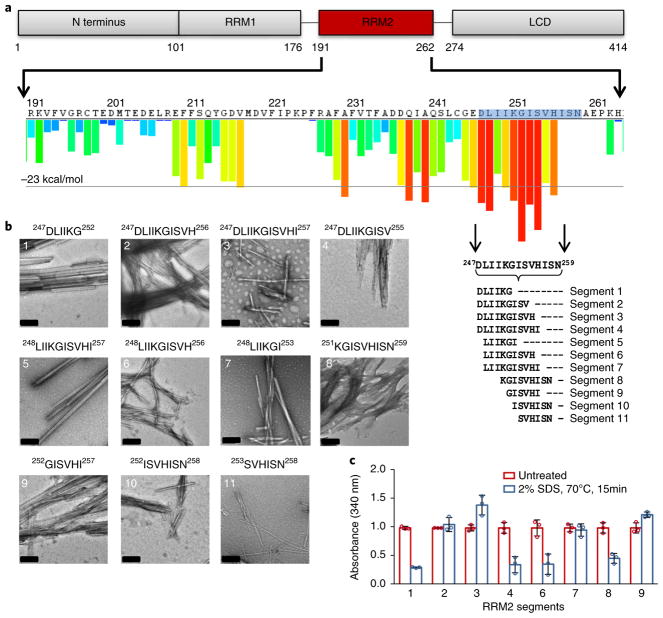
On the other hand, two other studies have shown that segments outside the C terminus are capable of forming fibrils. Peptide segments from the RRM2, 246–255 and 247–251, were proven to be fibrillogenic46,47. Segment 246–255 did not initiate nucleation; however, it was shown to assist in the promotion of amyloid fibril formation47. Additionally, when we threaded the TDP-43 sequence through the 3D profile method ZipperDB, which predicts segments likely to form amyloid steric zippers, five of the top 13 hits were localized to a 13-residue segment, 247DLIIKGISVHISN259, of the RRM2 (ref. 48) (Fig. 1a). This finding is significant, as there are 408 possible six-residue zippers that the 3D profile method evaluates for the 414-residue TDP-43. The presence of five different putative zipper-forming segments within a short residue span indicated that this segment could form a number of different structures or polymorphs. With the results of the ZipperDB output, and previous literature indicating the high propensity of this region for fibril formation, we sought to structurally characterize segment 247–259 to gain insight into polymorphic amyloid assemblies and the aggregation of TDP-43.
Fig. 1. Segments from the RRM2 domain of TDP-43 exhibit characteristic amyloid properties.
a, Schematic of TDP-43 domains (top) showing the sequence of RRM2 (middle) and ZipperDB predictions (histogram); predicted amyloid-promoting segments are shown as red bars)48. Four of the top ten potential zipper-forming peptide segments lie between residues 247 and 259, the RRMcore. The 11 segments chosen for further characterization are displayed below. b, Negatively stained EM images of 5 mM fibril samples of segments in the 247DLIIKGISVHISN259 region grown in PBS. Segments range 6–11 residues in length and span the entire 13-residue RRMcore. Fibril morphology differs for many of the segments, indicating the potential for polymorphic assembly in this region. Scale bars, 200 nm. c, Segments from the RRM2 exhibit moderate to high stability under heat and SDS denaturation. Fibril samples were grown then treated with 2% SDS and heated to 70 °C for 15 min. Data were normalized with untreated samples at an initial value of 1. Graph shows mean ± s.d. for experiments done in triplicate for distinct samples; individual data points are shown as circles. Samples that did not yield a high enough load for turbidity assays were not used. Additionally, some peptide segments exhibit greater stability when shaken for different time points or in higher-speed shakers. Data reported here are for samples prepared in the same shaker for the same duration of time.
We decided to biochemically characterize 11 segments within this region, ranging 6–11 residues in length, with average hydropathicity values ranging from 0.07 to 2.17 (ref. 49) (Fig. 1a and Supplementary Table 1). To determine whether these segments exhibit amyloid properties, we tested their propensity to aggregate, their stability under denaturing conditions, and their fibril diffraction pattern. We found that all 11 segments formed fibrils when shaken for one week (Fig. 1b). On the basis of previous studies demonstrating that amyloid fibrils are stable when incubated with SDS and heat, we subjected the fibrils to a 2% SDS solution at 70 °C for 15 min50. We found that under these extremely harsh denaturing conditions, the fibrils exhibited moderate to high stability. Segments 247DLIIKGISV255, 247DLIIKGISVH256, 248LIIKGISVHI257, and 252GISVHI257 all exhibited the highest stability, showing almost no dissociation of fibrils (Fig. 1c and Supplementary Fig. 1). Finally, we checked whether the dried fibril samples exhibit the characteristic cross-β diffraction pattern indicative of amyloid fibrils51. For the nine samples on which we were able to study diffraction, all exhibited reflections at spacings of 4.6–4.7 Å and 8.8–11.5Å, which is consistent with the presence of a steric-zipper-based amyloid fibril (Supplementary Fig. 2).
248LIIKGI253 and 247DLIIKGISVHI257 form distinct steric zippers
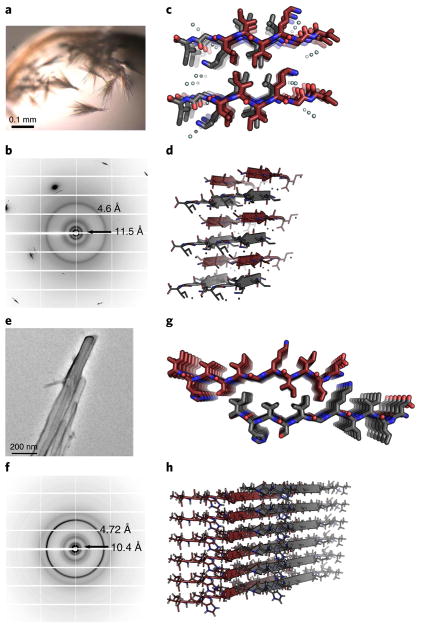
Following biochemical characterization of peptide segments from the 13-residue range within the RRM2, we attempted crystallization of all 11 segments. We successfully determined the structures of two constructs, 248LIIKGI253 and 247DLIIKGISVHI257, through the use of X-ray diffraction and MicroED, respectively.
Crystals of 248LIIKGI253 were grown in hanging-drop 96-well trays and were visible by light microscopy (Fig. 2a). The fibril diffraction pattern revealed rings at 4.6 Å and 11.5 Å, which are consistent with amyloid structure (Fig. 2b). Initial processing of the diffraction data from a microfocus X-ray beamline provided unit cell dimensions of 11.54 Å, 9.59 Å, and 21.18 Å, indicating an antiparallel β-sheet. Molecular replacement and refinement revealed a face = back, up-up, in-register antiparallel class 7 steric zipper9 (Table 1 and Fig. 2c). The hydrogen bond network is such that the sheet comprises only the first four residues, LIIK (Fig. 2d), thus leaving two-residue overhangs and a ragged appearance to the sheet edges. Owing to its ragged edges, the buried surface area is relatively low at 117 Å2; however, the shape complementarity is high at 0.78.
Fig. 2. Atomic structures of 248LIIKGI253 and 247DLIIKGISVHI257, determined by diffraction methods, illustrate different steric zipper interfaces.
a, Microcrystals of 248LIIKGI253 were grown in hanging-drop experiments at pH 8.5. Crystals were visible by light microscopy. b, Fibril diffraction of the 248LIIKGI253 sample exhibits reflections consistent with the characteristic cross-β pattern, showing 4.6-Å and 11.5-Å diffraction rings. c, The structure of 248LIIKGI253 was determined from X-ray diffraction data. The segment forms an antiparallel, face = back (class 7) steric zipper interface9, viewed here down the fibril axis. d, View of 248LIIKGI253, perpendicular to the fibril axis, illustrates the antiparallel stacking of the strands. e, Nanocrystals of the 247DLIIKGISVHI257 were grown in a shaker at 37 °C at a pH of 8.5. Crystals were invisible by light microscopy and successfully visualized using an electron microscope. f, Fibril diffraction of the 247DLIIKGISVHI257 segment exhibits reflections consistent with the characteristic cross-β pattern with 4.7-Å and 10.4-Å diffraction rings. g, The structure of 247DLIIKGISVHI257 was determined by MicroED. The segments form a parallel face-to-back class 2 steric zipper interface, viewed here down the fibril axis. h, Orthogonal view of the 247DLIIKGISVHI257 structure illustrates the stacking of β-strands.
