ABSTRACT
Distant homology relationships among proteins with many transmembrane regions (TMs) are difficult to detect as they are clouded by the TMs’ hydrophobic compositional bias and mutational divergence in connecting loops. In the case of several GPI lipid anchor biosynthesis pathway components, the hidden evolutionary signal can be revealed with dissectHMMER, a sequence similarity search tool focusing on fold-critical, high complexity sequence segments. We find that a sequence module with 10 TMs in PIG-W, described as acyl transferase, is homologous to PIG-U, a transamidase subunit without characterized molecular function, and to mannosyltransferases PIG-B, PIG-M, PIG-V and PIG-Z. We conclude that this new, membrane-embedded domain named BindGPILA functions as the unit for recognizing, binding and stabilizing the GPI lipid anchor in a modification-competent form as this appears the only functional aspect shared among all proteins. Thus, PIG-U's likely molecular function is shuttling/presenting the anchor in a productive conformation to the transamidase complex.
KEYWORDS: glycosylphosphatidylinositol, GPI lipid anchor, GPI biosynthesis, GPI mannosyltransferase, GPI transamidase complex, inositol acylase, transmembrane protein function, dissectHMMER
Introduction
Function annotation transfer within the homologous gene concept due to statistically significant sequence similarity of encoded protein sequences has revolutionized life science research [1–3]. Laborious experimental work for function characterization of many orthologous genes could be omitted at all or, at least, more precisely targeted. Yet, proteins with many transmembrane helices (TMs), low complexity regions or other non-globular segments remain difficult sequence-analytic targets as significance criteria break down due to the amino acid compositional bias [4]. For some functionally uncharacterized sequences, focusing the analysis on fold-critical segments [5,6] or on complex TMs (that carry an evolutionary signature versus simple, merely hydrophobic TMs) [7,8] can expand the reach of sequence similarity searches to previously not seen functionally annotated homologues.
As described below, these methodical innovations led to the discovery of a membrane-embedded protein sequence domain consisting of ten TMs and the inter-connecting loops in the GPI anchor biosynthesis pathway components PIG-B, PIG-M, PIG-U, PIG-W, PIG-V, and PIG-Z (Figure 1). As we find these six proteins evolutionarily related despite of their dissimilar molecular and cellular functions, we conclude that the common theme determining the function of this new membrane-embedded domain is the recognition/binding/shuttling of the GPI lipid anchor, a very complex lipid-sugar moiety, and its presentation to the various catalytic centers along the biosynthesis pathway.
Figure 1.
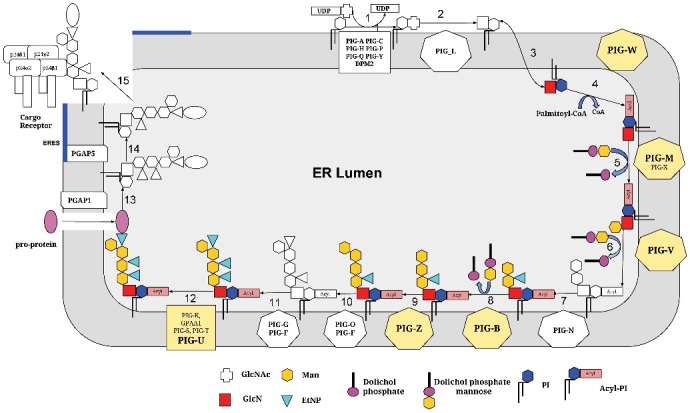
The GPI lipid anchor biosynthesis pathway – overview. The various steps of the pathway (from the N-acetylglucosaminyltransferase complex with PIG-A etc. to the step of leaving the ER where further anchor remodeling happens) are schematically depicted. All reactions and translocations are shown in white and black except for the six steps involving PIG-B, PIG-M, PIG-U, PIG-W, PIG-V, and PIG-Z that are highlighted in colors. Please note that PIG-U is a subunit of the transamidase complex and not all proteins, for example PIG-Z, are available in all organisms.
