Abstract
The death of several black bears at the black bear breeding base in Yunnan Pingbian Daweishan is a matter of concern. Multiple black bears exhibited decreased appetite or unusual waste, and some were soporific or suffered from vomiting and anhelation. In order to ascertain the cause of death, 16S rDNA gene sequencing and phylogenetic analysis was performed on bacteria isolated from tissue samples obtained from dead bears. The biochemical characteristics of the isolated bacteria were subsequently analyzed using different biochemical test systems. The bacteria can decompose glucose, but it cannot produce gas. The fermentation study of sucrose, lactose, trehalose, glycerol and mannitol yielded positive results; while it was unable to decompose urea or ODC (ornithine decarboxylase). Basic Local Alignment Search Tool (BLAST) analysis of a ~1500-bp DNA product amplified from the 16S rDNA of the bacterial isolate revealed that Enterococcus faecium from black bears is highly similar to other E. faecium isolates in the National Center for Biotechnology Information (NCBI) database, and the highest sequence similarity (99%) was with the reference strain. In addition, mice infected with the E. faecium isolate succumbed to severe damage to the lungs, liver, spleen, myocardium, and kidney tissues. In summary, the isolated E. faecium from dead black bears induced pathological changes in mice.
Key Words: Black bear, Characterization, Enterococcus faecium, Isolation
Introduction
Enterococci are part of the natural microbiota present in gastrointestinal tracts of animals, insects, and humans; however, they are also notorious hospital-acquired pathogens (Garcia-Migura et al., 2007 ▶; Leavis et al., 2007 ▶; Palmer et al., 2010 ▶). Enterococci have traditionally been used as indicators of faecal contamination, because they are abundant in the faeces of warm-blooded animals and are capable of surviving outside the body for prolonged periods of time (Lleò et al., 2005 ▶). Enterococcus faecium and Enterococcus faecalis are major opportunistic pathogens and can cause bacteremia, endocarditis, intra-abdominal and urinary tract wounds, as well as other infections (Fisher and Phillips, 2009 ▶). Enterococcus faecium strains have been isolated from various sources such as shellfish (Wilson et al., 2002 ▶), the intestine of fish from integrated fish farming (Petersen et al., 2003 ▶), Italian goat milk (Cocolin et al., 2007 ▶), varieties of cheese (Abriouel et al., 2005 ▶; Abeijón et al., 2006 ▶; Liu et al., 2011 ▶), Tunisian fermented meat (Belgacem et al., 2009 ▶), Italian rye grass (Izquierdo et al., 2008 ▶), Chungkukjang (Yoon et al., 2008 ▶), and chicken (Strompfová et al., 2007 ▶). In 2010, E. faecium was isolated from seafood (Koch et al., 2004 ▶; Sood et al., 2012 ▶).
Despite this, there have been few reports on the pathogenic characteristics of E. faecium. In the present study, we report the first systemic examination of E. faecium isolated from black bears exhibiting disease symptoms. Several black bears at the black bear breeding base in Yunnan Pingbian Daweishan died. Pingbian Daweishan is the provincial natural reserve in Honghe of Yunnan province.
Daweishan is a rich resource for wildlife including black bears, black apes, long hip cats, clouded leopards, sambars, lizards, gallus, golden pheasants and peacock pheasants, all listed as national protected animals. The black bear breeding base is a wildlife protection center which holds about 50 sick and injured black bears. The findings of this study may provide the basis for future studies on the treatment of infections caused by E. faecium.
Materials and Methods
Ethics statement
All sampling was carried out according to the Animal Ethics Procedures and Guidelines of the People’s Republic of China, and the Animal Care and Use Committee of the Centre for Animal Disease Control and Prevention (CADC), Yunnan province. All procedures carried out on animals were permitted by the Animal Care and Use Committee of the CADC, Yunnan province, and the Institutional Animal Care and Use Committee of Yunnan Agricultural University.
