ABSTRACT
Recent findings suggest that ephrinA5 (Efna5) has a novel role in female mouse fertility, in addition to its well-defined role as a neurogenesis factor. Nevertheless, its physiological roles in ovarian granulosa cells (GC) have not been determined. In this study, mouse GC were cultured and transfected with ephrin A5 siRNA and negative control to determine the effects of Efna5 on GC apoptosis, proliferation, cell cycle progression, and related signaling pathways. To understand the mode signaling, the mRNA expression levels of Efna5 receptors (Eph receptor A5, Eph receptor A3, Eph receptor A8, and Eph receptor B2) were examined. Both mRNA and protein expressions of apoptosis-related factors (Bax, Bcl-2, Caspase 8, Caspase 3, and Tnfα) and a proliferation marker, Pcna, were investigated. Additionally, the role of Efna5 on paracrine oocyte-secreted factors and steroidogenesis hormones were also explored. Efna5 silencing suppressed GC apoptosis by downregulating Bax and upregulating Bcl-2 in a Caspase 8-dependent manner. Efna5 knockdown promoted GC proliferation via p-Akt and p-ERK pathway activation. The inhibition of Efna5 enhanced BMH15 and estradiol expression, but suppressed GDF9, while progesterone level remained unaltered. These results demonstrated that Efna5 is a pro-apoptotic agent in GC and plays important role in folliculogenesis by mediating apoptosis, proliferation, and steroidogenesis in female mouse. Therefore Efna5 might be potential therapeutic target for female fertility disorders.
KEYWORDS: Apoptosis, EhrinA5, Ephrin-Eph signaling, Granulosa cells, Proliferation
Introduction
Folliculogenesis, a complex and core reproductive phenomenon, occurs spontaneously during the reproductive life of mammals. This process involves functional, morphological, and physiological changes in the theca cells (TC), cumulus cells (CC), and granulosa cells (GC), which are the main ovarian operational units. It is governed by paracrine [1,2], endocrine [3], and autocrine [4] factors. Follicular growth initiation, maturation, ovulation, steroidogenesis, and atresia are tightly regulated in an orchestrated manner by several regulatory molecules [5–9].
In a single cohort of growing follicles, several ovarian follicules begin to grow and proliferate; depending on the species, either a single or a few of these follicles complete the developmental process and reach fertilization, while the vast majority are lost due to atresia or degenerative processes [10,11]. However, it is unclear why such large proportions of ovarian follicules and oocytes are removed by degenerative processes during the reproductive life of mammals. As such, gaining an insight into how ovarian follicular growth, proliferation, and apoptosis are dynamically regulated is imperative. Additionally, identifying new regulatory molecules and cell signaling pathways involved in GC functions is crucial for understanding the complex processes involved in folliculogenesis. This mechanistic understanding would ultimately pave the way for new molecular diagnostic and therapeutic approaches to improve the ever-increasing issue of mammalian infertility.
Erythropoietin-producing hepatocellular (Eph) receptors, with 14 members, represent the largest family of receptor tyrosine kinase (RTKs) [12–15]. Eph and their eight ligands, Ephrins (Efns), are categorized into subclasses A and B based on their binding affinity to ligands and structural integration into the cell membrane respectively, according to the unified nomenclature of Eph receptors and ligands [16]. The ephrin–Eph family is ubiquitously expressed in several cell types and organs during embryonic development and regulates a wide range of early developmental processes, including tissue remodeling, skeletal and cardiac development, axon guidance, vascularization, and apoptosis [17,18]. Likewise, in adults, several normal cellular physiological functions have been attributed to ephrin–Eph signaling, such as angiogenesis [19], axon guidance [20], adhesion [21,22], morphogenesis [14,17], and neurogenesis [19,23]. In addition to its role in normal cellular functions, ephrin–Eph signaling has also been implicated in the regulation of cancer development and progression [14,24,25], bone disease [26], and diabetes [27].
Recent studies of the ephrin–Eph gene family in ovarian cells have revealed robust expressions of ephrin B1 in human luteinizing GC [28] as well as ephrinA5 (Efna5) in GC of bovines [29,30] and mouse [31], indicating a potential physiological role of ephrins and Eph receptors in the regulation of mammalian reproductive function. Buensuceso and Deroo [31] examined the expression of Efna5 and showed that its three cognate receptors, Eph receptor A5 (EphA5), Eph receptor A3 (EphA3), and Eph receptor A8 (EphA8), are regulated by follicle-stimulating hormones (FSH) via the protein kinase A (PKA) pathway in mouse primary GC and rat GC cell lines. They further highlighted that Efna5–EphA5 regulates mouse GC morphology and adhesion in vitro [31]. Furthermore, global Efna5 knockout results in sub-fertility phenotypes associated with ovarian histological disorders in mouse [32]. Moreover, based on bovine GC transcriptome profiling, Efna5 expression is elevated in large atretic follicles [30], indicating a role of Efna5 in follicular atresia. These limited studies support the roles of eph–Eph signaling, and particularly Efna5-Eph signaling, in the regulation of folliculogenesis; this is a novel emerging physiological role of the ephrin–Eph family, which is generally known for its involvement in neurogenesis landscape, prompting questions regarding its precise functions in ovarian function.
Despite the high expression levels of Efna5 in GC of various mammalian species, its physiological roles during folliculogenesis are barely elucidated. In this study, the biological role of Efna5 was examined by the treatment of mouse primary GC with ephrinA siRNAs in vitro. It was hypothesized that Efna5 regulate GC apoptosis, proliferation, apoptosis, cell cycle progression, oocyte development, and steroidogenesis. Furthermore, the mechanisms underlying the roles of Efna5 in these processes were evaluated.
Results
Efna5 expression is efficiently downregulated by ephrinA5 short interfering RNA (siRNA)
The biological roles of Efna5 in mammalian reproductive cells are largely unknown. Hence, to explore the physiological functions of Efna5 in the regulation of GC functions, a pool of ephrinA5 siRNA (siEfna5) was used to disrupt the expression of Efna5. Briefly, the transcriptional and translational silencing capacities of siEfna5 were determined by RT- qPCR and western blotting, respectively. The results are presented in Figure 1A, B, and C. These results indicated that siEfna5 transfection in mouse GC significantly inhibited the expression of Efna5 as compared to that of the NC at the protein level (0.3 ± 0.02 vs. 0.7 ± 0.029, P < 0.01, Figure 1A and B) and at the mRNA level (0.2 ± 0.03 vs. 1.1 ± 0.1, P < 0.001, Figure 1C).
Figure 1.