PIG-B, PIG-M, PIG-U, PIG-W, PIG-V, and PIG-Z occupy diverse positions in the GPI lipid anchor biosynthesis pathway (Figure 1). The oliogosaccharide chain of protein-linked GPI lipid anchors contains three or, in the case of yeast, some protozoa and mammalians, four mannose units (Figure 1). Up to four ER-membrane associated mannosyltransferases are known to catalyze the corresponding reactions [9–11]. PIG-M (in complex with auxiliary protein PIG-X [12] attaches the first mannose (via α1-4 bond) to the GlcN unit [13,14]. PIG-V (together with the auxiliary protein Pga1 reported only for fungi [15] is responsible for adding the second mannose (via α1-6 bond) [16]. The third and fourth mannose residues are added to the anchor (via α1-2 bond) by PIG-B [17,18] and PIG-Z [19,20], respectively. To some extent, the manosyltransferases are promiscuous; for example, PIG-B in Trypanosoma cruzi robustly complements deletions of PIG-B and PIG-Z in yeast [9].
Human PIG-U/yeast CdC91p was discovered as the fifth, most loosely attached subunit of the GPI transamidase complex in 2003 [21]. PIG-U has been reported as a potential oncogene [22–24] but the proteins’ molecular function and its exact role within the transamidase complex remain unclear [22].
The gene PIG-W/GWT1 was found to be required for inositol acylation of GPI anchors in rat [25] and yeast [26], respectively. Sagane et al. analyzed the topology of yeast Gwt1 and found that Gwt1 is a membrane protein containing 13 TM regions [27]. They also confirmed that the N- and C-termini of Gwt1 were oriented towards the ER lumen and cytoplasm, respectively.
Below, we provide arguments showing the evolutionary relatedness of the six proteins described above and conclusions with regard to the function of the newly derived membrane-embedded sequence domain that we suggest to name BindGPILA(“bind GPI lipid anchor).
Results and discussion
There is a homology relationship between the four mannosyltransferases PIG-M, PIG-V, PIG-B, and PIG-Z
It is known that all four mannosyltransferases share the same sequence architecture. An N-terminal transmembrane (TM) region is followed by a luminal domain which contains a functionally important signature motif of negatively charged amino acid residues and a stretch of C-terminal TM regions [28]. However, HMMER [29] searches with the mannosyltransferases as query sequences against the Pfam domain database (version 30) [30] reveal that they belong to different protein families: PIG-M to PIG-M mannosyltransferases (Mannosyl_trans, PF05007), PIG-V to PIG-V mannosyltransferases (Mannosyl_trans2, PF04188), and PIG-B and PIG-Z to Alg9-like mannosyltransferases (Glyco_transf_22, PF03901) (see Table 1 for detail). But all three protein families are members of the glycosyl transferase GT-C superfamily (clan CL0111) and, therefore, are thought to share a common fold and to be evolutionary related (see also Supplementary Table S1).
Table 1.
Sequence architecture of four human mannosyltransferases PIG-M, PIG-V, PIG-B, PIG-Z as well as PIG-U and PIG-W.
| Protein | Sequence length (AAs) | Total number of TMs | TMs involved in domain BindGPILA | Conserved catalytic motif between 1st and 2nd TM in the domain | Best Pfam Domain hit |
|---|---|---|---|---|---|
| PIGM_HUMAN (Q9H3S5) | 423 | 10 | 1–10 | DxD | Mannosyl_trans (PF05007) E-value = 4.0e-109 |
| PIGV_HUMAN (Q9NUD9) | 493 | 10 | 1–10 | D/ExE | Mannosyl_trans2 (PF04188) E-value = 4.0e-216 |
| PIGB_HUMAN (Q92521) | 554 | 11 | 1–10 | DE | Glyco_transf_22 (PF03901) E-value = 6.0e-123 |
| PIGZ_HUMAN (Q86VD9) | 579 | 11 | 1–10 | DE | Glyco_transf_22 (PF03901) E-value = 6.4e-52 |
| PIGU_HUMAN (Q9H490) | 435 | 10 | 1–10 | – | PIG-U (PF06728) E-value = 6.0e-115 |
| PIGW_HUMAN (Q7Z7B1) | 504 | 13 | 2–11 | – | GWT1 (PF06423) E-value = 1.2e-35 |
For each of the six human proteins each representing an orthologous protein family, total sequence lengths, total number of TMs and the range of TMs within the newly discovered membrane-embedded domain BindGPILA are listed. We also provide the conserved catalytically relevant motif in the luminal loop between the 1st and the 2nd TMs within the domain. Further, we list the best Pfam hit found with a plain HMMER search. To note, we provide a full list of hits from searches with sequences of one orthologous family into any of the other 5 families with HHPRED or dissectHMMER in Suplementary Table S1.
