Abstract
Sepsis, a life-threatening immune response to blood infections (bacteremia), has a ~30% mortality rate and is the 10th leading cause of US hospital deaths. The typical bacterial loads in adult septic patients are ≤100 bacterial cells (colony forming units, CFU) per mL blood, while pediatric patients exhibit only ~1000 CFU/mL. Due to the low numbers, bacteria must be propagated through ~24-hr blood cultures to generate sufficient CFUs for diagnosis and further analyses. Herein, we demonstrate that, unlike other rapid post-blood culture antibiotic susceptibility tests (ASTs), our phenotypic approach can drastically accelerate ASTs for the most common sepsis-causing gram-negative pathogens by circumventing long blood culture-based amplification. For all blood isolates of multi-drug resistant pathogens investigated (Escherichia coli, Klebsiella pneumoniae, and Acinetobacter nosocomialis), effective antibiotic(s) were readily identified within the equivalent of 8 hours from initial blood draw using <0.5mL of adult blood per antibiotic. These methods should drastically improve patient outcomes by significantly reducing time to actionable treatment information and reduce the incidence of antibiotic resistance.
Key terms: multidrug-resistant bacteria, antibiotic susceptibility test, flow cytometry, pre-blood culture, multidimensional statistics
Introduction
Leading to many deaths worldwide, sepsis can result from <100 CFU of bacteria/mL blood. Such low bacterial counts limits determinations of appropriate treatments, even in hospitals with advanced clinical diagnostics available. Early appropriate antibiotic treatment for bacteremia patients not only shortens hospitalizations and reduces antibiotic resistance proliferation, but it also lowers the incidence of septic shock and halves the fatality rate (2–6). As sepsis can be caused by any number of bacteria, effective treatment relies on the combination of bacterial identification and sensitivity profile determinations. While pathogen identification has been hastened to just a few hours post positive blood culture (7–11), antibiotic sensitivity tests (ASTs) still require an additional ~36–44 hrs, post blood culture (12). Although many flow cytometric-based ASTs have been proposed (13–22), development of general phenotypic ASTs has been elusive, due to the wide range of bacteria and antibiotics interactions, biovariability, noisy fluorescence background, and lack of reliable multidimensional statistical analyses to interpret small changes within the heterogeneous populations (17,19,23). These challenges have forced reliance on slow, but reliable blood cultures for amplifying populations, followed by multiple purification, growth, and antibiotic challenge cycles to guide treatment.
Developing ASTs directly from blood and circumvent the initial ~24 hr blood culture delay promise to drastically improve patient outcomes and impact public health efforts. Even though the ~109 mammalian blood cells/mL overwhelm any low-level bacteria signals (100~1000 CFU/mL) (24,25), bacterial presence determinations within blood samples have been reported by flow cytometry (26), microfluidics (27–29), and PCR (30,31). While most of these schemes detected the presence of bacterial genetic material, Hou et al. was able to detect mRNAs after pathogens were separated from blood in a microfluidic device (28). Like other molecular diagnosis approaches, however, they can only target known mRNA signatures for individual antibiotic resistance genetic markers for each bacterium-antibiotic pair. A phenotype-detecting flow cytometry-based AST specific for Y. pestis was proposed that relies on post-growth recovery of bacteria from a gel matrix and viability dye detection (32). Generalization, however, is problematic as careful bacterial recovery, significant post collection growth to reach ~106 CFU/mL, and user-dependent data gating were all needed to overcome the high scatter and fluorescence background. Additionally, viability dyes are known to produce false signals with various important bacteria/antibiotic pairs (17,19,23), and gating is highly subject to variations in day-to-day instrument fluctuations, alignment, and parameters, limiting application of this approach.
Recently, we developed a rapid, flow cytometry-based AST based on rigorous multidimensional statistical metrics (1) that matches the timescale of emerging post blood culture identification (~4hrs) (8–10). Our adaptive, multidimensional Probability Binned-signature Quadratic Form (PB-sQF) statistical distances (1) are ideal for quantifying small, but statistically significant changes relative to paired controls, even within broad, multidimensional flow cytometry datasets. PB-sQF calculates the true linear distance between any two multidimensional histograms, thereby enabling rapid direct comparisons of changes within heterogeneous populations, relative to their paired controls. Our prior order-of-magnitude improvement in post-blood culture time-to-result can in most cases be done label-free, and works with bacteria-antibiotic combinations that had failed with previous cytometric tests (1).
Without blood culture-based amplification, the highly disadvantageous bacteria:mammalian cell ratio, even in patients with bacteremia, demands that phenotypic ASTs remove nearly all mammalian cell background, without killing the bacteria. Additionally, sufficient bacteria must be recovered to allow assaying with multiple antibiotics at various concentrations, suggesting that at least some amplification, or a higher volume of blood (at ~100 CFU/mL), is needed. Because time is critical in ensuring appropriate treatment for patient survival (33) and reducing antibiotic resistance proliferation (6), we avoid the need for lengthy blood culture by utilizing saponin to complex with cholesterols and induce hemolysis (34,35), without affecting bacterial growth or morphology (36). This selective blood cell lysis enables even very small numbers of bacteria to be directly collected from the blood and enriched in blood-free growth medium for cytometric detection. Our robust statistics then enable quantification of very few bacterial counts, such that much shorter growth and antibiotic sensitivity times can be achieved.
Materials and Methods
Bacterial strains and antibiotics
All clinical isolates (E. coli strains Mu14S and Mu890, K. pneumoniae strains Mu670 and Mu55, A. nosocomialis strains M2) were obtained from the Georgia Emerging Infection Program (GEIP). The MIC of each isolate was determined by a clinical microbiology laboratory using post blood culture automated AST and confirmed using broth microdilution in our laboratory. The MICs of each strain were measured to be as follows. For A. nosocomialis strain M2, 1 μg/mL tetracycline, 2 μg/mL gentamicin, and > 1024 μg/mL ampicillin. For E. coli strain Mu890, 1 μg/mL tetracycline, 8 μg/mL gentamicin, and > 1024 μg/mL ampicillin. For E. coli strain Mu14S, > 64 μg/mL tetracycline, 8 μg/mL gentamicin, and > 1024 μg/mL ampicillin. For K. pneumoniae strain Mu55, > 64 μg/mL tetracycline, 1 μg/mL gentamicin, and > 1024 μg/mL ampicillin. For K. pneumoniae strain Mu670, 2 μg/mL tetracycline, 4 μg/mL gentamicin, and > 1024 μg/mL ampicillin.
