Abstract
Fluorogenic probes, small-molecule sensors that unmask brilliant fluorescence upon exposure to specific stimuli, are powerful tools for chemical biology. Those probes that respond to enzymatic catalysis illuminate the complex dynamics of biological processes at a level of spatiotemporal detail and sensitivity unmatched by other techniques. Here, we review recent advances in enzyme-activated fluorogenic probes for biological imaging. We organize our survey by enzyme classification, with emphasis on fluorophore masking strategies, modes of enzymatic activation, and the breadth of current and future applications. Key challenges such as probe selectivity and spectroscopic requirements are described alongside of therapeutic, diagnostic, and theranostic opportunities.
Graphical Abstract
Fluorogenic probes are latent fluorophores that reveal their signal in response to environmental changes, interactions with analytes, or specific chemical reactions.1 Fluorogenic probes are prepared by chemically modulating the fluorescence of a parent fluorophore, rendering it nonfluorescent until activation by a specific triggering event. Because of their high sensitivity and ability to monitor diverse events selectively, fluorogenic probes are important components in the toolkit of chemical biology.2–4
Enzyme-activated fluorogenic probes, which invoke enzymatic catalysis to trigger the generation of fluorescence, provide a versatile platform for monitoring biological processes in live cells and in vivo. Early enzyme-activated probes were based on xanthene dye scaffolds and detected galactosidases, phosphatases, lipases, and esterases.5–8 Later, rudimentary live-cell imaging was demonstrated with cell-permeable probes,9 leading to modern probe applications including cell-viability assays,10 diagnostic tests,11 and immunoassay technologies such as ELISA.12 Innovations in probe design continue to drive the development of companion techniques and applications.
The two overarching themes for fluorogenic probe design and applications are the spectroscopic properties of the parent fluorophore and the method employed to mask its fluorescence. Key properties of the parent fluorophore include brightness (which is the product of quantum yield and extinction coefficient), wavelengths and shapes of both excitation and emission peaks, pH effects on fluorescence, and resistance to photobleaching. Recent trends in fluorophore scaffolds include the reduction of phototoxicity and autofluorescence background with far-red,13 near-infrared,14 and two-photon excited probes15 and the fine-tuning of spectroscopic properties and brightness.16,17 Whereas parent fluorophore properties determine the post-activation performance of a probe, the fluorescence masking strategy governs its enzymic target and responsiveness. Accordingly, we focus this review on enzyme-catalyzed unmasking strategies that have been used for imaging in live cells and in vivo. We also constrain our survey to the last five years, and abstain from extensive discussion of parent fluorophore chemistry and spectroscopic properties covered elsewhere.3,16–19
PROBE DESIGN
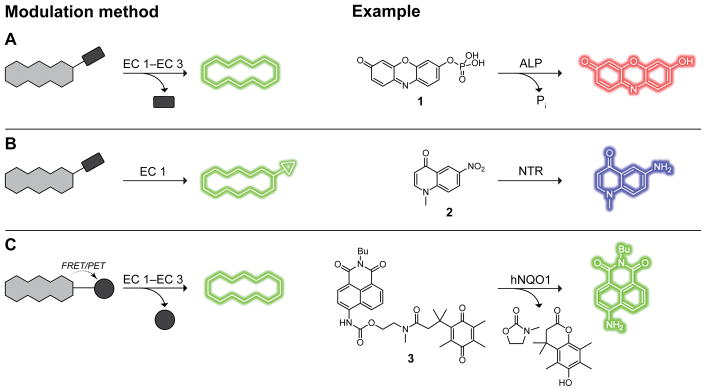
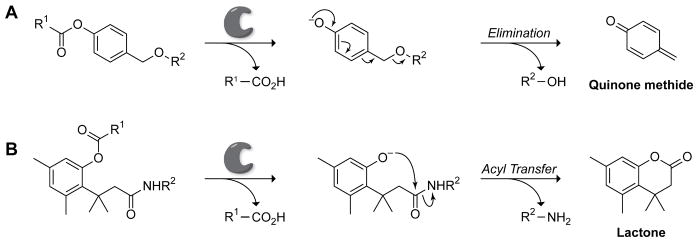
Optimal fluorescence masking methods are chemically stable, respond selectively to the desired event, and completely eliminate fluorescence and absorption at the excitation wavelength via quenching, masking groups, or other chemical modifications. Three main methods to modulate fluorescence in enzyme-activated fluorogenic probes are depicted in Scheme 1. Although masking groups to block constitutive fluorescence (Schemes 1A and 1B) are employed frequently, quenching strategies based on Förster resonance energy transfer (FRET) or photoinduced electron transfer (PET) effects can provide high modularity and, in some cases, ratiometric imaging (Schemes 1A and 1C). Other common themes for enzyme activation include fluorophore precipitation and auto-immolative linkers that can improve probe stability and performance.20 Two most frequently encountered auto-immolative motifs—elimination and acyl transfer—are used to release a fluorophore payload rapidly and spontaneously (Scheme 2).
Scheme 1.
Common fluorescence modulation methods in enzyme-activated fluorogenic probes, with examples for each method. (A) Enzymatic cleavage of blocking groups. (B) Enzymatic conversion of blocking groups into other functional groups. (C) Enzymatic release of FRET or PET quenchers.
Scheme 2.
Representative examples of two predominant mechanisms of activation in auto-immolative linkers: (A) elimination and (B) acyl transfer. Enzymatic catalysis releases a masking group (R1–CO2H). Then, elimination to form a quinone methide (A)27 or acyl transfer driven the action of a trimethyl lock (B)25 rapidly releases a fluorophore (R2–OH or R2–NH2).
Because the intracellular space is a dense heterogeneous mixture of molecules, organelles, and other subcellular structures, the complexity of biological systems is more faithfully represented by live-cell and in vivo models than by fixed-cell or isolated enzyme experiments.21,22 The rich trove of dynamic cellular processes that can be studied in live cells is not accessible to fixed-cell imaging or other disruptive methods. As a result, probe technologies developed using live-cell and in vivo models translate more readily to therapeutic, diagnostic, and clinical applications.23,24 Indeed, fluorogenic probes and masking groups are often inspired by prodrug and inhibitor design strategies.15,25,26
Operating in the crowded cellular environment poses additional challenges to the design of enzyme-activated probes for biological imaging applications. Constraints include optimizing the rates and specificities of enzymatic activation, directing probe uptake and localization, enhancing probe stability, and minimizing toxicity. Probe stability, enzyme specificity, and rate of activation are heavily influenced by the method of fluorescence modulation. In probes that employ a masking or blocking strategy (Schemes 1A and 1B), covalently attached group serve as both the enzyme-responsive moiety and fluorescence masking group. In contrast, probes utilizing quenching techniques require the addition of a separate enzyme-responsive group (Scheme 1C). Conversely, the modularity of masking and enzyme-responsive groups provides a convenient method of adapting fluorophore scaffolds to target different enzymes.
ENZYME TARGETS
To frame our review, we employ the enzyme classification system established by the International Union of Biochemistry and Molecular Biology (IUBMB). This system relies on the type of reaction catalyzed by the enzyme: oxidoreductases (EC 1), transferases (EC 2), hydrolases (EC 3), lyases (EC 4), isomerases (EC 5), and ligases (EC 6).28 The majority of enzyme-activated probes target enzymes in classes EC 1–EC 3. The sparsity of probes targeting EC 4–EC 6 can be attributed, at least partially, to the inaccessibility of fluorescence modulation mechanisms for reactions catalyzed by these enzymes. Probes targeting oxidoreductases in EC 1 employ the widest variety of fluorescence modulation strategies, whereas probes targeting EC 2 and EC 3 enzymes primarily rely on FRET/PET quenching or masking groups (Scheme 1C). Although probes for the three main enzyme classes EC 1–EC 3 have existed for several decades, recent advances in probe chemistry and design have rejuvenated the field with methods to tune spectroscopic properties and enzyme specificity.15,17,26
Below, we describe advances in enzyme-activated probes for biological imaging, organized by enzyme type. Each section contains descriptions of targeted enzyme classes or subclasses, modes of enzyme activation, and applications to live-cell and in vivo imaging. A summary of enzymes, probes, applications, and model cell lines and organisms can be found in Table 1.
Table 1.
Targeted enzymes and imaging applications of enzyme-activated probes.
