Abstract
While tuberculosis (TB) remains a global disease, the WHO estimates that 62% of the incident TB cases in 2016 occurred in the WHO South-East Asia and Western Pacific regions. TB in the Pacific is composed predominantly of two genetic families of Mycobacterium tuberculosis (Mtb): Beijing and Manila. The Manila family is historically under-studied relative to the families that comprise the majority of TB in Europe and North America (e.g. lineage 4), and it remains unclear why this lineage has persisted in Filipino populations despite the predominance of more globally successful Mtb lineages in most of the world. The Beijing family is of particular interest as it is increasingly associated with drug resistance throughout the world. Both of these lineages are important to the State of Hawaii, where they comprise over two-thirds of TB cases. Here, we performed whole genome sequencing on 82 Beijing family, Manila family, and outgroup clinical Mtb isolates from Hawaii to identify lineage-specific SNPs (SNPs found in all isolates from their respective families, and exclusively in those families) in established virulence factor genes. Six non-silent lineage-specific virulence factor SNPs were found in the Beijing family, including mutations in alternative sigma factor sigG and polyketide synthases pks5 and pks7. The Manila family displayed more than eleven non-silent lineage-specific and characteristic virulence factor mutations, including in genes coding for MCE-family protein Mce1B, two mutations in fatty-acid-AMP ligase FadD26, and virulence-regulating transcriptional regulator VirS. This study further identified an ancient clade that shared some virulence factor mutations with the Manila family, and investigated the relationship of those and other “Manila-like” spoligotypes to the Manila family with this SNP dataset. This work identified a set of virulence genes that are worth pursuing to determine potential differences in transmission or virulence displayed by these two Mtb families.
Introduction
Despite sustained control efforts, tuberculosis (TB) remains the ninth leading cause of death worldwide [1]. The World Health Organization (WHO) estimated that 62% of the incident TB cases in 2016 occurred in the WHO South-East Asia and Western Pacific Regions [1]. Within those regions, TB in both China and the Philippines in characterized by distinct families of TB–the Beijing family in China and the Manila family in the Philippines [2, 3]. These families fall into global genetic lineages 1 (the Manila family) and 2 (the Beijing family), and are distinct from the lineage 4 clades that are dominant in Europe and North America [4, 5].
The WHO’s Global Tuberculosis Report 2017 indicates these regions and Mtb families as areas of concern [1]. The Philippines was only surpassed in number of incident cases by India, Indonesia, and China. As noted in the report, the Fourth National Survey of the Prevalence of TB Disease in the Philippines was conducted from March to December of 2016, and the survey revealed that tuberculosis was not well controlled in the country [1]. Comparing the prevalence of smear-positive TB among participants aged 15 years or more to the case notification rate for smear-positive TB cases in 2016 (142 per 100,000 population) in the same age group gave a prevalence-to-notification ratio of 3.1. From that, the group’s adjusted prevalence of culture positive TB became 463 per 100,000 population (95% CI: 333−592) in 2007 and 512 per 100,000 population (95% CI: 420−603) from that study in 2016. The probability that prevalence did not decline over the period 2007–2016 was estimated at 75%. Following the survey, the estimate of TB incidence in the Philippines was revised to 554 cases per 100,000 (95% CI: 311–866), or 573,000 cases. That was well above the incidence previously estimated by the WHO (322 cases per 100,000). In contrast to the steady or increasing incidence of TB in the Philippines, TB incidence in China is thought to be declining, with the WHO’s estimate being 895,000 cases (64 per 100,000) in 2016.
However, as also noted in the WHO’s report, 4.1% of new TB cases and 19% of re-treated TB cases globally are multidrug-resistant (MDR) TB cases. China is joined by India and the Russian Federation to account for 47% of the world’s total incident MDR TB cases. China saw an estimated 73,000 incident MDR TB cases in 2016, compared to 30,000 for the Philippines. Considerably more concerning is China’s estimated percentage of new TB cases that are MDR in 2016, at 7.1%,compared to 2.6% for the Philippines, though the Philippines’ total MDR rate is expected to continue to increase [6]. Approximately 70% of TB cases in China are of the Beijing family, which also displayed the highest MDR rate of all lineages in the country [7]. Beyond China, it has further been proposed that the population structure of Mtb in areas of high drug resistance is shifting towards Beijing family strains [8].
While the Beijing family has been studied with increasing intensity, especially in the context of drug resistance, considerably less work has focused on the Manila family. Both families are of importance to TB control in the United States, where immigration from the WHO South-East Asia and Western Pacific Regions continually presents TB controllers with new TB cases. The State of Hawaii, centrally located in the Pacific and experiencing a high level of immigration from throughout both of those WHO Regions, accordingly has a composition of TB lineages that resembles the United States Affiliated Pacific Islands more than the continental United States [9]. As expected, immigration from Asia imports primarily Beijing family strains to Hawaii, while immigration from the Philippines brings primarily Manila family strains [2, 3]. Those two families combined are responsible for over two-thirds of the TB cases in Hawaii, a ratio that has remained constant since the introduction of the use of spoligotyping to type Hawaii’s Mtb in 1997 [10, 11]. Furthermore, high levels of immigration from throughout the Pacific into Hawaii result in the state perennially displaying either the highest or second-highest rates of TB in the United States, with Hawaii’s TB rate remaining steady even as TB rates throughout the US continue to decrease [12].
Thus, the need to further understand the characteristics of the Beijing and Manila families of Mtb is of considerable importance not only to countries throughout the Pacific, but also to the United States. Investigating unique differences in virulence factors that are characteristic of each of these families may help explain those families’ success in their originating environments, and may aid in developing improved control strategies if differences affecting latency, reactivation, or host preference are identified. The Manila family, for example, continues to predominate in Filipino hosts, where other, more globally successful lineages have been unable to displace the Manila family and rise to high incidence in the Philippines. Considerably less research has been performed on the Manila family than on the Beijing family or on lineage 4, thus which attributes may contribute to the Manila family’s persistence remains an open question.
This study seeks to utilize next-generation Illumina whole-genome sequencing on a diverse set of Beijing and Manila family-focused clinical Mtb isolates from Hawaii in order to identify lineage-specific virulence factor differences of these two families. Here, we examine the Mtb genomes to find non-silent (i.e. non-synonymous), single-nucleotide polymorphisms (SNPs) specific to each family in virulence genes as well as other genes of interest. These SNPs are alleles that are unique to either the Beijing or Manila family isolates among the 82 that we studied. In order for a SNP to be described here as a Beijing family-specific SNP, that SNP must have been found in all Beijing family isolates from this study, but not have been found in any of this study’s non-Beijing family isolates. Similarly, SNPs described as Manila family-specific must have been found in all Manila family isolates from this study, but not have been found in any of this study’s non-Manila family isolates.
Additionally, Hawaii and the Pacific region frequently see Mtb isolates with spoligotyping fingerprints that resemble the Manila family, but are less commonly encountered than isolates possessing spoligotyping patterns that have been established as “Manila family.” This study will utilize phylogenetics from full-genome sequences to better describe the relationship of such patterns and isolates to the Manila family.
Materials and methods
Selection of Mtb isolates for study
A comprehensive database of all 1,076 M. tuberculosis isolates processed for genetic fingerprinting for the State of Hawaii from 2002 through 2016 (and including a partial set from 2017) was obtained from the State of Hawaii Department of Health Tuberculosis Control Program for this study. Spoligotype octal codes are presented as recorded by the Centers for Disease Control and Prevention (CDC) contracted reference laboratory that performed the typing. Lineage or family names were assigned to spoligotype octal codes using the SpolDB4 database, with “EAI2_MANILLA” entries being considered “Manila family.” [13]. Eighty-two Mtb isolates from this set were selected for use in this study (Table 1). Additionally, a Mycobacterium bovis BCG P3 isolate was used as an outgroup for phylogenetic analysis, as M. bovis has been shown to have diverged from the M. tuberculosis ancestor prior to the differentiation of the TB lineages examined in this study [5].
Table 1. Full list of sequenced Mycobacterium tuberculosis isolates.