FAST
To simulate a blood specimen from a patient with bacteremia, the isolates were grown, diluted to the desired CFU/mL in blood (ZenBio, Research Triangle Park, NC) and saponin was added to achieve the final diluted sample. Initial bacterial cultures were prepared using Luria-Bertani (LB) broth for (E. coli) or cation-adjusted Mueller-Hinton broth (CAMHB). The fresh bacterial cultures started from ~0.05 optical density (OD) by inoculating a 6-mL fresh growth medium with overnight culture were grown in an incubator shaker (MaxQ 4000, Thermal Fisher Scientific, Waltham, MA) at 37°C and 225 rpm. After the culture reached mid-log phase, bacteria were collected and diluted into ~10 CFU/mL through serial 10-fold dilutions. Bacterial densities were determined by plating onto LB plates. The final 10-fold dilution was performed by adding 500μL of 100 CFU/mL into 4500 μL of 10% human blood in medium.
A 2.5% (w/v) of saponin solution was prepared, sonicated (Branson 2510, Emerson, St. Louis, MO) for 20 minutes and spun down with a clinical centrifuge (Centrific Model 228, Fisher Scientific, Waltham, MA) for 4 minutes. The supernatant cleared of particulates was collected. 500 μL of 2.5% saponin was then added to the 5 mL of 10% human blood sample with ~10 CFU/mL, described above and the mixtures were shaken at 300 rpm for 15 minutes at 37°C. After the saponin treatment, the bacteria were pelleted and washed with 2 mL of phosphate buffered saline (PBS) (Life Technologies, Carlsbad, CA) using a clinical centrifuge for 2 minutes. Bacterial growth medium (2.5 mL) was then added to the tube and incubated for 2 hours at 37°C and 225 rpm.
After the 2-hour incubation, 500-μL aliquots of the suspension were added to each of 4 wells in a 12-well microtiter plate that contained 500 μL of growth medium with or without antibiotic at 2-fold of specified concentrations. The plate was then incubated at 37°C for 3 hours. Bacteria were collected by centrifugation (Centrifuge 5417R, Eppendorf) and resuspended in 200 μL of PBS for flow cytometry detection. To ensure each clinical isolate was tested at its MIC values of the tested antibiotics, the initial bacterial cultures at the 1000 CFU/mL dilution were also tested for each experiment (Supporting Figs. 1, 2, 4, 5, and 6.), confirming that the antibiotic concentrations indeed inhibited bacterial growth.
Flow cytometry
Cytometric data were collected on a BD LSRFortessa flow cytometer (Becton Dickinson, Franklin Lake, NY) equipped with a 100 mW, 488 nm laser for the scatter signal. Set such that low scatter signals are observed when running pure PBS buffer, high-pass thresholds were set on both forward and side scatter to exclude instrumental noise. Data were recorded with FACSDiVa provided by BD. For FAST data, either 100,000 events were collected or collection was stopped when the sample volume was nearly depleted. Flow cytometry data were exported into .fcs files for further analysis and display in MATLAB 2016a (MathWorks, Natick, MA).
PB-sQF test statistics
The statistical tests were performed in MATLAB 2016a on an Intel® Core™ i7-4790 CPU (3.60GHz) machine, equipped with 12.0GB RAM running MS Windows 10. The PB-sQF procedure was as described in our previous publication (1). For each data set, thresholds were applied to both dimensions to include data points that lie within the range from 5 to 105 to exclude outliers. PB-sQF starts with the probability binning approach developed by Roederer et al. (37,38), but then uses a different and linear statistical distance metric related to quadratic form (39). The probability binning approach treats the original data as the initial bin. The variances of all dimensions (here, forward scatter and side scatter) were calculated and the initial bin was divided into 2 daughter bins at the median of the highest variance dimension. Data points equal to the median were randomly assigned to the 2 daughter bins. The same procedure was recursively applied to split each daughter bin into its daughter bins, until the designated number of bins were generated. All data were analyzed with 128 bins. The number of bins was determined by the calculation time and the accuracy of test results. For datasets with sufficient counts to avoid Poisson counting noise/bin from increasing uncertainty in centroid position, better accuracy can be obtained with more bins. With the low bacterial CFUs measured in these studies, 128 bins offered the best compromise of calculation time and counting noise minimization, while maintaining excellent convergence accuracy for bootstrapped confidence limit distances. Although any number of bins can be used in probability binning, with the weights taking care of the different counts/bin, using a power of two for the number of bins generates irregularly shaped bins containing similar numbers of counts (and nearly equal bin weights). The binning patterns are adaptive and represent the data with more (and smaller) bins in regions where the data is most highly concentrated. The same adaptive binning procedure is applied to both no-antibiotic controls and the antibiotic-treated samples. The centroids of the data within each bin and weights (normalized numbers of counts in each bin, relative to the total counts) were then calculated and stored for calculation. In this manner, data with ~100,000 bacterial counts in any number of dimensions is reduced to 128 adaptive bins/data set for fast distance calculations among data.
Bins are numbered for control (c) and sample (s) centroids in the order that the bins are created in the probability binning process, with N total bins. The weight for each bin is the number of counts per bin divided by the total number of counts. As with non-adaptively binned quadratic form statistics (39), the weight vector is the collection of the weights from the control and sample data:
The negative sign for the sample weights ensures that the difference between the control and sample is calculated in the following matrix multiplication.
The centroid is the multidimensional median of the data in each bin and the calculation is described below in the “Geometric quantile” section. The centroid matrix is written as:
The notation is the same as in the weight vector. “Cent” is a matrix listing all the centroids from control and sample, with each column representing a centroid and each row representing one dimension.