Table 1.
Data collection and refinement statistics
| LIIKGI (PDB 5W50) | DLIIKGISVHI (EMD-8765, PDB 5W52) | |
|---|---|---|
| Data collection | ||
| Space group | P1 | P1 |
| Cell dimensions | ||
| a, b, c (Å) | 11.54, 9.59, 21.18 | 24.81, 4.73, 15.83 |
| α, β, γ (°) | 905.63, 98.21, 76.50 | 80.88, 86.37, 89.78 |
| Resolution (Å) | 20.91–1.40 (1.45–1.40)a | 15.60–1.40 (1.43–1.40) |
| Rsym | 12.7 (36.1) | – |
| Rmerge | – | 20.3 (145.1)b |
| I/σ(I) | 11.5 (5.05) | 3.34 (0.92) |
| CC1/2 | 0.979 (0.879) | 0.992 (0.333) |
| Completeness (%) | 94.5 (94.6) | 73.4 (62.1) |
| Redundancy | 4.6 (4.5) | 9.0 (6.7) |
| Refinement | ||
| Resolution (Å) | 20.91–1.40 | 15.60–1.40 |
| No. reflections | 1,401 | 1,036 |
| Rwork/Rfree | 16.8 / 20.8 | 26.2 / 30.6 |
| No. atoms | ||
| Protein | 92 | 84 |
| Water | 9 | 0 |
| B factors | ||
| Protein | 7.97 | 33.41 |
| Water | 19.52 | – |
| R.m.s. deviations | ||
| Bond lengths (Å) | 0.013 | 0.009 |
| Bond angles (°) | 1.643 | 1.211 |
Values in parentheses are for highest-resolution shell.
Data are from 13 crystals.
In contrast to the microcrystals formed by segment 248LIIKGI253, crystals of 247DLIIKGISVHI257 were not visible by light microscopy, owing to their small size. The nanocrystals of 247DLIIKGISVHI257, which we term the RRMcore, were formed by shaking in pH 8.5 buffer and, at ~100 nm in width, were visible only by electron microscopy (Fig. 2e). Fibril diffraction of the sample illustrated characteristic amyloid reflections at spacings of 4.72 Å and 10.4 Å (Fig. 2f). We decided to use the cryo-EM method MicroED52,53 on the basis of its previous success in determining key structures from three other amyloid proteins: α-synuclein, tau and IAPP23,54–56. With this technique, an electron microscope is used to collect data in diffraction mode. The structure of 247DLIIKGISVHI257 was determined to be a parallel, face-to-back class 2 steric zipper9 (Table 1 and Fig. 2g,h). The steric zipper interface is formed by the interdigitation of residues DLIIKGIS from one strand with KGISVHI from the mating strand. The tight interface results in a buried surface area of 350 Å2 and a shape complementarity of 0.67.
The structures of 248LIIKGI253 and 247DLIIKGISVHI257 reveal two possible ways that segments from the RRMcore can associate to form an amyloid spine. The heterotypic nature of the two structures suggests that the RRMcore can form polymorphic assemblies.
Cryo-EM structure of 247DLIIKGISVHI257 fibril
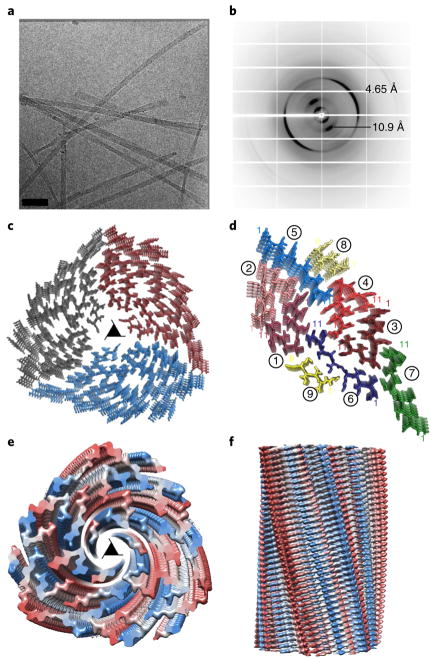
During biochemical characterization of segments within the RRMcore, we noticed that aggregates of 247DLIIKGISVHI257 grown at pH 8.5 differed in morphology from those grown at pH 7.5 or in water, of which the sample pH is 4. The crystals formed at pH 8.5 revealed the parallel class 2 zipper described above, whereas the aggregates grown at pH 7.5 and in water displayed fibrillar morphology, but not crystalline. By electron microscopy, the fibrils exhibited a regular helical twist (Fig. 3a and Supplementary Fig. 3). Fibril diffraction of this sample revealed cross-β diffraction at 4.65 Å and 10.5 Å, indicating the presence of steric zipper interfaces formed by parallel β-sheets (Fig. 3b). Following studies that used cryo-EM to determine structures of helical fibrils of amyloid-β and light chain amyloid16,17, we applied this method to our fibrils.
Fig. 3. Single-particle cryo-EM structure of the 247DLIIKGISVHI257 fibril illustrates a 27-filament, polymorphic three-start helix.
a, Cryo-EM image of 247DLIIKGISVHI257 fibrils. Scale bar, 60 nm. b, X-ray fibril diffraction of the 247DLIIKGISVHI257 sample grown in water exhibits a characteristic cross-β pattern with 4.65-Å and 10.9-Å reflections. c, The structure of 247DLIIKGISVHI257 fibrils is described as a three-start helix with a left-handed twist. The fibril is composed of 27 β-sheets running the full length of the fibril, with nine β-sheets per asymmetric unit, related by three-fold screw symmetry; view shown is down the fibril axis, through the center pore of the helical fibril. The three asymmetric units are colored gray, blue and red. The three-fold screw axis is represented by a triangle. d, The asymmetric unit of this structure is composed of nine β-sheets, each exhibiting a left-handed twist moving down the fibril axis. Each sheet is labeled 1–9. The β-sheets are classified by their relative conformation: kinked, red; straight, blue; curved, green and partial, yellow. Each strand is represented by a unique color within its designated family, with the first and last amino acid residues indicated by number in corresponding color. Two of the β-sheets, at the center and on the periphery of the helix, show density for only the first six residues: 247DLIIKG252. e, Top view of the density of the three-start helix viewed down the fibril axis. Density for each chain is color ramped blue to red from N terminus to C terminus. f, Density of the fibril viewed perpendicular to the fibril axis. Density color scheme is the same as that in e.
High-contrast images of the 247DLIIKGISVHI257 sample obtained by cryo-EM movie recording with a direct electron detector revealed eight classes of fibrils, including cylinders, twists, sheets and a three-start helix (Supplementary Fig. 3). The most dominant species, comprising ~60% of the sample, was the three-start helix, a common organization pattern of biological fibrils57. Subsequent 2D image classification of the three-start-helix produced 2D class averages that showed multiple faces of the fibril, generating enough data for single-particle 3D reconstruction (Supplementary Fig. 4).
We obtained a 3D structure at 3.7-Å resolution (Table 2, Fig. 3c and Supplementary Figs. 5 and 6) by single-particle 3D reconstruction, with helical parameters determined from the 2D class averages (Methods). The 247DLIIKGISVHI257 three-start helix appeared as a many-filament parallel β-sheet fibril, exhibiting three-fold screw symmetry (Supplementary Video 1). A single-layer cross-section of the fibril showed 27 peptide strands, with nine strands in the asymmetric unit (Fig. 3d and Supplementary Video 2). Each of the nine strands displays either a kinked, straight, or curved conformation, thereby contributing to the complex polymorphism of the structure (Supplementary Fig. 7). The asymmetric units are related by a left-handed three-fold screw with a rise of 1.60 Å and a rotation of 120.4° per layer. The individual protofilament β-sheets, 27 in total, exhibit a left-handed twist, as is common for β-sheets. The density map indicates that the β-sheets at the center and outer perimeter of the fibril have disordered termini (Fig. 3e,f). Thus, we modeled only the six ordered residues for these innermost and outermost β-sheets (displayed in yellow in Fig. 3d). At the center of the structure, the five disordered residues will occupy the apparently empty space, making it unlikely that this is a water channel.