| # | Accession Number |
Spoligotype Name |
Spoligotype Octal Code |
MIRU-VNTR Loci 1–12 | MIRU-VNTR Loci 13–24 | Comments |
|---|---|---|---|---|---|---|
| 7 | 09L1688 | Beijing | 000000000003771 | 223315175533 | 444444423348 | Beijing Diversity Selection |
| 10 | 09L2178 | Beijing | 000000000003771 | 222325173533 | 445644423328 | Beijing Clustered |
| 16 | 10L8643 | Beijing | 000000000003771 | 222325173533 | 445644423326 | Beijing Clustered |
| 18 | 10L9505 | Beijing | 000000000003771 | 223325173432 | 345544423539 | Beijing Diversity Selection |
| 22 | 11L1883 | Beijing | 000000000003771 | 222325173533 | 445644423326 | Beijing Clustered |
| 28 | 11L0426 | Beijing | 000000000003771 | 222325173533 | 445644423328 | Beijing Clustered |
| 29 | 11L0427 | Beijing | 000000000003771 | 222325173533 | 445644423328 | Beijing Clustered |
| 35 | 12L6290 | Beijing | 000000000003771 | 222325173533 | 445644423326 | Beijing Clustered |
| 38 | 12L7610 | Beijing | 000000000003771 | 223325173533 | 545534223328 | Beijing Diversity Selection |
| 39 | 12L7621 | Beijing | 000000000003771 | 223325173521 | 343544423339 | Beijing Diversity Selection |
| 40 | 12L7991 | Beijing | 000000000003771 | 222325173533 | 445644423328 | Beijing Clustered |
| 52 | 15RF4481 | Beijing | 000000000003771 | 223325163533 | 448544423328 | Beijing Clustered |
| 56 | 15RF6753 | Beijing | 000000000003771 | 222325173533 | 445644423328 | Beijing Clustered |
| 58 | 16RF0102 | Beijing | 000000000003771 | 222325173533 | 445644423328 | Beijing Clustered |
| 63 | 16RF2356 | Beijing | 000000000003771 | 222325173533 | 445644423328 | Beijing Clustered |
| 64 | 16RF4738 | Beijing | 000000000003771 | 223325163533 | 448544423328 | Beijing Clustered |
| 75 | 08L7125 | Beijing | 000000000003771 | 222325173533 | - | Beijing Clustered |
| 79 | 10L8839 | Beijing | 000000000003771 | 222325173533 | 445644423326 | Beijing Clustered |
| 80 | 10L9812 | Beijing | 000000000003771 | 222325173533 | 445644423326 | Beijing Clustered |
| 81 | 11L3224 | Beijing | 000000000003771 | 222325173533 | 445644423326 | Beijing Clustered |
| 82 | 12L6284 | Beijing | 000000000003771 | 222325173533 | 445644423326 | Beijing Clustered |
| 85 | 13L2956 | Beijing | 000000000003771 | 222325173533 | 445644423328 | Beijing Clustered |
| 86 | 16RF7084 | Beijing | 000000000003771 | 222325173533 | 445644423328 | Beijing Clustered |
| 74 | 08L0161 | Beijing | 000000000003751 | 222325173533 | - | Uncommon Beijing |
| 77 | 09L2141 | Beijing | 000000000003751 | 222325173533 | 445644423328 | Uncommon Beijing |
| 2 | 05L3477 | Manila | 677777477413771 | 254326223432 | 14a843263217 | Manila Clustered |
| 3 | 05L3487 | Manila | 677777477413771 | 254326223432 | 14a943263217 | Manila Clustered |
| 4 | 08L7789 | Manila | 677777477413771 | 254326223432 | 14a923273217 | Manila Diversity Selection |
| 6 | 09L1025 | Manila | 677777477413771 | 265326223432 | 148443263216 | Manila Diversity Selection |
| 9 | 09L1898 | Manila | 677777477413771 | 254326223432 | 14a843263217 | Manila Clustered |
| 11 | 09L2431 | Manila | 677777477413751 | 254316223432 | 14a743263217 | Manila Diversity Selection |
| 14 | 10L0115 | Manila | 677777477413771 | 254326223432 | 149843263217 | Manila Diversity Selection |
| 15 | 10L5331 | Manila | 677777477413671 | 254326223432 | 147743263217 | Manila Diversity Selection |
| 17 | 10L9503 | Manila | 677777477413771 | 244326223432 | 149443263217 | Manila Diversity Selection |
| 20 | 11L1563 | Manila | 677777477413771 | 254326222432 | 245843263217 | Manila Diversity Selection |
| 23 | 11L3658 | Manila | 677777477413771 | 254326223332 | 148943273217 | Manila Diversity Selection |
| 24 | 11L4599 | Manila | 677777477413771 | 254326223432 | 14a843243215 | Manila Diversity Selection |
| 34 | 12L6289 | Manila | 677777477413771 | 234326223432 | 176843263217 | Manila Diversity Selection |
| 41 | 12L9260 | Manila | 677777477413771 | 254326223432 | 13A843263217 | Manila Diversity Selection |
| 43 | 13L1313 | Manila | 677777477413771 | 244326223442 | 14A943263215 | Manila Diversity Selection |
| 45 | 13L2435 | Manila | 677777477413771 | 254326223432 | 14A943263217 | Manila Clustered |
| 46 | 13L2950 | Manila | 677777477413771 | 254326223432 | 14A843263217 | Manila Clustered |
| 48 | 14RF3136 | Manila | 677777477413771 | 254326223432 | 14a943263217 | Manila Clustered |
| 49 | 14RF3267 | Manila | 677777477000771 | 254326223432 | 149743263217 | Manila Clustered |
| 50 | 14RF5521 | Manila | 677777477413771 | 254326223432 | 14a843253217 | Manila Clustered |
| 51 | 15RF0460 | Manila | 677777477413771 | 255326223432 | 147843263217 | Manila Clustered |
| 53 | 15RF5637 | Manila | 677777477413771 | 254326223432 | 14a923263217 | Manila Clustered |
| 59 | 16RF0108 | Manila | 677777477413771 | 255326223432 | 147843263217 | Manila Clustered |
| 60 | 16RF1671 | Manila | 677777477413771 | 254326223432 | 14a943263217 | Manila Clustered |
| 61 | 16RF1679 | Manila | 677777477413771 | 254326223432 | 14a943253217 | Manila Clustered |
| 62 | 16RF1689 | Manila | 677777477413700 | 254326223432 | 14a943263217 | Manila Clustered |
| 65 | 16RF5423 | Manila | 677777477413771 | 254326223422 | 247843263217 | Manila Clustered |
| 69 | 09L0735 | Manila | 677777477413771 | - | - | Singleton, Deletions Study |
| 70 | 09L1111 | Manila | 677777477413771 | - | - | Singleton, Deletions Study |
| 89 | 96128 | Manila | 677777477413701 | - | - | Uncommon Manila |
| 90 | 20034 | Manila | 677777477413751 | - | - | Uncommon Manila |
| 5 | 09L0897 | EAI5 | 777777776413771 | 364225223533 | 245273243416 | For Manila-like Comparison |
| 33 | 11L5411 | EAI5 | 777777776413771 | 275225223532 | 245273243415 | For Manila-like Comparison |
| 84 | 13L2584 | no match* | 677774477413771 | 244326223432 | 14A843263217 | Manila-like Clustered |
| 21 | 11L1565 | no match** | 600777477413771 | 254326223432 | 146743263217 | Manila-like Cluster 1 |
| 30 | 11L2683 | no match** | 677777402003771 | 251326223432 | 14b843233217 | Manila-like Cluster 2 |
| 37 | 12L7424 | no match** | 677777402003771 | 251326223432 | 14B843233217 | Manila-like Cluster 2 |
| 44 | 13L1569 | no match** | 600777477413771 | 254326223432 | 146743263217 | Manila-like Cluster 1 |
| 8 | 09L1897 | no match | 777777607360371 | 123326143326 | 342224123235 | No Match Diversity Selection |
| 42 | 13L0560 | no match | 703717740003771 | 2254251b3533 | 524244223345 | No Match Diversity Selection |
| 71 | 04L0177 | no match | 737777377413771 | 264225223532 | - | No Match Diversity Selection |
| 72 | 06L6898 | no match | 737777377413771 | 264225223532 | - | No Match Diversity Selection |
| 73 | 06L7696 | no match | 737777377413771 | 264225223532 | - | No Match Diversity Selection |
| 13 | 09L4214 | H3 | 777777777720771 | 225325153323 | 232634423337 | Lineage 4 Diversity Selection |
| 27 | 10L8137 | H3 | 777777770020771 | 222225153322 | 143334213328 | Lineage 4 Diversity Selection |
| 76 | 09L2140 | H3 | 777777770020771 | 122325143323 | 233334213326 | Lineage 4 Diversity Selection |
| 78 | 10L6616 | H3 | 777777770020771 | 122325143323 | 233334213326 | Lineage 4 Diversity Selection |
| 1 | 04L0176 | LAM9 | 777777607760771 | 123326153326 | - | Lineage 4 Diversity Selection |
| 19 | 11L0415 | T1 | 777777777760771 | 242325142222 | 241544223124 | Lineage 4 Diversity Selection |
| 31 | 11L2831 | T1 | 777777777760771 | 2233151433–3 | 343544223326 | Lineage 4 Diversity Selection |
| 36 | 12L7116 | T1 | 777777777760771 | 233326153311 | 232334222523 | Lineage 4 Diversity Selection |
| 12 | 09L3451 | T3 | 377737777760771 | 223326153323 | 242334223425 | Lineage 4 Diversity Selection |
| 47 | 13RF4795 | U | 777777760000000 | 223325143322 | 242324223422 | U Diversity Selection |
| 54 | 15RF5640 | U | 777777760000000 | 223325143322 | 242324223422 | U Diversity Selection |
| 55 | 15RF6749 | U | 777777760000000 | 223325143322 | 242334223322 | U Diversity Selection |
| 57 | 15RF6755 | U | 777777760000000 | 223325143322 | 242324223422 | U Diversity Selection |
| 83 | 13L2363 | U | 777777760000000 | 223325143322 | 242324223422 | U Diversity Selection |
| 66 | BCG P3 | BOVIS1_BCG | 676773777777600 | - | - | M. bovis outgroup |
| - | Reference | H37Rv | 777777477760771 | 2x3226133321 | 242534233525 | Reference for alignment |
Complete list of Mycobacterium tuberculosis isolates selected for sequencing for this study. Isolates are identified by one or two digit University of Hawaii DNA extraction numbers and Centers for Disease Control and Prevention (CDC) genome accession numbers. Spoligotypes are named as presented in SpolDB4.
*No match in SpolDB4, but strongly suspected to be Manila family based on similarity.
**Manila-like.
Selection of Mtb isolates for identification of lineage-specific virulence factor differences
From the database, 25 Beijing family (spoligotype 000000000003771) isolates covering a chronological range from 2005 to 2016 were selected for whole genome sequencing. Of those, 19 isolates were selected from putative transmission clusters, four were selected to increase our represented Beijing family diversity (based on their MIRU-VNTR [Mycobacterial Interspersed Repetitive Unit-Variable Number Tandem Repeat] fingerprints), and two were selected due to possessing an uncommon Beijing family spoligotype (000000000003751). Thirty-one Manila family (representative spoligotyping pattern: 677777477413771) isolates were selected, of which 15 were selected from putative transmission clusters and 12 were selected based on uniqueness of MIRU-VNTR fingerprints to maximize this study’s Manila family diversity. To further increase the diversity of Manila family isolates examined in this study, two additional Manila family isolates with uncommon Manila family spoligotyping patterns (isolates 96128 and 20034) and two Manila family isolates with unique large-sequence-polymorphism (LSP) deletion patterns from a prior study (09L0735 and 09L1111) were selected from our laboratory’s Mycobacterial DNA Bank at the University of Hawaii, where their extracted DNA was archived [14]. Fourteen outgroup isolates from non-Beijing, non-Manila lineages from Hawaii during the 2002–2016 time period were selected for comparison, primarily on the basis of uniqueness of spoligotyping and MIRU-VNTR patterns. These isolates included four with the H3 spoligotype, one LAM9, three T1, one T3, and five from a suspected outbreak cluster with an undefined spoligotyping pattern (777777760000000). An additional three-isolate suspected transmission cluster with no SpolDB4 spoligotype match (737777377413771) was also selected.
Selection of Mtb isolates for comparison of “Manila-like” strains
Two putative two-isolate transmission clusters whose spoligotypes had no match in SpolDB4, but whose spoligotyping patterns otherwise exhibited substantial similarity to the Manila family, were selected for whole-genome comparison to the Manila family (Manila-like Cluster 1: isolates 21 and 44; Manila-like Cluster 2: isolates 30 and 37). Two EAI5 isolates were also selected (isolates 5 and 33) for sequencing due to the EAI5 clade’s similarity to the Manila family. An additional clustered isolate with no spoligotype match in SpolDB4 (isolate 84) that we considered to be Manila family based on the similarity of its spoligotype and MIRU-VNTR fingerprints was also included. Finally, two isolates with no spoligotype match in SpolDB4 and no near-matches with MIRU-VNTR patterns from other isolates (isolates 8 and 42) were selected to represent lineage-agnostic outgroups.
Whole genome sequencing
Recall of state of Hawaii Mtb isolates
All required isolates were requested from the laboratories where they had been previously sent by the State of Hawaii for contracted fingerprinting, and where they were archived (California Department of Public Health State Laboratory and Michigan Department of Community Health). Spoligotyping and 24-loci MIRU-VNTR fingerprints are displayed as provided by those laboratories. The Michigan Department of Community Health returned extracted DNA from their isolates, while isolates from the California Department of Public Health State Laboratory were returned as “double-killed” sample preps using a treatment of immersion in 70% ethanol and then heating at 80°C for one hour.
DNA extraction and whole genome sequencing
DNA extraction was performed as previously described by the National Institute of Public Health and Environmental Protection (RIVM), Bilthoven, The Netherlands (Isolation of Genomic DNA from Mycobacteria Protocol), or according to the source state laboratory’s internal protocol. DNA was quantified with the Qubit 2.0 dsDNA Broad Range Assay. Genomic libraries were prepared using the Illumina Nextera XT DNA Library Kit using manual normalization and sequenced on the Illumina MiSeq Platform with v3 Chemistry for 300 bp paired-end reads.