Interestingly, the sequences of the three mannosyltransferase protein families slightly differ in their conserved signature motif located in the first extracytoplasmic loop. It is DxD for PIG-M, D/ExE for PIG-V, and DE for PIG-B and PIG-Z. To note, all signature motifs are followed by an aromatic amino acid residue (Supplementary Figure S1). There is evidence for the motif to have a role in catalysis of PIG-M [14,28]. Most likely, the respective motif is also catalytically important for the other mannosyltransferases.
PIG-U is a member of the GPI lipid anchor mannosyltransferase family but lacks the functionally important DxD/DE motif in the first ER lumenal domain
Sequence analysis with the ANNOTATOR software suite [31] reveals that PIG-U has the same sequence architecture as the mannosyltransferases (Table 1). An N-terminal TM region is followed by a globular segment and multiple (we predicted another 9) TM regions towards the C-terminus. In some cases, e.g. human PIG-U (Q9H490), the predicted N-terminal TM region is at the very N-terminus of the sequence followed by a couple of small amino acid residues. Under these circumstances, a signal peptide is, as we suggest, incorrectly predicted with SIGNALP [32].
To note, the Trypanosoma brucei protein TTA2 (Q7YTW3) sequence is actually a PIG-U orthologue. TTA2 was originally described as a new transamidase complex component [33] but a PSI-Blast search [34] with its sequence as query clearly collects the PIG-U protein family (e.g., 3rd iteration lists Drosophila melanogaster PIG-U with E-value = 2e-104).
We observed that a simple Blast search [34] with human PIG-U (Q9H490) against Homo sapiens finds back its own sequence but, in addition, hits to human PIG-M (Q9H3S5) with a significant E-value = 3e-05. Interestingly, the Blast hit covers the most part of the C-terminal TM segment, namely the sequence range TM2- TM8. This evidence is further supported by numerous significant HHpred [35] and dissectHMMER hits with sequences of different members of the PIG-U protein family (finding mannosyltransferases) or mannosyltransferases (finding PIG-U) as queries, e.g., HHPred search with human PIG-U against Pfam (Version 31) hits to PF05007 (Mannosyl_trans, PIG-M, E-value = 3.5-31), PF04188 (Mannosyl_trans2, PIG-V, E-value = 0.0013) and PF03901 (Glyco_transf_22, E-value = 9.8e-5, see also Supplementary Table S1). These findings extend the similarity range to TM1-TM10 and clearly support that PIG-U is a member of the GPI mannosyltransferase family and the glycosyl transferase GT-C superfamily clan.
Therefore, the N-terminus of PIG-U is predicted to be cytoplasmic, which is consistent with the sequence architecture of the other four GPI mannosyltransferases. Subsequently, we have to assume that the globular segment between TM1 and TM2 is localized in the ER lumen. Members of the glycosyl transferase GT-C superfamily are known to be integral membrane proteins with a modified DxD motif in the first extracytoplasmic loop. Motif variations include DxD, ExD, DxE, DDx, or DEx residues [36]. In the case of PIG-M, change of either of the aspartic residues in the DxD motif to alanine abolishes its mannosyltransferase activity [14]. Inspection of the PIG-U sequences regarding the TM1-TM2 luminal segment reveals that there is no conserved DxD signature motif, not in any form described above.
To conclude, PIG-U shows sequence similarity to all four GPI mannosyltransferases and, thus, seems to be part of the glycosyl transferase GT-C superfamily but lacks the catalytically important DxD motif in the first extracytoplasmic loop. PIG-U is probably an inactive/non-functioning mannosyltransferase but still capable to recognize and to bind the substrate that all four GPI mannosyltransferases have in common: the GPI lipid anchor moiety. This predicted functionality makes PIG-U a perfect fit for the GPI transamidase complex as the function of GPI lipid anchor binding has not been assigned to any other transamidase subunit so far [10,37].
PIG-W is a member of the membrane acyl transferase superfamily but shares sequence features with Alg9-like mannosyltransferases (Glyco_transf_22, PF03901).
An HHpred search with human PIG-W (Q7Z7B1) as query sequence against Pfam (version 31) results in numerous hits: (1) The “own” domain GWT1 (PF06423, E-value = 1.6e-19, Table 1) is found. (2) Four domains with relatively weak but significant E-values are listed, namely OpgC_C (PF10129, 8.6e-9), DUF1624 (PF07786, 1.2e-09), DUF5009 (PF16401, 1.4e-08), and Acyl_transf_3 (PF01757, 0.0001). (3) The hit list is completed by three additional domains reported with non-significant E-values: Cas1_AcylT (PF07779, E-value = 12), DUF418 (PF04235, E-value = 14), and TraX (PF05857, E-value = 250). Strikingly, the domains listed under (2) and (3) have in common that they all belong to the membrane acyl transferase superfamily (Acyl_transf_3, CL0316).