| EC | Enzymes | Applications | Tested Cell Lines & Tissues | Model Organisms | Probes |
|---|---|---|---|---|---|
| 1.4 | Monoamine oxidase | Parkinson’s disease diagnosis; inhibitor screening | Hep-G2, SH-SY5Y | D. melanogaster, Mice | 4,5 |
|
| |||||
| 1.6 | NAD(P)H:quinone oxidoreductase | Rapid cancer cell screening; tissue resection | A549, HT29, H446, H596, OVCAR-3 | — | 3,6,7 |
|
| |||||
| Nitroreductases | Antibiotic-resistant pathogen identification | — | E. faecium, S. aureus, K. pneumoniae, A. baumannii | 8 | |
| Organelle-specific imaging in hypoxic tumor cells | A549, HEK293, HeLa, HTC116, liver | — | 2,9–14,16 | ||
| Selective mitochondrial imaging and drug delivery | A549, BT474, DU145, WI38 | — | 15 | ||
|
| |||||
| 1.7 | Azoreductase | Orthogonal reporter system | A549, HEK293T, HeLa, NIH-3T3 | — | 17 |
|
| |||||
| 1.8 | Thioredoxin reductase | Thioredoxin reductase-selective imaging in live cells | Hep-G2 | — | 18 |
|
| |||||
| 1.14 | ALKBH3 | Prostate cancer targeted therapy and inhibitor screening | B16, HeLa | Zebrafish | 19 |
| Tyrosinase | Vitiligo and Parkinson’s disease diagnosis | PC3, U2OS | — | 20 | |
|
| |||||
| 2.3 | γ-Glutamyltranspeptidase | Intraoperative fluorescence imaging | SHIN3, SKOV3, colon | — | 21,22 |
| Mycolyltransferases | Study of mycobacterial cell growth and division | — | M. smegmatis, C. glutamicum | 23 | |
|
| |||||
| 2.5 | Glutathione S-transferase | Isoform-selective glutathione transferase imaging | HL60 | — | 24 |
|
| |||||
| 2.7 | Bruton’s tyrosine kinase | Single-step selective kinase imaging | Jurkat, Namalwa | — | 25 |
|
| |||||
| 3.1.1 | Carboxylesterases | Super-resolution study of enzymatic activity | CHO, Drosophila S2, HEK293, HeLa, WBF344, neurons, brain | — | 26 |
| Orthogonal enzyme–probe pair; improved probe properties | HeLa | — | 27,28 | ||
| Theranostic agents | hiPSC neurons, neuro-2A | — | 29 | ||
| Pathogen profiling and detection | — | M. tuberculosis | 30 | ||
| Prodrug activation and multicolor imaging of ER esterases | HeLa, HT1080, SK-N-SH | — | 31 | ||
| Study of endocytic processes in cancer cells | HeLa, HTB125, HTB126 | — | 32 | ||
|
| |||||
| 3.1.2 | Acyl-protein thioesterase (APT) | Study of cellular responses to lipid stress, ratiometric imaging, visualization of mitochondrial APT | A549, HeLa, HEK293T, Hep-G2, MCF-7 | Human colon organoids | 33–35 |
|
| |||||
| 3.1.3 | Alkaline phosphatase (ALP) | Monitoring excreted phosphatases; near-infrared imaging of ALP | HeLa, Hep-G2, U-2OS tissue, Saos-2 tissue | D. melanogaster, Mice | 36,37 |
| Protein tyrosine phosphatase (PTP) | Two-photon visualization of PTP | HEK293, HeLa, Hep-G2 | S. saprophyticus, E. faecalis, A. baumannii, S. aureus | 1,38 | |
|
| |||||
| 3.2 | β-Galactosidase | Tracking of cell senescence, quantification of cytosolic delivery | C6, HeLa, SK-MEL-103 | Mice | 39,40 |
| Glucocerebrosidase | Study of lysosomal storage disorders | fibroblasts | — | 41 | |
| β-Glucuronidase | Deep-tissue tumor imaging | — | Mice | 42 | |
|
| |||||
| 3.4 | β-Alanyl aminopeptidase | Pathogen detection | — | P. aeruginosa, S. marcescens, B. cepacia | 43 |
| Hepsin matriptase | Prostate cancer imaging | DU145, LNCaP, PC3, PrEC | Mice | 44 | |
| Cathepsin B and S | Deep-tissue tumor imaging of lyososomal cathepsins | HEK293, HeLa, KB, MCF-7, MDA-MB-231, NIH-3T3, dendritic cells, U87 | Mice | 45–47 | |
| Caspase-3 | Visualization of pre-apoptotic enzymatic activity | HeLa | — | 48 | |
| ER aminopeptidase | Two-photon ER-targeted deep-tissue redox imaging | HeLa | — | 49 | |
|
| |||||
| 3.5 | β-Lactamases | Detection and labelling of antibiotic-resistant pathogens | — | E. coli, M. tuberculosis | 50,51 |
OXIDOREDUCTASES (EC 1)
Oxidoreductases are a highly diverse class of enzymes that oxidize or reduce a wide variety of substrates, often with cofactors NAD(P)H and flavin mononucleotide (FMN). These enzymes are classified further as oxidases or dehydrogenases depending on the nature of the redox reaction and the final electron acceptor. Oxidases transfer electrons to molecular oxygen as the acceptor, whereas dehydrogenases remove hydrogens from a donor in an NAD+- or FAD-dependent manner. Of the 22 different types of oxidoreductases, most fluorogenic probes target those classes relevant to pathogen detection and tumor imaging, diagnosis, and treatment. Key foci of recent work involve creating tools to investigate cancer biomarkers, tumor-cell hypoxia, and neurodegenerative diseases.
Oxidoreductases that act on the CH–NH2 group of donors (EC 1.4)
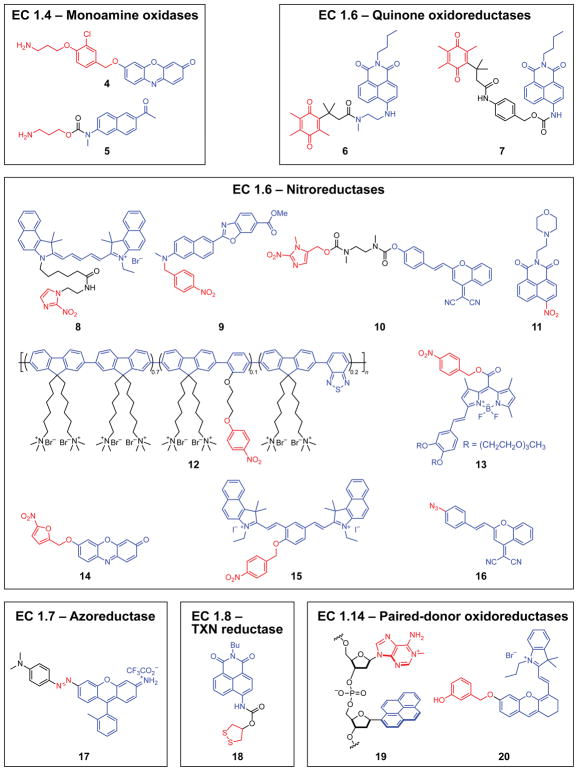
Oxidoreductases of subclass EC 1.4 include enzymes such as monoamine oxidase (MAO) that are associated with the outer mitochondrial membrane and catalyze the oxidative deamination of amines to aldehydes.29 Although both MAO-A and MAO-B isoforms are bound to mitochondria and abundant in the brain, they are differentially localized at a cellular and tissue level and have distinct substrate preferences. Isoforms MAO-A and MAO-B are targets for the treatment of Parkinson’s disease, as both MAO enzymes are important to maintain hormone and neurotransmitter homeostasis.29 Two-photon excited probes 4 and 5 were developed recently to monitor MAO-A and MAO-B activity selectively in deep-tissue imaging.15,26 Probe 4 is based on a resorufin scaffold with a linker connecting the fluorophore and propylamine enzyme-reactive moiety. MAO-A specificity is conferred by the ortho-halogenated linker, which is inspired by the MAO-A inhibitor clorgiline and also serves as an auto-immolative linker that undergoes elimination to release a quinone methide. Probe 5 is based on the acedan scaffold and derives its selectivity for MAO-B from a carbamate linker moiety, inspired by the MAO-B inhibitor pargyline and optimized by in silico molecular docking. Probes 4 and 5 demonstrate the utility of drug- and inhibitor-inspired probe design and enable selective imaging of MAO-A and MAO-B in live cells as well as in drosophila and mouse models of Parkinson’s disease.
Oxidoreductases acting on NADH or NADPH (EC 1.6)
The most frequently encountered enzymes in EC 1.6 are quinone oxidoreductases and nitroreductases (NTR). Although nitroreductases have also been assigned to the EC 1.5 and 1.7 subclass depending on their mechanism of action, most nitroreductases targeted by probes in Table 1 are of the EC 1.6 subclass. The characteristic feature of quinone oxidoreductases and nitroreductases is their dependence on NAD(P)H as an electron source.
Quinone oxidoreductases such as NAD(P)H:quinone oxidoreductase isozyme 1 (hNQO1) are upregulated in many tumors and constitute promising therapeutic targets.30 hNQO1 regulates the degradation of p53, p73α, and p33 tumor suppressors in breast, lung, liver, stomach, and kidney tumors, among others.30 Probes 3, 6, and 7 target hNQO1 via a quinone propionic acid motif, whose redox potential was tuned to quench the naphthalimide fluorophore by PET.31–33 Upon two-electron reduction by hNQO1, the resultant hydroquinone undergoes lactonization spurred by the gem-dimethyl substituents, in a manner akin to the trimethyl lock moiety (cf: EC 3.1.1, probe 32). In probes 3 and 6, the lactonization directly restores fluorescence, whereas in probe 7, an additional auto-immolative rearrangement step is necessary. Nonetheless, the hNQO1-mediated activation of probe 7 was found to be two orders of magnitude faster than that of 3 or 6, due largely to steric factors. Probe 7 also benefits from reduced phototoxicity and background autofluorescence. As a result of the favorable toxicity profiles and enzymic response of the naphthalimide probes, rapid identification of hNQO1-positive cells was achieved with probe 7 in <10 min with positive to negative ratios >500. More recently, a cyanine-based probe with a similar masking group strategy was applied to three-dimensional tumor spheroids and ovarian cancer mouse models.34
Similar to quinone oxidoreductases, two-electron nitroreductases are homodimers that employ NAD(P)H to reduce nitrogen-containing functional groups with aid from FMN as a cofactor. NTR activity is largely absent in most non-cancerous human tissue but highly prevalent in bacteria, with Escherichia coli and Enterobacter cloacae nitroreductases being of particular interest.35,36 Interestingly, E. cloacae NTR was first isolated from bacteria that metabolize TNT and were discovered in a munitions factory.35 Recent work with bacterial nitroreductases has focused on the rapid identification of pathogens, an application that benefits greatly from the low background and rapid response of fluorogenic probes. Probe 8 consists of a Cy5.5 fluorophore linked to an NTR-responsive nitroimidazole quencher.11 As a result of its cationic lipophilic nature, probe 8 readily penetrates both Gram-positive and Gram-negative bacteria and undergoes NTR-responsive regeneration of fluorescence. Probe 8 was utilized to identify and distinguish between key antibacterial-resistant pathogens Enterococcus faceium, Staphylococcus aureus, Klebsiella pneumonia, Acinetobacter baumannii, and Pseudomonas aeruginosa.