Data analysis
SNP matrices were produced using a modification of the NASP pipeline, with Bowtie2 used for alignment, GATK used for SNP-calling, and SNPs being filtered for a minimum of ten-fold read coverage and 90% read consensus [15, 16, 17]. Repetitive regions were removed by the NASP pipeline. A M. tuberculosis type strain H37Rv genome (GenBank NC_000962.3) was used as the scaffold for read alignment for identification of virulence factor SNPs, and our de novo sequenced representative Manila family genome “96121” (GenBank CP009427) was used as the scaffold for read alignment for construction of phylogenies and comparison of “Manila-like” strains. The number and percentage of mapped reads was determined using SAMtools flagstat. Percent coverage of the 4,411,532 base pair H37Rv reference genome NC_000962.3 was determined using SAMtools mpileup.
Identification of lineage-specific virulence factor differences
Output matrices from the NASP pipeline were imported into a Microsoft Access database and SQL queries were utilized to identify SNPs relative to H37Rv that were shared by all members of either the Beijing or Manila families that we sequenced. SQL queries were further used to search all non-Beijing isolates for those SNPs in order to exclude them as non-specific to the Beijing family if found. For the Manila family, SNPs that were shared by all Manila family isolates were searched for in all non-Manila family isolates–however, their presence in Manila-like isolates or isolates with no spoligotype match in SpolDB4 did not automatically exclude them, as the relationships of those isolates to the Manila family was examined separately and individually. SNPs in genes described in Forrellad et al.’s 2013 review of Mtb virulence were designated as “virulence factor SNPs” [18]. The strength of the association between lineage-specific and lineage-characteristic SNPs and their respective groups versus outgroups was calculated using the Fisher Exact method using R Statistics version 3.4.1 [19].
Exploring the relationship of Manila-like isolates to the Manila family
Spoligotypes of isolates (presented in this work as their CDC octal codes) were compared in order to identify the number of gained or lost spacers relative to the representative Manila family spoligotype (677777477413771). All isolates examined were individually aligned against Manila family do novo sequenced genome 96121 using the NASP pipeline to determine the numbers of SNPs separating those isolates from 96121. Putative Manila family-specific non-silent virulence factor SNPs were examined for all Manila-like, EAI5, and no-match isolates, with Mtb type strain H37Rv and the Beijing family serving as known outgroups.
A phylogenetic tree was constructed using MEGA7 utilizing maximum parsimony, obtaining the most parsimonious tree using the Subtree-Pruning-Regrafting algorithm with search level 2 where the initial trees were obtained by the random addition of sequences with ten replicates, and branch lengths were calculated using the average pathway method [20]. The maximum parsimony phylogeny was supported by 1,000 bootstrap repetitions, and rooted with Mycobacterium bovis BCG P3. To further support that phylogeny, a model averaged phylogeny was generated using jModelTest 2.1.10 with likelihood scores computed using a maximum likelihood optimized base tree with NNI base tree search and 11 substitution schemes [21]. Akaike Information Criterion (AIC), Akaike Information Criterion corrected for small sample sizes (AICc), and Bayesian Information Criterion (BIC) (all with model averaging and 100% confidence intervals) were evaluated to find the best-fit. The model-averaged phylogeny was created using AICc as the criterion for tree weights and with majority rule consensus and a 100% confidence interval.
Isolates were designated as “ancestral” or “modern” on the basis of the presence or absence of the TbD1 deletion region [22]. The presence of the TbD1 deletion was identified in the sequenced isolates by a gap in the bases aligned to de novo sequenced genome 96121 (GenBank CP009427) in the range of nucleotide positions 1,759,200 to 1,761,320.
Results
Whole genome sequencing
Of the 82 sequenced and analyzed Mtb genomes, 71 provided 90% or greater coverage of the H37Rv reference genome at 10X depth (Table 2). Eight more showed intermediate (40–90%) coverage, and three showed low coverage. Remarkably, despite showing less than 10% coverage of the reference genome, Manila family Isolate 53/15RF5637 still demonstrated 200-300X coverage depth at all of the Manila specific and characteristic SNP loci. Two isolates from a putative outbreak cluster with an undefined “U” spoligotype displayed extremely low coverage. However, the three other identically fingerprinting isolates from the same putative cluster displayed >95% coverage, ensuring that the putative cluster was still able to provide data for this study.
Table 2. Whole genome sequencing and SNP data.
| # | Accession Number | Spoligo- type |
Total SNPs | Unique SNPs |
Bases with 10X Coverage | Percent Coverage |
Reads Passed QC | Reads Mapped | % Reads Mapped |
|---|---|---|---|---|---|---|---|---|---|
| 7 | 09L1688 | Beijing | 1214 | 150 | 4366749 | 99.0% | 8000000 | 7864232 | 98.3% |
| 10 | 09L2178 | Beijing | 1259 | 5 | 4356699 | 98.8% | 2157898 | 2122913 | 98.4% |
| 16 | 10L8643 | Beijing | 1263 | 0 | 4377042 | 99.2% | 8000000 | 7616623 | 95.2% |
| 18 | 10L9505 | Beijing | 1199 | 106 | 4369709 | 99.1% | 1607256 | 1548473 | 96.3% |
| 22 | 11L1883 | Beijing | 1263 | 0 | 4297662 | 97.4% | 912052 | 886821 | 97.2% |
| 28 | 11L0426 | Beijing | 1254 | 0 | 4287003 | 97.2% | 912966 | 897819 | 98.3% |
| 29 | 11L0427 | Beijing | 1254 | 0 | 4370299 | 99.1% | 3370522 | 3268479 | 97.0% |
| 35 | 12L6290 | Beijing | 1263 | 0 | 4342168 | 98.4% | 1049434 | 1015433 | 96.8% |
| 38 | 12L7610 | Beijing | 1236 | 92 | 4338152 | 98.3% | 1302504 | 1274025 | 97.8% |
| 39 | 12L7621 | Beijing | 1219 | 126 | 4329373 | 98.1% | 1001768 | 967945 | 96.6% |
| 40 | 12L7991 | Beijing | 1236 | 5 | 4348892 | 98.6% | 1587788 | 1559313 | 98.2% |
| 52 | 15RF4481 | Beijing | 1224 | 0 | 4295503 | 97.4% | 1019258 | 987630 | 96.9% |
| 56 | 15RF6753 | Beijing | 1239 | 8 | 4269559 | 96.8% | 868080 | 838900 | 96.6% |
| 58 | 16RF0102 | Beijing | 1230 | 1 | 4298317 | 97.4% | 1066562 | 1033295 | 96.9% |
| 63 | 16RF2356 | Beijing | 1237 | 0 | 4216800 | 95.6% | 1316206 | 1276147 | 97.0% |
| 64 | 16RF4738 | Beijing | 1224 | 0 | 4311600 | 97.7% | 1271118 | 1211113 | 95.3% |
| 75 | 08L7125 | Beijing | 1231 | 0 | 4083094 | 92.6% | 1234036 | 1182148 | 95.8% |
| 79 | 10L8839 | Beijing | 1242 | 0 | 4284326 | 97.1% | 1887444 | 1811105 | 96.0% |
| 80 | 10L9812 | Beijing | 1242 | 0 | 4341917 | 98.4% | 2928112 | 2806986 | 95.9% |
| 81 | 11L3224 | Beijing | 1242 | 0 | 4038783 | 91.6% | 1208578 | 1159426 | 95.9% |
| 82 | 12L6284 | Beijing | 1264 | 0 | 4298709 | 97.4% | 2068176 | 1988761 | 96.2% |
| 85 | 13L2956 | Beijing | 1253 | 2 | 4339004 | 98.4% | 3256308 | 3158130 | 97.0% |
| 86 | 16RF7084 | Beijing | 1260 | 1 | 4340781 | 98.4% | 2406912 | 2312842 | 96.1% |
| 74 | 08L0161 | Beijing | 1256 | 1 | 4308269 | 97.7% | 1918020 | 1858140 | 96.9% |
| 77 | 09L2141 | Beijing | 1255 | 0 | 4325345 | 98.0% | 3012910 | 2903739 | 96.4% |
| 2 | 05L3477 | Manila | 1898 | 42 | 4382063 | 99.3% | 1391250 | 1361361 | 97.9% |
| 3 | 05L3487 | Manila | 1886 | 63 | 4399741 | 99.7% | 8000000 | 7401374 | 92.5% |
| 4 | 08L7789 | Manila | 1873 | 61 | 4381481 | 99.3% | 1868494 | 1826719 | 97.8% |
| 6 | 09L1025 | Manila | 1898 | 105 | 4392248 | 99.6% | 1694468 | 1656631 | 97.8% |
| 9 | 09L1898 | Manila | 1885 | 28 | 4375342 | 99.2% | 1906012 | 1854219 | 97.3% |
| 11 | 09L2431 | Manila | 1914 | 95 | 4383925 | 99.4% | 2261522 | 2216486 | 98.0% |
| 14 | 10L0115 | Manila | 1877 | 52 | 4374642 | 99.2% | 2365498 | 2325801 | 98.3% |
| 15 | 10L5331 | Manila | 1896 | 24 | 4368029 | 99.0% | 2555006 | 2505891 | 98.1% |
| 17 | 10L9503 | Manila | 1886 | 95 | 4396221 | 99.7% | 8000000 | 7576871 | 94.7% |
| 20 | 11L1563 | Manila | 1897 | 41 | 4355894 | 98.7% | 1132140 | 1047235 | 92.5% |
| 23 | 11L3658 | Manila | 1740 | 72 | 4386744 | 99.4% | 3066566 | 3006616 | 98.0% |
| 24 | 11L4599 | Manila | 1723 | 58 | 4373544 | 99.1% | 2375178 | 2323296 | 97.8% |
| 34 | 12L6289 | Manila | 1724 | 0 | 3096482 | 70.2% | 506266 | 497032 | 98.2% |
| 41 | 12L9260 | Manila | 1721 | 48 | 4352622 | 98.7% | 2009858 | 1977338 | 98.4% |
| 43 | 13L1313 | Manila | 1770 | 135 | 4366415 | 99.0% | 2166864 | 2126405 | 98.1% |
| 45 | 13L2435 | Manila | 1730 | 69 | 4346445 | 98.5% | 1431348 | 1406436 | 98.3% |
| 46 | 13L2950 | Manila | 1738 | 64 | 1952034 | 44.2% | 395948 | 348056 | 87.9% |
| 48 | 14RF3136 | Manila | 1735 | 49 | 4064968 | 92.1% | 587692 | 563574 | 95.9% |
| 49 | 14RF3267 | Manila | 1753 | 74 | 4210902 | 95.5% | 719036 | 682961 | 95.0% |
| 50 | 14RF5521 | Manila | 1755 | 70 | 2381270 | 54.0% | 897166 | 279096 | 31.1% |
| 51 | 15RF0460 | Manila | 1104 | 0 | 4255224 | 96.5% | 747934 | 720277 | 96.3% |
| 53 | 15RF5637 | Manila | 1125 | 37 | 401340 | 9.1% | 1424204 | 140153 | 9.8% |
| 59 | 16RF0108 | Manila | 1104 | 0 | 4274093 | 96.9% | 876532 | 838126 | 95.6% |
| 60 | 16RF1671 | Manila | 1121 | 32 | 3879987 | 88.0% | 551846 | 531611 | 96.3% |
| 61 | 16RF1679 | Manila | 1131 | 43 | 4175829 | 94.7% | 686132 | 658483 | 96.0% |
| 62 | 16RF1689 | Manila | 1133 | 64 | 4275638 | 96.9% | 899476 | 865810 | 96.3% |
