Abstract
Coenzyme Q (ubiquinone or CoQ) is an essential lipid that plays a role in mitochondrial respiratory electron transport and serves as an important antioxidant. In human and yeast cells, CoQ synthesis derives from aromatic ring precursors and the isoprene biosynthetic pathway. Saccharomyces cerevisiae coq mutants provide a powerful model for our understanding of CoQ biosynthesis. This review focusses on the biosynthesis of CoQ in yeast and the relevance of this model to CoQ biosynthesis in human cells. The COQ1–COQ11 yeast genes are required for efficient biosynthesis of yeast CoQ. Expression of human homologs of yeast COQ1–COQ10 genes restore CoQ biosynthesis in the corresponding yeast coq mutants, indicating profound functional conservation. Thus, yeast provides a simple yet effective model to investigate and define the function and possible pathology of human COQ (yeast or human gene involved in CoQ biosynthesis) gene polymorphisms and mutations. Biosynthesis of CoQ in yeast and human cells depends on high molecular mass multisubunit complexes consisting of several of the COQ gene products, as well as CoQ itself and CoQ intermediates. The CoQ synthome in yeast or Complex Q in human cells, is essential for de novo biosynthesis of CoQ. Although some human CoQ deficiencies respond to dietary supplementation with CoQ, in general the uptake and assimilation of this very hydrophobic lipid is inefficient. Simple natural products may serve as alternate ring precursors in CoQ biosynthesis in both yeast and human cells, and these compounds may act to enhance biosynthesis of CoQ or may bypass certain deficient steps in the CoQ biosynthetic pathway.
Keywords: coenzyme Q, mitochondrial dysfunction, Saccharomyces cerevisiae, ubiquinone
Introduction
Coenzyme Q (ubiquinone or CoQ) is a vital lipid component in mitochondrial energy metabolism. It is a two-part molecule containing a long polyisoprenyl tail of n isoprene units positioning the molecule in the mid-plane of membrane bilayer, and a fully substituted benzoquinone ring that undergoes reversible reduction and oxidation. The redox chemistry of CoQ and CoQH2 (ubiquinol, a hydroquinone) allows it to play its best-known role in mitochondrial respiration, accepting electrons and protons from Complex I or Complex II and donating them to Complex III, thereby establishing a proton gradient across the mitochondrial inner membrane. CoQ also serves as an essential electron and proton acceptor in other aspects of metabolism including fatty acid β-oxidation, uridine biosynthesis, and oxidation of sulphide, proline, glycerol-3-phosphate, choline, dimethylglycine, and sarcosine [1,2]. CoQH2 also serves a crucial antioxidant function, protecting membranes as a chain terminator of lipid peroxidation reactions, and in the maintenance of reduced forms of vitamin E [1,3]. CoQ/CoQH2 is a component of lipoproteins and is present in all cellular membranes including the plasma membrane where it functions in cellular redox regulation as part of the plasma membrane oxidoreductase system [1].
The focus of this review is on the biosynthesis of CoQ6 in the yeast Saccharomyces cerevisiae and the relevance of this model to the biosynthesis of CoQ10 in human cells. Readers are directed to other recent reviews that discuss the biosynthesis of CoQ in prokaryotes such as Escherichia coli [4], and in eukaryotes including Schizosaccharomyces pombe, plants, Caenorhabditis elegans, Mus musculus, and humans [5–7]. For an in-depth discussion of the effects of CoQ10 deficiencies and the clinical syndromes associated with these deficiencies, readers are directed to the article by Brea-Calvo and colleagues [8] in this issue of Essays in Biochemistry.
Overview of CoQ biosynthesis
S. cerevisiae is an extraordinarily useful model for understanding the biosynthesis of CoQ. Early yeast classic and molecular genetics combined with subcellular fractionation, biochemical assays, and lipid chemistry have helped to identify many of the steps required for CoQ biosynthesis. In particular, the collection of respiratory deficient coq mutants identified by Tzagoloff [9,10] set the stage for isolation and characterization of the yeast COQ genes. A particular advantage is that the CoQ-less coq mutants are viable when cultured on growth medium containing a fermentable carbon source, but are incapable of growth on medium containing a non-fermentable carbon source. In most cases, expression of the human COQ ( human polypeptide involved in CoQ10 biosynthesis) homolog restores function in the corresponding yeast coq mutant. This rescue of yeast coq mutants by human COQ genes is a powerful and simple functional assay still being used to ascertain the effects of human mutations or polymorphisms on human COQ gene function. Thus, what we have learned about the biosynthesis of CoQ6 in the yeast model is highly relevant to the biosynthesis of CoQ10 in humans (Figure 1).
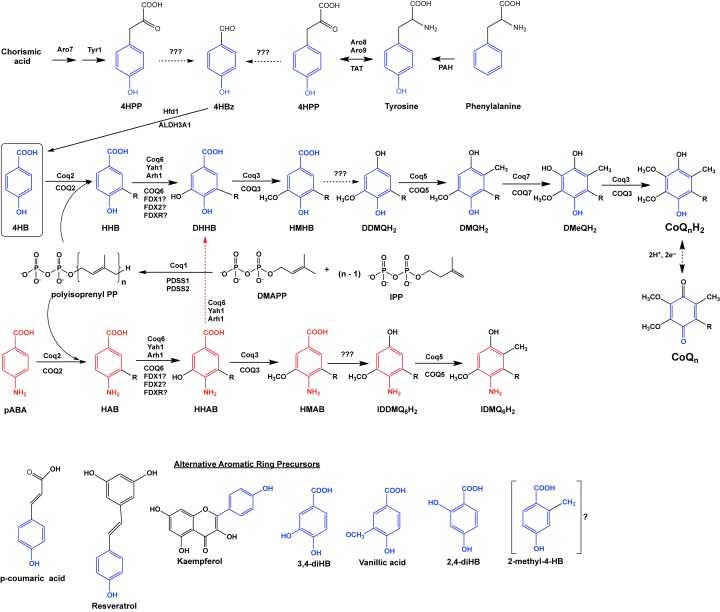
Figure 1. CoQ biosynthetic pathways in the yeast S. cerevisiae and in humans.
The CoQ biosynthetic pathway has been shown to involve at least 14 nuclear-encoded proteins that are necessary for mitochondrial CoQ biosynthesis in S. cerevisiae. Black dotted arrows denote more than one step. Solid arrows denote a single step attributed to the corresponding yeast polypeptide named above each arrow. The corresponding human homologs are named below each arrow. The main ring precursor used by both yeast and humans is 4-hydroxybenzoic acid (4HB). Yeasts synthesize 4HB de novo from chorismate or may obtain it from the metabolism of tyrosine. Humans rely on tyrosine to produce 4HB (or on phenylalanine and phenylalanine hydroxylase to produce tyrosine). Yeast and human cells produce isopentenyl pyrophosphate (IPP) and dimethylally pyrophosphate (DMAPP) as precursors to form hexaprenyl diphosphate (n=6) via Coq1 in yeast or decaprenyl diphosphate (n=10) via PDSS1/PDSS2 in humans. Yeast Coq2 and human COQ2 attach the polyisoprenyl tail to 4HB. Subsequent to this step, the next three intermediates are identified as yeast hexaprenyl-intermediates: HHB, 3-hexaprenyl-4HB; DHHB, 3-hexaprenyl-4,5-dihydroxybenzoic acid; HMHB, 3-hexaprenyl-4-hydroxy-5-methoxybenzoic acid. The next three intermediates are hydroquinones: DDMQH2, 2-hexaprenyl-6-methoxy-1,4-benzenediol; DMQH2, 2-hexaprenyl-3-methyl-6-methoxy-1,4-benzenediol; DMeQH2, 2-hexaprenyl- 3-methyl-6-methoxy-1,4,5-benzenetriol; to ultimately produce the final reduced product (CoQnH2). Red text identifies para-aminobenzoic acid (pABA) as an alternate ring precursor utilized by yeast (but not by humans). The next three intermediates are identified as yeast hexaprenyl-intermediates: HAB, 4-amino-3-hexaprenylbenzoic acid; HHAB, 4-amino-3-hexaprenyl-5-hydroxybenzoic acid; HMAB, 4-amino-3-hexaprenyl-5-methoxybenzoic acid. The next two intermediates are: IDDMQH2, 4-amino-3-hexaprenyl-5-methoxyphenol; IDMQH2, 4-amino-3-hexaprenyl-2-methyl-5-methoxyphenol. The step denoted by the red dotted arrow depends on yeast Coq6 and converts HHAB into DHHB. Interconversion of (CoQnH2) and (CoQn) is shown via a reversible two-electron reduction and oxidation. Steps indicated by ‘???’ are catalyzed by as yet unknown enzymes. Alternative compounds that may serve as ring precursors in CoQ biosynthesis are shown at the bottom of the panel: p-coumaric acid, resveratrol, and kaempferol. Analogs of 4HB that can function to bypass certain deficiencies in the CoQ biosynthetic pathway include: 3,4-dihydroxybenzoic acid (3,4-diHB), vanillic acid and 2,4-dihydroxybenzoic acid (2,4-diHB). It is not yet known whether 2-methyl-4HB (2-methyl-4HB) may also serve a bypass function.
The yeast model also provided early evidence that the eukaryotic CoQ biosynthetic pathway was localized to mitochondria. The Coq (denotes S. cerevisiae polypeptide involved in CoQ6 biosynthesis) polypeptides are nuclear encoded, and amino-terminal mitochondrial targetting sequences are needed to direct their transport to the mitochondrial matrix (Coq1, Coq3–Coq11) or to the inner mitochondrial membrane (Coq2). Assembly of Coq3–Coq9 plus Coq11 polypeptides into a high molecular mass complex termed the CoQ synthome in yeast (Figure 2) and Complex Q in human cells is another conserved feature of CoQ biosynthesis [7,11]. These complexes are essential for the biosynthesis of CoQ in yeast and human cells, and may serve to enhance catalytic efficiency and to minimize the escape of intermediates that may be toxic due to their redox or electrophilic properties. The CoQ-intermediates are quite hydrophobic and at least some of them appear to be essential partners in the assembly of the membrane-bound CoQ synthome [12] and Complex Q [7,13].
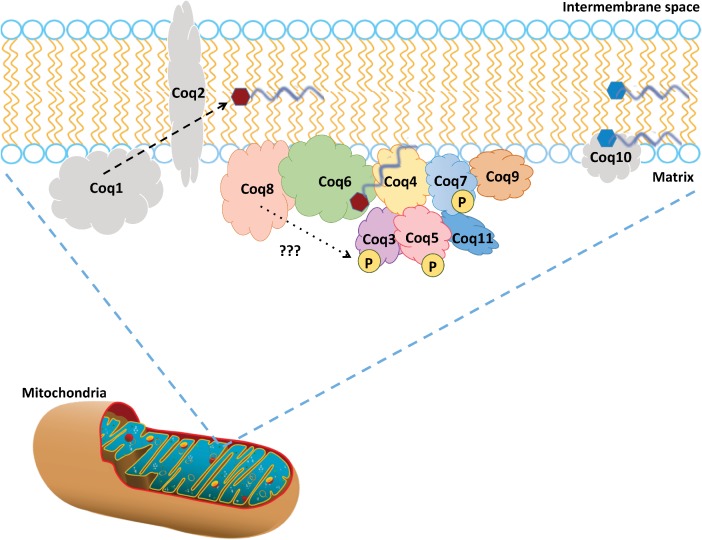
Figure 2. A model of the CoQ Synthome in the yeast S. cerevisiae.
