Abstract
Heterosis has been widely used to increase grain quality and yield, but its genetic mechanism remains unclear. In this study, the genetic basis of heterosis for four maize kernel traits was examined in two test populations constructed using a set of 184 chromosome segment substitution lines (CSSLs) and two inbred lines (Zheng58 and Xun9058) in two environments. 63 and 57 different heterotic loci (HL) were identified for four kernel traits in the CSSLs × Zheng58 and CSSLs × Xun9058 populations, respectively. Of these, nine HL and six HL were identified for four kernel traits in the CSSLs × Zheng58 and CSSLs × Xun9058 populations, at the two locations simultaneously. Comparative analysis of the HL for the four kernel traits identified only 21 HL in the two test populations simultaneously. These results showed that most HL for the four kernel traits differed between the two test populations. The common HL were important loci from the Reid × Tangsipingtou heterotic model, and could be used to predict hybrid performance in maize breeding.
Introduction
Heterosis is used to describe the superiority of heterozygous genotypes over parental homozygotes with respect to one or more characteristics1. Hybrid varieties in many crop species exhibit high fertility rates, improved nutrient quality and content, and an increased resilience under abiotic and biotic stress conditions2,3. The use of hybrid seed has significantly increased crop yield, with hybrid rice and maize currently accounting for over 50% of global production4,5. A credible approach to estimate hybrid phenotypes resulting from a better comprehension of the genetic basis of heterosis would substantially benefit hybrid breeding programs. In previous studies, dominance, epistasis, and overdominance have been the main genetic models employed to explain heterosis6–9. The dominance hypothesis supports the expression at multiple loci of beneficial dominant alleles from both parents that are combined in the hybrid6,7,10,11. In contrast, the overdominance hypothesis gives special attention to the presence of loci where the state of heterozygosis surpasses either homozygote1,12,13. Last, the interaction among favourable alleles from the given parents at different loci forms the foundation of the epistasis hypothesis14–18.
In previous investigations, three types of genetic populations have been used to identify quantitative trait loci (QTL) or heterotic loci (HL). The first population design, which is based on the North Carolina Design III (NCIII), uses F2, F3, or recombinant inbred lines (RILs) that are backcrossed with their parental lines. Such a design was once used to identify QTLs with overdominant or dominant effects13,19,20. Using a design of an immortalised F2 (IF2) population developed from paired crosses of RILs in rice, Hua et al.16 detected 44 HL for grain yield and its components in rice, while Tang et al.21 identified 13 HL for grain yield and its components in maize in the same population. Recently, chromosome segment substitution line (CSSL) backcross populations have been used to identify HL in tomato22, rice23, and cotton3. For instance, Meyer et al.24 reported a QTL for early stage heterosis related to biomass using testcross hybrids developed from 140 introgression line populations in Arabidopsis. Additionally, Shen et al.25 identified 15 dominance HL for plant height using a test population comprising a set of CSSLs in rice.
Maize has long served as a model species for dichotomising the genetic foundation of heterosis9. Despite an extensive and dramatic history of success, a striking discordance still exists, especially in maize, between the widespread agricultural utilisation of hybrid vigour and acknowledging the basis of heterosis5,8. Such disharmony blocks the effective exploitation of heterosis. The production of new hybrids is time-consuming26, so a better understanding of the underlying genetic basis of heterosis would improve the reliability of predicting hybrid phenotypes for use in hybrid breeding programs.
Grain yield, a complicated trait comprising several major components in different crops, is affected by many genetic and non-genetic factors. In maize, kernels per row, row number, and 100-kernel weight are the three major components of grain yield, with 100-kernel weight showing lower heterosis than the other two21. Kernel weight also consists of three secondary traits: kernel length, kernel width, and kernel thickness. Kernel weight and size are characterised as key components of grain yield in different hybrids and their parents27,28, and several reports have suggested that kernel length, width, and depth greatly influence kernel weight29–31. In the genetic basis of kernel weight and its related traits, several QTL mapping studies have been conducted for maize21,32–34, and the QTL for secondary traits of kernel weight, including kernel length, width, depth or thickness, volume, and ratio have also been identified in previous studies35,36.
For the heterosis of kernel weight and its related traits, Tang et al.21 identified two HL for 100-kernel weight in maize, and Wei et al.37 also reported five HL for 100-kernel weight using a single segment substitution lines testcross population. However, the heterosis of secondary traits of kernel weight is unclear. In this study, we dissected the genetic basis of heterosis for four kernel traits using two test populations constructed from a CSSL population and two test inbred lines, Zheng58 and Xun9058. The HL for the measured traits were identified by comparing single test crosses with the corresponding control hybrid (CK). The objectives of this study were to: (1) detect HL underlying the heterosis for kernel traits, and (2) analyse the genetic basis of heterosis for kernel traits in maize. These HL associated with kernel traits and their associated molecular markers may be used to predict hybrid performance in future maize breeding experiments.
Results
Performance of kernel traits and its mid-parent heterosis in the test populations
Average kernel lengths of the CSSLs × Zheng58 population were 10.21 mm and 10.62 mm at Changge and Hebi locations, respectively (Table 1), with corresponding mid-parent heterosis values of 12.31% and 11.46%. These values at the two locations were similar to those of the hybrid Zheng58 × lx9801. Mean kernel widths for the hybrid Zheng58 × lx9801 were 9.23 mm and 9.60 mm at the two locations, with mid-parent heterosis values of 7.88% and 6.27%, respectively. The CSSLs × Zheng58 population mean kernel width was similar to that of the control hybrid at the two locations. Mid-parent heterosis values for kernel width at the two locations were 8.46% and 7.69%, respectively. Regarding kernel thickness, average values of the CSSLs × Zheng58 population were 4.47 mm and 4.59 mm at the two locations, with mid-parent heterosis values of −9.32% and −4.09%, respectively. These test population values were similar to those of the control hybrid. The mean 100-kernel weights for the population were 30.23 g and 34.86 g at the two locations, with 18.61% and 24.34% mid-parent heterosis, respectively.
Table 1.
Performance of kernel traits in CSSLs × Zheng58 and CSSLs × Xun9058 populations.
| Locations | Traits | Parents | Zheng58 × lx9801 | CSSLs × Zheng58 | |||||
|---|---|---|---|---|---|---|---|---|---|
| lx9801 | Zheng58 | Mean | Mid-parent heterosis (%) | Mean | Mid-parent heterosis (%) | Ske. | Kur. | ||
| Changge | KL(mm) | 8.93 ± 0.08 | 9.36 ± 0.02 | 10.27 ± 0.03 | 12.28 | 10.21 ± 0.04 | 12.31 | −0.44 | 0.19 |
| KW(mm) | 8.50 ± 0.03 | 8.62 ± 0.07 | 9.23 ± 0.01 | 7.88 | 9.10 ± 0.04 | 8.46 | 0.12 | −0.68 | |
| KT(mm) | 4.57 ± 0.02 | 5.29 ± 0.04 | 4.48 ± 0.03 | −9.22 | 4.47 ± 0.01 | −9.32 | −0.48 | 0.36 | |
| HKW(g) | 21.24 ± 0.19 | 28.95 ± 0.71 | 29.75 ± 0.48 | 18.56 | 30.23 ± 0.17 | 18.61 | −0.09 | −0.57 | |
| Hebi | KL(mm) | 9.21 ± 0.05 | 9.76 ± 0.04 | 10.58 ± 0.02 | 11.58 | 10.62 ± 0.06 | 11.46 | −0.15 | 0.39 |
| KW(mm) | 9.13 ± 0.03 | 8.93 ± 0.04 | 9.60 ± 0.04 | 6.27 | 9.66 ± 0.06 | 7.69 | 0.37 | −0.21 | |
| KT(mm) | 4.57 ± 0.01 | 5.14 ± 0.06 | 4.66 ± 0.04 | −3.99 | 4.59 ± 0.01 | −4.09 | −0.31 | −0.30 | |
| HKW(g) | 24.97 ± 1.24 | 30.7 ± 1.13 | 34.54 ± 0.42 | 24.07 | 34.86 ± 0.18 | 24.34 | −0.14 | 0.86 | |
| Locations | Traits | Parents | Xun9058 × lx9801 | CSSLs × Xun9058 | |||||
| Xun9058 | CSSLs | Mean | Mid-parent heterosis (%) | Mean | Mid-parent heterosis (%) | Ske | kur | ||
| Changge | KL(mm) | 9.80 ± 0.04 | 8.92 ± 0.06 | 10 ± 0.03 | 6.78 | 9.94 ± 0.03 | 6.2 | −0.16 | 0.01 |
| KW(mm) | 8.55 ± 0.02 | 8.90 ± 0.05 | 9.1 ± 0.02 | 6.74 | 9.06 ± 0.03 | 3.84 | −0.51 | 0.49 | |
| KT(mm) | 5.29 ± 0.06 | 4.58 ± 0.04 | 4.37 ± 0.01 | −11.35 | 4.41 ± 0.01 | −10.64 | 0.02 | 0.24 | |
| HKW(g) | 24.17 ± 0.56 | 21.22 ± 0.25 | 24.91 ± 0.47 | 9.71 | 25.73 ± 0.25 | 13.37 | 0.01 | 0.60 | |
| Hebi | KL(mm) | 10.77 ± 0.06 | 9.23 ± 0.06 | 10.69 ± 0.04 | 7.01 | 10.63 ± 0.08 | 6.91 | −0.47 | 0.85 |
| KW(mm) | 8.93 ± 0.03 | 8.90 ± 0.07 | 10.01 ± 0.03 | 10.82 | 9.87 ± 0.09 | 12.30 | −0.18 | −0.62 | |
| KT(mm) | 5.14 ± 0.07 | 4.58 ± 0.03 | 4.57 ± 0.04 | −5.85 | 4.54 ± 0.02 | −5.93 | 0.46 | 0.47 | |
| HKW(g) | 30.23 ± 0.82 | 24.85 ± 0.29 | 34.35 ± 0.67 | 24.46 | 34.41 ± 0.29 | 24.73 | 0.01 | −0.01 | |
KL, kernel length; KW, kernel width; KT, kernel thickness; HKW, 100-kernel weight; ± , standard deviation.
Mean kernel lengths for the hybrid Xun9058 × lx9801 were 10.00 mm and 10.69 mm, with 6.78% and 7.01% mid-parent heterosis values at the two locations, respectively. The CSSLs × Xun9058 population had the closest kernel length and mid-parent heterosis to its control hybrid. Regarding kernel width, average values for the CSSLs × Xun9058 population were 9.06 mm and 9.87 mm, with 3.84% and 12.30% mid-parent heterosis at the two locations, respectively. The CSSLs × Xun9058 population had the closest values of kernel thickness and its mid-parent heterosis to the control hybrid. The mean 100-kernel weights for the CSSLs × Xun9058 population were 25.73 g and 34.41 g, and mid-parent heterosis values were 13.37% and 24.73% at the two locations, respectively.