| 65 | 16RF5423 | Manila | 1132 | 88 | 2048030 | 46.4% | 859888 | 311073 | 36.2% |
| 69 | 09L0735 | Manila | 1110 | 30 | 4356000 | 98.7% | 1655260 | 1608766 | 97.2% |
| 70 | 09L1111 | Manila | 1114 | 39 | 2757572 | 62.5% | 323612 | 310787 | 96.0% |
| 89 | 96128 | Manila | 1790 | 52 | 4370752 | 99.1% | 4774208 | 4632407 | 97.0% |
| 90 | 20034 | Manila | 1790 | 58 | 4360828 | 98.9% | 2170986 | 2085847 | 96.1% |
| 5 | 09L0897 | EAI5 | 1880 | 190 | 4342065 | 98.4% | 1479306 | 1428183 | 96.5% |
| 33 | 11L5411 | EAI5 | 1865 | 8 | 3068579 | 69.6% | 507294 | 497774 | 98.1% |
| 84 | 13L2584 | no match* | 1803 | 112 | 4335626 | 98.3% | 2260038 | 2178054 | 96.4% |
| 21 | 11L1565 | no match** | 1815 | 0 | 4361217 | 98.9% | 1318272 | 1279928 | 97.1% |
| 30 | 11L2683 | no match** | 1792 | 0 | 4365958 | 99.0% | 6147928 | 5990291 | 97.4% |
| 37 | 12L7424 | no match** | 1797 | 5 | 4352197 | 98.7% | 2665532 | 2352390 | 88.3% |
| 44 | 13L1569 | no match** | 1818 | 3 | 4389277 | 99.5% | 8000000 | 6583222 | 82.3% |
| 8 | 09L1897 | no match | 738 | 49 | 4372374 | 99.1% | 2008844 | 1977757 | 98.5% |
| 42 | 13L0560 | no match | 1242 | 574 | 4396157 | 99.7% | 6411876 | 5888007 | 91.8% |
| 71 | 04L0177 | no match | 1784 | 0 | 2273101 | 51.5% | 550312 | 525881 | 95.6% |
| 72 | 06L6898 | no match | 1785 | 0 | 4285043 | 97.1% | 1880038 | 1799285 | 95.7% |
| 73 | 06L7696 | no match | 1785 | 0 | 4229398 | 95.9% | 1585002 | 1518077 | 95.8% |
| 13 | 09L4214 | H3 | 820 | 348 | 4361423 | 98.9% | 2175416 | 2141283 | 98.4% |
| 27 | 10L8137 | H3 | 757 | 139 | 4097690 | 92.9% | 615790 | 598982 | 97.3% |
| 76 | 09L2140 | H3 | 767 | 0 | 4286721 | 97.2% | 1786962 | 1725623 | 96.6% |
| 78 | 10L6616 | H3 | 770 | 2 | 4336213 | 98.3% | 2394498 | 2321661 | 97.0% |
| 1 | 04L0176 | LAM9 | 808 | 76 | 4376679 | 99.2% | 2094096 | 2062342 | 98.5% |
| 19 | 11L0415 | T1 | 751 | 325 | 4358306 | 98.8% | 1089628 | 1063640 | 97.6% |
| 31 | 11L2831 | T1 | 725 | 310 | 4385415 | 99.4% | 8000000 | 7197683 | 90.0% |
| 36 | 12L7116 | T1 | 325 | 123 | 4393768 | 99.6% | 1499892 | 1186380 | 79.1% |
| 12 | 09L3451 | T3 | 507 | 191 | 4382703 | 99.3% | 2509032 | 2469187 | 98.4% |
| 47 | 13RF4795 | U | 62 | 0 | 4294777 | 97.4% | 891672 | 866742 | 97.2% |
| 54 | 15RF5640 | U | 86 | 5 | 585165 | 13.3% | 1128496 | 146887 | 13.0% |
| 55 | 15RF6749 | U | 57 | 0 | 4224552 | 95.8% | 796968 | 756386 | 94.9% |
| 57 | 15RF6755 | U | 181 | 101 | 92161 | 2.1% | 904078 | 55734 | 6.2% |
| 83 | 13L2363 | U | 60 | 2 | 4358815 | 98.8% | 2559816 | 2473275 | 96.6% |
Isolates are identified by one or two digit University of Hawaii DNA extraction numbers and Centers for Disease Control and Prevention (CDC) genome accession numbers. Spoligotypes are named as presented in SpolDB4. Total SNPs are those identified versus the Mtb type strain genome H37Rv GenBank NC_000962.3 (length: 4,411,532 base pairs). Unique SNPs are alleles not found at that locus in any other isolate in this study.
*No match in SpolDB4, but strongly suspected to be Manila family based on similarity.
**Manila-like.
The numbers of SNPs displayed by each isolate are generally determined by the isolates’ families, with ancestral Manila family isolates, as expected, displaying the most SNPs against the modern H37Rv genome. The number of unique SNPs for each isolate was mostly consistent with that isolate’s clustering. Isolates from direct transmission clusters displayed zero to six unique SNPs, while isolates that shared a MIRU-VNTR fingerprint, but were not actually transmission linked (common for Beijing and Manila family isolates) displayed dozens or more.
Identification of lineage-specific virulence factor differences
Multiple Beijing family specific virulence factor SNPs were identified in this study. In addition to identifying virulence factor differences specific to the Manila family, we also identified “lineage characteristic” virulence factor differences that were also found in a relatively uncommon non-Manila clade, but which may regardless be important for charactering the Manila family’s virulence.
Beijing family-specific virulence factor SNPs
Six SNPs in five previously described virulence factor genes were identified as lineage-specific non-silent virulence factor SNPs for the Beijing family (Table 3). Four additional Beijing-specific mutations in four genes possibly contributing to virulence or drug resistance, or otherwise of interest, were also identified.
Table 3. Beijing family-specific virulence factor SNP mutations, non-silent.
| Published Virulence Factor Mutations | |||||||||||
| Nucleotide Locus | Reference | Variant | Rv # | Nucleotide # | Strand | Old Codon | Old AA | New Codon | New AA | Gene Title | p-value |
| 213147 | C | A | Rv0182c | 994 | R | GAC | D | TAC | F | alternative RNA polymerase sigma factor SigG | <0.005 |
| 213281 | C | T | Rv0182c | 860 | R | GGC | G | GAC | D | alternative RNA polymerase sigma factor SigG | <0.005 |
| 1722228 | A | C | Rv1527c | 6182 | R | CTG | L | CGG | R | polyketide synthase Pks5 | <0.005 |
| 1877744 | A | C | Rv1661 | 2441 | F | GAA | E | GCA | A | polyketide synthase Pks7 | <0.005 |
| 2673818 | C | G | Rv2383c | 2020 | R | GTG | V | CTG | L | phenyloxazoline synthase MbtB | <0.005 |
| 3979990 | C | G | Rv3540c | 670 | R | GTG | V | CTG | L | lipid-transfer protein or keto acyl-CoA thiolase Ltp2 | <0.005 |
| Possible Virulence Factor Mutations and Genes of Interest | |||||||||||
| Nucleotide Locus | Reference | Variant | Rv # | Nucleotide # | Strand | Old Codon | Old AA | New Codon | New AA | Gene Title | p-value |
| 1361285 | C | T | Rv1217c | 517 | R | GCT | A | AGT | S | antibiotic transporter permease | <0.005 |
| 1643864 | T | C | Rv1458c | 397 | R | ACC | T | GCC | A | antibiotic transporter ATP-binding protein | <0.005 |
| 1955941 | G | C | Rv1730c | 1305 | R | GAC | D | GAG | E | penicillin-binding protein | <0.005 |
| 3127931 | T | A | Rv2820 | 342 | R | AAA | K | AAT | N | CRISPR type III-a/mtube-associated ramp protein csm4 | <0.005 |
List of non-silent SNPS in possible virulence factor genes or genes of interest that were exclusively found in all Beijing family isolates. None of these alleles were found in any non-Beijing-family isolates in this study. Rv numbers and gene titles are those provided by TubercuList [23]. Nucleotide loci are genomic nucleotide positions in Mtb H37Rv genome NC_000962.3. P-values were calculated using the Fisher Exact method. F: forward strand. R: reverse strand. AA: amino acid.
Manila family-specific virulence factor SNPs
Seven SNPs in six previously described virulence factor genes were identified as non-silent lineage-specific virulence factor SNPs for the Manila family (Table 4). Two of those mutations were located several bases upstream of the start codons, possibly in the promotor region or ribosome binding site. Four additional virulence factor mutations found in all Manila family isolates, but also in one epidemiological cluster with no SpoldDB4 spoligotype match and in one phylogenetically distinct EAI5 isolate, are also listed in Table 4.
Table 4. Family specific and characteristic virulence factor SNP mutations of the Manila family of Mtb.Family specific virulence factor mutations of the Manila family, non-silent.
| Nucleotide Locus | Reference | Variant | Rv # | Nucleotide # | Strand | Old Codon | Old AA | New Codon | New AA | Gene Title | p-value | # Non-Manila |
| 200379 | C | T | Rv0170 | 485 | F | GCC | A | GTC | V | MCE-family protein Mce1B | <0.005 | - |
| 236512 | C | T | Rv0198c -5 | -5 | - | - | - | - | - | 5 bases upstream Rv0198c zinc metalloprotease Zmp1, possibly in the ribosome binding site | <0.005 | - |
| 235681 | G | A | Rv0191c | 827 | R | GGC | A | GTG | V | zinc metalloprotease Zmp1 | <0.005 | - |
| 495198 | A | G | Rv0410c | 2117 | R | TTC | F | TCC | S | serine/threonine-protein kinase PknG | <0.005 | - |
| 1038500* | T | G | Rv0931c | 1415 | R | CAG | Q | CCG | P | transmembrane serine/threonine-protein kinase D PknD | <0.005 | - |
| 1875295* | C | T | Pks7–9 | -9 | - | - | - | - | - | 9 bases upstream of Rv1661 polyketide synthase Pks7, possibly in the promotor | <0.005 | - |
| 3244126 | G | A | Rv2930 | 430 | F | GTT | V | ATT | I | fatty-acid-AMP ligase FadD26 | <0.005 | - |
| Family characteristic virulence factor mutations of the Manila family, non-silent | ||||||||||||
| Nucleotide Locus | Reference | Variant | Rv # | Nucleotide # | Strand | Old Codon | Old AA | New Codon | New AA | Gene Title | p-value | # Non-Manila |
| 2745739 | G | A | Rv2445c -15 | -15 | - | - | - | - | - | 15 bases upstream of Rv2445c nucleoside diphosphate kinase NdkA, possibly in the promotor | <0.005 | 2 (4) |
| 3244414 | A | G | Rv2930 | 718 | F | ATG | M | GTG | V | fatty-acid-AMP ligase FadD26 | <0.005 | 2 (4) |
| 3447480 | A | C | Rv3082c | 947 | R | CTC | L | CGC | R | virulence-regulating transcriptional regulator VirS | <0.005 | 2 (4) |
| 4290827 | C | G | Rv3823c | 703 | R | GGG | G | CGG | R | membrane transporter MmpL8 | <0.005 | 2 (4) |
List of family-specific and family-characteristic non-silent SNPs in virulence factor genes. All family-specific SNPs were exclusively found in all Manila family isolates. None of these Manila-specific SNPs were found in any non-Manila-family isolates in this study. Family-characteristic SNPs are non-silent SNPs in virulence factor genes that were found in all Manila family isolates, as well as a limited set of similar isolates in this study.