Studies in S. cerevisiae have provided evidence for a high-molecular mass multisubunit protein and lipid complex, the CoQ synthome (see text for references). The Coq3–Coq9 and Coq11 polypeptides, designated in color, co-purify, and are members of this complex that is peripherally associated with the matrix-side of the inner mitochondrial membrane. Coq1, Coq2, and Coq10 are individual polypeptides that do not associate with the complex (indicated in gray). Coq1 and Coq2 synthesize the early intermediates HHB and HAB (denoted by red hexagon with a gray hexaprenyl tail). Coq10 binds CoQ (and also late-stage CoQ-intermediates denoted as blue hexagons with a gray tail), and functions as a chaperone for this hydrophobic lipid that normally resides at the mid-plane of the membrane bilayer. The Coq3, Coq5, and Coq7 polypeptides are phosphorylated in a Coq8-dependent manner (shown by ‘???’). The function of Coq8 is still under investigation; although part of a family of atypical kinases, Coq8 has been shown to autophosphorylate, but not yet shown to phosphorylate any other proteins, in vitro or in vivo. It is speculated to have ATPase function and potentially has the ability to phosphorylate lipids or other small molecules. Hence the phosphorylation of Coq3, Coq5, and Coq7 may be from Coq8 or be produced via another kinase that is recruited to the CoQ synthome to act upon those particular polypeptides. In yeast, it has been shown that the phosphatase that dephosphorylates Coq7 is Ptc7s, the product of the spliced form of PTC7 (not shown).
Ring precursors utilized in biosynthesis of CoQ
Origin of 4-hydroxybenzoic acid
In yeast and human cells, the primary precursor molecule that leads to the biosynthesis of CoQ is 4-hydroxybenzoic acid (4HB). Yeast cells generate 4HB via the shikimate pathway, but also utilize tyrosine as a ring precursor [14]. Unlike yeast, human cells contain phenylalanine hydroxylase, and so either phenylalanine or tyrosine may be utilized as precursors for the biosynthesis of 4HB. Many steps involved in the generation of 4HB from tyrosine are yet to be characterized [15,16]. However, two recent studies have shed light on the first and the last steps involved in yeast 4HB biosynthesis [17,18]. The first step involves the deamination of tyrosine to 4-hydroxyphenylpyruvate (4-HPP), catalyzed by either of the aminotransferases Aro8 or Aro9 [17]. Payet et al. [17] also identified 4-hydroxybenzaldehyde (4HBz) as the final intermediate leading to the biosynthesis of 4HB. The oxidation of 4HBz to 4HB is catalyzed by the aldehyde dehydrogenase Hfd1. Hfd1 is a mitochondrial outer membrane protein [19] indicating that 4HB is synthesized in the cytosol, and must be imported into the mitochondrial matrix, where it is incorporated into CoQ. Hence, there should be a mitochondrial transporter for 4HB that remains to be identified [7], and is responsible for use of exogenously added 4HB. Inactivation of HFD1 results in CoQ6 deficiency that may be complemented by the addition of exogenous 4HB. Expression of the human homolog ALDH3A1 restored CoQ6 biosynthesis in the hfd1 yeast mutant, and was shown to oxidize 4HBz to 4HB [17]. In an independent study Stefely et al. [18] confirmed these findings; MS was used to characterize the proteomes, lipidomes, and metabolomes of a large selection of yeast strains, each lacking a distinct gene related to mitochondrial biology. This multi-omic approach revealed that yeast Hfd1 and human ALDH3A1 serve as the aldehyde dehydrogenases responsible for the oxidation of 4HBz to 4HB. It will be important to determine whether human ALDH3A1 is required for CoQ10 biosynthesis in human cells; if so, it may be a potential target gene that should be considered when screening for CoQ10 deficiencies in patients.
Other aromatic ring precursors of CoQ
In addition to 4HB, yeast utilize para-aminobenzoic acid (pABA) as a ring precursor of CoQ6 [20,21]. Yeast Coq6 and Coq9 polypeptides are required for this metabolism. Yeast Coq6 is required for the oxidative deamination of the ring nitrogen substituent [22]. Coq9 is also required for Coq6 activity, including the Coq6-mediated deamination function [23,24]. Analogs of 4HB, including 2,4-dihydroxybenzoic acid (2,4-diHB), 3,4-dihydroxybenzoic acid (3,4-diHB) and vanillic acid may be incorporated into CoQ6 [16] (Figure 1). These analogs may allow for the bypass of CoQ6 biosynthetic defects in certain yeast coq6 and coq7 mutants [16,24,25], as discussed in ‘Yeast and human genes essential for CoQ biosynthesis’. Additional aromatic ring precursors incorporated into CoQ6 in yeast include p-coumarate and the polyphenols resveratrol and kaempferol [26,27] (Figure 1), although the use of kaempferol by yeast is very marginal.
In contrast, pABA is not utilized for CoQ synthesis by human or mouse cells [26], and instead it acts to inhibit the incorporation of 4HB into CoQ [16,28,29]. In mammalian cells, p-coumarate, vanillic acid, 3,4-diHB, resveratrol, and kaempferol also serve as CoQ ring precursors, with the difference that in this case kaempferol is a very efficient precursor that is even able to up-regulate CoQ9 and CoQ10 levels in human and mouse kidney cells [27]. While the mechanism underlying the use of polyphenols is still unknown, it is clear that a highly specific process occurs because other polyphenols with similar structures, such as piceatannol or apigenin, are not used for CoQ synthesis in mammalian cells [27].
A hypothesis for the use of p-coumarate, resveratrol, and kaempferol as ring precursors of CoQ is that they are metabolized to produce 4HB. The set of reactions that allow this conversion is not yet identified but, independent of the metabolic route involved, an increase in alternative CoQ ring precursors in cells will only turn into higher CoQ levels if cells have a low availability of endogenous 4HB, which is the primary precursor of CoQ. The fact that the availability of 4HB is a rate-limiting step in the CoQ biosynthetic pathway has been previously described in yeast and in mammalian kidney cells [21,27]. Supplementing mammalian kidney cells with exogenous 4HB resulted in an increase in CoQ levels four- to six-fold higher as compared with the non-supplemented control [27]. This observation led the authors to propose the possibility that increasing the availability of CoQ precursors in cells could move the metabolic flux in favor of the biosynthesis of CoQ, helping to ameliorate the phenotype associated with certain Q deficiencies.
Yeast and human genes essential for CoQ biosynthesis
Yeast COQ1; human PDSS1 and PDSS2
In yeast and human cells, the synthesis of the polyisoprenyl diphosphate tail derives from a non-sterol branch of the mevalonate pathway [7]. Yeast Coq1 is responsible for the synthesis of the hexaprenyl diphosphate tail moiety from the precursors dimethylallyl diphosphate and isopentenyl diphosphate [30]. The Coq1 polypeptide is peripherally associated with the matrix side of the inner mitochondrial membrane [31]. The analogous polyprenyl diphosphate synthases in other species determine the tail length (n) of the CoQn (ubiquinone-n or coenzyme Qn (refers to a specific isoform, where n is number of isoprenyl units in the tail of CoQn, e.g. CoQ10 in humans, CoQ6 in S. cerevisiae)) produced [32], and when expressed in yeast direct the synthesis of corresponding isoforms of CoQn [33]. The yeast Coq1 polypeptide is not associated with the CoQ synthome, but its lipid product is essential for the formation and/or stabilization of this complex [31,34].
PDSS1 and PDSS2 form a heterotetramer responsible for the synthesis of the decaprenyl-diphosphate tail precursor used to synthesize CoQ10 in human cells [35]. Patients with partial deficiencies in PDSS1 [36] and PDSS2 [37] show severe disruptions in multiple organ systems. As reviewed in this volume [8], the complexity of phenotypes is a hallmark of mitochondrial deficiency diseases.
Yeast COQ2; human COQ2
The yeast Coq2 polypeptide is required for the attachment of the polyisoprenyl ‘tail’ to 4HB [38]. In yeast, Coq2 generates 3-hexaprenyl-4-hydroxy benzoic acid (HHB; Figure 1), the first polyisoprenylated ring CoQ-intermediate in the biosynthetic pathway. This early hydrophobic CoQ-intermediate was found to accumulate in many of the yeast coq null mutants, including the coq3-coq9 null mutants [39,40]. Coq2 is imported into mitochondria via the Tim23 pathway [41], and is an integral membrane protein of the inner mitochondrial membrane [34]. It was originally hypothesized that Coq2 might serve to anchor the CoQ-synthome to the inner mitochondrial membrane [11], however there is no evidence that Coq2 is associated with the other Coq polypeptides that assemble into the CoQ-synthome [12]. Instead it appears that polyisoprenylated CoQ-intermediates produced by Coq1 and Coq2 are important for the stabilization of the CoQ synthome [34] (Figure 2).
Forsgren et al. [42] isolated human COQ2 cDNA and showed its expression in a yeast coq2 null mutant restored CoQ6 biosynthesis. Recently, Desbats et al. [43] have defined the 5′ transcription start sites of the human COQ2 transcript, indicating that of four potential upstream ATG translation initiation codons, the first two are rarely (if ever) used and that it is the fourth ATG that is in fact predominant. All isoforms of COQ2 were shown to co-localize to mitochondria. This finding argues against the previous hypothesis that the shorter COQ2 isoforms may represent non-mitochondrial polypeptides that may mediate cytoplasmic prenylation of 4HB [42]. The predominant use of the fourth ATG results in a shorter COQ2 polypeptide and the authors suggest new numbering that should be used to designate mutations of human COQ2 [43]. Desbats et al. [43] found a good correlation between disease severity in patients and the effect of COQ2 mutations on the decreased production of CoQ6 in a yeast complementation assay. Patients who harbored two alleles that markedly impair CoQ biosynthesis manifested multisystem severe clinical symptoms at birth or infancy, while patients who had a least one allele with residual CoQ biosynthesis manifested isolated steroid resistant nephrotic syndrome (SRNS) or adult onset encephalopathy [43].
Based on two structures determined for prokaryotic homologs of COQ2 (UbiA family aromatic prenyltransferases), human COQ2 is proposed to contain nine transmembrane helices [43]. The C-terminus of human COQ2 resides in the intermembrane space in mitochondria of HEK293 cells [43]. A recent structural model is compatible with this suggestion and predicts that the active site of human COQ2 faces the matrix [44]. Two of the disease-related mutations (given in the old nomenclature) are posited to interfere with the binding of the polyisoprenyl diphosphate (R197H) or to clash with the Mg2+ ions that participate in catalysis (A302V) [44].
A recent study by Herebian et al. [45] demonstrated that supplementation with 4HB fully restores endogenous CoQ10 biosynthesis in partially deficient COQ2 human fibroblasts harboring homozygous mutant alleles, including the A302V severe allele. Based on an in silico model of human COQ2, the authors identified several binding sites for 4HB and posited a channel for 4HB transport across the inner mitochondrial membrane. The authors proposed that the rescue of CoQ10 synthesis in fibroblasts from COQ2-deficient patients by treatment with 4HB may represent amelioration of a 4HB transport deficit and/or an enhancement of activity by increased supply of the ring substrate [45]. It will be important to experimentally determine whether the COQ2 polypeptide also functions as a 4HB transporter. In addition to restoring CoQ10 levels, the 4HB treatment also increased the steady state levels of COQ4 and COQ7 proteins involved in CoQ biosynthesis, and enhanced cell viability in response to stress conditions. This finding makes sense in light of the important role that CoQ and CoQ-intermediates play in stabilizing the CoQ-synthome in yeast and Complex Q in human cells [7,12,34]. This rescue of COQ2-deficient cells by 4HB treatment is quite striking and deserves further testing as a potential therapy. Even a small enhancement in the biosynthesis of CoQ is able to restore a wide array of phenotypes associated with CoQ deficiency [46].