The centroids and weights were then used to calculate the test statistics as described in sQF (40,41). First, a similarity matrix is constructed. The matrix elements at ith row and jth column, Aij, in the similarity matrix, A, is defined as:
The first term is a 2Nx2N matrix of 1’s and the second term is the dissimilarity matrix. The numerator, L[Cent(i),Cent(j)], calculates all pairwise Euclidean distances between multidimensional centroids i and j, for both the control and the sample. The denominator is the maximum distance to normalize the calculated distance. Lmax, is the maximum distance in one dimension. Since we have the same maximum range for each dimension, the maximum distance for n dimensions is . When the two centroids are identical, the numerator goes to 0 and thus no dissimilarity exists. Conversely, when the maximum difference occurs, the 2nd term goes to 1 due to normalization. The similarity matrix, A, is the logical opposite of the dissimilarity matrix so the dissimilarity matrix is subtracted from the 2Nx2N matrix of 1’s. Note that the diagonal elements, the similarity of centroid i and i, is always one. Each test statistic (statistical distance, D) were then calculated in quadratic form:
WeightT is the transpose of the Weight vector.
Confidence level estimation and fold distance
The bootstrap method was used to estimate the 99% confidence level of the no-antibiotic control and the 1/16x MIC data. By calculating the 99% confidence levels from small sample size sub-distributions, bootstrapping can estimate the confidence level at the data sample size accurately. 70 sub-distributions with the sample size ranging from 4*(number of bins) to (1/10 of sample size) with 20 steps, were randomly sampled from the 1/16x MIC data and the paired no-antibiotic control. The lower bound, 4*(number of bins), was chosen to prevent zero count per bin. The upper bound, (1/10 of sample size), was set to ensure the sampling process was random. All 140 sub-distributions were binned and the centroids and weights were calculated as described in “PB-sQF test statistics”. Test statistics were calculated between all 70 sub-distributions from the 1/16x MIC data and the 70 paired controls. Since all the sub-distributions were sampled randomly thus were different from each other, the test statistics yield a distribution (biological variability) and the 99% confidence level for each sample size was determined. The confidence levels decreased as a function of sub-sample size since all the sub-distributions come from the same mother distribution and the larger the sample size, the better the estimation of the mother distribution. The distribution of the 99% confidence level should approximate a Gaussian distribution at large sample size according central limit theory, thus the uncertainty in estimating the confidence level can be described with an equation used to estimates the standard error of the sample mean: , where n is the sample size and a1 is the standard deviation of the population. The 99% confidence level at sample size n, Conf(n), can then be described by the following equation:
where a0 is the confidence level of the population. The confidence level at sample size n converges to the population’s confidence level as the uncertainty decreases. From the fitting, we can get the confidence level of the mother distribution with sample size n. For our pure culture control, n = 100,000 counts. While in FAST, n varies. The test statistic of each antibiotic-treated sample was then normalized with the calculated confidence level and turned into fold distance relative to its paired control for direct comparison among different samples and replicates.
Error bar determination
Two different uncertainties contribute to the error bars. The first is biological variability and was determined by the standard deviation of the triplicate data. The second is the uncertainty in centroid position associated with the dispersion of data points in each bin. This uncertainty is determined by the median absolute deviation (MAD) of data within each bin. MAD was chosen over the standard deviation of each bin because MAD is more robust toward outliers. The MAD is calculated as follows:
The median of the absolute distance between each data point, Xi, and the centroid of each bin (42,43). The standard deviation can be estimated from MAD by:
where φ−1 is the inverse of the cumulative distribution function or the quantile function (43). Thus, the standard deviation (without the influence of outliers) can be calculated by dividing the MAD with the 75% quantile (See section “Geometric quantile”, below). The final binning uncertainty for each replicate i, , was estimated by propagating the uncertainty from each bin, .
Since all triplicate data were sub-samples from the same unknown population, the uncertainty from each replica was further pooled together to estimate the uncertainty of binning for the population as follows:
in which k = 3 for triplicate data. n is the sample size of each replicate.
The biological variation uncertainties from triplicate data and binning centroid estimation were propagated together to get the final uncertainty.
The error bars in the bar charts are one standard deviation above and below the test statistic value.
Geometric quantiles
Geometric quantiles were used for calculating the multidimensional median of each bin as the centroid and for estimating the standard deviation from MAD. The geometric quantile, Q, is defined as the data point that minimizes the following target function as described by Chaudhuri (44):
in which n is the number of data points in each bin; X⃗i is the data point, and Q⃗(m) is the quantile of the mth iteration; u = 2α − 1, where α is fractional quantile. For example, α = 0.5 for median (50% quantile) and the target function reduces to . The 50% quantile is the Q that minimizes the sum of distances between each data point to Q. For other quantiles, the second term in the target function is not zero and takes the deviation from the median into account. To minimize the target function, Newton’s method (with a quasi-Newton modification, if needed) was used to solve the unconstrained minimization problem, as detailed below.
Our initial guess, Q⃗(0), is the 1-D quantile in each dimension. Q⃗(1) is estimated using the following equations:
in which s⃗(m) is the increment determined by the first- and second-order derivative of the target function at the current iteration. For each step, f(Q⃗(m+1)) < f(Q⃗(m)) must be satisfied (Newton’s method). If not satisfied, a better Q⃗(m+1) is estimated by a backtracking line search (quasi-Newton method) (45). It has been shown that Newton’s method converges quadratically while the quasi-Newton method, with adequate line search, converges superlinearly (45). Convergence for all multidimensional medians for each bin was readily achieved in ≪50 iterations (usually <5).
The iteration stops when either (1) the iteration has been carried out 50 times or (2) the relative gradient in Q is smaller than the stopping criteria we set. The relative gradient is:
In this work, the iteration was stopped when relgrad(Q) is smaller than 10−4.