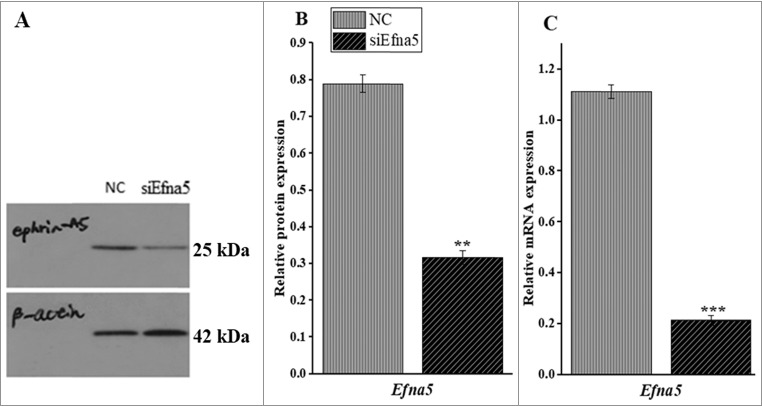
siEfna5 knocked down the protein and mRNA expression of Efna5 in mouse GCs in vitro. (A and B) siEfna5 effectively downregulated the protein expression of Efna5 in mouse GCs; (C) siEfna5 inhibited the expression of Efna5 at the mRNA level in mouse GCs. Protein and mRNA levels were determined by western blotting and RT-qPCR, respectively. Values represent Efna5 expression levels relative to β-actin levels and are presented as means ± SEM of 3 independent experiments. **P < 0.01, ***P < 0.001. NC, negative control; Efna5, EphrinA5; GC, granulosa cells; siEfna5; EphrinA5 siRNA.
mRNA expression levels of Efna5 receptors are downregulated by siEfna5
Like most members of the ephrin/Eph family, Efna5 associates with multiple receptors and functions via forward, backward, and/or bidirectional signaling. Therefore, in order to understand the signaling direction of Efna5 in mouse GC physiological function we determined the mRNA expression levels of a subclass of Efna5 receptors in siEfna5-transfected mouse by RT- qPCR. Of the eight known Efna5 receptors, we focused on the expression levels of four (Epha5, Epha3, Ephb2, and Epha8) reported to be regulated by FSH in mouse GC [31]. These receptors were downregulated by siEfna5. A RT-qPCR analysis of siEfna5-transfected GC indicated that the expression levels of the Ephb2, Epha5, Epha3, Epha8 receptors were lower than those in the NC (Figure 2A and B, P < 0.001 and Figure 2C and D, P < 0.01).
Figure 2.
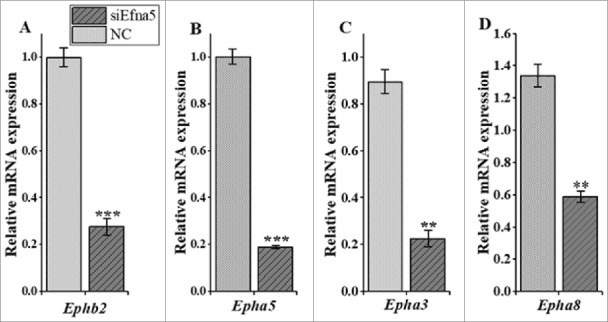
The mRNA expressions levels of four Efna5 receptors following siEfna5 transfection in GC. (A, B, C, and D) The mRNA expression levels of Ephab2, Epha5, Epha3, and Epha8 were significantly reduced following Efna5 silencing. The expression values are expressed relative to β-actin and are presented as means ± SEM. Compared to the NC group, **P < 0.01 and ***P < 0.001. NC, negative control, Epha3, Eph receptor A3; Epha5, Eph receptor A5; Epha8, Eph receptor A8; Ephb2, Eph receptor B2; Efna5; ephrinA5.
Silencing of Efna5 suppresses mouse GC apoptosis in vitro
Downregulation of Efna5 suppresses mouse GC apoptosis in vitro. The GC were transfected with siEfna5 and NC for 48 h and evaluated by flow cytometry to determine the rate of apoptosis. In this assay, the rate of apoptosis was lower in siEfna5-transfected cells than in NC (10.28 ± 0.45 vs. 22.54 ± 0.63, P < 0.001) (Figure 3A).
Figure 3.
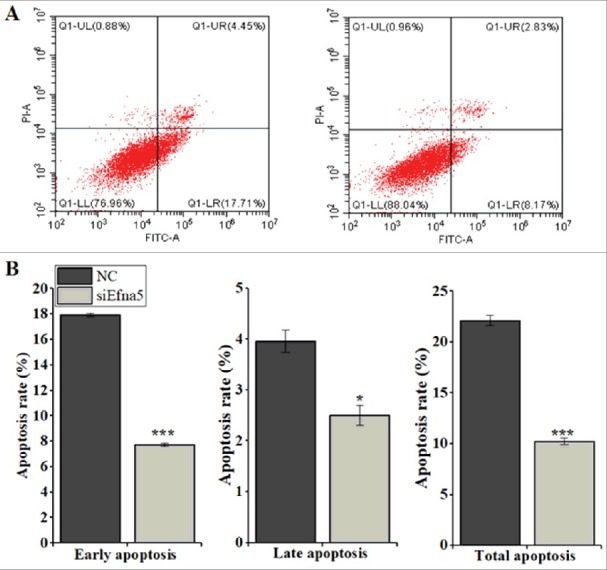
Downregulation of Efna5 inhibits mouse GC apoptosis in vitro. (A and B) Following transfection (5 × 105 cells per 6-well plate), annexin V-APC staining was used to detect dead cells by flow cytometry. The rates of apoptosis were analyzed separately for early, late, and total apoptosis and the results are expressed as means ± SEM for three (n = 3) independent biological replicates. *P < 0.05, ***P < 0.001. Quadrants: lower right (LR) = early apoptosis; upper right (UR) = late apoptosis; lower left (LL) = viable; Upper left (UL) = necrotic cells. NC, negative control; FITC, v-fluorescein isothiocyanate; PI, Propidium iodine.
Downregulation of Efna5 dysregulated the expression of apoptotic regulators in mouse GC in vitro
Briefly, in order to explore the underlying molecular mechanisms involved in the Efna5-mediated regulation of mouse GC apoptosis, the expression levels of key downstream apoptotic effector genes were analyzed at the mRNA and protein levels. The siRNA-mediated silencing of Efna5 resulted in significantly higher (P < 0.01) expression of the anti-apoptotic factor Bcl-2 at the mRNA and protein levels in comparison to that of the NC (Figure 4A), while the mRNA and protein expression levels of Caspase 3 (P < 0.01 and 0.05), Caspase 8 (P < 0.01), Bax (P < 0.01), and Tnf α (P < 0.01) were inhibited (Figure 4A and B).
Figure 4.
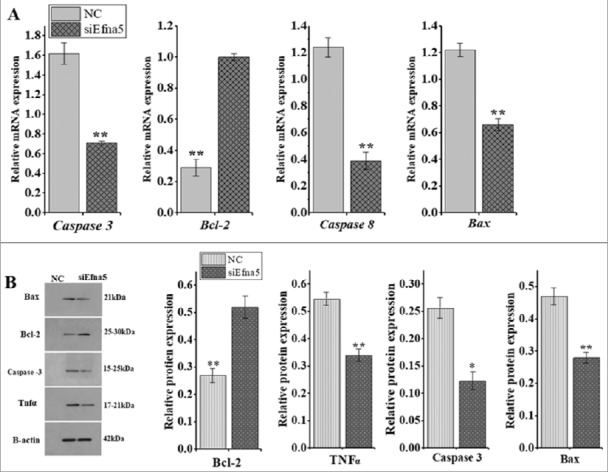
Silencing of Efna5 altered the expression levels of apoptosis regulators. (A) The effects of Efna5 silencing on the mRNA expression levels of Caspase 3, Caspase 8, Bcl-2, and Bax were determined by RT-qPCR and are presented as means ± SEM. (B) The effects of Efna5 silencing on the protein expression levels of Caspase 3, Caspase 8, Bcl-2, and Bax were analyzed by western blotting. β -actin signal intensity was used as an endogenous control for normalization. All results are presented as means ± SEM for three replicates, compared to the NC group *P < 0.05, **P < 0.01. Efna5, ephrinA5; GC granulosa cells; Bcl-2, B-cell lymphoma 2-associated x protein; Bax, BCL-2, B-cell lymphoma 2 family.