Yeast COQ3; human COQ3
Yeast Coq3 is an S-adenosylmethionine (AdoMet)-dependent methyltransferase required for the two O-methylation steps of CoQ biosynthesis [47–49]. Coq3 is peripherally associated with the matrix-side of the mitochondrial inner membrane [49]. Recent studies reveal that E. coli UbiG, a functional homolog of Coq3, binds to liposomes containing cardiolipin [50]. Structural determination of UbiG identifies it as a seven β-strand AdoMet-dependent methyltransferase that contains an unusual insertion sequence that mediates UbiG binding to membranes, and is required for CoQ biosynthesis [50].
Assays with farnesylated analogs of CoQ-intermediates provided early evidence that a complex of yeast Coq polypeptides is required to observe the Coq3 O-methyltransferase activity and hence CoQ biosynthesis [39,51]. Recovery of the yeast Coq3-consecutive non-denaturing affinity purification (CNAP) tagged polypeptide from digitonin-solubilized mitochondrial extracts showed that it co-purified with Coq4, Coq5, Coq6, Coq7, Coq9, and Coq11 polypeptides, in a high molecular mass complex that contained CoQ6 and several CoQ6-intermediates [12]. Thus Coq3 is an integral member of the CoQ synthome in yeast. The phosphorylation state of Coq3 may modulate the stability of the Coq3 polypeptide and that of the CoQ synthome [52,53]. Overexpression of Coq8, an atypical putative protein kinase, has been shown to stabilize several Coq polypeptides and the CoQ synthome in certain yeast coq null mutants [24,34]. Indeed, overexpression of Coq8 in the yeast coq3 null mutant increased steady state levels of the Coq4, Coq6, Coq7, and Coq9 polypeptides, and stabilized the CoQ synthome. Treatment of coq3 null mutants overexpressing Coq8 with vanillic acid (a 4HB analog that should bypass the first hydroxylation and first methylation steps) resulted in the production of the late stage CoQ-intermediate DMQ6 [24] (Figure 1). This finding indicates the potential difficulties in using analogs of 4HB to bypass deficiencies in Coq3, due to its involvement in two O-methylation steps, and to the apparent absence of Coq7 hydroxylase activity. It is tempting to speculate that treatment with 2,3-dimethoxy-4HB might serve to bypass both O-methyltransferase deficient steps in the coq3 null mutant. Such bypass would require that this analog could still be prenylated by Coq2, and subjected to decarboxylation, hydroxylation and C-methylation steps.
Expression of human COQ3 in yeast coq3 null mutants rescued growth on a non-fermentable source and partially restored the biosynthesis of CoQ6 [54]. Assays with farnesylated analogs of CoQ-intermediates showed that mitochondria prepared from coq3 null mutant yeast expressing human COQ3 performed both O-methylation steps [54]. Many lines of evidence indicate a similar Complex Q containing the COQ3–COQ9 polypeptides is involved in human CoQ10 biosynthesis [7]. So far, no mutations causing primary CoQ10 deficiency have been reported for the human COQ3 gene.
Yeast COQ4; human COQ4
Yeast Coq4 is required for CoQ6 biosynthesis in yeast, and is peripherally associated with the inner mitochondrial membrane on the matrix side [55]. It is thought to serve as a scaffold or organizer for the CoQ synthome [34,56], as it is associated with Coq3, Coq6, and Coq9 [12,51,57]. No enzyme activity or exact function has been associated with the Coq4 polypeptide. A known crystal structure of the Coq4 domain, determined as part of the structural genomics effort (PDB: 3KB4, Northeastern structural genomics program) identified long hydrophobic α helices bound to a geranylgeranyl monophosphate lipid. A conserved HDxxHx10–13E motif [56] chelated a magnesium ion (Mg2+) near to the phosphate head group [57]. From this structure, the function of Coq4 is speculated to bind the long polyisoprenyl tail of CoQ-intermediates and/or CoQ and to organize the enzymes that perform the ring modifications [34,57] (Figure 2).
Human COQ4 was shown to be a functional ortholog of yeast Coq4, and is capable of restoring CoQ6 biosynthesis in the yeast coq4 null mutant [58]. Distinct COQ4 RNA transcripts indicated the potential for two different isoforms of the human COQ4 polypeptide; the longest isoform was shown to possess a mitochondrial targetting sequence, was localized to mitochondria in HeLa cells, and restored CoQ6 biosynthesis in the yeast coq4 null mutant. The functional significance of the shorter isoform is not known; it lacks the mitochondrial targetting sequence and failed to rescue coq4 mutant yeast.
Patients who harbor two recessive COQ4 mutant alleles exhibit a broad spectrum of mitochondrial disorders associated with CoQ10 deficiencies [59]. Intriguingly, haploinsufficiency of COQ4 also causes CoQ10 deficiency in both human and yeast diploid cells [60]. Recently a heterozygous missense E161D mutation in COQ4 was reported in a patient with lethal rhabdomyolysis; introduction of the missense mutation was introduced to iPSCs, and recapitulated the muscle-specific CoQ10 deficiency [61].
Yeast COQ5; human COQ5
The yeast Coq5 polypeptide is an AdoMet-dependent methyltransferase required for the C-methylation step of CoQ biosynthesis [62,63]. It is peripherally associated with the matrix-side of the mitochondrial inner membrane [64]. Dai et al. [65] determined the structure of yeast Coq5; it has a typical seven β-strand AdoMet methyltransferase structure, and the protein was crystallized both in the presence and absence of AdoMet. The catalytic mechanism is yet to be determined; based on modeling the authors proposed an active site highly conserved Arg201 or Tyr78 act to deprotonate a water molecule that then acts as the base to deprotonate the C5-ring H from DDMQ6H2 [65]. Yeast coq5 point mutants that harbor mutations in the Class I methyltransferase motifs result in a loss of C-methyltransferase function, but retain steady state levels of the Coq5 polypeptide, and of the Coq polypeptide partner proteins of the CoQ synthome. In contrast, these CoQ synthome partner proteins are destabilized in the coq5 null mutant [64]. Overexpression of Coq8 in the coq5 null yeast mutant results in the increased steady-state levels of the Coq4, Coq7, Coq9 polypeptides, the stabilization of the CoQ synthome, and the accumulation of DDMQ6H2, the substrate of Coq5 [24,34] (Figure 1). It is possible that the 4HB analog 2-methyl-4-BH might function to bypass the defect in coq5 point mutants with stable Coq5 polypeptide, however, this has not yet been tested.
Regulated expression of yeast Coq5 is necessary for the correct assembly of the CoQ synthome. Recently, two mechanisms of COQ5 post-transcriptional regulation have been elucidated. The RNA binding protein Puf3 regulates the translation of appropriate amounts of Coq5 so the CoQ synthome can be assembled [66]. Oct1 is a mitochondrial matrix-localized protease that removes eight residues from the amino-terminal mitochondrial targetting sequence of Coq5 and is essential for formation of the mature amino-terminus of Coq5 and its stability [67]. There is also evidence that yeast Coq5 is phosphorylated in a Coq8-dependent manner [53].
Expression of the human COQ5 polypeptide was found to rescue the CoQ6 biosynthetic defect of the coq5 point mutants or in a coq5 null mutant overexpressing Coq8, but not a coq5 null mutant [68]. Thus, human COQ5 is an ortholog of yeast Coq5, but can rescue yeast only when the other yeast Coq partner proteins are present and the CoQ synthome is assembled. Primary CoQ10 deficiency has been recently diagnosed due to a partial loss of function of COQ5 [69]. The deficiency is shown to be due to a duplication of the COQ5 gene, and that due to alternative splicing appears to generate an unstable COQ5 mRNA with a long 3′-UTR. Steady state levels of the COQ5 polypeptide were dramatically decreased in fibroblasts from the affected homozygous patients as compared with controls. The affected patients had variable degrees of cerebellar ataxia, and showed a modest decrease in the levels of CoQ10 in peripheral blood leukocytes, and a more dramatic decrease in CoQ10 levels in a skeletal muscle biopsy [69]. The reduction in CoQ10 levels is consistent with the observation that decreases in COQ5-containing mitochondrial protein complex impairs the production of CoQ10 [70].
Yeast COQ6; human COQ6
The yeast Coq6 polypeptide is characterized as a flavin-dependent monooxygenase [71]. Conserved catalytic regions in Coq6 include the ADP binding fingerprint, the NAD(P)H/FAD binding motif, and the ribityl binding region [71]. Yeast Coq6 co-purifies with a tightly bound FAD, and modeling studies are consistent with its proposed activity as a ring hydroxylase [72]. Yeast Coq6 is responsible for the first hydroxylation step (ring C5) in CoQ biosynthesis [73]. It is also necessary for the deamination of ring C4 in S. cerevisiae when pABA is used an aromatic ring precursor [22]. The yeast Coq6 polypeptide is peripherally associated with inner mitochondrial membrane on the matrix side [71], and associates with Coq4, Coq5, Coq7, Coq8, and Coq9 polypeptides of the CoQ synthome [12]. Recent investigations discovered a physical association of Coq6 with Coq8 [12]. Yeast coq6 point mutants that affect the active site but preserve steady state levels of the Coq6 polypeptide and assembly of the CoQ synthome may be rescued by providing alternate ring precursors such as 3,4-diHB and vanillic acid (Figure 1). These alternate ring precursors, once prenylated by Coq2, allow the defective coq6 step to be bypassed [73]. Such bypass is also effective in coq6 null yeast mutants provided yeast COQ8 is overexpressed [24].
The human homolog COQ6, is able to rescue a yeast coq6 null mutant [25,74], and interacts with human COQ8B (ADCK4) and COQ7 [75]. Yeast coq6 null mutants expressing certain hypomorphic mutations of human COQ6 are rescued by treatment with either 3,4-diHB or vanillic acid [25]. The effectiveness of such bypass therapies as treatments for patients with mutations in COQ6 remains to be explored. It seems possible that these alternate ring precursors might serve to restore endogenous CoQ10 biosynthesis in patients with COQ6 deficiencies.
Mutations in human COQ6 have been implicated in an autosomal recessive disease characterized by severe progressive nephrotic syndrome and deafness [74]. It has been suggested that kidney biopsy should be performed on young children present with SRNS and sensorineural hearing loss [76]. The rationale for this suggestion is that the abnormal mitochondria in podocytes may provide an early diagnostic clue of mutations in CoQ biosynthetic genes. Supplementation with high doses of CoQ10 can stop the progression of kidney disease, and this therapy should be started immediately at first suspicion of CoQ10 deficiency [76,77].
Yeast YAH1 and ARH1; human FDX1, FDX2, and FDXR
Unlike most flavin-dependent monooxygenases that utilize NAD(P)H directly as a source of electrons, the electrons from NAD(P)H are funneled indirectly to yeast Coq6 via the coupled system of ferredoxin (Yah1) an iron–sulphur protein, and ferredoxin reductase (Arh1) [73]. Yeast engineered to be transiently depleted in Yah1 or Arh1 were shown to be defective in the same C5 ring-hydroxylation step, and to accumulate the same polyisoprenylated ring-intermediates as yeast mutants harboring inactive coq6 alleles [21,73]. YAH1 and ARH1 are essential genes in yeast, and in addition to CoQ biosynthesis, are also required for iron–sulphur cluster biosynthesis [78].