Results
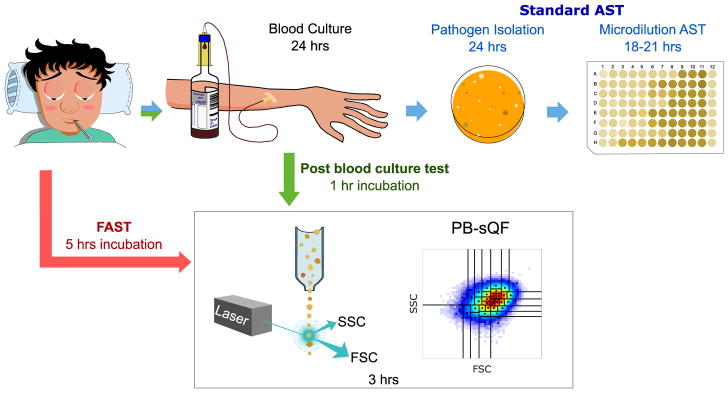
To demonstrate acceleration of the timeline for our Fast AST (i.e. FAST, Fig. 1), we obtained multidrug resistant, blood isolates of common bacteremia-causing pathogens (E. coli, K. pneumoniae, and A. nosocomialis). Varied counts of bacteria were mixed with human blood and diluted 1:9 (v/v) with bacterial growth medium to desired bacterial concentrations (< 10 CFU/mL final solution) and plated to independently confirm CFU/mL blood. Saponin was immediately added and the samples shaken for 15 minutes to achieve selective blood cell lysis. After hemolysis, bacteria were spun down and washed with PBS. The pellets were resuspended in bacterial growth medium and incubated at 37°C for 2 hours, followed by another 3-hour incubation with antibiotics at various fractions of the minimum inhibition concentration (MIC, as independently determined by microdilution from the pure starting culture, Table 1) for sensitive strains or at the CLSI MIC resistant breakpoints (12) for the highly resistant strains. Changes in scattered light signals largely resulting from bacterial growth inhibition were monitored by flow cytometry, and the difference in the 2-D scatter histograms with and without antibiotic treatment was quantified with PB-sQF statistics (1). Using PB-sQF, distance is calculated for each antibiotic concentration relative to its own no-antibiotic control and expressed as “fold distance” relative to the 99% confidence limit distance between the no-antibiotic control and the sensitive breakpoint. This allows comparison of fold distances among all samples as each is individually paired to its own control, as is each replicate. More detailed procedures are given in Materials and Methods.
Figure 1. Antibiotic susceptibility test (AST) timelines.
(Top, blue arrows) The standard clinical microbiology workflow requires >60 hours from initial blood draw. (Green arrows) Timeline for the post-blood culture cytometric AST using PB-sQF distances (1). (Red arrows) Timeline from initial blood draw for Fast AST (i.e. FAST). FSC: forward scatter. SSC: side scatter.
Table 1.
MIC (μg/mL) for each antibiotic/bacteria combination
| MIC (S/I/R) | E. coli Mu14S | E. coli Mu890 | K. pneumoniae Mu670 | K. pneumoniae Mu55 | A. nosocomialis M2 |
|---|---|---|---|---|---|
| Tetracycline | > 64 (R) | 1 (S) | 2 (S) | > 64 (R) | 1 (S) |
| Gentamicin | 8 (I) | 8 (I) | 4 (S) | 1 (S) | 2 (S) |
| Ampicillin | > 1024 (R) | > 1024 (R) | > 1024 (R) | > 1024 (R) | > 1024 (R) |
The MICs were determined from microdilution AST. S, I, and R represent sensitive, intermediate and resistant according to the 2016 Clinical & Laboratory Standards Institute (CLSI) handbook (12).
Multidrug-resistant E. coli isolates
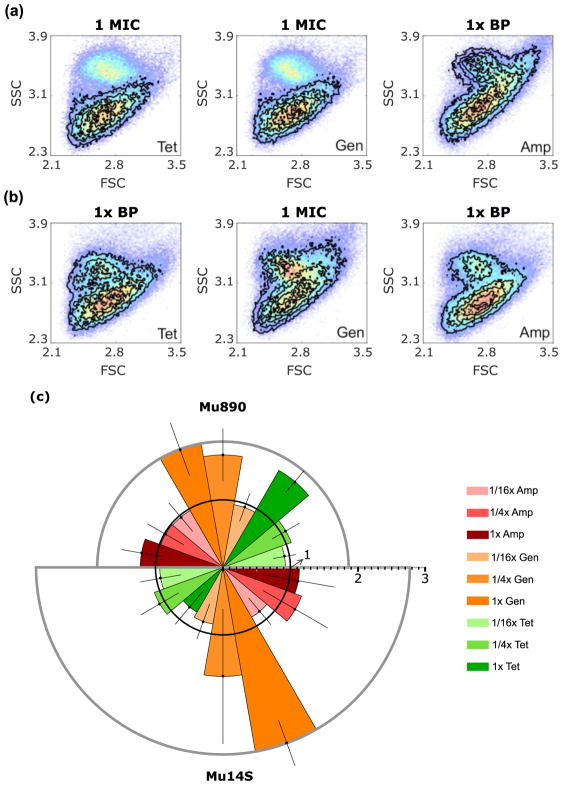
Two multidrug-resistant E. coli clinical isolates were tested, Mu890 and Mu14S. Following the hemolysis and growth procedure outlined above, flow cytometry was used to collect forward and side scattered light signals. Statistical comparison of these histograms (Fig. 2) demonstrates that susceptibility testing is readily performed by immediate hemolysis of 0.5 mL blood, followed by 2-hrs pre-incubation, and 3-hrs AST. When treated at the MICs of tetracycline or gentamicin, to which Mu890 is sensitive and intermediate, respectively, the Mu890 signals disappeared, indicating effective growth inhibition (Fig. 2a). When treated with 32 μg/mL ampicillin (the Enterobacteriaceae resistance breakpoint), however, scatter signals were indistinguishable from those of the no-antibiotic control (Fig. 2a). The complete cytometric data over 16-fold ranges encompassing the sensitive to resistant breakpoints of ineffective antibiotics or from 1/16x MIC to 1x MIC for antibiotics to which Mu890 are sensitive are in Supporting Figure S1. PB-sQF fold distance-based FAST beyond the 99% confidence levels match the much slower microscan AST data, demonstrating that tetracycline and gentamicin are indeed effective for Mu890 (Fig. 2c). The actual starting CFU/mL of Mu890 were confirmed with overnight plating to be 3, 3, and 5 CFU/mL for tetracycline, gentamicin, and ampicillin experiments, respectively. Since the E. coli/human blood was diluted 10-fold, the real concentrations before dilution corresponded to ~30, 30 and 50 CFU/mL of whole blood.
Figure 2. FAST antibiotic-induced scatter signals for E. coli isolates Mu890 and Mu14S.