Silencing of Efna5 promotes in vitro mouse GC proliferation without affecting cell cycle progression
Following the in vitro siEfna5-mediated downregulation of Efna5, the proliferation and cell cycle progression of GC were examined. A CCK-8 assay demonstrated that the depletion of Efna5 leads to greater mouse GC proliferation than that of the NC group (P < 0.001, Figure 5D). Additionally, RT-qPCR and western blot analyses showed increased (P < 0.01) expression levels of the proliferation marker, Pcna, at the mRNA (Figure 5C) and protein levels (Figure 5A and B). No significant differences were observed between siEfna5- and NC-transfected GC with respect to cell cycle progression (P < 0.05), as determined by Annexin V-FITC/PI staining (Figure 5E).
Figure 5.
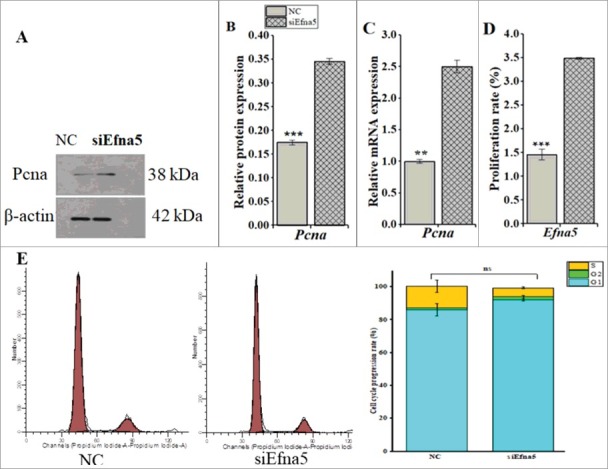
Downregulation of Efna5 promotes mouse GC proliferation in vitro, without affecting cell cycle progression. (A, B, C, D) The proliferation capacity of siEfna5-transfected mouse GC was evaluated by staining with the CCK-8 kit and determination using a multimode plate reader and was further confirmed by analyzing the protein and mRNA expression levels of Pcna. (E) Mouse GC were transfected with siEfna5 and incubated for 48 h, cells were saturated in PI, and the GC cycle was determined by FACS. All results are shown as means ± SEM and error bars with asterisks indicate significance (**P < 0.01 and ***P < 0.001). Efna5, ephrinA5; GC, granulosa; NC, negative control; ns, non-significant.
Inhibition of Efna5 regulates mouse GC proliferation by activating the ERK and Akt phosphorylation pathways
We next explored the pathways by which Efna5 influences GC proliferation. To investigate the downstream signaling pathways affecting cell proliferation, western blotting was conducted to assess the phosphorylation levels of extracellular kinase regulated protein 1 and 2 (ERK1/2) and Akt Serine/Threonine Kinase (Akt). The knockdown of Efna5 elevated the phosphorylation levels of Akt and ERK1/2 as compared to those of the NC (P < 0.001 and P < 0.01, respectively; Figure 6A).
Figure 6.
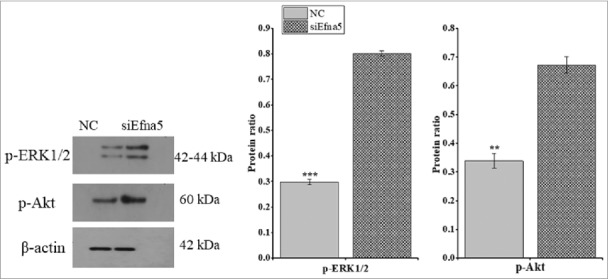
Downregulation of Efna5 activated Akt and ERK1/2 phosphorylation in mouse GCs in vitro. Protein extracts were collected 48-h post-transfection with siEfna5 and NC, and the phosphorylated protein levels were determined by western blotting and normalized against β-actin levels. The results are presented as means ± SEM and error bars with asterisks indicate significance (**P < 0.01 and ***P < 0.001). Efna5, ephrinA5; GC, granulosa; ERK1/2, extracellular kinase regulated protein; Akt, Akt Serine/Threonine Kinase.
Knockdown of Efna5 alters the concentration of steroidogenesis and expression of oocyte-secreted factors in mouse GC in vitro
Basal estradiol production was increased (P < 0.01) at 48 h after siEfna5 transfection in cultured medium within mouse GC, while progesterone levels were unaltered (Figure 7A and B). To assess the role of efna5 in ovarian function, the mRNA expression levels of growth differentiation factor 9 (GDF9) and bone morphogenetic (BMP15) genes, oocyte-related factors, were quantified by RT-qPCR. The suppression of Efna5 by siRNA significantly upregulated and downregulated the expression levels of BMP15 and GDF9 (P < 0.001 and P < 0.01, respectively; Figure 7), suggesting that role of ephrin: Ephrin signaling in oocyte development and intraovarian function.
Figure 7.
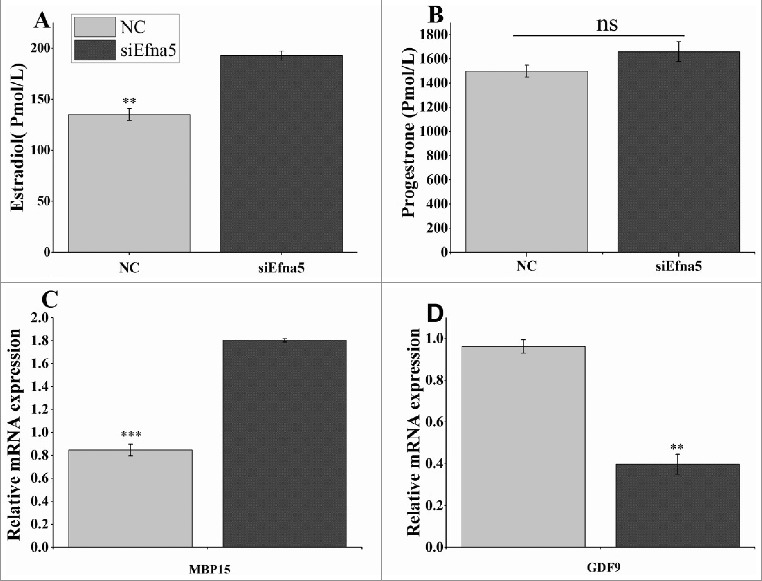
Effect of Efna5 knockdown on the mRNA expression of genes related to oocyte development and steroidogenesis in mouse GC in vitro. (A and B) The mRNA expression levels of BMP15 and GDF9 were quantified by real-time PCR following siEna5 transfection. (C and D) At 48 h after the transfection of siEfna5 and NC, serum was collected and the concentrations of estradiol and progesterone were determined using the mlbio kit. Results were obtained for triplicates and are presented as means ± SEM. **P < 0.01, ns, non-significant. NC, negative control; siEfna5, EphrinA5 siRNA; GCs, granulosa cells; GDF9, growth differentiation factor 9; BMP15, bone morphogenetic protein-15.
3. Discussion
Ephrin-Eph signaling has been implicated in several normal and pathological functions and most of its working principles are scientifically matured in cellular functions such as axon guidance [20], neurogenesis [19,33,34], and carcinogenesis [24,35]. However, knowledge on the biological functions of ephrin–Eph signaling in nearly all aspects of female reproduction is still in its infancy, despite its remarkable interplays and expressions in the mammalian ovary. In the global Efna5-knockout female mouse the mRNA expression levels of key genes involved in folliculogenesis were altered [32]. Paralleling to this study in a bovine transcriptome analysis, higher mRNA expression of Efna5 was reported in atretic follicles [30]. Consequently, we hypothesized that Efna5 regulates critical GC functions, such as apoptosis, proliferation, cell cycle progression, and intraovarian functions.