Sampling procedure and bacterial isolation
Samples were collected from the liver and spleen tissues of dead black bears, cut into 0.6 cm × 0.6 cm sections, and immersed in sterilized saline solution. The samples were inoculated into nutrient broth and incubated at 37°C for 18 h. After incubation, the bacterial culture was plated onto blood agar, MRS medium (De Man, Rogosa and Sharpe), BBL agar (Bifidobacterium agar medium) and MacConkey agar by streaking. The plates were then incubated at 37°C for 18 h. Single colonies on the blood agar plates were further investigated by smear tests and Gram stains. Culture conditions for the single colony isolates were optimized. Biochemical tests were then carried out in order to characterize the isolated bacteria by evaluating its ability to grow inurea, ODC (ornithine decarboxylase), maltose, sucrose, lactose, trehalose, glucoseandmannitol.
PCR amplification and sequencing of bacterial 16S rDNA
Total DNA was extracted from the bacterial isolate which was cultured on solid MRS medium. A 1-ml suspension of the bacterial isolate in phosphate-buffered saline was transferred to an Eppendorf tube, centrifuged at 10,000 rpm for 5 min, and then suspended in 100 µL of Tris-EDTA buffer. The DNA was then extracted using a TIANamp Bacteria DNA Kit according to the manufacturer’s instructions. The DNA was used directly for polymerase chain reaction (PCR) amplification of the 16S rDNA region. The coding region of 16S rDNA was amplified by PCR using the primers P1 (5´-AGA GTT TGA TCC TGG CTCA-3´) and P2 (5´-AGG AGG TGA TCC AGC CGA C-3´).
PCRs were carried out in a total volume of 25 µL, containing 2.5 µL of ×10 reaction buffer, 2.0 µL dNTPs (2.5 mM of each dNTP), 1.0 µL of each primer (20 pM each), 2.0 µL DNA template, 2.0 µL MgCl2 (25 mM) and 0.25 µL Taq DNA polymerase (5 U/µL). Sterile water was added in order to bring the reaction volume to 25 µL. Amplification was carried out in a Bio-Rad thermocycler with a cycling program consisting of an initial denaturing step at 95°C for 5 min, followed by 30 cycles of 94°C for 45 s, 55°C for 45 s and 72°C for 1.5 min, followed by 72°C for 10 min. The amplified product was then verified by electrophoresis on a 1% agarose gel, which was analyzed using a gel imaging system (Bio-Rad, USA). The PCR product was then sequenced by BGI Company. Analysis of the 16S rDNA sequence was conducted using the conserved domain architecture retrieval tool (Basic Local Alignment Search Tool, BLAST) at the National Center for Biotechnology Information (NCBI) repository. An aphylogenetic tree was constructed using the neighbor-joining (NJ) method and related 16S rDNA sequences obtained from the NCBI database.
Experimental infection and histopathological tissue examination
Infected tissues (spleen, liver, kidney, lung, and myocardium) from dead black bears were fixed in 4% paraformaldehyde and embedded in paraffin for histopathological examinations. To assess their pathogenicity, the bacteria were evaluated using small animal inoculations in a murine model, and bacterial plate counts. Ten BALB/c mice were inoculated intraperitoneally with 0.5 ml of bacteria in a solution containing 108 CFU/ml. All mice were examined for gross lesions on the day of death. After gross examinations, tissues from the spleen, liver, intestine, kidney, lung, and myocardium were removed aseptically and processed using established routines and methods. The samples were then cultured in an MRS medium to isolate bacteria, after which they were fixed in 4% paraformaldehyde and embedded in paraffin for histopathological examinations (Strompfová et al., 2007 ▶).