(a and b) Antibiotic induced scatter histograms (black contours) overlaid on paired no-antibiotic control (color dots, red indicating highest occurrence). BP: break point. (a) Mu890 treated with tetracycline (Tet) at 1 μg/mL (MIC), gentamicin (Gen) at 8 μg/mL (MIC). and ampicillin (Amp) at 32 μg/mL (resistance breakpoint). (b) Mu14S treated with tetracycline at 16 μg/mL (resistance breakpoint), gentamicin at 8 μg/mL (MIC) and ampicillin at 32 μg/mL (resistance breakpoint). (c) PB-sQF distances for (a) and (b). The radius is the fold distance, (the test statistics normalized by the 99% confidence distance to be different from the control). The 99% confidence fold-distance is represented by the inner black circle with radius equal to 1. Any test result exceeding the 99% confidence level (error bar included) is statistically different from the control and is an effective antibiotic treatment. Mu890 results: upper semicircle. Mu14S results: lower semicircle. Error bar is one standard deviation above and below the average fold distance obtained from triplicate trials. Details of test statistics and error bar calculation are presented in the Methods.
Also matching its standard AST data, FAST shows that Mu14S is intermediate to gentamicin and when treated at the MIC, exhibits growth inhibition (Fig. 2b and Supporting Fig. S2). When treated with tetracycline or ampicillin at each resistant breakpoint (16 μg/mL and 32 μg/mL), however, Mu14S signals remained statistically unchanged. The PB-sQF fold distance average from triplicate data shows clear differences between the 1x MIC gentamicin data versus the paired-control (Fig. 2c). This confirms that the gentamicin sensitivity of Mu14S observed after blood culture (1) can also be observed on much shorter timescales with FAST. Overnight plating confirms that initial Mu14S counts were 3, 2, 5 and CFU/mL for tetracycline, gentamicin and ampicillin data after 10-fold dilution of the blood/bacteria mixture, corresponding to FAST being performed on whole blood samples containing ~30, ~20 and ~50 CFU/mL. Controls of identical treatment of 10% human blood samples without bacteria inoculation were also tested, yielding scattered light histograms that do not significantly change around the resistance breakpoints of Enterobacteriaceae under gentamicin, ampicillin, or tetracycline treatment (Supporting Fig. S3). Overnight plating confirmed that no bacteria were found in the blood, except when specifically added. Thus, diluting bacterimic blood specimens 1:9 (v:v) directly into saponin-containing growth medium provides a path to ASTs within 8 hours from initial blood draw, with excellent results matching independent (36–44 hrs) MIC determinations from pure, overnight cultures (~108 CFU/mL) that could only be initiated after (~24hr) positive blood culture.
Multidrug resistant K. pneumoniae isolates
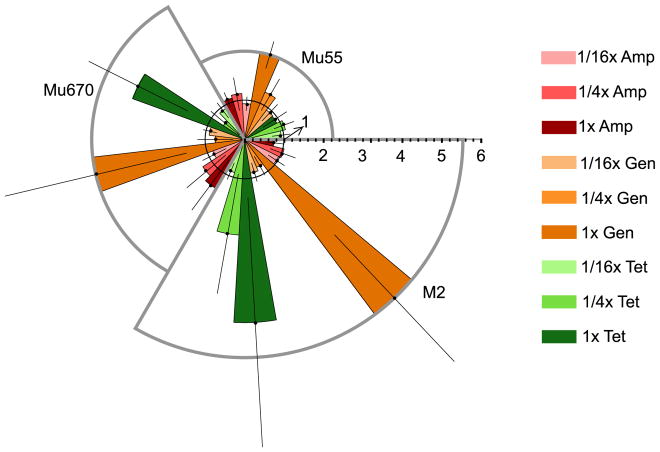
The same FAST procedure was applied to two multidrug-resistant K. pneumoniae clinical isolates, Mu55 and Mu670. Analogous to the E. coli data, K. pneumoniae growth inhibition is directly quantified with PB-sQF upon effective antibiotic treatment, and sensitivities are accurately determined. Importantly, when treated with antibiotics to which Mu55 or Mu670 were resistant, the scatter data (black contours) were not statistically different from each experiment’s paired control (pseudo-color plots) as shown in Supporting Figure S4 (Mu55), and Supporting Figure S5 (Mu670). PB-sQF confirms that tetracycline is effective toward Mu670 and gentamicin is appropriate for both Mu55 and Mu670. The initial bacterial concentrations post 10-fold dilution were confirmed by plating to be ~8 CFU/mL for Mu55 and ~9 CFU/mL for Mu670, demonstrating that FASTs can be readily completed within 8 hours of initial blood draw on blood samples exhibiting <100 CFU/mL.
Multidrug resistant A. nosocomialis isolates
Obtainable in <1/8th the time of standard AST results, FAST on A. nosocomialis clinical isolate M2 spiked in 10% human blood (Supporting Fig. S6) enabled its susceptibility profile to be similarly quickly determined. As with other species, PB-sQF reveals that M2 is resistant to ampicillin but susceptible to both tetracycline and gentamicin when assayed within 8 hrs of initial simulated blood draw (Fig. 3). Different from the ~105 CFU/mL E. coli and K. pneumoniae strains resulting from 2-hr preincubation, the final A. nosocomialis concentrations were ~104 CFU/mL, as confirmed by plating. Even with an order of magnitude fewer bacterial counts, the clear growth inhibition was readily quantified with the same procedure. As initial ~10 CFU/mL samples incubated for 5 hours are sufficient for analysis, ~10 doubling events are ideal to generate sufficient sample for FAST antibiotic panels to be performed on any bacteria. For the most common sepsis-inducing strains, including gram-negative E. coli, P. aeruginosa, and Klebsiella spp. (46,47), 10 doubling events should be achieved within the total 2-hr pre-incubation and 3-hr incubation with antibiotics in 1:9 whole blood:standard bacterial media.
Figure 3. FAST antibiotic-induced scatter signal changes for K. pneumoniae stain Mu55 and Mu670 and A. nosocomialis strain M2 reveal different susceptibilities.