Table 2.
Cryo-EM data collection, refinement and validation statistics
| DLIIKGISVHI (EMD-8781, PDB 5W7V) | |
|---|---|
| Data collection and processing | |
| Magnification | 130,000× |
| Voltage (kV) | 300 |
| Electron exposure (e−/Å2) | 30 |
| Defocus range (μm) | 2.5–4 |
| Pixel size (Å) | 1.07 |
| Symmetry imposed | Helical, 120.44° turn/ 1.60 Å rise |
| Initial particle images (no.) | 33,800 |
| Final particle images (no.) | 18,800 |
| Map resolution (Å) | 3.8 |
| FSC threshold | (0.5) |
| Map resolution range (Å) | 200–3.8 |
| Refinement | |
| Initial model used | De novo |
| Model resolution (Å) | 3.6 |
| FSC threshold | (0.5) |
| Model resolution range (Å) | 200–3.6 |
| Map-sharpening B factor (Å2) | –120 |
| Model composition | |
| Nonhydrogen atoms | 20,434 |
| Protein residues | 2,640 |
| B factors (Å2) | |
| Protein | 107.2 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.01 |
| Bond angles (°) | 0.77 |
| Validation | |
| MolProbity score | 1.62 |
| Clashscore | 12.75 |
| Poor rotamers (%) | 0 |
| Ramachandran plot | |
| Favored (%) | 97.22 |
| Allowed (%) | 2.78 |
| Disallowed (%) | 0 |
This complex helical fibril structure illustrates a tight packing of the 247DLIIKGISVHI257 segment into a polymorphic structure, the first amyloid molecular structure of its kind. Although this structure could not form with full-length TDP-43, the packing shows how the RRMcore of TDP-43, and amyloid proteins in general, can use the flexibility of their peptide backbones to adopt multiple conformations and form diverse steric-zipper interfaces, even in a single protofibril. This expanded repertoire of geometric shapes reveals the diverse mechanisms by which protofilaments can fit together to fill space, exclude water, and bury hydrophobic surfaces, as discussed below (Supplementary Fig. 8). Because of this complexity, the structure provides some of the first direct evidence for the ability of amyloid fibers to display positional polymorphism.
RRMcore structures and amyloid polymorphism
One of the interesting findings in determining these structures is that the same segment, RRMcore, does not form the same structure under two different conditions. Fibrils grown in water at pH 4 exhibited much stronger electrostatic interactions than the crystals grown at pH 8.5. At pH 4, both the histidine and N termini are protonated, allowing favorable electrostatic interactions between the histidine and aspartate residues as well as the N and C termini in the single-particle cryo-EM fibril structure (Supplementary Fig. 8). In the case of the crystals of RRMcore formed at pH 8.5, we did not observe strong electrostatic interactions, but rather, the stability of the structure appears to be generated by burying hydrophobic or apolar side chains. The difference in these two structures offered us our first insight into the polymorphic nature of this segment.
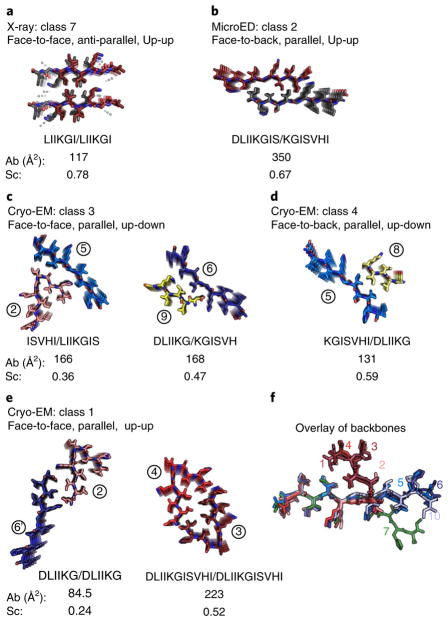
Our detailed analysis of the RRMcore polymorphs reveals that there are seven distinct interfaces that are formed among the three structures (Fig. 4 and Supplementary Table 2). For our analysis of the single-particle cryo-EM structure, we considered only interfaces with at least three interdigitating side chains. Each distinct steric zipper interface features different interacting amino acid side chains. Some of the interfaces feature homotypic interactions, whereas others feature heterotypic interactions, thus creating interfaces that have varying values of buried surface area and shape complementarity as well as different symmetries. These seven interfaces belong to five symmetry classes of steric zippers (Supplementary Fig. 9). Two of these classes were revealed by the X-ray and electron-diffraction structures. The 248LIIKGI253 X-ray structure is an antiparallel class 7 zipper (the two outer faces of the pair of the β-sheet are equivalent, and both β-sheets have the same strand edge facing up) (Fig. 4a). Here, up and down refer to opposite directions along the fibril axis. The 247DLIIKGISVHI257 MicroED structure is a parallel class 2 zipper (the two β-sheets are face to back, again with the same strand edge facing up) (Fig. 4b). This steric zipper has the greatest buried surface area of 350 Å2 (Fig. 4b). The cryo-EM structure accounts for the other five distinct interfaces that belong to three classes of zippers: classes 1, 3 and 4 (Fig. 4c–e). Class 3 zippers had been postulated (sheets face-to-face, with the strand edge facing up in one sheet and down in its mate), but not previously observed; we found class 3 zippers in our cryo-EM structure at the interfaces of ISVHI–LIIKGIS and DLIIKG–KGISVH (Fig. 4c). These various distinct interfaces are achieved because of the varied conformations adopted by the RRMcore backbone (conformations displayed as an overlay in Fig. 4f).
Fig. 4. Seven unique sheet-to-sheet interfaces, seen in the three structures presented.
These interfaces represent five classes of amyloid steric zippers9. The buried solvent-accessible surface area (Ab) and steric complementarity (Sc) are shown for each sheet-to-sheet interface. a, The 248LIIKGI253 structure, determined by X-ray diffraction, is the only example from all seven interfaces of an antiparallel zipper. This class 7 structure exhibits the highest shape complementarity at 0.78. b, The 247DLIIKGISVHI257 electron diffraction structure illustrates a class 2 steric zipper interface. This structure exhibits the tightest packing of all DLIIKGISSVHI interfaces, with a buried surface area of 350 Å2. c, Two interfaces from the 247DLIIKGISVHI257 cryo-EM structure exhibit class 3 morphology. These interfaces are the first two examples of class 3 zippers determined at molecular resolution. d, The third interface in the 247DLIIKGISVHI257 cryo-EM structure is a class 4 steric zipper formed at the periphery of the three-start helix. e, Two interfaces from the 247DLIIKGISVHI257 cryo-EM structure exhibit class 1 morphology. The kinked interface is one of the most distorted interfaces found to date for amyloid steric zippers. The segment β-strand appears to pack against itself as well as against a mating sheet. The DLIIKG–DLIIKG interface represents the weakest interface with the smallest area buried, 85 Å2, and the poorest shape complementarity at 0.24. f, The backbones of all full RRMcore strands have been superimposed for residues LIIK to illustrate their differences. Only the seven sheets formed from full-length 247DLIIKGISVHI257 strands of the cryo-EM structure are assigned numbers. Strand 10 is from the MicroED structure.
Within the single-particle cryo-EM structure, we see that the β-sheet interfaces exhibit both up-up and up-down backbone directionality58. This diversity is achieved by a kink that sharply reverses the up-down directionality of the backbone at the central isoleucine, the eighth amino acid of the 11-residue segment (Supplementary Fig. 10). That is, the carbonyl of the glycine residue faces down, and, at the kink, the carbonyl of the adjacent isoleucine also faces down, thereby creating a strand in which half of the segment exhibits ‘up’ directionality and the other half exhibits ‘down’ directionality. Here, the 247DLIIKGISVHI257 strand can have both an up-up interface on the first half of its sequence and an up-down interface on the last half. Because the RRMcore can switch directionality, the interactions of strands 6′, 2 and 5 show how only three copies of the peptide segment are needed to create two entirely distinct interfaces with different directionalities of the backbone (Fig. 5a,b). This enables polymorphic lateral growth of the fibril perpendicular to the fibril axis and suggests how a propagating fibril could be composed of two directionally distinct steric zipper cores within a localized region.