# Non-Manila is the number of clusters that are neither Manila family nor Manila-like that possess this allele (number of contained isolates is in parenthesis). Rv numbers and gene titles are those provided by TubercuList [23]. Nucleotide loci are genomic nucleotide positions in Mtb H37Rv genome NC_000962.3. P-values were calculated using the Fisher Exact method. F: forward strand. R: reverse strand. AA: amino acid. *One two-isolate Manila-like cluster did not display either of these two SNPs.
Manila family-specific possible virulence factor or gene-of-interest SNPs
Five Manila-specific SNPs in genes possibly contributing to virulence or drug resistance, or otherwise of interest, were identified (Table 5). Six additional such mutations found in all Manila family isolates, but also in one epidemiological cluster with no SpoldDB4 spoligotype match and in one phylogenetically distinct EAI5 isolate, are also listed in Table 5.
Table 5. Manila family specific and characteristic SNP mutations in possible virulence factor genes and genes of interest.
| Family specific possible virulence factor mutations and genes of interest of the Manila family, non-silent | ||||||||||||
| Nucleotide Locus | Reference | Variant | Rv # | Nucleotide # | Strand | Old Codon | Old AA | New Codon | New AA | Gene Title | p-value | # Non-Manila |
| 27199 | G | A | Rv0022c | 244 | R | CAG | Q | TAG | STOP | transcriptional regulator WhiB-like WhiB5 | <0.005 | - |
| 2239160 | G | C | Rv1996 | 157 | F | GGG | G | CGG | R | universal stress protein | <0.005 | - |
| 2654696 | G | A | Rv2374c | 398 | R | TCG | S | TTG | L | heat shock protein transcriptional repressor HrcA | <0.005 | - |
| 3129675 | C | T | Rv2823c | 2099 | R | CGC | R | CAC | H | CRISPR-associated protein cas10/csm1, subtype III-a/mtube | <0.005 | - |
| 27473 | G | T | WhiB5–31 | -31 | - | - | - | - | - | 31 bp upstream of transcriptional regulator WhiB-like WhiB5, possibly in the promotor | <0.005 | - |
| Family characteristic possible virulence factor mutations and genes of interest related to the Manila family, non-silent | ||||||||||||
| Nucleotide Locus | Reference | Variant | Rv # | Nucleotide # | Strand | Old Codon | Old AA | New Codon | New AA | Gene Title | p-value | # Non-Manila |
| 412280 | T | G | Rv0342 | 1443 | F | CAT | H | CAG | Q | isoniazid inducible protein IniA | <0.005 | 2 (4) |
| 1097023 | G | A | Rv0981 | 202 | F | GGC | G | AGC | S | mycobacterial persistence regulator MRPA | <0.005 | 2 (4) |
| 2239055 | C | T | Rv1996 | 52 | F | CCC | P | TCC | S | universal stress protein | <0.005 | 2 (4) |
| 2574598 | C | T | Rv2303c | 422 | R | AGC | S | AAC | N | antibiotic-resistance protein | <0.005 | 2 (4) |
| 2726051 | G | A | Rv2427ac | 37 | - | - | - | - | - | pseudogene OxyR protein | <0.005 | 2 (4) |
| 27469 | A | G | WhiB5–27 | -27 | - | - | - | - | - | 27 bp upstream of transcriptional regulator WhiB-like WhiB5, possibly in the promotor | <0.005 | 2 (4) |
List of family-specific and family-characteristic non-silent SNPs in possible virulence factor genes or genes of interest. All family-specific SNPs were exclusively found in all Manila family isolates. None of these Manila-specific SNPs were found in any non-Manila-family isolates in this study. Family-characteristic SNPs are non-silent SNPs in virulence factor genes that were found in all Manila family isolates, as well as a limited set of similar isolates in this study.
# Non-Manila is the number of clusters that are neither Manila family nor Manila-like that possess this allele (number of contained isolates is in parenthesis). Rv numbers and gene titles are those listed in TubercuList [23]. Nucleotide loci are genomic nucleotide positions in Mtb H37Rv genome NC_000962.3. P-values were calculated using the Fisher Exact method. F: forward strand. R: reverse strand. AA: amino acid
Relationship of Manila-like isolates to the Manila family
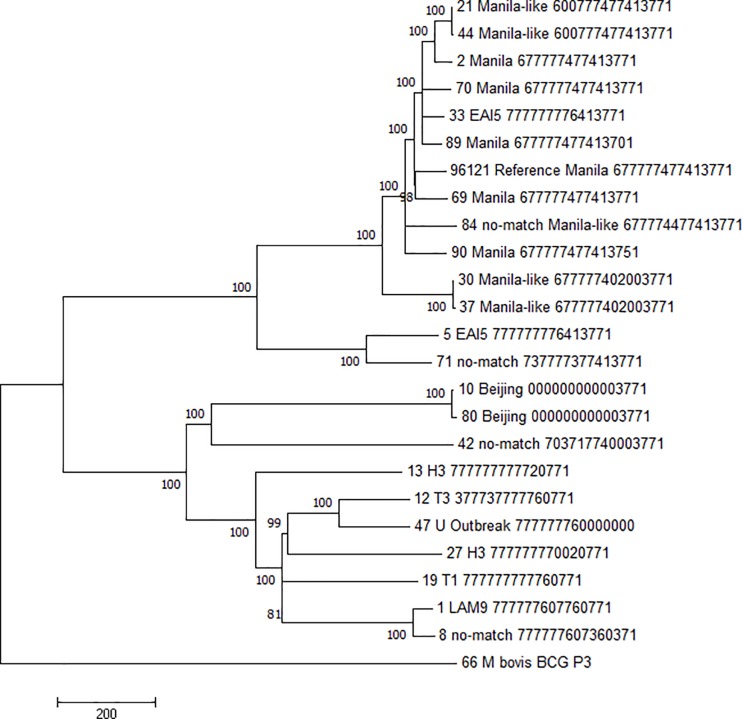
Phylogenetic analysis based on all genomic SNPs among selected isolates was conducted to further investigate the relationships of “Manila-like” and other strains to the Manila family (Fig 1). This phylogeny showed that the Manila family isolates that were selected due to their uncommon (but still Manila family) spoligotyping patterns (isolates 89 and 90) grouped within Manila family isolates that displayed the common Manila pattern. Likewise, the isolates selected based on previous deletion analysis to maximize the Manila family diversity in this study (isolates 69 and 70) grouped with other Manila family isolates as expected.
Fig 1. Maximum parsimony phylogeny of Manila-like isolates.
Evolutionary history inferred by MEGA7 using the maximum parsimony method and rooted by Mycobacterium bovis BCG P3. Branch support was determined by 1000 bootstrap repetitions (bootstrap values shown above each node). A model averaged phylogeny further supported this result (data not shown).
Other isolates displayed diverse relationships with the Manila family. One cluster of “Manila-like” isolates whose spoligotypes had no match in SpolDB4 (isolates 21 and 44) were shown to group inside the Manila family branch. However, one no-match Manila-like cluster (isolates 30 and 37), which was also observed to exhibit pigmentation when grown on Löwenstein–Jensen medium and differed from the Manila family at only two virulence factor SNP loci, occupies a branch of the phylogeny immediately divergent to the Manila family. Interestingly, one EAI5 isolate (5) and a three-isolate no SpolDB4 match epidemiological cluster (71–73) occupy a distinct branch between lineage 1 (Manila family and other EAI lineages) and the modern lineages that include lineages 2 (including the Beijing family) and 4 (including the LAM lineages, H lineages, and others). This was also an expected result, as those isolates matched the Manila-characteristic virulence factor SNP alleles and deviated from them in roughly equal number (Table 6). Other isolates with no SpolDB4 match (isolates 8 and 42, selected for diversity due to appearing to belong to no obvious lineage) grouped with lineage 4 isolates.
Table 6. Virulence factor SNPs specific to Manila and Manila-like isolates.
| Spoligotype Name | Manila Reference | Uncommon Manila Spoligotype | EAI5 | Uncommon Manila Spoligotype | Manila-like* | Manila-like** | Manila-like** † | No-SpolDB4 Match‡ | EAI5 | No SpolDB4 Match | Beijing (Outgroup) | No SpolDB4 Match | Mtb Type Strain |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Isolate # | 96121 | 89 | 33 | 90 | 21 | 84 | 37 | 71 | 5 | 8 | 10 | 42 | H37Rv |
| Spacers Lost | - | 3 | 1 | 1 | 6 | 2 | 7 | 2 | 1 | 8 | 26 | 13 | 3 |
| Spacer Gained | - | - | 3 | - | - | - | - | 3 | 3 | 6 | 0 | 1 | 5 |
| SNPs vs. 96121 | - | 239 | 239 | 278 | 281 | 299 | 437 | 872 | 1020 | 1920 | 1964 | 1970 | NA |
| 27,469 | G | G | G | G | G | G | G | G | G | A | A | A | A |
| 412,280 | G | G | G | G | G | G | G | G | G | T | T | T | T |
| 1,097,023 | A | A | A | A | A | A | A | A | A | G | G | G | G |
| 2,239,055 | T | T | T | T | T | T | T | T | T | C | C | C | C |
| 2,574,598 | T | T | T | T | T | T | T | T | T | C | C | C | C |
| 2,726,051 | A | A | A | A | A | A | A | A | A | G | G | G | G |
| 2,745,739 | A | A | A | A | A | A | A | A | A | G | G | G | G |
| 3,244,414 | G | G | G | G | G | G | G | G | G | A | A | A | A |
| 3,447,480 | C | C | C | C | C | C | C | C | C | A | A | A | A |
| 4,290,827 | G | G | G | G | G | G | G | G | G | C | C | C | C |
| 27,199 | A | A | A | A | A | A | A | G | G | G | G | G | G |
| 27,473 | T | T | T | T | T | T | T | G | G | G | G | G | G |
| 200,379 | T | T | T | T | T | T | T | C | C | C | C | C | C |
| 235,681 | A | A | A | A | A | A | A | G | G | G | G | G | G |
| 236,512 | T | T | T | T | T | T | T | C | C | C | C | C | C |
| 495,198 | G | G | G | G | G | G | G | A | A | A | A | A | A |
| 2,239,160 | C | C | C | C | C | C | C | G | G | G | G | G | G |
| 2,654,696 | A | A | A | A | A | A | A | G | G | G | G | G | G |
| 2,673,701 | T | T | T | T | T | T | T | C | C | C | C | C | C |
| 3,129,675 | T | T | T | T | T | T | T | C | C | C | C | C | C |
| 3,244,126 | A | A | A | A | A | A | A | G | G | G | G | G | G |
| 1,038,500 | G | G | G | G | G | G | T | T | T | T | T | T | T |
| 1,875,295 | T | T | T | T | T | T | C | C | C | C | C | C | C |
Comparison of SNP alleles between Manila family, Manila-like, unknown, and outgroup isolates. Shaded alleles match those displayed by Mtb type strain H37Rv, while non-shaded alleles match representative Manila family isolate 96121. Loci displayed in the lower two groups are found in Tables 4 and 5 as family-specific SNPs. Loci in the upper group are found in Tables 4 and 5 as family-characteristic SNPs; although not exclusive to the Manila family, these SNPs shared only with isolates from outside of existing major spoligotyping lineages may be important for defining virulence of the Manila family. All Manila family diversity selection isolates present the same alleles, as do two Manila-like clusters and one EAI5 isolate. One Manila-like cluster differed from the Manila family at two of these selected loci. One other EAI5 isolate and one cluster with no SpolDB4 match diverged from the Manila family at thirteen selected loci. These differences suggest possible differences in virulence for this small clade.