In the present study, using an in vitro model, we demonstrated the biological function and regulatory network of Efna5 in mouse primary GC, extending its roles beyond its well-known biological boundary, axon guidance [20,36]. In this experiment, siRNA was employed to disrupt the transcriptional and post-transcriptional abundance of Efna5 and to examine its physiological role in mouse GC. The expression of Efna5 was effectively inhibited by siEfna5 both at mRNA and protein levels. Concomitantly, its downstream cognate receptors (Epha5, Epha3, Epha8, and Ephb2) were also downregulated. Therefore, it is likely that Efna5 is involved in GC apoptosis and proliferation via forward signaling (ephrin A5–Eph receptor).
Apoptosis and proliferation are crucial cellular physiological processes during ovarian follicular development [9,37–39]. Notably, the crosstalk between cell death and survival signals is vital in regulating follicular cellular development and ultimately determines whether the follicles undergo atresia or ovulation, which is regulated by several ovarian factors [9,40]. In this regard, the potency of Efna5 and its receptors in regulating GC are not investigated. This study demonstrated, for the first time, that Efna5 is capable of inducing massive apoptosis, not only in neural tissue [41] but also in mouse GC and therefore it is a pro-apoptotic factor in immature mouse GC. In the present experiment, the downregulation of Efna5 promoted the survival of mouse GC by inhibiting apoptosis in vitro (Figure 3A and B). In support of this result, higher numbers of follicles were observed in Efna5 knockout immature superovulated mouse compared to wild-type mouse [32], leading to the ovulation of more follicles. In another study, elevated expression of Efna5 in early mouse embryonic neural cells [42]and recombinant Efna5-induced activation of Eph receptor A7 in mouse cortical progenitors cells [34] resulted in the induction of apoptosis. Furthermore, higher Efna5 expression was observed by transcriptome profiling of GC in bovine atretic follicles, indicating the involvement of ephrin-Eph signaling in GC apoptosis.
To further understand the mechanism underlying Efna5-mediated apoptosis, the mRNA and protein expression levels of key apoptosis regulators were analyzed. The downregulation of Efna5 suppressed the mRNA and protein expression levels of Caspase 8, Caspase 3, and Bax, though promoted Bcl-2 expression (Figure 3A and B). Moreover, the protein level of Tnfα was also inhibited (Figure 3B), suggesting that ephrin–Ephrin signaling cues the activation of cell death receptors concomitantly with Caspase 8, thereby promoting apoptosis. From the cellular membrane localization of Efna5 and its promiscuous interactions with Eph receptors, the observed extrinsic or Caspase 8-dependent apoptosis pathway and massive death of GC are plausible.
Our results provide evidence that ephrin–Eph signaling promotes apoptosis in a Caspase 8-dependent manner in ovarian follicular cells. In support of this observation, UV-induced apoptosis is mediated by Epha2 in a Caspase 8-dependent manner in mouse fibroblast cells [43]. However, Park et al. demonstrated that the ectopic overexpression of Efna5 induces massive apoptosis in mouse embryo neural epithelial cells by activation of Caspase 3, although failed to delineate the causal apoptotic pathway [18].
Granulosa cells proliferation was examined following the downregulation of Efna5 in vitro. In the current experiment 48 h incubation time was used to evaluate the GC proliferation based on our earlier in vitro proliferation result for mouse [44] and bovine [45] GC, suggesting that the time required for proliferation of cells depend on inherent biochemical and physiological property of the cells. The proliferation capacity of siEfna5-transfected mouse GC was significantly greater than that of the NC group, suggesting anti-proliferative behavior of Efna5 in mouse GC. The mRNA and protein expression levels of the proliferation marker Pcna [46] also increased in response to the downregulation of Efna5 (Figure 5 A, B,C and D), reinforcing the results of our proliferation assay. Currently, the molecular mechanism by which ephrin–Eph signaling influences mouse GC proliferation is not understood; accordingly, we further examined how Efna5 regulates GC proliferation. ERK and Akt are expressed in almost all cells and play essential roles in cell proliferation, cell death, and differentiation [47]. Our results demonstrated that the knockdown of Efna5 activates the phosphorylation of ERK 1/2 and Akt, thereby prompting mouse GC proliferation. In agreement with our findings, Efna5 regulates neurogenesis and angiogenesis via the phosphorylation of ERK and Akt in a non-ovarian cellular model, i.e., a temporal lobe epilepsy mouse model. Additionally, insulin-like growth factor 1 induces the activation and phosphorylation of Akt and thereby promotes porcine GC proliferation [48] and luteinizing human GC [49]. Earlier studies have reported that ephrin–Eph signaling regulates the proliferation of non-reproductive cells via the activation of ERK1/2 phosphorylation [50] and inhibition of phosphorylation [51]. These findings indicate that ephrin–Eph signaling is a negative or positive regulator of the phosphorylated ERK pathway, depending on the cell context. Surprisingly, in this experiment despite cell apoptosis and proliferation were influenced by downregualtion of Efna5 this did not affect cell cycle progression (Figure 5E). In line with our result ectopic over expression of Efna5 in mouse embryo led to cause excessive apoptosis neural epithelial cell leading to brain malformation while cell cycle progression rate was remained intact [18]. The current downregulation of Efna5 may be insufficient to disrupt cell cycle progressions or the forwarding signaling (efna5-Eph signaling) may not transmit the signals to cell cycle progression, thus further investigation is warranted for clarification.
Regulation of ovarian estradiol and progesterone levels by ephrin–Eph signaling is not well recognized in the current scientific literature. In the current study, the downregulation of Efna5 promoted the release of estradiol without influencing the progesterone level in the culture media of GC in vitro (Figure 7C and D). Presumably, the increased basal level of estradiol is in agreement with the fact that the silencing of Efna5 suppressed apoptosis, sustaining the follicular growth (Figure 3A). This finding agrees with those of a recent study in which the progesterone concentration remained intact in the blood samples of Efna5-knockout female mouse, though not with estradiol levels [32]. GDF9 and BMP15 are a subset of growth factors of oocyte origin [52]. They are involved in ovarian function [1,53] and are important in the subsequent growth of primary follicles. As depicted in Figure 7A and B, the mRNA expression levels of BMP15 and GDF9 were up- and downregulated in Efna5-silenced GC respectively. Since Efna5 is a neural gene with recently-identified functions in reproductive cells, the observed dysregulation of BMP15 and GDF9 can be the mechanism by which of Efna5 functions to maintain proper GC proliferation and effective female fertility [54]. The inhibition of BMP15 disrupts CC expansion, leading to impaired ovulation [55]. In our study, the elevated expression of BMP15 is in agreement with previous results, where the higher number of oocytes were recorded in mouse lacking Efna5 could be linked to BMP15 expression levels [32].
Conclusions
Our results provide the first components of in vitro evidence that ephrin–Eph signaling novel regulator of GC apoptosis in a Caspase 8-dependent manner in mouse GC. We confirmed that the siRNA-mediated inhibition of Efna5 suppressed GC apoptosis, supporting its pro-apoptotic function in female mouse ovarian cells. Furthermore, Efna5 mediated mouse GC proliferation through the activation of the p-Akt and p-ERK1/2 pathways without inducing significant changes in cell cycle progression.
In our study, the expression levels of paracrine factors (GDF9 and BMP15) and steroidogenesis hormones (progesterone and estradiol) were also altered following the downregulation of Efna5. Although our study provide novel role of Efna5 beyond its recognized biological border, the study has limitation firstly, the result is not confirmed in vivo study; secondly, eph: Eph signaling is mainly known to act through bidirectional signaling thus our study did not address whether the observed result is through bidirectional or only forward signaling thus further study will be required
Collectively, these results demonstrated the physiological role of efna5: Eph signaling in an ovarian cell, suggesting that additional studies should focus on its potential to protect infertility in female mammals as well as its use for the development of new therapeutic approaches.