Fig. 5. The conformational flexibility of the 247DLIIKGISVHI257 backbone provides insight into amyloid fibril formation and polymorphism.
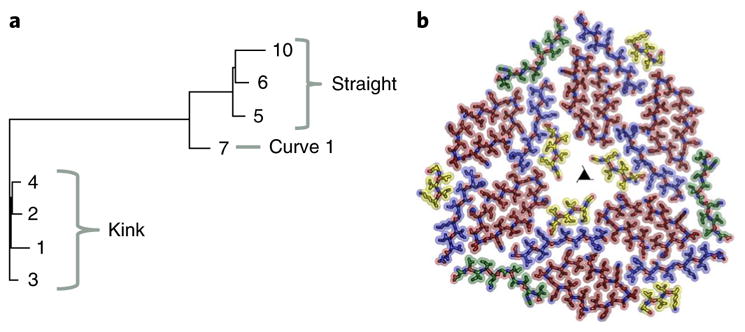
a, Structural relationships among the eight distinct β-sheet backbones illustrated by a pseudo-phylogenetic tree. The lengths of the branches correspond to r.m.s. deviation values between pairs of backbones. The two principal branches correspond to kinked and straight sheets. The straight branch forks into two sub-branches: straight and curve. b, A view down the fibril axis of one layer of the fibril represented by sticks and spheres. The strands are colored by relative conformation: kinked, red; straight, blue; curved, green; partial, yellow.
We also analyzed the flexibility of the 247DLIIKGISVHI257 backbone. To do this, we applied the LSQ superimposition feature from the program COOT on eight distinct β-strands of RRMcore in two of our structures59. We compared the backbone conformations of the seven full-length strands from the cryo-EM structure, strands 1–7, and the backbone of the MicroED structure, strand 10 (Fig. 4f). Comparison of r.m.s. deviation values reveal that the eight strands are divided into three subclasses, kinked, straight, and curved, with small but significant variations within each subclass (Supplementary Table 3). The cryo-EM structure contains four kinked segments in the asymmetric unit, with r.m.s. deviation values ranging from 0.41 to 0.72 Å. The other four strands of 247DLIIKGISVHI257 are more linear in conformation, with r.m.s. deviation values ranging from 0.59 to 2.34 Å. The structural relationships among the eight strand backbones are represented by a pseudo-phylogenic tree in Fig. 5a.
Finally, we investigated the possible modes of formation of this complex fibril from 27 distinct sheets. We noticed that a pseudo-two-fold axis runs between strands 1, 2, 5 and 8 and 3, 4, 6 and 9 (Supplementary Fig. 10). To visualize this, we have displayed all kinked strands as red, straight strands as blue, partial strands as yellow and curved strands as green in Fig. 5b. In this representation, one can see how two red strands associate with a blue and a yellow strand to form a core component of the asymmetric unit that is then repeated through two-fold symmetry. The green, curved strands serve to stabilize the asymmetric unit or protofibril, resulting in three-fold symmetry of the mature fibril.
Thus, the three structures provide us with information on how the RRMcore can form fibrils through polymorphic assemblies. The 247DLIIKGISVHI257 backbone can adopt at least eight different conformations, allowing the segment to associate with seven unique interfaces comprising five different classes of steric zippers. The polymorphic features of this region offer the first example of how an amyloid-forming segment can adopt multiple conformations and possibly contribute to unique fibril strains.
Discussion
Role of RRMcore in TDP-43 pathological aggregation
As described previously, the C terminus is widely believed to be responsible for nucleation of TDP-43 fibrils44. Recent findings have shown that the dimerization of TDP-43 through the N-terminal domain helps block TDP-43 aggregation by separating the LCD from neighboring copies of the protein, thus preventing amyloid formation60–62. Additionally, the structure of the mouse RRM2 domain, which shares 88% sequence identity with the human RRM2, is folded and highly stable. Within this RRM2 structure, the RRMcore forms two β-sheets, limiting the likelihood that it would randomly initiate aggregation63. This therefore elicits the question of the relevance of the RRMcore to TDP-43 pathological aggregation.
It is well supported that a segment from the RRM2 is unlikely to play a role in seeding pathological aggregation with full-length, folded TDP-43. However, patients with ALS and FTLD have aggregates in the brain and spinal cord that are primarily composed of C-terminal fragments ranging in size from 15 to 35 kDa37,64,65. The shorter segments include only part of the RRM2 and the C-terminal tail. Two of these C-terminal fragments from patients with ALS and FTLD result from cleavage of the RRM2 at residues 208 and 220 (refs 37,66). On the basis of RRM2 structure, cleavage at residue 208 would eliminate one α-helix and one β-sheet, and cleavage at residue 220 would eliminate an additional β-sheet, features that probably contribute to the folding and secondary structure of the RRM2. These cleavage products could result in destabilization of the RRM2 and partial, if not complete unfolding of the domain. In other amyloid proteins, such as superoxide dismutase 1, partial unfolding of the protein has already been shown to result in fibrillary amyloid aggregation. This unfolding could expose the RRMcore and thus allow it to contribute to TDP-43 aggregation. With a segment so polymorphic and capable of adopting many backbone conformations, it is possible it could form heterotypic interactions characteristic of mature amyloid fibrils and stabilize other fibril-forming segments of the C terminus. However, it is important to note that these assumptions are speculative and require further studies to validate the role of the RRM2 in TDP-43 C-terminal fragment aggregation.
Relevance of peptides for amyloid fibril studies
One of the caveats of studying small 6–11-residue segments of amyloid proteins is that their structural characterization results in homotypic interactions. However, recent studies using cryo-EM and solid-state NMR have revealed that the structures of full-length amyloid fibrils of α-synuclein, tau and amyloid-β are composed of much longer (~25–70 residues) heterotypic interactions6,11,67. In the case of tau, a key segment, VQIVYK, forms a staggered homotypic zipper as a short peptide but was revealed to form a non-staggered heterotypic zipper in the context of the longer protein by cryo-EM9,67. In the case of α-synuclein, the differences between peptide structure and fibril structure are even more pronounced. Here, a key 11-residue segment termed NACore forms a relatively linear homotypic steric zipper as a peptide; however, in the full-length protein, this segment bends twice, forming an S shape and a heterotypic zipper11,56. These findings bring into question the relevance of studying these small segments as they probably do not represent the full-length mature fibril structure.
Nevertheless, the information garnered from these homotypic structures of peptides has proven to be valuable. Previous work from our lab has shown that inhibitors rationally designed to block homo-typic amyloid structures of p53, transthyretin, and tau are effective at inhibiting fibril formation of the full-length protein, in some cases alleviating toxicity40,68,69. Specifically, an inhibitor rationally designed from the homotypic VQIVYK structure inhibits aggregation of the K12 tau construct68. This finding indicates that although the homotypic peptide structure does not fully describe the mature full-length fibril, some of the features of the segment structure, such as backbone conformation and side chain interdigitation, are present in the full-length protein. For the goal of designing inhibitors to block aggregation and alleviate toxicity of the full-length protein, structures of short peptide segments help achieve this goal through rational structure-based design, as demonstrated through our work with p53 (ref. 40).
Packing polymorphism of RRMcore
Understanding how amyloid proteins form polymorphic assemblies is a critical aspect of therapeutic development in neurodegenerative research. Here, we present evidence that an amyloid-forming segment can display two distinct types of packing polymorphism, positional and modal, concepts introduced by Caspar and Cohen70. Positional polymorphism is defined as the different disposition of identical parts within a single assembly. To date, this has been observed only once: a segment from amyloid-β, MVGGVV, formed a steric zipper with two different backbone conformations9. In contrast, modal polymorphism is defined as the different dispositions of identical parts in different modes of assembly, the customary usage of the term packing polymorphism in amyloid structures. The structures of RRMcore reported here show both positional and modal polymorphism. Positional polymorphism, which we report for the first time on such a vast scale for amyloid, is evident in the 27-β-sheet fibril, in which the seven β-sheets composed of RRMcore assume several distinctly different structures, as discussed above. Another different RRMcore β-sheet is the straight polymorph of the MicroED structure, which qualifies RRMcore as also exhibiting modal polymorphism. This remarkable variation in conformation of the β-sheets formed by RRMcore leads to prodigious packing polymorphism.
