Abstract
Diagnosing primary mitochondrial diseases is challenging in clinical practice. Although, defective oxidative phosphorylation (OXPHOS) is the common final pathway, it is unknown why different mtDNA or nuclear mutations result in largely heterogeneous and often tissue -specific clinical presentations. Mitochondrial tRNA (mt-tRNA) mutations are frequent causes of mitochondrial diseases both in children and adults. However numerous nuclear mutations involved in mitochondrial protein synthesis affecting ubiquitously expressed genes have been reported in association with very tissue specific clinical manifestations suggesting that there are so far unknown factors determining the tissue specificity in mitochondrial translation. Most of these gene defects result in histological abnormalities and multiple respiratory chain defects in the affected organs. The clinical phenotypes are usually early-onset, severe, and often fatal, implying the importance of mitochondrial translation from birth. However, some rare, reversible infantile mitochondrial diseases are caused by very specific defects of mitochondrial translation. An unbiased genetic approach (whole exome sequencing, RNA sequencing) combined with proteomics and functional studies revealed novel factors involved in mitochondrial translation which contribute to the clinical manifestation and recovery in these rare reversible mitochondrial conditions.
Keywords: mitochondrial translation, mitochondrial tRNA processing, mitochondrial tRNA modifications, mitochondrial tRNA synthetases
Introduction
All eukaryotic cells contain both genomic and mtDNA and two separate protein synthesis machineries [1]. Mitochondria are essential eukaryotic organelles with the main function to produce the majority of cellular energy by oxidative phosphorylation (OXPHOS). While the majority of OXPHOS components (complexes I–IV), the ATP synthase (complex V), and various factors required for mtDNA maintenance (replication, transcription, copy number control) are encoded within the nucleus, 13 polypeptides, two ribosomal RNAs (mt-rRNAs), and 22 transfer RNAs (mt-tRNAs) are encoded within the mtDNA [1]. The expression of these molecules is fundamental for cellular functioning and is closely co-ordinated with nuclear gene expression. Mutations in some nuclear genes can cause secondary instability of the mitochondrial genome in the form of depletion (decreased number of mtDNA molecules in the cell), multiple deletions or accumulation of point mutations, which in turn leads to mitochondrial diseases inherited in a Mendelian fashion [2]. Expression of the mitochondrial genome is initiated by transcription of the mtDNA from bidirectional heavy and light strand promoters to produce two polycistronic transcripts [3]. Instead of initiating at individual gene-specific promoters, transcription of mammalian mtDNA initiates from single promoters for H- and L-strand transcription, and progresses around almost the entire length of the genome [4]. Following endonucleolytic processing individual mitochondrial mRNA (mt-mRNA), mitochondrial rNA (mt-rRNA), and mitochondrial tRNA (mt-tRNA) transcripts undergo post-transcriptional modifications [5,6]. The transcription machinery of the mtDNA is regulated by several transcription factors TFAM, TEFM and TFB2M and mitochondrial RNA polymerase POLRMT [7]. The 13 mtDNA encoded components of the OXPHOS machinery using the mitochondrial translation mechanism are synthesized within the mitochondria, with the participation of the mitoribosome [8,9]. The assembled mitoribosome translates the mt-mRNAs and synthesizes proteins that are rapidly inserted into the inner mitochondrial membrane and integrated into their relevant complexes to form the OXPHOS system [10].
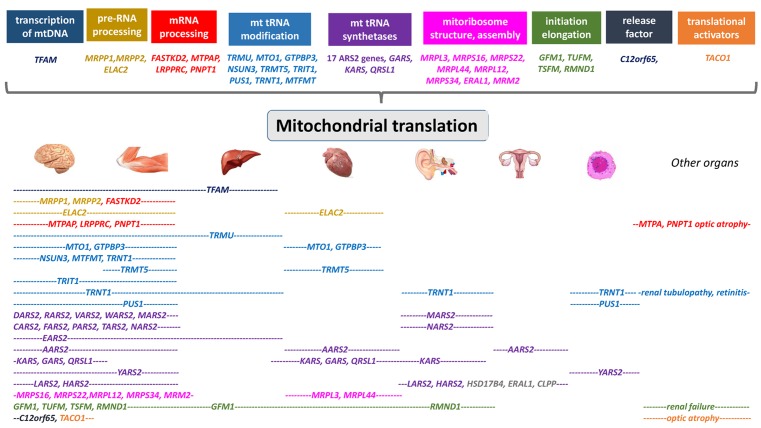
Approximately one-third of mitochondrial disorders have a presumed nuclear genetic defect of mitochondrial transcription and translation [11]. The identification of the molecular basis of this group has been particularly challenging and the recent availability of massively parallel sequencing have revealed several new disease genes, and unraveled new pathogenic mechanisms. Here, we present an overview of these tissue specific diseases (Figure 1).
Figure 1. Summary of the genes and disease mechanisms implicated in mitochondrial translation deficiencies with associated clinical phenotypes.
Defects of mitochondrial transcription
There are several genes involved in the initiation (POLRMT, TFAM, TFB2B) and elongation (TEFM, MTERF1) in transcription of mtDNA, however only mutations in TFAM have been shown to cause human diseases to date (Table 1) [12].
Table 1. Defects of mitochondrial transcription, pre-RNA, mRNA processing and stabilization.
| Gene | Protein | Clinical presentation | Age of onset | Mode of inheritance | OMIM | References |
|---|---|---|---|---|---|---|
| TFAM | Transcription factor A | Mitochondrial DNA depletion syndrome 15 | Infancy | AR | 617156 | Stiles et al. (2016) [12] |
| TRMT10C | tRNA methyltransferase 10 | Combined OXPHOS deficiency 30 | Infancy | AR | 616974 | Metodiev et al. (2016) [13] |
| HSD17B10 (MRPP2) | NAD(P)(H)-dependent short-chain dehydrogenase/reductases | Global developmental delay, epilepsy, and cardiac involvement | Early childhood | AR | 300256 | Oerum et al. (2017) [14], Falk et al. (2016) [15] |
| ELAC2 | RNase Z | Hypertrophic cardiomyopathy, hypotonia, lactic acidosis, delayed psychomotor development | Early childhood | AR | 605367 | Haack et al. (2013) [16], Shinwari et al. (2017) [17], Akawi et al. (2016) [18] |
| FASTKD2 | fas activated serine-threonine kinase domain 2 protein | Later onset, milder MELAS (mitochondrial encephalomyopathy, lactic acidosis and stroke-like episode)-like syndrome with seizures, stroke-like episodes and optic atrophy. Mitochondrial encephalomyopathy with developmental delay, hemiplegia, convulsions, asymmetrical brain atrophy | Childhood | AR | 612322 | Ghezzi et al. (2008) [19], Yoo et al. 2017 [20] |
| MTPAP | Mitochondrial poly-A polymerase | Progressive spastic ataxia with optic atrophy | Juvenile or early childhood | AR | 613672 | Crosby et al. (2010) [21] |
| LRPPRC | Leucine-rich PPR-motif containing protein | Leigh syndrome French–Canadian variant (LSFC) | Infantile | AR | 220111 | Mootha et al. (2003) [22], Olahova et al. (2015) [23], Han et al. (2017) [24] |
OMIM, Online Mendelian Inheritance in Man; AR, autosomal recessive.
TFAM
The TFAM gene encodes a mitochondrial transcription factor essential for initiating mtDNA transcription, replication, and nucleoid packaging [25]. Pathogenic mutations in TFAM are linked to an autosomal recessive disorder with infantile-onset progressive fatal liver failure. Infants were born with intrauterine growth restriction and developed hepatopathy with elevated transaminases, conjugated hyperbilirubinemia, and hypoglycemia. Liver failure and death occurred in early infancy [12]. The mtDNA copy number has been shown to be decreased in patient liver, muscle, and fibroblasts. Liver biopsy shows cirrhosis, micro- and macrovesicular steatosis and cholestasis and abnormal mitochondrial morphology on electron microscopy. Biochemical enzymology in muscle showed increased citrate synthase activity and borderline reduced RC enzyme activities [12]. Based on these findings, it is likely, that mutations in other genes involved in mitochondrial transcription also result in low mtDNA copy numbers and a combined defect of the enzymes of the respiratory chain.
It has been implicated that TFAM-mediated alterations may be an important mechanism in neurodegeneration in Alzheimer, Huntington, Parkinson, and other neurodegenerative diseases [13]. Because altered TFAM and mtDNA levels have been detected in multiple models of neurodegeneration, we suggest that the regulation of TFAM may be a key mechanism in disease pathomechanism or progression [13].
Maturation of the primary transcript: pre-RNA processing
Transcription of the mitochondrial genome generates large polycistronic transcripts punctuated by the 22 mt-tRNAs that are conventionally cleaved by the RNase P-complex and the RNase Z activity of ELAC2 at 5′ and 3′ ends, respectively (Table 1) [5,6].
TRMT10C/MRPP1
Mutations in TRMT10C (encoding the mitochondrial RNase P protein 1 (MRPP1)) were reported in infants presenting at birth with lactic acidosis, hypotonia, feeding difficulties, and deafness [13]. Both individuals died at 5 months after respiratory failure. MRPP1, along with MRPP2 and MRPP3, form the mitochondrial ribonuclease P (mt-RNase P) complex that cleaves the 5′ ends of mt-tRNAs from polycistronic precursor transcripts. Analyses of fibroblasts from affected individuals harboring TRMT10C missense variants revealed decreased protein levels of MRPP1 and an increase in mt-RNA precursors indicative of impaired mt-RNA processing and defective mitochondrial protein synthesis [13].
HSD17B10/MRPP2
MRPP2 (also known as HSD10/SDR5C1) belongs to the short-chain dehydrogenase/reductases (SDR) family and is involved in the catabolism of isoleucine and steroid metabolism [14]. MRPP2 also interacts in a complex with MRPP1 (TRMT10C) and MRPP3 (also known as PRORP), proteins involved in 5′-end processing of mitochondrial precursor tRNA [5].
A Caucasian boy with intractable epilepsy and global developmental delay carried a novel p.(Lys212Glu) mutation in the X-linked gene, HSD17B10 encoding for mitochondrial SDR5C1 [15]. Mutations in HSD17B10 lead to a metabolic disorder of fatty and amino acid metabolism, and affect an essential subunit of human mitochondrial RNase P, the enzyme responsible for 5′-processing and methylation of purine-9 of mt-tRNAs. The pathogenicity of the mutation is due to a general mitochondrial dysfunction caused by reduction in maturation of mt-tRNAs [15].
Two additional patients were reported with variable severity of developmental delay, epilepsy, and cardiac involvement. As a hallmark of the disease, urinary organic acid analysis showed elevated levels of 2-methyl-3-hydroxybutyric acid and tiglylglycine, and abnormalities were also detected in the acyl-carnitine spectrum in some cases [14].
ELAC2 (RNase Z)
Mt-tRNAs are cleaved by the RNase Z activity of ELAC2 at their 3′ ends [17]. Mutations in ELAC2 have been originally identified in five individuals with infantile hypertrophic cardiomyopathy and complex I deficiency and accumulation of mt-tRNA precursors in skeletal muscle and fibroblasts of the affected individuals, associated with impaired mitochondrial translation [17]. The association of severe, infantile cardiomyopathy and ELAC2 mutations was supported by 16 additional cases, suggesting that it is a relatively frequent cause of severe infantile-onset hypertrophic or dilated cardiomyopathy. The p.(Phe154Leu) variant has a severe effect with poor prognosis [18]. Affected children in a consanguineous Pakistani family with a homozygous splice-site mutation in ELAC2 presented with intellectual disability and minimal cardiac involvement [19].
Maturation of the primary transcript: mRNA processing and stability
FASTKD2
Mitochondrial encephalomyopathy with developmental delay, hemiplegia, convulsions, asymmetrical brain atrophy, and low cytochrome c oxidase (COX) activity in skeletal muscle were reported in patients with mutations in FASTKD2, encoding the fas activated serine-threonine kinase domain 2 protein [20]. FASTKD2 has a role in the assembly of the large ribosomal subunit and is required for 16S rRNA stability [26,27]. The tagged recombinant FASTKD2 protein co-localized with mitochondrial markers, and membrane potential-dependent mitochondrial import was demonstrated in isolated mitochondria in vitro. Later onset, milder mitochondrial encephalomyopathy, lactic acidosis and stroke-like episode (MELAS)-like syndrome with seizures, stroke-like episodes, and optic atrophy has been described in a Korean family with compound heterozygous mutations in FASTKD2 [28]. FASTKD2 has been also implicated as a target for modulating neurodegeneration and memory loss in ageing and dementia [29]. Furthermore, FASTKD2 has been also shown to mediate apoptosis in breast and prostate cancers [21].
MTPAP
In human mitochondria, polyadenylation of mRNA, undertaken by the nuclear-encoded mitochondrial poly(A) RNA polymerase, is essential for maintaining mitochondrial gene expression. An autosomal-recessive mutation has been identified in the MTPAP gene causing spastic ataxia with optic atrophy in the Old Order Amish population. Mt-mRNAs from affected individuals were shown to have severely truncated poly(A) tails [21]. Both mutated and wild-type MTPAP localized to the mitochondrial RNA-processing granules but the mutant protein generated only short oligo(A) extensions on RNA substrates, causing dysregulation of post-transcriptional expression leading to the reduction in respiratory chain complexes [30].
LRPPRC
LRPPRC is a mt-mRNA chaperone that relaxes secondary structures [31] enabling polyadenylation and co-ordinated translation of mitochondrially encoded proteins [32,33]. In addition, LRPPRC has been documented in various tumors, contributing to the apoptosis resistance of human cancer cells [34] and it has been identified as an inhibitor of autophagy and mitophagy via interaction with the mitophagy initiator Parkin [35].
