Abstract
Background
Delamanid, bedaquiline, and linezolid have recently been approved for the treatment of multidrug- and extensively drug-resistant (MDR and XDR, respectively) tuberculosis (TB). To use these drugs effectively, drug susceptibility tests, including rapid molecular techniques, are required for accurate diagnosis and treatment. Furthermore, mutation analyses are needed to assess the potential for resistance. We evaluated the minimum inhibitory concentrations (MICs) of these three anti-TB drugs for Korean MDR and XDR clinical strains and mutations in genes related to resistance to these drugs.
Methods
MICs were determined for delamanid, bedaquiline, and linezolid using a microdilution method. The PCR products of drug resistance-related genes from 420 clinical Mycobacterium tuberculosis strains were sequenced and aligned to those of M. tuberculosis H37Rv.
Results
The overall MICs for delamanid, bedaquiline, and linezolid ranged from ≤0.025 to >1.6 mg/L, ≤0.0312 to >4 mg/L, and ≤0.125 to 1 mg/L, respectively. Numerous mutations were found in drug-susceptible and -resistant strains. We did not detect specific mutations associated with resistance to bedaquiline and linezolid. However, the Gly81Ser and Gly81Asp mutations were associated with resistance to delamanid.
Conclusions
We determined the MICs of three anti-TB drugs for Korean MDR and XDR strains and identified various mutations in resistance-related genes. Further studies are needed to determine the genetic mechanisms underlying resistance to these drugs.
Keywords: Mycobacterium tuberculosis, Delamanid, Bedaquiline, Linezolid, Mutation, Minimum inhibitory concentration
INTRODUCTION
The spread of drug-resistant tuberculosis (TB) is a global concern. Compared with drug-susceptible TB, treatment of multidrug-resistant (MDR) TB takes longer and requires more expensive drugs with greater toxicity [1]. Therefore, new anti-TB compounds have been developed, and combination regimens, including these novel molecules, are being tested in clinical trials [2]. The WHO has amended its policy recommendations for treating drug-resistant TB, describing the use of new drugs such as delamanid, bedaquiline, and linezolid [2].
Delamanid is a nitro-dihydro-imidazooxazole derivative that inhibits the biosynthesis of mycolic acids [3,4]. Studies aimed at elucidating the structural basis for delamanid resistance have revealed the importance of a mutation in ddn (Rv3547) [5]. Moreover, five genes (ddn, fgd1, fbiA, fbiB, and fbiC) associated with either prodrug activation or the tangential F420 biosynthetic pathway have been identified in several resistant strains [5,6].
Bedaquiline, an ATP synthase inhibitor, is effective against MDR TB [7]. It acts as a broad-spectrum anti-mycobacterial compound, affecting drug-susceptible and drug-resistant TB as well as a wide range of nontuberculous mycobacteria [8]. Mutations in a transcriptional repressor (Rv0678) of genes encoding the MmpS5-MmpL5 efflux pump are involved in non-target-based resistance to bedaquiline and cross-resistance to clofazimine [9]. Furthermore, mutations in the atpE gene, which encodes the transmembrane oligomeric C subunit of ATP synthase, confer resistance to bedaquiline by preventing the drug from binding to the C subunit [10].
Linezolid is highly active against gram-positive bacteria and can be effectively used to treat MDR strains [11,12]. Linezolid resistance is associated primarily with a mutation at nucleotide position 2061 in rrl and the T460C mutation in rplC [13,14].
As delamanid, bedaquiline, and linezolid have only recently been approved, resistance to these drugs is expected to be low among patients with TB. However, drug resistance may occur spontaneously, even in the absence of antimicrobial exposure. Consequently, drug susceptibility tests, including rapid molecular techniques, are required for accurate diagnosis and treatment.
The minimum inhibitory concentrations (MICs) of these drugs have been evaluated [15,16,17], but studies on Korean clinical strains are lacking. Therefore, we investigated the MICs of delamanid, bedaquiline, and linezolid among MDR and extensively drug-resistant (XDR) Korean clinical strains and identified mutations in genes related to resistance to these drugs.