There are two human homologs of yeast Yah1 – FDX1 and FDX2. Human FDX2 was shown to complement the iron–sulphur cluster biosynthetic defect of Yah1 depleted yeast [79]. However, neither human FDX1 nor FDX2 were able to complement the CoQ6 biosynthetic defect of Yah1 depleted yeast [73]. The human homolog of Arh1 is termed as FDXR, which functions as an electron transfer protein in cholesterol biosynthesis and overall steroid metabolism, as well as iron–sulphur cluster biosynthesis [78]. Whether FDX1, FDX2, or FDXR function to assist human COQ6 catalytic activity in the biosynthesis of CoQ10 is not yet known.
Yeast COQ7 (CAT5) and PTC7; human COQ7 (CAT5, CLK-1) and PPTC7
Yeast Coq7 is a hydroxylase responsible for catalyzing the penultimate step of the CoQ biosynthetic pathway [80–82]. The hydroxylase activity depends on a carboxylate-bridged diiron binding motif, first identified as a highly conserved sequence across a wide array of organisms, and predicted to mediate hydroxylation similar to other members of the carboxylate-bridged diiron protein family, such as methane monoxygenase, ribonucleotide reductase, and phenolhydroxylase [82]. Modeling predicted Coq7 to be a four-helix bundle protein with an additional helix mediating an interfacial association with the membrane [82]. Experiments with isolated mitochondria and mitoplasts show yeast Coq7 polypeptide is peripherally associated with inner mitochondrial membrane on the matrix side [34].
Expression of human COQ7 rescues the CoQ6 deficiency of yeast coq7 null mutants [83], indicating profound conservation of function. Overexpression of a soluble fusion protein containing human COQ7 polypeptide fused to an immunoglobulin-binding domain of protein G was purified (termed as GB1-hCLK-1) and spectroscopic and kinetic methods provided evidence for the presence of the diiron center [84]. Binding of the substrate analogs DMQ0 or DMQ2 to GB1-hCLK1 mediated the reduction in the diiron site by NADH and in the presence of O2 the hydroxylation step was catalyzed [84].
Yeast coq7 null mutants accumulate the early CoQ-intermediates HHB and HAB, while coq7 point mutants [80] and coq7 null mutants overexpressing Coq8 accumulate DMQ6 the penultimate intermediate in the pathway [24,85]. Expression of the unrelated E. coli UbiF hydroxylase rescued a coq7 point mutant, but failed to rescue the coq7 null mutant [86]. These findings indicated Coq7 is an important polypeptide partner of the CoQ synthome; indeed, Coq7 co-purifies with tagged forms of Coq3, Coq6, and Coq9 polypeptides [12].
A mutation in the human COQ7 gene is associated with a primary ubiquinone deficiency, and results in multiple organ involvement [87]. Interestingly, the deficiencies in CoQ10 content and mitochondrial respiratory activities in fibroblasts isolated from this patient were improved following treatment with 2,4-diHB, a 4HB analog that bypasses the COQ7-dependent hydroxylase step [87]. The effectiveness of treatment with 2,4-diHB depends on the type of COQ7 mutation(s) present in patients [88]. It is likely that the success of these bypass therapies will depend on the stable presence of other COQ polypeptides and their ability to form the CoQ synthome (or complex Q) [16,24]. Regulated expression of COQ7 has been shown to impact the rates of CoQ10 biosynthesis, both at the level of NF-κB transcriptional up-regulation of COQ7 gene expression [89], and at the level of RNA binding proteins that mediate stability of COQ7 mRNA [90]. The regulated expression of COQ7 and the other component polypeptides of the CoQ synthome seem likely to influence its assembly and function, and so impact biosynthesis of CoQ10.
Yeast Coq7 is modified by phosphorylation [12,53]. Predictive algorithms suggested that the phosphorylation status of Coq7 was regulatory for CoQ6 biosynthesis, with at least three predicted phosphorylation sites on Ser20, Ser28, and Thr32 [91]. When these residues are replaced with alanine, phosphorylation was abolished and CoQ6 levels were significantly increased in yeast expressing Coq7 with the triple-Alanine substitution. In contrast, yeast expressing the Coq7 with substitution of acidic residues at these residues (Asp20, Glu28, and Asp32) had decreased levels of CoQ6 and accumulated DMQ6, indicating that the non-phosphorylatable form of Coq7 is the active form that catalyzes the penultimate pathway step [91]. Other sites of phosphorylation may also influence Coq7 activity; yeast expressing Coq7 harboring the phosphomimetic Ser114Glu substitution also produced lower amounts of CoQ6 and accumulated DMQ6 [92].
The phosphatase responsible for Coq7 dephosphorylation is Ptc7, a bona fide mitochondrial serine/threonine protein phosphatase belonging to the PPM family of phosphatases [93]. It was recently discovered that two distinct forms of PTC7 RNA exist, spliced and non-spliced forms, displaying a rare case of alternative splicing in yeast that results in two viable isoforms of a spliced protein [94,95]. The previously reported Ptc7 phosphatase was shown to be the spliced form (Ptc7s) that resides in the mitochondria, while the non-spliced form (Ptc7ns) is a nuclear membrane localized protein that contains a transmembrane helix that anchors it to the nuclear membrane [94]. Exclusive expression of Ptc7s showed significantly higher de novo CoQ biosynthesis, as compared with Ptc7ns. These findings suggest that the mitochondrial targetting of the Ptc7s results in Coq7 dephosphorylation, and allows Coq7 to catalyze the penultimate step of the CoQ biosynthetic pathway [95]. Ptc7s acts to dephosphorylate other mitochondrial proteins, and deletion of ptc7 perturbs mitochondrial function [96]. PPTC7 is the human serine/threonine phosphatase homolog of yeast Ptc7, however it is not known whether phosphorylation regulates human COQ7, and if so, whether PPTC7 recognizes it as a substrate.
Yeast COQ8; human COQ8A (ADCK3) and COQ8B (ADCK4)
The S. cerevisiae Coq8 polypeptide is identified as a putative kinase in the biosynthetic pathway of CoQ6. Coq8 harbors six of twelve motifs present in protein kinases and is required to observe the presence of phosphorylated forms of Coq3, Coq5, and Coq7 polypeptides [53]. Coq8 co-purifies with the CoQ synthome [12]. Further, overexpression of Coq8 in certain of the coq null strains restores steady state levels of Coq4, Coq7, and Coq9, and stabilizes the formation of the CoQ synthome [24,34].
Expression of human COQ8A (ADCK3) in yeast coq8 mutant strains restored CoQ6 biosynthesis and the phosphorylation state of several of the yeast Coq polypeptides, indicating a profound conservation of function [53]. Rescue of yeast coq8 mutants by human COQ8A depended on fusion to a yeast mitochondrial targetting sequence [53]. Yeast Coq8 and human COQ8A are homologs of atypical protein kinases. These proteins are able to autophosphorylate and show a surprising affinity and selectivity for ADP, as opposed to ATP [97]. Human COQ8A lacks in vitro protein kinase activity and instead shows ATPase activity that is essential for CoQ biosynthesis [98]. The ATPase activity is strongly activated by cardiolipin and small molecule mimics of CoQ intermediates [99]. Thus, the ATPase function of yeast Coq8 and human COQ8A is proposed to function in a chaperone-like activity to facilitate the assembly of the CoQ synthome and de novo [65] biosynthesis of CoQ [99].
Expression of human COQ8B (ADCK4) also rescues CoQ6 biosynthesis in yeast coq8 mutants [100]. Although the amino terminus of COQ8B has a typical mitochondrial matrix targetting sequence, rescue of coq8 mutant yeast by COQ8B depends on the addition of a yeast mitochondrial targetting sequence [100]. The effect of a COQ8B polymorphism present in 50% of the European population (COQ8B-H174R) was tested in the yeast expression model. Yeast coq8 mutants expressing the human COQ8B-His174 polypeptide had decreased steady state levels of the COQ8B polypeptide, decreased growth on medium containing a non-fermentable carbon source, and decreased CII + CIII activity as compared with mutants expressing the COQ8B-Arg174 polypeptide [100]. Thus, it is possible that this common COQ8B polymorphism may represent a risk factor for secondary CoQ10 deficiencies. Various human diseases are directly associated with mutations in the COQ8A and COQ8B genes. Most prevalent are recessive ataxia and childhood-onset cerebellar ataxia associated with mutated COQ8A [101,102], and a steroid-resistant nephrotic disease related to mutated COQ8B [75,100].
Yeast COQ9; human COQ9
In S. cerevisiae, Coq9 is required for CoQ biosynthesis, is a member of the CoQ synthome, and is peripherally associated with the inner mitochondrial membrane, on the matrix side [34,103]. A temperature-sensitive coq9 mutant (coq9-ts19) shifted to the non-permissive temperature results in the disassembling of the CoQ synthome, demonstrating that Coq9 is essential for the formation and stabilization of the high-molecular mass complex [23]. Coq9 is required for the deamination of Carbon 4 on CoQ-intermediates when pABA is utilized as the ring precursor in yeast [23]. The removal of the ring nitrogen substituent depends on the function of Coq6, and yeast with coq9 mutations accumulate 3-hexaprenyl-4-aminophenol (4-AP), an intermediate that has also been shown to build up in a coq6 null mutant overexpressing COQ8 [24]. It is therefore likely that both Coq6 and Coq9 are needed for the 5-hydroxylation and 4-deamination steps of CoQ-intermediates. Additionally, an accumulation of late-stage intermediates suggests Coq7 is not active in the absence of Coq9 [6,24]. In summary, the yeast Coq9 polypeptide is required for both Coq6 and Coq7 hydroxylation steps, via an indirect or supportive role.
Attempts to rescue yeast coq9 null mutants by expression of human COQ9 have so far failed [104–106]. However, expression of human COQ9 rescued the yeast coq9-ts19 mutant [106]. Under these conditions, a small amount of the human COQ9 polypeptide enhanced the synthesis of CoQ6 from 4HB (but not from pABA) and co-purified with the yeast Coq6-CNAP tagged polypeptide, indicating that human COQ9 is able to interact with the yeast CoQ synthome.
The presence of both human COQ9 and human COQ7 are needed for the hydroxylation step catalyzed by COQ7, and the two polypeptides interact [13]. Human cells with deficiencies in COQ9 accumulate DMQ10, the same intermediate that accumulates in COQ7 deficient cells [107]. Both COQ7 and COQ9 deficient cell lines respond to treatment with 2,4-diHB, another example of bypass therapy [108,109]. In fact, human fibroblasts with mutations in COQ9 show decreased steady state levels of the COQ7 polypeptide [109]. Interestingly, treatment with vanillic acid also restored function in the COQ9 deficient cells, but not the COQ7 deficient cells [108]. Perhaps human COQ9 and COQ6 function may also be linked, similar to the situation in yeast. In humans, COQ9 mutations result in various disease states, including predominant encephalomyopathy and an autosomal-recessive neonatal-onset CoQ deficiency [104,107].
Yeast and human genes required for efficient CoQ biosynthesis
Yeast COQ10; human COQ10A, COQ10B
Unlike the completely CoQ-less coq1–coq9 null mutants, the yeast coq10 null mutant produces near wild-type levels of CoQ at stationary phase, but synthesizes CoQ less efficiently during log phase growth [110,111]. While CoQ is eventually produced at near normal levels, the coq10 null mutant still has severe defects in respiratory electron transport and is sensitive to treatment with polyunsaturated fatty acids, phenotypes that are hallmarks of the coq1–coq9 null mutants [110]. Thus, even though CoQ6 content is similar to that of wild-type yeast, Coq10 is required for efficient function and biosynthesis of CoQ6. The Coq10 polypeptide contains a steroidogenic acute regulatory (StAR)-related lipid transfer (StART) domain, and binds CoQ and late-stage CoQ-intermediates both in vitro and in vivo, suggesting Coq10 may function as a lipid chaperone involving delivery of CoQ from site of synthesis to sites of function [110–114].
