Abstract
Listeria monocytogenes is a Gram-positive pathogen occurring in many refrigerated ready-to-eat foods. It is responsible for foodborne listeriosis, a rare but severe disease with a high mortality rate (20–30%). Lactococcus piscium CNCM I-4031 has the capacity to prevent the growth of L. monocytogenes in contaminated peeled and cooked shrimp and in a chemically defined medium using a cell-to-cell contact-dependent mechanism. To characterize this inhibition further, the effect of L. piscium was tested on a collection of 42 L. monocytogenes strains. All strains were inhibited but had different sensitivities. The effect of the initial concentration of the protective and the target bacteria revealed that the inhibition always occurred when L. piscium had reached its maximum population density, whatever the initial concentration of the protective bacteria. Viewed by scanning electron microscopy, L. monocytogenes cell shape and surface appeared modified in co-culture with L. piscium CNCM I-4031. Lastly, L. monocytogenes virulence, evaluated by a plaque-forming assay on the HT-29 cell line, was reduced after cell pre-treatment by the protective bacteria. In conclusion, the bioprotective effect of L. piscium toward L. monocytogenes growth and virulence was demonstrated, and a hypothesis for the inhibition mechanism is put forward.
Keywords: biopreservation, Lactococcus piscium, Listeria monocytogenes, co-culture, cell ratio, scanning electron microscopy, virulence
Introduction
Listeria monocytogenes is a human pathogenic Gram-positive bacterium, which is responsible for foodborne listeriosis generally associated with a high mortality rate (20–30%). All human population groups can be infected and particularly newborn infants, pregnant women, elderly people, and immuno-compromised patients (Lecuit et al., 2015). This species constitutes a major problem in refrigerated ready-to-eat (RTE) foods (Rocourt et al., 2003). According to the Codex Alimentarius Commission, RTE products are “any food which is normally eaten in its raw state or any food handled, processed, mixed, cooked, or otherwise prepared into a form which is normally eaten without further listericidal steps" (CAC, 2007). L. monocytogenes differs from most other food-borne pathogens in that it is ubiquitous and can grow or survive in most conditions encountered in the food chain and food-processing procedures (Buchanan et al., 2017). Among foodstuffs, fishery products recorded the highest level of non-compliance with EU safety criteria in 2015 (EFSA, 2016) while seafood products were implicated in 15% of L. monocytogenes outbreaks reported in Europe in the last 3 years. Its survival ability makes the control of this microorganism in food products, especially RTE foods, a major challenge. The lactic acid bacteria (LAB), generally recognized as safe (Salminen et al., 1998), are also present and dominant in RTE foods such as meat and seafood products stored under vacuum or modified atmosphere packaging. Selected LAB can limit the development of L. monocytogenes and are thus recognized as efficient bioprotective agents in food systems (Brillet et al., 2004; Nilsson et al., 2005; Tomé et al., 2006; Vermeiren et al., 2006a; Unlu et al., 2015). The competition between LAB and L. monocytogenes involves various bactericidal or bacteriostatic mechanisms such as (i) competition for nutrients (Nilsson et al., 2005); (ii) production of one or more antimicrobial active metabolites such as bacteriocins (Richard et al., 2003; Dortu et al., 2008; Martinez et al., 2015), reuterin (El-Ziney et al., 1999), organic acids (Amezquita and Brashears, 2002), and hydrogen peroxide (Ito et al., 2003; Batdorj et al., 2007). Lactococcus piscium CNCM I-4031 is an efficient bioprotective strain for seafood products isolated from raw salmon stored under modified atmosphere packaging (Matamoros et al., 2009a). It improves the sensory quality of cooked shrimp by preventing the growth of Brochothrix thermosphacta (Fall et al., 2012). L. piscium CNCM I-4031 can also limit the growth of L. monocytogenes RF191 during the storage of cooked shrimp (Fall et al., 2010). The inhibition mechanism has not yet been entirely elucidated but using a chemically defined medium (MSMA) to mimic the shrimp matrix, it has been suggested that cell contact is required for the inhibition of L. monocytogenes RF191 by L. piscium CNCM I-4031 (Saraoui et al., 2016).
The aim of this study was to characterize the further inhibitory effect of L. piscium CNCM I-4031 against L. monocytogenes species. First, the inhibitory activity of the CNCM I-4031 strain was evaluated on a collection of 42 L. monocytogenes strains. Then, the effect of the protective strain CNCM I-4031 on the growth, morphological shape, and virulence of L. monocytogenes was investigated.
Materials and Methods
Bacterial Strains, Culture Media, and Conditions
Lactococcus piscium CNCM-I 4031 was isolated from fresh salmon steak packed under modified atmosphere packaging (Matamoros et al., 2009b). The L. monocytogenes strains used in this study are listed in Table 1. All strains were stored in aliquots of 250 μl at −80°C in a final concentration of 10% (v/v) of glycerol. For all experiments, an aliquot of the strain was subcultured in Elliker broth (Biokar Diagnostic, Beauvais, France) for 24 h at 26°C for L. piscium, and in Brain Heart Infusion supplemented by 2% NaCl (mBHI) (Biokar Diagnostic, Beauvais, France) for 24 h at 20°C for L. monocytogenes. The cultures were diluted in their culture medium to obtain appropriate initial cell concentrations. The chemically defined medium MSMA used for the bacterial interaction observation was prepared as previously described by Saraoui et al. (2016). L. piscium was enumerated by spreading 100 μl of 10-fold serial dilutions on Elliker agar plates incubated at 8°C for 5 days under anaerobiosis (co-cultures) or at 26°C for 48 h (pure cultures). L. monocytogenes was enumerated by spread-plating 100 μl (classic enumeration) or 5 μl (microenumeration method) of 10-fold serial dilutions on mBHI agar incubated at 37°C for 24 h.
Table 1.