The four kernel traits of the two test populations exhibited significant variation between locations and genotypes (P < 0.01; Table 2), and only kernel thickness showed significant location × genotype interaction variation at the P < 0.05 level. The heritability (H2B) of kernel length, width, thickness, and 100-kernel weight was 73.50%, 73.18%, 78.58%, and 67.62%, respectively, in the CSSLs × Zheng58 population, with a relative lower heritability of the four measured traits (65.22%, 64.85%, 67.35%, and 64.89%); kernel thickness showed the highest heritability and negative mid-parent heterosis of all four kernel traits.
Table 2.
Analysis of variance for the four kernel traits in CSSLs × Zheng58 and CSSLs × Xun9058 populations.
| Traits | Populations | CSSLs × Zheng58 | CSSLs × Xun9058 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| DF | SS | MS | F | P | DF | SS | MS | F | P | ||
| Kernel length | Location | 1 | 40.42 | 40.42** | 289.29 | <0.0001 | 1 | 89.11 | 89.11** | 527.35 | <0.0001 |
| Repetition | 4 | 1.77 | 0.44 | 3.17 | 0.014 | 4 | 0.76 | 0.19 | 1.13 | 0.34 | |
| Genotype | 175 | 43.30 | 0.25** | 1.77 | <0.0001 | 164 | 41.29 | 0.25** | 1.49 | 0.0006 | |
| Location × Genotype | 171 | 28.62 | 0.17 | 1.20 | 0.07 | 159 | 26.81 | 0.17 | 1.00 | 0.49 | |
| Error | 507 | 70.84 | 0.14 | 466 | 78.74 | 0.17 | |||||
| Heritability (H2B) | 73.50% | 65.22% | |||||||||
| Kernel width | Location | 1 | 69.68 | 69.68** | 300.37 | <0.0001 | 1 | 114.95 | 114.95** | 393.68 | <0.0001 |
| Repetition | 4 | 9.94 | 2.49** | 10.71 | <0.0001 | 4 | 1.27 | 0.32 | 1.09 | 0.36 | |
| Genotype | 175 | 57.85 | 0.33** | 1.43 | 0.002 | 164 | 44.28 | 0.27** | 0.93 | 0.005 | |
| Location × Genotype | 171 | 39.08 | 0.23 | 0.99 | 0.54 | 159 | 53.49 | 0.34 | 1.15 | 0.13 | |
| Error | 507 | 117.62 | 0.23 | 466 | 136.07 | 0.29 | |||||
| Heritability (H2B) | 73.18% | 64.85% | |||||||||
| Kernel thickness | Location | 1 | 346.67 | 346.67** | 97.27 | <0.0001 | 1 | 338.07 | 338.07** | 81.32 | <0.0001 |
| Repetition | 4 | 12.76 | 3.19 | 0.89 | 0.47 | 4 | 10.02 | 2.50 | 0.60 | 0.66 | |
| Genotype | 175 | 1291.76 | 7.38** | 2.07 | <0.0001 | 164 | 1539.45 | 9.39** | 2.26 | <0.0001 | |
| Location × Genotype | 174 | 754.52 | 4.34* | 1.22 | 0.04 | 164 | 861.48 | 5.25* | 1.26 | 0.03 | |
| Error | 610 | 2174.06 | 3.56 | 572 | 2377.90 | 4.16 | |||||
| Heritability (H2B) | 78.58% | 67.35% | |||||||||
| 100-kernel weight | Location | 1 | 3038.61 | 3038.61** | 683.33 | <0.0001 | 1 | 10502.4 | 10502.4** | 1654.41 | <0.0001 |
| Repetition | 4 | 280.60 | 70.15** | 15.78 | <0.0001 | 4 | 19.21 | 4.80 | 0.76 | 0.55 | |
| Genotype | 175 | 1210.44 | 6.92** | 1.56 | 0.0002 | 164 | 1616.15 | 9.85** | 1.55 | 0.0003 | |
| Location × Genotype | 157 | 719.88 | 4.59 | 1.03 | 0.4 | 149 | 1281.23 | 8.6* | 1.35 | 0.01 | |
| Error | 404 | 1796.49 | 4.45 | 373 | 2367.85 | 6.35 | |||||
| Heritability (H2B) | 67.62% | 64.89% | |||||||||
DF, degrees of freedom; SS, Sum of square; MS, mean square; *, **p < 0.05 and p < 0.01, respectively.
Correlations between phenotype and heterosis for the four kernel traits
For the CSSLs × Zheng58 population, kernel length was significantly correlated with kernel width (P < 0.01; Table 3), while 100-kernel weight was significantly correlated with kernel length, width, and thickness (P < 0.01). The mid-parent heterosis for kernel length was also significantly correlated with kernel width and 100-kernel weight (P < 0.01); however, kernel length was negatively correlated with kernel thickness (P < 0.01). Additionally, kernel width was significantly correlated with 100-kernel weight (P < 0.01).
Table 3.
Correlation coefficients for four kernel traits in CSSLs × Zheng58 and CSSLs × Xun9058 populations.
| Populations | Trait | Kernel length | Kernel width | Kernel thickness | 100-kernel weight |
|---|---|---|---|---|---|
| CSSLs × Zheng58 | Kernellength | 0.25** | −0.09 | 0.19** | |
| Kernelwidth | 0.25** | 0.05 | 0.35** | ||
| Kernel thickness | −0.24** | 0.06 | 0.27** | ||
| 100-kernels weight | 0.20** | 0.31** | 0.04 | ||
| CSSLs × Xun9058 | Kernellength | 0.25** | −0.10 | 0.21** | |
| Kernelwidth | 0.16 | 0.05 | 0.36** | ||
| Kernel thickness | −0.14 | 0.03 | 0.27** | ||
| 100-kernel weight | 0.03 | 0.21** | 0.16* |
Correlation coefficients for phenotype and mid-parent heterosis of each trait are listed above and below the diagonal. *, **p < 0.05 and p < 0.01, respectively.
In the CSSLs × Xun9058 population, significant correlations were observed between kernel length and kernel width (P < 0.01), and between 100-kernel weight and kernel length, width, and thickness (P < 0.01). Regarding mid-parent heterosis, the 100-kernel weight was significantly correlated with kernel width and thickness (P < 0.01 and P < 0.05, respectively).
QTL detected for the four kernel traits in the CSSL population
For kernel length, 10 QTL were identified using the average data of each CSSL in the same location over 2 years (Table 4). Among them, the QTL qKL4 was detected in two locations simultaneously, making a 14.69% and 3.09% phenotypic contribution, and increasing kernel length by 0.13 mm and 0.03 mm at Changge and Hebi, respectively. Nine QTL for kernel width were also identified in the CSSL population at the two locations, and QTL qKW6b accounted for −3.90% and −3.83% phenotypic variation at the two locations. Seventeen QTL were identified for kernel thickness, with QTL qKT3b, qKT6d, and qKT9b detected at the two locations simultaneously. For 100-kernel weight, 20 QTL were detected, of which qHKW6f accounted for −6.86% and −5.07% phenotypic contribution at the two locations, and QTL qHKW9b made contributions of −6.11% and −4.03% in the CSSL population.
Table 4.
QTL detected for the four kernel traits in the CSSLs population.
| Location | Trait | QTL | Bin | Chromosomal region | Additive | Contribution (%) | P value |
|---|---|---|---|---|---|---|---|
| Changge | Kernel length | qKL1 | 1.06 | umc1035-umc1335-umc2396 | 0.13 | 14.69 | 0.019 |
| qKL2 | 2.04 | umc2088-umc1485-bnlg1861 | −0.02 | −2.39 | 0.036 | ||
| qKL4 | 4.01 | umc1017-umc1757-umc2280 | −0.03 | −3.23 | 0.009 | ||
| qKL6a | 6.05 | umc1614-umc2141-umc1805 | −0.03 | −3.51 | 0.013 | ||
| qKL9a | 9.02 | umc1170-umc1037-umc1033 | −0.05 | −5.75 | 0.045 | ||
| Kernel width | qKW1a | 1.06 | umc1035-umc1335-umc2396 | 0.10 | 12.09 | 0.009 | |
| qKW2 | 2.04 | umc2088-umc1485-bnlg1861 | −0.03 | −2.92 | 0.011 | ||
| qKW4 | 4.01 | umc1017-umc1757-umc2280 | 0.03 | 2.97 | 0.011 | ||
| qKW6a | 6.04 | umc2006-umc1614-umc2141 | 0.03 | 2.97 | 0.014 | ||
| qKW6a | 6.05 | umc1614-umc2141-umc1805 | 0.02 | 2.62 | 0.035 | ||
| qKW6b | 6.07 | umc1433-bnlg1380-bnlg1792 | −0.03 | −3.90 | 0.049 | ||
| Kernel thickness | qKT1a | 1.07 | umc1356-umc1278-umc1013 | −1.80 | −3.86 | 0.040 | |
| qKT1b | 1.08 | bnlg2228-dupssr12-umc2047 | −2.13 | −4.59 | 0.006 | ||
| qKT1d | 1.11 | umc2047-umc1538-bnlg131 | 2.21 | 4.77 | 0.006 | ||
| qKT2 | 2.04 | bnlg1064-umc1024-umc1465 | −0.54 | −1.17 | 0.030 | ||
| qKT2 | 2.04 | bnlg1064-umc1024-umc1465 | −1.48 | −3.19 | 0.027 | ||
| qKT3a | 3.07 | umc1489-umc1825-phi046 | −1.51 | −3.25 | 0.017 | ||
| qKT3b | 3.08 | umc1844-umc2275-umc2081 | −3.55 | −7.65 | 0.004 | ||
| qKT6a | 6.00 | phi075-bnlg238 | −1.58 | −3.41 | 0.000 | ||
| qKT6d | 6.08 | phi123-umc1127 | 2.08 | 4.48 | 0.001 | ||
| qKT7 | 7.03 | bnlg2271-umc1112-bnlg1805 | 1.64 | 3.54 | 0.003 | ||
| qKT9b | 9.02 | umc1170-umc1037-umc1033 | −2.14 | −4.61 | 0.009 | ||
| 100-kernel weight | qHKW1 | 1.03 | umc1403-umc1397-bnlg182 | −0.71 | −3.55 | 0.007 | |
| qHKW2b | 2.04 | umc2088-umc1485-bnlg1861 | −1.41 | −7.03 | 0.031 | ||