Although the entire TDP-43 protein cannot be modeled into the 27-β-sheet fibril determined here by single-particle cryo-EM, the structure offers an example at the molecular scale of the prodigious polymorphism of which amyloid assemblies are capable, and the great range of structures that amyloid fibrils can form, with implications for research in neurodegeneration. Distinct amyloid polymorphs may seed distinct ‘strains’ of amyloid, which can lead to different clinical conditions, as has been observed with both prion and tau protein, although the degree of polymorphism in those strains of full-length protein remains to be determined19,20. Each polymorph may require its own drug inhibitor. In cases in which multiple polymorphs form, inhibiting one structure may not be sufficient to block disease formation or progression. Thus, understanding amyloid polymorphism is likely to be important for drug development against amyloid diseases. We suggest that the structures presented in this paper offer insight not only about TDP-43 aggregation, but also about the challenges presented by amyloid aggregation in general.
Methods
Selection and generation of TDP-43 peptide targets
All targeted peptide segments were chosen based on the Rosetta Energy 3D Profile Method Zipper DB48. After the selection of peptide targets ranging in size from 6–11 residues, all peptides were purchased from Genscript at a purity of 95% or higher.
Peptide fibril formation
For initial biochemical characterization, all peptide segments were dissolved in PBS at pH 7.5. Peptide segments 247DLIIKG252, 247DLIIKGISV255, 247DLIIKGISVH256, 248LIIKGISVH256, 248LIIKGISVHI257, 250KGISVHISN259, 252GISVHI257 and 252ISVHISN259 were dissolved to a final concentration of 20 mM. Peptide segments 248LIIKGI253 and 253SVHISN259 were dissolved to a final concentration of 40 mM because of their high solubility. The last peptide segment, 247DLIIKGISVHI257, was dissolved to a final concentration of 1 mM because of its low solubility. All peptides were then incubated at 37 °C in a Torrey Pine Scientific shaker at level 9 for 1 week. Samples were examined by TEM following fibril formation.
Negative-stain transmission electron microscopy (TEM)
Negatively stained samples for TEM were prepared by applying 5 μL of sample on hydrophilic 400 mesh carbon-coated formvar support films mounted on copper grids (Ted Pella, Inc.). The fibrils were allowed to adhere for 4 min and washed once with 2% uranyl acetate. The samples were then stained for 2 min with 2% uranyl acetate, leaving a few nanoliters of stain on the grid. Grids were allowed to dry for 5 min before storage. Each grid was inspected on a T12 (FEI) microscope. Each fibril sample was replicated at least two times.
Fibril denaturation studies
Peptide fibrils were generated as described above. Untreated and treated samples were then prepared. For untreated samples, 6 μl of water was added to 54 μl of fibril preparation. For treated samples, 6 μl of 20% SDS solution was added to 54 μl of fibril preparation for a final 2% SDS concentration. The treated samples were then quiescently incubated at 70 °C for 15 min. Turbidity experiments were subsequently performed as untreated and treated samples were aliquoted into black 384-well plates (Nunc) covered with optical tape. Values were recorded at a wavelength of 340 nm in a Spectramax M5 (Molecular Devices). Experiments reported here were performed in triplicate. Data were normalized against untreated fibril samples so that the starting value was 1.00. Error bars represent s.d. Of note, final fibril load and stability can vary depending on shaking time and relative speed of shaker, which itself can vary based on frequency of shaker use. Here, we display samples that were shaken in the same Torrey Pines shaker for the same amount of time, 7 d.
Fibril diffraction
Fibrils prepared in PBS were spun down at 15,000 r.p.m. for 30 min in a tabletop microcentrifuge and washed with water a total of three times. Fibrils of 247DLIIKGISVHI257 prepared in water did not undergo this step. Concentrated fibrils were diluted in 20 μl of water. 3 μl of fibril sample was applied to the middle of two glass capillaries for alignment and allowed to dry overnight. For data collection, the glass capillaries, with fibril sample, were mounted on brass pins. Diffraction was collected at beamline 24-ID-E of the Advanced Photon Source, Argonne National Laboratory, Argonne, IL, USA, with a wavelength of 0.9792 Å. All patterns were collected at a distance of 300 mm at 100 K and analyzed using the Adxv software package71. Fibril diffraction patterns were tested once at the APS.
Crystallization
248LIIKGI253 was dissolved in 10 mM LiOH to a final concentration of 18.7 mg/mL. Crystals were grown by 24-well hanging-drop vapor diffusion over a reservoir containing 0.1 M CHES, pH 9.5, and 17.5% w/v PEG 8000. Each hanging drop contained 1.5 μl of peptide solution mixed with 1.5 μl of reservoir solution. Crystals were mounted in loops and stored in liquid nitrogen before data collection.
247DLIIKGISVHI257 was dissolved in CHES buffer at pH 8.5 to a final concentration of 1 mM. Crystals were formed by shaking the peptide at 37 °C for 1 week in a Torrey Pines Scientific Shaker at level 9.
X-ray data collection and structure determination
The X-ray diffraction data for segment 248LIIKGI253 was collected at beamline 24-ID-E of the Advanced Photon Source, Argonne National Laboratory, Argonne, IL, USA. Detector ADSC HF -4M was used. Data were collected at a wavelength of 0.9792 Å, temperature of 100 K, at 5° oscillations and 175 mm detector distance. Indexing was done using XDS, integration was done with DENZO and datasets from two crystals were merged in P1 and scaled with SCALEPACK to 1.4-Å resolution72. The data were imported using the CCP4 suite73. Molecular replacement was done using Phaser74. Initial models of different idealized polyalanine β-strands did not pack correctly in the cell. The final model used was a sheet composed of two antiparallel strands with the sequence AIIAGI, based on the structure of VEAVYL. The model was manually adjusted over 13 cycles to the correct sequence and rotamer orientation in Coot59. The atomic model was refined by Refmac with a final Rfactor/Rfree of 20.7/22.6 (ref. 75) and 100% of Ramachandran angles favored.
MicroED data collection and structure determination
The MicroED data for segment 247DLIIKGISVHI257 were collected using the same protocol as Rodriguez et al.56 with minor differences detailed below56,76,77. Data were collected at the HHMI Janelia Research Campus using the Tecnai F20 TEM operating at 200 kV and recorded using a bottom-mount TVIPS F416 CMOS camera. Individual image frames were exposed for 3 s per image. Diffraction data were collected in rolling-shutter mode. During each exposure, crystals were continuously rotated unidirectionally within the beam at a rate of 0.3° per second. Indexing and integration was done using XDS, and datasets from 13 crystals were merged in P1 and scaled with SCALEPACK to 1.4-Å resolution78. The data were imported using the CCP4 suite73. Molecular replacement was done using Phaser73. An idealized 11-residue β-strand model of DLIIKGISVHI was used for molecular replacement. The model was further refined in Refmac, accounting for twinning operations and adjusting rotamer orientations in Coot59,75. The twinning operator is –H, K, K-L with a fraction of 0.5. The final model had an Rfactor/Rfree of 26.2/30.6 and 100% of Ramachandran angles favored.
Cryo-EM and reconstruction
247DLIIKGISVHI257 peptide was dissolved in water to a final concentration of 1 mM at pH 4. The sample was shaken for 1 week at 37 °C in a Torrey Pine shaker on level 9. The fibril sample was subsequently spun down at 15,000 r.p.m. for 30 min. The supernatant, with contaminant monomer, was disposed of, and the fibril sample was resuspended in 20 μl of water.