METHODS
1. Clinical strains
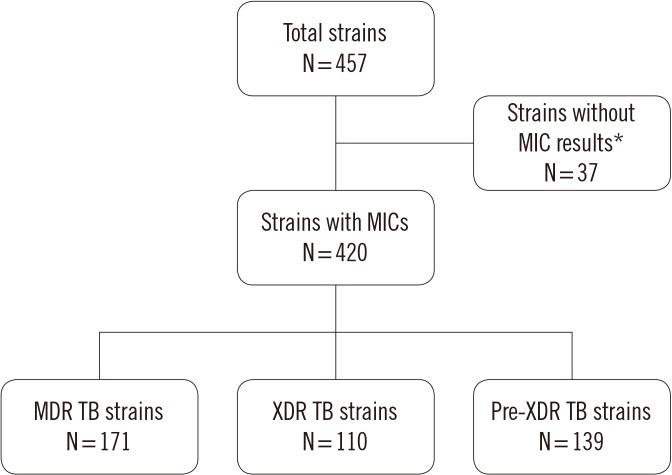
In total, 457 M. tuberculosis clinical strains isolated from patients were obtained from the culture collection at the Korean Institute of Tuberculosis, Cheongju, Korea. Of the 457 clinical strains, 37 were excluded because of contamination or lack of growth. Thus, 420 clinical strains were included in this study (Fig. 1). The study was exempt from ethical approval by the Institutional Review Board of the Korea National Institute for Bioethics Policy, as clinical strains were used.
Fig. 1. Drug resistance patterns of the clinical strains used in this study, as assessed by phenotypic drug susceptibility testing.
*Failed test isolates (N=26) and contaminated isolates (N=11).
Abbreviations: MIC, minimum inhibitory concentration; MDR TB, multidrug-resistant tuberculosis; XDR TB, extensively drug-resistant tuberculosis; Pre-XDR, pre-extensively drug-resistant tuberculosis (MDR TB with resistant to either a fluoroquinolone and second-line injectable drug but not both).
2. Anti-TB drugs
Delamanid powder was obtained from Otsuka Pharmaceutical (Tokyo, Japan), bedaquiline powder was obtained from Janssen Pharmaceutica (Beerse, Belgium), and linezolid was supplied by Yungjin Pharmaceutical (Seoul, Korea). Delamanid and bedaquiline were each dissolved in dimethyl sulfoxide; linezolid was dissolved in methanol. Stock solutions were stored at −70℃.
3. Drug concentration ranges and phenotypic drug susceptibility testing
A range of concentrations was prepared for each anti-TB drug using serial two-fold dilutions, as previously described with slight modifications [15,16,18]: delamanid, 0.0125–1.6 mg/L; bedaquiline, 0.03125–4 mg/L; and linezolid, 0.125–16 mg/L.
Each 2× antibiotic working solution (50 µL) was added to a microtiter plate well, except for the growth control well. M. tuberculosis clinical strains were grown on Lowenstein-Jensen medium. The turbidity of the resulting bacterial suspension was adjusted with phosphate-buffered saline (PBS) to an equivalent of McFarland standard no. 1 (~5×107 colony forming units [CFU]/mL) and then diluted 1:100 (~5×105 CFU/mL) in 2×7H9 broth medium (Becton Dickinson, Sparks, MD, USA) with 100 mg/L 2,3-diphenyl-5-(2-thienyl)-tetrazolium chloride (STC; Tokyo Kasei Kogyo Co., Ltd., Tokyo, Japan). The 2× inoculum was poured into a disposable reservoir, and 50 µL was added to each well containing the 2× antibiotic working solutions. The inoculum was added to all wells (including the growth control wells) except the negative control well, which was inoculated with 50 µL of 2×7H9 broth medium. The final inoculum concentration in each well was 2.5×105 CFU/mL. All plates were incubated at 37℃ for 14 days, and the MIC was then determined as the lowest concentration of the antibiotic that prevented visual growth. Phenotypic drug susceptibility testing (DST) using the broth microdilution method was performed twice.