List of L. monocytogenes strains used in this study.
| Strain | Origin | Country | Collection |
|---|---|---|---|
| ScottA | Milk (incriminated in listeriosis) | United States | CIP 103575 |
| EGD-e | Rabbit (incriminated in listeriosis) | England | ATCC BAA-679 |
| EU2208 | Cod croquette | Spain | AZTI |
| EU2169 | Fresh trout | Spain | AZTI |
| EU2170 | Fresh trout | Spain | AZTI |
| EU2171 | Fresh trout | Spain | AZTI |
| EU2148 | Shrimp | Iceland | MATIS |
| EU2209 | Smoked cod | Spain | AZTI |
| EU2158 | Smoked salmon production plant | France | ASEPT |
| EU2159 | Smoked salmon production plant | France | ASEPT |
| EU2160 | Smoked salmon production plant | France | ASEPT |
| EU2161 | Smoked salmon production plant | France | ASEPT |
| EU2162 | Smoked salmon production plant | France | ASEPT |
| EU2163 | Smoked salmon production plant | France | ASEPT |
| EU2164 | Smoked salmon production plant | France | ASEPT |
| RF101 | Smoked salmon production plant | France | ASEPT |
| RF102 | Smoked salmon production plant | France | ASEPT |
| RF103 | Smoked salmon production plant | France | ASEPT |
| RF104 | Smoked salmon production plant | France | ASEPT |
| RF105 | Smoked salmon production plant | France | ASEPT |
| RF106 | Smoked salmon production plant | France | ASEPT |
| RF96 | Smoked salmon production plant | France | ASEPT |
| RF97 | Smoked salmon production plant | France | ASEPT |
| RF98 | Smoked salmon production plant | France | ASEPT |
| RF99 | Smoked salmon production plant | France | ASEPT |
| RF113 | Smoked salmon production plant | France | ASEPT |
| RF115 | Smoked salmon production plant | France | ASEPT |
| RF116 | Smoked salmon production plant | France | ASEPT |
| RF118 | Smoked salmon production plant | France | ASEPT |
| RF120 | Smoked salmon production plant | France | ASEPT |
| RF122 | Smoked salmon production plant | France | ASEPT |
| RF123 | Smoked salmon production plant | France | ASEPT |
| RF124 | Smoked salmon production plant | France | ASEPT |
| RF125 | Smoked salmon production plant | France | ASEPT |
| RF133 | Smoked salmon production plant | France | ASEPT |
| RF135 | Smoked salmon production plant | France | ASEPT |
| RF138 | Smoked salmon production plant | France | ASEPT |
| RF142 | Smoked salmon production plant | France | ASEPT |
| RF152 | Smoked salmon production plant | France | ASEPT |
| RF92 | Smoked trout | France | Aqualande |
| RF93 | Smoked trout | France | Aqualande |
| RF166 | Taramasalata | France | Biocéane |
| RF191 | Tropical cooked peeled shrimp | France | PFI Nouvelles Vagues |
ATCC, American Type Culture Collection; AZTI, Derio, Spain; MATIS, Reykjavik, Iceland; ASEPT, Le Mans, France; Aqualande, Roquefort, France; Biocéane, Nantes, France; PFI Nouvelles Vagues, Boulogne-sur-Mer, France.
Biodiversity of L. monocytogenes Sensitivity to L. piscium CNCM I-4031
After subculture, L. piscium CNCM I-4031 and each of the 42 L. monocytogenes strains (Table 1) were co-cultured in a 96-well microplate filled with 200 μl of MSMA at an initial concentration of 106 and 103 CFU/ml, respectively. The microplate was incubated at 26°C for 30 h without shaking. Controls consisted of monocultures of each L. monocytogenes strain inoculated in the same conditions. L. monocytogenes strains were enumerated in CFU/ml using the microenumeration method described in section “Bacterial Strains, Culture Media, and Conditions.” The inhibition was calculated by the difference in the log-concentration of L. monocytogenes strain in pure culture and in co-culture. The standard deviation was estimated according to five independent replicates of the inhibition tests for L. monocytogenes RF191 and L. piscium CNCM-4031.
Effect of Co-culture Ratios on the Inhibition of L. monocytogenes by L. piscium CNCM I-4031
Lactococcus piscium CNCM I-4031 and L. monocytogenes RF191 were co-inoculated in 250-ml flasks of MSMA medium, without shaking. Depending on the experiments, the initial ratios between L. piscium and L. monocytogenes were 103/103 (A), 105/103 (B), and 106/106 (C) CFU/ml, respectively. The co-cultures were incubated at 26°C, and the growth of both strains was monitored during 48 to 72 h by the classic enumeration method described in section “Bacterial Strains, Culture Media, and Conditions.” Controls consisted of monocultures of each strain in MSMA at 26°C at the same inoculation levels. All the cultures were performed in triplicate.
Observation of Cells in Co-culture by Scanning Electron Microscopy
Pure cultures and co-cultures (ratio 106/103 CFU/ml) of L. piscium CNCM I-4031 and L. monocytogenes RF191 were cultivated in 10 ml of MSMA medium at 26°C for 24 h. Then, 1 ml (108 cells) of the suspension was filtered on a Nuclepore® polycarbonate membrane with a 0.22-μm pore size and 13-mm diameter (Whatman International Ltd., Maidstone, United Kingdom). In order to observe the bacterial interaction on a solid medium (Dubey and Ben-Yehuda, 2011), another filter membrane was placed on an MSMA agar (15 g/l) plate and spotted with 10 μl of co-culture and incubated for 6 h at 26°C. All membranes containing the cells were fixed with 2.5% (v/v) glutaraldehyde (diluted in sodium cacodylate 0.1 M, pH 7.2) (Sigma Aldrich, Saint-Quentin Fallavier, France) for 48 h at 4°C. The fixing solution was renewed twice. Then, the samples were washed using a solution of sodium cacodylate (0.2 M, pH 7.2) and dehydrated in serial concentrations of ethanol (60, 70, 80, 90, 95%), 10 min for each concentration, followed by three times/20 min in 100% ethanol. The membranes were transferred to a critical point dryer, and the samples were subsequently sputter-coated and observed with a scanning electron microscope (Jeol JSM 6301F) at the CMEBA platform (Rennes, France).
Analysis of the Virulence of L. monocytogenes Co-cultivated With L. piscium
Lactococcus piscium CNCM I-4031, L. monocytogenes RF191, and L. monocytogenes Scott A strains were cultivated in MSMA medium at 26°C for 24 h in triplicates. The cultures were centrifuged, re-suspended in phosphate buffered-saline (PBS, Eurobio, Courtaboeuf, France), and then diluted to obtain appropriate cell concentrations for the cell line infection.
The human adenocarcinoma cells (line HT-29) (European Collection of Animal Cell Cultures, Salisbury, United Kingdom) were routinely grown in 75-cm2 flasks (Sigma) in a complete medium, DMEM (Dulbecco’s modified Eagle’s medium), with 10% (v/v) fetal calf serum (SCF), and 1% amphotericin B 250 μg/ml (Eurobio). Gentamicin 100 μg/ml (Sigma) was added to the culture medium. Cells were kept in the humidified atmosphere of a 5% CO2 incubator at 37°C. One hundred microliter of HT-29 cell suspension (2 × 104 cells) was deposited per well in a 96-well tissue culture plate (Sigma). The plates were incubated for 4 days with antibiotics followed by incubation for 24 h without antibiotics to obtain confluent monolayers.