Supplementary Figure S2 shows an alignment of four PIG-W sequences (sequence range TM4-TM13) with the seed alignment of the OpgC_C (PF10129) domain. There are quite a few conserved residues in the alignment, e.g. a strongly conserved Arginine in TM4 (here and further PIG-W numbering), a strongly conserved Glycine in TM5, and a conserved aromatic residue (mainly tyrosine) at the beginning of TM6. To note, the two sequence positions D145 and K155 reported to be of some functional importance for PIG-W in yeast GWT1 (P47026) [27] are not conserved in the total alignment.
A dissectHMMER search [6] with PIG-W_HUMAN (Q7Z7B1) as query sequence results in more domain hits with significant fold-critical E-values based on HMMER2: (1) GWT1 (PF06423, 1.11e-18), (2) Acyl_transf_3 (PF01757, 4.65e-4), (3) Baculo_11_kDa (PF06143, 3.e-4), and (4) Glyco_transf_22 (PF03901, 6.16e-4). The first two hits (1) and (2) are consistent with the HHpred prediction results. Unfortunately, the alignment for hit (3) is short covering only PIG-W's positions 317–374 and the Baculovirus 11 kDa protein family is functionally uncharacterized; there is no further data available. The very surprising result is that sequence similarity between PIG-W_HUMAN and Alg9-like mannosyltransferases (Glyco_transf_22, PF03901, hit (4)) was detected. The sequence range for the alignment with PIG-W_HUMAN (Q7Z7B1) covers positions 64–429 and it corresponds to the sequence segment TM2 – TM11 including both TM2 as domain-starting TM and TM11 as final TM (Table 1).
Thus, we conclude on the one hand that PIG-W/GWT1 is a member of the membrane acyl transferase Acyl_transf_3 superfamily (clan CL0316). This finding complements prior knowledge that PIG-W/GWT1 is required for inositol acylation of GPI anchors and it was proposed to be the acyl transferase itself [25,26]. On the other hand, PIG-W shows sequence similarity to Alg9-like mannosyltransferases over a sequence stretch of 10 TMs (TM2- TM11). The fact that PIG-W is an acyltransferase and not a mannosyltransferase suggests that the similarity is not due to the catalysis. Most likely, this extended membrane embedded region is necessary for the productive binding of the enzyme's substrate, namely the GPI lipid anchor moiety.
PIG-B, PIG-M, PIG-U, PIG-W, PIG-V, and PIG-Z harbor a common sequence module most likely involved in recognition and binding of the GPI moiety
All four mannosyltransferase protein families PIG-M, PIG-V, PIG-B and PIG-Z and, despite missing the functionally important signature motif in the first extracytoplasmic loop, also PIG-U are members of the glycosyl transferase GT-C superfamily. The application of different sequence homology search methods like Blast/PSI-Blast [34], HHPred [35], and dissectHMMER [6] demonstrates that the members of one protein family are similar to the members of the other four protein families (see Supplementary Table S1 for more detail). The sequence range where the similarity is observed covers ten TM regions: All 10 TMs of PIG-M, PIG-V, and PIG-U are included, for PIG-B and PIG-Z, the first 10 TMs (excluding the last 11th TM) contribute to sequence similarity.
Most interestingly, PIG-W, an enzyme with a different molecular function – an acyl transferase and not a mannosyltransferase but part of the same GPI lipid anchor biosynthesis pathway – also shows sequence similarity to Alg9-like mannosyltransferases (Glyco_transf_22, PF03901), which are represented by the PIG-B and PIG-Z protein families (Table 1). For PIG-W, the sequence stretch TM2-TM11 (excluding the 1st TM and TMs 12 and 13) is responsible for the similarity to mannosyltransferase sequences (Figure 2). To note, the N-terminus of PIG-W is oriented towards ER lumen [27]. Therefore, the sequence segment between TM1 and TM2 is cytoplasmic, PIG-W's TM2 crosses the membrane in direction cytoplasm –> ER lumen. This is consistent with the direction of TM1 of the four mannosyltransfease protein families because their N-terminus is cytoplasmic. PIG-U's N-terminus is also predicted to be cytoplasmic, which would fit into the overall context.
Figure 2.