Results
Necropsy observations
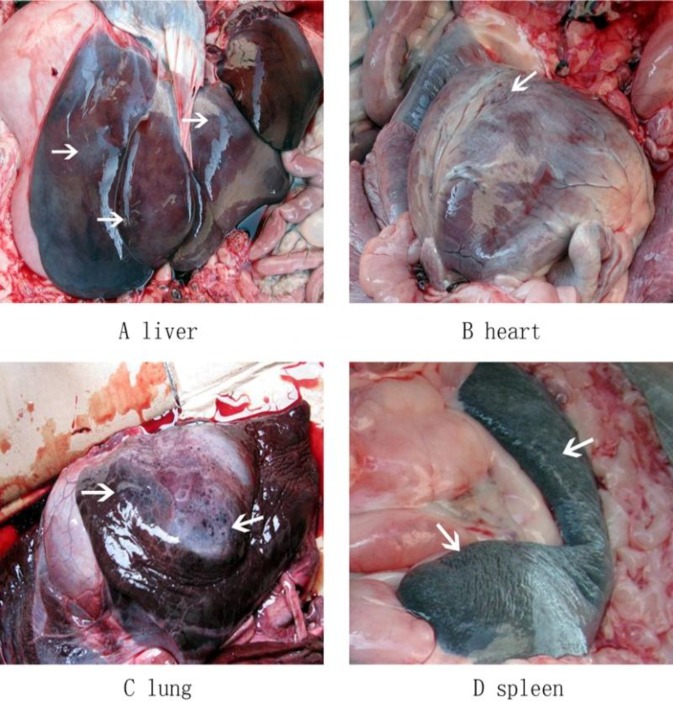
The diseased black bears showed clinical manifestations including loss of appetite and mental depression, and some exhibited vomiting and respiratory difficulties. The disease is characterized by an acute onset, hyperthermia (40.5-41.5°C), visible mucous membrane cyanosis, diarrhea (black loose stool), and convulsions before death. There were about 50 black bears, 28 of which showed similar clinical symptoms, and 4 that died. The morbidity rate was 56%, and the mortality rate was 8%. Necropsy revealed similar lesions and enlarged fragile and brown or yellowish-brown livers in all dead bears (Fig. 1A). Diffuse, off-white or gray-yellow necrosis was visible in the subcapsular and liver parenchyma. Cardiac dilatation was observed; the hearts were filled with crass amentum and some pinpoint hemorrhages were spotted on the cardiac tissue (Fig. 1B). In addition, further examination revealed congestion and edema in the lungs (Fig. 1C) and the parenchyma, and tracheal hyperemiaas well as swelling and off-white necrotic foci on the spleen (Fig. 1D).
Fig. 1.
Gross lesions observed in dead black bears. Livers were fragile and brown/yellowish-brown (A), hearts were filled with crass amentum with some pinpoint hemorrhages spotted on the cardiac tissue (B), lungs had edema and were congested (C), spleens were swellen with an off-white necrotic foci (D)
Enterococcus faecium isolation and biochemical characteristics
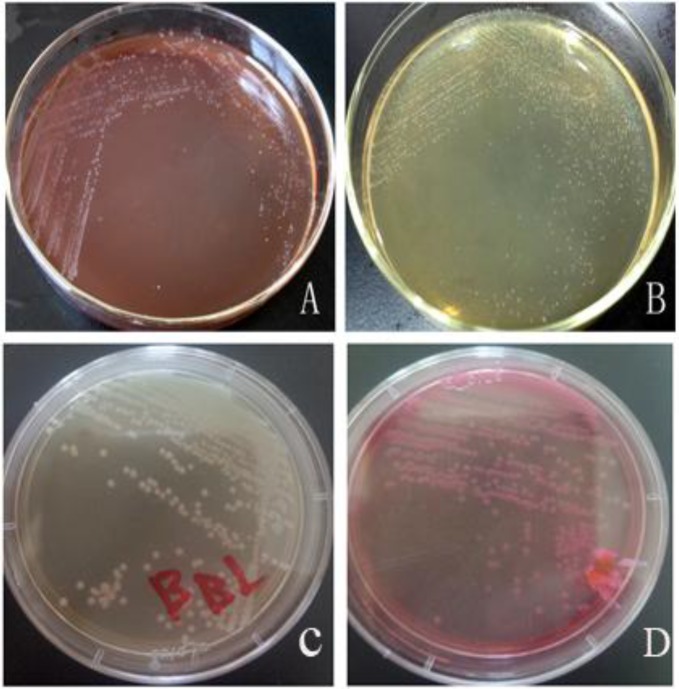
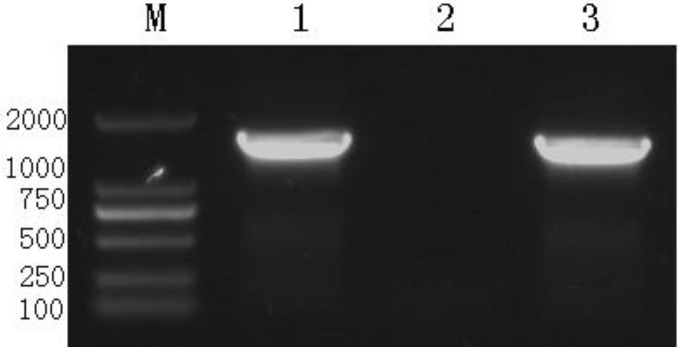
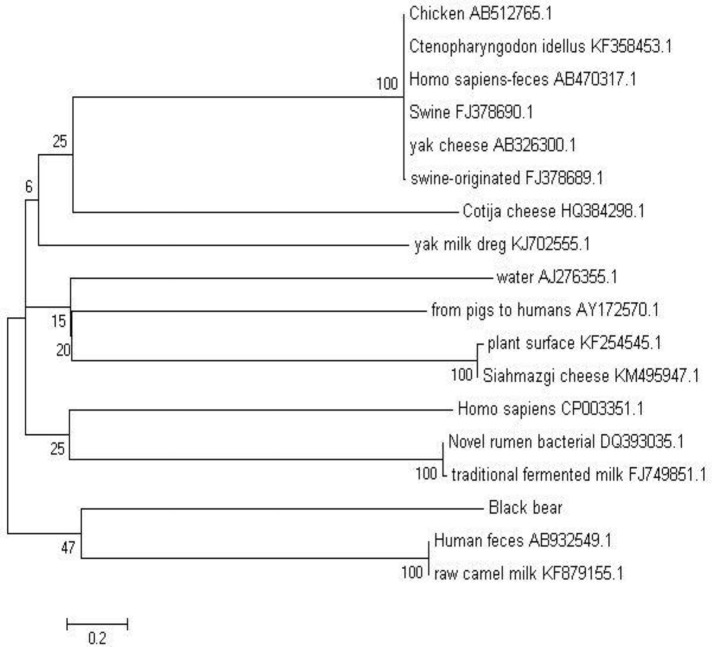