In each well, HT-29 cells were infected with 108 L. piscium CNCM I-4031 and incubated for 1 h at 37°C. L. piscium cells were then removed by suction, and the HT-29 cells were infected with 100-μl suspensions from 108 to 103 CFU/ml of L. monocytogenes per well and incubated for 2 h at 37°C. L. monocytogenes cells were then removed by suction. The HT-29 cells were covered with 100 μl of DMEM 10% SCF with 100 μg/ml of gentamicin and incubated for 1.5 h at 37°C to eliminate bacterial cells from the plates. Each well was then overlaid with DMEM 10% SCF with 100 μg/ml of gentamicin containing 0.47% of indubiose to prevent cell starvation. Incubation was carried out at 37°C for 24 to 48 h. The virulence of the bacterial cells was evaluated by plaque-forming assay (PFA) in the cell monolayers using an optical microscope (VWR, Pennsylvania, United States). Controls consisted of HT-29 cells infected with serial dilutions of 108 to 102 UFC of L. piscium CNCM I-4031 or L. monocytogenes RF191 or ScottA per well. The whole experiment was repeated three times (different weeks) corresponding to nine independent tests.
Statistical Analyses
Statistical analyses on bacterial counts concentrations were performed using R software (R Core Team, 2014) by the analysis of one-way analysis of factor variance (ANOVA) followed by least significant difference (LSD) test.
Results and Discussion
Inhibition Capacity of L. piscium CNCM I-4031 Strain Toward Various L. monocytogenes Strains
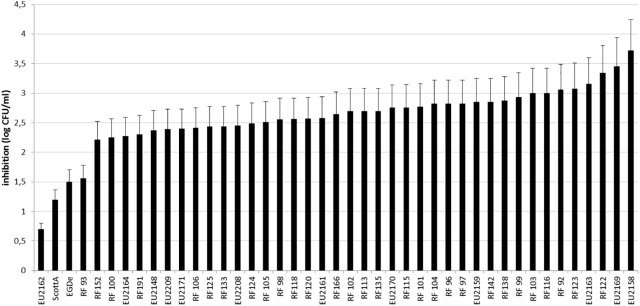
A total of 42 L. monocytogenes strains from diverse sources, and geographical origins were selected (Table 1). Most of them were isolated from seafood except the reference strains ScottA (Fleming et al., 1985) and EGD-e (Murray et al., 1926) isolated during a listeriosis outbreak from human and animal tissue, respectively. The inhibition of L. monocytogenes strains was measured after 30 h of co-culture on microplates with L. piscium CNCM I-4031, in MSMA liquid medium. In these miniaturized experimental conditions, L. monocytogenes RF191 displayed an inhibition of 2.40 ± 0.35 log CFU/ml, which is lower than that previously described in larger culture volumes with an inhibition of 3–4 log CFU/ml (Saraoui et al., 2016). However, this screening test showed that L. piscium CNCM I-4031 could inhibit all L. monocytogenes strains, whatever their origin (Figure 1). The inhibition yield was strain-dependent, varying between 0.69 log CFU/ml for EU2162 and 3.72 log CFU/ml for EU2158. These values can be correlated with the low growth in MSMA medium of EU2262 in pure culture (7.3 log CFU/ml) and, in contrast, the strong growth of EU2158 (9.3 log CFU/ml). In mixed culture, after 30 h of incubation, the concentration of L. monocytogenes ranged between 5.6 and 7.1 log CFU/ml when cultivated alone and 7.3 and 9.5 in co-culture (Figure 2).
FIGURE 1.
Inhibition of 42 L. monocytogenes strains after a 30-h culture with L. piscium CNCM I-4031 in MSMA.
FIGURE 2.
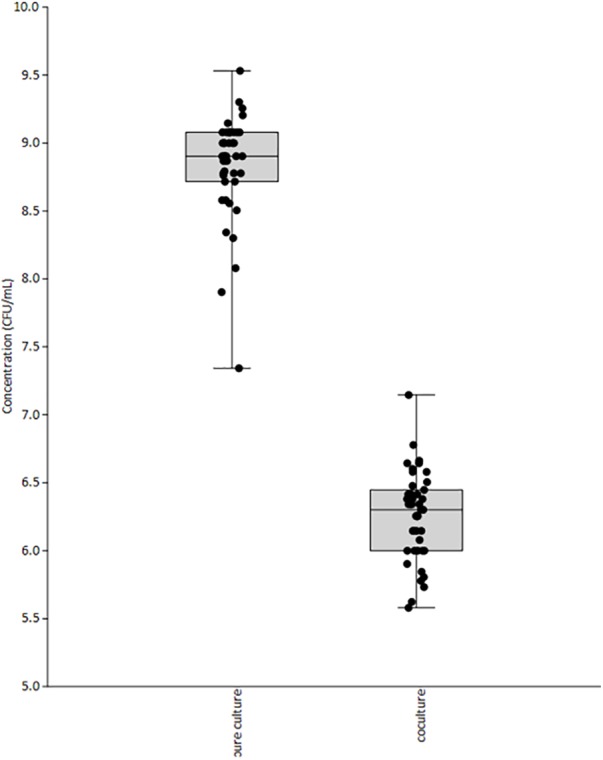
Final concentration of 42 L. monocytogenes strains in pure and mixed culture with L. piscium CNCM I-4031, after 30 h in MSMA medium.