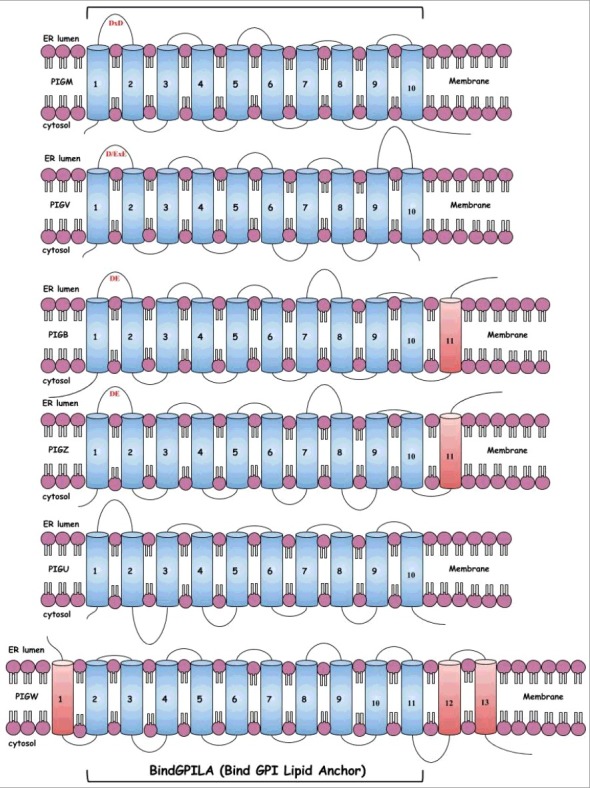
Scheme of the sequence architecture of PIG-B, PIG-M, PIG-U, PIG-W, PIG-V, and PIG-Z. The figure shows the schematic architecture of the transmembrane domains from PIGB, PIGM, PIGU, PGW, PIGV and PIGZ. The TMs colored in blue forms the membrane embedded sequence domain, BindGPILA, comprising of 10 TMs. The extracellular loops are shown by curved loops and the length of the loop shows the distance between two TMs. The conserved catalytic motif, which is present in the first extracellular loop between the first and second TM, are labelled in red.
It is important to emphasize that all ten TMs in the newly defined BindGPILA domain are complex (as determined with TMSOC) and, thus, carry an evolutionary signal beyond the hydrophobic sequence bias [7,8]. Supplementary Figure S1 shows an alignment of the sequences of all six protein families over the relevant 10 TMs [39,40]. Each family is represented by human, mouse, fruit fly, yeast and, if available, Trypanosoma brucei sequences. The first TM in the new domain harbors a strongly conserved charged amino acid residue, a positively charged arginine in the case of the mannosyltransferases and PIG-U, and a negatively charged residue in the case of PIG-W. Apparently, the first TM might not be absolutely essential for function in all proteins with this domain or in all species. It was reported that GPI14, PIG-M in yeast, can still function somehow with the first TM impaired by an amino terminal deletion [13]. As binding of a substrate is an additive effect provided from several sites, this functionality might not be fully disrupted by losing one site.
The linker sequence segment between the BindGPILA domain's TM1 and TM2 contains the functionally important DXD/DE signature motifs for the mannosyltransferases but there is no such motif for PIG-U and PIG-W. Further C-terminally, we observe quite a few conserved residues within protein families but also shared ones by different protein families. E.g., there are a conserved proline and a conserved aspartic acid in the motif TM3. Whereas we see the conserved proline in PIG-M, PIG-U, PIG-B and PIG-Z, the conserved aspartic acid is found in PIG-M, PIG-B, PIG-Z, PIG-V, and PIG-W.
The domain's TM6 contains a conserved proline. Similar to TM1, the domain's TM7 harbors a conserved charged amino acid residue, in this case, a positively charged one for PIG-M and PIG-U, and a negatively charged one for PIG-B, PIG-Z, PIG-V, and PIG-W. At the C-terminus of TM6, there is an enrichment of aromatic residues and/or amino acids with long hydrophobic sidechains. Apparently, the exact identity of these residues in the species-specific context is critical for function. Mutations F274L and W275L in human PIG-U tested in CHOPA16.1 cells show no activity in restoring the surface expression of CD59 [21]. The TM8 is glycine rich but not for PIG-M, PIG-U, and PIG-V.