Quality control was performed under conditions equivalent to those of the experiment using M. tuberculosis strain H37Rv (ATCC 27294). The MICs were defined as 0.002–0.016 mg/L for delamanid [15], 0.015–0.06 mg/L for bedaquiline [16], and 0.5 mg/L for linezolid [19].
4. PCR and sequencing
DNA was extracted from the clinical strains by heating [20] and was stored at −20℃ until used. The primers used to amplify and sequence genes associated with drug resistance are shown in Table 1. The primers were designed in-house for this study. PCR amplification was performed according to a standard protocol under the following conditions: pre-denaturation at 95℃ for 5 minutes, followed by 35 cycles of denaturation at 95℃ for 30 seconds; annealing at 62℃ (fbiA), 66℃ (ddn), or 52℃ (Rv0678, atpE, rrl, and rplC) for 30 seconds; extension at 72℃ for 30 seconds (fbiA, Rv0678, atpE, rrl, and rplC) or 1 minute (ddn); and a final extension at 72℃ for 10 minutes. The PCR products were sequenced using a BigDye Terminator Cycle Sequencing kit and an ABI Prism 3730xl DNA analyzer (Applied Biosystems, Foster City, CA, USA). The results were analyzed using the BioEdit software, and sequences were aligned to those of M. tuberculosis H37Rv (GenBank accession no. AL123456.3).
Table 1. Primer sequences used for PCR.
| Antimicrobial agent | Primer | Sequence (5′ to 3′) |
|---|---|---|
| Delamanid | fbiA_F | GCG GTT CTG TTG TGG TTG GG |
| fbiA_R | CCG ATG ACG GGC AGG ATC TCG ATG G | |
| ddn_F | CGA GCG CAC CGA CCA GAG C | |
| ddn_R | GCA TGG CCC GCA GGT GGA CAA | |
| Bedaquiline | Rv0678_F | GTA TCC AGG CAC GCT TGA |
| Rv0678_R | CCC CAC AAT CGA TAA CC | |
| atpE_F | GTA CTT CAG CCA AGC GAT GG | |
| atpE_R | CCG TTG GGA ATG AGG AAG TTG | |
| Linezolid | rrl_F | TAC CAA GGC GTA CGA GAT |