Influence of the Initial Ratio of L. piscium CNCM I-4031/L. monocytogenes
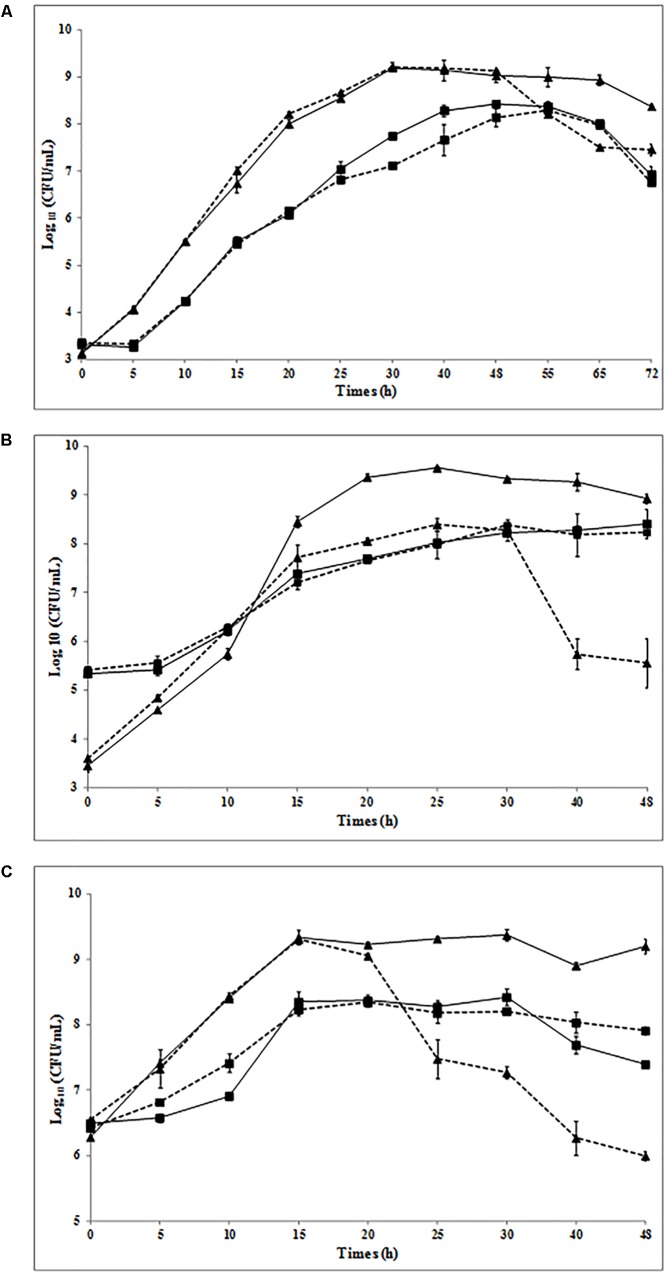
Previous studies have shown that L. piscium CNCM I-4031 can inhibit the growth of L. monocytogenes RF191 from 3 to 4 log units with an inoculum ratio of 106/103 CFU/ml (L. piscium/L. monocytogenes) in shrimp at 8°C and in MSMA medium at 26°C (Fall et al., 2010; Saraoui et al., 2016). Considering that the concentration of pathogenic bacteria in food at the beginning of storage is low, protective bacteria are usually added at high concentrations to food products (Ananou et al., 2005; Brillet et al., 2005). In order to determine whether the inhibition was linked with these initial concentrations, three different initial ratios of L. piscium/L. monocytogenes RF191 were analyzed (Figure 3): 103/103 CFU/ml (A), 105/103 CFU/ml (B), and 106/106 CFU/ml (C).
FIGURE 3.
Growth of L. piscium CNCM I-4031 (■) and L. monocytogenes RF191 ( ) in pure culture (solid line) and in co-culture (dotted line) in MSMA at 26°C. Initial concentrations of L. piscium/L. monocytogenes: (A) 103/103 CFU/ml, (B) 105/103 CFU/ml, and (C) 106/106 CFU/ml.
) in pure culture (solid line) and in co-culture (dotted line) in MSMA at 26°C. Initial concentrations of L. piscium/L. monocytogenes: (A) 103/103 CFU/ml, (B) 105/103 CFU/ml, and (C) 106/106 CFU/ml.
The growth kinetics of L. piscium in pure and co-culture were similar whatever the initial ratios. The maximum population density (MPD) of approximately 108 CFU/ml was reached after 40–48 h in (A), 25 h in (B), and 15 h in (C) with a growth rate between 0.32 to 0.45 h−1 (Table 2). L. monocytogenes growth started with an exponential phase and reached the MPD (∼109 CFU/ml) at 30 h in (A), 20 h in (B), and 15 h in (C) (Table 2). In each co-culture, the L. monocytogenes population increased to reach 108 to 109 CFU/ml until L. piscium achieved the MPD, then decreased (Figure 3). These results indicate that, regardless of the inoculum ratio of the two strains, L. monocytogenes was always significantly inhibited in co-cultures when L. piscium reached its MPD. This inhibition was proportional to the initial concentration of L. piscium and ranged from 1.42 to 3.37 log CFU/g (Table 2) with a higher inhibitory effect when the protective strain was inoculated at 105–106 CFU/ml. These observations are in accordance with previous studies with other bioprotective LAB isolated from seafood, such as a non-bacteriocin-producing Carnobacterium piscicola A9b (Nilsson et al., 2005) and Lactobacillus sakei 10A (Vermeiren et al., 2006b). Nevertheless, contrary to L. sakei 10A, which required an initial concentration of up to 105 CFU/g to inhibit L. monocytogenes effectively, L. piscium showed an inhibition with a low initial concentration of 103 CFU/ml.
Table 2.
Maximal population density and growth rate of L. piscium CNCM I-4031 (Lp) and L. monocytogenes RF191 (Lm) in pure (PC) or co-culture (CC) in MSMA medium at 26°C.
| Lactococcus piscium CNCM I-4031 | Listeria monocytogenes RF191 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Initial ratio UFC/ml (Lp/Lm) | (A) 103/103 | (B) 105/103 | (C) 106/106 | (A) 103/103 | (B) 105/103 | (C) 106/106 | ||||||
| Culture specifications | PC | CC | PC | CC | PC | CC | PC | CC | PC | CC | PC | CC |
| Maximal population density (log CFU/ml) ± SD | 8.28 ± 0.12 | 8.14 ± 0.2 | 8.01 ± 0.01 | 7.98 ± 0.27 | 8.35 ± 0.15 | 8.23 ± 0.09 | 9.19 ± 0.04 | 9.21 ± 0.09 | 9.36 ± 0.03 | 8.05 ± 0.07 | 9.33 ± 0.11 | 9.31 ± 0.05 |
| Growth rate (h-1) | 0.43 | 0.40 | 0.45 | 0.37 | 0.40 | 0.32 | 0.60 | 0.63 | 0.76 | 0.66 | 0.51 | 0.63 |
| Final population density (log CFU/ml) ± SD | 6.92 ± 0.02a | 6.75 ± 0.02a | 8.40 ± 0.29a | 8.23 ± 0.03a | 7.39 ± 0.04b | 7.91 ± 0.05a | 8.37 ± 0.11a | 7.46 ± 0.04b | 8.92 ± 0.09a | 5.55 ± 0.50b | 9.20 ± 0.10a | 5.99 ± 0.06b |
SD, standard deviation. a,bRepresent groups determined with LSD test, a same letter indicates values not significantly different (p-value < 0.05 by one-way ANOVA) through one condition.
The competition between LAB species and other populations in food and mixed cultures by one single “dominant” strain and when LAB have reached their maximum level is described in the literature as the “Jameson effect” (Jameson, 1962; Gimenez and Dalgaard, 2004). The Jameson effect is considered a race between species in order to maximize their growth by exploiting the environmental nutrients. Thus, the species that first reaches its MPD inhibits the growth of the other species (Mellefont et al., 2008; Cornu et al., 2011). However, in our experiments, we showed that the inhibition of L. monocytogenes by L. piscium occurred even when the concentration of the pathogenic bacteria was higher than that of the protective bacteria (Figures 3A,C). In these conditions, L. monocytogenes reached its MPD before L. piscium but no inhibition of L. piscium occurred. The same results were found when L. monocytogenes ScottA was co-inoculated with Escherichia coli whereas in co-culture with Lactobacillus plantarum or Pseudomonas fluorescens, the first strain reaching its MPD stopped the growth of the other one (Mellefont et al., 2008).