In order to characterize the pathogen, the bacterial isolate was incubated on plates containing various media (blood agar plate, MRS agar plate, BBL agar plate, MacConkey agar plate) after which white colonies were observed on the blood agar plate (Fig. 2A), off-white colonies on the MRS agar plate (Fig. 2B), white colonies on the BBL agar plate (Fig. 2C), and pink colonies on the MacConkey agar plate (Fig. 2D). Gram staining methods indicated that the bacterial isolate was gram-positive. The bacteria could decompose glucose, but could not produce gas. The fermentation study of sucrose, lactose, trehalose, glycerol and mannitol yielded positive results, but urea or ODC were not decomposed, hence negative results were obtained. PCR amplification yielded an amplicon of approximately 1540 bp (Fig. 3) from the bacterial isolate. BLAST analysis revealed that the sequence had maximum similarity (99%) with 17 reference strains. A phylogenetic tree was generated using the NJ algorithm for the 16s rDNA amplicon of the isolate and sequences obtained from the GenBank database. The phylogenetic analysis revealed that the 16S rDNA gene of E. faecium isolated from black bears formed an independent cluster (Fig. 4).
Fig. 2.
Growth of the bacterial isolate on different media with colony morphologies after incubation for 18 h. The bacterial isolate was incubated on plates containing various media; white colonies were observed on the blood agar plate (A), off-white colonies were observed on the MRS agar plate (B), white colonies on the BBL agar plate (C), and pink colonies on the MacConkey agar plate (D)
Fig. 3.
PCR amplification for 16S rDNA identification of E. faecium. M: DNA marker 2000. Lane 1: Amplified product of 16S rDNA of E. faecium from black bears (about 1540 bp), Lane 2: Negative control (physiological saline), and Lane 3: Amplified product of 16S rDNA of E. faecium isolated from infected mice
Fig. 4.
