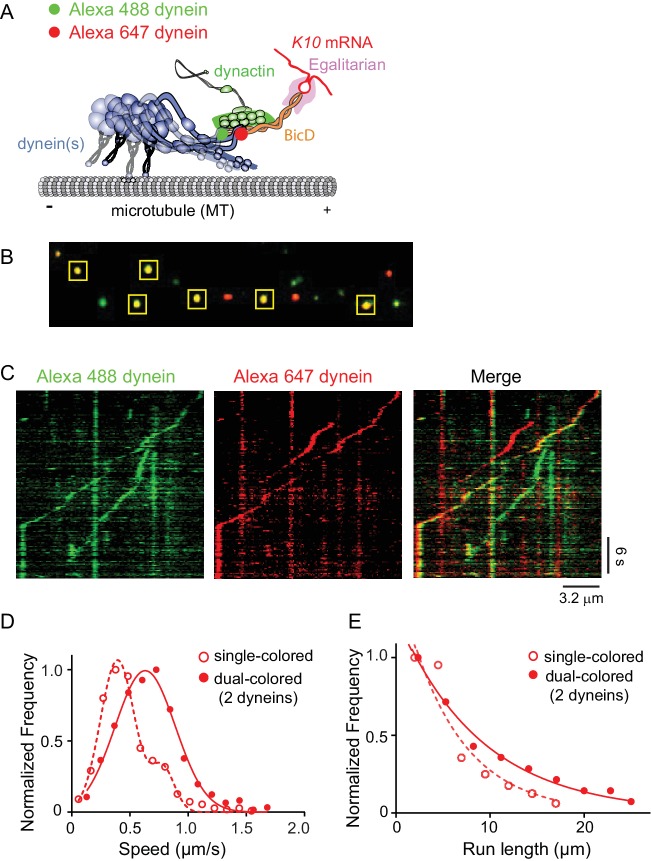
Figure 7. Recruitment of two dynein motors to the mRNP results in faster and longer runs.
(A) Schematic of the two-color experiment. Dynein was either labeled with Alexa 488 (green) or Alexa 647 (red) for the single-molecule pulldowns shown in panel B. mRNPs were formed by incubating with two dyneins per dynactin (figure adapted from [Reck-Peterson et al., 2018]). (B) Single-molecule pulldowns in the presence of AMP-PNP showed that 22% of complexes were dual-colored, implying that 44% of complexes had two dynein motors bound. (C) Kymographs showing single- and dual-colored runs, with dynein labeled with either a 525 or a 655 nm Qdot. Speed and run length are quantified in panels D and E. (D) The speed of dual-colored complexes (filled red circles) (0.63 ± 0.26 μm/s, n = 40), were compared to that of single-colored complexes (open red circles) (0.40 ± 0.14 μm/s and 0.74 ± 0.13 μm/s, n = 44). (E) Run lengths of dual-colored runs (filled red circles) (8.9 ± 0.5 μm, n = 40) were 53% longer than the single-colored complexes (open red circles) (5.8 ± 1.1 μm, n = 44, p=0.036, Kolmogorov–Smirnov test, mean ± SE of the fit). See Figure 7—source data 1.