Materials and methods
Management of experimental animals
Wild type female kumming mouse (KM) of three weeks old was used for the study. Experimental mouse were purchased from Liaoning Longevity Biotechnology Co. Ltd. (Liaoning, China) and were managed in the experimental center of animal breeding genetics and reproduction laboratory of Huazhong Agricultural University (Wuhan, China). The experimental mouse were housed at 22°C with 12 h alternating light/dark conditions and was provided food and water ad libitum. Experiments were conducted in accordance with the guidelines established by the ethics committee of the Hubei Research Center of Experimental Animals (Approval ID: SCXK (Hubei) 2008-0005).
Mouse GC isolation and culture
Ovaries from 21-day post-natal female KM were collected and pooled in a 65-mm cell culture dish containing phosphate-buffered saline (PBS) (Hyclone, Logan, UT, USA). The GC samples were isolated using needle puncture methods. Finally, cells were filtered by removing follicular impurities and debris using a 70-µm nylon mesh cellular strainer (Life Sciences, Corning, Inc., Durham, NC, USA). Separated GC were centrifuged at 15,100 rpm for 5 min three times, followed by washing with Dulbecco's modified Eagle's medium, F12 (DMEM/F12) (Hyclone, Inc., UT, USA). The supernatants were discarded and the pellets were rescued for culturing. Eventually, the cells were cultured into 6-well plates (Thermo Scientific, Jiangsu, P.R. China) in DMEM/F12 (culture medium) supplemented with 10% fetal bovine serum (FBS; Hyclone) and 1% penicillin/streptomycin (Hyclone). The seeded GC were incubated at 1 × 106 cells/mL at 37°C, 5%CO2, and 95% O2 for 48 h, and the medium was replaced with fresh medium at 24 h.
EphrinA5 siRNA transfection
EphrinA5 siRNA, referred as siEfna5 in this manuscript, contained a pool of three target-specific siRNAs designed to down-regulate mouse Efna5 gene expression (Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA) (Table 1). The negative control (NC) scrambled siRNA did not target any sequences (sense 5′-uucuccgaacgugucacgutt-3′ and antisense 5′-acgugacacguucggagaatt-3′) and was purchased from Shanghai Gene Pharma Co., Ltd. (Shanghai, China). Lipofectamine® RNAiMAX reagent was purchased from Invitrogen (Life Technology, Inc., Carlsbad, CA, USA). Both siEfna5 and RNAiMAX were diluted in OPTI-MEM® (Gibco, USA) following the recommendations of the manufacturer and used for transfection.
Table 1.
Pool of EphrinA5 siRNAs used in the experiment.
| Cat No | Sense | Antisense |
|---|---|---|
| Sc-39435A | GGAAGAAGGUCCUGUCUAAtt | UUAGACAGGACCUUCUUCCtt |
| sc-39435B | CCUUCAGCCUGAUUUCUAAtt | UUAGAAAUCAGGCUGAAGGtt |
| sc-39435C | GAUCGGAAAUUGCUUAGAAtt | UUCUAAGCAAUUUCCGAUCtt |
RNA extraction and reverse-transcription polymerase chain Reaction (RT-PCR)
Total RNA was extracted from cultured GC after 48 h incubation using the E.Z.N.A. Total RNA Kit I (R6834-02; Omega Bio-tek, Norcross, GA, USA) according to the manufacturer's protocol. The purity and concentration of RNA were determined by spectrophotometry using the Nanodrop 2000 Analyzer (Thermo Scientific, Wilmington, DE, USA) to determine absorbance at 260/280. Total RNA with absorbance at 260/280 of 1.8–2.1 was chosen for complementary DNA (cDNA) synthesis. Consequently, 1 µg of total RNA was used to perform reverse transcription using the FastKing RT Kit (Tiangen Biotech, Co., Ltd., Beijing, China) based on the manufacturer's guidelines.
Quantitative real-time PCR
For all target genes, primers were generated using Primer 5.00 (Primer Biosoft, Palo Alto, CA, USA) (Table 2). Gene expression levels were determined by quantitative real-time PCR (qRT-PCR) using SYBR-Green Master Mix (Qiagen, Hilden, Germany). QRT-PCR was performed using 96-well plates (Light Cycler 480 Multiwell Plate 96; Roche, Indianapolis, IN, USA) on the Bio-Rad CFX96 Real-Time system C100 Touch Thermal cycler (Bio-Rad, Inc., Hercules, CA, USA). In each reaction, a 10 µL volume was used, consisting of cDNA, RNase-free water, sense and antisense primers, and SYBR-Green Master Mix (QIAGEN, Germany). All reactions were performed in triplicate. The fold change in target gene expression was calculated based on earlier described model [56] and expression levels were normalized against β-actin, as an endogenous control.
Table 2.
List of primer sequences for quantitative real-time PCR.
| Gene symbol | AC/No | Primer sequences | Product length (bp) | Annealing Temp. (°C) |
|---|---|---|---|---|
| Efna5 | NM_207654.2 | F: 5′-TAGTTACAGGTTTGTGGGTTCG-3′ R: 5′- GGTGTTTCATCATTGGTTAGAG-3′ |
230 | 61 |
| Epha5 | NM_007937.3 | F: 5′- GCTGGTTCCCATTGGGAAAT-3′ R: 5′- CACATTGGTGACTGGAGAAGGA-3′ |
95 | 57.1 |
| Epha3 | NM_010140.3 | F: 5′-TCACACTGCGGGACTGTAAC-3′ R: 5′-TTCAGAATGCGATCCCCGAG-3′ |
187 | 58.7 |
| Epha8 | NM_007939.2 | F: 5′- CTCAAACCAGGCACTCGCTA-3′ R: 5′- CGATGGTCCGAGTGTCGTAG-3′ |
127 | 60.5 |
| Ephb2 | NM_001290753.2 | F: 5′-ACCTCAGTTCGCCTCTGTGAA-3′ R: 5′- GGCTCACCTGGTGCATGAT-3′ |
77 | 59 |
| Pcna | NM_011045.2 | F: 5′- GTGGATAAAGAAGAGGAGGCG-3′ R: 5′-TGTAGGAGACAGTGGAGTGGC-3′ |
111 | 60.5 |
| Bax | NM_007527 | F: 5′- GCCTCCTCTCCTACTTCGG-3′ R: 5′-AAAAATGCCTTTCCCCTTC-3′ |
187 | 58.5 |
| Bcl2 | NM_009741 | F: 5′-TCTCTCGTCGCTACCGTCG-3′ R: 5′- CCCAGTTCACCCCATCCCT-3′ |
123 | 58 |
| Caspase8 | NM_009812.2 | F: 5′-ACTCGGCCACAGGTTACGG-3′ R: 5′-TGTGTGGGATGTAGTCCAAGC-3′ |
137 | 61.6 |
| Gdf9 | NM_008110 | F: 5′- CGGCTCCATCGCTTACAAA-3′ R: 5′- CTTCCCCCGCTCACACAGT-3′ |
187 | 55.1 |
| Bmp15 | NM_009757 | F: 5′- GAAAATGGTGAGGCTGGTAA-3′ R: 5′- GATGAAGTTGATGGCGGTAA-3′ |
152 | 57.7 |
| β-actin | NM_ 007393.5 | F: 5′- CACGATGGAGGGGCCGGACTCATC-3′ R:5′-TAAAGACCTCTATGCCAACACAGT-3′ |
241 | 59 |
Protein extraction and western blotting
Following 48 h of incubation of siEfna5-transfected GC (5 × 105 cells per 6-well plate), total protein was extracted. The GCs were washed on ice with PBS and lysed with the protein extraction reagent RIPA buffer (100 µL; 25 mM Tris HCl, pH 7.6, 150 mM NaCl, 1% NP-40, 1% sodium deoxycholate, 0.1% SDS) supplemented with 1% phenylmethanesulfonyl fluoride (PMSF), 1% protease inhibitor (P8340; Sigma, St. Louis, MO, USA), and 1% phosphatase inhibitor (P0040; Sigma). The lysed cells were scrambled and centrifuged at 12,000 rpm for 5 min at 4°C and the supernatants were maintained at -80°C until use. The protein concentration was determined using the BCA Protein Assay Kit (Pierce, Rockford, IL, USA).