Nucleotide loci are genomic nucleotide positions in Mtb H37Rv genome NC_000962.3.
Spacers gained or lost are relative to the Manila family reference spoligotype 677777477413771.
* No match in SpolDB4, but Manila-like; representative of a two-isolate epidemiological cluster with both isolates sharing the same alleles at these loci.
**No match in SpolDB4, but almost certainly Manila family; representative of a two-isolate epidemiological cluster with both isolates sharing the same alleles at these loci.
† Representative of a two-isolate epidemiological cluster with both isolates sharing the same alleles at these loci.
‡ Representative of a three-isolate epidemiological cluster with all isolates sharing the same alleles at these loci.
Table 6 further illustrates the distribution of loci that were designated as either Manila-specific (found only in Manila family isolates) or Manila-characteristic (found in all Manila family isolates as well as certain related groups). Notably, two SNPs (1,038,500 and 1,875,295) that were designated as Manila-specific were not shared by one two-isolate Manila-like cluster (isolates 30 and 37), though the cluster matched the Manila family at the other virulence loci. Additionally, since octal coding might not intuitively convey how many spacers are gained or lost with a change from one digit to another, Table 7 presents the numbers of spacers gained or lost by each isolate relative to Manila family reference genome 96121. This comparison of SNPs versus 96121 and spacers gained or lost clearly illustrates that the number of spacers lost by an isolate relative to 96121 fails to predict an isolate or lineage’s closeness to the Manila family.
Table 7. Clustering and spoligotype details of selected Manila and Manila-like isolates.
| 96121 vs isolate # | SNPs vs. 96121 | Isolate Cluster | Spoligotyping Pattern (96121: 677777477413771) |
Spacers Lost | Spacers Gained |
|---|---|---|---|---|---|
| 70 | 193 | Manila Diversity Selection (Deletions) | 677777477413771 | - | - |
| 69 | 210 | Manila Diversity Selection (Deletions) | 677777477413771 | - | - |
| 89 | 239 | Manila Diversity Selection | 677777477413701 | 3 | - |
| 33 | 239 | EAI5 | 777777776413771 | 1 | 3 |
| 2 | 278 | Manila Cluster A | 677777477413771 | - | - |
| 90 | 278 | Manila Diversity Selection | 677777477413751 | 1 | - |
| 21 | 281 | Manila-like Cluster 1 | 600777477413771 | 6 | - |
| 44 | 293 | Manila-like Cluster1 | 600777477413771 | 6 | - |
| 84 | 299 | Manila-like Cluster B | 677774477413771 | 2 | - |
| 37 | 437 | Manila-like Cluster 2 (Pigmented) | 677777402003771 | 7 | - |
| 30 | 438 | Manila-like Cluster 2 (Pigmented) | 677777402003771 | 7 | - |
| 71 | 872 | 02–06 No SpolDB4 Match | 737777377413771 | 2 | 3 |
| 72 | 873 | 02–06 No SpolDB4 Match | 737777377413771 | 2 | 3 |
| 73 | 873 | 02–06 No SpolDB4 Match | 737777377413771 | 2 | 3 |
| 5 | 1020 | EAI5 | 777777776413771 | 1 | 3 |
| 8 | 1920 | Diversity Selection | 777777607360371 | 8 | 6 |
| 80 | 1925 | Beijing Family (Outgroup) | 000000000003771 | 26 | 0 |
| 10 | 1964 | Beijing Family (Outgroup) | 000000000003771 | 26 | 0 |
| 42 | 1970 | Diversity Selection | 703717740003771 | 13 | 1 |
Details of the clusters represented by isolates in Fig 1. Spoligotyping octal elements that differ from the standard Manila family spoligotyping pattern 677777477413771 are marked in bold and underlined, with the numbers of CRISPR spacers gained or lost (versus the standard Manila family pattern) also presented. Isolates are ordered by the total number of genomic SNPs against de novo sequenced representative Manila family isolate 96121. Note that the number of spacers lost relative to the Manila family reference fails to indicate the closeness of the isolate’s relationship to 96121, as determined by the number of SNPs between them.
Discussion
Identification of lineage-specific virulence factor differences
Lineage-specific SNPs in virulence factor genes were identified for both the Beijing and Manila families. These were defined as non-silent mutations that were shared by all isolates in that family, but could not be found outside of that family in this study.
Lineage-specific Beijing family virulence factor SNPs
This study identified several virulence factor mutations specific to the Beijing family. Two mutations were found in alternative RNA polymerase sigma factor gene sigG, which has been shown to be upregulated during macrophage infection as well as by DNA damage, despite not regulating DNA repair genes. Accordingly, its deletion has been shown to impair survival in a macrophage infection model [24, 25]. Mutations were also found in polyketide synthase genes pks5 and pks7. The contribution of these genes to Mtb virulence has been investigated by multiple studies. For example, disruption of Pks5 showed no difference in cell envelope lipid composition, but resulted in severe growth defects in a mouse infection model. In contrast, disruption of Pks7 produced a strain that was deficient in the production of phthiocerol dimycocerosates (PDIMs), which has been known to attenuate growth during in vivo infection, and in this case attenuated growth in mice infected by respiratory inoculation [18, 26, 27]. In addition, W-Beijing family strains have been shown to produce a polyketide synthase-derived phenolic glycolipid and demonstrated hypervirulence in a murine model, associated with a reduction in production of pro-inflammatory cytokines TNF-α, IL-6, and IL-12 [28]. One mutation was found in phenyloxazoline synthase MbtB, which is involved in iron acquisition, an important virulence factor for growth inside a macrophage, where iron levels can be 1/1000th those outside the leukocyte [18]. The final Beijing family virulence factor found to display a lineage-specific mutation was lipid-transfer protein or keto acyl-CoA thiolase Ltp2, which may be involved in cholesterol transport [18].
The Beijing family’s lineage-specific mutations in other genes of interest included one in antibiotic transporter permease Rv1217c, which has been shown to actively transport the ionophore antibiotic tetronasin across the cell membrane, and one in antibiotic transporter ATP-binding protein Rv1458c, which is similar to other described antibiotic transporters [29]. A mutation was also found in a penicillin-binding protein (Rv1730c). Penicillin is widely regarded as ineffective against Mtb, but the rise of extensively drug-resistant TB has renewed interest in the use of beta-lactam drugs against it [30]. Additionally, studies combining beta lactam drugs with beta lactamase inhibitors have shown promise [31]. Finally, CRISPR type III-a/mtube-associated ramp protein Csm4 showed one mutation. Although the exact role of Csm4 is still being deduced, CRISPR-related changes in the Beijing family are of interest due to its lack of many of the CRISPR spacers from the set of 43 utilized by spoligotyping that are present in many other lineages.
The Beijing family has been shown to be split into ancient (atypical) and modern (typical) sublineages on the basis of the presence or absence of one to two IS6110 insertion sequences in the NTF region, with the modern sublineage being shown to display increased virulence relative to the ancestral sublineage [32]. Depending on their sublineage, Beijing family isolates display mutations in two DNA repair genes (mutT4 and mutT2) as well as ogg, which are thought to result in an increased mutation rate, allowing for quicker adaptation to new host pressures or environments [32, 33]. The genetic difference between ancient and modern Beijing strains has been further explored, and while mutations have been identified, their phenotypic results still require investigation [34]. Our present study, however, did not seek to include isolates of the less clinically important ancient (atypical) clade. A previous study identified a set of 81 SNPs that characterized all modern Beijing family strains [35]. That study identified virulence factor SNPs in the mce and vapBC gene families, but our study indicated that those SNPs are not specific to the Beijing family (data not shown). Another study that sought to characterize the modern/typical Beijing lineage identified 51 SNPs that defined the lineage and were not found in a search of 29 non-Beijing strains [36]. Notably, their set of SNPs found regulatory network mutations to be over-represented. As our study only searched for mutations in established virulence factor genes, mutations in regulatory genes that might be responsible for major differences in virulence would not have been captured by this study unless those regulatory genes had already been characterized as virulence factors.
Lineage-specific Manila family virulence factor SNPs
The Manila family exhibited a similar number of lineage-specific virulence factor mutations as the Beijing family. However, we also identified a group of isolates (EAI5 isolate 5 and a cluster with no SpolDB4 match composed of isolates 71–73) that showed virulence factor SNPs matching the Manila family at some loci, but matching an outgroup at others. Those SNPs that were found only in the Manila family and in this small, ancient clade were presented as Manila family characteristic SNPs rather than Manila family specific SNPs. While not 100% specific to the family, these mutations may still be important for understanding the virulence or transmission characteristics of the Manila family.
Among the notable virulence factor genes hosting the Manila family’s lineage specific SNPs, one mutation was found in mce1B. One study showed that when the mce1 operon was knocked out, the resulting infection was unable to enter a state of persistent infection in mouse lungs, instead exhibiting hypervirulence and killing the mice more rapidly than wild-type Mtb. From ex vivo infection of murine macrophages, this was proposed to have resulted from the mutant failing to stimulate a Th-1 mediated immune response that would have induced protective granuloma formation [37]. One Manila family-specific mutation was found five bases upstream of the zinc metalloprotease zmp1 start codon, which might place it within the ribosome binding site (RBS). Mycobacterial ribosome binding sites are typically G and A-rich regions of six to eight base pairs in length, located four to seven base pairs upstream of the start codon [38]. Zmp1 is further of interest as it contains a second Manila family specific mutation, and zmp1’s deletion has been shown to result in hypervirulence in a C57BL/6 mouse model [39]. One mutation was found in the gene encoding PknG, which is secreted into the macrophage cytosol upon mycobacterial infection, where it is required for inhibition of phago-lysosomal fusion and mediates persistence under stressful conditions [40, 41]. Another mutation was found in pknD, which is required for invasion of brain endothelia, but not lung epithelia or macrophages [42]. While the Beijing family displayed a family-specific mutation in pks7, the Manila family displayed a family-specific mutation nine bases upstream of the gene, possibly affecting the promotor or ribosome binding site. Finally, the Manila family exhibits a mutation in fadD26, whose disruption results in impaired synthesis of phthiocerol dimycocerosates and attenuation in a mouse model [43]. Interestingly, one potential Mtb vaccine that has now completed a phase I clinical trial, MTBVAC, was produced from a double deletion mutant that removed phoP and fadD26 [44].