| qHKW3 | 3.03 | umc2259-phi036-umc1495 | −1.62 | −8.09 | 0.007 | ||
| qHKW4a | 4.01 | umc1017-umc1757-umc2280 | 1.11 | 5.56 | 0.027 | ||
| qHKW5a | 5.00 | umc2302-umc1990-umc1482 | 0.71 | 3.53 | 0.024 | ||
| qHKW6f | 6.07 | umc1433-bnlg1380-bnlg1792 | −1.37 | −6.86 | 0.013 | ||
| qHKW9a | 9.00 | umc1037-umc1033-bnlg1082 | −1.09 | −5.43 | 0.016 | ||
| qHKW9b | 9.02 | umc1170-umc1037-umc1033 | −1.22 | −6.11 | 0.002 | ||
| qHKW9c | 9.04 | umc1522-umc1492-umc1519 | −1.60 | −8.02 | 0.037 | ||
| qHKW9d | 9.06 | bnlg1191-umc2345-umc1310 | 1.94 | 9.7 | 0.033 | ||
| Hebi | Kernel length | qKL4 | 4.01 | umc1017-umc1757-umc2280 | 0.03 | 3.09 | 0.049 |
| qKL5 | 5.00 | umc2302-umc1990-umc1482 | 0.06 | 5.67 | 0.018 | ||
| qKL6b | 6.05 | bnlg1732-umc1424-umc1296 | 0.05 | 5.5 | 0.012 | ||
| qKL9b | 9.04 | umc1492-umc1519-umc1375 | 0.04 | 3.78 | 0.058 | ||
| qKL9c | 9.06 | bnlg1191-umc2345-umc1310 | 0.04 | 4.38 | 0.047 | ||
| qKL10 | 10.04 | umc1336-umc2163-umc2350 | 0.06 | 6.36 | 0.026 | ||
| Kernel width | qKW1b | 1.09 | umc2047-umc1538-bnlg131 | −0.05 | −5.11 | 0.033 | |
| qKW3 | 3.08 | umc1844-umc2275-umc2081 | −0.07 | −7.57 | 0.037 | ||
| qKW6b | 6.07 | umc1433-bnlg1380-bnlg1792 | −0.04 | −3.83 | 0.012 | ||
| qKW9a | 9.04 | umc1492-umc1519-umc1375 | −0.04 | −4.2 | 0.026 | ||
| qKW9b | 9.06 | bnlg1191-umc2345-umc1310 | 0.06 | 6.11 | 0.015 | ||
| Kernel thickness | qKT1b | 1.08 | bnlg2228-dupssr12-umc2047 | −1.81 | −3.95 | 0.011 | |
| qKT1c | 1.09 | umc2047-umc1538-bnlg131 | 2.53 | 5.54 | 0.005 | ||
| qKT2 | 2.04 | umc2088-umc1485-bnlg1861 | −0.78 | −1.7 | 0.008 | ||
| qKT3b | 3.08 | umc1844-umc2275-umc2081 | −3.22 | −7.05 | 0.008 | ||
| qKT5 | 5.00 | umc1496-umc1097-bnlg1006 | 1.97 | 4.3 | 0.002 | ||
| qKT6b | 6.00 | umc1178-phi389203-umc2316 | −1.26 | −2.75 | 0.002 | ||
| qKT6c | 6.07 | umc1433-bnlg1380-bnlg1792 | −1.75 | −3.84 | 0.003 | ||
| qKT6d | 6.08 | phi123-umc1127 | 0.87 | 1.9 | 0.033 | ||
| qKT9a | 9.00 | bnlg1272-bnlg1810-umc1809 | 2.47 | 5.4 | 0.027 | ||
| qKT9b | 9.02 | umc1170-umc1037-umc1033 | −1.82 | −3.97 | 0.015 | ||
| qKT9c | 9.04 | umc1492-umc1519-umc1375 | 1.11 | 2.44 | 0.019 | ||
| qKT9d | 9.05 | bnlg1091-bnlg1191-umc2345 | 2.90 | 6.34 | 0.005 | ||
| 100-kernel weight | qHKW2a | 2.03 | bnlg1064-umc1024-umc1465 | −1.55 | −4.97 | 0.038 | |
| qHKW4b | 4.00 | umc1232-phi072-umc1228 | 0.57 | 1.82 | 0.036 | ||
| qHKW5b | 5.05 | umc1729-bnlg118-umc1792 | 1.53 | 4.91 | 0.000 | ||
| qHKW6a | 6.02 | umc1979-nc009-umc1014 | 1.21 | 3.9 | 0.005 | ||
| qHKW6b | 6.04 | umc2006-umc1614-umc2141 | 0.73 | 2.34 | 0.018 | ||
| qHKW6c | 6.04 | nc009-umc1014-mmc0523 | 1.19 | 3.83 | 0.027 | ||
| qHKW6d | 6.04 | nc012-umc1020-bnlg1732 | −1.86 | −5.98 | 0.008 | ||
| qHKW6e | 6.05 | bnlg1732-umc1424-umc1296 | 1.74 | 5.58 | 0.000 | ||
| qHKW6f | 6.07 | umc1433-bnlg1380-bnlg1792 | −1.58 | −5.07 | 0.000 | ||
| qHKW6g | 6.07 | phi123-umc1127 | 0.55 | −0.77 | 0.039 | ||
| qHKW9b | 9.02 | umc1170-umc1037-umc1033 | −1.26 | −4.03 | 0.005 | ||
| qHKW10 | 10.04 | umc1336-umc2163-umc2350 | 0.79 | 2.53 | 0.014 |
Heterotic loci identified for kernel traits in the CSSLs × Zheng58 population
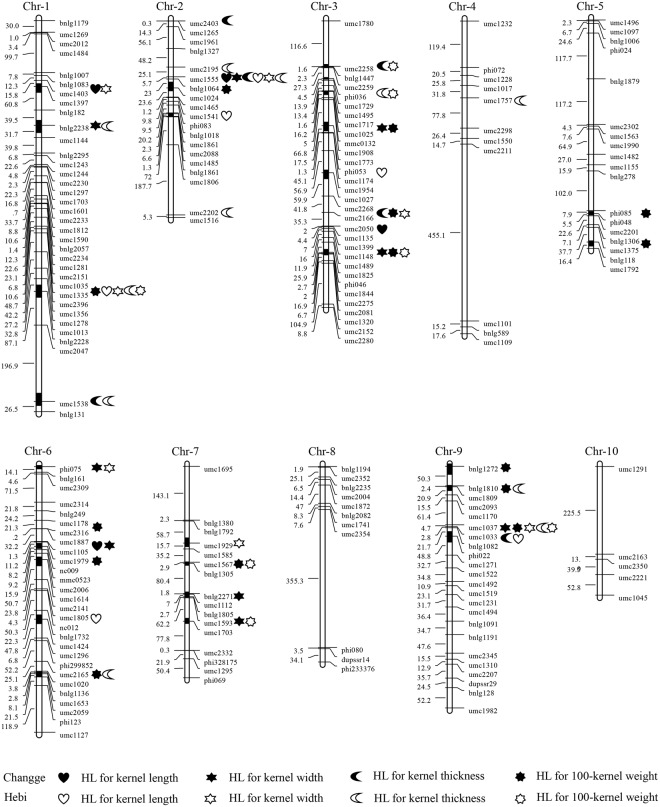
A total of 63 different HL were identified for the four kernel traits in the CSSLs × Zheng58 population at the two locations (Table 5; Fig. 1). Of these, 14 different HL for kernel length were detected in the test populations; hKL9b, identified at the two locations simultaneously, had −4.21% and −4.57% control heterosis at Changge and Hebi, respectively.
Table 5.
Heterotic loci detected for the four kernel traits in the CSSLs × Zheng58 population.
| Locations | Traits | HL | Bin | Chromosomal region | Control heterosis (%) | P value |
|---|---|---|---|---|---|---|
| Changge | Kernel length | hKL1b | 1.03 | umc1397-bnlg182-bnlg2238 | −7.12 | 0.004 |
| hKL1c | 1.06 | umc1035-umc1335-umc2396 | −7.28 | 0.028 | ||
| hKL3c | 3.07 | umc1135-umc1399-umc1148 | −7.28 | 0.016 | ||
| hKL3d | 3.07 | umc1844-umc2275-umc2081 | −7.77 | 0.032 | ||
| hKL4 | 4.01 | umc1228-umc1017-umc1757 | −6.31 | 0.024 | ||
| hKL5a | 5.06 | bnlg278-umc1680-phi085 | −6.8 | 0.045 | ||
| hKL6a | 6.04 | mmc0523-umc2006-umc1614 | −7.44 | 0.005 | ||
| hKL7b | 7.03 | umc1567-bnlg1305-bnlg2271 | 6.80 | 0.045 | ||
| hKL9b | 9.03 | bnlg1082-phi022-umc1271 | −4.21 | 0.023 | ||
| Kernel width | hKW1b | 1.04 | bnlg182-bnlg2238-umc1144 | 5.54 | 0.006 | |
| hKW1d | 1.08 | bnlg2228-dupssr12-umc2047 | 10.15 | 0.010 | ||
| hKW2 | 2.03 | umc2195-umc1555-bnlg1064 | 5.54 | 0.028 | ||
| hKW3a | 3.04 | umc2259-phi036-umc1495 | 5.54 | 0.042 | ||
| hKW3b | 3.05 | umc1174-bnlg1035-umc2127 | 6.27 | 0.016 | ||
| hKW3d | 3.08 | umc1844-umc2275-umc2081 | −5.9 | 0.029 | ||
| hKW3d | 3.08 | phi046-umc1844-umc2275 | 6.27 | 0.043 | ||
| hKW6b | 6.04 | umc1979-nc009-umc1014 | 6.83 | 0.014 | ||
| hKW7a | 7.02 | bnlg1792-umc1929-umc1585 | 5.90 | 0.005 | ||
| hKW7c | 7.03 | umc1567-bnlg1305-bnlg2271 | 5.17 | 0.026 | ||
| hKW7d | 7.04 | umc2332-phi328175-umc1295 | 8.12 | 0.036 | ||
| hKW9a | 9.02 | umc1037-umc1033-bnlg1082 | 4.06 | 0.049 | ||
| hKW9d | 9.06 | umc1310-umc2207-dupssr29 | 4.61 | 0.040 | ||
| Kernel thickness | hKT1d | 1.11 | bnlg2228-dupssr12-umc2047 | −5.53 | 0.024 | |
| hKT2a | 2.03 | umc2195-umc1555-bnlg1064 | −3.66 | 0.026 | ||
| hKT3a | 3.03 | phi374118-umc2258-bnlg1447 | −4.15 | 0.021 | ||
| hKT3c | 3.05 | phi053-umc1174-bnlg1035 | 6.05 | 0.034 | ||
| hKT3d | 3.05 | umc2127-umc1954-umc2166 | −4.51 | 0.039 | ||
| hKT5 | 5.04 | umc2302-umc1990-umc1482 | 3.43 | 0.041 | ||
| hKT7b | 7.03 | umc1567-bnlg1305-bnlg2271 | 10.56 | 0.037 | ||
| hKT8 | 8.03 | bnlg2082-umc1741-umc2354 | 5.28 | 0.015 | ||
| hKT9c | 9.03 | bnlg1082-phi022-umc1271 | −10.19 | 0.028 | ||
| 100-kernel weight | hHKW1b | 1.04 | bnlg182-bnlg2238-umc1144 | 8.95 | 0.001 | |
| hHKW1c | 1.07 | umc1335-umc2396-umc1356 | 7.98 | 0.005 | ||
| hHKW1c | 1.07 | umc1356-umc1278-umc1013 | 6.81 | 0.008 | ||
| hHKW1d | 1.08 | umc1278-umc1013-bnlg2228 | 7.39 | 0.005 | ||
| hHKW2b | 2.03 | umc2195-umc1555-bnlg1064 | 6.81 | 0.026 | ||
| hHKW3a | 3.03 | phi374118-umc2258-bnlg1447 | 6.81 | 0.008 | ||
| hHKW3b | 3.04 | umc2259-phi036-umc1495 | 12.06 | 0.000 | ||
| hHKW3d | 3.05 | umc2127-umc1954-umc2166 | −13.62 | 0.002 | ||
| hHKW7b | 7.03 | umc1567-bnlg1305-bnlg2271 | 11.48 | 0.006 | ||
| hHKW9a | 9.00 | phi233376-bnlg1272-bnlg1810 | −6.61 | 0.046 | ||
| hHKW9c | 9.02 | umc1170-umc1037-umc1033 | 12.06 | 0.035 | ||
| hHKW9d | 9.03 | bnlg1082-phi022-umc1271 | 12.65 | 0.004 | ||
| hHKW9e | 9.06 | umc1310-umc2207-dupssr29 | 8.56 | 0.024 | ||
| hHKW10 | 10.04 | umc2163-umc2350-umc1272 | 6.23 | 0.018 | ||
| Hebi | Kernel length | hKL1d | 1.08 | umc1013-bnlg2228-dupssr12 | 5.20 | 0.002 |
| hKL2a | 2.03 | umc2195-umc1555-bnlg1064 | 4.88 | 0.043 | ||
| hKL2b | 2.04 | umc2088-umc1485-bnlg1861 | 7.24 | 0.001 | ||
| hKL3a | 3.06 | umc1593-umc1027-umc2268 | 4.57 | 0.002 | ||
| hKL6b | 6.06 | bnlg1732-umc1424-umc1296 | −4.09 | 0.003 | ||
| hKL9b | 9.03 | bnlg1082-phi022-umc1271 | −4.57 | 0.01 | ||
| Kernel width | hKW1a | 1.03 | umc1397-bnlg182-bnlg2238 | 9.03 | 0.021 | |
| hKW1c | 1.07 | umc1335-umc2396-umc1356 | 6.94 | 0.034 | ||
| hKW2 | 2.03 | umc2195-umc1555-bnlg1064 | 5.73 | 0.037 | ||
| hKW3c | 3.05 | umc1954-umc2166-umc1593 | −5.90 | 0.027 | ||
| hKW6a | 6.00 | phi075-bnlg238 | 5.21 | 0.038 | ||
| hKW7a | 7.02 | bnlg1792-umc1929-umc1585 | 4.51 | 0.023 | ||
| hKW9a | 9.02 | umc1037-umc1033-bnlg1082 | 10.76 | 0.003 | ||
| Kernel thickness | hKT1a | 1.04 | bnlg182-bnlg2238-umc1144 | −5.06 | 0.034 | |
| hKT1c | 1.08 | umc1013-bnlg2228-dupssr12 | 6.55 | 0.047 | ||
| hKT1d | 1.11 | umc2047-umc1538-bnlg131 | −4.09 | 0.023 | ||