The PB-sQF result for each strain is shown in each segment. Mu55 (upper right segment) was treated with tetracycline at 16 μg/mL (resistance breakpoint) or gentamicin at 1 μg/mL (MIC) or ampicillin at 32 μg/mL (resistance breakpoint). Mu670 (left segment) was treated with tetracycline at 2 μg/mL (MIC) or gentamicin at 4 μg/mL (MIC) or ampicillin at 32 μg/mL (resistance breakpoint). M2 (lower right segment) was treated with either tetracycline at 1 μg/mL (MIC), gentamicin at 2 μg/mL (MIC), or ampicillin at 128 μg/mL (resistance breakpoint for penicillin type antibiotic). As in Figure 2c, the radius is the fold distance normalized by the 99% confidence level (inner black circle with radius equals to 1). The error bar is one standard deviation of each test result above and below the average.
Discussion
Most cytometric-based bacterial viability tests utilize fluorescent dyes to assay live/dead populations after positive blood cultures, but have problems with many antibiotic/bacteria combinations. Since there are no statistically significant scattered-light signal changes in the blood-only sample with antibiotic exposure, and the antibiotic breakpoint is related to growth inhibition, the scattered-light signal changes in the bacteria-spiked samples with inhibitory antibiotic concentration observed here result from antibiotic-induced growth inhibition. This growth inhibition is observed only in the presence of antibiotic concentrations that matched the independently determined MICs, and was confirmed by overnight plating for each sample. Colony counts for the prototypical example, Mu14S (Supporting Table S1) correlate with the independently determined resistance profile. This supports that the primary contribution to the changes in scattered light signals indeed reflect the inhibition/reduction of viable bacterial counts in the sample. Here, we show that by using PB-sQF to characterize the differences between dataset, the changes in 2D scatter patterns are sufficient for a robust growth inhibition-based phenotypic AST directly from bacteria-containing blood. A scatter-only AST is more general to different bacterial strains since there is no staining problem as seen in the previous studies (17,19).
As described in our previous study (1), PB-sQF combines the advantages from Quadratic Form (QF) (39), which uses rectangular binning, and PB-χ2 (37,38), which uses chi-squared statistics. QF calculates linear distances that can be compared directly. Rectangular binning, however, does not scale well with high data dimensionality. PB-χ2 addresses the dimensionality problem with probability binning, by significantly reducing the number of bins required to represent the data, thereby reducing the computation burden. The chi-squared statistics, however, give non-linear distances which cannot be quantitatively compared among samples. This non-linear distance also prevents the test results of different machines or different days from being compared directly. By using probability binning, calculating the signatures (centroids) of each bin, and performing Quadratic Form calculations of linear distances, PB-sQF enables direct similarity comparison for multidimensional data through fold distance comparisons, each of which is individually normalized to its own paired control for more quantitative comparisons.
The data shown in Figures 2 and 3 fluctuate from run to run due to the varying ratio between bacterial cells and blood debris resulting from sample processing. Also, when the blood debris counts are high, the signals can partially obscure the bacteria signal. Additionally, bacteria with different size and shape produce different scatter patterns that can increase overlap with the blood debris background. As a result, setting an artificial gate to discriminate bacteria counts is problematic when the bacterial identity is unknown. Without robust multidimensional statistics, unbiased by artificial gating, determining statistical distances between any two data sets that have different background signals is an outstanding challenge. PB-sQF calculates similar levels of fold distances of the triplicate data from their own paired controls with different levels of background noise. With triplicate error bars included in the PB-sQF analysis, the 1x MIC data were statistically significantly different from the 99% confidence level.
To rapidly determine appropriate treatments for gram-negative bacteria, we developed FAST to minimize time to result from initial blood draw. By selectively removing blood cells, FAST requires a total of only 8 hours to complete susceptibility testing. Using flow cytometry to acquire the entire distribution of bacterial responses to antibiotic exposure, PB-sQF statistical metrics directly quantify the differences between antibiotic-treated data and no-antibiotic paired controls. Consistent results are obtained even when data vary among different replicates, or if performed on different instruments. This procedure, without time-consuming overnight incubation and multiple serial subculturing, reduces the time to result from >60 hours to <8 hours total time from initial blood draw, with identical susceptibility determinations. Since rapid identification of the correct antibiotic treatment is crucial in treating suspected bacterial infections, FAST has the potential to greatly improve patient outcomes, while minimizing antibiotic resistance proliferation. While both bacterial identity and susceptibility profile are currently needed for appropriate treatment, CLSI breakpoints for the most common bacteremia-causing bacteria differ by <4-fold (12). Thus, testing additional antibiotic concentrations to encompass the full susceptibility ranges of the most common bacteremia-causing pathogens could rapidly provide susceptibility information without waiting for much slower, post blood culture bacterial identity determinations. As the majority of blood stream infections are caused by gram-negative bacteria (46,47), this approach offers a path to drastically improved patient outcomes, while still allowing for subsequent confirmation from standard post blood culture pathogen identification and ASTs.
Supplementary Material
Acknowledgments
The authors gratefully acknowledge support of these studies from the National Institutes of Health, Award number R01AI107116 and the Vasser-Woolley Foundation.
Footnotes
Data and Code availability. All data will be made freely available on request. PB-sQF codes used for flow cytometry analysis are available upon request for non-commercial purposes.
Competing Financial Interests. The authors declare no competing financial interests.