Inspired by recent reports of NTR activity in hypoxic tumor cells, probes 2 and 9–14 were created to study hypoxia-dependent nitroreductase activity in cancerous cells.37–43 Probes 2, 9, 10, were highly selective for nitroreductase in the presence of other biologically relevant reducing agents, and were used in the imaging of live A549, HCT116, and HeLa cells. Spatial differences in nitroreductase activity between different cellular compartments can be studied using targeted probes such as 12, a cationic conjugated polymer that accumulates in the nucleus, or 11, which contains a lysosome-targeting morpholine moiety. These probes are selective and highly responsive to nitroreductases, enable targeted imaging, and span several regions of the visible spectrum, enabling further studies of hypoxic tumor masses and other hypoxia-related diseases such as stroke and cardiac ischemia.
In consideration of the putative prokaryotic origin of mitochondria and nitroreductases, probe 15 was created to search for intramitochondrial NTR activity in normoxic cancer cells.24 The cationic and lipophilic nature of probe 15 enabled selective mitochondrial accumulation in live A549 cells and selective NTR response over other background reductases. Probe 15 revealed intramitochondrial nitroreductase activity that was attenuated by bacterial NTR inhibitors. This strategy was then expanded to prepare a prodrug version of Antimycin A for targeted release in mitochondria, which showed enhanced biological activity in WI38, BT474, and DU145 cells.
Unlike the nitroaromatic enzyme-reactive moieties in probes 8–15, probe 16 incorporates an aryl azido group as a mask. Surprisingly, probe 16 was selective for CYP450 enzymes (EC 1.14) rather than the expected cytochrome P450 reductases.44 This orthogonal response could be exploited for imaging and delivery applications. On the other hand, the effective reduction of probe 16 in several different cancer-cell lines suggests instability of the aryl azido groups commonly used for proximity proteomics and photoaffinity labeling.45
Oxidoreductases acting on other nitrogenous compounds as donors (EC 1.7)
As an alternative to nitroreductase-responsive probes, probe 17 is responsive to E. coli azoreductase (EC 1.7.1.6), which reduces the azo group that links the rhodamine scaffold and the dimethylaniline auto-immolative linker.46 Reduction of the azo group not only reverses PET quenching, but also enables elimination of the parent rhodamine green dye. The azoreductase–probe pair can be used as an orthogonal reporter in any cell line amenable to transfection, including HeLa, A549, HEK293T, and NIH3T3. The slight susceptibility of the azo bond to non-specific bioreduction under hypoxic conditions is, however, a potential limitation to the scope of applications.
Enzymes acting on a sulfur group of donors (EC 1.8)
Thioredoxin reductase (TrxR) is a cornerstone of the thioredoxin pathway as the only known reductase of thioredoxin.47 Similar to the nitroreductases described above, TrxR is a homodimer requiring NADPH and FMN cofactors. Probe 18 enables selective and rapid imaging of TrxR activity in live cells.48 The selectivity arises from the five-membered cyclic disulfide attached to a naphthalimide fluorophore via a carbamate linker. Reduction by TxrR generates a thiolate that cleaves the carbamate linker to form a stable cyclic carbonothioate, releasing the naphthalimide fluorophore. Probe 18 resists nonspecific activation by reducing agents and closely related enzymes, as demonstrated by in vitro assays and live-cell imaging in Hep-G2 cells.
Enzymes acting on paired donors, with incorporation or reduction of molecular oxygen (EC 1.14)
Probe 19 enables the direct measurement of the activity of α-ketoglutarate-dependent dioxygenase alkB homolog 3 (ALKBH3), which is also known as prostate cancer antigen-1.49 ALKBH3 demethylates 1-methyladenine in single-stranded DNA or RNA, and elevated levels of ALKBH3 are correlated with increased invasiveness and cell survival in cancer cells.50 Probe 19 incorporates an electron-deficient 1-methyladenine quencher and adjacent pyrene fluorophores into a single-strand of DNA. Enzymatic demethylation by ALKBH3 attenuates 1-methyladenine PET quenching, thus restoring pyrene fluorescence. Enzyme-optimized probe 19 possesses two pyrene nucleosides on the 3′ end of 1-methyladenine with symmetric poly(A) tails flanking both 3′ and 5′ ends for a total length of 10–12 nucleotides. The combination of length and positioning imbues probe 19 with steady-state kinetic parameters that mirror those of native substrates and higher selectivity for ALKBH3 than for nine other homologs. As evidenced by flow cytometry and live-cell imaging with prostate cancer line PC3, probe 19 enables direct measurement of ALKBH3 activity in lieu of laborious immunohistochemistry or in vitro assays of enzymatic activity.
Another target of fluorogenic probes in subclass EC 1.14 is tyrosinase, an enzyme that converts phenols into ortho-quinones and that limits the rate of melanin biosynthesis.51 Although abnormal tyrosinase levels have been implicated in Parkinson’s disease and vitiligo, detection and quantification of these enzymes is often complicated by cross-reactivity of probes with reactive oxygen species (ROS). To circumvent this limitation, near-infrared probe 20 incorporates a tyrosinase-responsive mask having an additional methylene group inserted between the hemi-cyanine fluorophore and aromatic ring, obviating ROS oxidation.51 Elevated tyrosinase levels in murine melanoma B16 cells relative to HeLa cells were demonstrated by live-cell imaging with cell-permeable 20 and corroborated by an ELISA. Imaging zebrafish with probe 20 revealed previously unknown asymmetric distributions of tyrosinase between the yolk sac and tail.
TRANSFERASES (EC 2)
Transferases facilitate the transfer of functional groups from donor to acceptor substrates, and accommodate an extensive array of groups such as sulfuryl, phosphoryl, methyl, amino acyl, and acetyl groups.28 Although transferases are essential to key biochemical pathways, fewer fluorogenic probes targeting transferases have been reported than those targeting the more frequently studied oxidoreductases (EC 1) or hydrolases (EC 3). This dichotomy is in part due to the difficulty of designing selective masking groups that can distinguish transferase activity from that of hydrolases and other enzymes. Indeed, early observations of transferase activity attributed the activity to a combination of hydrolases and other enzymes instead of a single transferase enzyme.52 Recent probes that have achieved selective activation by transferases rely on transfer-group mimetics (probes 21–23),53,54 specific acceptors (probe 24),55 or recognition moieties (probe 25).56
Acyltransferases (EC 2.3)
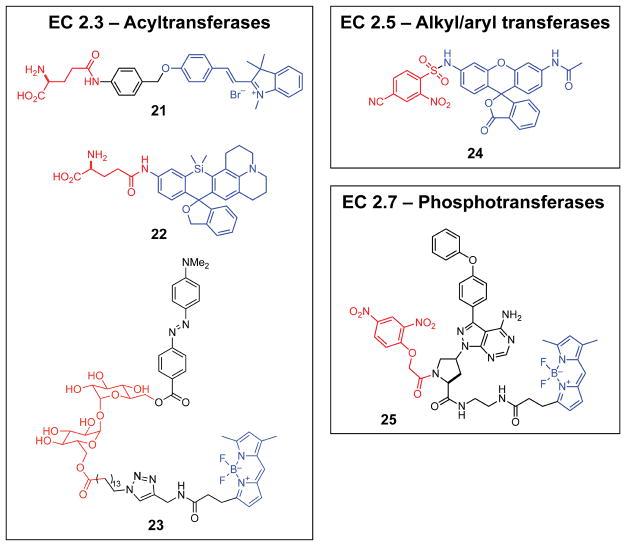
γ-Glutamyltransferase is one of approximately 32 aminoacyltransferases that act on amines to transfer peptide bonds.28 γ-Glutamyltransferase-activated probes are of particular interest for oncologic surgeries because γ-glutamyltransferase is highly expressed in a variety of cancers.57 Fluorogenic probes for such intraoperative applications require rapid activation and low background to be effective. Probes 21 and 22 are activated by the enzyme-catalyzed hydrolysis of a glutamyl amide (which is a side reaction of γ–glutamyltransferase), releasing indocyanine or silarhodamine fluorophores. These probes, which share the same rapid-response masking group and minimize autofluorescence via red or near-infrared emission, have been used to image tumors in mouse intestines.
In mycobacteria, such as Mycobacterium tuberculosis, acyltransferases play vital roles in cell-envelope biosynthesis.58 FRET-quenched fluorogenic probe 2359 has been developed to study the spatiotemporal dynamics of mycolyltransferases that are essential for the construction of mycobacterial cell envelopes. Enzyme–probe specificity in probe 23 was achieved by using a substrate-mimetic linker between a BODIPY fluorophore and DABCYL quencher. Images of Mycobacterium smegmatis obtained with this probe revealed that mycolyltransferase activity is asymmetric. Notably, fluorescence was generated by a hydrolytic side reaction catalyzed by mycolyltransferases, thereby leaving the cell envelope intact (cf: cytotoxic β-lactamase probes). The ability of this enzyme-activated probe to visualize mycobacteria selectively and with high sensitivity (>104-fold versus E. coli or Bacillus subtilis) suggests utility in the diagnosis of tuberculous.
Alkyl- or aryltranferases (EC 2.5)
Glutathione S-transferase (GST) is an important enzyme for the detoxification of xenobiotic substances through transfer of glutathione for subsequent metabolic decomposition.60 The structural variety of encountered xenobiotics exerts evolutionary pressure towards either a few enzymes with high substrate promiscuity or many substrate-specific enzymes. Ultimately, multiple isoforms of human GST evolved, with at least eight subclasses present in varied cellular locations including the cytosol, mitochondria, and microsomes.60 These defensive enzymes are also frequently commandeered by cancer cells to acquire drug-resistance. GST catalyzes nucleophilic aromatic substitution of glutathione into probe 24 to form a Meisenheimer complex, which collapses to release the parent fluorophore.55 Selectivity for the α and μ isoforms is achieved by tuning aryl substituents, and probe 24 was effective for imaging GST in HL60 cells.