| hKT2a | 2.03 | bnlg1327-umc2195-umc1555 | −4.89 | 0.010 | ||
| hKT2a | 2.03 | umc2195-umc1555-bnlg1064 | −14.08 | 0.017 | ||
| hKT2b | 2.08 | umc1806-umc2202-umc1516 | −5.71 | 0.036 | ||
| hKT3b | 3.04 | umc1717-umc1025-mmc0132 | −5.52 | 0.033 | ||
| hKT4b | 4.03 | umc1757-umc2280-umc1550 | 5.37 | 0.029 | ||
| hKT6e | 6.07 | bnlg1136-umc1653-umc2059 | 6.42 | 0.025 | ||
| hKT9a | 9.01 | bnlg1272-bnlg1810-umc1809 | −7.75 | 0.025 | ||
| hKT9b | 9.02 | umc1170-umc1037-umc1033 | 6.21 | 0.001 | ||
| 100-kernel weight | hHKW1d | 1.08 | umc1013-bnlg2228-dupssr12 | 5.20 | 0.043 | |
| hHKW3a | 3.03 | phi374118-umc2258-bnlg1447 | 6.17 | 0.014 | ||
| hHKW3b | 3.04 | umc2259-phi036-umc1495 | 10.03 | 0.039 | ||
| hHKW3f | 3.08 | umc1844-umc2275-umc2081 | 12.6 | 0.024 | ||
| hHKW7a | 7.02 | umc1666-umc1703-umc1433 | 15.82 | 0.007 | ||
| hHKW7b | 7.03 | umc1567-bnlg1305-bnlg2271 | 6.49 | 0.013 | ||
| hHKW9b | 9.02 | umc1037-umc1033-bnlg1082 | 10.99 | 0.025 |
HL, heterotic loci.
Figure 1.
Integrated heterotic loci (HL) on genetic linkage maps for kernel traits in two environments in the CSSLs × Zheng58 population. The heterotic chromosome ranges are represented by black color.
Sixteen HL for kernel width were detected in the CSSLs × Zheng58 population at the two locations, including hKW2, hKW7a, and hKW9a, which were detected at both locations simultaneously. hKW2 increased kernel width by 5.54% and 5.73% compared with the control hybrid at Changge and Hebi, respectively, compared with 5.90% and 4.51% for hKW7a, and 4.06% and 10.76% for hKW9a.
We identified 17 different HL for kernel thickness in the CSSLs × Zheng58 population at the two locations, including nine detected at Changge and 10 at Hebi. hKT1d decreased the kernel thickness by −5.53% and −4.09% compared with the control hybrid, while hKT2a made −3.66% and −4.89% contributions to heterosis for kernel thickness at Changge and Hebi, respectively.
For 100-kernel weight, 16 HL were detected in the CSSLs × Zheng58 populations at the two locations, including 13 at Changge and seven at Hebi. Four of these, hHKW1d, hHKW3a, hHKW3b, and hHKW7b were identified at both locations simultaneously. hHKW1d increased the 100-kernel weight by 7.39% and 5.20%, and hHKW3b increased it by 12.60% and 10.30% compared with the control hybrid at the two locations, respectively. HL hHKW7b made 7.03% and 6.49% contributions to heterosis of the 100-kernel weight at the two locations, respectively.
Heterotic loci identified for kernel traits in the CSSLs × Xun9058 population
A total of 57 different HL for the four kernel traits were identified in the CSSLs × Xun9058 population at the two locations (Table 6; Fig. 2), including 36 detected at Changge and 26 at Hebi. For kernel length, a total of nine HL were detected in the populations at the two locations. hKL9a decreased the kernel length by 7.00% and 3.17% compared with the control hybrid at the two locations, respectively.
Table 6.
Heterotic loci detected for kernel-related traits in the CSSLs × Xun9058 population.
| Locations | Traits | HL | Bin | Chromosomal region | Control heterosis(%) | P value |
|---|---|---|---|---|---|---|
| Changge | Kernel length | hKL1a | 1.02 | umc2191-bnlg1007-bnlg1083 | 5.67 | 0.009 |
| hKL3b | 3.07 | umc1489-umc1825-phi046 | −4.33 | 0.041 | ||
| hKL5b | 5.09 | umc1792-umc1153 | −7.33 | 0.005 | ||
| hKL6c | 6.04 | umc1979-nc009-umc1014 | −7.50 | 0.027 | ||
| hKL7a | 7.02 | umc1666-umc1703-umc1433 | −10.00 | 0.017 | ||
| hKL9a | 9.01 | umc1809-umc2093-umc1170 | −7.00 | 0.045 | ||
| hKL9b | 9.06 | bnlg1091-bnlg1191-umc2345 | −8.00 | 0.013 | ||
| Kernel width | hKW1a | 1.03 | umc1403-umc1397-bnlg182 | −13.74 | 0.04 | |
| hKW3a | 3.05 | umc1954-umc2166-umc1593 | −6.04 | 0.026 | ||
| hKW3d | 3.08 | phi046-umc1844-umc2275 | −4.95 | 0.044 | ||
| hKW3e | 3.09 | umc1320-umc2152-umc2277 | −10.99 | 0.007 | ||
| hKW4 | 4.03 | umc1757-umc2280-umc1550 | −13.55 | 0.016 | ||
| hKW6c | 6.06 | bnlg1732-umc1424-umc1296 | 7.69 | 0.038 | ||
| hKW7b | 7.03 | bnlg2271-umc1112-bnlg1805 | −8.79 | 0.027 | ||
| hKW7d | 7.04 | umc2332-phi328175-umc1295 | 6.23 | 0.006 | ||
| hKW9b | 9.05 | umc1519-umc1375-umc1231 | 4.40 | 0.026 | ||
| hKW9c | 9.06 | bnlg1091-bnlg1191-umc2345 | −7.69 | 0.012 | ||
| Kernel thickness | hKT1a | 1.04 | bnlg182-bnlg2238-umc1144 | −4.64 | 0.001 | |
| hKT1b | 1.07 | umc1356-umc1278-umc1013 | 4.87 | 0.001 | ||
| hKT3f | 3.09 | umc1320-umc2152-umc2277 | 4.35 | 0.005 | ||
| hKT6a | 6.03 | umc1178-phi389203-umc2316 | 8.17 | 0.001 | ||
| hKT6b | 6.04 | umc1979-nc009-umc1014 | 7.28 | 0.002 | ||
| hKT6c | 6.05 | mmc0523-umc2006-umc1614 | 6.66 | 0.004 | ||
| hKT6d | 6.06 | bnlg1732-umc1424-umc1296 | 3.52 | 0.002 | ||
| hKT6e | 6.07 | umc2165-bnlg1136-umc1653 | 7.30 | 0.000 | ||
| hKT7a | 7.02 | bnlg1792-umc1929-umc1585 | 2.35 | 0.003 | ||
| hKT7c | 7.04 | bnlg1805-umc2332-phi328175 | 2.32 | 0.001 | ||
| hKT7d | 7.04 | bnlg2271-umc1112-bnlg1805 | 2.73 | 0.003 | ||
| hKT9b | 9.02 | umc1170-umc1037-umc1033 | 5.90 | 0.002 | ||
| hKT9e | 9.06 | bnlg1191-umc2345-umc1310 | −3.98 | 0.000 | ||
| 100-kernel weight | hHKW1a | 1.01 | bnlg1179-umc1269-umc2012 | 21.36 | 0.007 | |
| hHKW1d | 1.08 | umc1278-umc1013-bnlg2228 | 21.14 | 0.001 | ||
| hHKW2a | 2.03 | bnlg1792-umc1929-umc1585 | −18.00 | 0.012 | ||
| hHKW3a | 3.04 | umc2259-phi036-umc1495 | 13.48 | 0.006 | ||
| hHKW3e | 3.07 | umc2050-umc1135-umc1399 | 15.01 | 0.046 | ||
| hHKW5b | 5.06 | phi085-phi048-umc2201 | 10.2 | 0.008 | ||
| hHKW5b | 5.06 | bnlg278-umc1680-phi085 | 7.58 | 0.041 | ||
| Hebi | Kernel length | hKL1b | 1.03 | umc1397-bnlg182-bnlg2238 | 3.53 | 0.008 |
| hKL1b | 1.03 | umc1403-umc1397-bnlg182 | 3.33 | 0.021 | ||
| hKL1c | 1.06 | umc1281-umc2151-umc1035 | −4.89 | 0.002 | ||
| hKL9a | 9.01 | umc1809-umc2093-umc1170 | −3.17 | 0.040 | ||
| hKL9a | 9.02 | umc1170-umc1037-umc1033 | −5.51 | 0.003 | ||
| Kernel width | hKW3c | 3.05 | umc2127-umc1954-umc2166 | −10.6 | 0.028 | |
| hKW3d | 3.08 | phi046-umc1844-umc2275 | −6.44 | 0.043 | ||
| hKW7a | 7.02 | bnlg1792-umc1929-umc1585 | −11.6 | 0.026 | ||
| hKW9a | 9.02 | umc1037-umc1033-bnlg1082 | 6.55 | 0.037 | ||
| hKW9b | 9.05 | umc1492-umc1519-umc1375 | 8.21 | 0.025 | ||
| Kernel thickness | hKT1b | 1.07 | umc1335-umc2396-umc1356 | 3.64 | 0.013 | |
| hKT1c | 1.08 | umc1013-bnlg2228-dupssr12 | 5.86 | 0.024 | ||
| hKT1d | 1.11 | umc2047-umc1538-bnlg131 | −5.87 | 0.005 | ||
| hKT3e | 3.07 | umc1135-umc1399-umc1148 | 7.31 | 0.050 | ||
| hKT4a | 4.01 | umc1228-umc1017-umc1757 | 4.42 | 0.003 | ||
| hKT4a | 4.01 | phi072-umc1228-umc1017 | 6.68 | 0.016 | ||
| hKT4c | 4.09 | umc2211-umc1101-bnlg589 | 5.41 | 0.000 | ||
| hKT6b | 6.04 | umc1979-nc009-umc1014 | 6.60 | 0.033 | ||
| hKT9a | 9.01 | umc1809-umc2093-umc1170 | 6.81 | 0.012 | ||
| hKT9d | 9.05 | umc1231-umc1494-bnlg1091 | −5.48 | 0.039 | ||
| 100-kernel weight | hHKW1b | 1.04 | bnlg182-bnlg2238-umc1144 | 5.12 | 0.035 | |
| hHKW2c | 2.04 | bnlg1064-umc1024-umc1465 | −4.80 | 0.033 | ||
| hHKW3c | 3.04 | umc1908-umc1773-phi053 | 5.88 | 0.029 | ||
| hHKW3d | 3.05 | umc2127-umc1954-umc2166 | 11.59 | 0.050 | ||
| hHKW5a | 5.01 | bnlg1006-phi024-bnlg1879 | 6.74 | 0.000 | ||
| hHKW6 | 6.05 | umc1614-umc2141-umc1805 | 5.77 | 0.047 | ||
| hHKW7b | 7.03 | umc1567-bnlg1305-bnlg2271 | 6.49 | 0.013 | ||
| hHKW7c | 7.04 | umc2332-phi328175-umc1295 | 13.21 | 0.015 | ||
| hHKW9c | 9.02 | umc1037-umc1033-bnlg1082 | 10.99 | 0.025 |
Figure 2.
Integrated heterotic loci (HL) on genetic linkage maps for kernel traits in two environments in the CSSLs × Xun9058 population. The heterotic chromosome ranges are represented by black color.