A homozygous founder mutation in the LRPPRC gene (c.1061C>T, p.(Ala354Val)) was identified as one of the first nuclear mitochondrial disease genes [22], associated with the French-Canadian variant of Leigh Syndrome (LSFC) and COX deficiency. LSFC is characterized by Leigh syndrome (a subacute neurodegeneration of the brainstem and basal ganglia), developmental delay, hypotonia, mild facial dysmorphism, and high mortality due to episodes of severe acidosis and coma that typically arise in the first year of life [22]. Subsequently, LSFC has also been described outside Quebec in ten patients from seven unrelated families of Caucasian, Pakistani, Indian, Turkish, and Iraqi origin [23] and in a Chinese boy with a milder phenotype [24]. The phenotype of these patients resembles LSFC, but in addition, neonatal cardiomyopathy or congenital malformations of the heart and the brain were reported. Decreased levels of mutant LRPPRC protein and impaired Complex IV enzyme activity were associated with abnormal COX assembly and reduced steady-state levels of numerous OXPHOS subunits in patients’ fibroblasts and skeletal muscle. In some patients complex I was also reduced, suggesting the role of LRPPRC in tissue-specific post-transcriptional regulation of mt-mRNAs [23].
Diseases caused by abnormal tRNA modifications
Mt-tRNA modifications play a crucial role in regulating cellular energy delivery in response to local needs, and dysfunctional modifications may participate in the pathomechanism of mt-tRNA-related disorders (Table 2) [36].
Table 2. Defects of mt-tRNA modification.
| Gene | Protein | Clinical presentation | Age of onset | Mode of inheritance | OMIM | References |
|---|---|---|---|---|---|---|
| TRMU | tRNA 5-methylamino-methyl-2- thiouridy-late methyl-transferase |
Reversible infantile liver failure | Infantile | AR | 613070 | Zeharia et al. (2009) [37] Schara et al. (2011) [38] Uusimaa et al. (2011) [39] Gaignard et al. (2013) [40] |
| MTO1 | Mitochondrial translation optimization 1 homolog | Hypertrophic cardiomyopathy and lactic acidosis | Infantile | AR | 614702 | Ghezzi et al. (2012) [41], Baruffini et al. (2013) [42] O’Byrne et al. (2018) [43] |
| GTPBP3 | GTP-binding protein 3 | Hypertrophic or dilated cardiomyopathy, encephalopathy (hypotonia, developmental delay, seizures, visual impairment), lactate↑ | Early childhood | AR | 608536 | Kopajtich et al. (2014) [44] |
| NSUN3 | 5-methylcytosine (m(5)C) methyltransferase | Developmental delay, microcephaly, failure to thrive, lactic acidosis, muscular weakness, external ophthalmoplegia, and nystagmus | Neonatal | AR | 617491 | van Haute et al. (2016) [45] |
| TRMT5 | tRNA methyltransferase 5 | Exercise intolerance, lactic acidosis, growth retardation, developmental delay, complex hereditary spastic paraparesis | Childhood neonatal | AR | 611023 | Powell et al. (2015) [46] Tarnopolsky et al. (2017) [47] |
| TRIT1 | tRNA isopentenyl-transferase | Encephalopathy and myoclonic epilepsy, brain abnormalities | Childhood | AR | Yarham et al. (2014) [48] Kernohan et al. (2017) [49] |
|
| TRNT1 | tRNA nucleotidyltransferase | Retinitis pigmentosa, erythrocitic microcytosis; sideroblastic anemia with B-cell immunodeficiency, periodic fevers, and developmental delay | Neonatal, juvenile | AR | 612907 | Chakraborty et al. (2014) [50] DeLuca et al. (2016) [51] |
| PUS1 | Pseudouridine synthase | Myopathy, lactic acidosis, and sideroblastic anemia (MLASA1) | Early childhood to adult age | AR | 608109 | Bykhovskaya et al. (2004) [52] Fernandez-Vizarra et al. (2007) [53] Metodiev et al. (2015) [54] |
| MTFMT | Methionyl-tRNA formyltransferase | Leigh encephalopathy, white matter lesions, microcephaly, mental retardation, ataxia, and muscular hypotonia | Childhood | AR | 611766 | Tucker et al. (2011) [55] Neeve et al. (2013) [56] Haack et al. (2014) [57] |
OMIM, Online Mendelian Inheritance in Man; AR, autosomal recessive.
Wobble base modifications (TRMU, MTO1, GTPBP3, NSUN3)
TRMU
Reversible infantile liver failure is caused by autosomal recessive mutations in the tRNA 5-methylaminomethyl-2-thiouridylate methyltransferase [37–40] and the majority of these patients show complete spontaneous recovery if they survive the first year of life [58]. TRMU is an enzyme responsible for the thiouridylation of mt-tRNAGlu, mt-tRNAGln, and mt-tRNALys, which requires cysteine. Cysteine is an essential amino acid in the first months of life, because of the physiologically low activity of the cystathionine γ-lyase (cystathionase) enzyme in infants [59]. The age-dependent, partially reversible clinical presentation of TRMU mutations resembles reversible infantile respiratory chain deficiency due to them. 14674T>C/G mutation in mt-tRNAGlu. Low dietary cysteine may be a common trigger of the clinical presentation of both diseases [60]. Mutations in TRMU have been also suggested to aggravate the deafness phenotype of the mitochondrial m.1555A>G 12S rRNA mutation [61], however the variants reported here were rather variants of unknown significance and had no involvement in liver disease.
MTO1
MTO1 (mt-tRNA Translation Optimization 1), an evolutionarily conserved gene encodes the enzyme that catalyzes the 5-taurinomethylation of the wobble uridine base in mt-tRNAGln, tRNAGlu, and tRNALys. This post-transcriptional modification increases the accuracy and efficiency of mtDNA translation [61].
The first patients carrying recessive mutations in the MTO1 gene were identified in 2012 [41]. The clinical presentation was severe infantile hypertrophic cardiomyopathy. Two patients died within the first days of life, while the third unrelated subject showed marked improvement of the cardiomyopathy in childhood, and at the age of 19 years he suffered a stable hypertrophic cardiomyopathy with normal ejection fraction and moderate bilateral optic atrophy. Five additional patients were presented with hypertrophic cardiomyopathy and lactic acidosis in association with encephalopathy and psychomotor delay [42]. All patients complained of first symptoms soon after birth and two of them died in their first days of life. More recently, in a large cohort of 35 cases of MTO1 deficiency [61], none of the patients had bi-allelic null variants suggesting that the complete loss of MTO1 is not viable. The most common features at presentation are lactic acidosis and hypertrophic cardiomyopathy with global developmental delay/intellectual disability (97%), feeding difficulties (49%), hypotonia (63%) failure to thrive (34%), seizures (34%), optic atrophy (52%), and ataxia (21%) and low activity of respiratory chain enzymes I, III, and IV. A subjective clinical improvement was observed in some patients on ketogenic diet and therapy with dichloroacetate [43].
GTPBP3
Mutations in GTPBP3 are associated with a severe mitochondrial translation defect, due to the abnormal formation of 5-taurinomethyluridine (τm(5)U) in the anticodon wobble position of mt-tRNAs [44]. Eleven individuals from nine families were reported with recessive mutations in GTPBP3, encoding the mitochondrial GTP-binding protein 3 [44]. All patients presented with lactic acidosis and nine developed hypertrophic cardiomyopathy, but in contrast with individuals with mutations in MTO1 (involved in the same modification), most individuals with GTPBP3 mutations developed neurological symptoms and MRI involvement of thalamus, putamen, and brainstem resembling Leigh syndrome [44]. Affected individuals from eight out of nine families presented with combined respiratory chain complex deficiencies in skeletal muscle.
NSUN3
The recently characterized 5-methylcytosine (m(5)C) methyltransferase, NSun3 links m(5)C RNA modifications with energy metabolism [45]. Loss of function mutations in NSUN3 a previously uncharacterized m(5)C methyltransferase, have been identified in a patient who developed combined developmental delay, microcephaly, failure to thrive, recurrent lactic acidosis, muscular weakness, external ophthalmoplegia, and nystagmus at 3 months of age with combined OXPHOS deficiency in skeletal muscle [45].
Position 37 modifications (TRMT5, TRIT1)
TRMT5
Autosomal recessive mutations in the TRMT5 gene (encoding tRNA methyltransferase 5) were reported in two patients with strikingly different clinical presentation [46]. While both affected individuals presented with lactic acidosis and evidence of multiple mitochondrial respiratory chain complex deficiencies in skeletal muscle, one presented with failure to thrive and hypertrophic cardiomyopathy in childhood, and the other was an adult with a life-long history of exercise intolerance. Recently, TRMT5 mutations were also linked to complex hereditary spastic paraparesis [47]. Mutations in TRMT5 were associated with the hypomodification of a guanosine residue at position 37 (G37) of mt-tRNA, predominantly in skeletal muscle.
TRIT1
The first pathogenic mutation in TRIT1 (encoding the tRNA isopentenyltransferase, responsible for i6A37 modification of some cytosolic and mt-tRNAs) has been identified in two siblings with encephalopathy and myoclonic epilepsy and severe combined mitochondrial respiratory chain defects [48]. It has been show that a previously reported pathogenic m.7480A>G mt-tRNASer(UCN) mutation also acts by causing a loss of i6A37 modification, demonstrating that mt-tRNASerUCN is the substrate for TRIT1 [48]. Four individuals from three unrelated families ‘matched’ by GeneMatcher and MatchMakerExchange confirmed the role of TRIT1 in human disease [49]. The patients had microcephaly, developmental delay, epilepsy, and decreased levels of selected mitochondrial proteins [49].
CCA adding: TRNT1
TRNT1 (CCA-adding transfer RNA nucleotidyl transferase) enzyme deficiency is a complex metabolic disease caused by defective post-transcriptional modification of mitochondrial and cytosolic tRNAs [62]. Mutations in TRNT1 cause congenital sideroblastic anemia, immunodeficiency, fevers, and developmental delay (SIFD) [50]. Further mutations in TRNT1 have been reported in patients with a combination of abnormal blood cells (sideroblastic anemia, B lymphocyte or combined B and T immunodeficiency), metabolic crisis, and multisystem mitochondrial disease (retinitis pigmentosa, hepatosplenomegaly, exocrine pancreatic insufficiency, and renal tubulopathy [62,63–65]. Other clinical features include sensorineural deafness, cerebellar atrophy, brittle hair, partial villous atrophy, and nephrocalcinosis. TRNT1 mutations cause a spectrum of symptoms ranging from a childhood-onset complex disease with manifestations in most organs to an adult-onset isolated retinitis pigmentosa presentation. Acute management of these patients includes transfusion for anemia, fluid and electrolyte replacement, immunoglobulin therapy, and potentially bone marrow transplantation. A defect of 3′-CCA addition to mt-tRNAs (tRNA(Cys), tRNA(LeuUUR) and tRNA(His)) demonstrates a novel pathomechanism [62].
Pseudouridylation: PUS1
Pseudouridylate synthase 1 (PUS1) is an enzyme located in both nucleus and mitochondria, which converts uridine into pseudouridine in several cytosolic and mt-tRNA positions and increases the efficiency of protein synthesis in both compartments [66,52]. Myopathy, lactic acidosis, sideroblastic anemia (MLASA) syndrome is a rare autosomal recessive disease caused by recessive mutations in PUS1 encoding the pseudouridine synthase 1 enzyme [52–54,66]. A similar phenotype has been observed in mutations in YARS2 encoding the mitochondrial tyrosyl-tRNA synthetase [67]. Patients in consanguineous families of Persian, Jewish, and Italian origins presented with mental retardation, dysmorphic features, lactic acidosis, myopathy, sideroblastic anemia, and low activity of complexes 1 and 4 of the respiratory chain in muscles [53,54,68]. Some patients were reported with a mild phenotype of sideroblastic anemia and muscle weakness in adult age [54,69]. A double localization of PUS1 has been demonstrated, the isoform localized to the nucleus is predicted to be shorter (isoform 2) than the mitochondrial isoform, which contains an N-terminal mitochondrial targetting sequence. The structural differences in nuclear compared with mitochondrial isoforms of PUS1 may be implicated in the variability of the clinical presentations in MLASA [53].
Formylation of the mitochondrial methionine tRNA (Met-tRNAMet)
The first mutations in the MTFMT gene in patients with Leigh syndrome and combined respiratory chain deficiency were reported by Tucker et al. [55]. In the past 5 years, several patients have been reported with MTFMT mutations and the clinical presentation is variable (Leigh encephalopathy, white matter lesions, microcephaly, mental retardation, ataxia, and muscular hypotonia) but often milder and later onset than other genetic forms of Leigh syndrome [56,57,70]. The mutations are usually loss-of-function mutations resulting in a severe decrease in MTFMT protein and reduced steady-state levels of complex I and IV subunits. The c.626C>T mutation has been detected in >80% of patients with MTFMT deficiency, and represents a relatively frequent cause of Leigh syndrome.
Diseases of tRNA aminoacylation: mt-tRNA synthetases
Defects in nuclear genes encoding mitochondrial aminoacyl-tRNA synthetases (mt-ARSs) are increasingly linked to a variety of pediatric and adult onset tissue specific disorders [71]. Several recent reviews [72–75] presented detailed information, therefore here, we only provide a short summary of the most common phenotypes of mt-tRNA synthetase-related diseases (Table 3).