| rrl_R | CGC CGT AAC TCT ATG CAA | |
| rplC_F | AAC GCA TAA GTA CAA GGA | |
| rplC_R | CGT CTT CGT CGA GTG GGT A |
RESULTS
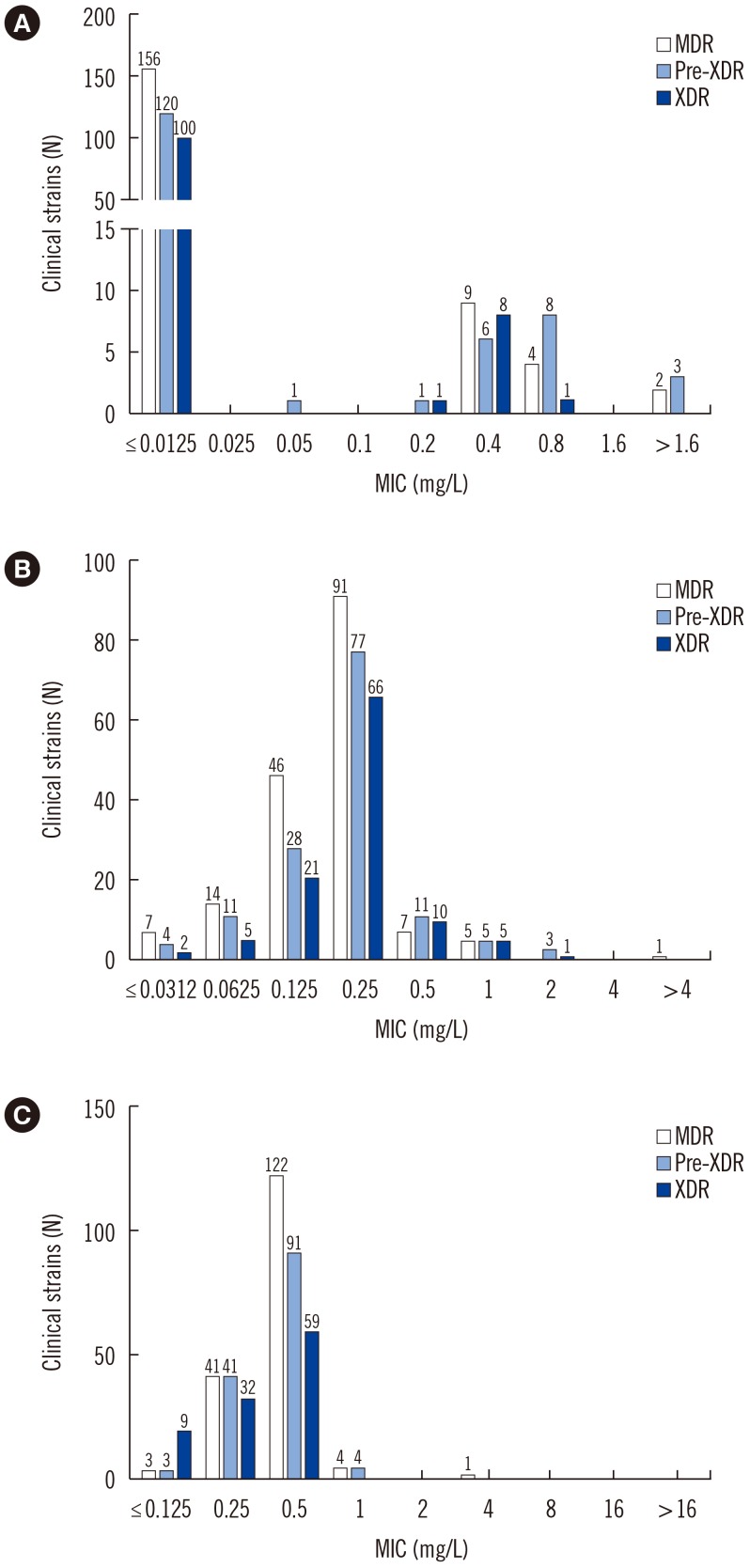
The MICs of delamanid, bedaquiline, and linezolid were ≤0.0125, 0.0625, and 0.5 mg/L, respectively, in the H37Rv control strain. The overall drug MIC distribution is shown in Fig. 2. Of the 420 strains, 379 (90%) were susceptible and 41 (10%) were resistant to delamanid based on the critical concentration of 0.2 mg/L, as suggested by Otsuka Pharmaceutical [21]. Of the latter, 15, 17, and 9 were MDR, pre-extensively drug resistant (pre-XDR), and XDR strains, respectively.
Fig. 2. Overall MIC distribution for 420 clinical strains: (A) Delamanid, (B) bedaquiline, and (C) linezolid MICs. The number above each bar indicates the number of strains.
Abbreviations: MIC, minimum inhibitory concentration; MDR, multidrug-resistant tuberculosis; XDR, extensively drug-resistant tuberculosis; Pre-XDR, pre-extensively drug-resistant tuberculosis.
For bedaquiline, 372 (89%) clinical strains were susceptible, and 48 (11%) were resistant, based on the European Committee on Antimicrobial Susceptibility Testing (EUCAST) breakpoint of 0.25 mg/L [22]. Thirteen MDR, 19 pre-XDR, and 16 XDR strains showed resistance to bedaquiline. One MDR strain (0.2%) was resistant to linezolid, based on the critical concentration of 1 mg/L [23].
We sought to identify mutations in delamanid resistance-related genes in the 379 delamanid-susceptible strains. We found three nonsynonymous and six silent mutations in fbiA. In addition, one non-silent and one silent mutation were identified in ddn. Among the 41 delamanid-resistant strains, two non-silent mutations were identified in ddn (Table 2).