Each aliquot of 2.5 μl of the fibril sample was applied onto a “baked”79 Quantifoil 1.2/1.3 μm, 200 mesh grid, blotted for 4 s at force 1, then vitrified in liquid ethane at liquid nitrogen temperature in a Vitrobot Mark IV (FEI). Cryo-EM data were collected in an FEI Titan Krios microscope equipped with a Gatan Quantum LS/K2 Summit direct electron detection camera (operated with a 300 kV acceleration voltage and a slit width 20 eV). Data collection was in counting mode driven by Leginon automation software package80. Defocus values were controlled with Leginon by applying a single 3.0-μm target defocus. Dose-fractionation movies were recorded at a frame duration of 200 ms and total duration of 10 s. The total dosage was 60 e−/Å2, with each frame 1.2 e−/Å2 (6 e−/Å2 per second) (measured on Digital Micrograph (Gatan) software).
Frames were aligned as described81, except that an iterative alignment scheme as previously described82 was implemented after the original alignment method. We used the summary of all frames to determine defocus and particle locations, and frames 3–20 for data processing. The focal parameters of the data were determined by CTFFIND4 (ref. 83). We selectively included images within a defocus range of 2.5–4 μm.
D11I fibrils were manually marked in EMAN84helixboxer. A total of 1,904 individual filaments were selected. These filaments were segmented by 90% overlap scheme into 33,758 boxes of 288 × 288 pixels. A series of Relion85 2D classifications were done to the dataset: (1) all boxes were subjected to a general (tau_fudge =2) 2D classification to eliminate bad particles and to separate different morphologies (morphological distributions described in main text); (2) we chose the group of fibrils with the most abundant morphology (20,138 boxes) for the next run (2. tau_fudge = 4) to further eliminate bad particles and misclassified particles. The second 2D classification resulted in 18,887 boxes; (3) we used a last 2D classification with a very high tau_fudge factor (16) to extend the working resolution to beyond 4.8 Å, making it possible to catch the layer line corresponding to the helical arrangement of β-strands.
A total of 18,818 boxes were included in the 3D analyses. We used EMAN-based IHRSR86 to reconstruct the 3D structure of the fibril. The resolution was slightly better than 4 Å, making it possible to trace the backbone and register the 11-peptide sequence. We built an atomic model for an asymmetric unit (with nine strands) with Coot59. We built 18 helically related copies of the asymmetric unit to account for the helix. We did a preliminary Fourier space pseudo-crystallographic model refinement86. We then carried out another model refinement with the phenix.real_space_refine command of the Phenix package87 using default settings.
We further refined the 3D model of the fibril in Relion with IHRSR implementation88 by Class3D with one class (owing to the fine feature of the β-strands, Refine3D was unable to adequately regulate working resolution to refine with high enough resolution factors). We tested the resolution by even-odd division of the dataset according to movie numbers. The FSC crosses 0.5 at 3.8 Å and 0.143 at 3.2 Å. The local resolution was also assessed by ResMap89 to be 3.5 Å. The side chain features of the map are consistent with a resolution of 3.5–3.6 Å. We further refined the atomic model in Phenix with phenix.real_space_ refine command with 30 asymmetric units.
Analysis of structural features
All images displaying the interfaces of the RRM2 structures were created in Pymol90. Area buried and shape complementarity were calculated based on established protocols91,92. Backbone r.m.s. deviation values were determined using the LSQ superimpose feature in COOT59. The pseudo-phylogeny tree comparing these r.m.s. deviation values was generated using the T-REX interface93 at http://www.trex.uqam.ca/.
Life Sciences Reporting Summary
Further information on experimental design is available in the Life Sciences Reporting Summary.
Data availability
All structural data have been deposited into the Worldwide Protein Data Bank (wwPDB) and the Electron Microscopy Data Bank (EMDB) with the following accession codes: LIIKGI (PDB 5W50), DLIIKGISVHI MicroED (PDB 5W52, EMD-8765) and DLIIKGISVHI Cryo-EM (EMD-8781, PDB 5W7V). All other data are available from the authors upon reasonable request.
Supplementary Material
Acknowledgments
We thank Q. Cao, L. Saelices, S. Sangwan, and P. Seidler for discussion, D. Shi at Janelia for microscope support, and M. Collazo at UCLA-DOE Macromolecular Crystallization Core Technology Center for crystallization support. We thank the staff at the Argonne Photon Source (APS) NECAT Beamline 24-ID-E, which is funded by National Institutes of Health (P41 GM103403). This research was supported in part by grants from the National Science Foundation (NSF MCB 1616265 to D.S.E.) and the National Institutes of Health (GM071940 to Z.H.Z.), Howard Hughes Medical Institute and the Janelia Research Campus visitor program. D.R.B. was supported by the National Science Foundation Graduate Research Fellowship. We acknowledge the use of instruments at the Electron Imaging Center for Nanomachines supported by UCLA and by instrumentation grants from NIH (1S10RR23057 and 1U24GM116792) and NSF (DBI-1338135).
Footnotes
Author contributions
E.L.G. and D.S.E. designed the project and wrote the manuscript with input from all other authors especially M.R.S. E.L.G. and H.T. conducted fibril growth experiments, stability assays and crystallization and structure determination of 248LIIKGI253. E.L.G. and M.R.S. did the fibril diffraction assays. E.L.G. grew crystals of 247DLIIKGISVHI257, while D.R.B. collected MicroED data of the sample. M.R.S., D.C. and E.L.G. processed data and solved the structure of 247DLIIKGISVHI257 crystal. E.L.G. prepared and optimized the 247DLIIKGISVHI257 fibril sample for cryo-EM while P.G. processed and solved the cryo-EM structure. E.L.G., M.R.S. and D.S.E. analyzed structures, conducted computational analysis such as area buried and designed models of fibril nucleation and elongation. T.G. and Z.H.Z. contributed to analysis of microED and cryoEM structures, respectively.
Competing interests
D.S.E. is an advisor and equity shareholder in ADDRx, Inc.
Supplementary information is available for this paper at https://doi.org/10.1038/s41594-018-0045-5.