For kernel width, 13 different HL were identified in the CSSLs × Xun9058 population, of which only hKW9b was detected at the two locations simultaneously. This contributed 4.40% and 8.21% to kernel width compared with the control hybrid at Changge and Hebi, respectively.
Twenty HL for kernel thickness were detected in the CSSLs × Xun9058 population. hKT1b, identified at both locations simultaneously, increased kernel thickness by 4.87% and 3.64% compared with the control hybrid at Changge and Hebi, respectively, while hKT6b made 7.28% and 6.60% contributions compared with the control hybrid at the same locations.
Fifteen different HL for 100-kernel weight were identified in the CSSLs × Xun9058 population at the two locations, including 10 detected at Changge and eight at Hebi.
Comparison of heterotic loci identified in the two test populations
When comparing common HL detected at the two test populations, only 21 (33.33% and 36.84%) for the four kernel traits were detected in this study simultaneously (Supplemental Table 1); most HL (42/63, 66.67%; 36/57, 63.16%) differed between the two test populations. This supports the notion that heterosis is controlled by multiple loci, and that the interaction of multiple loci affects heterosis for a given trait in different hybrids. The HL detected in the two test populations simultaneously, such as hKL9b, hKW9a, hHKW1d, hHKW3a, and hHKW7b, may be common for the measured traits between the Reid × Tangsipingtou (TSPT) heterotic pattern, so could be used to predict hybrid performance in future maize breeding.
Discussion
The use of CSSL test populations with different parents
Because parental lines of maize commercial hybrids belong to different heterotic groups, the HL involved in different hybrids are always diverse. hQTL and HL mapping research has usually been carried out with biparental populations such as F2:3, DH, and RIL test populations13,18,38–40, IF2 populations2,16, and CSSLs or SSSLs test populations23,37,41,42. Although each type of segregating population has its own merits and corresponding shortcomings, none of them can be used to compare HL derived from different crosses. Larièpe et al.43 adopted an extension of design III derived from three initial inbred parents. The NCIII extension enabled the study of heterosis in families derived from both unrelated and related parents, and comparisons of contrasts between homozygous and heterozygous genotypes and between heterozygous genotypes to be made at each locus.
In this study, out of the 56 QTL and 120 HL for kernel traits identified using a CSSL population and its two test populations, only four QTL and HL (qKT1 vs hKT1c, qKT1a vs hKT1b, qKT9b vs hKT9b, qHKW9b vs hHKW9c) were detected at the same or overlapping introgression region lines at the same locations (Tables 4–6); many QTL (52/56, 92.86%) and HL (117/120, 97.50%) were not located on the same chromosomal region. This result indicated that heterosis and performance are controlled by different genetic mechanisms37, and that the HL for kernel construction traits detected by comparing each CSSL test hybrid with its corresponding CK have different genetic effects in the inbred lines lx9801 and Chang7-2 compared with the two test parents. Additionally, the CSSL population was tested further with multiple inbred lines belonging to the Reid heterotic group to identify HL between Reid and TSPT heterotic group. This type of test population can therefore not only identify specific HL of multiple inbred lines but can also be used to screen for common HL between Reid × TSPT heterotic model systems.
The advantage of heterotic loci identification using CSSLs test populations
Heterosis is a complicated characteristic controlled by minor additive and dominant multigenes. It is also readily affected by environmental factors such as soil fertility, sunlight, rainfall, and plant density44. Several important factors have limited dissection of the heterosis genetic basis. First, many agronomic traits and economical characters are compound traits involving several secondary traits, so trait heterotic values are the concurrent results of secondary traits. Second, it is debatable whether mid-parent heterosis or over-parent heterosis data represent the real heterotic expression, although mid-parent heterosis has been previously used to identify heterotic loci16,21. Third, the accuracy of phenotypic values of heterosis in different environments is uncertain when using mid-parent heterosis to dissect the genetic basis of heterosis. This is because inbred lines are easily affected by environmental factors, so mid-parent heterosis values vary in different environments. Finally, because the heterotic gene always has a distinctly different genetic effect among different heterozygous alleles, it is important to identify common heterotic loci between different test parents.
In previous studies, several types of segregated populations have been used to dissect the genetic basis of heterosis; these include F2 populations, DH and RIL test populations, IF2 populations, triple testcross populations, and SSSL backcross populations3,14,16,17,38,45–49. In comparison with these, the CSSLs test populations with different test parents in this study has several merits for dissecting the genetic basis of heterosis. First, the heterotic loci identified by comparing the test population and its corresponding CK have little influence in different environments, so it is easy to obtain accurate phenotypic values. Second, the genetic effect of heterotic loci in different test backgrounds can be analysed to identify the main heterotic loci between different groups. Theoretically, if the significant loci identified by comparing test hybrids and corresponding CK are true HL, many will not be simultaneously detected in the CSSL population. In this study, only four QTL and HL were identified simultaneously in the same CSSLs, and most HL (97.50%) were not detected in the CSSL population. Thus, the significant loci identified were real HL, which showed different heterotic genetic effects between the two inbred lines lx9801 and Chang7-2 when tested against the inbred Zheng 58 and Xun 9058.
In a previous study, Wang et al.50 identified 169 HL associated with grain yield and its five components using two test populations. Additionally, we identified several common HL of kernel-related traits over the same and overlapping introgression region lines. These included hKW3d for kernel width, hHKW3b and hHKW5b for 100-kernel weight, hKL1a and hKL9a for kernel length, and hKT4a for kernel thickness in the CSSLs × Zheng58 population (Tables 5, 6).
Hybrid performance production
The identification of high-performing hybrids is an integral part of every maize breeding program. However, because the field evaluation of all potential hybrids is too resource-intensive, only a small subset are tested in field trials, from which only a few elite hybrids are selected51. Therefore, it is important to estimate hybrid performance52. In a previous study, hybrid vigour was predicted using molecular markers to estimate genetic distances among parents53; several studies have uncovered a direct correlation between superior hybrid performance and the genetic distance of parental lines54,55. Recent investigations have used molecular markers and QTL for the genomic prediction of hybrid performance in maize56–58, sunflowers8, and wheat26. An important component of hybrid performance is the specific combining ability between the parental lines of a hybrid. As a consequence, both additive and dominance effects of markers must be estimated to account for the entire genetic variance.