In a previous study, we demonstrated that the amount of lactic acid produced by L. piscium CNCM I-4031 in co-culture conditions was not responsible for the inhibition (Saraoui et al., 2016). Moreover, the supplementation of the co-culture with nutrients did not restore the ability of L. monocytogenes to grow, and no inhibition was observed when both cultures were separated by a 0.45-μm membrane. The present study evidenced that L. piscium could limit the growth of L. monocytogenes when it reached its maximum density, whatever its initial level. Inhibition phenomena linked to maximum cellular concentration have been shown in other LAB, and some authors have suggested that they could involve quorum sensing (Kuipers et al., 2000; Risøen et al., 2000; Rohde and Quadri, 2006; Moslehi-Jenabian et al., 2011; Rizzello et al., 2014). In our case, the mechanism remains unknown and has still to be investigated.
Observation of Cells in Co-culture by Scanning Electron Microscopy
With the aim of investigating the behavior of the pathogenic bacteria in co-culture at the microscopic scale, the cell morphology of L. piscium CNCM I-4031 and L. monocytogenes RF191 was compared in monoculture in MSMA medium with co-culture conditions at the time of inhibition. The results presented in Figure 4 show that L. piscium in mono-cultures were spherical (Elliker) or ovoid (MSMA) cells between 0.5 and 1 μm in diameter and appeared in pairs or short chains. L. monocytogenes were rod-shaped cells between 0.5 μm (MSMA) and 1.5 μm (mBHI) in length and ∼0.5 μm in width, appearing individually or in pairs (Figure 4B). For co-cultures, strains were observed in either liquid or solid media (Figures 4C,D). Co-cultures on solid media have already revealed the presence of nanotubes, which are extensions of the cytoplasmic membrane used by bacteria to exchange their cellular compounds in cell contact interactions (Dubey and Ben-Yehuda, 2011). These authors described intra-species nanotubes between cells of Bacillus subtilis, as well as inter-species connections between cells of B. subtilis, S. aureus, and E. coli using scanning electron microscopy. In the conditions of our experiments, such nanotubes were not observed between the cells. However, the pictures show that in the presence of L. piscium, L. monocytogenes cells appeared to be more elongated (Figures 4C,D). The elongation of Listeria cells in stress conditions (low pH, high salt concentration) has been reported in previous studies (Besnard et al., 2000; Bereksi et al., 2002). Listeria cells can divide without septation, leading to the modification of their surface properties with the presence of filamentous structures. Our observations also suggest that the surface of L. monocytogenes cells was damaged or completely altered in the presence of LAB (Figures 4C,D). These effects on L. monocytogenes morphology have been reported in previous studies dealing with the mechanism of action of anti-Listeria components (Dieuleveux et al., 1998).
FIGURE 4.
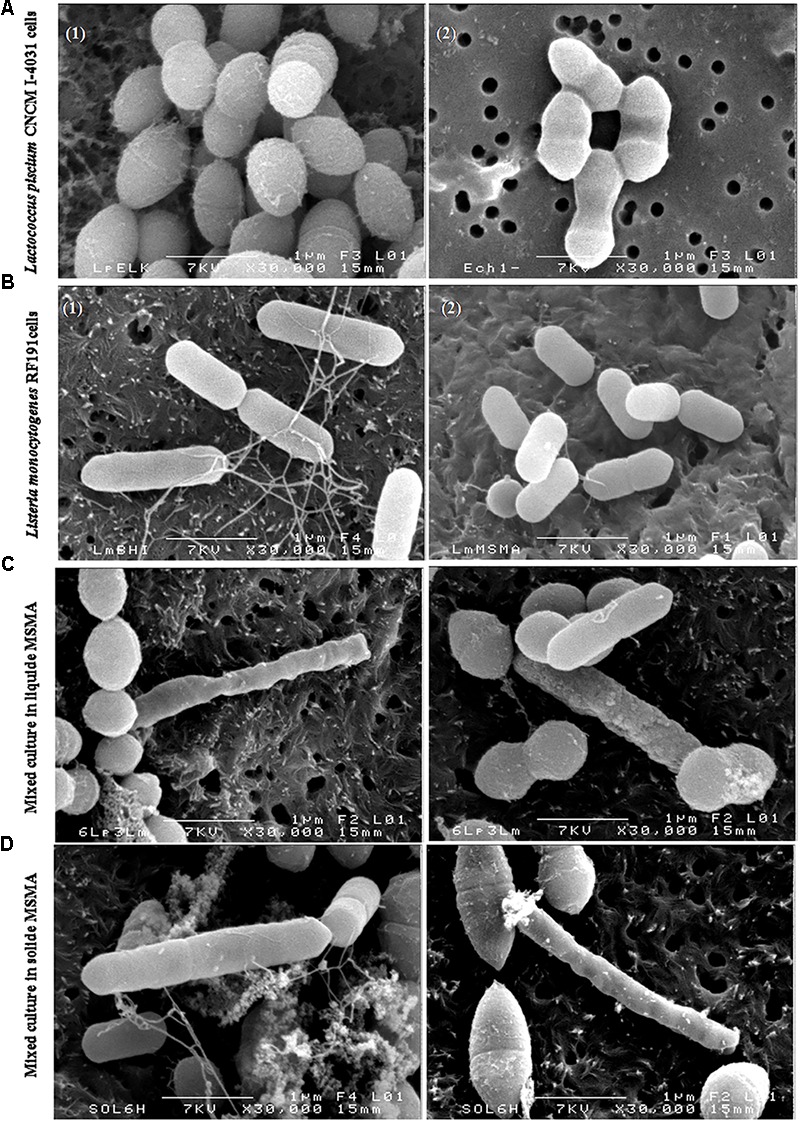
L. piscium CNCM I-4031 and L. monocytogenes RF191 cells in pure and co-culture viewed using scanning electron microscopy on polycarbonate membranes. Magnification: × 30,000. (A) L. piscium in pure culture (1) in Elliker; (2) in MSMA, 24 h at 26°C. (B) L. monocytogenes RF191 in pure culture (1) in mBHI; (2) in MSMA, 24 h at 26°C. (C) Co-culture of L. piscium CNCM I-4031 and L. monocytogenes RF191 in liquid MSMA. (D) Co-culture of L. piscium CNCM I-4031 and L. monocytogenes RF191 on MSMA plate.