References
- 1.Huang T-H, Ning X, Wang X, Murthy N, Tzeng Y-L, Dickson RM. Rapid Cytometric Antibiotic Susceptibility Testing Utilizing Adaptive Multidimensional Statistical Metrics. Analytical Chemistry. 2015;87:1941–1949. doi: 10.1021/ac504241x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kreger BE, Craven DE, McCabe WR. Gram-negative bacteremia. IV. Re-evaluation of clinical features and treatment in 612 patients. Am J Med. 1980;68:344–55. doi: 10.1016/0002-9343(80)90102-3. [DOI] [PubMed] [Google Scholar]
- 3.Kollef MH. Broad-Spectrum Antimicrobials and the Treatment of Serious Bacterial Infections: Getting It Right Up Front. Clinical Infectious Diseases. 2008;47:S3–S13. doi: 10.1086/590061. [DOI] [PubMed] [Google Scholar]
- 4.Fraser A, Paul M, Almanasreh N, Tacconelli E, Frank U, Cauda R, Borok S, Cohen M, Andreassen S, Nielsen AD, et al. Benefit of appropriate empirical antibiotic treatment: thirty-day mortality and duration of hospital stay. Am J Med. 2006;119:970–6. doi: 10.1016/j.amjmed.2006.03.034. [DOI] [PubMed] [Google Scholar]
- 5.Ibrahim EH, Sherman G, Ward S, Fraser VJ, Kollef MH. The Influence of Inadequate Antimicrobial Treatment of Bloodstream Infections on Patient Outcomes in the ICU Setting*. Chest. 2000;118:146–155. doi: 10.1378/chest.118.1.146. [DOI] [PubMed] [Google Scholar]
- 6.Kohanski MA, DePristo MA, Collins JJ. Sublethal Antibiotic Treatment Leads to Multidrug Resistance via Radical-Induced Mutagenesis. Molecular Cell. 2010;37:311–320. doi: 10.1016/j.molcel.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Christner M, Rohde H, Wolters M, Sobottka I, Wegscheider K, Aepfelbacher M. Rapid identification of bacteria from positive blood culture bottles by use of matrix-assisted laser desorption-ionization time of flight mass spectrometry fingerprinting. J Clin Microbiol. 2010;48:1584–91. doi: 10.1128/JCM.01831-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sauer S, Kliem M. Mass spectrometry tools for the classification and identification of bacteria. Nat Rev Microbiol. 2010;8:74–82. doi: 10.1038/nrmicro2243. [DOI] [PubMed] [Google Scholar]
- 9.Carey JR, Suslick KS, Hulkower KI, Imlay JA, Imlay KRC, Ingison CK, Ponder JB, Sen A, Wittrig AE. Rapid Identification of Bacteria with a Disposable Colorimetric Sensing Array. Journal of the American Chemical Society. 2011;133:7571–7576. doi: 10.1021/ja201634d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barczak AK, Gomez JE, Kaufmann BB, Hinson ER, Cosimi L, Borowsky ML, Onderdonk AB, Stanley SA, Kaur D, Bryant KF, et al. RNA signatures allow rapid identification of pathogens and antibiotic susceptibilities. Proc Natl Acad Sci U S A. 2012;109:6217–22. doi: 10.1073/pnas.1119540109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huletsky A, Giroux R, Rossbach V, Gagnon M, Vaillancourt M, Bernier M, Gagnon F, Truchon K, Bastien M, Picard FJ, et al. New Real-Time PCR Assay for Rapid Detection of Methicillin- Resistant Staphylococcus aureus Directly from Specimens Containing a Mixture of Staphylococci. Journal of Clinical Microbiology. 2004;42:1875–1884. doi: 10.1128/JCM.42.5.1875-1884.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.CLSI. M100-S26 Performace Standards for Antimicrobial Susceptibility Testing. 2016 [Google Scholar]
- 13.Mason DJ, Allman R, Stark JM, Lloyd D. Rapid estimation of bacterial antibiotic susceptibility with flow cytometry. Journal of Microscopy. 1994;176:8–16. doi: 10.1111/j.1365-2818.1994.tb03494.x. [DOI] [PubMed] [Google Scholar]
- 14.Walberg M, Gaustad P, Steen HB. Rapid flow cytometric assessment of mecillinam and ampicillin bacterial susceptibility. Journal of Antimicrobial Chemotherapy. 1996;37:1063–1075. doi: 10.1093/jac/37.6.1063. [DOI] [PubMed] [Google Scholar]
- 15.Walberg M, Gaustad P, Steen HB. Rapid assessment of ceftazidime, ciprofloxacin, and gentamicin susceptibility in exponentially-growing E. coli cells by means of flow cytometry. Cytometry. 1997;27:169–178. [PubMed] [Google Scholar]
- 16.Suller MTE, Lloyd D. Fluorescence monitoring of antibiotic-induced bacterial damage using flow cytometry. Cytometry. 1999;35:235–241. doi: 10.1002/(sici)1097-0320(19990301)35:3<235::aid-cyto6>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- 17.Mortimer FC, Mason DJ, Gant VA. Flow Cytometric Monitoring of Antibiotic-Induced Injury in Escherichia coli Using Cell-Impermeant Fluorescent Probes. Antimicrobial Agents and Chemotherapy. 2000;44:676–681. doi: 10.1128/aac.44.3.676-681.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Walberg M, Steent HB. Zbigniew Darzynkiewicz HACJPR, editor. Methods in Cell Biology. Part B. Vol. 64. Academic Press; 2001. flow cytometric monitoring of bacterial susceptibility to antibiotics; pp. 553–566. [DOI] [PubMed] [Google Scholar]
- 19.Gauthier C, St-Pierre Y, Villemur R. Rapid antimicrobial susceptibility testing of urinary tract isolates and samples by flow cytometry. Journal of Medical Microbiology. 2002;51:192–200. doi: 10.1099/0022-1317-51-3-192. [DOI] [PubMed] [Google Scholar]
- 20.Assunção P, Antunes NT, Rosales RS, Poveda C, De La Fe C, Poveda JB, Davey HM. Application of flow cytometry for the determination of minimal inhibitory concentration of several antibacterial agents on Mycoplasma hyopneumoniae. Journal of Applied Microbiology. 2007;102:1132–1137. doi: 10.1111/j.1365-2672.2006.03170.x. [DOI] [PubMed] [Google Scholar]
- 21.Faria-Ramos I, Espinar MJ, Rocha R, Santos-Antunes J, Rodrigues AG, Cantón R, Pina-Vaz C. A novel flow cytometric assay for rapid detection of extended-spectrum beta-lactamases. Clinical Microbiology and Infection. 2013;19:E8–E15. doi: 10.1111/j.1469-0691.2012.03986.x. [DOI] [PubMed] [Google Scholar]
- 22.Nuding S, Zabel TL. Detection, Identification and Susceptibility Testing of Bacteria by Flow Cytometry. J Bacteriol Parasitol. 2013;S5:005. [Google Scholar]