Kinases (EC 2.7)
Kinases are integral to signaling pathways and exhibit extraordinary substrate variety. The quantification and spatiotemporal tracking of a particular kinase activity has required multistep protocols involving the introduction of non-native proteins into cells.61,62 As alternatives, probe 2556 and analogs63 have been developed for the single-step imaging of specific kinases. Probe 25 consists of three components—a Bruton’s tyrosine kinase (Btk) inhibitor-based recognition moiety, a fluorophore–quencher pair, and a kinase-cleavable linker. A cysteine residue (Cys481) in the active site of Btk attaches covalently to the α-carbon of the amide linker in probe 25, inducing elimination of the dinitrophenol quencher and enabling real-time imaging of Btk in live Namalwa cells. Because the kinase specificity of probe 25 is derived from the inhibitor mimic, this strategy is generally applicable to any kinase with accessible, selective small-molecule inhibitors.
HYDROLASES (EC 3)
Hydrolases, which catalyze the hydrolytic cleavage of chemical bonds, are preeminent targets of enzyme-activated fluorogenic probes. Early work with enzyme-activated probes was predominately on hydrolase-activated probes, which continue to be important tools for intracellular drug delivery, 64 imaging of dynamic cell processes,20 as well as diagnostics and therapeutics.65–67
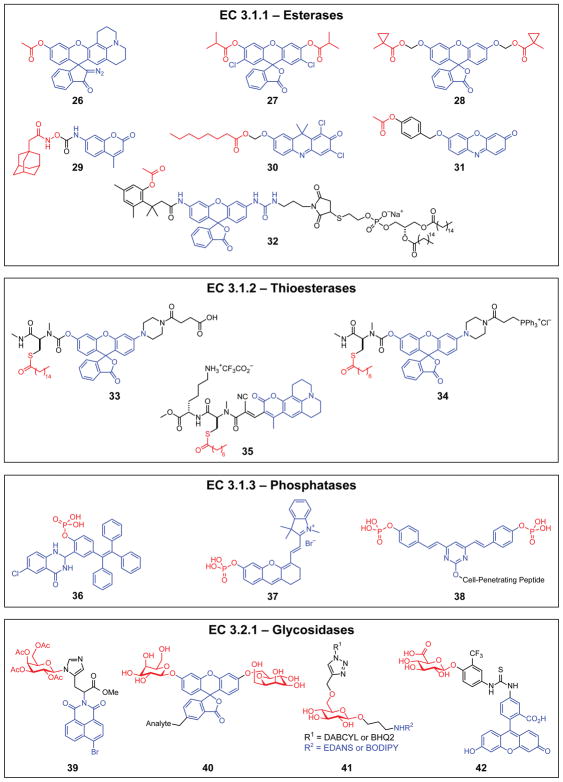
Esterases (EC 3.1.1)
Esterases are particularly amenable to repurposing for drug delivery and diagnostic applications. Masking negatively charged carboxylic acids with esters is a proven technique to enhance intracellular delivery of sensors, biomolecules,68 and therapeutic agents.64 Recent advances in esterase-activated probes include improvements in performance and utility (probes 26–28) and new applications in therapeutics, pathogen detection, and cell physiology (probes 29–32).
Probe 26 is a dual-input probe for the super-resolution imaging of enzymatic activity. Many super-resolution microscopy techniques require photoactivatable probes, which is achieved with probe 26 by replacing the traditional xanthene lactone with a diazo moiety.69 Because the esterase-reactive masking group is installed independently of the diazo moiety, this strategy can also be used to target other classes of enzymes in live-cell imaging. Probe 27 provides high stability, brightness, and photostability due to generally applicable electronic and steric optimization of the fluorophore and esterase-labile masking group.70,71 Probe 28 along with exogenous pig liver esterase form an enzyme–probe pair that is orthogonal to esterases in human cells. This pair enables an additional dimension of selectivity and demonstrates the feasibility of overlaying multiple enzymes catalyzing similar reactions in a single biological system while also preserving neatly discernable enzymatic responses.
In recent applications, esterase-activated fluorogenic probes 29 and 30 were shown to target bacterial enzymes relevant to botulism and tuberculosis. Probe 29 is a combination therapeutic and diagnostic agent, or “theranostic”,72 with a dual-purpose masking group that is also a potent inhibitor of botulinum neurotoxins.23 A hydroxamate linker enables 29 to undergo esterase-catalyzed activation of the prodrug and fluorogenic probe after freely diffusing across the plasma membrane, with enhanced intracellular delivery and effectiveness in live neurons. Probe 30 is a far-red sensor with variable lipid tails for profiling bacterial esterase and lipase activities.73 By testing samples with a battery of probes containing different lipid moieties, a characteristic esterase fingerprint can be used to distinguish M. tuberculosis from similar bacterial strains. In contrast to bacterial probes 29 and 30, probes 31 and 32 function in human cells to provide refined spatiotemporal control and detailed physiological information. Probe 31 is one of a family of probes tuned to report esterase activity in the endoplasmic reticulum selectively, with potential for application as a drug release trigger.74 Probe 32 contains a phosphatidylglycerol moiety, allowing it to embed in the outer surface of cells and monitor endocytic events.20 The internalization of probe 32 allows intracellular esterases to unmask fluorescence, a feature that was used to demonstrate fundamental differences in rates of endocytosis in matched cancer and non-cancer cell lines.
Thioesterases (EC 3.1.2). The S-acylation of cysteine residues is a dynamic post-translational modification prevalent in mammalian cells. The degree of protein S-acylation is tuned by the careful balance between S-acyltransferases and thioesterases, which are themselves regulated by a variety of cell processes including lipid signaling cascades, neuronal activation, and growth factor signaling.75 Reversible S-palmitoylation is of particular interest given its association with disease and the importance of palmitoylation for proper protein location and function.76
Acyl-protein thioesterase 1 and 2 (APT1 and APT2) are depalmitoylases essential for the regulation of S-palmitoylation in the cytosol. Probes 33–35 target APT1 and APT2 via a substrate mimic that, upon activation, undergoes intramolecular cleavage of a carbamate to release coumarin or rhodol fluorophores.77–79 The carboxylic acid in probe 33 increases aqueous solubility, enabling inclusion of a native S-palmitoyl trigger substrate compared to the abbreviated S-octanoyl triggers in probes 34 and 35.79 Discovery of previously unknown mitochondrial depalmitoylase activity was accomplished by appending a triphenylphosphine moiety to yield mitochondria-targeted probe 34.77 Further, imaging of live HEK293T cells with an APT1-preferential probe variant showed that APT1 is the primary enzyme responsible for mitochondrial depalmitoylation. To improve quantification of APT activity, ratiometric probe 35 was prepared from a substituted aminocoumarin and used to visualize cellular responses to lipid stress and APT activity in human colon organoids.78 Probes 33–35 constitute a portfolio of tools to study the spatiotemporal dynamics of APT1 and APT2 activity.
Phosphatases (EC 3.1.3)
Minimizing background and deep-tissue imaging are common themes of probes 36–38, which target alkaline phosphatases (ALP) or protein tyrosine phosphatases (PTP)—enzymes that play key roles in disease pathogenesis and cell regulation.80 ALP probe 36 was designed to detect secreted phosphatases, triggering excited state intramolecular proton transfer (ESIPT) fluorescence enhancement and irreversibly staining the surrounding area via fluorophore precipitation. Probe 36 was able to distinguish cells with different physiological profiles in heterogeneous tumor tissues.81 In contrast to the sedimentary nature of probe 36, near-infrared probe 37 was designed for dynamic imaging of ALP in live mice.14 Probe 38, which targets PTP, is a two-photon acyloxymethyl ketone probe conjugated to cell-penetrating peptides to facilitate organelle-specific detection at tissue depths of up to 100 μm.82 Far-red PTP probe 1 provides reduced phototoxicity and background autofluorescence along with enhanced cellular uptake compared to existing PTP probes. Probe 1 was used to image PTP activity in HeLa cells and to identify S. aureus from a panel of similar human pathogens.13
Glycosidases (EC 3.2.1)
Glycosidase-activated probes typically incorporate monosaccharides as masking groups or cleavable linkers for FRET quenching. Probes 39 and 40 are activated by β-galactosidase and are based on naphthalimide and xanthene scaffolds, respectively.83,84 Two-photon excited probe 39 is designed to identify cell senescence in live SK-MEL-103 cells and xenograft tumor-imaging experiments.84 Bioconjugable probe 40, together with E. coli β-galactosidase installed in the cytosol, forms an enzyme–probe pair for quantification of exogenous protein entry into the cytosol of HeLa cells.83 Similar pairs with near-infrared emission profiles have been developed for general imaging applications in other cell lines.85 Probe 41 detects lysosomal glucocerebrosidase activity, which is deficient in Gaucher’s disease,86 via cleavage of the glycosidic bond to release the parent fluorophore. The modularity of the linkers, quenchers, and fluorophores provides access to a variety of wavelengths for imaging fibroblasts. Probe 42 is similarly modular but instead employs masking groups activated by β–glucuronidase,87 a promising enzyme for prodrug-activation.88,89 The linker between masking group and near-infrared fluorophore contains a latent ortho-quinone methide electrophile that covalently binds the activated probe to the β-glucuronidase enzyme, a trapping motif also in β–lactamase probe 51 (EC 3.5.2). Trappable probe 42 has been used to visualize both subcutaneous and deep-tissue liver tumors in mice.