Manila family characteristic mutations were found in several additional genes. Nucleoside diphosphate kinase NdkA inactivates GTPase Rac1 in macrophages (which reduces production of reactive oxygen species), and knock down of ndkA resulted in increased susceptibility to intracellular killing and reduced persistence in the lungs of infected mice [45]. Another mutation was also observed for fadD26 (in addition to the family-specific mutation), suggesting that FadD26 may be important in directing the virulence of the Manila family. VirS is a transcriptional regulator controlling the mymA operon, which is induced by infection in macrophages, and may be involved in remodeling Mtb’s cellular envelope under acidic, intracellular conditions [46]. Finally, a mutation was found in mmpL8, disruption of which resulted in attenuation presenting as significantly increased time to death in a mouse aerosol infection model, corresponding to less efficient suppression of cytokines directing a Th1-type immune response [47].
SNPs in genes of interest were likewise split between lineage-specific and lineage-characteristic for the Manila family. The most notable lineage-specific gene was transcriptional regulator whiB5, which influences the expression of 58 genes, and which our study revealed is truncated due to a premature stop codon in the Manila family [48]. Both chronic and progressive infections have been found to be attenuated in a mouse model when whiB5 was deleted. The mutant was also shown to be unable to resume growth after reactivation from chronic infection. However, as previously noted, human infections with Manila family strains do not appear to display any deficiency in reactivation [49]. Furthermore, two mutations were observed upstream of whiB5’s start codon (-27 and -31). These mutations may be located in the -35 promoter sequence, but that is difficult to determine as mycobacteria have little homology in their -35 promoter sequences [38]. Although studies have shown that the -35 region is not essential for promoter function in mycobacteria, manipulation of the region can vary the wild-type promotor activity from as little as 6% to as much as 200% of its original activity [38]. Regardless, it is interesting that while the whiB5–31 mutation was observed only in Manila family isolates, a whiB5–27 mutation was observed in the Manila family, both EAI5 clades, and the ancient three-isolate epidemiological cluster with no SpolDB4 match. Another gene of interest is the oxyR pseudogene. oxyR nucleotide 37 T has been used as a marker for the Manila family and EAI5 lineage [50]. However, this study revealed that oxyR 37 T is not fully exclusive to the Manila family, as the newly identified ancient clade with an EIA5 isolate and a no SpolDB4 match cluster also exhibited the mutation. Finally, a Manila family characteristic mutation was observed in mycobacterial persistence regulator MRPA, which of Mtb’s ~214 transcriptional regulators, was one of seven shown to be upregulated during nonreplicating persistent stage stage 2 (NRP2) [51]. MRPA had been previously shown to be required for entrance into and maintenance of persistent infection [52].
Relationship of Manila-like isolates to the Manila family
Full genome sequencing provides insight here into the connection of isolates with putative “Manila-like” spoligotypes to the Manila family. A phylogenetic tree determined from those full genome SNP sets has further established their relationships (Fig 1).
In general, the number of spoligotyping/CRISPR spacers lost by an isolate relative to the Manila family did not serve as a predictor of the number of total SNP differences between that isolate and reference Manila family isolate 96121 (Table 7). Multiple examples demonstrating this are presented below. Manila family isolate 89, with its three missing spacers relative to the Manila family’s prototypical pattern, displayed the same number of SNPs from 96121 as EAI5 isolate 33 (which lost one spacer and gained three). Furthermore, both of those isolates had fewer SNPs versus 96121 than isolate 2 (chosen to represent a large Manila family epidemiological cluster). The epidemiological cluster represented by isolates 21 and 41 was missing six spacers, but did not display many more SNPs versus 96121. However, other than for EAI5, gain of spacers relative to the Manila family appeared to be a better indication of distance from the Manila family than loss of spacers. EAI5 isolate 5 and a three-isolate no SpolDB4 match epidemiological cluster (isolates 71–73) occupy a branch together, with both gaining the same three spacers, but the one spacer lost by isolate 5 and the two spacers lost by the 71–73 cluster do not overlap. Other isolates whose spoligotypes had no match in SpolDB4 differed from Manila by the largest numbers of SNPs, similar to the Beijing family outgroup isolates. Of those no-match isolates, isolate 8 was selected due to having a large number of spacers gained versus 96121, and it clustered closest to LAM9 isolate 1, while isolate 42, selected to represent a large number of spacers lost, clustered closest to the Beijing family isolates (despite the apparent difference in spoligotypes).
The numbers of SNPs between individual isolates and our Manila family reference genome further clarify the relationships of isolates displaying various spoligotyping patterns to the Manila family. Isolates 69 and 70, selected from a prior study (which used deletion-based analysis) in order to maximize our present study’s diversity within the Manila family, expectedly clustered within the other Manila family isolates, and displayed the fewest SNPs versus 96121 (210 and 193 SNPs, respectively). Manila family isolate 89, selected due to losing three of the Manila family’s characteristic spoligotyping spacers, likewise clustered with the rest of the Manila family, while Manila family isolate 90 (which lost only a single spacer) was more distinct, and clustered closer to isolate 84 (which lost two spacers and had no SpolDB4 match). Another un-matched, but seemingly Manila-like spoligotype displayed by epidemiologically clustered isolates 21 and 44 (missing seven spacers) clustered within the Manila family. An additional un-matched, but seemingly Manila-like, epidemiological cluster of isolates 30 and 37 (missing seven spacers) clustered furthest from the Manila family, yet still on a distinct branch from the Manila family’s known outgroups. Despite this, it was the only “Manila-like” group not to match the Manila family in all of the virulence factor mutations that characterized the family in this study, differing in a SNP at transmembrane serine/threonine-protein kinase D pknD and a SNP 9 bases upstream of Rv1661 polyketide synthase pks7, possibly in the promotor. Furthermore, isolates from this cluster were also observed to exhibit pigmentation when grown on Löwenstein–Jensen medium.
The EAI5 spoligotype revealed interesting results. We have previously identified in other EAI5 isolates that this spoligotype covers both Principle Genetic Groups 1 and 2 [53]. Thus, this EAI5 spoligotype spans evolutionarily distinct clades, possibly as the result of convergent evolution of CRISPR regions due to phage pressure. Both EAI5 isolates in this study were found to be ancient, despite both phylogenetics and virulence SNPs showing them to be quite distinct. While EAI5 isolate 33 clustered with the Manila family, EAI5 isolate 5 clustered with a three-isolate epidemiological cluster with no SpolDB4 match (isolates 71–73). This sub-clade of ancient isolates was roughly split between matching the Manila family and matching outgroup H37Rv at the Manila family’s lineage specific and characteristic SNP loci (10 to 13). Some notable virulence factor differences for those isolates include losing the Manila family’s truncated transcriptional regulator WhiB5 and one of its two upstream mutations, SNPs in MCE-family protein Mce1B, zinc metalloprotease Zmp1 and its upstream mutation, and one of the two fatty-acid-AMP ligase FadD26 mutations, among others (Table 6). These differences suggest that this ancient sub-clade displays virulence characteristics different from the Manila family.
Conclusions
This study identified distinct sets of non-silent virulence factor SNPs that were specific to the Beijing and Manila families of Mtb. Further study of these mutations could lead to greater understanding of differences in virulence in these two families. Future work investigating the effects of these SNPs in animal or macrophage infection models, which is beyond the scope of this present study, will be required to fully determine their phenotypes. However, future studies should not be limited to investigating only single SNPs. Mtb lacks singular genomic features that account for host preference, and epistatic interactions and coordination of transcriptional regulation must be considered together to account for Mtb’s transmission and virulence [5]. Thus, investigating only single SNPs from a specific family alone (in the absence of the family’s other mutations) may be insufficient to produce a phenotype with altered virulence. Additionally, this study utilized only a small (82-isolate) set of Hawaii-focused Mtb isolates. Future work will further establish the lineage-specificity of these mutations by significantly expanding the number of genomes investigated. Although repositories such as the National Center for Biotechnology Information (NCBI) GenBank already hold a large number of Mtb genomes, the lack of paired spoligotyping data attached to those genomes limits their use in this study. A tool for in silico spoligotyping of Mtb genomes, SpolPred, enables identification of spoligotypes from FASTQ sequencing read files, allowing data from repositories such as the NCBI Sequence Read Archive (SRA) to be investigated [54]. However, while Beijing family sequencing data is abundant, Manila family sequencing data currently remains uncommon.
Data Availability
All FASTQ Illumina read files from this project are available from the NCBI SRA repository under BioProject PRJNA480117: http://www.ncbi.nlm.nih.gov/bioproject/480117.
Funding Statement
This study was funded in part by the University of Hawaii Office of the Vice Chancellor for Research. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.World Health Organization. Global Tuberculosis Report 2017. 2018. Available from: http://www.who.int/entity/tb/publications/global_report/MainText_13Nov2017.pdf?ua=1
- 2.Douglas JT, Qian L, Montoya J, Musser J, Van Embden J, Van Soolingen D, et al. Characterization of the Manila Family of Mycobacterium tuberculosis. J Clin Microbiol. 2003;41: 2723–2726. 10.1128/JCM.41.6.2723-2726.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van Soolingen D, Qian L, de Haas PE, Douglas JT, Traore H, Portaels F, et al. Predominance of a single genotype of Mycobacterium tuberculosis in countries of east Asia. J Clin Microbiol. 1995;33: 3234–3238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Comas I, Homolka S, Niemann S, Gagneux S. Genotyping of Genetically Monomorphic Bacteria: DNA Sequencing in Mycobacterium tuberculosis Highlights the Limitations of Current Methodologies. PLoS One. 2009;4–11: e7815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gagneux S. Ecology and evolution of Mycobacterium tuberculosis. 2018. Nat Rev Microbiol. 2018;16:202–213. 10.1038/nrmicro.2018.8 [DOI] [PubMed] [Google Scholar]
- 6.Sharma A, Hill A, Kurbatova E, van der Walt M, Kvasnovsky C, Tupasi TE, et al. Estimating the future burden of multidrug-resistant and extensively drug-resistant tuberculosis in India, the Philippines, Russia, and South Africa: a mathematical modelling study. Lancet Infect Dis. 2017;S1473-3099(17): 30247–30255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou Y, van den Hof S, Wang S, Pang Y, Zhao B, Xia H, et al. Association between genotype and drug resistance profiles of Mycobacterium tuberculosis strains circulating in China in a national drug resistance survey. PLoS ONE. 12(3): e0174197 10.1371/journal.pone.0174197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Steenwinkel J, ten Kate M, de Knegt G, Kremer K, Aarnoutse R, Boeree M, et al. Drug Susceptibility of Mycobacterium tuberculosis Beijing Genotype and Association with MDR TB. Emerg Infect Dis. 2012;18(4): 660–663. 10.3201/eid1804.110912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bamrah S, Desmond E, Ghosh S, France AM, Kammerer JS, Cowan LS, et al. Molecular Epidemiology of Mycobacterium tuberculosis in the United States–Affiliated Pacific Islands. Asia Pac J Public Health. 2014;26(1): 77–84. 10.1177/1010539512469249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koster K, Qian L, Flores C, Desmond E, Brostrom R, Foster J, et al. Cluster Identification and Analysis of Clinical Mycobacterium tuberculosis Isolates in the State of Hawaii. 2015;Abstr 115th Gen Meet Am Soc Microbiol. American Society for Microbiology, Washington, DC.
- 11.Qian L, Yamamoto S, Zheng X, Douglas JT. Movement and Analysis of Spoligotypes of M. tuberculosis found in Hawaii. 2001;Abstr 101th Gen Meet Am Soc Microbiol. American Society for Microbiology, Washington, DC.