Table 2. Mutations identified in delamanid resistance-related genes.
| Delamanid | Gene | Nucleotide substitution | Amino acid change | Strains (N) | MIC (mg/L) |
|---|---|---|---|---|---|
| Susceptible | fbiA | C110A, C883T | Ala37Asp, His295Tyr | 1 | ≤0.0125 |
| C189T | Asp63Asp | 1 | ≤0.0125 | ||
| C337T | Leu113Leu | 3 | ≤0.0125 | ||
| C337T, G542A | Leu113Leu, Pro181Pro | 1 | 0.2 | ||
| C582T | Ser194Ser | 1 | ≤0.0125 | ||
| C801T | Ser267Ser | 1 | ≤0.0125 | ||
| T659G | I220Ser | 1 | ≤0.0125 | ||
| ddn | C117A | Gly39Gly | 5 | ≤0.0125 | |
| G241A | Gly81Ser | 4 | ≤0.0125 | ||
| 1 | 0.2 | ||||
| Resistant | ddn | G241A | Gly81Ser | 21 | 0.4 |
| 9 | 0.8 | ||||
| 1 | >1.6 | ||||
| G242A | Gly81Asp | 2 | >1.6 |
Abbreviation: MIC, minimum inhibitory concentration.
Among the 372 bedaquiline-susceptible strains, five non-silent and four silent mutations were identified in Rv0678. In addition, one non-silent mutation was identified in atpE. Among the 48 bedaquiline-resistant strains, seven non-silent mutations and one silent mutation were identified in Rv0678 (Table 3).
Table 3. Mutations identified in bedaquiline resistance-related genes.
| Bedaquiline | Gene | Nucleotide substitution | Amino acid change | Strains (N) | MIC (mg/L) |
|---|---|---|---|---|---|
| Susceptible | rv0678 | A199G | Ile67Val | 1 | 0.25 |
| A263G | Asp88Gly | 1 | 0.0625 | ||
| C270A | Arg90Arg | 1 | 0.25 | ||
| C33T | Gly11Gly | 2 | 0.125 | ||
| 12 | 0.25 | ||||
| C33T, A63C | Gly11Gly, Glu21Asp | 1 | 0.25 | ||
| C9A | Val3Val | 1 | 0.0625 | ||
| G368A | Arg123Lys | 1 | ≤ 0.03125 | ||
| 1 | 0.125 | ||||
| 2 | 0.25 | ||||
| T437C | Met146Thr | 1 | 0.125 | ||
| atpE | C124T | Gln42stop | 1 | 0.25 | |
| Resistant | rv0678 | A292G | Asn98Asp | 1 | 1 |
| A63C | Glu21Asp | 1 | 0.5 | ||
| 1 | 1 | ||||
| A91C | Ser31Arg | 1 | 0.5 | ||
| C268T | Arg90cys | 1 | 1 | ||
| C33T | Gly11Gly | 1 | 0.5 | ||
| 1 | 2 | ||||
| C33T, T236C | Gly11Gly, Phe79Ser | 1 | 1 | ||
| C98G | Thr33Ser | 1 | 2 | ||
| G368A | Arg123Lys | 1 | 1 |
Abbreviation: MIC, minimum inhibitory concentration.
One non-silent mutation in rplC and seven substitution mutations in rrl were identified in the 419 linezolid-susceptible strains. The single linezolid-resistant strain did not have any mutations in these genes (Table 4).
Table 4. Mutations identified in linezolid resistance-related genes.
| Linezolid | Gene | Nucleotide substitution | Amino acid change | Strains (N) | MIC (mg/L) |
|---|---|---|---|---|---|
| Susceptible | rrl | C2223T, C2232T | 1 | 0.25 | |
| G2324A | 1 | ≤ 0.125 | |||
| 1 | 0.25 | ||||
| A2526C | 1 | ≤ 0.125 | |||
| 1 | 0.25 | ||||
| C2990T | 1 | 0.5 | |||
| C2993G | 1 | 0.5 | |||
| A3094G | 1 | 0.5 | |||
| rplC | A328G | Ser110Gly | 1 | 0.25 |
Abbreviation: MIC, minimum inhibitory concentration.