Table 3. Mutations in aminoacyl-tRNA synthetases.
| Gene | Protein | Clinical presentation | Age of onset | Mode of inheritance | OMIM | References |
|---|---|---|---|---|---|---|
| DARS2 | Aspartyl-tRNA sythetase 2 | - Leukoencephalopathy with brainstem and spinal cord involvement (LBSL) - Paroxysmal exercise-induced gait ataxia |
Childhood or adulthood | AR | 610956 | Scheper et al. (2007) [76] Isohanni et al. (2010) [77] Miyake et al. (2011) [78] van Berge et al. (2014) [79] Shimojima et al. (2017) [80] Pinto et al. (2014) [81] Synofzik et al. (2011) [82] |
| RARS2 | Arginyl-tRNA synthetase 2 |
Pontocerebellar hypoplasia type 6 (PCHD-6) | Neonatal or early childhood |
AR | 611523 | Edvardson et al. (2007) [83] Rankin et al. (2010) [84] Cassandrini et al. (2013) [85] Li et al. (2015) [86] Lühl et al. (2016) [87] |
| EARS2 | Glutamyl-tRNA synthetase 2 | Leukoencephalopathy with thalamus and brainstem involvement and high lactate (LTBL); multiple congenital anomalies and multisystem dysfunction dysgenesis of corpus callosum | Congenital or infantile |
AR | 612799 | Steenweg et al. (2012) [88] Talim et al. (2013) [89] Kevelam et al. (2016) [90] Güngör et al. (2016) [91] Şahin et al. (2016) [92] |
| MARS2 | Methionyl-tRNA synthetase 2 | Autosomal recessive spastic ataxia with leukoencephalopathy | Juvenile or adulthood | AR | 609728 | Bayat et al. (2012) [93] Webb et al. (2015) [94] |
| FARS2 | Phenylalanyl-tRNA synthetase 2 | Alpers syndrome, encephalopathy, epilepsy, lactic acidosis, spastic paraplegia | Neonatal or infantile | AR | 611592 | Elo et al. (2012) [95] Shamseldin et al. (2012) [96] Yang et al. (2016) [97] |
|
AARS2 |
Alanyl-tRNA synthetase 2 | - Hypertrophic cardiomyopathy - Ovario-leukodystrophy - Leukoencephalopathy with axonal spheroids and pigmented glia (ALSP) |
Infantile to adulthood | AR | 614096 | Götz et al. (2011) [98] Taylor et al. (2014) [99] Dallabona et al. (2014) [100] Lynch et al. (2016) [101] Szpisjak et al. (2017) [102] |
| YARS2 | Tyrosyl-tRNA synthetase | MLASA2, gastrointestinal difficulties, cardiomyopathy | Infantile | AR | 613561 | Riley et al. (2010) [67] Sasarman et al. (2012) [103] Shahni et al. (2013) [104] Riley et al. (2013) [105] Nakajima et al. (2014) [106] |
| SARS | Seryl-tRNA synthetase 2 | - HUPRA syndrome (hyperuricemia, pulmonary hypertension, renal failure in infancy, and alkalosis) - Progressive spastic paresis |
Infantile | AR | 613845 | Belostotsky et al. (2011) [107] Linnankivi et al. (2016) [108] |
| HARS2 | Histidyl-tRNA synthetase 2 | Perrault syndrome (sensorineural deafness, ovarian dysgenesis) | Juvenile or adulthood | AR | 600783 | Pierce et al. (2011) [109] |
| LARS2 | Leucyl-tRNA synthetase | Perrault syndrome (sensorineural deafness, ovarian dysgenesis) hydrops, lactic acidosis, and sideroblastic anemia |
Juvenile neonatal |
AR | 604544 | Pierce et al. (2013) [110] Soldà et al. (2016) [111] Demain et al. (2017) [112] Riley et al. (2016) [113] |
| TARS2 | Threonyl-tRNA synthetas | Mitochondrial encephalomyopathy Axial hypotonia and limb hypertonia, psychomotor delay, and high levels of blood lactate |
Infantile | AR | 612805 | Diodato et al. (2014) [114] |
| NARS2 | Asparginyl-tRNA synthetase | Non-syndromic deafness, Leigh syndrome, Alpers syndrome, infantile onset neurodegenerative disorder | Infantile | AR | 612803 | Sofou et al. (2015) [115] Vanlander et al. (2015) [116] Simon et al. (2015) [117] Mizuguchi et al. (2017) [118] |
| CARS2 | Cysteinyl-tRNA synthetas | Combined oxidative phosphorylation deficiency-27 (COXPD27); severe epileptic encephalopathy and complex movement disorders | Juvenile | AR | 612800 | Coughlin et al. (2015) [119] |
| IARS2 | Ileucyl-tRNA synthetase | - Skeletal dysplasia, infantile cataract, congenital neurotrophic keratitis, orbital myopathy, Leigh syndrome - CAGSSS syndrome |
Adulthood or infantile |
AR | 616007 612801 |
Schwartzentruber et al. (2014) [120] Moosa et al. (2017) [121] |
| VARS2 | Valyl-tRNa synthetase | Mitochondrial encephalomyopathy: psychomotor delay, epilepsy, mental retardation, growth hormone deficiency, hypogonadism | Juvenile | AR | 612802 | Diodato et al. (2014) [114] Baertling et al. (2017) [122] Alsemari et al. (2017) [123] |
| WARS2 | Tryptophanyl-tRNA synthetase | - Autosomal recessive intellectual disability - Mitochondrial encephalopathy - Infantile-onset Parkinsonism |
Infantile or juvenile |
AR | 604733 | Musante et al. (2017) [124] Wortmann et al. (2017) [125] Theisen et al. (2017) [126] Burke et al. (2017) [127] |
| PARS2 | Prolyl-tRNA synthetase | Non-syndromic hearing loss, Leigh syndrome, intellectual disability with epilepsy and severe myopathy, seizure | Infantile | AR | 612036 | Sofou et al. (2015) [128] Mizuguchi et al. (2017) [118] |
| GARS | Glycil-tRNA synthetase | - Charcot-Marie-Tooth disease, type 2D - Neuropathy, distal hereditary motor, type VA - Multisystem developmental delay, growth retardation- Lactic acidosis, cardiomyopathy, exercise intolerance |
Adulthood, early childhood |
AD AR |
601472 600794 |
Antonellis et al. (2003) [129] Oprescu et al. (2017) [130] Nafisinia et al. (2017) [131] McMillan et al. (2014) [132] |
| KARS | Lysyl-tRNA synthetases | - Charcot-Marie-Tooth disease, recessive intermediate, B - Deafness, autosomal recessive 89 - Visual impairment and progressive microcephaly - Hypertrophic cardiomyopathy and combined mitochondrial respiratory chain defect |
Adult, infantile, childhood |
AR | 613641 613916 |
Kohda et al. (2016) [133] Verrigini et al. (2017) [134] McMillan et al. (2015) [135] Santos-Cortez et al. (2013) [136] McLaughlin et al. (2010) [137] |
OMIM, Online Mendelian Inheritance in Man; AR, autosomal recessive; AD, autosomal dominant.
Mutations in each of the 19 human mt-ARS genes have been reported in human disease [74]. Glycyl-(GARS) and lysyl tRNA (KARS) synthetase genes encode both cytosolic and mitochondrial ARS enzymes, suggesting links between protein syntheses in these two distinct cellular compartments. Other cytosolic ARSs are encoded by a set of genes distinct from those encoding mt-ARSs [138]. All mt-ARSs genes are located in the nucleus, synthesized in the cytosol, imported into the mitochondria by an N-terminal pre-sequence (mitochondrial targetting sequence, MTS), which is cleaved upon entry into the mitochondria [139].
Despite being ubiquitously expressed, mutations in these genes show an unexpected variety of phenotypes, including many neurological disorders affecting the white matter (DARS2, EARS2, MARS2, AARS2) or causing epileptic encephalopathy (CARS2, FARS2, PARS2, TARS2, VARS2), pontocerebellar hypoplasia (RARS2), or intellectual disability (RARS2, WARS2). While other characteristic phenotypes are sensori-neuronal hearing loss and ovarian failure (Perrault syndrome: HARS2, LARS2), mitochondrial myopathy, MLASA: YARS2, hyperuricemia, pulmonary hypertension, renal failure, alkalosis (HUPRA: SARS2), cardiomyopathy (AARS2), or sensori-neural hearing loss (MARS2, NARS2). Besides the fact that new mutations are continuously discovered, neither the cause of the selective vulnerability, nor the exact molecular mechanisms leading to the diseases, are well understood. Degeneration of the central nervous system is speculated with early impairment of mitochondrial energy production that is crucial for myelination and maintenance of compact myelin [140]. Mutations in DARS2 and EARS2 result in very characteristic MRI phenotypes of leukoencephalopathy with brainstem and spinal cord involvement and lactate elevation (LBSL) [76] and leukoencephalopathy with thalamus and brainstem involvement and high lactate (LTBL) [141]. LBSL caused by mutations in DARS2 is clinically characterized by slowly progressive pyramidal, cerebellar and dorsal column impairment, variably associated with delayed intellectual and/or motor development, cognitive impairment, epilepsy and peripheral neuropathy. The severity is ranging from early-onset severe disease, which can be fatal within the first years of life, to adult-onset forms [79,80]. The majority of patients carry a splice site mutation in intron 2, upstream of exon 3 [79]. Subgroups of patients with similar mutations (the common variants c.228-21_-20delTTinsC together with c.455G>T and c.492+2T>C) and a mild disease progression were identified. MRI abnormalities were correlated with the severity of the phenotype in mildly affected patients [81].
LTBL due to EARS2 mutations is characterized by a biphasic clinical course [141,89,90]. Approximately one-third of patients suffered from hypotonia soon after birth, followed by spastic tetraparesis, dystonia, visual impairment, and seizures. The majority (two-third) of patients had normal or mildly delayed early development, disease-onset in the second half of the first year of life with clinical regression, spasticity, loss of milestones, sometimes seizures and irritability, and an improvement in symptoms and MRI abnormalities from the second year of life.
A founder mutation, p.(Arg590Trp) in AARS2, encoding the mt alanyl tRNA synthetase may predominantly affect the heart (infantile cardiomyopathy) [98,99], while other AARS2 mutations are characterized by childhood- to adulthood-onset ataxia, spasticity, and dementia with frontal lobe dysfunction with leukoencephalopathy, cerebellar atrophy, and involvement of the corpus callosum on MRI [100]. Notably, all female patients also had ovarian failure. None of these cases suffered from a cardiomyopathy. Cardiomyopathy-associated mutations severely compromise aminoacylation, whereas partial activity is retained by the mutation combinations found in the leukodystrophy patients [142]. Similar molecular mechanisms may underlie the tissue specific manifestations of the other mt tRNA synthetases.
A few patients presented severe infantile multisystem disease predominantly affecting the heart and brain associated with combined OXPHOS enzyme deficiency have been reported recently with autosomal recessive mutations in the QRSL1 gene [143,144]. No mitochondrial glutaminyl-tRNA synthetase (GlnRS) has been known and Gln-tRNAGln synthesis occurs via an indirect pathway involving QRSL1 (GatA). In this pathway, mt tRNAGln is first misaminoacylated by mt glutamyl-tRNA synthetase (GluRS) to form Glu-tRNAGln, which is then followed by transamidation to Gln-tRNAGln. This transamidation is processed by the hGatCAB heterotrimer. It has been shown that mutations in QRSL1 (GatA), a component of hGatCAB were associated with severe transamidation activity defects [143].
Perrault syndrome: LARS2, HARS2 (HSD17B4, CLLP, ERAL1)
Perrault syndrome is characterized by sensorineural hearing loss (SNHL) in males and females, and ovarian dysfunction in females. The SNHL is bilateral and ranges in severity from moderate with early-childhood onset to profound with congenital onset. Ovarian dysfunction ranges from gonadal dysgenesis (absent or streak gonads) manifesting as primary amenorrhea to primary ovarian insufficiency (POI) defined as cessation of menses before the age of 40. Fertility in affected males is reported as normal. Neurological features described in some affected women include developmental delay or intellectual disability, cerebellar ataxia, and motor and sensory peripheral neuropathy [145]. The diagnosis is confirmed by the presence of biallelic pathogenic variants in the genes HARS2, HSD17B4, LARS2, ERAL1, or CLPP. The fact that these seemingly different molecular mechanisms of mitochondrial translation can result in very similar, characteristic phenotypes raise the possibility of some common mechanisms.
Mitoribosomal structure and assembly: MRPL3, MRPS16, MRPS22, MRPL44, MRPL12, MRPS34, ERAL1
Autosomal recessive mutations in nuclear encoded mitochondrial ribosomal proteins are rare and cause severe, infantile onset disease with growth retardation, neurological phenotypes (MRPL3, MRPS16, MRPS22, MRPL12, MRPS34) and cardiac involvement (MRPL3, MRPL44) (Table 4) [10]. Autosomal recessive mutations in the ribosomal assembly factor ERAL1 have been associated with Perrault syndrome [145].
Table 4. Mutations in mitochondrial ribosomal proteins and ribosome assembly proteins.
| Gene | Protein | Clinical presentation | Age of onset | Mode of inheritance | OMIM | References |
|---|---|---|---|---|---|---|
| MRPL | Mitochondrial ribosomal protein L3 | Hypertrophic cardiomyopathy and psychomotor retardation | Infantile | AR | 614582 | Galmiche et al. (2011) [146] |
| MRPS16 | Mitochondrial ribosomal protein S16 | Corpus callosum agenesia, hypothonia, and fatal neonatal lactic acidosis | Neonatal | AR | 610498 | Miller et al. (2004) [147] |
| MRPS22 | Mitochondrial ribosomal protein S22 | Cornelia de Lange-like syndrome Edema, cardiomyopathy and tubulopathy |
Neonatal | AR | 611719 | Saada et al. (2007) [148] Smits et al. (2011) [149] |
| MRPL44 | Mitochondrial ribosomal protein L44 | Hypertrophic cardiomyopathy | Neonatal | AR | 611849 | Carroll et al. (2013) [150] Distelmaier et al. (2015) [151] |
| MRPL12 | Mitochondrial ribosomal protein L12 | Growth retardation and neurological deterioration | Neonatal | AR | 602375 | Serre et al. (2013) [152] |
| MRPS34 | Mitoribosomal ribosomal protein S34 | Leigh syndrome and combined OXPHOS defects | Neonatal | AR | 611994 | Richman et al. (2015) [153] Lake et al. (2017) [154] |
| ERAL1 | mt-rRNA chaperone | Perrault syndrome | Childhood or adult | AR | 607435 | Newman et al. (2014) [143] |
OMIM, Online Mendelian Inheritance in Man; AR, autosomal recessive.