Effect of L. piscium CNCM I-4031 on the Virulence of L. monocytogenes
As an important pathogen, L. monocytogenes encompasses a large spectrum of strains with varying virulence effects (Buchanan et al., 2017). In order to examine another aspect of the bioprotective effect of L. piscium, the impact on L. monocytogenes virulence was investigated. For this purpose, the virulence of two selected L. monocytogenes strains, RF191 isolated from seafood and ScottA known to be a highly virulent strain (Lindback et al., 2010), were tested using an HT-29 cell PFA, in the presence or absence of L. piscium. On an HT-29 monolayer pre-treated with 108 L. piscium, no lysis plates were detected after 24 or 48 h confirming that L. piscium has no pathogenic activity. Plaques were counted in the well containing HT-29 cells infected by 5 log CFU/ml of L. monocytogenes ScottA or RF191 strains. The ScottA strain formed large deep plaques whereas the RF191 strain formed small shallow ones. The mean log PFA values were 3.93 ± 0.06 and 3.68 ± 0.08, respectively (Figure 5). According to the study of Roche et al. (2001), the RF191 strain that forms more than 3.34 log plaques should be considered virulent, even though it remains less virulent than the ScottA strain. When HT-29 cells were infected by L. monocytogenes strains after being treated by L. piscium, the mean log PFA values were 3.40 ± 0.36 and 1.25 ± 0.29 for ScottA and RF191 strains, respectively (Figure 5). A significant effect was found for the RF191 strain that had lost its virulence after pre-treatment with L. piscium CNCM I-64031. A slight but a significant decrease in virulence was also observed for the ScottA strain, suggesting that the effect of the protective bacteria on L. monocytogenes virulence is strain-dependent. The effect on virulence reduction by LAB has already been described for foodborne pathogens such as L. monocytogenes (Garriga et al., 2015; Pilchova et al., 2016) or Campylobacter (Alemka et al., 2010). The effect is usually investigated as one of the probiotic properties of bacteria; however, such additional properties also increase the bioprotective value of strains and their safety assessment.
FIGURE 5.
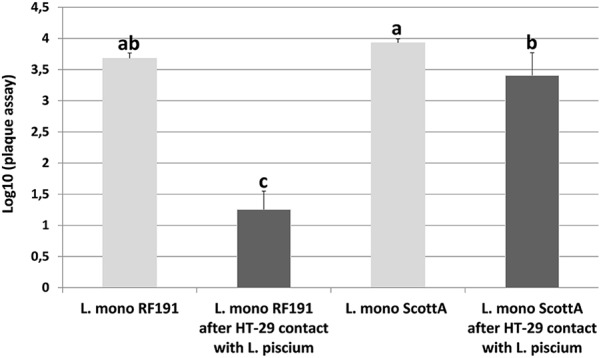
L. monocytogenes plaque-forming assay (PFA) using HT-29 epithelial cells infected with strains ScottA and RF191 directly or after 1 h of pre-incubation with L. piscium CNCM I-4031. Plaques were counted in HT-29 cells infected with 105 UFC/ml of L. monocytogenes. No plaque was observed when L. piscium CNCM I-4031 was inoculated alone. Error bars indicate standard deviations from at least three independent experiments. a,b,cRepresent groups determined with LSD test, a same letter indicates values not significantly different (p-value < 0.05 by one way-ANOVA).
Conclusion
In this study, we have demonstrated the ability of L. piscium CNCM I-4031 to inhibit a large collection of L. monocytogenes strains. This inhibition occurs whatever the initial concentration of the protective strain when L. piscium has reached its MPD. The inhibition mechanism is still under investigation but our different sets of results suggest that it requires the proximity of cells and affects the cellular surface of the targeted bacteria. In addition to inhibiting L. monocytogenes growth, L. piscium decreased L. monocytogenes virulence with a variable effect according to the strain. This study provides additional knowledge about the inhibitory activity of a non-bacteriocin-producing LAB toward L. monocytogenes and significant information for the potential use of L. piscium CNCM I-4031 to control L. monocytogenes in seafood products.
Author Contributions
M-FP and FL designed the work. TS, DP, and FC performed the experiments. TS and FL assisted with the scanning electron microscopy analysis. TS, M-FP, FL, DP, and J-MC analyzed the data. TS drafted the paper. MF-P, FL, and DP wrote the final version of the manuscript, which was read and approved by all authors.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank the staff of the CMEBA platform (Rennes, France) for their technical assistance with the scanning electron microscopy images. The authors also thank MATIS, AZTI, PFI Nouvelle Vague, ASEPT, Biocéane, and Aqualande for providing Listeria monocytogenes strains.
Footnotes
Funding. TS was the recipient of a Ph.D. fellowship from the French Ministry of Higher Education and Research. This work was financially supported by the project COM-BACT from the Région des Pays de la Loire, France.