Using a simulation study, Technow et al.51 demonstrated a strong alternative to genomic best linear unbiased prediction, in the form of the Bayesian whole-genome regression method, BayesB59. This method was first used by Maenhout et al.57 for the genomic estimation of hybrid performance. In terms of heterosis optimum manipulation, the parental inbred lines of different hybrids have been extracted from genetically distant germplasm pools, called heterotic groups53, and have been widely used by maize breeders. As for hybrid prediction, a key question is how many hybrids per inbred line, i.e., a cross with lines from the opposite heterotic group, should be included in the training set. Technow et al.58 followed the Dent and Flint heterotic pattern to analyse the genomic and phenotypic data of 1,254 hybrids in a typical maize hybrid breeding program. They found that the estimated accuracy of untested hybrids was highest if both parents were parents of other hybrids in the training set, and that the prediction accuracy of untested hybrids was lowest if neither parents were involved in any training set hybrid. Technow et al.51 also showed that prediction accuracies increased with marker density and the number of tested parents. They reported that under low linkage disequilibrium the modelling of marker effects as population-specific was the most beneficial. In China, Reid and TSPT are components of the first heterotic pattern that has been widely used in maize breeding60. In the present study, two test populations, constructed from representative inbred lines derived from the Reid and TSPT heterotic group, were used to detect HL for kernel traits in maize. We detected 21 HL in the two test populations simultaneously, and suggest that these HL for kernel traits and their linked molecular markers could be used to predict hybrid performance in future maize breeding programs.
Genetic dissection of heterosis for kernel traits in maize
Grain yield is the ultimate product of multiple processes that occur throughout the growing season61. Grain yield in maize is a function of many harvested kernels and their individual weights. In these two components, the number of kernels usually explains most of the variation62, while kernel weight has a high heritability63,64 and varies significantly among genotypes65. Several QTL mapping studies for maize kernel weight have been conducted, but results are inconsistent in terms of effect size and localisation32–34. Such inconsistencies may be linked to the complexity of the trait, which thus requires further dissection into simpler components. Regarding the secondary traits of kernel weight, Zhang et al.35 detected three QTL for kernel depth using a 229-line F2:3 population derived from inbred lines Yu 82 and Shen 137. Zhang et al.36 also identified 54 unconditional main QTL for five kernel-related traits, namely kernel thickness, weight, length, volume, and width using an IF2 population in maize. In the present study, 63 HL for four kernel traits were identified in the CSSLs × Zheng58 population, with nine (13.8%), namely hKL9a, hKW2, hKW9b, hKT1d, hKT2a, hHKW1d, hHKW3a, hHKW3b, and hHKW7b, identified at two locations simultaneously. Fifty-seven HL were identified for four kernel traits in the CSSLs × Xun9058 population, of which seven (12.18%), hKL9a, hKW3c, hKW3d, hKW7a, hKW9b, hKT1b, and hKT6b, were identified for four kernel traits at two locations simultaneously. Several HL for different kernel traits sharing common chromosomal regions were also detected. For example, HL for 100-kernel weight, length, thickness, and width were detected in chromosomal bin 2.03 in the same chromosomal region, umc2195-umc1555-bnlg1064, in the CSSLs × Zheng58 population at the two locations simultaneously. Moreover, the HL for kernel length and thickness were detected in chromosomal bin 9.02 at the same chromosomal region, bnlg1170-umc1037-umc1033, in the CSSLs × Xun9058 population at the two locations.
Many HL detected for grain yield and its components in a previous study50, as well as HL for kernel traits located on common chromosomal regions in the same test populations have been found on the same chromosomal region. For example, in the CSSLs × Xun9058 population, HL hlEL1a for ear length, hlEW1b for ear width, hlGY1a for grain yield, and hKW1a for kernel width were detected on umc1403-umc1397-bnlg182. Similarly, HL hlEL1b for ear length, hlRN1a for row number, hlKPR1b for kernels per row, hKT1a for kernel thickness, and hHKW1b for 100-kernel weight were detected on the chromosomal region bnlg182-bnlg2238-umc1144, while HL hlEW9b for ear width, hlRN9c for row number, hlKPR9b for kernels per row, hKT9b for kernel thickness, and hKL9a for kernel length were located on umc1170-umc1037-umc1033. In the CSSLs × Zheng58 population, HL hlEL1b for ear length, hlRN1a for row number, hlGY1b for grain yield, hKW1b for kernel width, hHKW1b for 100-kernel weight, and hKT1a for kernel thickness were found on bnlg182-bnlg2238-umc1144. Moreover, on the chromosomal region umc1844-umc2275-umc2081, HL hlEL3e for ear length, hlEW3e for ear width, hlRN3e for row number, hKL3d for kernel length, hKW3d for kernel width, and hHKW3f for 100-kernel weight were detected simultaneously. These results show that the HL for grain yield and its components form clusters, and that the regions containing these clusters may be useful for future maize breeding.
Combining ability and heterosis in maize
It can be difficult to predict the yield performance of hybrids by that of their parents per se in breeding practice66. Therefore, selecting inbred lines with a high combining ability becomes important. Griffing67 first carried out diallel tests to comprehend the genetic basis of combining ability, and proposed that SCA and GCA were mainly respectively correlated with nonadditive and additive genetic effects. In a previous study, Qi et al.68 identified 56 significant loci for GCA and 21 loci for SCA of five yield-related traits of introgression lines test populations in maize, and only five loci for GCA and SCA simultaneously; this indicated a different genetic basis for GCA and SCA.
Significant positive correlations were also reported for SCA with high parent heterosis (HPH), mid parent heterosis (MPH), and hybrid performance69,70. Hence, improved selection for SCA would indirectly increase MPH and HPH for hybrids in maize breeding70. In this study, 63 and 57 different HL were identified for four kernel traits in the CSSLs × Zheng58 and CSSLs × Xun9058 populations, of which only 21 (33.33% and 36.84%) were detected in the two test populations simultaneously. The results are consistent with different SCA for different crosses in previous studies69–71. Moreover, our work suggests that common HL identified from different tests can be used to select elite inbred lines with high SCA, and contribute to predicting hybrid performance in the Reid × TSPT heterotic model in maize breeding.
Materials and Methods
CSSL and test population construction and field experiments
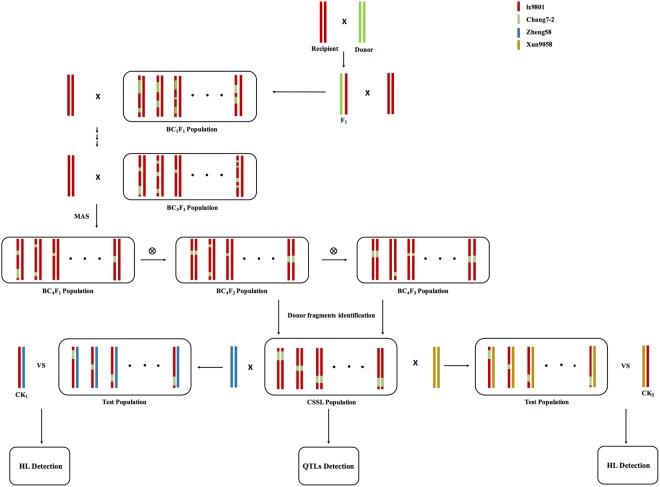
A maize 184 CSSL population constructed using two elite inbred lines, lx9801 and Chang7-2, was used in this study. The two elite inbred lines belonged to the TSPT heterotic group, an important germplasm widely used in China. The inbred line Chang7-2, used as the donor parent, is one parent of the elite hybrid Zhengdan958 (Zheng58 × Chang7-2), the first commercial hybrid widely used in China (from 2005 to 2014). The recipient parent was lx9801, a parent of Ludan9002 (Zheng58 × lx9801), another elite commercial hybrid. The two hybrids, Zhengdan958 and Ludan9002, have a common female parent, Zheng58. Based on the simple sequence repeat (SSR) molecular marker linkage map integrated by IBM 2008 Neighbors (http://www.maizegdb.org), 700 paired SSR markers were used to screen for polymorphisms between the two parents, and a total of 225 SSR polymorphic markers were used to screen donor fragments of the inbred line Chang7-2 at different backcross populations.
In the winter of 2009, 929 BC3F1 lines were planted in Sanya (N18°15′, E109°30′) China. Five plants of each BC3F1 line were screened using the polymorphic markers, and 875 plants with fewer (<5) donor chromosomal regions were backcrossed with recipient parents to produce the BC4F1 population. BC4F1 plants with one or two donor chromosomal regions were screened using polymorphic markers, and selfed in the summer of 2010 in Zhengzhou (N34°16′, E112°42′) China. In the winter of 2011, 1718 BC4F2 lines were selected and planted in Sanya, and plants with single chromosomal segment substitutions were verified and selfed to produce 102 homozygous CSSLs. Additionally, some single heterozygous CSSLs were also selfed, and 84 were selected from the BC4F3 population. Finally, we achieved a population of 184 CSSLs with a single segment of donor parent Chang7-2 in the recipient parent lx9801 background50.
Test populations were constructed using the CSSL population and two inbred lines, Zheng58 and Xun9058 (Fig. 3). These inbred lines belong to the Reid heterotic group, which are a pair of the heterotic model Reid × TSPT, and are widely used in China.
Figure 3.
Group construction pattern map. Each genome is represented by a specific color. Red and green represents lx9801 as recipient parent and Chang7-2 as donor parent respectively; Blue and brown represents test parent Zheng58 and Xun9058. HL, heterotic loci; QTLs, quantitative traits loci; CK1, Zheng58 × lx9801; CK2, Xun9058 × lx9801; MAS, marker-assisted selection.
Field experiments
Because test hybrids are taller than the CSSL population, the planting of test hybrids adjacent to their parents in the field would affect the normal growth of inbred lines. To avoid interactions of test hybrids and the CSSL population, they were divided into two individual experiments in this study. First, the two test populations and their corresponding control hybrids (Zheng58 × lx9081 and Xun9058 × lx9081) were evaluated on farms of the Hebi Agricultural Institute (Hebi; E 114° 33′, N 35° 41′) and Changge (E 113° 29′, N 34° 1′), respectively. Materials were planted in June 2012 and 2013 following the wheat harvest. The experimental design of the test population consisted of a randomised complete block with three replicates of corresponding hybrids (Zheng58 × lx9801 and Xun9058 × lx9801; CK) added between the 10 test crosses. Each plant material was planted in one plot per field. The plots constituted of rows 4 m long, separated by 0.66 m between each row. A total population density of 67,500 plants per hectare was maintained. The CSSL population and the four inbred lines (lx9801, Chang7-2, Zheng58, and Xun9058) were planted in the adjacent field using the same field design to identify the additive QTL and analyse mid-parent heterosis. Fields were managed according to local maize cultivation practices.
Performance measurement
After maturity, 10 ears were harvested from successive plants in each plot, and air-dried to a grain moisture level of 13%. Measured traits were kernel length (mm), kernel thickness (mm), kernel width (mm), and 100-kernel weight (g). Phenotypic data were recorded as follows: (1) KL (mm kernel−1) = (ear diameter − cob diameter)/2, for which cob and ear diameters were measured at the middle of the ear; (2) kernel width in the middle of a kernel (KW, mm kernel−1) = [cob diameter + (ear diameter − cob diameter)/2] π/(ear row number); (3) KT (mm kernel−1), judged from the thickness of 10 kernels in the middle of an ear36,72,73; (4) HKW (g), the average value of the three measurements of the weight of 100-kernels randomly selected74.