DISCUSSION
To investigate the MIC distributions of delamanid, bedaquiline, and linezolid among Korean MDR and XDR clinical strains, we performed phenotypic DST using the broth microdilution method and sequencing of drug resistance-related genes for 420 clinical M. tuberculosis strains. In the case of delamanid, based on a critical concentration of 0.2 mg/L, we found that most susceptible strains exhibited an MIC of ≤0.0125 mg/L. This MIC distribution among susceptible strains is similar to previous estimates [15,17,21]. Keller et al [17] reported three resistant strains exhibiting MICs for delamanid of >0.32 mg/L, while others reported resistant strains with MICs of >8 mg/L [15,21]. Unlike previous studies, most delamanid-resistant strains in the present study exhibited MICs of 0.4–0.8 mg/L and showed different MIC distribution profiles.
A genetic analysis of the 379 delamanid-susceptible strains revealed four nonsynonymous and seven silent mutations in fbiA and ddn. The positions of nucleotide substitutions were different from those reported by Schena et al [15], except for Leu113Leu. Of the 41 resistant strains, 33 possessed mutations in fbiA or ddn. Although various mutations in fbiA and ddn were reported in resistant strains, there was little overlap between these previously reported mutations and those identified in our study. Schena et al [15] reported four mutations in fbiA and ddn, all of which were nonsense mutations that resulted in a premature stop codon. Another study reported a resistant strain with an Asp49Thr mutation in fbiA [18]. Haver et al [5] reported 19 mutations, including stop mutations, at various positions in ddn; the Gly81Asp mutation (MIC=1 mg/L), which is associated with drug activation, was particularly notable. This was the only mutation that was also identified in the present study (MIC ≥1.6 mg/L), and it showed a high resistance.
Of note, the Gly81Ser mutation appeared at a high frequency (75.6%) in the resistant strains. In an additional experiment, we confirmed that this mutation occurred at an even higher frequency (81%) in resistant strains with MIC >0.4 mg/L (data not shown). However, the same mutation was found in susceptible strains (MIC ≤0.0125 mg/L); therefore, it is unclear whether it is involved in drug resistance.
As the critical concentration of bedaquiline has yet to be determined by Johnson & Johnson, we used an MIC of 0.25 mg/L, which was approved by EUCAST [22]. We confirmed that the MIC for most susceptible strains was 0.125–0.25 mg/L and that for most resistant strains was 0.5–1 mg/L. A genetic analysis of bedaquiline resistance-related genes identified nonsynonymous and silent mutations in both bedaquiline-susceptible and -resistant strains. The mutations we identified in Rv0678 and atpE differed from those previously reported [9,24]. There was no single mutation harbored by a large proportion of the resistant strains.
Based on a critical concentration of 1 mg/L for linezolid, the MIC for the most susceptible strains was 0.25–0.5 mg/L. Although nucleotide substitutions in rrl and rplC were detected in susceptible strains, no mutations were found in these genes in the single linezolid-resistant strain. Several previous studies have reported mutations that confer resistance to linezolid. Hillemann et al [13] reported nucleotide substitutions at positions 2016 and 2576 in rrl among in vitro-selected linezolid-resistant M. tuberculosis mutants. Beckert et al [14] identified a T460C mutation in rplC in in vitro-selected mutants and clinical isolates, and this mutation was associated with higher linezolid MIC values (>2 mg/L) [25].
We used MDR and XDR strains that have presumably not been exposed to these three drugs. Therefore, it is difficult to confirm the relationship between the identified mutations and resistance, as high MIC values due to acquired resistance were not observed. However, we identified resistant strains with high MIC values (>1.6 mg/L) in the case of delamanid, most of which carried the Gly81Ser mutation in ddn. We hypothesize that this is due to a mechanism other than acquired resistance.
The emergence of drug-resistant TB is accompanied by low treatment success, especially in patients with MDR and XDR TB. The three anti-TB drugs we examined have been used to treat patients with MDR and XDR TB, with improved success rates. Continued phenotypic DST for these drugs is imperative for mitigating the inappropriate use of antimicrobial agents.
A limitation of this study is that we did not evaluate clinical strains with acquired resistance to the three drugs. However, we were able to determine the MICs for three anti-TB drugs in Korean MDR and XDR strains and isolated a few strains resistant to these drugs. Further studies are needed to confirm the genetic basis and determine the underlying mechanisms of resistance to these anti-TB drugs.