Translation initiation and elongation factors: GFM1, TUFM, TSFM, RMND1
The diseases caused by mutations in these factors are severe neonatal or infantile onset rare diseases affecting the brain (GFM1, TUFM, TSFM, RMND1), liver (GFM1), heart (TSFM), and other organs (RMND1) (Table 5) [11]. There are no diseases linked to mutations in translation termination factors to date. The most frequent gene defect in this group is caused by mutations in RMND1 leading to a severe defect of mitochondrial translation in all tissues. The RMND1 gene encodes an integral inner membrane mitochondrial protein that assembles into a large 240-kDa complex to support translation of the 13 polypeptides encoded on mtDNA [155,156]. Clinical and genetic features of 32 RMND1 patients from 21 pedigrees are hypotonia and developmental delay (75%), sensori-neural hearing loss (72%), nephropathy (53%), failure to thrive (53%), seizures (44%), microcephaly (41%), and spasticity (19%) [157]. The disease usually starts early, before 2 years of life, but patients with renal involvement show a later onset, better prognosis, and longer survival [157]. Four patients were successfully treated with kidney transplantation with a good clinical response.
Table 5. Mitochondrial translation initiation, elongation, termination, and release factors and translational activators.
| Gene | Protein | Clinical presentation | Age of onset | Mode of inheritance | OMIM | References |
|---|---|---|---|---|---|---|
| GFM1 | Elongation factor G 1, mitochondrial (EFG1mt) | Encephalopathy with or without liver involvement | Neonatal | AR | 609060 | Coenen et al. (2004) [158] Valente et al. (2007) [159] Smits et al. (2011) [160] |
| TUFM | Elongation factor Tu, mitochondrial (EF-TUmt) | Lactic acidosis, leukoencephalopathy, and polymicrogyria | Neonatal | AR | 610678 | Valente et al. (2007) [159] Kohda et al. (2016) [161] |
| TSFM | Elongation factor Ts, mitochondrial (EF-Tsmt) | Encephalomyopathy, hypertrophic cardiomyopathy | Neonatal or childhood | AR | 610505 | Smeitink et al. (2006) [162] Smits et al. (2011) [160] Shamseldin et al. (2012) [163] Ahola et al. (2014) [164] |
| RMND1 | Regulator of microtubule dynamics 1 | Deafness, myopathy, renal involvement, cardiomyopathy and a severe biochemical defect Combined oxidative phosphorylation deficiency -11 |
neonatal | AR | 614917 614922 |
Janer et al. (2012) [144] Garcia-Diaz et al. (2012) [145] Taylor et al. (2014) [99] Janer et al. (2015) [165] Gupta et al. (2016) [166] Ravn et al. (2016) [159] Vinu et al. (2018) [167] |
| C12orf65 | Chromosome 12 ORF 65 | Leigh syndrome, optic atrophy, ophthalmoplegia Spastic paraplegia with optic atrophy and axonal neuropathy (SPG55) |
Infantile | AR | 613559 | Antonicka et al. (2010) [156] Pyle et al. (2014) [168] Shimazaki et al. (2012) [157] Spiegel et al. (2014) [169] |
| TACO1 | Translational activator of COX1 | Leigh syndrome | Juvenile | AR | 612958 | Weraarpachai et al. (2009) [170] Makrythanasis et al.(2014) [171] |
OMIM, Online Mendelian Inheritance in Man; AR, autosomal recessive.
Release factors: C12orf65
The C12orf65 gene encodes a protein that is critical for the release of newly synthesized proteins from mitochondrial ribosomes and its deficiency was reported in patients with Leigh syndrome and optic atrophy [172], in autosomal recessive hereditary spastic paraplegia 55 (SPG55) [168] or Charcot-Marie-Tooth disease type 6 [169], or Behr’s syndrome (optic atrophy, spastic paraparesis, motor neuropathy, ataxia, ophthalmoparesis) [170]. The spectrum of C12orf65-related phenotypes includes the triad of early-onset optic atrophy, axonal neuropathy, and spastic paraparesis as key clinical features [170,173].
Translational activators: TACO1
As mammalian mt-mRNAs do not have significant 5′ UTRs, alternate mechanisms exist to promote their translation. A defect in the translational activator of the mtDNA-encoded COX I subunit has been identified in a pedigree segregating late-onset Leigh syndrome and cytochrome c oxidase (COX) deficiency [174]. A single homozygous one-base-pair insertion has been identified in one large consanguineous Turkish family with teenage onset Leigh syndrome, cognitive decline, dystonia, and optic atrophy in TACO1 for translational activator of COX I [174,175]. No other mutations have been reported to date worldwide to confirm the phenotype. However, our group has detected the previously described TACO1 mutation in an additional consanguineous Turkish family (unpublished). The clinical phenotype in patients has been supported by the Taco1 mutant mice, which develop a late-onset visual impairment, motor dysfunction, and cardiac hypertrophy [176].
Mutation in PNPT1, which encodes a polyribonucleotide nucleotidyltransferase, impairs RNA import into mitochondria and causes respiratory-chain deficiency.
Other mechanisms affecting mitochondrial translation
Abnormal import of RNA into the mitochondria: PNPT1
PNPT1 encodes the mitochondrial polynucleotide phosphorylase (PNPase), which is predominantly localized in the mitochondrial intermembrane space and is a 3′–5′ exoribonuclease which acts together with SUV3 to form the RNA degradosome within the mitochondrial matrix [177]. Two siblings with severe encephalomyopathy, choreoathetotic movements, and combined respiratory-chain defects carried a homozygous PNPT1 missense mutation (c.1160A>G), which disrupts the trimerization of the protein. A defect of mitochondrial translation has been detected in the patient’s fibroblasts. Recently additional patients have been reported with recessive PNPT1 mutations and the clinical presentation of early onset of severe axonal neuropathy, optic atrophy, intellectual disability, auditory neuropathy, and chronic respiratory and gut disturbances [178], and severe Leigh syndrome [179]. Specific RNA processing intermediates derived from mitochondrial transcripts of the ND6 subunit of Complex I, as well as small mRNA fragments, accumulated in the subject’s myoblasts indicates that PNPase activity is essential for the correct maturation of the ND6 transcript [179].
Modification of rRNAs: MRM2
A homozygous missense mutation (c.567G>A; p.Gly189Arg) has been identified in a 7-year-old Italian boy with the clinical presentation of childhood-onset rapidly progressive encephalomyopathy and stroke-like episodes with multiple OXPHOS deficiency in skeletal muscle. MRM2 encodes an enzyme responsible for 2′-O-methyl modification at position U1369 in the human mitochondrial 16S rRNA. Although a confirmation of the clinical phenotype in a second independent patient is still lacking, it is possible that mutations in MRM2 cause a MELAS-like phenotype, and suggests the genetic screening of MRM2 in patients with a m.3243 A > G negative MELAS-like disease [180].
Summary
Here we have illustrated the large variety of clinical presentations caused by defects of mitochondrial translation. More detailed understanding of the molecular mechanisms involved in mitochondrial translation may reveal some insights on the tissue specific phenotypes. Processing and modifications of mt-tRNAs may provide novel approaches to develop treatment to defects of mitochondrial translation.
It has been recently shown that supplementation with cysteine (l-cysteine and N-acetyl-cysteine) improves mitochondrial translation in patients with reversible mitochondrial disease (TRMU, mt-tRNAGlu) and with m.3243A>G and m.8344A>G frequent mt-tRNA mutations [181], as absence of post-transcriptional modifications at the wobble positions of mt-tRNAs for LeuUUR and Lys has been related to MELAS and myoclonic epilepsy with ragged-red fiber (MERRF), respectively.
As another novel approach, leucyl tRNA synthetase is able to partially rescue defects caused by mutations in non-cognate mt-tRNAs and furthermore, a C-terminal peptide alone can enter mitochondria and interact with the same spectrum of mt-tRNAs as the entire synthetase in intact cells [182,183]. These data support the possibility that a small peptide may correct the biochemical defect associated with many mt-tRNA mutations, inferring a novel therapy for these disorders.
Abbreviations
- COX
cytochrome c oxidase
- LBSL
leukoencephalopathy with brainstem and spinal cord involvement and lactate elevation
- LSFC
French-Canadian variant of Leigh syndrome
- LTBL
leukoencephalopathy with thalamus and brainstem involvement and high lactate
- MLASA
myopathy, lactic acidosis, and sideroblastic anemia
- MELAS
mitochondrial encephalomyopathy, lactic acidosis and stroke-like episode
- mt-ARS
mitochondrial aminoacyl-tRNA synthetase
- mt-mRNA
mitochondrial mRNA
- mt-rRNA
mitochondrial rRNA
- mt-tRNA
mitochondrial tRNA
- OXPHOS
oxidative phosphorylation
- PNPase
polynucleotide phosphorylase
- SDR
short-chain dehydrogenase/reductase
Competing interests
The authors declare that there are no competing interests associated with the manuscript.
Funding
This work was supported by the Wellcome Centre for Mitochondrial Research [grant number 203105/Z/16/Z] which provides support to the Wellcome Trust Investigator [grant number 109915/Z/15/Z (to R.H.)]; the Medical Research Council (U.K.) [grant number MR/N025431/1]; the European Research Council [grant number 309548]; the Wellcome Trust Pathfinder Scheme [grant number 201064/Z/16/Z]; the Newton Fund (U.K./Turkey) [grant number MR/N027302/1]; the European Union H2020—Research and Innovation Actions (Solve-RD) [grant number SC1-PM-03-2017]; the Rotary Foundation (TRF) Global Grant Scholarship [grant number GG1862130].
Author contribution
V.B., G.R. and R.H. were equally involved in data collection and drafting of the manuscript.