Humans have two homologs of yeast COQ10, namely COQ10A and COQ10B. Each isoform has several transcript variants as a result of alternative transcription initiation and/or alternative splicing [6]. The function of the COQ10 polypeptide is widely conserved across different organisms; expression of homologs from either Caulobacter crescentus or human COQ10A rescue the impaired growth of yeast coq10Δ mutant yeast [110]. Currently, there is no known disease phenotype associated with mutations in human COQ10A or COQ10B. However, the postulated lipid chaperone function of COQ10A and COQ10B makes these polypeptides intriguing targets for study of the movement of CoQ between mitochondrial membranes and the respiratory complexes.
Yeast COQ11; human NDUFA9
COQ11 (YLR290C) was recently identified to be required for efficient de novo CoQ biosynthesis in S. cerevisiae. Affinity purification of CNAP-tagged Coq11 showed Coq11-CNAP co-purified with Coq4, Coq5, and Coq7 – members of the high molecular mass CoQ synthome [12]. A separate high throughput study also identified Coq11 as a mitochondrial protein, confirming its localization to the portion of the cell where CoQ is synthesized [115]. Due to its novelty, the functional roles, organization, and stoichiometry of Coq11 within the CoQ-synthome are not yet fully understood. However, numerous features of Coq11 and its homologs solidify its link to CoQ biosynthesis. In five fungal genomes, the existence of Coq11-Coq10 fusion proteins suggests these proteins may have a functional relationship [12]. High throughput studies found COQ11 to have a genetic correlation with both COQ2 and COQ10, which further supports this hypothesis [115]. Sequence analyses establish Coq11 as a member of the atypical short-chain dehydrogenase/reductase superfamily of oxidoreductases (SDR). SDR superfamily proteins contain a conserved Rossmann fold, a protein structural motif used to bind nucleotide co-factors such as FAD, FMN, and NAD(P) [116]. This bound co-factor is then used to assist the protein in its catalysis of different chemical reactions including isomerization, decarboxylation, epimerization, imine reduction, and carbonyl-alcohol oxidoreduction [116,117]. It is therefore tempting to speculate that Coq11 may use its Rossmann fold to perform enzymatic reactions within the CoQ biosynthetic pathway.
Interestingly, a protein similarity network analysis reveals that the taxonomy of YLR290C-like proteins includes the SDR subfamily protein NDUFA9, an auxiliary subunit of Complex I in humans important for complex stability [12,118–120]. Patients with decreased levels of NDUFA9 are unable to assemble Complex I properly and may develop a degenerative infancy respiratory disorder known as Leigh syndrome, which is often fatal in the first years of life [121,122]. Differences in NDUFA9 deficiency produce phenotypic variations in patients [46]. It will be challenging to evaluate whether NDUFA9 deficiencies impact CoQ biosynthesis directly, because the deficiencies resulting from Complex I defects (and other mitochondrial defects) may secondarily influence CoQ biosynthesis [123,124]. Further exploration of Coq11 homology with NDUFA9 will help define their functional relationship.
Summary
The COQ1–COQ11 genes identified in the S. cerevisiae yeast model are required for efficient biosynthesis of CoQ6.
Expression of human homologs of yeast COQ1–COQ10 genes restore CoQ biosynthesis in the corresponding yeast coq mutants, indicating profound functional conservation.
Yeast provides a simple yet powerful model to investigate and define the function and possible pathology of human COQ gene polymorphisms and mutations.
Simple natural products may serve as alternate ring precursors in CoQ biosynthesis, and these compounds may act to enhance biosynthesis of CoQ or may bypass select deficient steps.
Biosynthesis of CoQ in yeast and human cells depends on high molecular mass multi-subunit complexes consisting of several of the COQ gene products, as well as CoQ itself and CoQ-intermediates.
Thus the CoQ synthome in yeast, or Complex Q in human cells is essential for de novo biosynthesis of CoQ.
Acknowledgments
We thank Dr A. Tzagoloff (Columbia University) for the original yeast coq mutant strains. We also thank the UCLA Molecular Instrumentation Core for the use of the QTRAP 4000.
Abbreviations
- AdoMet
S-adenosylmethionine
- CNAP
consecutive non-denaturing affinity purification
- CoQ
ubiquinone or coenzyme Q
- CoQH2
ubiquinol or CoQ hydroquinone
- CoQn
ubiquinone-n or coenzyme Qn (refers to a specific isoform, where n is number of isoprenyl units in the tail of CoQn, e.g. CoQ10 in humans, CoQ6 in Saccharomyces cerevisiae)
- Coq
Saccharomyces cerevisiae polypeptide involved in CoQ6 biosynthesis
- COQ
human polypeptide involved in CoQ10 biosynthesis
- COQ
yeast or human gene involved in CoQ biosynthesis
- pABA
para-aminobenzoic acid
- SDR
short-chain dehydrogenase/reductase superfamily of oxidoreductases
- SRNS
steroid resistant nephrotic syndrome
- 2,4-diHB
2,4-dihydroxybenzoic acid
- 3,4-diHB
3,4-dihydroxybenzoic acid
- 4HB
4-hydroxybenzoic acid
- 4HBz
4-hydroxybenzaldehyde
Competing interests
The authors declare that there are no competing interests associated with this manuscript.
Funding
This work was supported in part by the National Science Foundation [grant number MCB-1330803 (to C.F.C.)]; and the National Institutes of Health [grant number T32 GM 008496 (to M.C.B. and H.T.)].
Author contribution
A.M.A., M.C.B., L.F.-d.-R., A.N., H.T., and C.F.C. contributed to drafting this review, revising it for intellectual content and approved the final version. A.M.A., A.N., L.F.-d.R., and C.F.C. contributed to preparation of the figures.
References
- 1.Turunen M., Olsson J. and Dallner G. (2004) Metabolism and function of coenzyme Q. Biochim. Biophys. Act. 1660, 171–199 10.1016/j.bbamem.2003.11.012 [DOI] [PubMed] [Google Scholar]
- 2.Crane F.L. (2001) Biochemical functions of coenzyme Q10. J. Am. Coll. Nutr. 20, 591–598 10.1080/07315724.2001.10719063 [DOI] [PubMed] [Google Scholar]
- 3.Bentinger M., Brismar K. and Dallner G. (2007) The antioxidant role of coenzyme Q. Mitochondrion 7S, S41–S50 10.1016/j.mito.2007.02.006 [DOI] [PubMed] [Google Scholar]
- 4.Aussel L., Pierrel F., Loiseau L., Lombard M., Fontecave M. and Barras F. (2014) Biosynthesis and physiology of coenzyme Q in bacteria. Biochim. Biophys. Acta 1837, 1004–1011 10.1016/j.bbabio.2014.01.015 [DOI] [PubMed] [Google Scholar]
- 5.Hayashi K., Ogiyama Y., Yokomi K., Nakagawa T., Kaino T. and Kawamukai M. (2014) Functional conservation of coenzyme Q biosynthetic genes among yeasts, plants, and humans. PLoS ONE 9, 14 10.1371/journal.pone.0099038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang Y. and Hekimi S. (2013) Molecular genetics of ubiquinone biosynthesis in animals. Crit. Rev. Biochem. Mol. Biol. 48, 69–88 10.3109/10409238.2012.741564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stefely J.A. and Pagliarini D.J. (2017) Biochemistry of mitochondrial coenzyme Q biosynthesis. Trends Biochem. Sci. 42, 824–843 10.1016/j.tibs.2017.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alcázar-Fabra M., Trevisson E., and Brea-Calvo G. (2018) Clinical syndromes associated with Coenzyme Q10 deficiency. Essays Biochem., 62, 377–398 10.1042/EBC20170107 [DOI] [PubMed] [Google Scholar]
- 9.Tzagoloff A., Akai A. and Needleman R.B. (1975) Assembly of the mitochondrial membrane system. Characterization of nuclear mutants of Saccharomyces cerevisiae with defects in mitochondrial ATPase and respiratory enzymes. J. Biol. Chem. 250, 8228–8235 [PubMed] [Google Scholar]
- 10.Tzagoloff A. and Dieckmann C.L. (1990) PET genes of Saccharomyces cerevisiae. Microbiol. Rev. 54, 211–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tran U.C. and Clarke C.F. (2007) Endogenous synthesis of coenzyme Q in eukaryotes. Mitochondrion 7S, S62–S71 10.1016/j.mito.2007.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Allan C.M., Awad A.M., Johnson J.S., Shirasaki D.I., Wang C., Blaby-Haas C.E. et al. (2015) Identification of Coq11, a new coenzyme Q biosynthetic protein in the CoQ-Synthome in Saccharomyces cerevisiae. J. Biol. Chem. 290, 7517–7534 10.1074/jbc.M114.633131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lohman D.C., Forouhar F., Beebe E.T., Stefely M.S., Minogue C.E., Ulbrich A. et al. (2014) Mitochondrial COQ9 is a lipid-binding protein that associates with COQ7 to enable coenzyme Q biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 111, E4697–E4705 10.1073/pnas.1413128111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clarke C.F. (2000) New advances in coenzyme Q biosynthesis. Protoplasma 213, 134–147 10.1007/BF01282151 [DOI] [Google Scholar]
- 15.Kawamukai M. (2016) Biosynthesis of coenzyme Q in eukaryotes. Biosci. Biotechnol. Biochem. 80, 23–33 10.1080/09168451.2015.1065172 [DOI] [PubMed] [Google Scholar]
- 16.Pierrel F. (2017) Impact of chemical analogs of 4-hydroxybenzoic acid on coenzyme Q biosynthesis: from inhibition to bypass of coenzyme Q deficiency. Front. Physiol. 8, 436 10.3389/fphys.2017.00436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Payet L.A., Leroux M., Willison J.C., Kihara A., Pelosi L. and Pierrel F. (2016) Mechanistic details of early steps in coenzyme Q biosynthesis pathway in yeast. Cell Chem. Biol. 23, 1241–1250 10.1016/j.chembiol.2016.08.008 [DOI] [PubMed] [Google Scholar]
- 18.Stefely J.A., Kwiecien N.W., Freiberger E.C., Richards A.L., Jochem A., Rush M. J.P. et al. (2016) Mitochondrial protein functions elucidated by multi-omic mass spectrometry profiling. Nat. Biotechnol. 34, 1191–1197 10.1038/nbt.3683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zahedi R.P., Sickmann A., Boehm A.M., Winkler C., Zufall N., Schonfisch B. et al. (2006) Proteomic analysis of the yeast mitochondrial outer membrane reveals accumulation of a subclass of preproteins. Mol. Biol. Cell 17, 1436–1450 10.1091/mbc.e05-08-0740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marbois B., Xie L.X., Choi S., Hirano K., Hyman K. and Clarke C.F. (2010) para-Aminobenzoic acid is a precursor in coenzyme Q6 biosynthesis in Saccharomyces cerevisiae. J. Biol. Chem. 285, 27827–27838 10.1074/jbc.M110.151894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pierrel F., Hamelin O., Douki T., Kieffer-Jaquinod S., Muhlenhoff U., Ozeir M. et al. (2010) Involvement of mitochondrial ferredoxin and para-aminobenzoic acid in yeast coenzyme Q biosynthesis. Chem. Biol. 17, 449–459 10.1016/j.chembiol.2010.03.014 [DOI] [PubMed] [Google Scholar]