Acknowledgment
This study was supported by a grant from the Korean Health Technology R&D Project, Ministry of Health & Welfare, Korea (HI16C1554).
Footnotes
Authors' Disclosures of Potential Conflicts of Interest: No potential conflicts of interest relevant to this article were reported.
References
- 1.Blair HA, Scott LJ. Delamanid: a review of its use in patients with multidrug-resistant tuberculosis. Drugs. 2015;75:91–100. doi: 10.1007/s40265-014-0331-4. [DOI] [PubMed] [Google Scholar]
- 2.World Health Organization. Global tuberculosis report 2016. [Updated on Jun 2016]. http://www.who.int/tb/publications/global_report/en.
- 3.Gler MT, Skripconoka V, Sanchez-Garavito E, Xiao H, Cabrera-Rivero JL, Vargas-Vasquez DE, et al. Delamanid for multidrug-resistant pulmonary tuberculosis. N Engl J Med. 2012;366:2151–2160. doi: 10.1056/NEJMoa1112433. [DOI] [PubMed] [Google Scholar]
- 4.Matsumoto M, Hashizume H, Tomishige T, Kawasaki M, Tsubouchi H, Sasaki H, et al. OPC-67683, a nitro-dihydro-imidazooxazole derivative with promising action against tuberculosis in vitro and in mice. PLoS Med. 2006;3:e466. doi: 10.1371/journal.pmed.0030466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Haver HL, Chua A, Ghode P, Lakshminarayana SB, Singhal A, Mathema B, et al. Mutations in genes for the F420 biosynthetic pathway and a nitroreductase enzyme are the primary resistance determinants in spontaneous in vitro-selected PA-824-resistant mutants of Mycobacterium tuberculosis. Antimicrob Agents Chemother. 2015;59:5316–5323. doi: 10.1128/AAC.00308-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Feuerriegel S, Köser CU, Baù D, Rüsch-Gerdes S, Summers DK, Archer JA, et al. Impact of fgd1 and ddn diversity in Mycobacterium tuberculosis complex on in vitro susceptibility to PA-824. Antimicrob Agents Chemother. 2011;55:5718–5722. doi: 10.1128/AAC.05500-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Andries K, Verhasselt P, Guillemont J, Göhlmann HW, Neefs JM, Winkler H, et al. A diarylquinoline drug active on the ATP synthase of Mycobacterium tuberculosis. Science. 2005;307:223–227. doi: 10.1126/science.1106753. [DOI] [PubMed] [Google Scholar]
- 8.Huitric E, Verhasselt P, Andries K, Hoffner SE. In vitro antimycobacterial spectrum of a diarylquinoline ATP synthase inhibitor. Antimicrob Agents Chemother. 2007;51:4202–4204. doi: 10.1128/AAC.00181-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Andries K, Villellas C, Coeck N, Thys K, Gevers T, Vranckx L, et al. Acquired resistance of Mycobacterium tuberculosis to bedaquiline. PLoS One. 2014;9:e102135. doi: 10.1371/journal.pone.0102135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huitric E, Verhasselt P, Koul A, Andries K, Hoffner S, Andersson DI. Rates and mechanisms of resistance development in Mycobacterium tuberculosis to a novel diarylquinoline ATP synthase inhibitor. Antimicrob Agents Chemother. 2010;54:1022–1028. doi: 10.1128/AAC.01611-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gu B, Kelesidis T, Tsiodras S, Hindler J, Humphries RM. The emerging problem of linezolid-resistant Staphylococcus. J Antimicrob Chemother. 2013;68:4–11. doi: 10.1093/jac/dks354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Birmingham MC, Rayner CR, Meagher AK, Flavin SM, Batts DH, Schentag JJ. Linezolid for the treatment of multidrug-resistant, gram-positive infections: experience from a compassionate-use program. Clin Infect Dis. 2003;36:159–168. doi: 10.1086/345744. [DOI] [PubMed] [Google Scholar]