Phylogenetic tree of E. faecium isolated from black bears in relation to different reference strains using MEGA
Histopathologic observations
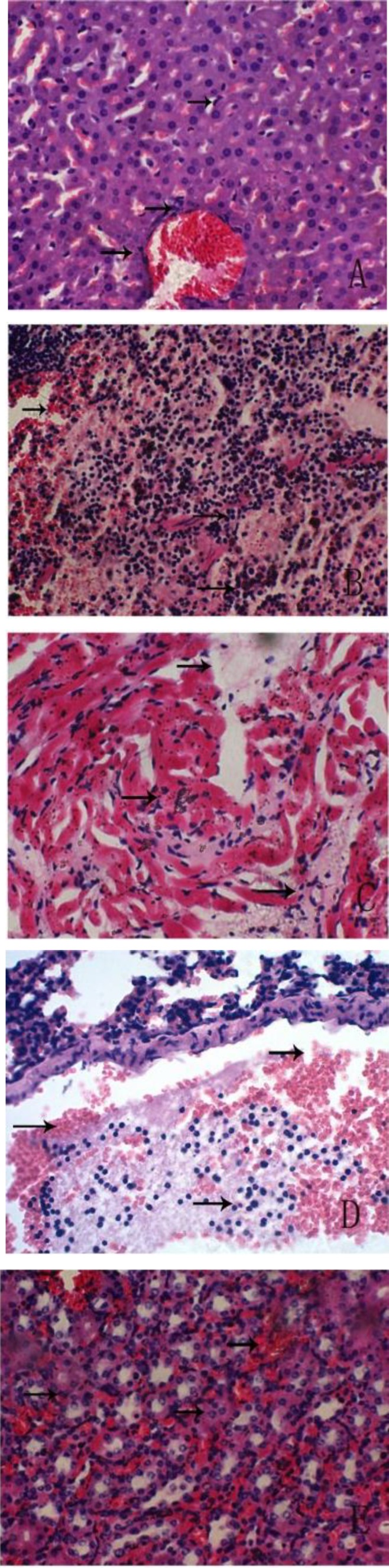
When the bacterial isolates were injected into mice, the infected mice were soporific and exhibited tousled fur and poor appetite. Four mice died at 8 h following infection, and the rest died 48 h after infection. The mice were later dissected and bleeding sites were found on the surface of the livers and lungs. The infected mice exhibited hepatic sinusoidal congestion, obvious degeneration and necrosis of hepatocytes and hepatic cord destruction in the liver (Fig. 5A), serious hemorrhaging in the spleen, necrosis of lymphocytes and splenic infarction (Fig. 5B), fracturing of myocardial fibers, hemorrhaging in the myocardium, and irregular, discrete, random hemosiderin rims in the myocardium (Fig. 5C). In addition, the mice showed obvious alveolar wall congestion, epithelial hyperplasia, interstitial thickening, and inflammatory exudation in the alveolar space of the lung (Fig. 5D), renal interstitial hemorrhaging, granular degeneration and necrosis in the renal tubular epithelia, and tumefaction and disappearance of the lumen (Fig. 5E).
Fig. 5.
Photomicrographs of histopathological lesions of E. faecium in the liver, spleen, myocardium, lung, and kidney of infected mice (A, C, B, D, E, H&E, ×400). (A) Congestion in hepatic sinusoid, obvious degeneration and necrosis of hepatocytes and destruction of the hepatic cord in the liver, (B) serious hemorrhage in spleen, necrosis of lymphocytes, splenic infarction, (C) myocardial fiber fracture and hemorrhage in the myocardium with discrete and random irregular rims of hemosiderin in the myocardium, (D) obvious congestion of the alveolar wall, epithelial hyperplasia, interstitial thickening, inflammatory exudation in the alveolar space in the lung, and (E) renal interstitial hemorrhage, granular degeneration and necrosis in renal tubular epithelia, tumefaction and disappearance of lumen (the positions of the lesion are indicated by arrows)
Discussion
Historically, Enterococci were not thought to be dangerous pathogens. However, these bacteria have subsequently been isolated from severe occurrences of disease and mortality, and have therefore been recognized as an important cause of nosocomial infections (Sood et al., 2012 ▶). Clinical infections caused by the Enterococcus species predominantly include bacteremia, and to a lesser extent urinary tract infections, bacterial endocarditis, and diverticulitis (Fisher et al., 2009 ▶). Enterococcus faecium is a gram-positive bacterium belonging to the family Enterococcaceae, and is an important opportunistic pathogen easily transmitted between diseased and healthy animals. As a result, it is known as a significant etiological agent of both acute and chronic infections (Koch et al., 2004 ▶).