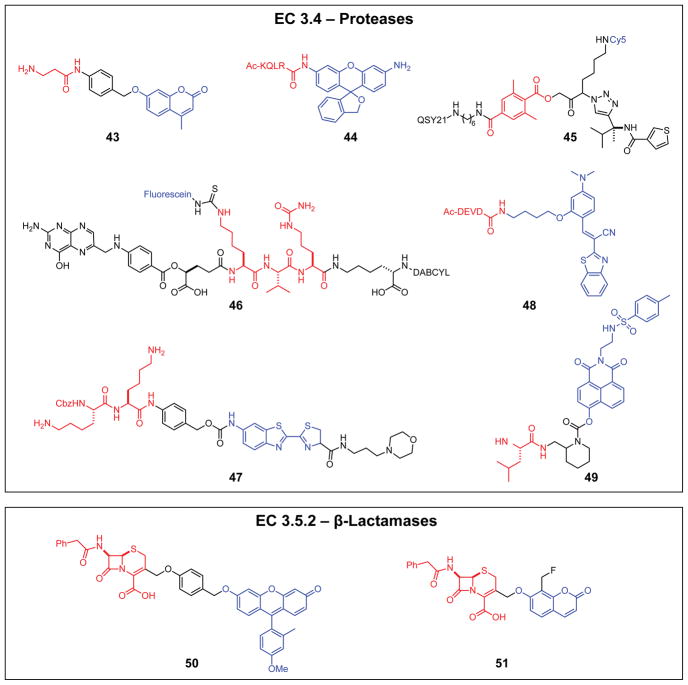
Proteases (EC 3.4)
Proteases, enzymes that cleave peptide bonds, act with varying degrees of substrate discrimination. Targeting highly specific proteases with enzyme-activated fluorogenic probes can be achieved by incorporating peptides or peptide mimics as masking groups or linkers. Depending on the desired enzyme target, these protease-recognition moieties can range from a single amino acid to more than 20 residues. Although activation mechanisms in protease-activated probes have been fairly constant, recent advances have enhanced probe performance, selectivity, and breadth of applications.
Probe 43 detects β-alanyl aminopeptidase activity in P. aeruginosa, a multi-drug resistant pathogen commonly found in hospital-acquired infections.90 The β-alanyl masking group of the probe was applied to resorufin, naphthalimide, and other fluorophore scaffolds to create panels of probes with clinical utility for identifying P. aeruginosa in culture.91
Probes 44–47 are designed for tumor visualization to improve diagnosis and intraoperative inspection during surgery. Probe 44 contains a single KQLR peptide masking group that is selectively cleaved by hepsin matriptase in prostate tumors.92 By leveraging the pH-dependent fluorescence of the masked probe relative to that of the parent hydroxymethyl rhodamine, high contrast ratios are achieved even with the use of only a single masking group. The diagnostic utility of probe 44 was established broadly in live-cell and mouse imaging experiments. Probes 45–47 target cathepsins, a group of proteases activated in the acidic environment of lysosomes that have become a centerpiece of prodrug strategies.93,94 Probe 45 is a non-peptide activity-based probe containing an electrophilic moiety that first selectively labels cathepsin S and then eliminates a quencher (QSY21), resulting in covalently labeling the enzyme with a fluorophore (Cy5).95 Probe 45, when deployed with nonspecific cathepsin-activated sister probes, has been instrumental for imaging syngeneic mammary tumors in vivo and profiling pathways of endolysosomal proteolysis in live dendritic cells. Probes 46, 47, and others incorporate short peptides targeting cathepsin B and have been used for selective imaging of cathepsin B in a wide variety of cell lines (Table 1).65,96 A discussion of protease-targeted activity-based probes and their clinical applications can be found elsewhere.97–100
Protease probes 48 and 49 rely on amino acid- or peptide-masking groups to target different proteases. Upon cleavage of the DEVD peptide in probe 48 by apoptotic protease caspase-3, the released monomer forms hydrophobic excimers with long-wavelength fluorescence.101 Excimer formation spurs precipitation of aggregates, enabling live-cell imaging of location-specific caspase activity without risk of fluorophore diffusion after cell apoptosis. Probe 49, which targets endoplasmic reticulum (ER) aminopeptidase 1, is a two-photon excited fluorescent probe that undergoes a bicyclic urea cyclization to release an ER-targeted naphthalimide.102 Selective enzymatic hydrolysis of the appropriate amide bond by the desired enzyme target, rather than nonspecific cleavage of thiourea and carbamate linkers in probes 46, 47, and 49, was confirmed by the absence of undesirable background signal upon incubation with non-target proteases.
β-Lactamases (EC 3.5.2)
Because of their clinical significance in antibiotic resistance, β–lactamases are frequent targets for fluorogenic probes in this class. With more than 890 β–lactamases having been identified to date, these cyclic amide hydrolases seriously challenge the efficacy of current antibiotics portfolios.103 Advances in β-lactamase-activated probes focus primarily on improvements to the mechanism of activation and enzymatic specificity, enabling clinically crucial identification of different antibiotic-resistant bacteria. A challenge in the development of meaningful probes for β-lactamases is the intrinsic cytotoxicity of β-lactams, which can limit the usable concentration of these probes for live-cell imaging.
Probe 50 selectively targets BlaC, a β-lactamase overexpressed in M. tuberculosis.66 Specificity was attained by tuning substituents to complement the substrate recognition loop in BlaC. As a result, probe 50 is 104-fold more responsive to BlaC than to homologous β–lactamases (e.g., TEM-1) and can identify M. tuberculosis in human sputum, though with a false-positive rate of 27%.
β-Lactamase generates a highly reactive Michael acceptor from probe 51, comparable to the action of β-glucaronidase probe 42.67 The nascent electrophile forms covalent bonds with nucleophilic residues of β-lactamase before probe diffusion occurs, enabling spatiotemporal tracking of the enzyme. The utility of this approach was demonstrated in E. coli cells that produce β-lactamase.
FUTURE DIRECTIONS
Enzyme-activated fluorogenic probes are highly sensitive tools for biological imaging applications. Advances in probe design and application have expanded the toolkit for assessing enzymatic activity and have established generalizable methods for imaging in live-cell and in vivo model systems.
Future work will likely focus on two primary areas—enhanced specificity of enzyme activation and improved probe chemistries. The attainable spatiotemporal and physiological information for imaging experiments is related directly to the degree of enzyme specificity. Expanding the palette of single enzyme-specific probes (i.e., 24 and 45) and isoform-specific probes (i.e., 4 and 5) would provide higher-resolution information to identify the location and function of a particular enzyme amidst the intracellular tapestry. Alternatively, new orthogonal enzyme–probe systems involving probes like 28 and 40 could be used to interrogate biological processes selectively while minimizing undesirable noise from endogenous enzymes. A logical extension of improved selectivity would be new theranostic agents such as probe 29, which deliver targeted therapeutic agents and illuminate disease sites simultaneously.
Improved probe chemistries would enhance the photophysical performance of parent fluorophore scaffolds and improve enzymatic responses.19 Tuning masking groups and enzyme-recognition moieties, as demonstrated in probes 27 and 45, could yield greater stability, reduced background, and enhanced activation kinetics. The incorporation of highly tunable17 and multi-input104,105 fluorophore scaffolds into new probes would facilitate sophisticated biological imaging and provide new insight into fundamental aspects of cell biology and physiology.
Figure 1.
Structures of probes activated by oxidoreductase enzymes EC 1.4–EC 1.6, EC 1.8, and EC 1.14. Enzyme-reactive moieties (red) and fluorophore scaffolds (blue) are highlighted.
Figure 2.
Structures of probes activated by transferase enzymes (EC 2). Enzyme-reactive moieties (red) and fluorophore scaffolds (blue) are highlighted.
Figure 3.
Structures of probes activated by hydrolases in classes EC 3.1 and EC 3.2. Enzyme-reactive moieties (red) and fluorophore scaffolds (blue) are highlighted.
Figure 4.
Structures of probes activated by hydrolases in classes EC 3.4 and EC 3.5. Enzyme-reactive moieties (red) and fluorophore scaffolds (blue) are highlighted.
Acknowledgments
W.C. was supported by an NSF Graduate Research Fellowship. Work on fluorogenic probes in the Raines laboratory is supported by grants R01 GM044783 and R01 CA073808 (NIH).
KEYWORDS
- enzyme-activated fluorogenic probe
a type of fluorogenic probe that uses the catalytic activity of an enzyme to generate fluorescence
- fluorogenic probe
a constitutively nonfluorescent small molecule that becomes fluorescent upon a change in environment (e.g., pH), its binding to an analyte, or its undergoing a chemical reaction, hydrolase, a class of enzyme (EC 3) that catalyzes the hydrolytic cleavage of specific chemical bonds
- live-cell imaging
imaging experiments conducted in proliferating cells without the use of chemical fixation
- oxidoreductase
a class of enzyme (EC 1) that catalyzes the transfer of electrons from a donor molecule to an acceptor molecule
- photoinduced electron transfer (PET)
nonradiative transfer of an excited state electron from a donor to an acceptor functional group, generating charge separation
- Förster resonance energy transfer (FRET)
nonradiative transfer of energy between dipole–dipole coupled chromophores
- transferase
a class of enzyme (EC 2) that catalyzes the transfer of specific functional groups from a donor molecule to an acceptor molecule
Footnotes
Notes
The authors declare no competing financial interest.