- 23.Shapiro HM. Multiparameter flow cytometry of bacteria: Implications for diagnostics and therapeutics. Cytometry. 2001;43:223–226. doi: 10.1002/1097-0320(20010301)43:3<223::aid-cyto1054>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- 24.Dietzman DE, Fischer GW, Schoenknecht FD. Neonatal escherichia coli septicemia—bacterial counts in blood. The Journal of Pediatrics. 1974;85:128–130. doi: 10.1016/s0022-3476(74)80308-2. [DOI] [PubMed] [Google Scholar]
- 25.Yagupsky P, Nolte FS. Quantitative aspects of septicemia. Clinical Microbiology Reviews. 1990;3:269–279. doi: 10.1128/cmr.3.3.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mansour JD, Robson JA, Arndt CW, Schulte TH. Detection of Escherichia coli in blood using flow cytometry. Cytometry. 1985;6:186–90. doi: 10.1002/cyto.990060303. [DOI] [PubMed] [Google Scholar]
- 27.Pitt WG, Alizadeh M, Husseini GA, McClellan DS, Buchanan CM, Bledsoe CG, Robison RA, Blanco R, Roeder BL, Melville M, et al. Rapid separation of bacteria from blood-review and outlook. Biotechnol Prog. 2016;32:823–39. doi: 10.1002/btpr.2299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hou HW, Bhattacharyya RP, Hung DT, Han J. Direct detection and drug-resistance profiling of bacteremias using inertial microfluidics. Lab Chip. 2015;15:2297–307. doi: 10.1039/c5lc00311c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tay A, Pavesi A, Yazdi SR, Lim CT, Warkiani ME. Advances in microfluidics in combating infectious diseases. Biotechnology Advances. 2016;34:404–421. doi: 10.1016/j.biotechadv.2016.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gosiewski T, Szala L, Pietrzyk A, Brzychczy-Wloch M, Heczko PB, Bulanda M. Comparison of methods for isolation of bacterial and fungal DNA from human blood. Curr Microbiol. 2014;68:149–55. doi: 10.1007/s00284-013-0451-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gosiewski T, Jurkiewicz-Badacz D, Sroka A, Brzychczy-Włoch M, Bulanda M. A novel, nested, multiplex, real-time PCR for detection of bacteria and fungi in blood. BMC Microbiology. 2014;14:144. doi: 10.1186/1471-2180-14-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Steinberger-Levy I, Zahavy E, Cohen S, Flashner Y, Mamroud E, Aftalion M, Gur D, Ber R. Enrichment of Yersinia pestis from blood cultures enables rapid antimicrobial susceptibility determination by flow cytometry. Adv Exp Med Biol. 2007;603:339–50. doi: 10.1007/978-0-387-72124-8_31. [DOI] [PubMed] [Google Scholar]
- 33.Doern GV, Vautour R, Gaudet M, Levy B. Clinical impact of rapid in vitro susceptibility testing and bacterial identification. Journal of Clinical Microbiology. 1994;32:1757–1762. doi: 10.1128/jcm.32.7.1757-1762.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gogelein H, Huby A. Interaction of saponin and digitonin with black lipid membranes and lipid monolayers. Biochim Biophys Acta. 1984;773:32–8. doi: 10.1016/0005-2736(84)90547-9. [DOI] [PubMed] [Google Scholar]
- 35.Francis G, Kerem Z, Makkar HP, Becker K. The biological action of saponins in animal systems: a review. Br J Nutr. 2002;88:587–605. doi: 10.1079/BJN2002725. [DOI] [PubMed] [Google Scholar]
- 36.Arabski M, Wegierek-Ciuk A, Czerwonka G, Lankoff A, Kaca W. Effects of Saponins against Clinical E. coli Strains and Eukaryotic Cell Line. Journal of Biomedicine and Biotechnology. 2012;2012:6. doi: 10.1155/2012/286216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roederer M, Moore W, Treister A, Hardy RR, Herzenberg LA. Probability binning comparison: a metric for quantitating multivariate distribution differences. Cytometry. 2001;45:47–55. doi: 10.1002/1097-0320(20010901)45:1<47::aid-cyto1143>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 38.Roederer M, Treister A, Moore W, Herzenberg LA. Probability binning comparison: A metric for quantitating univariate distribution differences. Cytometry. 2001;45:37–46. doi: 10.1002/1097-0320(20010901)45:1<37::aid-cyto1142>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 39.Bernas T, Asem EK, Robinson JP, Rajwa B. Quadratic form: A robust metric for quantitative comparison of flow cytometric histograms. Cytometry Part A. 2008;73A:715–726. doi: 10.1002/cyto.a.20586. [DOI] [PubMed] [Google Scholar]
- 40.Beecks C, Uysal MS, Seidl T. Signature quadratic form distances for content-based similarity. Proceedings of the 17th ACM international conference on Multimedia; Beijing, China: ACM; 2009. pp. 697–700. [Google Scholar]
- 41.Beecks C, Uysal MS, Seidl T. Signature Quadratic Form Distance. Proceedings of the ACM International Conference on Image and Video Retrieval; Xi’an, China: ACM; 2010. pp. 438–445. [Google Scholar]
- 42.Zeitschrift für Astronomie und verwandte Wissenschaften: J. G. Cotta; 1816.
- 43.Ruppert D. Statistics and Data Analysis for Financial Engineering. Springer; 2010. [Google Scholar]
- 44.Chaudhuri P. On a Geometric Notion of Quantiles for Multivariate Data. Journal of the American Statistical Association. 1996;91:862–872. [Google Scholar]
- 45.Dennis JE, Jr, Schnabel RB. Siam. 1996. Numerical methods for unconstrained optimization and nonlinear equations. [Google Scholar]
- 46.Deen J, von Seidlein L, Andersen F, Elle N, White NJ, Lubell Y. Community-acquired bacterial bloodstream infections in developing countries in south and southeast Asia: a systematic review. Lancet Infect Dis. 2012;12:480–7. doi: 10.1016/S1473-3099(12)70028-2. [DOI] [PubMed] [Google Scholar]
- 47.Vincent JL, Rello J, Marshall J, Silva E, Anzueto A, Martin CD, Moreno R, Lipman J, Gomersall C, Sakr Y, et al. International study of the prevalence and outcomes of infection in intensive care units. Jama. 2009;302:2323–9. doi: 10.1001/jama.2009.1754. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.