- 13.Hillemann D, Rüsch-Gerdes S, Richter E. In vitro-selected linezolid-resistant Mycobacterium tuberculosis mutants. Antimicrob Agents Chemother. 2008;52:800–801. doi: 10.1128/AAC.01189-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beckert P, Hillemann D, Kohl TA, Kalinowski J, Richter E, Niemann S, et al. rplC T460C identified as a dominant mutation in linezolid-resistant Mycobacterium tuberculosis strains. Antimicrob Agents Chemother. 2012;56:2743–2745. doi: 10.1128/AAC.06227-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schena E, Nedialkova L, Borroni E, Battaglia S, Cabibbe AM, Niemann S, et al. Delamanid susceptibility testing of Mycobacterium tuberculosis using the resazurin microtiter assay and the BACTEC™ MGIT™ 960 system. J Antimicrob Chemother. 2016;71:1532–1539. doi: 10.1093/jac/dkw044. [DOI] [PubMed] [Google Scholar]
- 16.Kaniga K, Cirillo DM, Hoffner S, Ismail NA, Kaur D, Lounis N, et al. A multilaboratory, multicountry study to determine bedaquiline minimal inhibitory concentration quality control ranges for phenotypic drug-susceptibility testing. J Clin Microbiol. 2016;54:2956–2962. doi: 10.1128/JCM.01123-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Keller PM, Hömke R, Ritter C, Valsesia G, Bloemberg GV, Bötteger EC. Determination of MIC distribution and epidemiological cutoff values for bedaquiline and delamanid in Mycobacterium tuberculosis using the MGIT 960 system equipped with TB eXiST. Antimicrob Agents Chemother. 2015;59:4352–4355. doi: 10.1128/AAC.00614-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bloemberg GV, Keller PM, Stucki D, Trauner A, Borrell S, Latshang T, et al. Acquired resistance to bedaquiline and delamanid in therapy for tuberculosis. N Engl J Med. 2015;373:1986–1988. doi: 10.1056/NEJMc1505196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang TS, Liu YC, Sy CL, Chen YS, Tu HZ, Chen BC. In vitro activities of linezolid against clinical isolates of Mycobacterium tuberculosis complex isolated in Taiwan over 10 years. Antimicrob Agents Chemother. 2008;52:2226–2227. doi: 10.1128/AAC.00414-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee SH, Choi HB, Yu SY, Chang UJ, Kim CK, Kim HJ. Detection of first-line anti-tuberculosis drug resistance mutations by allele-specific primer extension on a microsphere-based platform. Ann Lab Med. 2015;35:487–493. doi: 10.3343/alm.2015.35.5.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stinson K, Kurepina N, Venter A, Fujiwara M, Kawasaki M, Timm J, et al. MIC of delamanid (OPC-67683) against Mycobacterium tuberculosis clinical isolates and a proposed critical concentration. Antimicrob Agents Chemother. 2016;60:3316–3322. doi: 10.1128/AAC.03014-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.European Committee on Antimicrobial Susceptibility Testing. Definitions of clinical breakpoints and epidemiological cut-off values. Clinical Breakpoints Table V.7.1. Valid From 2017-03-10. [Updated on Mar 2017]. http://www.eucast.org/clinical_breakpoints/
- 23.CLSI. Susceptibility testing of mycobacteria, nocardiae, and other aerobic actinomycetes. 2nd edition. Wayne, PA: Clinical Laboratory Standards Institute; 2011. CLSI M24-A2. [PubMed] [Google Scholar]
- 24.Segala E, Sougakoff W, Chauffour AN, Jarller V, Petrella S. New mutations in the mycobacterial ATP synthase: new insights into the binding of the diarylquinoline TMC207 to the ATP synthase C-ring structure. Antimicrob Agents Chemother. 2012;56:2326–2334. doi: 10.1128/AAC.06154-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang S, Chen J, Cui P, Shi W, Shi X, Niu H, et al. Mycobacterium tuberculosis mutations associated with reduced susceptibility to linezolid. Antimicrob Agents Chemother. 2016;60:2542–2544. doi: 10.1128/AAC.02941-15. [DOI] [PMC free article] [PubMed] [Google Scholar]