References
- 1.Lakowicz JR. Principles of Fluorescence Spectroscopy. 3. Springer; New York: 2006. [Google Scholar]
- 2.Chan J, Dodani SC, Chang CJ. Reaction-based small-molecule fluorescent probes for chemoselective bioimaging. Nat Chem. 2012;4:973–984. doi: 10.1038/nchem.1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grimm JB, Heckman LM, Lavis LD. The chemistry of small-molecule fluorogenic probes. Prog Mol Biol Transl. 2013;113:1–34. doi: 10.1016/B978-0-12-386932-6.00001-6. [DOI] [PubMed] [Google Scholar]
- 4.Nadler A, Schultz C. The power of fluorogenic probes. Angew Chem Int Ed. 2013;52:2408–2410. doi: 10.1002/anie.201209733. [DOI] [PubMed] [Google Scholar]
- 5.Rotman B. Measurement of activity of single molecules of β-D-galactosidase. Proc Natl Acad Sci USA. 1961;47:1981–1991. doi: 10.1073/pnas.47.12.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rotman B, Zderic JA, Edelstein M. Fluorogenic substrates for β-D-galactosidases and phosphatases derived from fluorescein (3,6-dihydroxyfluoran) and its monomethyl ether. Proc Natl Acad Sci USA. 1963;50:1–6. doi: 10.1073/pnas.50.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kramer DN, Guilbault GG. A substrate for the fluorometric determination of lipase activity. Anal Chem. 1963;35:588–589. [Google Scholar]
- 8.Guilbault GG, Kramer DN. Resorufin butyrate and indoxyl acetate as fluorogenic substrates for cholinesterase. Anal Chem. 1965;37:120–123. doi: 10.1021/ac60220a031. [DOI] [PubMed] [Google Scholar]
- 9.Rotman B, Papermaster BW. Membrane properties of living mammalian cells as studied by enzymatic hydrolysis of fluorogenic esters. Proc Natl Acad Sci USA. 1966;55:134–141. doi: 10.1073/pnas.55.1.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jones KH, Senft JA. An improved method to determine cell viability by simultaneous staining with fluorescein diacetate–propidium iodide. J Histochem Cytochem. 1985;33:77–79. doi: 10.1177/33.1.2578146. [DOI] [PubMed] [Google Scholar]
- 11.Xu S, Wang Q, Zhang Q, Zhang L, Zuo L, Jiang JD, Hu HY. Real time detection of ESKAPE pathogens by a nitroreductase-triggered fluorescence turn-on probe. Chem Commun. 2017;53:11177–11180. doi: 10.1039/c7cc07050k. [DOI] [PubMed] [Google Scholar]
- 12.Coutlee F, Viscidi RP, Yolken RH. Comparison of colorimetric, fluorescent, and enzymatic amplification substrate systems in an enzyme-immunoassay for detection of DNA–RNA hybrids. J Clin Microbiol. 1989;27:1002–1007. doi: 10.1128/jcm.27.5.1002-1007.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Biswas S, McCullough BS, Ma ES, LaJoie D, Russell CW, Garrett Brown D, Round JL, Ullman KS, Mulvey MA, Barrios AM. Dual colorimetric and fluorogenic probes for visualizing tyrosine phosphatase activity. Chem Commun. 2017;53:2233–2236. doi: 10.1039/c6cc09204g. [DOI] [PubMed] [Google Scholar]
- 14.Tan Y, Zhang L, Man KH, Peltier R, Chen G, Zhang H, Zhou L, Wang F, Ho D, Yao SQ, Hu Y, Sun H. Reaction-based off–on near-infrared fluorescent probe for imaging alkaline phosphatase activity in living cells and mice. ACS Appl Mater Interfaces. 2017;9:6796–6803. doi: 10.1021/acsami.6b14176. [DOI] [PubMed] [Google Scholar]
- 15.Li L, Zhang CW, Chen GYJ, Zhu B, Chai C, Xu QH, Tan EK, Zhu Q, Lim KL, Yao SQ. A sensitive two-photon probe to selectively detect monoamine oxidase B activity in Parkinson’s disease models. Nat Commun. 2014;5:3276. doi: 10.1038/ncomms4276. [DOI] [PubMed] [Google Scholar]
- 16.Zheng Q, Lavis LD. Development of photostable fluorophores for molecular imaging. Curr Opin Chem Biol. 2017;39:32–38. doi: 10.1016/j.cbpa.2017.04.017. [DOI] [PubMed] [Google Scholar]
- 17.Grimm JB, Muthusamy AK, Liang Y, Brown TA, Lemon WC, Patel R, Lu R, Macklin JJ, Keller PJ, Ji N, Lavis LD. A general method to fine-tune fluorophores for live-cell and in vivo imaging. Nat Methods. 2017;14:987–994. doi: 10.1038/nmeth.4403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lavis LD, Raines RT. Bright ideas for chemical biology. ACS Chem Biol. 2008;3:142–155. doi: 10.1021/cb700248m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lavis LD, Raines RT. Bright building blocks for chemical biology. ACS Chem Biol. 2014;9:855–866. doi: 10.1021/cb500078u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Levine MN, Hoang TT, Raines RT. Fluorogenic probe for constitutive cellular endocytosis. Chem Biol. 2013;20:614–618. doi: 10.1016/j.chembiol.2013.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ellis RJ. Macromolecular crowding: Obvious but underappreciated. Trends Biochem Sci. 2001;26:597–604. doi: 10.1016/s0968-0004(01)01938-7. [DOI] [PubMed] [Google Scholar]
- 22.Minton AP. The influence of macromolecular crowding and macromolecular confinement on biochemical reactions in physiological media. J Biol Chem. 2001;276:10577–10580. doi: 10.1074/jbc.R100005200. [DOI] [PubMed] [Google Scholar]
- 23.Šilhár P, Eubanks LM, Seki H, Pellett S, Javor S, Tepp WH, Johnson EA, Janda KD. Targeting botulinum A cellular toxicity: A prodrug approach. J Med Chem. 2013;56:7870–7879. doi: 10.1021/jm400873n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chevalier A, Zhang Y, Khdour OM, Kaye JB, Hecht SM. Mitochondrial nitroreductase activity enables selective imaging and therapeutic targeting. J Am Chem Soc. 2016;138:12009–12012. doi: 10.1021/jacs.6b06229. [DOI] [PubMed] [Google Scholar]
- 25.Levine MN, Raines RT. Trimethyl lock: A trigger for molecular release in chemistry, biology, and pharmacology. Chem Sci. 2012;3:2412–2420. doi: 10.1039/C2SC20536J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu X, Shi W, Li X, Ma H. A strategy for specific fluorescence imaging of monoamine oxidase A in living cells. Angew Chem, Int Ed. 2017;56:15319–15323. doi: 10.1002/anie.201708428. [DOI] [PubMed] [Google Scholar]
- 27.Rokita SE, editor. Quinone Methides. John Wiley & Sons; Hoboken, NJ: 2009. [Google Scholar]
- 28.Webb EC, editor. Enzyme Nomenclature 1992: Recommendations of the Nomenclature Committee of the International Union of Biochemistry and Molecular Biology on the Nomenclature and Classification of Enzymes. Academic Press; San Diego, CA: 1992. [Google Scholar]
- 29.Shih JC, Chen K, Ridd MJ. Monoamine oxidase: From genes to behavior. Annu Rev Neurosci. 1999;22:197–217. doi: 10.1146/annurev.neuro.22.1.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ross D, Kepa JK, Winski SL, Beall HD, Anwar A, Siegel D. NAD(P)H:quinone oxidoreductase 1 (NQO1): Chemoprotection, bioactivation, gene regulation and genetic polymorphisms. Chem Biol Interact. 2000;129:77–97. doi: 10.1016/s0009-2797(00)00199-x. [DOI] [PubMed] [Google Scholar]
- 31.Silvers WC, Prasai B, Burk DH, Brown ML, McCarley RL. Profluorogenic reductase substrate for rapid, selective, and sensitive visualization and detection of human cancer cells that overexpress NQO1. J Am Chem Soc. 2013;135:309–314. doi: 10.1021/ja309346f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hettiarachchi SU, Prasai B, McCarley RL. Detection and cellular imaging of human cancer enzyme using a turn-on, wavelength-shiftable, self-immolative profluorophore. J Am Chem Soc. 2014;136:7575–7578. doi: 10.1021/ja5030707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Prasai B, Silvers WC, McCarley RL. Oxidoreductase-facilitated visualization and detection of human cancer cells. Anal Chem. 2015;87:6411–6418. doi: 10.1021/acs.analchem.5b01615. [DOI] [PubMed] [Google Scholar]
- 34.Shen Z, Prasai B, Nakamura Y, Kobayashi H, Jackson MS, McCarley RL. A near-infrared, wavelength-shiftable, turn-on fluorescent probe for the detection and imaging of cancer tumor cells. ACS Chem Biol. 2017;12:1121–1132. doi: 10.1021/acschembio.6b01094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bryant C, DeLuca M. Purification and characterization of an oxygen-insensitive NAD(P)H nitroreductase from Enterobacter cloacae. J Biol Chem. 1991;266:4119–4125. [PubMed] [Google Scholar]
- 36.Williams EM, Little RF, Mowday AM, Rich MH, Chan-Hyams JV, Copp JN, Smaill JB, Patterson AV, Ackerley DF. Nitroreductase gene-directed enzyme prodrug therapy: Insights and advances toward clinical utility. Biochem J. 2015;471:131–153. doi: 10.1042/BJ20150650. [DOI] [PubMed] [Google Scholar]
- 37.Su J, Guise CP, Wilson WR. FSL-61 is a 6-nitroquinolone fluorogenic probe for one-electron reductases in hypoxic cells. Biochem J. 2013;452:79–86. doi: 10.1042/BJ20121695. [DOI] [PubMed] [Google Scholar]