- 22.Ozeir M., Pelosi L., Ismail A., Mellot-Draznieks C., Fontecave M. and Pierrel F. (2015) Coq6 is responsible for the C4-deamination reaction in coenzyme Q biosynthesis in Saccharomyces cerevisiae. J. Biol. Chem. 290, 24140–24151 10.1074/jbc.M115.675744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.He C.H., Black D.S., Nguyen T.P., Wang C., Srinivasan C. and Clarke C.F. (2015) Yeast Coq9 controls deamination of coenzyme Q intermediates that derive from para-aminobenzoic acid. Biochim. Biophys. Acta 1851, 1227–1239 10.1016/j.bbalip.2015.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xie L.X., Ozeir M., Tang J.Y., Chen J.Y., Jaquinod S.K., Fontecave M. et al. (2012) Overexpression of the Coq8 kinase in Saccharomyces cerevisiae coq null mutants allows for accumulation of diagnostic intermediates of the coenzyme Q6 biosynthetic pathway. J. Biol. Chem. 287, 23571–23581 10.1074/jbc.M112.360354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Doimo M., Trevisson E., Airik R., Bergdoll M., Santos-Ocana C., Hildebrandt F. et al. (2014) Effect of vanillic acid on COQ6 mutants identified in patients with coenzyme Q10 deficiency. Biochim. Biophys. Acta 1842, 1–6 10.1016/j.bbadis.2013.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xie L.X., Williams K.J., He C.H., Weng E., Khong S., Rose T.E. et al. (2015) Resveratrol and para-coumarate serve as ring precursors for coenzyme Q biosynthesis. J. Lipid Res. 56, 909–919 10.1194/jlr.M057919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fernandez-Del-Rio L., Nag A., Gutierrez Casado E., Ariza J., Awad A.M., Joseph A.I. et al. (2017) Kaempferol increases levels of coenzyme Q in kidney cells and serves as a biosynthetic ring precursor. Free Radic. Biol. Med. 110, 176–187 10.1016/j.freeradbiomed.2017.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alam S.S., Nambudiri A.M. and Rudney H. (1975) J-Hydroxybenzoate: polyprenyl transferase and the prenylation of 4-aminobenzoate in mammalian tissues. Arch. Biochem. Biophys. 171, 183–190 10.1016/0003-9861(75)90022-3 [DOI] [PubMed] [Google Scholar]
- 29.Gonzalez-Aragon D., Buron M.I., Lopez-Lluch G., Herman M.D., Gomez-Diaz C., Navas P. et al. (2005) Coenzyme Q and the regulation of intracellular steady-state levels of superoxide in HL-60 cells. Biofactors 25, 31–41 10.1002/biof.5520250105 [DOI] [PubMed] [Google Scholar]
- 30.Ashby M.N. and Edwards P.A. (1990) Elucidation of the deficiency in two yeast coenzyme Q mutants. Characterization of the structural gene encoding hexaprenyl pyrophosphate synthetase. J. Biol. Chem. 265, 13157–13164 [PubMed] [Google Scholar]
- 31.Gin P. and Clarke C.F. (2005) Genetic evidence for a multi-subunit complex in coenzyme Q biosynthesis in yeast and the role of the Coq1 hexaprenyl diphosphate synthase. J. Biol. Chem. 280, 2676–2681 10.1074/jbc.M411527200 [DOI] [PubMed] [Google Scholar]
- 32.Okada K., Suzuki K., Kamiya Y., Zhu X., Fujisaki S., Nishimura Y. et al. (1996) Polyprenyl diphosphate synthase essentially defines the length of the side chain of ubiquinone. Biochim. Biophys. Acta 1302, 217–223 10.1016/0005-2760(96)00064-1 [DOI] [PubMed] [Google Scholar]
- 33.Okada K., Kainou T., Matsuda H. and Kawamukai M. (1998) Biological significance of the side chain length of ubiquinone in Saccharomyces cerevisiae. FEBS Lett. 431, 241–244 10.1016/S0014-5793(98)00753-4 [DOI] [PubMed] [Google Scholar]
- 34.He C.H., Xie L.X., Allan C.M., Tran U.C. and Clarke C.F. (2014) Coenzyme Q supplementation or over-expression of the yeast Coq8 putative kinase stabilizes multi-subunit Coq polypeptide complexes in yeast coq null mutants. Biochim. Biophys. Acta 1841, 630–644 10.1016/j.bbalip.2013.12.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saiki R., Nagata A., Kainou T., Matsuda H. and Kawamukai M. (2005) Characterization of solanesyl and decaprenyl diphosphate synthases in mice and humans. FEBS J. 272, 5606–5622 10.1111/j.1742-4658.2005.04956.x [DOI] [PubMed] [Google Scholar]
- 36.Mollet J., Giurgea I., Schlemmer D., Dallner G., Chretien D., Delahodde A. et al. (2007) Prenyldiphosphate synthase, subunit 1 (PDSS1) and OH-benzoate polyprenyltransferase (COQ2) mutations in ubiquinone deficiency and oxidative phosphorylation disorders. J. Clin. Invest. 117, 765–772 10.1172/JCI29089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lopez L.C., Schuelke M., Quinzii C.M., Kanki T., Rodenburg R.J., Naini A. et al. (2006) Leigh syndrome with nephropathy and CoQ10 deficiency due to decaprenyl diphosphate synthase subunit 2 (PDSS2) mutations. Am. J. Hum. Genet. 79, 1125–1129 10.1086/510023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ashby M.N., Kutsunai S.Y., Ackerman S., Tzagoloff A. and Edwards P.A. (1992) COQ2 is a candidate for the structural gene encoding para-hydroxybenzoate:polyprenyltransferase. J. Biol. Chem. 267, 4128–4136 [PubMed] [Google Scholar]
- 39.Hsu A.Y., Do T.Q., Lee P.T. and Clarke C.F. (2000) Genetic evidence for a multi-subunit complex in the O-methyltransferase steps of coenzyme Q biosynthesis. Biochim. Biophys. Acta 1484, 287–297 10.1016/S1388-1981(00)00019-6 [DOI] [PubMed] [Google Scholar]
- 40.Johnson A., Gin P., Marbois B.N., Hsieh E.J., Wu M., Barros M.H. et al. (2005) COQ9, a new gene required for the biosynthesis of coenzyme Q in Saccharomyces cerevisiae. J. Biol. Chem. 280, 31397–31404 10.1074/jbc.M503277200 [DOI] [PubMed] [Google Scholar]
- 41.Leuenberger D., Bally N.A., Schatz G. and Koehler C.M. (1999) Different import pathways through the mitochondrial intermembrane space for inner membrane proteins. EMBO J. 18, 4816–4822 10.1093/emboj/18.17.4816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Forsgren M., Attersand A., Lake S., Grunler J., Swiezewska E., Dallner G. et al. (2004) Isolation and functional expression of human COQ2, a gene encoding a polyprenyl transferase involved in the synthesis of CoQ. Biochem. J. 382, 519–526 10.1042/BJ20040261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Desbats M.A., Morbidoni V., Silic-Benussi M., Doimo M., Ciminale V., Cassina M. et al. (2016) The COQ2 genotype predicts the severity of coenzyme Q10 deficiency. Hum. Mol. Genet. 25, 4256–4265 10.1093/hmg/ddw257 [DOI] [PubMed] [Google Scholar]
- 44.Li W. (2016) Bringing bioactive compounds into membranes: the UbiA superfamily of intramembrane aromatic prenyltransferases. Trends Biochem. Sci. 41, 356–370 10.1016/j.tibs.2016.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Herebian D., Seibt A., Smits S. H.J., Rodenburg R.J., Mayatepek E. and Distelmaier F. (2017) 4-Hydroxybenzoic acid restores CoQ10 biosynthesis in human COQ2 deficiency. Ann. Clin. Transl. Neurol. 4, 902–908 10.1002/acn3.486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Baertling F., Sanchez-Caballero L., van den Brand M. A.M., Fung C.W., Chan S.H., Wong V.C. et al. (2018) NDUFA9 point mutations cause a variable mitochondrial complex I assembly defect. Clin. Genet. 93, 111–118 10.1111/cge.13089 [DOI] [PubMed] [Google Scholar]
- 47.Clarke C.F., Williams W. and Teruya J.H. (1991) Ubiquinone biosynthesis in Saccharomyces cerevisiae. Isolation and sequence of COQ3, the 3,4-dihydroxy-5-hexaprenylbenzoate methyltransferase gene. J. Biol. Chem. 266, 16636–16644 [PubMed] [Google Scholar]
- 48.Hsu A.Y., Poon W.W., Shepherd J.A., Myles D.C. and Clarke C.F. (1996) Complementation of coq3 mutant yeast by mitochondrial targeting of the Escherichia coli UbiG polypeptide: evidence that UbiG catalyzes both O-methylation steps in ubiquinone biosynthesis. Biochemistry 35, 9797–9806 10.1021/bi9602932 [DOI] [PubMed] [Google Scholar]
- 49.Poon W.W., Barkovich R.J., Hsu A.Y., Frankel A., Lee P.T., Shepherd J.N. et al. (1999) Yeast and rat Coq3 and Escherichia coli UbiG polypeptides catalyze both O-methyltransferase steps in coenzyme Q biosynthesis. J. Biol. Chem. 274, 21665–21672 10.1074/jbc.274.31.21665 [DOI] [PubMed] [Google Scholar]
- 50.Zhu Y., Wu B., Zhang X., Fan X., Niu L., Li X. et al. (2015) Structural and biochemical studies reveal UbiG/Coq3 as a class of novel membrane-binding proteins. Biochem. J. 470, 105–114 10.1042/BJ20150329 [DOI] [PubMed] [Google Scholar]
- 51.Marbois B., Gin P., Faull K.F., Poon W.W., Lee P.T., Strahan J. et al. (2005) Coq3 and Coq4 define a polypeptide complex in yeast mitochondria for the biosynthesis of coenzyme Q. J. Biol. Chem. 280, 20231–20238 10.1074/jbc.M501315200 [DOI] [PubMed] [Google Scholar]
- 52.Tauche A., Krause-Buchholz U. and Rodel G. (2008) Ubiquinone biosynthesis in Saccharomyces cerevisiae: the molecular organization of O-methylase Coq3p depends on Abc1p/Coq8p. FEMS Yeast Res. 8, 1263–1275 10.1111/j.1567-1364.2008.00436.x [DOI] [PubMed] [Google Scholar]
- 53.Xie L.X., Hsieh E.J., Watanabe S., Allan C.M., Chen J.Y., Tran U.C. et al. (2011) Expression of the human atypical kinase ADCK3 rescues coenzyme Q biosynthesis and phosphorylation of Coq polypeptides in yeast coq8 mutants. Biochim. Biophys. Acta 1811, 348–360 10.1016/j.bbalip.2011.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jonassen T. and Clarke C.F. (2000) Isolation and functional expression of human COQ3, a gene encoding a methyltransferase required for ubiquinone biosynthesis. J. Biol. Chem. 275, 12381–12387 10.1074/jbc.275.17.12381 [DOI] [PubMed] [Google Scholar]
- 55.Belogrudov G.I., Lee P.T., Jonassen T., Hsu A.Y., Gin P. and Clarke C.F. (2001) Yeast COQ4 encodes a mitochondrial protein required for coenzyme Q synthesis. Arch. Biochem. Biophys. 392, 48–58 10.1006/abbi.2001.2448 [DOI] [PubMed] [Google Scholar]