- 12.Schmit KM, Wansaula Z, Pratt R, Price SF, Langer AJ. Tuberculosis—United States, 2016. MMWR Morb Mortal Wkly Rep. 2017;66:289–294. 10.15585/mmwr.mm6611a2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brudey K, Driscoll JR, Rigouts L, Prodinger WM, Gori A, Al-Hajoj SA, et al. Mycobacterium tuberculosis complex genetic diversity: mining the fourth international spoligotyping database (SpolDB4) for classification, population genetics and epidemiology. BMC Microbiol. 2006;6(23). 10.1186/1471-2180-6-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frink S, Qian L, Yu S, Cruz L, Desmond E, Douglas JT. Rapid Deletion-Based Subtyping System for the Manila Family of Mycobacterium tuberculosis. J Clin Microbiol. 2011;49(5): 1951–1955. 10.1128/JCM.01338-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sahl JW, Lemmer D, Travis J, Schupp JM, Gillece JD, Aziz M, et al. NASP: an accurate, rapid method for the identification of SNPs in WGS datasets that supports flexible input and output formats. Microbial Genomics. 2016;2: e000074 10.1099/mgen.0.000074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Langmead B, Salzberg S. Fast gapped-read alignment with Bowtie 2. Nature Methods. 2012;9:357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20: 1297–303. 10.1101/gr.107524.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Forrellad MA, Klepp LI, Gioffré A, Sabio y García J, Morbidoni HR, de la Paz Santangelo M, et al. Virulence factors of the Mycobacterium tuberculosis complex. Virulence. 2013;4(1): 3–66. 10.4161/viru.22329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.R Development Core Team (2008). R: A language and environment for statistical computing R Foundation for Statistical Computing, Vienna, Austria: ISBN 3-900051-07-0, URL http://www.R-project.org. [Google Scholar]
- 20.Kumar S, Stecher G, Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33: 1870–1874. 10.1093/molbev/msw054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Darriba D, Taboada GL, Doallo R, Posada D. jModelTest 2: more models, new heuristics and parallel computing. Nature Methods. 2012;9(8): 772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brosch R, Gordon SV, Marmiesse M, Brodin P, Buchrieser C, Eiglmeier K, et al. A new evolutionary scenario for the Mycobacterium tuberculosis complex. Proc Natl Acad Sci U S A. 2002;99(6): 3684–3689. 10.1073/pnas.052548299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lew JM, Kapopoulou A, Jones LM, Cole ST. TubercuList—10 years after. Tuberculosis (Edinb). 2011;91(1):1–7. 10.1016/j.tube.2010.09.008 [DOI] [PubMed] [Google Scholar]
- 24.Gaudion A, Dawson L, Davis E, Smollett K. Characterisation of the Mycobacterium tuberculosis alternative sigma factor SigG: Its operon and regulon. Tuberculosis (Edinb). 2013;93(5): 482–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee JH, Geiman DE, Bishai WR. Role of stress response sigma factor SigG in Mycobacterium tuberculosis. J Bacteriol. 2008;190: 1128–1133. 10.1128/JB.00511-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rousseau C, Sirakova TD, Dubey VS, Bordat Y, Kolattukudy PE, Gicquel B, et al. Virulence attenuation of two Mas-like polyketide synthase mutants of Mycobacterium tuberculosis. Microbiology. 2003;149: 1837–1847. 10.1099/mic.0.26278-0 [DOI] [PubMed] [Google Scholar]
- 27.Domenech P, Reed MB. Rapid and spontaneous loss of phthiocerol dimycocerosate (PDIM) from Mycobacterium tuberculosis grown in vitro: implications for virulence studies. Microbiology. 2009;155: 3532–3543. 10.1099/mic.0.029199-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reed MB, Domenech P, Manca C, Su H, Barczak AK, Kreiswirth BN, et al. A glycolipid of hypervirulent tuberculosis strains that inhibits the innate immune response. Nature. 2004;431: 84–87. 10.1038/nature02837 [DOI] [PubMed] [Google Scholar]
- 29.Tuberculist. [cited 14 Feb 2018]. Available from: http://tuberculist.epfl.ch/.
- 30.Chambers HF, Moreau F, Yajko D, Miick C, Wagner C, Hackbarth C, et al. Can penicillins and other beta-lactam antibiotics be used to treat tuberculosis?. Antimicrob Agents Chemother. 1995;39(12): 2620–2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Keener A. Oldie but goodie: Repurposing penicillin for tuberculosis. Nat Med. 2014;20(9): 976–978. 10.1038/nm0914-976 [DOI] [PubMed] [Google Scholar]
- 32.Ribeiro SCM, Gomes LL, Amaral EP, Andrade MRM, Almeida FM, Rezende AL, et al. Mycobacterium tuberculosis Strains of the Modern Sublineage of the Beijing Family Are More Likely To Display Increased Virulence than Strains of the Ancient Sublineage. J Clin Microbiol. 2014;52(7): 2615–2624. 10.1128/JCM.00498-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mestre O, Luo T, Dos Vultos T, Kremer K, Murray A, Namouchi A, et al. Phylogeny of Mycobacterium tuberculosis Beijing Strains Constructed from Polymorphisms in Genes Involved in DNA Replication, Recombination and Repair. PLoS ONE. 6(1): e16020 10.1371/journal.pone.0016020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu Q, Luo T, Dong X, Sun G, Liu Z, Gan M, et al. Genetic features of Mycobacterium tuberculosis modern Beijing sublineage. Emerg Microbes Infect. 2016;5(14). 10.1038/emi.2016.14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Merker M, Blin C, Mona S, Duforet-Frebourg N, Lecher S, Willery E, et al. Evolutionary history and global spread of the Mycobacterium tuberculosis Beijing lineage. Nat Genet. 2015;47(3):242–249. 10.1038/ng.3195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schurch AC, Kremer K, Warren RM, Hung NV, Zhao Y, Wan K, et al. Mutations in the regulatory network underlie the recent clonal expansion of a dominant subclone of the Mycobacterium tuberculosis Beijing genotype. Infect Genet Evol. 2011;11: 587–597. 10.1016/j.meegid.2011.01.009 [DOI] [PubMed] [Google Scholar]
- 37.Shimono N, Morici L, Casali N, Cantrell S, Sidders B, Ehrt S, et al. Hypervirulent mutant of Mycobacterium tuberculosis resulting from disruption of the mce1 operon. Proc Natl Acad Sci U S A. 2003;100(26): 15918–15923. 10.1073/pnas.2433882100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Newton-Foot M, van Pittius NCG. The complex architecture of mycobacterial promoters. Tuberculosis. 2013;93(1): 60–74. 10.1016/j.tube.2012.08.003 [DOI] [PubMed] [Google Scholar]
- 39.Muttucumaru DG, Smith DA, McMinn EJ, Reese V, Coler RN, Parish T. Mycobacterium tuberculosis Rv0198c, a putative matrix metalloprotease is involved in pathogenicity. Tuberculosis (Edinb). 2011;91: 111–116. [DOI] [PubMed] [Google Scholar]
- 40.Scherr N, Muller P, Perisa D, Combaluzier B, Jeno P, Pieters J. Survival of Pathogenic Mycobacteria in Macrophages Is Mediated through Autophosphorylation of Protein Kinase G. J Bacteriol. 2009;191(14): 4546–4554. 10.1128/JB.00245-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Khan MZ, Bhaskar A, Upadhyay S, Kumari P, Rajmani RS, Jain P, et al. Protein Kinase G confers survival advantage to Mycobacterium tuberculosis during latency like conditions. J Biol Chem. 2017;292: 16093–16108. 10.1074/jbc.M117.797563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Be NA, Bishai WR, Jain SK. Role of Mycobacterium tuberculosis pknD in the pathogenesis of central nervous system tuberculosis. BMC Microbiol. 2012;12(7): 10.1186/1471-2180-12-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Infante E, Aguilar LD, Gicquel B, Pando RH. Immunogenicity and protective efficacy of the Mycobacterium tuberculosis fadD26 mutant. Clin Exp Immunol. 2005;141(1): 21–28. 10.1111/j.1365-2249.2005.02832.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kaufmanna SHE, Weinera J, von Reyn CF. Novel approaches to tuberculosis vaccine development. Int J Infect Dis. 2017;56: 263–267. 10.1016/j.ijid.2016.10.018 [DOI] [PubMed] [Google Scholar]
- 45.Sun J, Singh V, Lau A, Stokes RW, Obregón-Henao A, Orme IM, et al. Mycobacterium tuberculosis Nucleoside Diphosphate Kinase Inactivates Small GTPases Leading to Evasion of Innate Immunity. PLoS Pathog. 9(7): e1003499 10.1371/journal.ppat.1003499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Singh R, Singh A, Tyagi AK. Deciphering the genes involved in pathogenesis of Mycobacterium tuberculosis. Tuberculosis (Edinb). 2005;85(5); 325–335. [DOI] [PubMed] [Google Scholar]
- 47.Domenech P, Reed MB, Dowd CS, Manca C, Kaplan G, Barry CE. The Role of MmpL8 in Sulfatide Biogenesis and Virulence of Mycobacterium tuberculosis. J Biol Chem. 2004;279(20): 21257–21265. 10.1074/jbc.M400324200 [DOI] [PubMed] [Google Scholar]
- 48.Casonato S, Cervantes Sánchez A, Haruki H, Rengifo González M, Provvedi R, Dainese E, et al. WhiB5, a Transcriptional Regulator That Contributes to Mycobacterium tuberculosis Virulence and Reactivation. Infect Immun. 80(9):3132–3144. 10.1128/IAI.06328-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wan X, Koster K, Qian L, Desmond E, Brostrom R, Hou S, et al. (2017) Genomic analyses of the ancestral Manila family of Mycobacterium tuberculosis. PLoS ONE. 12(4): e0175330 10.1371/journal.pone.0175330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Douglas JT, Qian L, Montoya JC, Musser JM, Van Embden JDA, Van Soolingen D, et al. Characterization of the Manila Family of Mycobacterium tuberculosis. J Clin Microbiol. 2003;41(6): 2723–2726. 10.1128/JCM.41.6.2723-2726.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gopinath V, Raghunandanan S, Lawrence Gomez R, Jose L, Surendran A, Ramachandran R, et al. Profiling the Proteome of Mycobacterium tuberculosis during Dormancy and Reactivation. Mol Cell Proteomics. 2015;14(8): 2160–2176. 10.1074/mcp.M115.051151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zahrt TC, Deretic V. Mycobacterium tuberculosis signal transduction system required for persistent infections. Proc Natl Acad Sci U S A. 2001;98(22):12706–12711. 10.1073/pnas.221272198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Koster K, Qian L, Hoshide M, Crudu V, Warren R, Victor T, et al. Analysis of Evolutionary Markers among Mycobacterium tuberculosis Spoligotyping Groups. 2012;Abstr 112th Gen Meet Am Soc Microbiol. American Society for Microbiology, Washington, DC.
- 54.Coll F, Mallard K, Preston MD, Bentley S, Parkhill J, McNerney R, et al. SpolPred: rapid and accurate prediction of Mycobacterium tuberculosis spoligotypes from short genomic sequences. Bioinformatics. 2012;28(22):2991–3. 10.1093/bioinformatics/bts544 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All FASTQ Illumina read files from this project are available from the NCBI SRA repository under BioProject PRJNA480117: http://www.ncbi.nlm.nih.gov/bioproject/480117.