References
- 1.Gorman G.S., Chinnery P.F., DiMauro S., Hirano M., Koga Y., McFarland R. et al. (2016) Mitochondrial diseases. Nat. Rev. Dis. Primers 2, 16080 10.1038/nrdp.2016.80 [DOI] [PubMed] [Google Scholar]
- 2.Viscomi C. and Zeviani M. (2017) MtDNA-maintenance defects: syndromes and genes. J. Inherit. Metab. Dis. 40, 587–599 10.1007/s10545-017-0027-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gustafsson C.M., Falkenberg M. and Larsson N.G. (2016) Maintenance and expression of mammalian mitochondrial DNA. Annu. Rev. Biochem. 85, 133–160 10.1146/annurev-biochem-060815-014402 [DOI] [PubMed] [Google Scholar]
- 4.Pearce S.F., Rebelo-Guiomar P., D’Souza A.R., Powell C.A., Van Haute L. and Minczuk M. (2017) Regulation of mammalian mitochondrial gene expression: recent advances. Trends Biochem. Sci. 42, 625–639 10.1016/j.tibs.2017.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Holzmann J., Frank P., Löffler E., Bennett K.L, Gerner C. and Rossmanith W. (2008) RNase P without RNA: identification and functional reconstitution of the human mitochondrial tRNA processing enzyme. Cell 135, 462–474 10.1016/j.cell.2008.09.013 [DOI] [PubMed] [Google Scholar]
- 6.Rackham O., Busch J.D., Matic S., Siira S.J., Kuznetsova I., Atanassov I. et al. (2016) Hierarchical RNA processing is required for mitochondrial ribosome assembly. Cell Rep. 16, 1874–1890 10.1016/j.celrep.2016.07.031 [DOI] [PubMed] [Google Scholar]
- 7.Rusecka J., Kaliszewska M., Bartnik E. and Tońska K. (2018) Nuclear genes involved in mitochondrial diseases caused by instability of mitochondrial DNA. J. Appl. Genet. 59, 43–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Greber B.J., Bieri P., Leibundgut M., Leitner A., Aebersold R., Boehringer D. et al. (2015) The complete structure of the 55S mammalian mitochondrial ribosome. Science 348, 303–308 10.1126/science.aaa3872 [DOI] [PubMed] [Google Scholar]
- 9.Amunts A., Brown A., Toots J., Scheres S.H.W. and Ramakrishnan V. (2015) The structure of the human mitochondrial ribosome. Science 348, 95–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Minczuk M. and D’Souza A.R., (2018) Mitochondrial transcription and translation: overview. Essays Biochem. 62, 309–320 10.1042/EBC20170102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boczonadi V. and Horvath R. (2014) Mitochondria: impaired mitochondrial translation in human disease. Int. J. Biochem. Cell. Biol. 48, 77–84 10.1016/j.biocel.2013.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stiles A.R, Simon M.T., Stover A., Eftekharian S., Khanlou N., Wang H.L. et al. (2016) Mutations in TFAM, encoding mitochondrial transcription factor A, cause neonatal liver failure associated with mtDNA depletion. Mol. Genet. Metab 119, 91–99 10.1016/j.ymgme.2016.07.001 [DOI] [PubMed] [Google Scholar]
- 13.Metodiev M.D., Thompson K., Alston C.L., Morris A.A., He L., Assouline Z. et al. (2016) Recessive mutations in TRMT10C cause defects in mitochondrial RNA processing and multiple respiratory chain deficiencies. Am. J. Hum. Genet. 99, 246 10.1016/j.ajhg.2016.06.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oerum S., Roovers M., Leichsenring M., Acquaviva-Bourdain C., Beermann F., Gemperle-Britschgi C. et al. (2017) Novel patient missense mutations in the HSD17B10 gene affect dehydrogenase and mitochondrial tRNA modification functions of the encoded protein. Biochim. Biophys. Acta 1863, 3294–3302 10.1016/j.bbadis.2017.09.002 [DOI] [PubMed] [Google Scholar]
- 15.Falk M.J., Gai X., Shigematsu M., Vilardo E., Takase R., McCormick E. et al. (2016) A novel HSD17B10 mutation impairing the activities of the mitochondrial RNase P complex causes X-linked intractable epilepsy and neurodevelopmental regression. RNA Biol. 13, 477–485 10.1080/15476286.2016.1159381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haack T.B., Kopajtich R., Freisinger P., Wieland T., Rorbach J., Nicholls T.J. et al. (2013) ELAC2 mutations cause a mitochondrial RNA processing defect associated with hypertrophic cardiomyopathy. Am. J. Hum. Genet. 93, 211–223 10.1016/j.ajhg.2013.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shinwari Z.M.A., Almesned A., Alakhfash A., Al-Rashdan A.M., Faqeih E., Al-Humaidi Z. et al. (2017) The phenotype and outcome of infantile cardiomyopathy caused by a homozygous ELAC2 mutation. Cardiology 137, 188–192 10.1159/000465516 [DOI] [PubMed] [Google Scholar]
- 18.Akawi N.A., Ben-Salem S., Hertecant J., John A., Pramathan T., Kizhakkedath P. et al. (2016) A homozygous splicing mutation in ELAC2 suggests phenotypic variability including intellectual disability with minimal cardiac involvement. Orphanet J. Rare Dis. 11, 139 10.1186/s13023-016-0526-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ghezzi D., Saada A., D’Adamo P., Fernandez-Vizarra E., Gasparini P., Tiranti V. et al. (2008) FASTKD2 nonsense mutation in an infantile mitochondrial encephalomyopathy associated with cytochrome c oxidase deficiency. Am. J. Hum. Genet. 83, 415–423 10.1016/j.ajhg.2008.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yoo D.H., Choi Y.C., Nam D.E., Choi S.S., Kim J.W., Choi B.O. et al. (2017) Identification of FASTKD2 compound heterozygous mutations as the underlying cause of autosomal recessive MELAS-like syndrome. Mitochondrion 35, 54–58 10.1016/j.mito.2017.05.005 [DOI] [PubMed] [Google Scholar]
- 21.Crosby A.H., Patel H., Chioza B.A., Proukakis C., Gurtz K., Patton M.A. et al. (2010) Defective mitochondrial mRNA maturation is associated with spastic ataxia. Am. J. Hum. Genet. 87, 655–660 10.1016/j.ajhg.2010.09.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mootha V.K., Lepage P., Miller K., Bunkenborg J., Reich M., Hjerrild M. et al. (2003) Identification of a gene causing human cytochrome c oxidase deficiency by integrative genomics. Proc. Natl. Acad. Sci. U.S.A. 100, 605–610 10.1073/pnas.242716699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oláhová M., Hardy S.A., Hall J., Yarham J., Haack T.B., Wilson W.C. et al. (2015) LRPPRC mutations cause early-onset multisystem mitochondrial disease outside of the French-Canadian population. Brain 138, 3503–3519 10.1093/brain/awv291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Han V.X., Tan T.S., Wang F.S. and Tay S.K. (2017) Novel LRPPRC mutation in a boy with mild Leigh syndrome, French-Canadian type outside of Québec. Child Neurol. Open 4, 10.1177/2329048X17737638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kang I., Chu C.T., Kaufman B.A.. The mitochondrial transcription factor TFAM in neurodegeneration: emerging evidence and mechanisms. FEBS Lett. 592, 793–811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Antonicka H. and Shoubridge E.A. (2015) Mitochondrial RNA granules are centers for posttranscriptional rna processing and ribosome biogenesis. Cell Rep. 1247, pii: S2211, 10.1016/j.celrep.2015.01.030 [DOI] [PubMed] [Google Scholar]
- 27.Popow J., Alleaume A.M., Curk T., Schwarzl T., Sauer S. and Hentze M.W. (2015) FASTKD2 is an RNA-binding protein required for mitochondrial RNA processing and translation. RNA 1873–1884 10.1261/rna.052365.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ramanan V.K., Nho K., Shen L., Risacher S.L., Kim S., McDonald B.C. et al. (2015) FASTKD2 is associated with memory and hippocampal structure in older adults. Mol. Psychiatry 20, 1197–1204 10.1038/mp.2014.142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Das S., Yeung K.T., Mahajan M.A. and Samuels H.H. (2014) Fas activated serine-threonine kinase domains 2 (FASTKD2) mediates apoptosis of breast and prostate cancer cells through its novel FAST2 domain. BMC Cancer 14, 852 10.1186/1471-2407-14-852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wilson W.C., Hornig-Do H.T., Bruni F., Chang J.H., Jourdain A.A., Martinou J.C. et al. (2014) A human mitochondrial poly(A) polymerase mutation reveals the complexities of post-transcriptional mitochondrial gene expression. Hum. Mol. Genet. 23, 6345–6355 10.1093/hmg/ddu352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Siira S.J, Spåhr H., Shearwood A.J., Ruzzenente B., Larsson N.G., Rackham O. et al. (2017) LRPPRC-mediated folding of the mitochondrial transcriptome. Nat. Commun. 8, 1532 10.1038/s41467-017-01221-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sasarman F., Brunel-Guitton C., Antonicka H., Wai T. and Shoubridge E.A. (2010) LRPPRC and SLIRP interact in a ribonucleoprotein complex that regulates posttranscriptional gene expression in mitochondria. Mol. Biol. Cell 21, 1315–1323 10.1091/mbc.e10-01-0047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ruzzenente B., Metodiev M.D., Wredenberg A., Bratic A., Park C.B., Camara Y. et al. (2012) LRPPRC is necessary for polyadenylation and coordination of translation of mitochondrial mRNAs. EMBO J. 31, 443–456 10.1038/emboj.2011.392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tian T., Ikeda J., Wang Y., Mamat S., Luo W., Aozasa K. et al. (2012) Role of leucine-rich pentatricopeptide repeat motif-containing protein (LRPPRC) for anti-apoptosis and tumourigenesis in cancers. Eur. J. Cancer 48, 2462–2473 10.1016/j.ejca.2012.01.018 [DOI] [PubMed] [Google Scholar]
- 35.Zou J., Yue F., Li W., Song K., Jiang X., Yi J. et al. (2014) Autophagy inhibitor LRPPRC suppresses mitophagy through interaction with mitophagy initiator Parkin. PLoS ONE 9, e94903 10.1371/journal.pone.0094903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Boczonadi V., Smith P.M., Pyle A., Gomez-Duran A., Schara U., Tulinius M. et al. (2013) Altered 2-thiouridylation impairs mitochondrial translation in reversible infantile respiratory chain deficiency. Hum. Mol. Genet. 22, 4602–4615 10.1093/hmg/ddt309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zeharia A., Shaag A., Pappo O., Mager-Heckel A.M., Saada A., Beinat M. et al. (2009) Acute infantile liver failure due to mutations in the TRMU gene. Am. J. Hum. Genet. 85, 401–407 10.1016/j.ajhg.2009.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schara U., von Kleist-Retzow J.C., Lainka E., Gerner P., Pyle A., Smith P.M. et al. (2011) Acute liver failure with subsequent cirrhosis as the primary manifestation of TRMU mutations. J. Inherit. Metab. Dis. 34, 197–201 10.1007/s10545-010-9250-z [DOI] [PubMed] [Google Scholar]
- 39.Uusimaa J., Jungbluth H., Fratter C., Crisponi G., Feng L., Zeviani M. et al. (2011) Reversible infantile respiratory chain deficiency is a unique, genetically heterogenous mitochondrial disease. J. Med. Genet. 48, 660–668 10.1136/jmg.2011.089995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gaignard P., Gonzales E., Ackermann O., Labrune P., Correia I., Therond P et al. (2013) Mitochondrial infantile liver disease due to TRMU gene mutations: three new cases. JIMD Rep. 11, 117–123 10.1007/8904_2013_230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ghezzi D, Baruffini E, Haack TB, Invernizzi F, Melchionda L, Dallabona C et al. (2012) Mutations of the mitochondrial-tRNA modifier MTO1 cause hyper- trophic cardiomyopathy and lactic acidosis. Am. J. Hum. Genet. 90, 1079–1087 10.1016/j.ajhg.2012.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Baruffini E., Dallabona C., Invernizzi F., Yarham J. W., Melchionda L., Blakely E. L. et al. (2013) MTO1 mutations are associated with hypertrophic cardiomyopathy and lactic acidosis and cause respiratory chain deficiency in humans and yeast. Hum. Mutat. 34, 1501–1509 10.1002/humu.22393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.O’Byrne J.J., Tarailo-Graovac M., Ghani A., Champion M., Deshpande C., Dursun A. et al. (2018) The genotypic and phenotypic spectrum of MTO1 deficiency. Mol. Genet. Metab. 123, 28–42 10.1016/j.ymgme.2017.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kopajtich R., Nicholls T.J., Rorbach J., Metodiev M.D., Freisinger P., Mandel H. et al. (2014) Mutations in GTPBP3 cause a mitochondrial translation defect associated with hypertrophic cardiomyopathy, lactic acidosis, and encephalopathy. Am. J. Hum. Genet. 95, 708–720 10.1016/j.ajhg.2014.10.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Van Haute L., Dietmann S., Kremer L., Hussain S., Pearce S.F., Powell C.A. et al. (2016) Deficient methylation and formylation of mt-tRNA(Met) wobble cytosine in a patient carrying mutations in NSUN3. Nat. Commun. 7, 12039 10.1038/ncomms12039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Powell C.A., Kopajtich R., D’Souza A.R., Rorbach J., Kremer L.S., Husain R.A. et al. (2015) TRMT5 mutations cause a defect in posttranscriptional modification of mitochondrial tRNA associated with multiple respiratory-chain deficiencies. Am. J. Hum. Genet. 97, 319–328 10.1016/j.ajhg.2015.06.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tarnopolsky M.A., Brady L., Tetreault M. and (2017) TRMT5 mutations are associated with features of complex hereditary spastic paraparesis. Neurology 89, 2210–2211 10.1212/WNL.0000000000004657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yarham J.W., Lamichhane T.N., Pyle A., Mattijssen S., Baruffini E., Bruni F. et al. (2014) Defective i6A37 modification of mitochondrial and cytosolic tRNAs results from pathogenic mutations in TRIT1 and its substrate tRNA. PLoS Genet. 10, e1004424 10.1371/journal.pgen.1004424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kernohan K.D., Dyment D.A., Pupavac M., Cramer Z., McBride A., Bernard G. et al. (2017) Matchmaking facilitates the diagnosis of an autosomal-recessive mitochondrial disease caused by biallelic mutation of the tRNA isopentenyltransferase (TRIT1) gene. Hum. Mutat. 38, 511–516 10.1002/humu.23196 [DOI] [PubMed] [Google Scholar]