Two hints in the literature are corroborative for our function assignment for PIG-U. (1) It is known that PIG-U is the subunit most loosely connected to the transamidase. PIG-S, PIG-T, GAA1 and GPI8 form a stable, active transamidase complex in the absence of PIG-U [21]. Thus, PIG-U is involved in an event independent of the cleavage of the GPI attachment signal sequence from the precursor protein and does not have a role in the recognition of the GPI attachment signal or the presentation of the precursor protein to the catalytic site of GPI8. (2) Further, Gab1 (PIG-U in yeast) forms a complex with GPI17 (PIG-S) in vivo; thus, PIG-U interacts with the transamidase possibly via PIG-S. A temperature-sensitive GAB1 mutant is defective in transfer of GPI precursors to proteins and accumulates complete GPI lipid anchors [40]. Thus, both findings (1) and (2) are agreeable with PIG-U (or GAB1) being involved in presenting free GPI lipid moieties in a competent conformation to the catalytic subunit GPAA1/GAA1 for establishing the peptide bond between the anchor and the C-terminus of the substrate protein.
As supplementary files with this article, we provide “BindGPILA.aln”, the domain alignment in aln-format, as well as the hidden Markov models in the HMMER2 (“BindGPILA-hmm2.hmm”) and HMMER3 (“BindGPILA-hmm3.hmm”) formats. The latter domain models have the sequences from the six families as top hits and can be used for automatically annotating uncharacterized protein sequences.
To conclude, there is sequence-analytic evidence that the six protein families PIG-B, PIG-M, PIG-U, PIG-W, PIG-V, and PIG-Z harbor a common sequence domain comprising 10 TMs: These are the first/all 10 TMs of PIG-B, PIG-Z/ PIG-M, PIG-V, and PIG-U and TM2-TM11 of PIG-W. Despite the very different molecular functionalities represented by those six protein families, they have one feature in common; namely, they are involved in the GPI lipid anchor biosynthesis and have to recognize/bind the “growing” GPI moiety. We conclude that the most likely function of the common module of 10 TMs, the domain BindGPILA, shared by the six protein families is the recognition/binding of the GPI moiety. Further, we implicate that presenting the GPI lipid anchor to the transamidase complex is the true molecular function of PIG-U.
Supplementary Material
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
References
- [1].Bork P, Dandekar T, Diaz-Lazcoz Y, et al. . Predicting function: from genes to genomes and back. J Mol Biol. 1998;283:707–725. doi: 10.1006/jmbi.1998.2144. PMID:9790834 [DOI] [PubMed] [Google Scholar]
- [2].Eisenhaber F. A decade after the first full human genome sequencing: when will we understand our own genome? J Bioinform Comput Biol. 2012;10:1271001. doi: 10.1142/S0219720012710011. PMID:22849370 [DOI] [PubMed] [Google Scholar]
- [3].Kuznetsov V, Lee HK, Maurer-Stroh S, et al. . How bioinformatics influences health informatics: usage of biomolecular sequences, expression profiles and automated microscopic image analyses for clinical needs and public health. Health Inform Sci Syst. 2013;1:2. doi: 10.1186/2047-2501-1-2. PMID:25825654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Wong WC, Maurer-Stroh S, Eisenhaber F. More than 1,001 problems with protein domain databases: transmembrane regions, signal peptides and the issue of sequence homology. PLoS Comput Biol. 2010;6:e1000867. doi: 10.1371/journal.pcbi.1000867. PMID:20686689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Wong WC, Maurer-Stroh S, Eisenhaber B, et al. . On the necessity of dissecting sequence similarity scores into segment-specific contributions for inferring protein homology, function prediction and annotation. BMC Bioinformatics. 2014;15:166. doi: 10.1186/1471-2105-15-166. PMID:24890864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Wong WC, Yap CK, Eisenhaber B, et al. . dissectHMMER: a HMMER-based score dissection framework that statistically evaluates fold-critical sequence segments for domain fold similarity. Biol Direct. 2015;10:39. doi: 10.1186/s13062-015-0068-3. PMID:26228544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Wong WC, Maurer-Stroh S, Eisenhaber F. Not all transmembrane helices are born equal: towards the extension of the sequence homology concept to membrane proteins. Biol Direct. 