References
- 1.Chiti F, Dobson CM. Protein misfolding, functional amyloid, and human disease. Annu Rev Biochem. 2006;75:333–366. doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- 2.Hardy J, Selkoe DJ. The amyloid hypothesis of Alzheimer’s disease: progress and problems on the road to therapeutics. Science. 2002;297:353–356. doi: 10.1126/science.1072994. [DOI] [PubMed] [Google Scholar]
- 3.Knowles TPJ, Vendruscolo M, Dobson CM. The amyloid state and its association with protein misfolding diseases. Nat Rev Mol Cell Biol. 2014;15:384–396. doi: 10.1038/nrm3810. [DOI] [PubMed] [Google Scholar]
- 4.Greenwald J, Riek R. Biology of amyloid: structure, function, and regulation. Structure. 2010;18:1244–1260. doi: 10.1016/j.str.2010.08.009. [DOI] [PubMed] [Google Scholar]
- 5.Goldsbury C, Frey P, Olivieri V, Aebi U, Müller SA. Multiple assembly pathways underlie amyloid-β fibril polymorphisms. J Mol Biol. 2005;352:282–298. doi: 10.1016/j.jmb.2005.07.029. [DOI] [PubMed] [Google Scholar]
- 6.Paravastu AK, Leapman RD, Yau WM, Tycko R. Molecular structural basis for polymorphism in Alzheimer’s β-amyloid fibrils. Proc Natl Acad Sci USA. 2008;105:18349–18354. doi: 10.1073/pnas.0806270105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wiltzius JJW, et al. Molecular mechanisms for protein-encoded inheritance. Nat Struct Mol Biol. 2009;16:973–978. doi: 10.1038/nsmb.1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nelson R, et al. Structure of the cross-β spine of amyloid-like fibrils. Nature. 2005;435:773–778. doi: 10.1038/nature03680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sawaya MR, et al. Atomic structures of amyloid cross-beta spines reveal varied steric zippers. Nature. 2007;447:453–457. doi: 10.1038/nature05695. [DOI] [PubMed] [Google Scholar]
- 10.Stroud JC. The zipper groups of the amyloid state of proteins. Acta Crystallogr D Biol Crystallogr. 2013;69:540–545. doi: 10.1107/S0907444912050548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tuttle MD, et al. Solid-state NMR structure of a pathogenic fibril of full-length human α-synuclein. Nat Struct Mol Biol. 2016;23:409–415. doi: 10.1038/nsmb.3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wälti MA, et al. Atomic-resolution structure of a disease-relevant Aβ(1-42) amyloid fibril. Proc Natl Acad Sci USA. 2016;113:E4976–E4984. doi: 10.1073/pnas.1600749113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Colvin MT, et al. Atomic resolution structure of monomorphic Aβ42 amyloid fibrils. J Am Chem Soc. 2016;138:9663–9674. doi: 10.1021/jacs.6b05129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Petkova AT, et al. A structural model for Alzheimer’s β-amyloid fibrils based on experimental constraints from solid state NMR. Proc Natl Acad Sci USA. 2002;99:16742–16747. doi: 10.1073/pnas.262663499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xiao Y, et al. Aβ(1-42) fibril structure illuminates self-recognition and replication of amyloid in Alzheimer’s disease. Nat Struct Mol Biol. 2015;22:499–505. doi: 10.1038/nsmb.2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schmidt A, Annamalai K, Schmidt M, Grigorieff N, Fändrich M. Cryo-EM reveals the steric zipper structure of a light chain-derived amyloid fibril. Proc Natl Acad Sci USA. 2016;113:6200–6205. doi: 10.1073/pnas.1522282113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schmidt M, et al. Peptide dimer structure in an Aβ(1-42) fibril visualized with cryo-EM. Proc Natl Acad Sci USA. 2015;112:11858–11863. doi: 10.1073/pnas.1503455112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bai XC, et al. An atomic structure of human γ-secretase. Nature. 2015;525:212–217. doi: 10.1038/nature14892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tanaka M, Chien P, Naber N, Cooke R, Weissman JS. Conformational variations in an infectious protein determine prion strain differences. Nature. 2004;428:323–328. doi: 10.1038/nature02392. [DOI] [PubMed] [Google Scholar]
- 20.Kaufman SK, et al. Tau prion strains dictate patterns of cell pathology, progression rate, and regional vulnerability in vivo. Neuron. 2016;92:796–812. doi: 10.1016/j.neuron.2016.09.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Riek R, Eisenberg DS. The activities of amyloids from a structural perspective. Nature. 2016;539:227–235. doi: 10.1038/nature20416. [DOI] [PubMed] [Google Scholar]
- 22.Eisenberg D, Jucker M. The amyloid state of proteins in human diseases. Cell. 2012;148:1188–1203. doi: 10.1016/j.cell.2012.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krotee P, et al. Atomic structures of fibrillar segments of hIAPP suggest tightly mated β-sheets are important for cytotoxicity. eLife. 2017;6:1–26. doi: 10.7554/eLife.19273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Soriaga AB, Sangwan S, Macdonald R, Sawaya MR, Eisenberg D. Crystal structures of IAPP amyloidogenic segments reveal a novel packing motif of out-of-register beta sheets. J Phys Chem B. 2016;120:5810–5816. doi: 10.1021/acs.jpcb.5b09981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Colletier JP, et al. Molecular basis for amyloid-beta polymorphism. Proc Natl Acad Sci USA. 2011;108:16938–16943. doi: 10.1073/pnas.1112600108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Neumann M, et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006;314:130–133. doi: 10.1126/science.1134108. [DOI] [PubMed] [Google Scholar]
- 27.Hasegawa M, et al. Phosphorylated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Ann Neurol. 2008;64:60–70. doi: 10.1002/ana.21425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schwab C, Arai T, Hasegawa M, Yu S, McGeer PL. Colocalization of transactivation-responsive DNA-binding protein 43 and huntingtin in inclusions of Huntington disease. J Neuropathol Exp Neurol. 2008;67:1159–1165. doi: 10.1097/NEN.0b013e31818e8951. [DOI] [PubMed] [Google Scholar]
- 29.Amador-Ortiz C, et al. TDP-43 immunoreactivity in hippocampal sclerosis and Alzheimer’s disease. Ann Neurol. 2007;61:435–445. doi: 10.1002/ana.21154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nakashima-Yasuda H, et al. Co-morbidity of TDP-43 proteinopathy in Lewy body related diseases. Acta Neuropathol. 2007;114:221–229. doi: 10.1007/s00401-007-0261-2. [DOI] [PubMed] [Google Scholar]
- 31.Higashi S, et al. Concurrence of TDP-43, tau and α-synuclein pathology in brains of Alzheimer’s disease and dementia with Lewy bodies. Brain Res. 2007;1184:284–294. doi: 10.1016/j.brainres.2007.09.048. [DOI] [PubMed] [Google Scholar]
- 32.Kraemer BC, et al. Loss of murine TDP-43 disrupts motor function and plays an essential role in embryogenesis. Acta Neuropathol. 2010;119:409–419. doi: 10.1007/s00401-010-0659-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sephton CF, et al. TDP-43 is a developmentally regulated protein essential for early embryonic development. J Biol Chem. 2010;285:6826–6834. doi: 10.1074/jbc.M109.061846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu LSS, et al. TDP-43, a neuropathosignature factor, is essential for early mouse embryogenesis. Genesis. 2010;48:56–62. doi: 10.1002/dvg.20584. [DOI] [PubMed] [Google Scholar]
- 35.Chiang PM, et al. Deletion of TDP-43 down-regulates Tbc1d1, a gene linked to obesity, and alters body fat metabolism. Proc Natl Acad Sci USA. 2010;107:16320–16324. doi: 10.1073/pnas.1002176107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhu L, et al. An ALS-mutant TDP-43 neurotoxic peptide adopts an anti-parallel β-structure and induces TDP-43 redistribution. Hum Mol Genet. 2014;23:6863–6877. doi: 10.1093/hmg/ddu409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Igaz LM, et al. Expression of TDP-43 C-terminal fragments in vitro recapitulates pathological features of TDP-43 proteinopathies. J Biol Chem. 2009;284:8516–8524. doi: 10.1074/jbc.M809462200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Iguchi Y, et al. TDP-43 depletion induces neuronal cell damage through dysregulation of Rho family GTPases. J Biol Chem. 2009;284:22059–22066. doi: 10.1074/jbc.M109.012195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Di Carlo V, et al. TDP-43 regulates the microprocessor complex activity during in vitro neuronal differentiation. Mol Neurobiol. 2013;48:952–963. doi: 10.1007/s12035-013-8564-x. [DOI] [PubMed] [Google Scholar]
- 40.Soragni A, et al. A designed inhibitor of p53 aggregation rescues p53 tumor suppression in ovarian carcinomas. Cancer Cell. 2016;29:90–103. doi: 10.1016/j.ccell.2015.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guo W, et al. An ALS-associated mutation affecting TDP-43 enhances protein aggregation, fibril formation and neurotoxicity. Nat Struct Mol Biol. 2011;18:822–830. doi: 10.1038/nsmb.2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jiang LL, et al. Structural transformation of the amyloidogenic core region of TDP-43 protein initiates its aggregation and cytoplasmic inclusion. J Biol Chem. 2013;288:19614–19624. doi: 10.1074/jbc.M113.463828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jiang LL, et al. Two mutations G335D and Q343R within the amyloidogenic core region of TDP-43 influence its aggregation and inclusion formation. Sci Rep. 2016;6:23928. doi: 10.1038/srep23928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Budini M, et al. Cellular model of TAR DNA-binding protein 43 (TDP-43) aggregation based on its C-terminal Gln/Asn-rich region. J Biol Chem. 2012;287:7512–7525. doi: 10.1074/jbc.M111.288720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mompeán M, et al. Structural characterization of the minimal segment of TDP-43 competent for aggregation. Arch Biochem Biophys. 2014;545:53–62. doi: 10.1016/j.abb.2014.01.007. [DOI] [PubMed] [Google Scholar]