References
- Alemka A., Clyne M., Shanahan F., Tompkins T., Corcionivoschi N., Bourke B. (2010). Probiotic colonization of the adherent mucus layer of HT29MTXE12 cells attenuates Campylobacter jejuni virulence properties. Infect. Immun. 78 2812–2822. 10.1128/IAI.01249-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amezquita A., Brashears M. M. (2002). Competitive inhibition of Listeria monocytogenes in ready-to-eat meat products by lactic acid bacteria. J. Food Prot. 65 316–325. 10.4315/0362-028X-65.2.316 [DOI] [PubMed] [Google Scholar]
- Ananou S., Garriga M., Hugas M., Maqueda M., Martinez-Bueno M., Galvez A., et al. (2005). Control of Listeria monocytogenes in model sausages by enterocin AS-48. Int. J. Food Microbiol. 103 179–190. 10.1016/j.ijfoodmicro.2004.12.024 [DOI] [PubMed] [Google Scholar]
- Batdorj B., Trinetta V., Dalgalarrondo M., Prevost H., Dousset X., Ivanova I., et al. (2007). Isolation, taxonomic identification and hydrogen peroxide production by Lactobacillus delbrueckii subsp. lactis T 31, isolated from Mongolian yoghurt: inhibitory activity on food-borne pathogens. J. Appl. Microbiol. 103 584–593. 10.1111/j.1365-2672.2007.03279.x [DOI] [PubMed] [Google Scholar]
- Bereksi N., Gavini F., Benezech T., Faille C. (2002). Growth, morphology and surface properties of Listeria monocytogenes Scott A and LO28 under saline and acid environments. J. Appl. Microbiol. 92 556–565. 10.1046/j.1365-2672.2002.01564.x [DOI] [PubMed] [Google Scholar]
- Besnard V., Federighi M., Cappelier J. M. (2000). Development of a direct viable count procedure for the investigation of VBNC state in Listeria monocytogenes. Lett. Appl. Microbiol. 31 77–81. 10.1046/j.1472-765x.2000.00771.x [DOI] [PubMed] [Google Scholar]
- Brillet A., Pilet M. F., Prévost H., Bouttefroy A., Leroi F. (2004). Biodiversity of Listeria monocytogenes sensitivity to bacteriocin-producing Carnobacterium strains and application in sterile cold-smoked salmon. J. Appl. Bacteriol. 97 1029–1037. 10.1111/j.1365-2672.2004.02383.x [DOI] [PubMed] [Google Scholar]
- Brillet A., Pilet M. F., Prévost H., Cardinal M., Leroi F. (2005). Effect of inoculation of Carnobacterium divergens V 41, a biopreservative strain against Listeria monocytogenes risk, on the microbiological, and sensory quality of cold-smoked salmon. Int. J. Food Microbiol. 104 309–324. 10.1016/j.ijfoodmicro.2005.03.012 [DOI] [PubMed] [Google Scholar]
- Buchanan R. L., Gorris L. G. M., Hayman M. M., Jackson T. C., Whiting R. C. (2017). A review of Listeria monocytogenes: an update on outbreaks, virulence, dose-response, ecology, and risk assessments. Food Control 75 1–13. 10.1016/j.foodcont.2016.12.016 [DOI] [Google Scholar]
- CAC (2007). CAC/GL 61-2007. Guidelines on the Application of General Principles of Food Hygiene to the Control of Listeria monocytogenes in Ready-to-Eat Foods. Geneva: Codex Alimentarius Commission, 1–28. [Google Scholar]
- Cornu M., Billoir E., Bergis H., Beaufort A., Zuliani V. (2011). Modeling microbial competition in food: application to the behavior of Listeria monocytogenes and lactic acid flora in pork meat products. Food Microbiol. 28 639–647. 10.1016/j.fm.2010.08.007 [DOI] [PubMed] [Google Scholar]
- Dieuleveux V., Lemarinier S., Gueguen M. (1998). Antimicrobial spectrum and target site of D-3-phenyllactic acid. Int. J. Food Microbiol. 40 177–183. 10.1016/S0168-1605(98)00031-2 [DOI] [PubMed] [Google Scholar]
- Dortu C., Huch M., Holzapfel W. H., Franz C. M., Thonart P. (2008). Anti-listerial activity of bacteriocin-producing Lactobacillus curvatus CWBI-B28 and Lactobacillus sakei CWBI-B1365 on raw beef and poultry meat. Lett. Appl. Microbiol. 47 581–586. 10.1111/j.1472-765X.2008.02468.x [DOI] [PubMed] [Google Scholar]
- Dubey G. P., Ben-Yehuda S. (2011). Intercellular nanotubes mediate bacterial communication. Cell 144 590–600. 10.1016/j.cell.2011.01.015 [DOI] [PubMed] [Google Scholar]
- EFSA (2016). The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2015. EFSA J. 14:4364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Ziney M. G., van den Tempel T., Debevere J., Jakobsen M. (1999). Application of reuterin produced by Lactobacillus reuteri 12002 for meat decontamination and preservation. J. Food Prot. 62 257–261. 10.4315/0362-028X-62.3.257 [DOI] [PubMed] [Google Scholar]
- Fall P. A., Leroi F., Chevalier F., Guerin C., Pilet M. F. (2010). Protective effect of a non-bacteriocinogenic Lactococcus piscium CNCM I-4031 strain against Listeria monocytogenes in sterilized tropical cooked peeled shrimp. J. Aquat. Food Prod. Technol. 19 84–92. 10.1080/10498850.2010.486910 [DOI] [Google Scholar]
- Fall P. A., Pilet M. F., Leduc F., Cardinal M., Duflos G., Guérin C., et al. (2012). Sensory and physicochemical evolution of tropical cooked peeled shrimp inoculated by Brochothrix thermosphacta and Lactococcus piscium CNCM I-4031 during storage at 8°C. Int. J. Food Microbiol. 152 82–90. 10.1016/j.ijfoodmicro.2011.07.015 [DOI] [PubMed] [Google Scholar]
- Fleming D. W., Cochi S. L., MacDonald K. L., Brondum J., Hayes P. S., Plikaytis B. D., et al. (1985). Pasteurized milk as a vehicle of infection in an outbreak of listeriosis. N. Engl. J. Med. 312 404–407. 10.1056/NEJM198502143120704 [DOI] [PubMed] [Google Scholar]
- Garriga M., Rubio R., Aymerich T., Ruas-Madiedo P. (2015). Potentially probiotic and bioprotective lactic acid bacteria starter cultures antagonise the Listeria monocytogenes adhesion to HT29 colonocyte-like cells. Benef. Microbes 6 337–343. 10.3920/BM2014.0056 [DOI] [PubMed] [Google Scholar]
- Gimenez B., Dalgaard P. (2004). Modelling and predicting the simultaneous growth of Listeria monocytogenes and spoilage micro-organisms in cold-smoked salmon. J. Appl. Microbiol. 96 96–109. 10.1046/j.1365-2672.2003.02137.x [DOI] [PubMed] [Google Scholar]
- Ito A., Sato Y., Kudo S., Sato S., Nakajima H., Toba T. (2003). The screening of hydrogen peroxide-producing lactic acid bacteria and their application to inactivating psychrotrophic food-borne pathogens. Curr. Microbiol. 47 231–236. 10.1007/s00284-002-3993-1 [DOI] [PubMed] [Google Scholar]
- Jameson J. E. (1962). A discussion of dynamics of Salmonella enrichment. J. Hyg. 60 193–207. 10.1017/S0022172400039462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuipers O. P., Buist G., Kok J. (2000). Current strategies for improving food bacteria. Res. Microbiol. 151 815–822. 10.1016/S0923-2508(00)01147-5 [DOI] [PubMed] [Google Scholar]