- 56.Marbois B., Gin P., Gulmezian M. and Clarke C.F. (2009) The yeast Coq4 polypeptide organizes a mitochondrial protein complex essential for coenzyme Q biosynthesis. Biochim. Biophys. Acta 1791, 69–75 10.1016/j.bbalip.2008.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rea S.L., Graham B.H., Nakamaru-Ogiso E., Kar A. and Falk M.J. (2010) Bacteria, yeast, worms, and flies: exploiting simple model organisms to investigate human mitochondrial diseases. Dev. Disabil. Res. Rev. 16, 200–218 10.1002/ddrr.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Casarin A., Jimenez-Ortega J.C., Trevisson E., Pertegato V., Doimo M., Ferrero-Gomez M.L. et al. (2008) Functional characterization of human COQ4, a gene required for Coenzyme Q10 biosynthesis. Biochem. Biophys. Res. Commun. 372, 35–39 10.1016/j.bbrc.2008.04.172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Brea-Calvo G., Haack T.B., Karall D., Ohtake A., Invernizzi F., Carrozzo R. et al. (2015) COQ4 mutations cause a broad spectrum of mitochondrial disorders associated with CoQ10 deficiency. Am. J. Hum. Genet. 96, 309–317 10.1016/j.ajhg.2014.12.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Salviati L., Trevisson E., Rodriguez Hernandez M.A., Casarin A., Pertegato V., Doimo M. et al. (2012) Haploinsufficiency of COQ4 causes coenzyme Q10 deficiency. J. Med. Genet. 49, 187–191 10.1136/jmedgenet-2011-100394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Romero-Moya D., Santos-Ocana C., Castano J., Garrabou G., Rodriguez-Gomez J.A., Ruiz-Bonilla V. et al. (2017) Genetic rescue of mitochondrial and skeletal muscle impairment in an induced pluripotent stem cells model of coenzyme Q10 deficiency. Stem Cells 35, 1687–1703 10.1002/stem.2634 [DOI] [PubMed] [Google Scholar]
- 62.Barkovich R.J., Shtanko A., Shepherd J.A., Lee P.T., Myles D.C., Tzagoloff A. et al. (1997) Characterization of the COQ5 gene from Saccharomyces cerevisiae. Evidence for a C-methyltransferase in ubiquinone biosynthesis. J. Biol. Chem. 272, 9182–9188 10.1074/jbc.272.14.9182 [DOI] [PubMed] [Google Scholar]
- 63.Dibrov E., Robinson K.M. and Lemire B.D. (1997) The COQ5 gene encodes a yeast mitochondrial protein necessary for ubiquinone biosynthesis and the assembly of the respiratory chain. J. Biol. Chem. 272, 9175–9181 10.1074/jbc.272.14.9175 [DOI] [PubMed] [Google Scholar]
- 64.Baba S.W., Belogrudov G.I., Lee J.C., Lee P.T., Strahan J., Shepherd J.N. et al. (2004) Yeast Coq5 C-methyltransferase is required for stability of other polypeptides involved in coenzyme Q biosynthesis. J. Biol. Chem. 279, 10052–10059 10.1074/jbc.M313712200 [DOI] [PubMed] [Google Scholar]
- 65.Dai Y.N., Zhou K., Cao D.D., Jiang Y.L., Meng F., Chi C.B. et al. (2014) Crystal structures and catalytic mechanism of the C-methyltransferase Coq5 provide insights into a key step of the yeast coenzyme Q synthesis pathway. Acta Crystallogr. Sec. D Biol. Crystallogr. 70, 2085–2092 10.1107/S1399004714011559 [DOI] [PubMed] [Google Scholar]
- 66.Lapointe C.P., Stefely J.A., Jochem A., Hutchins P.D., Wilson G.M., Kwiecien N.W. et al. (2018) Multi-omics reveal specific targets of the RNA-binding protein Puf3p and its orchestration of mitochondrial biogenesis. Cell Syst. 6, 125–135.e126, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Veling M.T., Reidenbach A.G., Freiberger E.C., Kwiecien N.W., Hutchins P.D., Drahnak M.J. et al. (2017) Multi-omic mitoprotease profiling defines a role for Oct1p in coenzyme Q production. Mol. Cell 68, 970–977.e911, 10.1016/j.molcel.2017.11.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nguyen T.P., Casarin A., Desbats M.A., Doimo M., Trevisson E., Santos-Ocana C. et al. (2014) Molecular characterization of the human COQ5 C-methyltransferase in Coenzyme Q biosynthesis. Biochim. Biophys. Acta, 10.1016/j.bbalip.2014.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Malicdan M. C.V., Vilboux T., Ben-Zeev B., Guo J., Eliyahu A., Pode-Shakked B. et al. (2018) A novel inborn error of the coenzyme Q10 biosynthesis pathway: cerebellar ataxia and static encephalomyopathy due to COQ5 C-methyltransferase deficiency. Hum. Mutat. 39, 69–79 10.1002/humu.23345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yen H.C., Liu Y.C., Kan C.C., Wei H.J., Lee S.H., Wei Y.H. et al. (2016) Disruption of the human COQ5-containing protein complex is associated with diminished coenzyme Q10 levels under two different conditions of mitochondrial energy deficiency. Biochim. Biophys. Acta 1860, 1864–1876 10.1016/j.bbagen.2016.05.005 [DOI] [PubMed] [Google Scholar]
- 71.Gin P., Hsu A.Y., Rothman S.C., Jonassen T., Lee P.T., Tzagoloff A. et al. (2003) The Saccharomyces cerevisiae COQ6 gene encodes a mitochondrial flavin-dependent monooxygenase required for coenzyme Q biosynthesis. J. Biol. Chem. 278, 25308–25316 10.1074/jbc.M303234200 [DOI] [PubMed] [Google Scholar]
- 72.Ismail A., Leroux V., Smadja M., Gonzalez L., Lombard M., Pierrel F. et al. (2016) Coenzyme Q biosynthesis: evidence for a substrate access channel in the FAD-dependent monooxygenase Coq6. PLoS Comput. Biol. 12, e1004690 10.1371/journal.pcbi.1004690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ozeir M., Muhlenhoff U., Webert H., Lill R., Fontecave M. and Pierrel F. (2011) Coenzyme Q biosynthesis: Coq6 is required for the C5-hydroxylation reaction and substrate analogs rescue Coq6 deficiency. Chem. Biol. 18, 1134–1142 10.1016/j.chembiol.2011.07.008 [DOI] [PubMed] [Google Scholar]
- 74.Heeringa S.F., Chernin G., Chaki M., Zhou W., Sloan A.J., Ji Z. et al. (2011) COQ6 mutations in human patients produce nephrotic syndrome with sensorineural deafness. J. Clin. Invest. 121, 2013–2024 10.1172/JCI45693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ashraf S., Gee H.Y., Woerner S., Xie L.X., Vega-Warner V., Lovric S. et al. (2013) ADCK4 mutations promote steroid-resistant nephrotic syndrome through CoQ10 biosynthesis disruption. J. Clin. Invest. 123, 5179–5189 10.1172/JCI69000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Park E., Ahn Y.H., Kang H.G., Yoo K.H., Won N.H., Lee K.B. et al. (2017) COQ6 mutations in children with steroid-resistant focal segmental glomerulosclerosis and sensorineural hearing loss. Am. J. Kidney Dis. 70, 139–144 10.1053/j.ajkd.2016.10.040 [DOI] [PubMed] [Google Scholar]
- 77.Gigante M., Diella S., Santangelo L., Trevisson E., Acosta M.J., Amatruda M. et al. (2017) Further phenotypic heterogeneity of CoQ10 deficiency associated with steroid resistant nephrotic syndrome and novel COQ2 and COQ6 variants. Clin. Genet. 92, 224–226 10.1111/cge.12960 [DOI] [PubMed] [Google Scholar]
- 78.Sheftel A., Stehling O. and Lill R. (2010) Iron-sulfur proteins in health and disease. Trends Endocrinol. Metab. 21, 302–314 10.1016/j.tem.2009.12.006 [DOI] [PubMed] [Google Scholar]
- 79.Sheftel A.D., Stehling O., Pierik A.J., Elsasser H.P., Muhlenhoff U., Webert H. et al. (2010) Humans possess two mitochondrial ferredoxins, Fdx1 and Fdx2, with distinct roles in steroidogenesis, heme, and Fe/S cluster biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 107, 11775–11780 10.1073/pnas.1004250107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Marbois B.N. and Clarke C.F. (1996) The COQ7 gene encodes a protein in Saccharomyces cerevisiae necessary for ubiquinone biosynthesis. J. Biol. Chem. 271, 2995–3004 10.1074/jbc.271.6.2995 [DOI] [PubMed] [Google Scholar]
- 81.Jonassen T., Proft M., Randez-Gil F., Schultz J.R., Marbois B.N., Entian K.D. et al. (1998) Yeast Clk-1 homologue (Coq7/Cat5) is a mitochondrial protein in coenzyme Q synthesis. J. Biol. Chem. 273, 3351–3357 10.1074/jbc.273.6.3351 [DOI] [PubMed] [Google Scholar]
- 82.Stenmark P., Grunler J., Mattsson J., Sindelar P.J., Nordlund P. and Berthold D.A. (2001) A new member of the family of di-iron carboxylate proteins. Coq7 (clk-1), a membrane-bound hydroxylase involved in ubiquinone biosynthesis. J. Biol. Chem. 276, 33297–33300 10.1074/jbc.C100346200 [DOI] [PubMed] [Google Scholar]
- 83.Vajo Z., King L.M., Jonassen T., Wilkin D.J., Ho N., Munnich A. et al. (1999) Conservation of the Caenorhabditis elegans timing gene clk-1 from yeast to human: a gene required for ubiquinone biosynthesis with potential implications for aging. Mamm. Genome 10, 1000–1004 10.1007/s003359901147 [DOI] [PubMed] [Google Scholar]
- 84.Lu T.T., Lee S.J., Apfel U.P. and Lippard S.J. (2013) Aging-associated enzyme human clock-1: substrate-mediated reduction of the diiron center for 5-demethoxyubiquinone hydroxylation. Biochemistry 52, 2236–2244 10.1021/bi301674p [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Padilla S., Tran U.C., Jimenez-Hidalgo M., Lopez-Martin J.M., Martin-Montalvo A., Clarke C.F. et al. (2009) Hydroxylation of demethoxy-Q6 constitutes a control point in yeast coenzyme Q6 biosynthesis. Cell. Mol. Life Sci. 66, 173–186 10.1007/s00018-008-8547-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tran U.C., Marbois B., Gin P., Gulmezian M., Jonassen T. and Clarke C.F. (2006) Complementation of Saccharomyces cerevisiae coq7 mutants by mitochondrial targeting of the Escherichia coli UbiF polypeptide. Two functions of yeast Coq7 polypeptide in coenzyme Q biosynthesis. J. Biol. Chem. 281, 16401–16409 10.1074/jbc.M513267200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Freyer C., Stranneheim H., Naess K., Mourier A., Felser A., Maffezzini C. et al. (2015) Rescue of primary ubiquinone deficiency due to a novel COQ7 defect using 2,4-dihydroxybensoic acid. J. Med. Genet. 52, 779–783 10.1136/jmedgenet-2015-102986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wang Y., Smith C., Parboosingh J.S., Khan A., Innes M. and Hekimi S. (2017) Pathogenicity of two COQ7 mutations and responses to 2,4-dihydroxybenzoate bypass treatment. J. Cell. Mol. Med. 21, 2329–2343 10.1111/jcmm.13154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Brea-Calvo G., Siendones E., Sanchez-Alcazar J.A., de Cabo R. and Navas P. (2009) Cell survival from chemotherapy depends on NF-kappaB transcriptional up-regulation of coenzyme Q biosynthesis. PLoS ONE 4, e5301 10.1371/journal.pone.0005301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Cascajo M.V., Abdelmohsen K., Noh J.H., Fernandez-Ayala D.J., Willers I.M., Brea G. et al. (2016) RNA-binding proteins regulate cell respiration and coenzyme Q biosynthesis by post-transcriptional regulation of COQ7. RNA Biol. 13, 622–634 10.1080/15476286.2015.1119366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Martin-Montalvo A., Gonzalez-Mariscal I., Padilla S., Ballesteros M., Brautigan D.L., Navas P. et al. (2011) Respiratory-induced coenzyme Q biosynthesis is regulated by a phosphorylation cycle of Cat5p/Coq7p. Biochem. J. 440, 107–114 10.1042/BJ20101422 [DOI] [PubMed] [Google Scholar]