Data analysis
Mid-parent heterosis (HMP) for the four kernel traits in the two test populations was evaluated in the two environments. The values of mid-parent heterosis were calculated as HMP (%) = (F1 − MP)/MP × 100%, for which HMP is the percentage of mid-parent heterosis75, F1 is the average data of the four kernel traits in each testcross population, and MP is the mean of the average values of the CSSL population and the corresponding test parent. Mid-parent heterosis values of the corresponding hybrids (Zheng58 × lx9801 and Xun9058 × lx9801) were calculated using the same formula.
SAS 9.2 was used to evaluate the four kernel traits of the two test populations exhibited significant variation between locations and genotypes. The model followed was: phenotypeijk = μ + locationi + genotypej + genotype*location + repetitionk + errorijk, where μ is overall mean76. H2B = σ2genotype/(σ2genotype + σ2genotype*location/e + σ2error/r*e), where e and r are the numbers of environments and replicates76.
A QTL was considered to exist in the CSSL population when there was a significant difference in the measured value between the CSSL and the recurrent inbred line lx9801 (P < 0.05) by one-way analysis of variance and Duncan’s multiple comparisons using SPSS 17.0 software. The additive effect was calculated using the following equation: A = (SSSL − lx9801)/2. The percentage of the additive effect (A%) was calculated using the following equation2: A% = A/lx9801 × 100%.
Heterotic genes usually have different genetic effects and phenotypes when heterozygosis with different alleles is observed50. We identified HL for kernel-related traits and compared the significance between single hybrids and the average value of two adjacent corresponding CK (hybrids Zheng58 × lx9801 or Xun9058 × lx9801). The value of a given trait of one test hybrid in the three replicates between two years at one location was compared between the test population and its corresponding hybrid using the Student’s t-test. If a significant difference was observed (P < 0.05), the corresponding chromosomal region was considered to be a HL between the CSSL and its test inbred line, and the HL showed a different heterotic genetic effect between inbred line Chang7-2 and lx9801. The heterotic effect was calculated as follows: HL% = (H − CK)/CK × 100%, where H represented the value of the trait in a single cross in the CSSL test populations and CK represented the value of the trait in the corresponding hybrid77.
Electronic supplementary material
Acknowledgements
This work gained support from the State Key Basic Research and Development Plan of China (2014CB138203), the National Natural Science Foundation of China (91335205) and the Major Science and Technology Programs of Henan Province (161100110500). We also thank Sarah Williams, PhD, from Liwen Bianji, Edanz Group China (www.liwenbianji.cn), for editing the English text of a draft of this manuscript.
Author Contributions
J.T. and G.Z. conceived this study. Experiments and data collection were conducted by W.Y., S.C. and T.R. S.X., H.X., J.J. and X.Z. analysed the data. The manuscript was written by J.T. and W.X. The final manuscript was read and approved by all authors.
Competing Interests
The authors declare no competing interests.
Footnotes
Yafei Wang and Xiangge Zhang contributed equally to this work.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-29338-1.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Zhanyong Guo, Email: gu0zhy@163.com.
Jihua Tang, Email: tangjihua1@163.com.
References
- 1.Shull GH. The composition of a field of maize. Journal of Heredity. 1908;1:296–301. doi: 10.1093/jhered/os-4.1.296. [DOI] [Google Scholar]
- 2.Zhou G, et al. Genetic composition of yield heterosis in an elite rice hybrid. Proc Natl Acad Sci USA. 2012;109:15847–15852. doi: 10.1073/pnas.1214141109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guo X, et al. Mapping heterotic loci for yield and agronomic traits using chromosome segment introgression lines in cotton. J Integr Plant Biol. 2013;55:759–774. doi: 10.1111/jipb.12054. [DOI] [PubMed] [Google Scholar]
- 4.Cheng SH, Zhuang JY, Fan YY, Du JH, Cao LY. Progress in research and development on hybrid rice: a super-domesticate in China. Ann Bot. 2007;100:959–966. doi: 10.1093/aob/mcm121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Duvick, D. N. Heterosis: feeding people and protecting natural resources, pp. 19–29. In the genetics and exploitation of heterosis in crops, edited by J. Coors, and S. Pandey. (CSSA, Madison, WI, 1999).
- 6.Bruce AB. The Mendelian theory of heredity and the augmentation of vigor. Science. 1910;32:627–628. doi: 10.1126/science.32.827.627-a. [DOI] [PubMed] [Google Scholar]
- 7.Jones DF. Dominance of linked factors as a means of accounting for heterosis. Genetics. 1917;2:466. doi: 10.1093/genetics/2.5.466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reif JC, Zhao YS, Würschum T, Gowda M, Hahn V. Genomic prediction of sunflower hybrid performance. Plant Breeding. 2013;132:107–114. doi: 10.1111/pbr.12007. [DOI] [Google Scholar]
- 9.Lippman ZB, Zamir D. Heterosis: revisiting the magic. Trends Genet. 2007;23:60–66. doi: 10.1016/j.tig.2006.12.006. [DOI] [PubMed] [Google Scholar]
- 10.Xiao J, Li J, Yuan L, Tanksley SD. Dominance is the major genetic basis of heterosis in rice as revealed by QTL analysis using molecular markers. Genetics. 1995;140:745–754. doi: 10.1093/genetics/140.2.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cockerham CC, Zeng ZB. Design III with marker loci. Genetics. 1996;143:1437–1456. doi: 10.1093/genetics/143.3.1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.East EM. Heterosis. Genetics. 1936;21:375. doi: 10.1093/genetics/21.4.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stuber CW, Lincoln SE, Wolff DW, Helentjaris T, Lander ES. Identification of genetic factors contributing to heterosis in a hybrid from two elite maize inbred lines using molecular markers. Genetics. 1992;132:823–39. doi: 10.1093/genetics/132.3.823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yu SB, et al. Importance of epistasis as the genetic basis of heterosis in an elite rice hybrid. Proc Natl Acad Sci USA. 1997;94:9226–9231. doi: 10.1073/pnas.94.17.9226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li ZK, et al. Overdominant epistatic loci are the primary genetic basis of inbreeding depression and heterosis in rice. I. Biomass and grain yield. Genetics. 2001;158:1737–1753. doi: 10.1093/genetics/158.4.1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hua JP, et al. Single-locus heterotic effects and dominance by dominance interactions can adequately explain the genetic basis of heterosis in an elite rice hybrid. Proc Natl AcadSci USA. 2003;100:2574–2579. doi: 10.1073/pnas.0437907100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Melchinger AE. Genetic basis of heterosis for growth-related traits in Arabidopsis investigated by testcross progenies of near-isogenic lines reveals a significant role of epistasis. Genetics. 2007;177:1827–1837. doi: 10.1534/genetics.107.080564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li LZ, et al. Dominance, overdominance and epistasis condition the heterosis in two heterotic rice hybrids. Genetics. 2008;180:1725–1742. doi: 10.1534/genetics.108.091942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu H, Romero-Severson J, Bernardo R. Genetic basis of heterosis explored by simple sequence repeat markers in a random-mated maize population. Theor Appl Genet. 2003;107:494–502. doi: 10.1007/s00122-003-1271-7. [DOI] [PubMed] [Google Scholar]
- 20.Frascaroli E, et al. Classical genetic and quantitative trait loci analyses of heterosis in a maize hybrid between two elite inbred lines. Genetics. 2007;176:625–644. doi: 10.1534/genetics.106.064493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tang JH, et al. Dissection of the genetic basis of heterosis in an elite maize hybrid by QTL mapping in an immortalized F2 population. Theor Appl Genet. 2010;120:333–340. doi: 10.1007/s00122-009-1213-0. [DOI] [PubMed] [Google Scholar]
- 22.Semel Y, et al. Overdominant quantitative trait locus for yield and fitness in tomato. Proc Natl AcadSci USA. 2006;103:12981–12986. doi: 10.1073/pnas.0604635103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang ZQ, et al. Identification of Indica rice chromosome segments for the improvement of Japonica inbreds and hybrids. Theor Appl Genet. 2012;124:1351–1364. doi: 10.1007/s00122-012-1792-z. [DOI] [PubMed] [Google Scholar]
- 24.Meyer RC, et al. QTL analysis of early stage heterosis for biomass in Arabidopsis. Theor Appl Genet. 2010;120:227–237. doi: 10.1007/s00122-009-1074-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shen GJ, Zhan W, Chen HX, Xing YZ. Dominance and epistasis are the main contributors to heterosis for plant height in rice. Plant Science. 2014;215:11–18. doi: 10.1016/j.plantsci.2013.10.004. [DOI] [PubMed] [Google Scholar]
- 26.Zhao YS, Zeng J, Fernando R, Reif JC. Genomic prediction of hybrid wheat performance. Crop Sci. 2013;53:802–810. doi: 10.2135/cropsci2012.08.0463. [DOI] [Google Scholar]
- 27.Messmer R, et al. Drought stress and tropical maize: QTL-by-environment interactions and stability of QTLs across environments for yield components and secondary traits. Theor Appl Genet. 2009;119:913–930. doi: 10.1007/s00122-009-1099-x. [DOI] [PubMed] [Google Scholar]
- 28.Doebley JF, Gaut BS, Smith BD. The molecular genetics of crop domestication. Cell. 2006;127:1309–1321. doi: 10.1016/j.cell.2006.12.006. [DOI] [PubMed] [Google Scholar]
- 29.Guo T, et al. Genetic basis of grain yield heterosis in an “immortalized F2” maize population. Theor Appl Genet. 2014;127:2149–2158. doi: 10.1007/s00122-014-2368-x. [DOI] [PubMed] [Google Scholar]
- 30.Li C, et al. Quantitative trait loci mapping for yield components and kernel-related traits in multiple connected RIL populations in maize. Euphytica. 2013;193:303–316. doi: 10.1007/s10681-013-0901-7. [DOI] [Google Scholar]
- 31.Zhang WQ, Ku LX, Zhang J, Han ZP, Chen YH. QTL analysis of kernel ratio, kernel depth, and 100-kernel weight in maize (Zea mays L.) Acta Agronomica Sinica. 2013;39:455–463. doi: 10.3724/SP.J.1006.2013.00455. [DOI] [Google Scholar]
- 32.Schön, C. C. et al. RFLP mapping in maize: quantitative trait loci affecting testcross performance of elite European flint lines. Crop Sci34, 379–389.