- 50.Chakraborty P. K., Schmitz-Abe K., Kennedy E. K., Mamady H., Naas T., Durie D. et al. (2014) Mutations in TRNT1 cause congenital sideroblastic anemia with immunodeficiency, fevers, and developmental delay (SIFD). Blood 124, 2867–2871 10.1182/blood-2014-08-591370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.DeLuca A. P., Whitmore S. S., Barnes J., Sharma T. P., Westfall T. A., Scott C. A. et al. (2016) Hypomorphic mutations in TRNT1 cause retinitis pigmentosa with erythrocytic microcytosis. Hum. Mol. Genet. 25, 44–56 10.1093/hmg/ddv446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bykhovskaya Y., Casas K., Mengesha E., Inbal A. and Fischel-Ghodsian N. (2004) Missense mutation in pseudouridine synthase 1 (PUS1) causes mitochondrial myopathy and sideroblastic anemia (MLASA). Am. J. Hum. Genet. 74, 1303–1308 10.1086/421530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fernandez-Vizarra E., Berardinelli A., Valente L., Tiranti V. and Zeviani M. (2007) Nonsense mutation in pseudouridylate synthase 1 (PUS1) in two brothers affected by myopathy, lactic acidosis and sideroblastic anaemia (MLASA). J. Med. Genet. 44, 173–180 10.1136/jmg.2006.045252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Metodiev M.D., Assouline Z., Landrieu P., Chretien D., Bader-Meunier B., Guitton C. et al. (2015) Unusual clinical expression and long survival of a pseudouridylate synthase (PUS1) mutation into adulthood. Europ. J. Hum. Genet. 23, 880–882 10.1038/ejhg.2014.192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tucker E.J., Hershman S.G., Köhrer C., Belcher-Timme C.A., Patel J., Goldberger O.A. et al. (2011) Mutations in MTFMT underlie a human disorder of formylation causing impaired mitochondrial translation. Cell Metab. 14, 428–434 10.1016/j.cmet.2011.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Neeve V.C., Pyle A., Boczonadi V., Gomez-Duran A., Griffin H., Santibanez-Koref M. et al. (2013) Clinical and functional characterisation of the combined respiratory chain defect in two sisters due to autosomal recessive mutations in MTFMT. Mitochondrion 13, 743–748 10.1016/j.mito.2013.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Haack T.B., Gorza M., Danhauser K., Mayr J.A., Haberberger B. and Wieland T. (2014) Phenotypic spectrum of eleven patients and five novel MTFMT mutations identified by exome sequencing and candidate gene screening. Mol. Genet. Metab. 111, 342–352 10.1016/j.ymgme.2013.12.010 [DOI] [PubMed] [Google Scholar]
- 58.Boczonadi V, Bansagi B and Horvath R (2015) Reversible infantile mitochondrial diseases. J. Inherit. Metab. Dis 38, 427–435 10.1007/s10545-014-9784-6 [DOI] [PubMed] [Google Scholar]
- 59.Sturman J.A., Gaull G. and Raiha N.C. (1970) Absence of cystathionase in human fetal liver: is cystine essential? Science 169, 74–76 10.1126/science.169.3940.74 [DOI] [PubMed] [Google Scholar]
- 60.Guan M.X., Yan Q., Li X., Bykhovskaya Y., Gallo-Teran J., Hajek P. et al. (2006) Mutation in TRMU related to transfer RNA modification modulates the phenotypic expression of the deafness-associated mitochondrial 12S ribosomal RNA mutations. Am. J. Hum. Genet. 79, 291–302 10.1086/506389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li R., Li X., Yan Q., Mo J. Q. and Guan M.X. (2003) Identification and characterization of mouse MTO1 gene related to mitochondrial tRNA modification. Biochim. Biophys. Acta 1629, 53–59 10.1016/S0167-4781(03)00160-X [DOI] [PubMed] [Google Scholar]
- 62.Wedatilake Y., Niazi R., Fassone E., Powell C.A., Pearce S., Plagnol V. et al. (2016) TRNT1 deficiency: clinical, biochemical and molecular genetic features. Orphanet J. Rare Dis. 11, 90 10.1186/s13023-016-0477-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lougaris V., Chou J., Baronio M., Gazzurelli L., Lorenzini T., Soresina A. et al. (2017) Novel biallelic TRNT1 mutations resulting in sideroblastic anemia, combined B and T cell defects, hypogammaglobulinemia, recurrent infections, hypertrophic cardiomyopathy and developmental delay. Clin. Immunol. 188, 20–22 [DOI] [PubMed] [Google Scholar]
- 64.Hull S., Malik A.N., Arno G., Mackay D.S., Plagnol V. and Michaelides M. (2016) Expanding the phenotype of TRNT1-related immunodeficiency to include childhood cataract and inner retinal dysfunction. JAMA Ophthalmol. 134, 1049–1053 10.1001/jamaophthalmol.2015.5833 [DOI] [PubMed] [Google Scholar]
- 65.Barton C., Kausar S., Kerr D., Bitetti S. and Wynn R. (2017) SIFD as a novel cause of severe fetal hydrops and neonatal anaemia with iron loading and marked extramedullary haemopoiesis. J. Clin. Pathol. 71, 275–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chen J. and Patton J. R. (1999) Cloning and characterization of a mammalian pseudouridine synthase. RNA 5, 409–419 10.1017/S1355838299981591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Riley L.G., Cooper S., Hickey P., Rudinger-Thirion J., McKenzie M., Compton A. et al. (2010) Mutation of the mitochondrial tyrosyl-tRNA synthetase gene YARS2, causes myopathy, lactic acidosis, and sideroblastic anemia—MLASA syndrome. Am. J. Hum. Genet. 87, 52–59 10.1016/j.ajhg.2010.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zeharia A., Fischel-Ghodsian N., Casas K., Bykhocskaya Y., Tamari H., Lev D. et al. (2005) Mitochondrial myopathy, sideroblastic anemia, and lactic acidosis: an autosomal recessive syndrome in Persian Jews caused by a mutation in the PUS1 gene. J. Child Neurol. 20, 449–452 10.1177/08830738050200051301 [DOI] [PubMed] [Google Scholar]
- 69.Cao M., Donà M., Valentino M.L., Valentino L., Semplicini C., Maresca A. et al. (2016) Clinical and molecular study in a long-surviving patient with MLASA syndrome due to novel PUS1 mutations. Neurogenetics 17, 65–70 10.1007/s10048-015-0465-x [DOI] [PubMed] [Google Scholar]
- 70.La Piana R., Weraarpachai W., Ospina L.H., Tetreault M., Majewski J. and (2017) Identification and functional characterization of a novel MTFMT mutation associated with selective vulnerability of the visual pathway and a mild neurological phenotype. Neurogenetics 18, 97–103 10.1007/s10048-016-0506-0 [DOI] [PubMed] [Google Scholar]
- 71.Boczonadi V., Jennings M.J. and Horvath R. (2017) The role of tRNA synthetases in neurological and neuromuscular disorders. FEBS Lett., 592, 703–717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sissler M., González-Serrano L.E. and Westhof E. (2017) Recent advances in mitochondrial aminoacyl-tRNA synthetases and disease. Trends Mol. Med. 23, 693–708 10.1016/j.molmed.2017.06.002 [DOI] [PubMed] [Google Scholar]
- 73.Oprescu S.N., Griffin L.B., Beg A.A. and Antonellis A. (2017) Predicting the pathogenicity of aminoacyl-tRNA synthetase mutations. Methods 113, 139–151 10.1016/j.ymeth.2016.11.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Meyer-Schuman R. and Antonellis A. (2017) Emerging mechanisms of aminoacyl-tRNA synthetase mutations in recessive and dominant human disease. Hum. Mol. Genet. 26, R114–R127 10.1093/hmg/ddx231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rajendran V., Kalita P., Shukla H., Kumar A. and Tripathi T. (2018) Aminoacyl-tRNA synthetases: Structure, function, and drug discovery. Int. J. Biol. Macromol. 111, 400–414 10.1016/j.ijbiomac.2017.12.157 [DOI] [PubMed] [Google Scholar]
- 76.Scheper G.C., van der Klok T., van Andel R.J., van Berkel C.G., Sissler M., Smet J. et al. (2007) Mitochondrial aspartyl-tRNA synthetase deficiency causes leukoencephalopathy with brain stem and spinal cord involvement and lactate elevation. Nat. Genet. 39, 534–539 10.1038/ng2013 [DOI] [PubMed] [Google Scholar]
- 77.Isohanni P., Linnankivi T., Buzkova J., Lönnqvist T., Pihko H., Valanne L. et al. (2010) DARS2 mutations in mitochondrial leucoencephalopathy and multiple sclerosis. J. Med. Genet. 47, 66–70 10.1136/jmg.2009.068221 [DOI] [PubMed] [Google Scholar]
- 78.Miyake N., Yamashita S., Kurosawa K., Miyatake S., Tsurusaki Y., Doi H. et al. (2011) A novel homozygous mutation of DARS2 may cause a severe LBSL variant. Clin. Genet. 80, 293–296 10.1111/j.1399-0004.2011.01644.x [DOI] [PubMed] [Google Scholar]
- 79.van Berge L., Hamilton E.M., Linnankivi T., Uziel G., Steenweg M.E., Isohanni P. et al. (2014) Leukoencephalopathy with brainstem and spinal cord involvement and lactate elevation: clinical and genetic characterization and target for therapy. Brain 137, 1019–1029 10.1093/brain/awu026 [DOI] [PubMed] [Google Scholar]
- 80.Shimojima K., Higashiguchi T., Kishimoto K., Miyatake S., Miyake N., Takanashi J.I. et al. (2017) A novel DARS2 mutation in a Japanese patient with leukoencephalopathy with brainstem and spinal cord involvement but no lactate elevation. Hum. Genome Var. 4, 17051 10.1038/hgv.2017.51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Pinto W.B. and de Souza P.V. (2014) DARS2 gene clinical spectrum: new ideas regarding an underdiagnosed leukoencephalopathy. Brain 137, e289 10.1093/brain/awu134 [DOI] [PubMed] [Google Scholar]
- 82.Synofzik M., Schicks J., Lindig T., Biskup S., Schmidt T., Hansel J. et al. (2011) Acetazolamide-responsive exercise-induced episodic ataxia associated with a novel homozygous DARS2 mutation. J. Med. Genet. 48, 713–715 10.1136/jmg.2011.090282 [DOI] [PubMed] [Google Scholar]
- 83.Edvardson S., Shaag A., Kolesnikova O., Gomori J.M., Tarassov I., Einbinder T. et al. (2007) Deleterious mutation in the mitochondrial arginyl-transfer RNA synthetase gene is associated with pontocerebellar hypoplasia. Am. J. Hum. Genet. 81, 857–862 10.1086/521227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Rankin J., Brown R., Dobyns W.B., Harington J., Patel J., Quinn M. et al. (2010) Pontocerebellar hypoplasia type 6: A British case with PEHO-like features. Am. J. Med. Genet. 152, 2079–2084 10.1002/ajmg.a.33531 [DOI] [PubMed] [Google Scholar]
- 85.Cassandrini D., Cilio M.R., Bianchi M., Doimo M., Balestri M., Tessa A. et al. (2013) Pontocerebellar hypoplasia type 6 caused by mutations in RARS2: definition of the clinical spectrum and molecular findings in five patients. J. Inherit. Metab. Dis. 36, 43–53 10.1007/s10545-012-9487-9 [DOI] [PubMed] [Google Scholar]
- 86.Li Z., Schonberg R., Guidugli L., Johnson A.K., Arnovitz S., Yang S. et al. (2015) A novel mutation in the promoter of RARS2 causes pontocerebellar hypoplasia in two siblings. J. Hum. Genet. 60, 363–369 10.1038/jhg.2015.31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lühl S., Bode H., Schlötzer W., Bartsakoulia M, Horvath R., Abicht A. et al. (2016) Novel homozygous RARS2 mutation in two siblings without pontocerebellar hypoplasia - further expansion of the phenotypic spectrum. Orphanet J. Rare Dis. 11, 140 10.1186/s13023-016-0525-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Steenweg M.E., Ghezzi D., Haack T., Abbink T.E., Martinelli D., van Berkel C.G. et al. (2012) Leukoencephalopathy with thalamus and brainstem involvement and high lactate ‘LTBL’ caused by EARS2 mutations. Brain 135, 1387–1394 10.1093/brain/aws070 [DOI] [PubMed] [Google Scholar]
- 89.Talim B., Pyle A., Griffin H., Topaloglu H., Tokatli A., Keogh M.J. et al. (2013) Multisystem fatal infantile disease caused by a novel homozygous EARS2 mutation. Brain 136, e228 10.1093/brain/aws197 [DOI] [PubMed] [Google Scholar]
- 90.Kevelam S.H., Klouwer F.C., Fock J.M., Salomons G.S., Bugiani M. and van der Knaap M.S. (2016) Absent thalami caused by a homozygous EARS2 mutation: expanding disease spectrum of LTBL. Neuropediatrics 47, 64–67 [DOI] [PubMed] [Google Scholar]
- 91.Güngör O., Özkaya A.K., Şahin Y., Güngör G., Dilber C. and Aydın K.A. (2016) compound heterozygous EARS2 mutation associated with mild leukoencephalopathy with thalamus and brainstem involvement and high lactate (LTBL). Brain Dev. 38, 857–861 10.1016/j.braindev.2016.04.002 [DOI] [PubMed] [Google Scholar]
- 92.Şahin S., Cansu A., Kalay E., Dinçer T., Kul S., Çakır İ.M. et al. (2016) Leukoencephalopathy with thalamus and brainstem involvement and high lactate caused by novel mutations in the EARS2 gene in two siblings. J. Neurol. Sci. 365, 54–58 10.1016/j.jns.2016.04.008 [DOI] [PubMed] [Google Scholar]
- 93.Bayat V., Thiffault I., Jaiswal M., Tétreault M., Donti T., Sasarman F. et al. (2012) Mutations in the mitochondrial methionyl-tRNA synthetase cause a neurodegenera- tive phenotype in flies and a recessive ataxia (ARSAL) in humans. PLoS Biol. 10, e1001288 10.1371/journal.pbio.1001288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Webb B.D., Wheeler P.G., Hagen J.J., Cohen N., Linderman M.D., Diaz G.A. et al. (2015) Novel, compound heterozygous, single-nucleotide variants in MARS2 associated with developmental delay, poor growth, and sensorineural hearing loss. Hum. Mutat. 36, 587–592 10.1002/humu.22781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Elo J.M., Yadavalli S.S., Euro L., Isohanni P., Götz A., Carroll C.J. et al. (2012) Mitochondrial phenylalanyl-tRNA synthetase mutations underlie fatal infantile Alpers encephalopathy. Hum. Mol. Genet. 21, 4521–4529 10.1093/hmg/dds294 [DOI] [PubMed] [Google Scholar]