2011;6:57. doi: 10.1186/1745-6150-6-57. PMID:22024092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Wong WC, Maurer-Stroh S, Schneider G, et al. . Transmembrane helix: simple or complex. Nucleic Acids Res. 2012;40:W370–W375. doi: 10.1093/nar/gks379. PMID:22564899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Cortes LK, Scarcelli JJ, Taron CH. Complementation of essential yeast GPI mannosyltransferase mutations suggests a novel specificity for certain Trypanosoma and Plasmodium PigB proteins. PLoS One. 2014;9:e87673. doi: 10.1371/journal.pone.0087673. PMID:24489949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Eisenhaber B, Maurer-Stroh S, Novatchkova M, et al. . Enzymes and auxiliary factors for GPI lipid anchor biosynthesis and post-translational transfer to proteins. Bioessays. 2003;25:367–385. doi: 10.1002/bies.10254. PMID:12655644 [DOI] [PubMed] [Google Scholar]
- [11].Ferguson MAJ, Hart GW, Kinoshita T. Glycosylphosphatidylinositol anchors. In: Varki A, Cummings RD, Esko JD, et al., editors. Essentials of glycobiology [Internet]. Cold Spring Harbor: (NY: ): Cold Spring Harbor Laboratory Press; 2017. p. 137–150. [Google Scholar]
- [12].Ashida H, Hong Y, Murakami Y, et al. . Mammalian PIG-X and yeast Pbn1p are the essential components of glycosylphosphatidylinositol-mannosyltransferase I. Mol Biol Cell. 2005;16:1439–1448. doi: 10.1091/mbc.E04-09-0802. PMID:15635094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Davydenko SG, Feng D, Jantti J, et al. . Characterization of GPI14/YJR013w mutation that induces the cell wall integrity signalling pathway and results in increased protein production in Saccharomyces cerevisiae. Yeast. 2005;22:993–1009. doi: 10.1002/yea.1286. PMID:16134120 [DOI] [PubMed] [Google Scholar]
- [14].Maeda Y, Watanabe R, Harris CL, et al. . PIG-M transfers the first mannose to glycosylphosphatidylinositol on the lumenal side of the ER. EMBO J. 2001;20:250–261. doi: 10.1093/emboj/20.1.250. PMID:11226175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Sato K, Noda Y, Yoda K. Pga1 is an essential component of Glycosylphosphatidylinositol-mannosyltransferase II of Saccharomyces cerevisiae. Mol Biol Cell. 2007;18:3472–3485. doi: 10.1091/mbc.E07-03-0258. PMID:17615295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Kang JY, Hong Y, Ashida H, et al. . PIG-V involved in transferring the second mannose in glycosylphosphatidylinositol. J Biol Chem. 2005;280:9489–9497. doi: 10.1074/jbc.M413867200. PMID:15623507 [DOI] [PubMed] [Google Scholar]
- [17].Sutterlin C, Escribano MV, Gerold P, et al. . Saccharomyces cerevisiae GPI10, the functional homologue of human PIG-B, is required for glycosylphosphatidylinositol-anchor synthesis. Biochem J. 1998;332(Pt 1):153–159. doi: 10.1042/bj3320153. PMID:9576863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Takahashi M, Inoue N, Ohishi K, et al. . PIG-B, a membrane protein of the endoplasmic reticulum with a large lumenal domain, is involved in transferring the third mannose of the GPI anchor. EMBO J. 1996;15:4254–4261. PMID:8861954 [PMC free article] [PubMed] [Google Scholar]
- [19].Grimme SJ, Westfall BA, Wiedman JM, et al. . The essential Smp3 protein is required for addition of the side-branching fourth mannose during assembly of yeast glycosylphosphatidylinositols. J Biol Chem. 2001;276:27731–27739. doi: 10.1074/jbc.M101986200. PMID:11356840 [DOI] [PubMed] [Google Scholar]
- [20].Taron BW, Colussi PA, Wiedman JM, et al. . Human Smp3p adds a fourth mannose to yeast and human glycosylphosphatidylinositol precursors in vivo. J Biol Chem. 2004;279:36083–36092. doi: 10.1074/jbc.M405081200. PMID:15208306 [DOI] [PubMed] [Google Scholar]
- [21].Hong Y, Ohishi K, Kang JY, et al. . Human PIG-U and yeast Cdc91p are the fifth subunit of GPI transamidase that attaches GPI-anchors to proteins. Mol Biol Cell. 2003;14:1780–1789. doi: 10.1091/mbc.E02-12-0794. PMID:12802054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Gamage DG, Hendrickson TL. GPI transamidase and GPI anchored proteins: oncogenes and biomarkers for cancer. Crit Rev Biochem Mol Biol. 2013;48:446–464. doi: 10.3109/10409238.2013.831024. PMID:23978072 [DOI] [PubMed] [Google Scholar]
- [23].Guo Z, Linn JF, Wu G, et al. . CDC91L1 (PIG-U) is a newly discovered oncogene in human bladder cancer. Nat Med. 2004;10:374–381. doi: 10.1038/nm1010. PMID:15034568 [DOI] [PubMed] [Google Scholar]