- 46.Saini A, Chauhan VS. Self-assembling properties of peptides derived from TDP-43 C-terminal fragment. Langmuir. 2014;30:3845–3856. doi: 10.1021/la404710w. [DOI] [PubMed] [Google Scholar]
- 47.Saini A, Chauhan VS. Delineation of the core aggregation sequences of TDP-43 C-terminal fragment. Chembiochem. 2011;12:2495–2501. doi: 10.1002/cbic.201100427. [DOI] [PubMed] [Google Scholar]
- 48.Goldschmidt L, Teng PK, Riek R, Eisenberg D. Identifying the amylome, proteins capable of forming amyloid-like fibrils. Proc Natl Acad Sci USA. 2010;107:3487–3492. doi: 10.1073/pnas.0915166107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kyte J, Doolittle RF. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 50.Kato M, et al. Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell. 2012;149:753–767. doi: 10.1016/j.cell.2012.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sunde M, et al. Common core structure of amyloid fibrils by synchrotron X-ray diffraction. J Mol Biol. 1997;273:729–739. doi: 10.1006/jmbi.1997.1348. [DOI] [PubMed] [Google Scholar]
- 52.Shi D, Nannenga BL, Iadanza MG, Gonen T. Three-dimensional electron crystallography of protein microcrystals. eLife. 2013;2:e01345. doi: 10.7554/eLife.01345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nannenga BL, Shi D, Leslie AGW, Gonen T. High-resolution structure determination by continuous-rotation data collection in MicroED. Nat Methods. 2014;11:927–930. doi: 10.1038/nmeth.3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu S, et al. Atomic resolution structure determination by the cryo-EM method MicroED. Protein Sci. 2017;26:8–15. doi: 10.1002/pro.2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.de la Cruz MJ, et al. Atomic-resolution structures from fragmented protein crystals with the cryoEM method MicroED. Nat Methods. 2017;14:399–402. doi: 10.1038/nmeth.4178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rodriguez JA, et al. Structure of the toxic core of α-synuclein from invisible crystals. Nature. 2015;525:486–490. doi: 10.1038/nature15368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sept D, Baker NA, McCammon JA. The physical basis of microtubule structure and stability. Protein Sci. 2003;12:2257–2261. doi: 10.1110/ps.03187503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Eisenberg DS, Sawaya MR. Structural studies of amyloid proteins at the molecular level. Annu Rev Biochem. 2017;86:69–95. doi: 10.1146/annurev-biochem-061516-045104. [DOI] [PubMed] [Google Scholar]
- 59.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 60.Jiang LL, et al. The N-terminal dimerization is required for TDP-43 splicing activity. Sci Rep. 2017;7:6196. doi: 10.1038/s41598-017-06263-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mompeán M, et al. Point mutations in the N-terminal domain of transactive response DNA-binding protein 43 kDa (TDP-43) compromise its stability, dimerization, and functions. J Biol Chem. 2017;292:11992–12006. doi: 10.1074/jbc.M117.775965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Afroz T, et al. Functional and dynamic polymerization of the ALS-linked protein TDP-43 antagonizes its pathologic aggregation. Nat Commun. 2017;8:45. doi: 10.1038/s41467-017-00062-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kuo PH, Doudeva LG, Wang YT, Shen CKJ, Yuan HS. Structural insights into TDP-43 in nucleic-acid binding and domain interactions. Nucleic Acids Res. 2009;37:1799–1808. doi: 10.1093/nar/gkp013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nonaka T, Kametani F, Arai T, Akiyama H, Hasegawa M. Truncation and pathogenic mutations facilitate the formation of intracellular aggregates of TDP-43. Hum Mol Genet. 2009;18:3353–3364. doi: 10.1093/hmg/ddp275. [DOI] [PubMed] [Google Scholar]
- 65.Li Q, Yokoshi M, Okada H, Kawahara Y. The cleavage pattern of TDP-43 determines its rate of clearance and cytotoxicity. Nat Commun. 2015;6:6183. doi: 10.1038/ncomms7183. [DOI] [PubMed] [Google Scholar]
- 66.Zhang YJ, et al. Aberrant cleavage of TDP-43 enhances aggregation and cellular toxicity. Proc Natl Acad Sci USA. 2009;106:7607–7612. doi: 10.1073/pnas.0900688106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fitzpatrick AWP, et al. Cryo-EM structures of tau filaments from Alzheimer’s disease. Nature. 2017;547:185–190. doi: 10.1038/nature23002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sievers SA, et al. Structure-based design of non-natural amino-acid inhibitors of amyloid fibril formation. Nature. 2011;475:96–100. doi: 10.1038/nature10154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Saelices L, et al. Uncovering the mechanism of aggregation of human transthyretin. J Biol Chem. 2015;290:28932–28943. doi: 10.1074/jbc.M115.659912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Caspar DLD, Cohen C. In: Engstrom A, Strandberg B, editors. Polymorphism of tropomyosin and a view of protein function; Nobel Symposium 11. Symmetry and Function of Biological Systems at the Macromolecular Level; Stockholm: New York: Almquist and Wiksell; John Wiley and Sons, Inc; 1969. pp. 393–414. [Google Scholar]
- 71.Arvai A. Adxv - A Program to Display X-ray Diffraction Images. 2015. [Google Scholar]
- 72.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 73.Winn MD, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.McCoy AJ, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Vagin AA, et al. REFMAC5 dictionary: organization of prior chemical knowledge and guidelines for its use. Acta Crystallogr D Biol Crystallogr. 2004;60:2184–2195. doi: 10.1107/S0907444904023510. [DOI] [PubMed] [Google Scholar]
- 76.Hattne J, et al. MicroED data collection and processing. Acta Crystallogr A Found Adv. 2015;71:353–360. doi: 10.1107/S2053273315010669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Shi D, et al. The collection of MicroED data for macromolecular crystallography. Nat Protoc. 2016;11:895–904. doi: 10.1038/nprot.2016.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kabsch W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J Appl Crystallogr. 1993;26:795–800. [Google Scholar]
- 79.Miyazawa A, Fujiyoshi Y, Unwin N. Structure and gating mechanism of the acetylcholine receptor pore. Nature. 2003;423:949–955. doi: 10.1038/nature01748. [DOI] [PubMed] [Google Scholar]
- 80.Suloway C, et al. Automated molecular microscopy: the new Leginon system. J Struct Biol. 2005;151:41–60. doi: 10.1016/j.jsb.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 81.Li X, et al. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nat Methods. 2013;10:584–590. doi: 10.1038/nmeth.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Banerjee S, et al. 2.3 Å resolution cryo-EM structure of human p97 and mechanism of allosteric inhibition. Science. 2016;351:871–875. doi: 10.1126/science.aad7974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Rohou A, Grigorieff N. CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J Struct Biol. 2015;192:216–221. doi: 10.1016/j.jsb.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ludtke SJ, Baldwin PR, Chiu W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J Struct Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- 85.Scheres SHW. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ge P, Zhou ZH. Hydrogen-bonding networks and RNA bases revealed by cryo electron microscopy suggest a triggering mechanism for calcium switches. Proc Natl Acad Sci USA. 2011;108:9637–9642. doi: 10.1073/pnas.1018104108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Clemens DL, Ge P, Lee BY, Horwitz MA, Zhou ZH. Atomic structure of T6SS reveals interlaced array essential to function. Cell. 2015;160:940–951. doi: 10.1016/j.cell.2015.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kucukelbir A, Sigworth FJ, Tagare HD. Quantifying the local resolution of cryo-EM density maps. Nat Methods. 2014;11:63–65. doi: 10.1038/nmeth.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.DeLano WL. The PyMOL Molecular Graphics System. DeLano Scientific; 2002. [Google Scholar]
- 91.Lawrence MC, Colman PM. Shape complementarity at protein/protein interfaces. J Mol Biol. 1993;234:946–950. doi: 10.1006/jmbi.1993.1648. [DOI] [PubMed] [Google Scholar]
- 92.Collaborative Computational Project, Number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 93.Boc A, Diallo AB, Makarenkov V. T-REX: a web server for inferring, validating and visualizing phylogenetic trees and networks. Nucleic Acids Res. 2012;40:W573–W579. doi: 10.1093/nar/gks485. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All structural data have been deposited into the Worldwide Protein Data Bank (wwPDB) and the Electron Microscopy Data Bank (EMDB) with the following accession codes: LIIKGI (PDB 5W50), DLIIKGISVHI MicroED (PDB 5W52, EMD-8765) and DLIIKGISVHI Cryo-EM (EMD-8781, PDB 5W7V). All other data are available from the authors upon reasonable request.