- 96.Shamseldin H.E., Alshammari M., Al-Sheddi T., Salih M.A., Alkhalidi H., Kentab A. et al. (2012) Genomic analysis of mitochondrial diseases in a consanguineous population reveals novel candidate disease genes. J. Med. Genet. 49, 234–241 10.1136/jmedgenet-2012-100836 [DOI] [PubMed] [Google Scholar]
- 97.Yang Y., Liu W., Fang Z., Shi J., Che F., He C. et al. . et al. , 2016. A newly identified missense mutation in FARS2 causes autosomal-recessive spastic paraplegia. Hum. Mutat. 37, 165–169 10.1002/humu.22930 [DOI] [PubMed] [Google Scholar]
- 98.Götz A., Tyynismaa H., Euro L., Ellonen P., Hyötyläinen T., Ojala T. et al. (2011) Exome sequencing identifies mitochondrial alanyl-tRNA synthetase mutations in infantile mitochondrial cardiomyopathy. Am. J. Hum. Genet. 88, 635–642 10.1016/j.ajhg.2011.04.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Taylor R.W., Pyle A., Griffin H., Blakely E.L., Duff J., He L. et al. (2014) Use of whole-exome sequencing to determine the genetic basis of multiple mitochondrial respiratory chain complex deficiencies. JAMA 312, 68–77 10.1001/jama.2014.7184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Dallabona C., Diodato D., Kevelam S.H., Haack T.B., Wong L.J., Salomons G.S. et al. (2014) Novel (ovario) leukodystrophy related to AARS2 mutations. Neurology 82, 2063–2071 10.1212/WNL.0000000000000497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lynch D.S., Zhang W.J., Lakshmanan R., Kinsella J.A., Uzun G.A., Karbay M. et al. (2016) Analysis of mutations in AARS2 in a series of CSF1R-negative patients with adult-onset leukoencephalopathy with axonal spheroids and pigmented glia. JAMA Neurol. 73, 1433–1439 10.1001/jamaneurol.2016.2229 [DOI] [PubMed] [Google Scholar]
- 102.Szpisjak L., Zsindely N., Engelhardt J.I., Vecsei L., Kovacs G.G. and Klivenyi P. (2017) Novel AARS2 gene mutation producing leukodystrophy: a case report. J. Hum. Genet. 62, 329–333 10.1038/jhg.2016.126 [DOI] [PubMed] [Google Scholar]
- 103.Sasarman F., Nishimura T., Thiffault I. and Shoubridge E.A. (2012) A novel mutation in YARS2 causes myopathy with lactic acidosis and sideroblastic anemia. Hum. Mutat. 33, 1201–1206 10.1002/humu.22098 [DOI] [PubMed] [Google Scholar]
- 104.Shahni R., Wedatilake Y., Cleary M.A., Lindley K.J., Sibson K.R. and Rahman S. (2013) A distinct mitochondrial myopathy, lactic acidosis and sideroblastic anemia (MLASA) phenotype associates with YARS2 mutations. Am. J. Med. Genet. 161, 2334–2338 10.1002/ajmg.a.36065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Riley L.G., Menezes M.J., Rudinger-Thirion J., Duff R., de Lonlay P., Rotig A. et al. (2013) Phenotypic variability and identification of novel YARS2 mutations in YARS2 mitochondrial myopathy, lactic acidosis and sideroblastic anaemia. Orphanet J. Rare Dis. 8, 193 10.1186/1750-1172-8-193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Nakajima J., Eminoglu T.F., Vatansever G., Nakashima M., Tsurusaki Y., Saitsu H. et al. (2014) A novel homozygous YARS2 mutation causes severe myopathy, lactic acidosis, and sideroblastic anemia 2. J. Hum. Genet. 59, 229–232 10.1038/jhg.2013.143 [DOI] [PubMed] [Google Scholar]
- 107.Belostotsky R., Ben-Shalom E., Rinat C., Becker-Cohen R., Feinstein S., Zeligson S. et al. (2011) Mutations in the mitochondrial seryl-tRNA synthetase cause hyperuricemia, pulmonary hypertension, renal failure in infancy and alkalosis, HUPRA syndrome. Am. J. Hum. Genet. 88, 193–200 10.1016/j.ajhg.2010.12.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Linnankivi T., Neupane N., Richter U., Isohanni P. and Tyynismaa H. (2016) Splicing defect in mitochondrial Seryl-tRNA synthetase gene causes progressive spastic paresis instead of hupra syndrome. Hum. Mutat. 37, 884–888 10.1002/humu.23021 [DOI] [PubMed] [Google Scholar]
- 109.Pierce S.B., Chisholm K.M., Lynch E.D., Lee M.K., Walsh T., Opitz J.M. et al. (2011) Mutations in mitochondrial histidyl tRNA synthetase HARS2 cause ovarian dysgenesis and sensorineural hearing loss of Perrault syndrome. Proc. Natl. Acad. Sci. U.S.A. 108, 6543–6548 10.1073/pnas.1103471108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Pierce S.B., Gersak K., Michaelson-Cohen R., Walsh T., Lee M.K., Malach D. et al. (2013) Mutations in LARS2, encoding mitochondrial leucyl-tRNA synthetase, lead to premature ovarian failure and hearing loss in Perrault syndrome. Am. J. Hum. Genet. A 92, 614–620 10.1016/j.ajhg.2013.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Soldà G., Caccia S., Robusto M., Chiereghin C., Castorina P., Ambrosetti U. et al. (2016) First independent replication of the involvement of LARS2 in Perrault syndrome by whole-exome sequencing of an Italian family. J. Hum. Genet. 61, 295–300 10.1038/jhg.2015.149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Demain L.A., Urquhart J.E., O’Sullivan J., Williams S.G., Bhaskar S.S. and Jenkinson E.M. (2017) Expanding the genotypic spectrum of Perrault syndrome. Clin. Genet. 91, 302–312 10.1111/cge.12776 [DOI] [PubMed] [Google Scholar]
- 113.Riley L.G., Rudinger-Thirion J., Schmitz-Abe K., Thorburn D.R., Davis R.L., Teo J. et al. (2016) LARS2 variants associated with hydrops, lactic acidosis, sideroblastic anemia, and multisystem failure. JIMD Rep. 28, 49–57 10.1007/8904_2015_515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Diodato D., Melchionda L., Haack T.B., Dallabona C., Baruffini E., Donnini C. et al. (2014) VARS2 and TARS2 mutations in patients with mitochondrial encephalomyopathies. Hum. Mutat. 35, 983–989 10.1002/humu.22590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Sofou K., Kollberg G., Holmstrom M., Davila M., Darin N., Gustafsson C. M. et al. (2015) Whole exome sequencing reveals mutations in NARS2 and PARS2, encoding the mitochondrial asparaginyl-tRNA synthetase and prolyl-tRNA synthetase in patients with Alpers syndrome. Mol. Genet. Genomic Med. 3, 59–68 10.1002/mgg3.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Vanlander A.V., Menten B., Smet J., De Meirleir L., Sante T., De Paepe B. et al. (2015) Two siblings with homozygous pathogenic splice-site variant in mitochondrial asparaginyl-tRNA synthetase (NARS2). Hum. Mutat. 36, 222–231 10.1002/humu.22728 [DOI] [PubMed] [Google Scholar]
- 117.Simon M., Richard E.M., Wang X., Shahzad M., Huang V.H., Qaiser T.A. et al. (2015) Mutations of human NARS2, encoding the mitochondrial asparaginyl-tRNA synthetase, cause nonsyndromic deafness and Leigh syndrome. PLoS Genet. 11, e1005097 10.1371/journal.pgen.1005097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Mizuguchi T., Nakashima M., Kato M., Yamada K., Okanishi T., Ekhilevitch N. et al. (2017) PARS2 and NARS2 mutations in infantile-onset neurodegenerative disorder. J. Hum. Genet. 62, 525–529 10.1038/jhg.2016.163 [DOI] [PubMed] [Google Scholar]
- 119.Coughlin C.R., Scharer G.H., Friederich M.W., Yu H.C., Geiger E.A., Creadon-Swindell G. et al. (2015) Mutations in the mitochondrial cysteinyl-tRNA synthase gene, CARS2, lead to a severe epileptic encephalopathy and complex movement disorder. J. Med. Genet. 52, 532–540 10.1136/jmedgenet-2015-103049 [DOI] [PubMed] [Google Scholar]
- 120.Schwartzentruber J., Buhas D., Majewski J., Sasarman F., Papillon-Cavanagh S., Thiffault I. et al. (2014) Mutation in the nuclear-encoded mitochondrial isoleucyl-tRNA synthetase IARS2 in patients with cataracts, growth hormone deficiency with short stature, partial sensorineural deafness, and peripheral neuropathy or with Leigh syndrome. Hum. Mutat. 35, 1285–1289 [DOI] [PubMed] [Google Scholar]
- 121.Moosa S., Haagerup A., Gregersen P.A., Petersen K.K., Altmüller J., Thiele H. et al. (2017) Confirmation of CAGSSS syndrome as a distinct entity in a Danish patient with a novel homozygous mutation in IARS2. Am. J. Med. Genet. A. 173, 1102–1108 10.1002/ajmg.a.38116 [DOI] [PubMed] [Google Scholar]
- 122.Baertling F., Alhaddad B., Seibt A., Budaeus S., Meitinger T., Strom T.M. et al. (2017) Neonatal encephalocardiomyopathy caused by mutations in VARS2. Metab. Brain Dis. 32, 267–270 10.1007/s11011-016-9890-2 [DOI] [PubMed] [Google Scholar]
- 123.Alsemari A., Al-Younes B., Goljan E., Jaroudi D., BinHumaid F., Meyer B.F. et al. (2017) Recessive VARS2 mutation underlies a novel syndrome with epilepsy, mental retardation, short stature, growth hormone deficiency, and hypogonadism. Hum. Genomics 11, 28 10.1186/s40246-017-0124-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Musante L., Püttmann L., Kahrizi K., Garshasbi M., Hu H., Stehr H. et al. (2017) Mutations of the aminoacyl-tRNA-synthetases SARS and WARS2 are implicated in the etiology of autosomal recessive intellectual disability. Hum. Mutat. 38, 621–636 10.1002/humu.23205 [DOI] [PubMed] [Google Scholar]
- 125.Wortmann S.B., Timal S., Venselaar H., Wintjes L.T., Kopajtich R., Feichtinger R.G. et al. (2017) Biallelic variants in WARS2 encoding mitochondrial tryptophanyl-tRNA synthase in six individuals with mitochondrial encephalopathy. Hum. Mutat. 38, 1786–1795 10.1002/humu.23340 [DOI] [PubMed] [Google Scholar]
- 126.Theisen B.E., Rumyantseva A., Cohen J.S., Alcaraz W.A., Shinde D.N., Tang S. et al. (2017) Deficiency of WARS2, encoding mitochondrial tryptophanyl tRNA synthetase, causes severe infantile onset leukoencephalopathy. Am. J. Med. Genet. A 173, 2505–2510 10.1002/ajmg.a.38339 [DOI] [PubMed] [Google Scholar]
- 127.Burke E.A., Frucht S.J., Thompson K., Wolfe L.A., Yokoyama T., Bertoni M. et al. (2017) Biallelic mutations in mitochondrial tryptophanyl-tRNA synthetase cause levodopa-rresponsive infantile-onset parkinsonism. Clin. Genet. 93, 712–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Sofou K., Kollberg G., Holmstrom M., Davila M., Darin N., Gustafsson C. M. et al. (2015) Whole exome sequencing reveals mutations in NARS2 and PARS2, encoding the mitochondrial asparaginyl-tRNA synthetase and prolyl-tRNA synthetase, in patients with Alpers syndrome. Mol. Genet. Genomic Med. 3, 59–68 10.1002/mgg3.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Antonellis A., Ellsworth R.E., Sambuughin N., Puls I., Abel A., Lee-Lin S.Q. et al. (2003) Glycyl tRNA synthetase mutations in Charcot-Marie-Tooth disease type 2D and distal spinal muscular atrophy type V. Am. J. Hum. Genet. 72, 1293–1299 10.1086/375039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Oprescu S.N., Chepa-Lotrea X., Takase R., Golas G., Markello T.C., Adams D.R. et al. (2017) Compound heterozygosity for loss-of-function GARS variants results in a multisystem developmental syndrome that includes severe growth retardation. Hum. Mutat. 38, 1412–1420 10.1002/humu.23287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Nafisinia M., Riley L.G., Gold W.A., Bhattacharya K., Broderick C.R., Thorburn D.R. et al. (2017) Compound heterozygous mutations in glycyl-tRNA synthetase (GARS) cause mitochondrial respiratory chain dysfunction. PLoS ONE 12, e0178125 10.1371/journal.pone.0178125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.McMillan H.J., Schwartzentruber J., Smith A., Lee S., Chakraborty P., Bulman D.E. et al. (2014) Compound heterozygous mutations in glycyl-tRNA synthetase are a proposed cause of systemic mitochondrial disease. BMC Med. Genet. 15, 36 10.1186/1471-2350-15-36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Kohda M., Tokuzawa Y., Kishita Y., Nyuzuki H., Moriyama Y., Mizuno Y. et al. (2016) A comprehensive genomic analysis reveals the genetic landscape of mitochondrial respiratory chain complex deficiencies. PLoS Genet. 12, e1005679 10.1371/journal.pgen.1005679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Verrigni D., Diodato D., Di Nottia M., Torraco A., Bellacchio E., Rizza T. et al. (2017) Novel mutations in KARS cause hypertrophic cardiomyopathy and combined mitochondrial respiratory chain defect. Clin. Genet. 91, 918–923 10.1111/cge.12931 [DOI] [PubMed] [Google Scholar]
- 135.McMillan H.J., Humphreys P., Smith A., Schwartzentruber J., Chakraborty P., Bulman D.E. et al. (2015) Congenital visual impairment and progressive microcephaly due to lysyl-transfer ribonucleic acid (RNA) synthetase (KARS) mutations: the expanding phenotype of aminoacyl-transfer RNA synthetase mutations in human disease. J. Child Neurol. 30, 1037–1043 10.1177/0883073814553272 [DOI] [PubMed] [Google Scholar]
- 136.Santos-Cortez R.L., Lee K., Azeem Z., Antonellis P.J., Pollock L.M., Khan S. et al. (2013) Mutations in KARS, encoding lysyl-tRNA synthetase, cause autosomal-recessive nonsyndromic hearing impairment DFNB89. Am. J. Hum. Genet. 93, 132–140 10.1016/j.ajhg.2013.05.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.McLaughlin H.M., Sakaguchi R., Liu C., Igarashi T., Pehlivan D., Chu K. et al. (2010) Compound heterozygosity for loss-of-function lysyl-tRNA synthetase mutations in a patient with peripheral neuropathy. Am. J. Hum. Genet. 87, 560–566 10.1016/j.ajhg.2010.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Bonnefond L., Fender A., Rudinger-Thirion J., Giegé R., Florentz C. and Sissler M, Toward the full set of human mitochondrial aminoacyl-tRNA synthetases: characterization of AspRS and TyrRS. Biochemistry 44, 4805–4816 10.1021/bi047527z [DOI] [PubMed] [Google Scholar]