- [24].Nagpal JK, Dasgupta S, Jadallah S, et al. . Profiling the expression pattern of GPI transamidase complex subunits in human cancer. Mod Pathol. 2008;21:979–991. doi: 10.1038/modpathol.2008.76. PMID:18487995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Murakami Y, Siripanyapinyo U, Hong Y, et al. . PIG-W is critical for inositol acylation but not for flipping of glycosylphosphatidylinositol-anchor. Mol Biol Cell. 2003;14:4285–4295. doi: 10.1091/mbc.E03-03-0193. PMID:14517336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Umemura M, Okamoto M, Nakayama K, et al. . GWT1 gene is required for inositol acylation of glycosylphosphatidylinositol anchors in yeast. J Biol Chem. 2003;278:23639–23647. doi: 10.1074/jbc.M301044200. PMID:12714589 [DOI] [PubMed] [Google Scholar]
- [27].Sagane K, Umemura M, Ogawa-Mitsuhashi K, et al. . Analysis of membrane topology and identification of essential residues for the yeast endoplasmic reticulum inositol acyltransferase Gwt1p. J Biol Chem. 2011;286:14649–14658. doi: 10.1074/jbc.M110.193490. PMID:21367863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Menon AK, Kinoshita T, Orlean P, et al. . Glycosylphosphatidylinositol (GPI) anchoring of proteins. Vol. 26. – series “The Enzymes”. New York: Academic Press; 2017. [Google Scholar]
- [29].Finn RD, Clements J, Arndt W, et al. . HMMER web server: 2015 update. Nucleic Acids Res. 2015;43:W30–W38. doi: 10.1093/nar/gkv397. PMID:25943547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Finn RD, Coggill P, Eberhardt RY, et al. . The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 2016;44:D279–D285. doi: 10.1093/nar/gkv1344. PMID:26673716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Eisenhaber B, Kuchibhatla D, Sherman W, et al. . The recipe for protein sequence-based function prediction and its implementation in the ANNOTATOR software environment. Methods Mol Biol. 2016;1415:477–506. doi: 10.1007/978-1-4939-3572-7_25. PMID:27115649 [DOI] [PubMed] [Google Scholar]
- [32].Petersen TN, Brunak S, von HG, et al. . SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods. 2011;8:785–786. doi: 10.1038/nmeth.1701. PMID:21959131 [DOI] [PubMed] [Google Scholar]
- [33].Nagamune K, Ohishi K, Ashida H, et al. . GPI transamidase of Trypanosoma brucei has two previously uncharacterized (trypanosomatid transamidase 1 and 2) and three common subunits. Proc Natl Acad Sci USA. 2003;100:10682–10687. doi: 10.1073/pnas.1833260100. PMID:12958211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Altschul SF, Koonin EV. Iterated profile searches with PSI-BLAST–a tool for discovery in protein databases. Trends Biochem Sci. 1998;23:444–447. doi: 10.1016/S0968-0004(98)01298-5. PMID:9852764 [DOI] [PubMed] [Google Scholar]
- [35].Soding J, Biegert A, Lupas AN. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005;33:W244–W248. doi: 10.1093/nar/gki408. PMID:15980461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Liu J, Mushegian A. Three monophyletic superfamilies account for the majority of the known glycosyltransferases. Protein Sci. 2003;12:1418–1431. doi: 10.1110/ps.0302103. PMID:12824488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Eisenhaber B, Eisenhaber S, Kwang TY, et al. . Transamidase subunit GAA1/GPAA1 is a M28 family metallo-peptide-synthetase that catalyzes the peptide bond formation between the substrate protein's omega-site and the GPI lipid anchor's phosphoethanolamine. Cell Cycle. 2014;13:1912–1917. doi: 10.4161/cc.28761. PMID:24743167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Larkin MA, Blackshields G, Brown NP, et al. . Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. PMID:17846036 [DOI] [PubMed] [Google Scholar]
- [39].Waterhouse AM, Procter JB, Martin DM, et al. . Jalview Version 2–a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. PMID:19151095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Grimme SJ, Gao XD, Martin PS, et al. . Deficiencies in the endoplasmic reticulum (ER)-membrane protein Gab1p perturb transfer of glycosylphosphatidylinositol to proteins and cause perinuclear ER-associated actin bar formation. Mol Biol Cell. 2004;15:2758–2770. doi: 10.1091/mbc.E04-01-0035. PMID:15075373 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