In this study, E. faecium was isolated from black bears, and the morphological and biochemical characteristics of the bacterium were identified. 16S rDNA sequencing analysis demonstrated that black bear E. faecium had maximum sequence similarity with the reference strains for E. faecium (99%), indicating an evolutionarily relationship between them. This was further confirmed by phylogenetic clustering, which revealed that black bear E. faecium clustered independently from other isolates, This finding suggests that the isolate may have undergone gene mutation or deletion; however, further research will be needed to verify this assumption. The black bear isolate was positive for sucrose, lactose, trehalose, glycerol and mannitol, and negative for urea and ODC, which matches the species description of E. faecium in Bergey’s Manual of Systematic Bacteriology.
Histopathological observations of infected mice revealed that the isolated strain severely damaged myocardium, liver, spleen, kidney and lung tissues. There was obvious degeneration and necrosis of hepatocytes and destruction of the hepatic cord in liver tissues, serious hemorrhaging in spleen tissues, granular degeneration in the renal tubular epithelia, hemorrhaging in kidney tissues, and congestion of the alveolar wall in lung tissues. In a previous study, Enterococci were found to be involved in pathogenic infections and were transmitted by contaminated food. Enterococcus faecium is part of the normal gut flora in humans, and usually does not cause any pathological symptoms. However, E. faecium is an opportunistic pathogen, and can cause disease under certain conditions. In the case of black bears, water shortage, underfeeding or sudden changes in feed and temperature, the prevalence of mosquitoes, flies, and rats and other factors increase the animals’ susceptibility to the disease. The bacterial community of the gut microflora may play an essential role in the developmental stages of pathogens. For example, the malaria transmission cycle may be affected by the presence of gram-negative bacteria, which promote the formation of malaria parasites. It should be noted, however, that although specific clinical manifestations of E. faecium in other animals are yet to be verified, the bacteria may be transmitted from food-producing animals to humans through the food supply chain.
Enterococcus faecium isolated from dead black bears were investigated in this study. The biochemical characteristics of the isolated bacteria were analyzed, and pathological biopsies revealed that E. faecium severely damaged hepatocytes, caused lymphocytes necrosis, splenic infarction, myocardial fiber fracture, epithelial hyperplasia, and renal interstitial hemorrhage in infected mouse tissues. Such findings can be used to control this pathogen.
Acknowledgements
The present research was supported by the Natural Science Foundation of Yunnan Agricultural University (2015ZR04), Monitoring Important Diseases of the Wild Animals in Yunnan (K2400033), and The Key Laboratory of Veterinary Public Health in Yunnan Higher Education (A3008366).
Conflict of interest
The authors declare that they have no competing interests, financial or otherwise.