- 38.Li Z, Li X, Gao X, Zhang Y, Shi W, Ma H. Nitroreductase detection and hypoxic tumor cell imaging by a designed sensitive and selective fluorescent probe, 7-[(5-nitrofuran-2-yl)methoxy]-3H-phenoxazin-3-one. Anal Chem. 2013;85:3926–3932. doi: 10.1021/ac400750r. [DOI] [PubMed] [Google Scholar]
- 39.Zhang J, Liu HW, Hu XX, Li J, Liang LH, Zhang XB, Tan W. Efficient two-photon fluorescent probe for nitroreductase detection and hypoxia imaging in tumor cells and tissues. Anal Chem. 2015;87:11832–11839. doi: 10.1021/acs.analchem.5b03336. [DOI] [PubMed] [Google Scholar]
- 40.Zhou J, Shi W, Li LH, Gong QY, Wu XF, Li XH, Ma HM. A lysosome-targeting fluorescence off–on probe for imaging of nitroreductase and hypoxia in live cells. Chem Asian J. 2016;11:2719–2724. doi: 10.1002/asia.201600012. [DOI] [PubMed] [Google Scholar]
- 41.Kim TI, Kim H, Choi Y, Kim Y. meso-Ester BODIPYs for the imaging of hypoxia in tumor cells. Sens Actuators B Chem. 2017;249:229–234. [Google Scholar]
- 42.Zhang X, Zhao Q, Li Y, Duan X, Tang Y. Multifunctional probe based on cationic conjugated polymers for nitroreductase-related analysis: Sensing, hypoxia diagnosis, and imaging. Anal Chem. 2017;89:5503–5510. doi: 10.1021/acs.analchem.7b00477. [DOI] [PubMed] [Google Scholar]
- 43.Jin C, Zhang Q, Lu W. Selective turn-on near-infrared fluorescence probe for hypoxic tumor cell imaging. RSC Adv. 2017;7:18217–18223. [Google Scholar]
- 44.O’Connor LJ, Mistry IN, Collins SL, Folkes LK, Brown G, Conway SJ, Hammond EM. CYP450 enzymes effect oxygen-dependent reduction of azide-based fluorogenic dyes. ACS Cent Sci. 2017;3:20–30. doi: 10.1021/acscentsci.6b00276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pham ND, Parker RB, Kohler JJ. Photocrosslinking approaches to interactome mapping. Curr Opin Chem Biol. 2013;17:90–101. doi: 10.1016/j.cbpa.2012.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shin N, Hanaoka K, Piao W, Miyakawa T, Fujisawa T, Takeuchi S, Takahashi S, Komatsu T, Ueno T, Terai T, Tahara T, Tanokura M, Nagano T, Urano Y. Development of an azoreductase-based reporter system with synthetic fluorogenic substrates. ACS Chem Biol. 2017;12:558–563. doi: 10.1021/acschembio.6b00852. [DOI] [PubMed] [Google Scholar]
- 47.Arnér ESJ, Holmgren A. Physiological functions of thioredoxin and thioredoxin reductase. Eur J Biochem. 2000;267:6102–6109. doi: 10.1046/j.1432-1327.2000.01701.x. [DOI] [PubMed] [Google Scholar]
- 48.Zhang L, Duan D, Liu Y, Ge C, Cui X, Sun J, Fang J. Highly selective off–on fluorescent probe for imaging thioredoxin reductase in living cells. J Am Chem Soc. 2014;136:226–233. doi: 10.1021/ja408792k. [DOI] [PubMed] [Google Scholar]
- 49.Beharry AA, Lacoste S, O’Connor TR, Kool ET. Fluorescence monitoring of the oxidative repair of DNA alkylation damage by ALKBH3, a prostate cancer marker. J Am Chem Soc. 2016;138:3647–3650. doi: 10.1021/jacs.6b00986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Camps M, Eichman BF. Unraveling a connection between DNA demethylation repair and cancer. Mol Cell. 2011;44:343–344. doi: 10.1016/j.molcel.2011.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wu X, Li L, Gong Q, Ma H. Near-infrared fluorescent probe with new recognition moiety for specific detection of tyrosinase activity: Design, synthesis, and application in living cells and zebrafish. Angew Chem, Int Ed. 2016;55:14728–14732. doi: 10.1002/anie.201609895. [DOI] [PubMed] [Google Scholar]
- 52.Morton RK. Transferase activity of hydrolytic enzymes. Nature. 1953;172:65–68. doi: 10.1038/172065a0. [DOI] [PubMed] [Google Scholar]
- 53.Park S, Lim S-Y, Bae SM, Kim S-Y, Myung S-J, Kim H-J. Indocyanine-based activatable fluorescence turn-on probe for γ-glutamyltranspeptidase and its application to the mouse model of colon cancer. ACS Sens. 2016;1:579–583. [Google Scholar]
- 54.Iwatate RJ, Kamiya M, Umezawa K, Kashima H, Nakadate M, Kojima R, Urano Y. Silicon rhodamine-based near-infrared fluorescence probe for γ-glutamyltransferase. Bioconjugate Chem. 2018;29:241–244. doi: 10.1021/acs.bioconjchem.7b00776. [DOI] [PubMed] [Google Scholar]
- 55.Shibata A, Nakano Y, Ito M, Araki M, Zhang J, Yoshida Y, Shuto S, Mannervik B, Mogenstern R, Ito Y, Abe H. Fluorogenic probes using 4-substituted-2-nitrobenzenesulfonyl derivatives as caging groups for the analysis of human glutathione transferase catalyzed reactions. Analyst. 2013;138:7326–7330. doi: 10.1039/c3an01339a. [DOI] [PubMed] [Google Scholar]
- 56.Zhang Q, Liu H, Pan Z. A general approach for the development of fluorogenic probes suitable for no-wash imaging of kinases in live cells. Chem Commun. 2014;50:15319–15322. doi: 10.1039/c4cc07429g. [DOI] [PubMed] [Google Scholar]
- 57.Corti A, Franzini M, Paolicchi A, Pompella A. Gamma-glutamyltransferase of cancer cells at the cossroads of tumor progression, drug resistance and drug targeting. Anticancer Res. 2010;30:1169–1181. [PubMed] [Google Scholar]
- 58.Röttig A, Steinbüchela A. Acyltransferases in bacteria. Microbiol Mol Biol Rev. 2013;77:277–321. doi: 10.1128/MMBR.00010-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hodges HL, Brown RA, Crooks JA, Weibel DB, Kiessling LL. Fluorogenic probe for imaging mycobacterial growth and division. Proc Natl Acad Sci USA. 2018;115:5271–5276. doi: 10.1073/pnas.1720996115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Salinas AE, Wong MG. Glutathione S-transferases—A review. Curr Med Chem. 1999;6:279–309. [PubMed] [Google Scholar]
- 61.Ng T, Squire A, Hansra G, Bornancin F, Prevostel C, Hanby A, Harris W, Barnes D, Schmidt S, Mellor H, Bastiaens PIH, Parker PJ. Imaging protein kinase Cα activation in cells. Science. 1999;283:2085–2089. doi: 10.1126/science.283.5410.2085. [DOI] [PubMed] [Google Scholar]
- 62.Zhang L, Lee KC, Bhojani MS, Khan AP, Shilman A, Holland EC, Ross BD, Rehemtulla A. Molecular imaging of Akt kinase activity. Nat Med. 2007;13:1114–1119. doi: 10.1038/nm1608. [DOI] [PubMed] [Google Scholar]
- 63.Zuo Y, Shi Y, Li X, Teng Y, Pan Z. A novel 2,5-diaminopyrimidine-based affinity probe for Bruton’s tyrosine kinase. Sci Rep. 2015;5:16136. doi: 10.1038/srep16136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Testa B, Mayer JM. Hydrolysis in Drug and Prodrug Metabolism: Chemistry, Biochemistry, and Enzymology. Verlag Helvetica Chimica Acta; Zürich, Switzerland: 2003. [Google Scholar]
- 65.Tian R, Li M, Wang J, Yu M, Kong X, Feng Y, Chen Z, Li Y, Huang W, Wu W, Hong Z. An intracellularly activatable, fluorogenic probe for cancer imaging. Org Biomol Chem. 2014;12:5365–5374. doi: 10.1039/c4ob00297k. [DOI] [PubMed] [Google Scholar]
- 66.Cheng Y, Xie H, Sule P, Hassounah H, Graviss EA, Kong Y, Cirillo JD, Rao J. Fluorogenic probes with substitutions at the 2 and 7 positions of cephalosporin are highly BlaC-specific for rapid Mycobacterium tuberculosis detection. Angew Chem, Int Ed. 2014;53:9360–9364. doi: 10.1002/anie.201405243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mao W, Xia L, Wang Y, Xie H. A self-immobilizing and fluorogenic probe for β-lactamase detection. Chem Asian J. 2016;11:3493–3497. doi: 10.1002/asia.201601344. [DOI] [PubMed] [Google Scholar]
- 68.Mix KA, Lomax JE, Raines RT. Cytosolic delivery of proteins by bioreversible esterification. J Am Chem Soc. 2017;139:14396–14398. doi: 10.1021/jacs.7b06597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Halabi EA, Thiel Z, Trapp N, Pinotsi D, Rivera-Fuentes P. A photoactivatable probe for super-resolution imaging of enzymatic activity in live cells. J Am Chem Soc. 2017;139:13200–13207. doi: 10.1021/jacs.7b07748. [DOI] [PubMed] [Google Scholar]
- 70.Chyan W, Kilgore HR, Gold B, Raines RT. Electronic and steric optimization of fluorogenic probes for biomolecular imaging. J Org Chem. 2017;82:4297–4304. doi: 10.1021/acs.joc.7b00285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chyan W, Kilgore HR, Raines RT. Cytosolic uptake of large monofunctionalized dextrans. Bioconjugate Chem. 2018;29:1942–1949. doi: 10.1021/acs.bioconjchem.8b00198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Thanou M, editor. Theranostics and Image Guided Drug Delivery. The Royal Society of Chemistry; London, UK: 2018. [Google Scholar]
- 73.Tallman KR, Levine SR, Beatty KE. Profiling esterases in Mycobacterium tuberculosis using far-red fluorogenic substrates. ACS Chem Biol. 2016;11:1810–1815. doi: 10.1021/acschembio.6b00233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hakamata W, Tamura S, Hirano T, Nishio T. Multicolor imaging of endoplasmic reticulum-located esterase as a prodrug activation enzyme. ACS Med Chem Lett. 2014;5:321–325. doi: 10.1021/ml400398t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zeidman R, Jackson CS, Magee AI. Protein acyl thioesterases (review) Mol Membr Biol. 2009;26:32–41. doi: 10.1080/09687680802629329. [DOI] [PubMed] [Google Scholar]