- 139.Carapito C., Kuhn L., Karim L., Rompais M., Rabilloud T., Schwenzer H. et al. (2017) Two proteomic methodologies for defining N-termini of mature human mitochondrial aminoacyl-tRNA synthetases. Methods 113, 111–119 10.1016/j.ymeth.2016.10.012 [DOI] [PubMed] [Google Scholar]
- 140.Ognjenović J. and Simonović M. (2017) Human aminoacyl-tRNA synthetases in diseases of the nervous system. RNA Biol. 23, 1–12 10.1080/15476286.2017.1330245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Steenweg M.E., Ghezzi D., Haack T., Abbink T.E., Martinelli D., van Berkel C.G. et al. (2012) Leukoencephalopathy with thalamus and brainstem involvement and high lactate ‘LTBL’ caused by EARS2 mutations. Brain 135, 1387–1394 10.1093/brain/aws070 [DOI] [PubMed] [Google Scholar]
- 142.Euro L., Konovalova S., Asin-Cayuela J., Tulinius M., Griffin H., Horvath R. et al. (2015) Structural modeling of tissue-specific mitochondrial alanyl-tRNA synthetase (AARS2) defects predicts differential effects on aminoacylation. Front. Genet. 6, 21 10.3389/fgene.2015.00021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Kohda M., Tokuzawa Y., Kishita Y., Nyuzuki H., Moriyama Y., Mizuno Y. et al. (2016) A Comprehensive genomic analysis reveals the genetic landscape of mitochondrial respiratory chain complex deficiencies. PLoS Genet. 12, e1005679 10.1371/journal.pgen.1005679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Kamps R., Szklarczyk R., Theunissen T.E., Hellebrekers D.M.E.I., Sallevelt S.C.E.H., Boesten I.B. et al. (2018) Genetic defects in mtDNA-encoded protein translation cause pediatric, mitochondrial cardiomyopathy with early-onset brain disease. Eur. J. Hum. Genet. 10.1038/s41431-017-0058-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Newman W.G., Friedman T.B. and Conway G.S. (2014) Perrault syndrome. In SourceGeneReviews® (Adam M.P., Ardinger H.H., Pagon R.A., Wallace S.E., Bean L.J.H., Stephens K. and Amemiya A., eds), pp. 1993–2018, University of Washington, Seattle, Seattle (WA) [Google Scholar]
- 146.Galmiche L., Serre V., Beinat M., Assouline Z., Lebre A.S., Chretien D. et al. (2011) Exome sequencing identifies MRPL3 mutation in mitochondrial cardiomyopathy. Hum. Mutat. 32, 1225–1231 10.1002/humu.21562 [DOI] [PubMed] [Google Scholar]
- 147.Miller C., Saada A., Shaul N., Shabtai N., Ben-Shalom E., Shaag A. et al. (2004) Defective mitochondrial translation caused by a ribosomal protein (MRPS16) mutation. Ann. Neurol. 56, 734–738 10.1002/ana.20282 [DOI] [PubMed] [Google Scholar]
- 148.Saada A., Shaag A., Arnon S., Dolfin T., Miller C., Fuchs-Telem D. et al. (2007) Antenatal mitochondrial disease caused by mitochondrial ribosomal protein (MRPS22) mutation. J. Med. Genet. 44, 784–786 10.1136/jmg.2007.053116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Smits P., Saada A., Wortmann S.B., Heister A.J., Brink M., Pfundt R. et al. (2011) Mutation in mitochondrial ribosomal protein MRPS22 leads to Cornelia de Lange-like phenotype, brain abnormalities and hypertrophic cardiomyopathy. Eur. J. Hum. Genet. 19, 39–399 10.1038/ejhg.2010.214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Carroll C.J., Isohanni P., Pöyhönen R., Euro L., Richter U., Brilhante V. et al. (2013) Whole- exome sequencing identifies a mutation in the mitochondrial ribosome protein MRPL44 to underlie mitochondrial infantile cardiomyopathy. J. Med. Genet. 50, 151–159 10.1136/jmedgenet-2012-101375 [DOI] [PubMed] [Google Scholar]
- 151.Distelmaier F., Haack T.B., Catarino C.B., Gallenmüller C., Rodenburg R.J., Strom T.M. et al. (2015) MRPL44 mutations cause a slowly progressive multisystem disease with childhood-onset hypertrophic cardiomyopathy. Neurogenetics 16, 319–323 10.1007/s10048-015-0444-2 [DOI] [PubMed] [Google Scholar]
- 152.Serre V., Rozanska A., Beinat M., Chretien D., Boddaert N., Munnich A. et al. (2013) Mutations in mitochondrial ribosomal protein MRPL12 leads to growth retardation, neuro- logical deterioration and mitochondrial translation deficiency. Biochim. Biophys. Acta 1832, 1304–1312 10.1016/j.bbadis.2013.04.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Richman T.R., Ermer J.A., Davies S.M., Perks K.L., Viola H.M., Shearwood A.M. et al. (2015) Mutation in MRPS34 compromises protein synthesis and causes mitochondrial dysfunction. PLoS Genet. 11, e1005089 10.1371/journal.pgen.1005089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Lake N.J., Webb B.D., Stroud D.A., Richman T.R., Ruzzenente B., Compton A.G. et al. (2017) Biallelic mutations in MRPS34 lead to instability of the small mitoribosomal subunit and Leigh syndrome. Am. J. Hum. Genet. 101, 239–254 10.1016/j.ajhg.2017.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Janer A., Antonicka H., Lalonde E., Nishimura T., Sasarman F., Brown G.K. et al. (2012) An RMND1 Mutation causes encephalopathy associated with multiple oxidative phosphorylation complex deficiencies and a mitochondrial translation defect. Am. J. Hum. Genet. 91, 737–743 10.1016/j.ajhg.2012.08.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Garcia-Diaz B., Barros M.H., Sanna-Cherchi S., Emmanuele V., Akman H.O., Ferreiro- Barros C.C. et al. (2012) Infantile encephaloneuromyopathy and defective mitochondrial translation are due to a homozygous RMND1 mutation. Am. J. Hum. Genet. 91, 729–736 10.1016/j.ajhg.2012.08.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Ng Y.S., Alston C.L., Diodato D., Morris A.A., Ulrick N., Kmoch S. et al. (2016) The clinical, biochemical and genetic features associated with RMND1-related mitochondrial disease. J. Med. Genet. 10.1136/jmedgenet-2016-103910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Coenen M.J.H., Antonicka H., Ugalde C., Sasarman F., Rossi R., Angelien Heister J.G.A.M. et al. (2004) Mutant mitochondrial elongation factor G1 and combined oxidative phosphorylation deficiency. N. Engl. J. Med. 351, 2080–2086 10.1056/NEJMoa041878 [DOI] [PubMed] [Google Scholar]
- 159.Ravn K., Neland M., Wibrand F., Duno M. and Ostergaard E. (2016) Hearing impairment and renal failure associated with RMND1 mutations. Am. J. Med. Genet. A. 170A, 142–147 10.1002/ajmg.a.37399 [DOI] [PubMed] [Google Scholar]
- 160.Smits P., Antonicka H., van Hasselt P.M., Weraarpachai W., Haller W., Schreurs M. et al. (2011) Mutation in subdomain G’ of mitochondrial elongation factor G1 is associated with combined OXPHOS deficiency in fibroblasts but not in muscle. Europ. J. Hum. Genet. 19, 275–279 10.1038/ejhg.2010.208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Kohda M., Tokuzawa Y., Kishita Y., Nyuzuki H., Moriyama Y., Mizuno Y. et al. (2016) A comprehensive genomic analysis reveals the genetic landscape of mitochondrial respiratory chain complex deficiencies. PLoS Genet. 12, e1005679 10.1371/journal.pgen.1005679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Smeitink J.A., Elpeleg O., Antonicka H., Diepstra H., Saada A., Smits P. et al. (2006) Distinct clinical phenotypes associated with a mutation in the mitochondrial translation elongation factor EFTs. Am. J. Hum. Genet. 79, 869–877 10.1086/508434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Shamseldin H.E., Alshammari M., Al-Sheddi T., Salih M.A., Alkhalidi H., Kentab A. et al. (2012) Genomic analysis of mitochondrial diseases in a consanguineous population reveals novel candidate disease genes. J. Med. Genet. 49, 234–241 10.1136/jmedgenet-2012-100836 [DOI] [PubMed] [Google Scholar]
- 164.Ahola S., Isohanni P., Euro L., Brilhante V., Palotie A., Pihko H. et al. (2014) Mitochondrial EFTs defects in juvenile-onset Leigh disease, ataxia, neuropathy, and optic atrophy. Neurology 83, 743–751 10.1212/WNL.0000000000000716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Janer A., van Karnebeek C.D., Sasarman F., Antonicka H., Ghamdi M., Shyr C. et al. (2015) RMND1 deficiency associated with neonatal lactic acidosis, infantile onset renal failure, deafness, and multiorgan involvement. Eur. J. Hum. Genet. 23, 1301–1307 10.1038/ejhg.2014.293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Gupta A., Colmenero I., Ragge N.K., Blakely E.L., He L., McFarland R. et al. (2016) Compound heterozygous RMND1 gene variants associated with chronic kidney disease, dilated cardiomyopathy and neurological involvement: a case report. BMC Res. Notes 9, 325 10.1186/s13104-016-2131-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Vinu N., Puri R.D., Anand K. and Verma I.C. (2018) Expanding the phenotype of the founder south asian mutation in the nuclear encoding mitochondrial RMND1 gene. Indian J. Pediatr. 85, 87–92 10.1007/s12098-017-2515-x [DOI] [PubMed] [Google Scholar]
- 168.Shimazaki H., Takiyama Y., Ishiura H., Sakai C., Matsushima Y., Hatakeyama H. et al. (2012) A homozygous mutation of C12orf65 causes spastic paraplegia with optic atrophy and neuropathy (SPG55). J. Med. Genet. 49, 777–784 10.1136/jmedgenet-2012-101212 [DOI] [PubMed] [Google Scholar]
- 169.Tucci A., Liu Y.T., Preza E., Pitceathly R.D., Chalasani A., Plagnol V. et al. (2014) Novel C12orf65 mutations in patients with axonal neuropathy and optic atrophy. J. Neurol. Neurosurg. Psychiatry 85, 486–492 10.1136/jnnp-2013-306387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Pyle A., Ramesh V., Bartsakoulia M., Boczonadi V., Gomez-Duran A., Herczegfalvi A. et al. (2014) Behr’s syndrome is typically associated with disturbed mitochondrial translation and mutations in the C12orf65 Gene. J. Neuromuscul. Dis. 1, 55–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Makrythanasis P., Nelis M., Santoni F. A., Guipponi M., Vannier A., Bena F. et al. (2014) Diagnostic exome sequencing to elucidate the genetic basis of likely recessive disorders in consanguineous families. Hum. Mutat. 35, 1203–1210 10.1002/humu.22617 [DOI] [PubMed] [Google Scholar]
- 172.Antonicka H., Ostergaard E., Sasarman F., Weraarpachai W., Wibrand F., Pedersen A.M. et al. (2010) Mutations in C12orf65 in patients with encephalomyopathy and a mitochondrial translation defect. Am. J. Hum. Genet. 87, 115–122 10.1016/j.ajhg.2010.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Spiegel R., Mandel H., Saada A., Lerer I., Burger A., Shaag A. et al. (2014) Delineation of C12orf65-related phenotypes: a genotype-phenotype relationship. Eur. J. Hum. Genet. 22, 1019–1025 10.1038/ejhg.2013.284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Weraarpachai W., Antonicka H., Sasarman F., Seeger J., Schrank B., Kolesar J.E. et al. (2009) Mutation in TACO1, encoding a translational activator of COX I, results in cytochrome c oxidase deficiency and late-onset Leigh syndrome. Nat. Genet. 41, 833–837 10.1038/ng.390 [DOI] [PubMed] [Google Scholar]
- 175.Seeger J., Schrank B., Pyle A., Stucka R., Lörcher U., Müller-Ziermann S. et al. (2010) Clinical and neuropathological findings in patients with TACO1 mutations. Neuromuscul. Disord. 20, 720–724 10.1016/j.nmd.2010.06.017 [DOI] [PubMed] [Google Scholar]
- 176.Richman T.R., Spåhr H., Ermer J.A., Davies S.M., Viola H.M. and Bates K.A. (2016) nLoss of the RNA-binding protein TACO1 causes late-onset mitochondrial dysfunction in mice. Nat. Commun. 7, 11884 10.1038/ncomms11884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Vedrenne V., Gowher A., De Lonlay P., Nitschke P., Serre V. and Boddaert N. (2012) Mutation in PNPT1, which encodes a polyribonucleotide nucleotidyltransferase, impairs RNA import into mitochondria and causes respiratory-chain deficiency. Am. J. Hum. Genet. 91, 912–918 10.1016/j.ajhg.2012.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Alodaib A., Sobreira N., Gold W.A., Riley L.G., Van Bergen N.J., Wilson M.J. et al. (2016) Whole-exome sequencing identifies novel variants in PNPT1 causing oxidative phosphorylation defects and severe multisystem disease. Eur. J. Hum. Genet. 25, 79–84 10.1038/ejhg.2016.128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Matilainen S., Carroll C.J., Richter U., Euro L., Pohjanpelto M. et al. (2017) Defective mitochondrial RNA processing due to PNPT1 variants causes Leigh syndrome. Hum. Mol. Genet. 26, 3352–3361 10.1093/hmg/ddx221 [DOI] [PubMed] [Google Scholar]
- 180.Garone C., D’Souza A.R., Dallabona C., Lodi T., Rebelo-Guiomar P., Rorbach J. et al. (2017) Defective mitochondrial rRNA methyltransferase MRM2 causes MELAS-like clinical syndrome. Hum. Mol. Genet. 26, 4257–4266 10.1093/hmg/ddx314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Bartsakoulia M., Mϋller J.S., Gomez-Duran A., Yu-Wai-Man P., Boczonadi V. and Horvath R. (2016) Cysteine supplementation may be beneficial in a subgroup of mitochondrial translation deficiencies. J. Neuromuscul. Dis. 3, 363–379 10.3233/JND-160178 [DOI] [PubMed] [Google Scholar]
- 182.Perli E., Giordano C., Pisano A., Montanari A., Campese A.F., Reyes A. et al. (2014) The isolated carboxy-terminal domain of human mitochondrial leucyl-tRNA synthetase rescues the pathological phenotype of mitochondrial tRNA mutations in human cells. EMBO Mol. Med. 6, 169–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Hornig-Do H.T., Montanari A., Rozanska A., Tuppen H.A., Almalki A.A., Abg-Kamaludin D.P. et al. (2014) Human mitochondrial leucyl tRNA synthetase can suppress non cognate pathogenic mt-tRNA mutations. EMBO Mol. Med. 6, 183–193 [DOI] [PMC free article] [PubMed] [Google Scholar]