References
- Abeijón, MC , Medina, RB , Katz, MB , González, SN Technological properties of Enterococcus faecium isolated from ewe’s milk and cheese with importance for flavour development. Can. J. Microbiol. 2006;52:237–245. doi: 10.1139/w05-136. [DOI] [PubMed] [Google Scholar]
- Abriouel, H , Lucas, R , Ben, ON , Valdivia, E , Maqueda, M , Martínez-Cañamero, M , Gálvez, A Enterocin AS-48RJ: a variant of enterocin AS-48 chromosomally encoded by Enterococcus faecium RJ16 isolated from food. Syst. Appl. Microbiol. 2005;28:383–397. doi: 10.1016/j.syapm.2005.01.007. [DOI] [PubMed] [Google Scholar]
- Belgacem, ZB , Abriouel, H , Omar, NB , Lucasb, R , Martínez-Canamerob, M , Gálvezb, A , Manaia, M Antimicrobial activity, safety aspects and some technological properties of bacteriocinogenic Enterococcus faecium from artisanal Tunisian fermented meat. Food Contr. 2009;21:462–470. [Google Scholar]
- Cocolin, L , Foschino, R , Comi, G , Grazia, FM Description of the bacteriocins produced by two strains of Enterococcus faecium isolated from Italian goat milk. Food Microbiol. 2007;24:752–758. doi: 10.1016/j.fm.2007.03.001. [DOI] [PubMed] [Google Scholar]
- Fisher, K , Phillips, C The ecology, epidemiology and virulence of Enterococcus. Microbiology. 2009;155:1749–1757. doi: 10.1099/mic.0.026385-0. [DOI] [PubMed] [Google Scholar]
- Garcia-Migura, L , Liebana, E , Jensen, LB Transposon characterization of vancomycin-resistant Enterococcus faecium (VREF) and dissemination of resistance associated with transferable plasmids. J. Antimicrob. Chemother. 2007;60:263–268. doi: 10.1093/jac/dkm186. [DOI] [PubMed] [Google Scholar]
- Izquierdo, E , Bednarczyk, A , Schaeffer, C , Cai, Y , Marchioni, E , Van, DAand Ennahar, S Production of enterocin L50A, L50B, and IT, a new enterocin, by Enterococcus faecium IT62, a strain isolated from Italian ryegrass in Japan. Antimicrob. Agents Chemother. 2008;52:1917–1923. doi: 10.1128/AAC.01409-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch, S , Hufnagel, M , Theilacker, Cand Huebner, J Enterococcal infections: host response, therapeutic, and prophylactic possibilities. 2004;Vaccine. 22:822–830. doi: 10.1016/j.vaccine.2003.11.027. [DOI] [PubMed] [Google Scholar]
- Leavis, HL , Willems, RJ , van, WWJ , Schuren, FH , Caspers, MP , Bonten, MJ Insertion sequence-driven diversification creates a globally dispersed emerging multiresistant subspecies of E-faecium. PLoS Pathog. 2007;3:e7. doi: 10.1371/journal.ppat.0030007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, G , Griffiths, MW , Wu, P , Wang, H , Zhang, X , Li, P Enterococcus faecium LM-2, a multi-bacteriocinogenic strain naturally occurring in “Byaslag”, a traditional cheese of inner Mongolia in China. Food Contr. 2011;22:283–289. [Google Scholar]
- Lleò, MM , Bonato, B , Benedetti, Dand Canepari, P Survival of enterococcal species in aquatic environments. FEMS Microbiol. Ecol. 2005;54:189–196. doi: 10.1016/j.femsec.2005.03.016. [DOI] [PubMed] [Google Scholar]
- Palmer, KL , Kos, VN , Gilmore, MS Horizontal gene transfer and the genomics of enterococcal antibiotic resistance. Curr. Opin. Microbiol. 2010;13:632–639. doi: 10.1016/j.mib.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen, A , Dalsgaard, A Antimicrobial resistance of intestinal Aeromonas spp and Enterococcus spp in fish cultured in integrated broiler-fish farms in Thailand. Aquaculture. 2003;219:71–82. [Google Scholar]
- Sood, S , Malhotra, M , Das, BK andKapil, A Enterococcus in wound infections: virulence and antimicrobial resistance. Acta Microbiol. Immunol. Hung. 2012;59:263–269. doi: 10.1556/AMicr.59.2012.2.11. [DOI] [PubMed] [Google Scholar]
- Strompfová, V , Lauková, A In vitro study on bacteriocin production of enterococci associated with chickens. Anaerobe. 2007;13:228–237. doi: 10.1016/j.anaerobe.2007.07.002. [DOI] [PubMed] [Google Scholar]
- Wilson, IG , McAfee, GG Vancomycin-resistant enterococci in shellfish, unchlorinated waters, and chicken. Int. J. Food Microbiol. 2002;79:143–151. doi: 10.1016/s0168-1605(02)00063-6. [DOI] [PubMed] [Google Scholar]
- Yoon, MY , Kim, YJ , Hwang, HJ Properties and safety aspects of Enterococcus faecium strains isolated from Chungkukjang, a fermented soy product. Food Sci. Technol. 2008;41:925–933. [Google Scholar]