- Lecuit M., Charlier-Woerther C., Leclerq A. (2015). Rapport Annuel D’activité du Centre national de Référence des Listeria – Année 2014. Paris: Institut Pasteur. [Google Scholar]
- Lindback T., Rottenberg M. E., Roche S. M., Rorvik L. M. (2010). The ability to enter into an avirulent viable but non-culturable (VBNC) form is widespread among Listeria monocytogenes isolates from salmon, patients and environment. Vet. Res. 41:8. 10.1051/vetres/2009056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez R. C., Staliano C. D., Vieira A. D., Villarreal M. L., Todorov S. D., Saad S. M., et al. (2015). Bacteriocin production and inhibition of Listeria monocytogenes by Lactobacillus sakei subsp. sakei 2a in a potentially synbiotic cheese spread. Food Microbiol. 48 143–152. 10.1016/j.fm.2014.12.010 [DOI] [PubMed] [Google Scholar]
- Matamoros S., Leroi F., Cardinal M., Gigout F., Kasbi Chadli F., Cornet J., et al. (2009a). Psychrotrophic lactic acid bacteria used to improve the safety and quality of vacuum-packaged cooked and peeled tropical shrimp and cold-smoked salmon. J. Food Prot. 72 365–374. [DOI] [PubMed] [Google Scholar]
- Matamoros S., Pilet M. F., Gigout F., Prévost H., Leroi F. (2009b). Selection and evaluation of seafood-borne psychrotrophic lactic acid bacteria as inhibitors of pathogenic and spoilage bacteria. Food Microbiol. 26 638–644. 10.1016/j.fm.2009.04.011 [DOI] [PubMed] [Google Scholar]
- Mellefont L. A., McMeekin T. A., Ross T. (2008). Effect of relative inoculum concentration on Listeria monocytogenes growth in co-culture. Int. J. Food Microbiol. 121 157–168. 10.1016/j.ijfoodmicro.2007.10.010 [DOI] [PubMed] [Google Scholar]
- Moslehi-Jenabian S., Vogensen F. K., Jespersen L. (2011). The quorum sensing luxS gene is induced in Lactobacillus acidophilus NCFM in response to Listeria monocytogenes. Int. J. Food Microbiol. 149 269–273. 10.1016/j.ijfoodmicro.2011.06.011 [DOI] [PubMed] [Google Scholar]
- Murray E. G. D., Webb R. A., Swann M. B. R. (1926). A disease of rabbits characterised by a large mononuclear leucocytosis, caused by a hitherto undescribed bacillus Bacterium monocytogenes (n.sp.). J. Pathol. Bacteriol. 29 407–439. 10.1002/path.1700290409 [DOI] [Google Scholar]
- Nilsson L., Hansen T. B., Garrido P., Buchrieser C., Glaser P., Knochel S., et al. (2005). Growth inhibition of Listeria monocytogenes by a non bacteriocinogenic Carnobacterium piscicola. J. Appl. Microbiol. 98 172–183. 10.1111/j.1365-2672.2004.02438.x [DOI] [PubMed] [Google Scholar]
- Pilchova T., Pilet M. F., Cappelier J. M., Pazlarova J., Tresse O. (2016). Protective effect of Carnobacterium spp. against Listeria monocytogenes during host cell invasion using in vitro HT29 model. Front. Cell. Infect. Microbiol. 6:88. 10.3389/fcimb.2016.00088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team (2014). A Language and Environment for Statistical Computing [Internet]. Vienna: R Foundation for Statistical Computing. [Google Scholar]
- Richard C., Brillet A., Pilet M. F., Prévost H., Drider D. (2003). Evidence on inhibition of Listeria monocytogenes by divercin V41 action. Lett. Appl. Microbiol. 36 288–292. 10.1046/j.1472-765X.2003.01310.x [DOI] [PubMed] [Google Scholar]
- Risøen P. A., Brurberg M. B., Eijsink V. G. H., Nes I. F. (2000). Functional analysis of promoters involved in quorum sensing-based regulation of bacteriocin production in Lactobacillus. Mol. Microbiol. 37 619–628. 10.1046/j.1365-2958.2000.02029.x [DOI] [PubMed] [Google Scholar]
- Rizzello C. G., Filannino P., Di Cagno R., Calasso M., Gobbetti M. (2014). Quorum-sensing regulation of constitutive plantaricin by Lactobacillus plantarum strains under a model system for vegetables and fruits. Appl. Environ. Microbiol. 80 777–787. 10.1128/AEM.03224-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roche S. M., Velge P., Bottreau E., Durier C., Marquet-van der Mee N., Pardon P. (2001). Assessment of the virulence of Listeria monocytogenes: agreement between a plaque-forming assay with HT-29 cells and infection of immucompetent mice. Int. J. Food Microbiol. 68 33–44. 10.1016/S0168-1605(01)00460-3 [DOI] [PubMed] [Google Scholar]
- Rocourt J., BenEmbarek P., Toyofuku H., Schlundt J. (2003). Quantitative risk assessment of Listeria monocytogenes in ready to eat foods: the FAO/WHO approach. FEMS Immunol. Med. Microbiol. 35 263–267. 10.1016/S0928-8244(02)00468-6 [DOI] [PubMed] [Google Scholar]
- Rohde B. H., Quadri L. E. (2006). Functional characterization of a three-component regulatory system involved in quorum sensing-based regulation of peptide antibiotic production in Carnobacterium maltaromaticum. BMC Microbiol. 6:93. 10.1186/1471-2180-6-93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salminen S., von Wright A., Morelli L., Marteau P., Brassart D., de Vos W. M., et al. (1998). Demonstration of safety of probiotics - a review. Int. J. Food Microbiol. 44 93–106. 10.1016/S0168-1605(98)00128-7 [DOI] [PubMed] [Google Scholar]
- Saraoui T., Fall P. A., Leroi F., Antignac J.-P., Chéreau S., Pilet M. F. (2016). Inhibition mechanism of Listeria monocytogenes by a bioprotective bacteria Lactococcus piscium CNCM I-4031. Food Microbiol. 53 70–78. 10.1016/j.fm.2015.01.002 [DOI] [PubMed] [Google Scholar]
- Tomé E., Teixeira P., Gibbs P. A. (2006). Anti-listerial inhibitory lactic acid bacteria isolated from commercial cold smoked salmon. Food Microbiol. 23 399–405. 10.1016/j.fm.2005.05.004 [DOI] [PubMed] [Google Scholar]
- Unlu G., Nielsen B., Ionita C. (2015). Production of antilisterial bacteriocins from lactic acid bacteria in dairy-based media: a comparative study. Probiotics Antimicrob. Proteins 7 259–274. 10.1007/s12602-015-9200-z [DOI] [PubMed] [Google Scholar]
- Vermeiren L., Devlieghere F., Vandekinderen I., Debevere J. (2006a). The interaction of the non-bacteriocinogenic Lactobacillus sakei 10A and lactocin S producing Lactobacillus sakei 148 towards Listeria monocytogenes on a model cooked ham. Food Microbiol. 23 511–518. [DOI] [PubMed] [Google Scholar]
- Vermeiren L., Devlieghere F., Vandekinderen I., Rajtak U., Debevere J. (2006b). The sensory acceptability of cooked meat products treated with a protective culture depends on glucose content and buffering capacity: a case study with Lactobacillus sakei 10A. Meat Sci. 74 532–545. 10.1016/j.meatsci.2006.05.003 [DOI] [PubMed] [Google Scholar]