- 76.Yeste-Velasco M, Linder ME, Lu YJ. Protein S-palmitoylation and cancer. Biochim Biophys Acta. 2015;1856:107–120. doi: 10.1016/j.bbcan.2015.06.004. [DOI] [PubMed] [Google Scholar]
- 77.Kathayat R, Cao Y, Elvira PD, Sandoz PA, Zaballa ME, Springer MZ, Drake LE, Macleod KF, van der Goot FG, Dickinson BC. Active and dynamic mitochondrial S-depalmitoylation revealed by targeted fluorescent probes. Nat Commun. 2017;9:334. doi: 10.1038/s41467-017-02655-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Beck MW, Kathayat RS, Cham CM, Chang EB, Dickinson BC. Michael addition-based probes for ratiometric fluorescence imaging of protein S-depalmitoylases in live cells and tissues. Chem Sci. 2017;8:7588–7592. doi: 10.1039/c7sc02805a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Qiu T, Kathayat RS, Cao U, Beck MW, Dickinson BC. A fluorescent probe with improved water solubility permits the analysis of protein S-depalmitoylation activity in live cells. Biochemistry. 2018;57:221–225. doi: 10.1021/acs.biochem.7b00835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Coleman JE. Structure and mechanism of alkaline-phosphatase. Annu Rev Biophys Biomol Struct. 1992;21:441–483. doi: 10.1146/annurev.bb.21.060192.002301. [DOI] [PubMed] [Google Scholar]
- 81.Liu HW, Li K, Hu XX, Zhu L, Rong Q, Liu Y, Zhang XB, Hasserodt J, Qu FL, Tan W. In situ localization of enzyme activity in live cells by a molecular probe releasing a precipitating fluorochrome. Angew Chem, Int Ed. 2017;56:11788–11792. doi: 10.1002/anie.201705747. [DOI] [PubMed] [Google Scholar]
- 82.Li L, Ge J, Wu H, Xu QH, Yao SQ. Organelle-specific detection of phosphatase activities with two-photon fluorogenic probes in cells and tissues. J Am Chem Soc. 2012;134:12157–12167. doi: 10.1021/ja3036256. [DOI] [PubMed] [Google Scholar]
- 83.Chao TY, Raines RT. Fluorogenic label to quantify the cytosolic delivery of macromolecules. Mol BioSyst. 2013;9:339–342. doi: 10.1039/c3mb25552b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lozano-Torres B, Galiana I, Rovira M, Garrido E, Chaib S, Bernardos A, Muñoz-Espín D, Serrano M, Martínez-Máñez R, Sancenón F. An OFF–ON two-photon fluorescent probe for tracking cell senescence in vivo. J Am Chem Soc. 2017;139:8808–8811. doi: 10.1021/jacs.7b04985. [DOI] [PubMed] [Google Scholar]
- 85.Han J, Han MS, Tung CH. A fluorogenic probe for β-galactosidase activity imaging in living cells. Mol BioSyst. 2013;9:3001–3008. doi: 10.1039/c3mb70269c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yadav AK, Shen DL, Shan X, He X, Kermode AR, Vocadlo DJ. Fluorescence-quenched substrates for live cell imaging of human glucocerebrosidase activity. J Am Chem Soc. 2015;137:1181–1189. doi: 10.1021/ja5106738. [DOI] [PubMed] [Google Scholar]
- 87.Cheng TC, Roffler SR, Tzou SC, Chuang KH, Su YC, Chuang CH, Kao CH, Chen CS, Harn IH, Liu KY, Cheng TL, Leu YL. An activity-based near-infrared glucuronide trapping probe for imaging β-glucuronidase expression in deep tissues. J Am Chem Soc. 2012;134:3103–3110. doi: 10.1021/ja209335z. [DOI] [PubMed] [Google Scholar]
- 88.de Graaf M, Boven E, Scheeren HW, Haisma HJ, Pinedo HM. Beta-glucuronidase–mediated drug release. Curr Pharm Des. 2002;8:1391–1403. doi: 10.2174/1381612023394485. [DOI] [PubMed] [Google Scholar]
- 89.Burke PJ, Hamilton JZ, Jeffrey SC, Hunter JH, Doronina SV, Okeley NM, Miyamoto JB, Anderson ME, Stone IJ, Ulrich ML, Simmons JK, McKinney EE, Senter PD, Lyon RP. Optimization of a PEGylated glucuronide-monomethylauristatin E linker for antibody–drug conjugates. Mol Cancer Ther. 2017;16:116–123. doi: 10.1158/1535-7163.MCT-16-0343. [DOI] [PubMed] [Google Scholar]
- 90.Váradi L, Hibbs DE, Orenga S, Babolat M, Perry JD, Groundwater PW. β-Alanyl aminopeptidase-activated fluorogenic probes for the rapid identification of Pseudomonas aeruginosa in clinical samples. RSC Adv. 2016;6:58884–58889. [Google Scholar]
- 91.Luo JL, Jin T, Váradi L, Perry JD, Hibbs DE, Groundwater PW. Evaluation of fluorogenic aminonaphthalenesulfonamides and 6-hydrazinobenz[de]isoquinoline-1,3-diones for the detection of bacteria. Dyes Pigm. 2016;125:15–26. [Google Scholar]
- 92.Yogo T, Umezawa K, Kamiya M, Hino R, Urano Y. Development of an activatable fluorescent probe for prostate cancer imaging. Bioconjugate Chem. 2017;28:2069–2076. doi: 10.1021/acs.bioconjchem.7b00233. [DOI] [PubMed] [Google Scholar]
- 93.Dubowchik GM, Firestone RA, Willner D, Hofstead SJ, Trail PA, Lasch SJ, Henderson AJ, Jure M, Mosure KW, Knipe JO. Peptide linkers for selective intralysosomal release of anticancer drugs from monoclonal antibody conjugates. In: Ramage R, Epton R, editors. Peptides 1996: Proceedings of the Twenty-Fourth European Peptide Symposium; Kingswinford, UK: Mayflower Scientific; 1998. pp. 347–348. [Google Scholar]
- 94.Doronina SO, Toki BE, Torgov MY, Mendelsohn BA, Cerveny CG, Chace DF, DeBlanc RL, Gearing RP, Bovee TD, Siegeall CB, Francisco JA, Wahl AF, Meyer DL, Senter PD. Development of potent monoclonal antibody auristatin conjugates for cancer therapy. Nat Biotechnol. 2003;21:778–784. doi: 10.1038/nbt832. [DOI] [PubMed] [Google Scholar]
- 95.Bender KO, Ofori L, van der Linden WA, Mock ED, Datta GK, Chowdhury S, Li H, Segal E, Lopez MS, Ellman JA, Figdor CG, Bogyo M, Verdoes M. Design of a highly selective quenched activity-based probe and its application in dual color imaging studies of cathepsin S activity localization. J Am Chem Soc. 2015;137:4771–4777. doi: 10.1021/jacs.5b00315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wang Y, Li J, Feng L, Yu J, Zhang Y, Ye D, Chen HY. Lysosome-targeting fluorogenic probe for cathepsin B imaging in living cells. Anal Chem. 2016;88:12403–12410. doi: 10.1021/acs.analchem.6b03717. [DOI] [PubMed] [Google Scholar]
- 97.Verdoes M, Bender KO, Segal E, van der Linden WA, Syed S, Withana NP, Sanman LE, Bogyo M. Improved quenched fluorescent probe for imaging of cysteine cathepsin activity. J Am Chem Soc. 2013;135:14726–14730. doi: 10.1021/ja4056068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Sanman LE, Bogyo M. Activity-based profiling of proteases. Annu Rev Biochem. 2014;83:249–273. doi: 10.1146/annurev-biochem-060713-035352. [DOI] [PubMed] [Google Scholar]
- 99.Ofori LO, Withana NP, Prestwood TR, Verdoes M, Brady JJ, Winslow MM, Sorger J, Bogyo M. Design of protease activated optical contrast agents that exploit a latent lysosomotropic effect for use in fluorescence-guided surgery. ACS Chem Biol. 2015;10:1977–1988. doi: 10.1021/acschembio.5b00205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Withana NP, Ma X, McGuire HM, Verdoes M, van der Linden WA, Ofori LO, Zhang R, Li H, Sanman LE, Wei K, Yao S, Wu P, Li F, Huang H, Xu Z, Wolters PJ, Rosen GD, Collard HR, Zhu Z, Cheng Z, Bogyo M. Non-invasive imaging of idiopathic pulmonary fibrosis using cathepsin protease probes. Sci Rep. 2016;6:19755. doi: 10.1038/srep19755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kim TI, Jin H, Bae J, Kim Y. Excimer emission-based fluorescent probe targeting caspase-3. Anal Chem. 2017;89:10565–10569. doi: 10.1021/acs.analchem.7b02790. [DOI] [PubMed] [Google Scholar]
- 102.Xu S, Liu HW, Hu XX, Huan SY, Zhang J, Liu YC, Yuan L, Qu FL, Zhang XB, Tan W. Visualization of endoplasmic reticulum aminopeptidase 1 under different redox conditions with a two-photon fluorescent probe. Anal Chem. 2017;89:7641–7648. doi: 10.1021/acs.analchem.7b01561. [DOI] [PubMed] [Google Scholar]
- 103.Bush K. Bench-to-bedside review: The role of β-lactamases in antibiotic-resistant Gram-negative infections. Crit Care. 2010;14:224. doi: 10.1186/cc8892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Grimm JB, Gruber TD, Ortiz G, Brown TA, Lavis LD. Virginia Orange: A versatile, red-shifted fluorescein scaffold for single- and dual-input fluorogenic probes. Bioconjugate Chem. 2016;27:474–480. doi: 10.1021/acs.bioconjchem.5b00566. [DOI] [PubMed] [Google Scholar]
- 105.Gonçalves CCS, da Costa BZ, Lima MLSO, Fiorito GF, Ruiz ALTG, de Oliveira SBP, Barbosa GO, de Carvalho HF, Marsaioli AJ. Enzymatic profiling in prostate and breast cancer cells: Phosphate hydrolysis and alcohol oxidation. Tetrahedron. 2016;72:7235–7240. [Google Scholar]