- 33.Austin DF, Lee M. Comparative mapping in F2:3 and F6:7 generations of quantitative trait loci for grain yield and yield components in maize. Theor Appl Genet. 1996;92:817–826. doi: 10.1007/BF00221893. [DOI] [PubMed] [Google Scholar]
- 34.Frova C, Krajewski P, di, Fonzo. N, Villa M, Sarli-Gorla M. Genetic analysis of drought tolerance in maize by molecular markers. I. Yield components. Theor Appl Genet. 1999;99:280–288. doi: 10.1007/s001220051233. [DOI] [Google Scholar]
- 35.Zhang G, et al. Fine mapping a major QTL for kernel number per row under different phosphorus regimes in maize (Zea mays L.) Theor Appl Genet. 2013;126:1545–1553. doi: 10.1007/s00122-013-2072-2. [DOI] [PubMed] [Google Scholar]
- 36.Zhang ZH, et al. QTL analysis of kernel-related traits in maize using an immortalized F2 population. PLoS One. 2014;9(2):e89645. doi: 10.1371/journal.pone.0089645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wei XY, et al. Genetic analysis of heterosis for maize grain yield and its components in a set of SSSL testcross populations. Euphytica. 2016;210:181–193. doi: 10.1007/s10681-016-1695-1. [DOI] [Google Scholar]
- 38.Melchinger AE, Utz HF, Schön CC. Quantitative trait locus (QTL) mapping using different testers and independent population samples in maize reveals low power of QTL detection and large bias in estimates of QTL effects. Genetics. 1998;149:383–403. doi: 10.1093/genetics/149.1.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Syed NH, Chen ZJ. Molecular marker genotypes, heterozygosity and genetic interactions explain heterosis in Arabidopsis thaliana. Heredity. 2005;94:295–304. doi: 10.1038/sj.hdy.6800558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Luo XJ, et al. Additive and over-dominant effects resulting from epistatic loci are the primary genetic basis of heterosis in rice. J Integr Plant Biol. 2009;51:393–408. doi: 10.1111/j.1744-7909.2008.00807.x. [DOI] [PubMed] [Google Scholar]
- 41.Yu CY, et al. Study on heterosis of inter-subspecies between indica and japonica rice (Oryza sativa L.) using chromosome segment substitution lines. Chin Sci Bull. 2005;50:131–136. doi: 10.1007/BF02897516. [DOI] [Google Scholar]
- 42.Liu GF, Zhu HT, Zhang GQ, Li LH, Ye GY. Dynamic analysis of QTLs on tiller number in rice (Oryza sativa L.) with single segment substitution lines. Theor Appl Genet. 2012;125:143–153. doi: 10.1007/s00122-012-1822-x. [DOI] [PubMed] [Google Scholar]
- 43.Larièpe A, et al. The genetic basis of heterosis: multiparental quantitative trait loci mapping reveals contrasted levels of apparent overdominance among traits of agronomical interest in maize (Zea mays L.) Genetics. 2012;190:795–811. doi: 10.1534/genetics.111.133447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li B, et al. Genetic effects and heterosis of yield and yield component traits based on Gossypium Barbadense chromosome segment substitution lines in two Gossypium Hirsutum backgrounds. PLoS One. 2016;11(6):e0157978. doi: 10.1371/journal.pone.0157978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Koester R, Sisco PH, Stuber CW. Identification of quantitative trait loci controlling days to flowering and plant height in two near isogenic lines of maize. Crop Sci. 1993;33(6):1209–1216. doi: 10.2135/cropsci1993.0011183X003300060020x. [DOI] [Google Scholar]
- 46.Guo JF, Su GQ, Zhang JP, Wang GY. Genetic analysis and QTL mapping of maize yield and associate agronomic traits under semi-arid land condition. Afr J Biotechnol. 2008;7(12):1829–1838. doi: 10.5897/AJB2008.000-5031. [DOI] [Google Scholar]
- 47.Ma LY, et al. Quantitative trait loci for panicle layer uniformity identified in doubled haploid lines of rice in two environments. J Integr Plant Biol. 2009;51:818–824. doi: 10.1111/j.1744-7909.2009.00854.x. [DOI] [PubMed] [Google Scholar]
- 48.Li ZK, et al. Analysis of plant height heterosis based on QTL mapping in wheat. ActaAgron Sin. 2010;36:771–778. [Google Scholar]
- 49.Song FW, et al. Heterosis for plant height and ear position in maize revealed by quantitative trait loci analysis with triple testcross design. Acta Agron Sin. 2011;37(7):1186–1195. doi: 10.3724/SP.J.1006.2011.01186. [DOI] [Google Scholar]
- 50.Wang HQ, et al. Identification of heterotic loci associated with grain yield and its components using two CSSL test populations in maize. Scientific Reports. 2016;6:38205. doi: 10.1038/srep38205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Technow F, Riedelsheimer C, Schrag TA, Melchinger AE. Genomic prediction of hybrid performance in maize with models incorporating dominance and population specific marker effects. Theor Appl Genet. 2012;125:1181–1194. doi: 10.1007/s00122-012-1905-8. [DOI] [PubMed] [Google Scholar]
- 52.Bernardo R. Best linear unbiased prediction of maize single-cross performance. Crop Sci. 1996;36:50–56. doi: 10.2135/cropsci1996.0011183X003600010009x. [DOI] [PubMed] [Google Scholar]
- 53.Melchinger, A. E. & Gumber, R. K. Overview of heterosis and heterotic groups in agronomic crops. Concepts and Breeding of Heterosis in Crop Plants, 29–44 (1998).
- 54.Liu XC, Ishikiand K, Wang WX. Identification of AFLP markers favorable to heterosis in hybrid rice. Breed Sci. 2002;52:201–206. doi: 10.1270/jsbbs.52.201. [DOI] [Google Scholar]
- 55.Barbosa AMM, et al. Relationship of intra- and inter population tropical maize single cross hybrid performance and genetic distances computed from AFLP and SSRmarkers. Euphytica. 2003;130:87–99. doi: 10.1023/A:1022381008721. [DOI] [Google Scholar]
- 56.Massman JM, Gordillo A, Lorenzana RE, Bernardo R. Genomewide predictions from maize single-cross data. Theor Appl Genet. 2012;126:13–22. doi: 10.1007/s00122-012-1955-y. [DOI] [PubMed] [Google Scholar]
- 57.Maenhout S, De, Baets. B, Haesaert G. Prediction of maize single-cross hybrid performance: support vector machine regression vs. best linear prediction. Theor Appl Genet. 2010;120:415–427. doi: 10.1007/s00122-009-1200-5. [DOI] [PubMed] [Google Scholar]
- 58.Technow F, et al. Genome properties and prospects of genomic prediction of hybrid performance in a breeding program of maize. Genetics. 2014;197:1343–55. doi: 10.1534/genetics.114.165860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Meuwissen THE, Hayes BJ, Goddard ME. Prediction of total genetic value using genome-wide dense marker maps. Genetics. 2001;157:1819–1829. doi: 10.1093/genetics/157.4.1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Teng WT, et al. Analysis of maize heterosis groups and patterns during past decade in China. Scientia Agricultura Sinica. 2004;37:1804–1811. [Google Scholar]
- 61.Slafer GA. Genetic basis of yield as viewed from a crop physiologist’s perspective. Ann Appl Biol. 2003;142:117–128. doi: 10.1111/j.1744-7348.2003.tb00237.x. [DOI] [Google Scholar]
- 62.Borrás L, Gambín BL. Trait dissection of maize kernel weight: towards integrating hierarchical scales using a plant growth approach. Field Crops Res. 2010;118:1–12. doi: 10.1016/j.fcr.2010.04.010. [DOI] [Google Scholar]
- 63.Sadras VO. Evolutionary aspects of the trade-off between seed size and number in crops. Field Crops Res. 2007;100:125–138. doi: 10.1016/j.fcr.2006.07.004. [DOI] [Google Scholar]
- 64.Alvarez PS, et al. Correlation between parental inbred lines and derived hybrid performance for grain filling traits in maize. Crop Sci. 2013;53:1636–1645. doi: 10.2135/cropsci2013.01.0035. [DOI] [Google Scholar]
- 65.Reddy VM, Daynard TB. Endosperm characteristics associated with rate of grain filling and kernel size in corn. Maydica. 1983;28:339–355. [Google Scholar]
- 66.Hallauer AR. Methods used in developing maize inbreds. Maydica. 1990;35:1–6. [Google Scholar]
- 67.Griffing B. Concept of general and specific combining ability in relation to diallel crossing systems. Aus J Biol Sci. 1956;9:463–493. doi: 10.1071/BI9560463. [DOI] [Google Scholar]
- 68.Qi H, et al. Identification of combining ability loci for five yield-related traitsin maize using a set of testcrosses with introgression lines. Theor Appl Genet. 2013;126:369–377. doi: 10.1007/s00122-012-1985-5. [DOI] [PubMed] [Google Scholar]
- 69.Makumbi D, Betrán JF, Bänziger M, Ribaut JM. Combining ability, heterosis and genetic diversity in tropical maize (Zea mays L.) under stress and non-stress conditions. Euphytica. 2011;180:143–162. doi: 10.1007/s10681-010-0334-5. [DOI] [Google Scholar]
- 70.Ndhlela T, et al. Relationships between heterosis, genetic distances and specific combining ability among CIMMYT and Zimbabwe developed maize inbred lines under stress and optimal conditions. Euphytica. 2015;204:635–647. doi: 10.1007/s10681-015-1353-z. [DOI] [Google Scholar]
- 71.Aminu D, Garba YM, Muhammad AS. Combining ability and heterosis for phenologic and agronomic traits in maize (Zea mays L.) under drought conditions in the Northern Guinea Savanna of BornoState, Nigeria. African Journal of Biotechnology. 2014;13(24):2400–2406. doi: 10.5897/AJB2013.13371. [DOI] [Google Scholar]
- 72.Li YX, et al. Correlation analysis and QTL mapping for traits of kernel structure and yield components in maize. Sci Agron Sin. 2009;42:408–418. [Google Scholar]
- 73.Peng B, et al. QTL analysis for yield components and kernel-related traits in maize across multi-environments. Theor Appl Genet. 2011;122:1305–1320. doi: 10.1007/s00122-011-1532-9. [DOI] [PubMed] [Google Scholar]
- 74.Peng B, et al. Correlations and comparisons of quantitative trait loci with family perse and testcross performance for grain yield and related traits in maize. Theor Appl Genet. 2013;126:773–789. doi: 10.1007/s00122-012-2017-1. [DOI] [PubMed] [Google Scholar]
- 75.Jiang L, Ge M, Zhao H, Zhang TF. Analysis of heterosis and quantitative trait loci for kernel shape related traits using triple testcross population in maize. PLoS One. 2015;10(4):e0124779. doi: 10.1371/journal.pone.0124779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Liu X, et al. Factors affecting genomic selection revealed by empirical evidence in maize. The Crop Journal. 2018;290:12–23. [Google Scholar]
- 77.Wei XY, et al. Heterotic loci for various morphological traits of maize detected using a single segment substitution lines test-cross population. Molecular Breeding. 2015;35(3):1–13. doi: 10.1007/s11032-015-0287-4. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.