- 92.Busso C., Ferreira-Junior J.R., Paulela J.A., Bleicher L., Demasi M. and Barros M.H. (2015) Coq7p relevant residues for protein activity and stability. Biochimie 119, 92–102 10.1016/j.biochi.2015.10.016 [DOI] [PubMed] [Google Scholar]
- 93.Martin-Montalvo A., Gonzalez-Mariscal I., Pomares-Viciana T., Padilla-Lopez S., Ballesteros M., Vazquez-Fonseca L. et al. (2013) The phosphatase Ptc7 induces coenzyme Q Biosynthesis by activating the hydroxylase Coq7 in yeast. J. Biol. Chem. 288, 28126–28137 10.1074/jbc.M113.474494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Juneau K., Nislow C. and Davis R.W. (2009) Alternative splicing of PTC7 in Saccharomyces cerevisiae determines protein localization. Genetics 183, 185–194 10.1534/genetics.109.105155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Awad A.M., Venkataramanan S., Nag A., Galivanche A.R., Bradley M.C., Neves L.T. et al. (2017) Chromatin-remodeling SWI/SNF complex regulates coenzyme Q6 synthesis and a metabolic shift to respiration in yeast. J. Biol. Chem. 292, 14851–14866 10.1074/jbc.M117.798397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Guo X., Niemi N.M., Hutchins P.D., Condon S.G., Jochem A., Ulbrich A. et al. (2017) Ptc7p dephosphorylates select mitochondrial proteins to enhance metabolic function. Cell Rep. 18, 307–313 10.1016/j.celrep.2016.12.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Stefely J.A., Reidenbach A.G., Ulbrich A., Oruganty K., Floyd B.J., Jochem A. et al. (2015) Mitochondrial ADCK3 employs an atypical protein kinase-like fold to enable coenzyme Q biosynthesis. Mol. Cell 57, 83–94 10.1016/j.molcel.2014.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Stefely J.A., Licitra F., Laredj L., Reidenbach A.G., Kemmerer Z.A., Grangeray A. et al. (2016) Cerebellar ataxia and coenzyme Q deficiency through loss of unorthodox kinase activity. Mol. Cell 63, 608–620 10.1016/j.molcel.2016.06.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Reidenbach A.G., Kemmerer Z.A., Aydin D., Jochem A., McDevitt M.T., Hutchins P.D. et al. (2018) Conserved lipid and small-molecule modulation of COQ8 reveals regulation of the ancient kinase-like UbiB family. Cell Chem. Biol. 25, 154–165.e111, 10.1016/j.chembiol.2017.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Vazquez Fonseca L., Doimo M., Calderan C., Desbats M.A., Acosta M.J., Cerqua C. et al. (2018) Mutations in COQ8B (ADCK4) found in patients with steroid-resistant nephrotic syndrome alter COQ8B function. Hum. Mutat. 39, 406–414 10.1002/humu.23376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lagier-Tourenne C., Tazir M., Lopez L.C., Quinzii C.M., Assoum M., Drouot N. et al. (2008) ADCK3, an ancestral kinase, is mutated in a form of recessive ataxia associated with coenzyme Q10 deficiency. Am. J. Hum. Genet. 82, 661–672 10.1016/j.ajhg.2007.12.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Mollet J., Delahodde A., Serre V., Chretien D., Schlemmer D., Lombes A. et al. (2008) CABC1 gene mutations cause ubiquinone deficiency with cerebellar ataxia and seizures. Am. J. Hum. Genet. 82, 623–630 10.1016/j.ajhg.2007.12.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Hsieh E.J., Gin P., Gulmezian M., Tran U.C., Saiki R., Marbois B.N. et al. (2007) Saccharomyces cerevisiae Coq9 polypeptide is a subunit of the mitochondrial coenzyme Q biosynthetic complex. Arch. Biochem. Biophys. 463, 19–26 10.1016/j.abb.2007.02.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Duncan A.J., Bitner-Glindzicz M., Meunier B., Costello H., Hargreaves I.P., Lopez L.C. et al. (2009) A nonsense mutation in COQ9 causes autosomal-recessive neonatal-onset primary coenzyme Q10 deficiency: a potentially treatable form of mitochondrial disease. Am. J. Hum. Genet. 84, 558–566 10.1016/j.ajhg.2009.03.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hayashi K., Ogiyama Y., Yokomi K., Nakagawa T., Kaino T. and Kawamukai M. (2014) Functional conservation of coenzyme Q biosynthetic genes among yeasts, plants, and humans. PLoS ONE 9, e99038 10.1371/journal.pone.0099038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.He C.H., Black D.S., Allan C.M., Meunier B., Rahman S. and Clarke C.F. (2017) Human COQ9 rescues a coq9 yeast mutant by enhancing coenzyme Q biosynthesis from 4-hydroxybenzoic acid and stabilizing the CoQ-synthome. Front. Physiol. 8, 463 10.3389/fphys.2017.00463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Danhauser K., Herebian D., Haack T.B., Rodenburg R.J., Strom T.M., Meitinger T. et al. (2016) Fatal neonatal encephalopathy and lactic acidosis caused by a homozygous loss-of-function variant in COQ9. Eur. J. Hum. Genet. 24, 450–454 10.1038/ejhg.2015.133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Herebian D., Seibt A., Smits S. H.J., Bunning G., Freyer C., Prokisch H. et al. (2017) Detection of 6-demethoxyubiquinone in CoQ10 deficiency disorders: Insights into enzyme interactions and identification of potential therapeutics. Mol. Genet. Metab. 121, 216–223 10.1016/j.ymgme.2017.05.012 [DOI] [PubMed] [Google Scholar]
- 109.Luna-Sanchez M., Diaz-Casado E., Barca E., Tejada M.A., Montilla-Garcia A., Cobos E.J. et al. (2015) The clinical heterogeneity of coenzyme Q10 deficiency results from genotypic differences in the Coq9 gene. EMBO Mol. Med. 7, 670–687 10.15252/emmm.201404632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Allan C.M., Hill S., Morvaridi S., Saiki R., Johnson J.S., Liau W.S. et al. (2013) A conserved START domain coenzyme Q-binding polypeptide is required for efficient Q biosynthesis, respiratory electron transport, and antioxidant function in Saccharomyces cerevisiae. Biochim. Biophys. Acta 1831, 776–791 10.1016/j.bbalip.2012.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Barros M.H., Johnson A., Gin P., Marbois B.N., Clarke C.F. and Tzagoloff A. (2005) The Saccharomyces cerevisiae COQ10 gene encodes a START domain protein required for function of coenzyme Q in respiration. J. Biol. Chem. 280, 42627–42635 10.1074/jbc.M510768200 [DOI] [PubMed] [Google Scholar]
- 112.Busso C., Bleicher L., Ferreira J.R. and Barros M.H. (2010) Site-directed mutagenesis and structural modeling of Coq10p indicate the presence of a tunnel for coenzyme Q6 binding. FEBS Lett. 584, 1609–1614 10.1016/j.febslet.2010.03.024 [DOI] [PubMed] [Google Scholar]
- 113.Cui T.Z. and Kawamukai M. (2009) Coq10, a mitochondrial coenzyme Q binding protein, is required for proper respiration in Schizosaccharomyces pombe. FEBS J. 276, 748–759 10.1111/j.1742-4658.2008.06821.x [DOI] [PubMed] [Google Scholar]
- 114.Shen Y., Goldsmith-Fischman S., Atreya H.S., Acton T., Ma L., Xiao R. et al. (2005) NMR structure of the 18 kDa protein CC1736 from Caulobacter crescentus identifies a member of the START domain superfamily and suggests residues mediating substrate specificity. Proteins 58, 747–750 10.1002/prot.20365 [DOI] [PubMed] [Google Scholar]
- 115.Perocchi F., Jensen L.J., Gagneur J., Ahting U., von Mering C., Bork P. et al. (2006) Assessing systems properties of yeast mitochondria through an interaction map of the organelle. PLoS Genet. 2, e170 10.1371/journal.pgen.0020170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Marchler-Bauer A., Zheng C., Chitsaz F., Derbyshire M.K., Geer L.Y., Geer R.C. et al. (2013) CDD: conserved domains and protein three-dimensional structure. Nucleic Acids Res. 41, D348–D352 10.1093/nar/gks1243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Rossmann M.G., Moras D. and Olsen K.W. (1974) Chemical and biological evolution of nucleotide-binding protein. Nature 250, 194–199 10.1038/250194a0 [DOI] [PubMed] [Google Scholar]
- 118.Pagliarini D.J., Calvo S.E., Chang B., Sheth S.A., Vafai S.B., Ong S.E. et al. (2008) A mitochondrial protein compendium elucidates complex I disease biology. Cell 134, 112–123 10.1016/j.cell.2008.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Hirst J. (2011) Why does mitochondrial complex I have so many subunits? Biochem. J. 437, e1–e3 10.1042/BJ20110918 [DOI] [PubMed] [Google Scholar]
- 120.Stroud D.A., Formosa L.E., Wijeyeratne X.W., Nguyen T.N. and Ryan M.T. (2013) Gene knockout using transcription activator-like effector nucleases (TALENs) reveals that human NDUFA9 protein is essential for stabilizing the junction between membrane and matrix arms of complex I. J. Biol. Chem. 288, 1685–1690 10.1074/jbc.C112.436766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Leshinsky-Silver E., Lev D., Tzofi-Berman Z., Cohen S., Saada A., Yanoov-Sharav M. et al. (2005) Fulminant neurological deterioration in a neonate with Leigh syndrome due to a maternally transmitted missense mutation in the mitochondrial ND3 gene. Biochem. Biophys. Res. Commun. 334, 582–587 10.1016/j.bbrc.2005.06.134 [DOI] [PubMed] [Google Scholar]
- 122.van den Bosch B.J., Gerards M., Sluiter W., Stegmann A.P., Jongen E.L., Hellebrekers D.M. et al. (2012) Defective NDUFA9 as a novel cause of neonatally fatal complex I disease. J. Med. Genet. 49, 10–15 10.1136/jmedgenet-2011-100466 [DOI] [PubMed] [Google Scholar]
- 123.Kuhl I., Miranda M., Atanassov I., Kuznetsova I., Hinze Y., Mourier A. et al. (2017) Transcriptomic and proteomic landscape of mitochondrial dysfunction reveals secondary coenzyme Q deficiency in mammals. Elife 6, 10.7554/eLife.30952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Yubero D., Montero R., Martin M.A., Montoya J., Ribes A., Grazina M. et al. (2016) Secondary coenzyme Q10 deficiencies in oxidative phosphorylation (OXPHOS) and non-OXPHOS disorders. Mitochondrion 30, 51–58 10.1016/j.mito.2016.06.007 [DOI] [PubMed] [Google Scholar]