Equal volumes (40 µg) of protein samples were separated by 12% sodium dodecyl sulfate polyacrylamide gel electrophoresis (Sigma Aldrich) transferred to 0.45-µm polyvinylidene difluoride membranes (Millipore, Billerica, MA, USA) and incubated with specific antibodies. In brief, the proteins on the membranes were blocked to avoid non-specific binding using 5% skimmed milk diluted in Tris-buffered saline at room temperature for 2 h. Consequently, the membrane was incubated at 4°C overnight with the following primary anti-bodies: Efna5 (1:200, 380400; Life Technology); Bax (1:1000, Ab32503; Abcam, Cambridge, UK), Pcna, Bcl-2, Tnfα (1:1000; San Ying Bio-Tech, Wuhan, China), p-Akt 1:2000 (13038S; CST, Danvers, MA, USA), p-ERK1/2 (1:1000, 4370S; CS, USA), and β-actin as a control (1:200, BM0627; BosterBio, Wuhan, China). Enhanced chemiluminescence (NCI5079; Bio-Rad, USA) was used to identify the target proteins in each sample according to the manufacturer's protocol. Gel-Pro Analyzer version 4 (Media Cybernetics, Rockville, MD, USA) and ImageJ software [57] were used to capture images and determine bands respectively and the results were normalized against β-actin.
Cell apoptosis detection
Primary mouse GC were seeded in 6-well plates at 5 × 105 cells/well for 48 h. Consequently, the cells were transfected with siEfna5 and NC, followed by incubation for 48 h at 37°C and 5%CO2. GC apoptosis was determined by flow cytometry using the Annexin V-fluorescein isothiocyanate (FITC)/iodine (PI) Apoptosis Detection Kit (KeyGen Biotech, Nanjing, China) following the manufacturer's instructions. The cells were lysed using trypsin, washed with PBS, and re-suspended in binding buffer solution. The cells were stained with FITC and PI for 15 min and incubated at room temperature. Finally, the apoptosis rate was determined by flow cytometry using the FACSVerse Calibur (BD Biosciences, San Jose, CA, USA) following the manufacturer's protocol. The results of the apoptosis assay were verified by analyzing the mRNA and protein expression levels of apoptosis-related molecules.
Cell proliferation assay
For proliferation studies, siEfna5 and NC-transfected GC were seeded at 2 × 104 cells/well in a 96-well plate and incubated in culture medium for 48 h. The incubation time used to evaluate GC proliferation was based on our previous laboratory result [44,45]. The GC proliferation capacity was assayed using the Cell Counting Kit- 8 (CCK- 8) (Donjindo Molecular Technologies, Inc., Rockville, MD, USA) and the viable cells were determined based on the quantity of fromazan dye, which is a cell viability indicator, following the manufacturer's instructions. A 100-µL CCK-8 solution was added to culture medium (10%) in each well and incubated for 2 h at 37°C and 5% CO2. Then, the GCs were loaded onto a multimode plate reader (PerkinElmer, EnSpire, Waltham, MA, USA) and the absorbance was measured at 450 nm for each experimental group. This result was further verified by determining the protein and mRNA expression levels of the proliferation marker Pcna.
Cell cycle assay
The siEfna5 – and NC-transfected mouse primary GCs were harvested at 48 h post-transfection using trypsin for three minutes at 37°C, followed by centrifugation at 15,000 rpm for five minutes. The harvested cells were washed in 70% ethanol overnight at 4°C and stained with 100 µL of RNAse and 400 µL of PI solution for 30 minutes. Cell cycle progression was determined using the FACSVerse Calibur (BD Biosciences, CA, USA) and the proportion of cells in differing phases of the cell cycle were analyzed using ModFit LT, DNA cell-cycle (version 2) for three independent experiments.
Enzyme-linked Immunosorbent assay for steroid hormones
After transfection with siEfna5 and NC, the GC were incubated for 48 h and culture medium was collected and subjected to centrifugation at 15,000 rpm for 5 min. The concentrations of oestradiol-17β (E2) and progesterone (P4) were measured using the Iodine Radioimmunoassay Mouse ELISA Kit purchased from MLBIO Biotechnology Co., Ltd. (Shanghai, China). Hormone assays were conducted according to the manufacturer's guidelines.
Statistical analysis
Data were obtained from three independent biological parameters, such as apoptosis, cell proliferation, cell cycle progression, and steroidogenesis levels with mean values used to determine statistical significance. All data were analyzed using Statistical Package Software for Social Science (version 23; SPSS, Inc., Cary, NC, USA). Mean values for the treatment and control groups were evaluated using Student's t-tests. A value of P < 0.05 was considered significant. All results are depicted on graphs as means ± SEM using Origin Pro version 17.
Funding Statement
This work was supported by Natural Science Foundation of China (31772602) and the Earmarked Fund for Modern Agro-industry Technology Research System (CARS-37-04B). Both them are highly appreciated.
Acknowledgments
Financial support from both the Natural Science Foundation of China (31772602) and the Earmarked Fund for Modern Agro-industry Technology Research System (CARS-37-04B) are highly appreciated.
Disclosure of Potential Conflicts of Interest
The authors declare no conflict of interest.
References
- [1].Emori C, Sugiura K. Role of oocyte-derived paracrine factors in follicular development. Anim Sci J.. 2014;85(6):627–633. doi: 10.1111/asj.12200. PubMed PMID: 24717179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Hillier SG. Paracrine support of ovarian stimulation. MHR: Basic Science of Reproductive Medicine. 2009;15(12):843–850. doi: 10.1093/molehr/gap086. [DOI] [PubMed] [Google Scholar]
- [3].Salustri A, Camaioni A, D'Alessandris C. Endocrine and paracrine regulation of cumulus expansion. Zygote. 1996;4(4):313–315. doi: 10.1017/S0967199400003312. PubMed PMID: 9153772. [DOI] [PubMed] [Google Scholar]
- [4].Reis FM, Cobellis L, Luisi S, et al. . Paracrine/autocrine control of female reproduction. Gynecol Endocrinol. 2000;14(6):464–475. doi: 10.3109/09513590009167720. PubMed PMID: 11228069. [DOI] [PubMed] [Google Scholar]
- [5].Rimon-Dahari N, Yerushalmi-Heinemann L, Alyagor L, et al. . Ovarian Folliculogenesis. Results Probl Cell Differ. 2016;58:167–190. doi: 10.1007/978-3-319-31973-5_7. PubMed PMID: 27300179. [DOI] [PubMed] [Google Scholar]
- [6].Worku T, Rehman ZU, Talpur HS, et al. . MicroRNAs: new insight in modulating follicular atresia: a review. Int J Mol Sci. 2017;18(2):333. doi: 10.3390/ijms18020333. PubMed PMID: 28208755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Wang N, Zhao F, Lin P, et al. . Knockdown of XBP1 by RNAi in Mouse Granulosa Cells Promotes Apoptosis, Inhibits Cell Cycle, and Decreases Estradiol Synthesis. Int J Mol Sci. 2017;18(6):1152. doi: 10.3390/ijms18061152. PubMed PMID: 28555054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Kranc W, Brazert M, Ozegowska K, et al. . Expression profile of genes regulating steroid biosynthesis and metabolism in human ovarian granulosa cells-a primary culture approach. Int J Mol Sci. 2017;18(12):2673. doi: 10.3390/ijms18122673. PubMed PMID: 29232835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Kaipia A, Hsueh AJ. Regulation of ovarian follicle atresia. Annu Rev Physiol. 1997;59:349–363. doi: 10.1146/annurev.physiol.59.1.349. PubMed PMID: 9074768. [DOI] [PubMed] [Google Scholar]
- [10].Matsuda F, Inoue N, Manabe N, et al. . Follicular growth and atresia in mammalian ovaries: regulation by survival and death of granulosa cells. J Reprod Dev. 2012;58(1):44–50. doi: 10.1262/jrd.2011-012. PubMed PMID: 22450284. [DOI] [PubMed] [Google Scholar]
- [11].Tiwari M, Prasad S, Tripathi A, et al. . Apoptosis in mammalian oocytes: a review. Apoptosis. 2015;20(8):1019–1025. doi: 10.1007/s10495-015-1136-y. PubMed PMID: 25958165. [DOI] [PubMed] [Google Scholar]
- [12].Manning G, Whyte DB, Martinez R, et al. . The protein kinase complement of the human genome. Science. 2002;298(5600):1912–1934. doi: 10.1126/science.1075762. PubMed PMID: 12471243. [DOI] [PubMed] [Google Scholar]
- [13].Flanagan JG, Vanderhaeghen P. The ephrins and Eph receptors in neural development. Annu Rev Neurosci. 1998;21:309–345. doi: 10.1146/annurev.neuro.21.1.309. PubMed PMID: 9530499. [DOI] [PubMed] [Google Scholar]
- [14].Andres AC, Ziemiecki A. Eph and ephrin signaling in mammary gland morphogenesis and cancer. J Mammary Gland Biol Neoplasia. 2003;8(4):475–485. doi: 10.1023/b:jomg.0000017433.83226.22. PubMed PMID: 14985642. [DOI] [PubMed] [Google Scholar]
- [15].Kullander K, Klein R. Mechanisms and functions of Eph and ephrin signalling. Nat Rev Mol Cell Biol. 2002;3(7):475–486. doi: 10.1038/nrm856. PubMed PMID: 12094214. [DOI] [PubMed] [Google Scholar]
- [16].Unified nomenclature for Eph family receptors and their ligands, the ephrins. Eph Nomenclature Committee. Cell. 1997;90(3):403–404. doi: 10.1016/S0092-8674(00)80500-0. PubMed PMID: 9267020. [DOI] [PubMed] [Google Scholar]
- [17].Palmer A, Klein R. Multiple roles of ephrins in morphogenesis, neuronal networking, and brain function. Genes Dev. 2003;17(12):1429–1450. doi: 10.1101/gad.1093703. PubMed PMID: 12815065. [DOI] [PubMed] [Google Scholar]
- [18].Park E, Kim Y, Noh H, et al. . EphA/ephrin-A signaling is critically involved in region-specific apoptosis during early brain development. Cell Death Differ. 2013;20(1):169–180. doi: 10.1038/cdd.2012.121. PubMed PMID: 22976838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Shu Y, Xiao B, Wu Q, et al. . The Ephrin-A5/EphA4 Interaction Modulates Neurogenesis and Angiogenesis by the p-Akt and p-ERK Pathways in a Mouse Model of TLE. Mol Neurobiol. 2016;53(1):561–576. doi: 10.1007/s12035-014-9020-2. PubMed PMID: 25502292. [DOI] [PubMed] [Google Scholar]
- [20].Egea J, Klein R. Bidirectional Eph-ephrin signaling during axon guidance. Trends Cell Biol. 2007;17(5):230–238. doi: 10.1016/j.tcb.2007.03.004.PubMed. PMID: 17420126. [DOI] [PubMed] [Google Scholar]
- [21].Lin S, Gordon K, Kaplan N, et al. . Ligand Targeting of EphA2 Enhances Keratinocyte Adhesion and Differentiation via Desmoglein 1. Mol Biol Cell. 2010;21(22):3902–3914. doi: 10.1091/mbc.E10-03-0242. PubMed PMID: 20861311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Pasquale EB. Eph receptors and ephrins in cancer: bidirectional signaling and beyond. Nat Rev Cancer. 2010;10(3):165–180.doi: 10.1038/nrc2806. PubMed PMID: PMC2921274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Laussu J, Khuong A, Gautrais J, et al. . Beyond boundaries–Eph:ephrin signaling in neurogenesis. Cell Adh Migr. 2014;8(4):349–359. doi: 10.4161/19336918.2014.969990. PubMed PMID: 25482631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Xi HQ, Wu XS, Wei B, et al. . Eph receptors and ephrins as targets for cancer therapy. J Cell Mol Med. 2012;16(12):2894–2909. doi: 10.1111/j.1582-4934.2012.01612.x. PubMed PMID: 22862837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Pasquale EB. Eph-ephrin bidirectional signaling in physiology and disease. Cell. 2008;133(1):38–52. doi: 10.1016/j.cell.2008.03.011. PubMed PMID: 18394988. [DOI] [PubMed] [Google Scholar]
- [26].Davy A, Bush JO, Soriano P. Inhibition of gap junction communication at ectopic Eph/ephrin boundaries underlies craniofrontonasal syndrome. PLoS Biol. 2006;4(10):e315. doi: 10.1371/journal.pbio.0040315. PubMed PMID: 16968134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Konstantinova I, Nikolova G, Ohara-Imaizumi M, et al. . EphA-Ephrin-A-mediated beta cell communication regulates insulin secretion from pancreatic islets. Cell. 2007;129(2):359–370. doi: 10.1016/j.cell.2007.02.044. PubMed PMID: 17448994. [DOI] [PubMed] [Google Scholar]
- [28].Egawa M, Yoshioka S, Higuchi T, et al. . Ephrin B1 is expressed on human luteinizing granulosa cells in corpora lutea of the early luteal phase: the possible involvement of the B class Eph-ephrin system during corpus luteum formation. J Clin Endocrinol Metab. 2003;88(9):4384–4392. doi: 10.1210/jc.2002-021910. PubMed PMID: 12970314. [DOI] [PubMed] [Google Scholar]
- [29].Hatzirodos N, Irving-Rodgers HF, Hummitzsch K, et al. . Transcriptome profiling of granulosa cells of bovine ovarian follicles during growth from small to large antral sizes. BMC Genomics. 2014;15:24. doi: 10.1186/1471-2164-15-24. PubMed PMID: 24422759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Hatzirodos N, Hummitzsch K, Irving-Rodgers HF, et al. . Transcriptome profiling of granulosa cells from bovine ovarian follicles during atresia. BMC Genomics. 2014;15:40. doi: 10.1186/1471-2164-15-40. PubMed PMID: 24438529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Buensuceso AV, Deroo BJ. The ephrin signaling pathway regulates morphology and adhesion of mouse granulosa cells in vitro. Biol Reprod. 2013;88(1):25. doi: 10.1095/biolreprod.112.100123. PubMed PMID: 23242526. [DOI] [PubMed] [Google Scholar]
- [32].Buensuceso AV, Son AI, Zhou R, et al. . Ephrin-A5 Is Required for Optimal Fertility and a Complete Ovulatory Response to Gonadotropins in the Female Mouse. Endocrinology. 2016;157(2):942–955. doi: 10.1210/en.2015-1216. PubMed PMID: 26672804. [DOI] [PubMed] [Google Scholar]
- [33].Jiao JW, Feldheim DA, Chen DF. Ephrins as negative regulators of adult neurogenesis in diverse regions of the central nervous system. Proc Natl Acad Sci U S A. 2008;105(25):8778–8783. doi: 10.1073/pnas.0708861105. PubMed PMID:18562299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Depaepe V, Suarez-Gonzalez N, Dufour A, et al. . Ephrin signalling controls brain size by regulating apoptosis of neural progenitors. Nature. 2005;435(7046):1244–1250. doi: 10.1038/nature03651. PubMed PMID: 15902206. [DOI] [PubMed] [Google Scholar]
- [35].Pasquale EB. Eph receptors and ephrins in cancer: bidirectional signalling and beyond. Nat Rev Cancer. 2010;10(3):165–180.doi: 10.1038/nrc2806. PubMed PMID: 20179713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Lim YS, McLaughlin T, Sung TC, et al. . p75(NTR) mediates ephrin-A reverse signaling required for axon repulsion and mapping. Neuron. 2008;59(5):746–758. doi: 10.1016/j.neuron.2008.07.032. PubMed PMID: 18786358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Iijima K, Jiang JY, Shimizu T, et al. . Acceleration of follicular development by administration of vascular endothelial growth factor in cycling female rats. J Reprod Dev. 2005;51(1):161–168. doi: 10.1262/jrd.51.161. PubMed PMID: 15750308. [DOI] [PubMed] [Google Scholar]
- [38].Sargent KM, Lu N, Clopton DT, et al. . Loss of vascular endothelial growth factor A (VEGFA) isoforms in granulosa cells using pDmrt-1-Cre or Amhr2-Cre reduces fertility by arresting follicular development and by reducing litter size in female mice. PloS One. 2015;10(2):e0116332. doi: 10.1371/journal.pone.0116332. PubMed PMID: 25658474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Quirk SM, Cowan RG, Harman RM, et al. . Ovarian follicular growth and atresia: the relationship between cell proliferation and survival. J Anim Sc. 2004;82(E-Suppl):E40–52. doi: 10.2527/2004.8213_supplE40x. PubMed PMID: 15471814. [DOI] [PubMed] [Google Scholar]
- [40].Hubbard CJ, Oxberry B. Follicular atresia. In: Familiari G, Makabe S, Motta PM, Ultrastructure of the ovary. Boston, MA: Springer US; 1991. p. 273–285. [Google Scholar]
- [41].Jin Z, El-Deiry WS. Overview of cell death signaling pathways. Cancer Biol Ther. 2005;4(2):139–163. doi: 10.4161/cbt.4.2.1508. PubMed PMID: 15725726. [DOI] [PubMed] [Google Scholar]
- [42].Noh H, Park S. Over-Expression of Ephrin-A5 in Mice Results in Decreasing the Size of Progenitor Pool through Inducing Apoptosis. Mol Cells. 2016;39(2):136–140. doi: 10.14348/molcells.2016.2245. PubMed PMID: 26674965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Zhang G, Njauw CN, Park JM, et al. . EphA2 is an essential mediator of UV radiation-induced apoptosis. Cancer Res. 2008;68(6):1691–1696. doi: 10.1158/0008-5472.can-07-2372. PubMed PMID: 18339848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Talpur HS, Worku T, Rehman ZU, et al. . Knockdown of melatonin receptor 1 and induction of follicle-stimulating hormone on the regulation of mouse granulosa cell function. Reproductivebiology. 2017;17(4):380–388. doi: 10.1016/j.repbio.2017.10.005. PubMed PMID: 29097083. [DOI] [PubMed] [Google Scholar]
- [45].Riaz H, Dong P, Shahzad M, et al. . Constitutive and follicle-stimulating hormone-induced action of somatostatin receptor-2 on regulation of apoptosis and steroidogenesis in bovine granulosa cells. J Steroid Biochem Mol Biol. 2014;141:150–159. doi: 10.1016/j.jsbmb.2014.02.001. PubMed PMID: 24530462. [DOI] [PubMed] [Google Scholar]
- [46].Iatropoulos MJ, Williams GM. Proliferation markers. Experimental and Toxicologic Pathology: Official Journal of the Gesellschaft fur Toxikologische Pathologie. 1996;48(2-3):175–181. doi: 10.1016/s0940-2993(96)80039-x. PubMed PMID: 8672872. [DOI] [PubMed] [Google Scholar]
- [47].Lefloch R, Pouyssegur J, Lenormand P. Single and combined silencing of ERK1 and ERK2 reveals their positive contribution to growth signaling depending on their expression levels. Mol Cell Biol. 2008;28(1):511–527. doi: 10.1128/mcb.00800-07. PubMed PMID: 17967895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Westfall SD, Hendry IR, Obholz KL, et al. . Putative role of the phosphatidylinositol 3-kinase-Akt signaling pathway in the survival of granulosa cells. Endocrine. 2000;12(3):315–321. doi: 10.1385/endo:12:3:315. PubMed PMID: 10963053. [DOI] [PubMed] [Google Scholar]
- [49].Goto M, Iwase A, Harata T, et al. . IGF1-induced AKT phosphorylation and cell proliferation are suppressed with the increase in PTEN during luteinization in human granulosa cells. Reproduction. 2009;137(5):835–842. doi: 10.1530/rep-08-0315. PubMed PMID: 19225041. [DOI] [PubMed] [Google Scholar]
- [50].Miao H, Wei BR, Peehl DM, et al. . Activation of EphA receptor tyrosine kinase inhibits the Ras/MAPK pathway. Nat Cell Biol. 2001;3(5):527–530. doi: 10.1038/35074604. PubMed PMID: 11331884. [DOI] [PubMed] [Google Scholar]
- [51].Aoki M, Yamashita T, Tohyama M. EphA receptors direct the differentiation of mammalian neural precursor cells through a mitogen-activated protein kinase-dependent pathway. J Biol Chem. 2004;279(31):32643–32650. doi: 10.1074/jbc.M313247200. PubMed PMID: 15145949. [DOI] [PubMed] [Google Scholar]
- [52].Paulini F, Melo EO. The role of oocyte-secreted factors GDF9 and BMP15 in follicular development and oogenesis. Reprod Domest Anim. 2011;46(2):354–361. doi: 10.1111/j.1439-0531.2010.01739.x. PubMed PMID:21198974. [DOI] [PubMed] [Google Scholar]
- [53].Diaz FJ, Wigglesworth K, Eppig JJ. Oocytes determine cumulus cell lineage in mouse ovarian follicles. J Cell Sci. 2007;120(Pt 8):1330–1340. doi: 10.1242/jcs.000968. PubMed PMID: 17389684. [DOI] [PubMed] [Google Scholar]
- [54].Peng J, Li Q, Wigglesworth K, et al. . Growth differentiation factor 9:bone morphogenetic protein 15 heterodimers are potent regulators of ovarian functions. Proc Natl Acad Sci U S A. 2013;110(8):E776–E785. doi: 10.1073/pnas.1218020110. PubMed PMID: PMC3581982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Yoshino O, McMahon HE, Sharma S, et al. . A unique preovulatory expression pattern plays a key role in the physiological functions of BMP-15 in the mouse. Proc Natl Acad Sci. 2006;103(28):10678–10683. doi: 10.1073/pnas.0600507103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Hellemans J, Mortier G, De Paepe A, et al. . qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol. 2007;8(2):R19.doi: 10.1186/gb-2007-8-2-r19. PubMed PMID: 17291332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Abramoff MD, Magalhães PJ, Ram SJ. Image processing with image. J Biophotonics Int. 2004;11(7):36–42. [Google Scholar]


