Abstract
BACKGROUND: The analysis of exosomes in blood obtained from the tumor-draining mesenteric vein (MV) can identify tumor biomarkers before they reach target organs and form the premetastatic niche where circulating tumor cells can anchor. Our group has recently shown that microRNAs in plasma from the MV—but not the peripheral vein (PV)—have been related to liver metastases in colon cancer (CC) patients. Here we examine the exosomal protein cargo in plasma from the MV and paired PV in 31 CC patients. PATIENTS AND METHODS: The study included patients who were initially diagnosed with stage I-III CC and 10 healthy controls. Exosomes from the MV and PV of all patients and controls were isolated by ultracentrifugation and confirmed by cryogenic transmission electron microscopy. High-throughput proteomic analysis by mass spectrometry was used to identify expression levels of exosomal proteins. Findings were confirmed by Western blot. RESULTS: Exosomal ECM1 protein was more highly expressed in patients than in controls and was 13.55 times higher in MV from relapsed than relapse-free patients. High exosomal ECM1 expression was associated with liver metastases. Patients with high exosomal ECM1 expression in MV—but not PV—plasma had shorter time to relapse than those with low ECM1 expression (P = .04). CONCLUSION: High levels of exosomal ECM1 protein can identify CC patients with a higher risk of relapse. The analysis of exosomes isolated from the tumor-draining MV is a promising method for the identification of biomarkers before they reach the target organ.
Introduction
Colon cancer (CC) is the third most frequent cancer and the second cause of death from cancer in the developed world [1]. Disease stage is the main prognostic factor for relapse and survival. Surgery is the standard treatment for stage I-III CC, followed by adjuvant therapy in stage III but not in stage I, while in stage II, the benefit of adjuvant therapy is still controversial [2]. However, approximately 30% of patients with stage I-III disease and up to 65% of stage IV patients will relapse after treatment [3], highlighting the need for prognostic biomarkers that can identify patients more likely to develop metastases.
In recent years, exosomes and their cargo have been shown to be promising markers of tumor growth and metastasis development. Exosomes are small vesicles (30-100 nm) involved in cell-to-cell communication. An exosome contains a small cytosol carrying coding and noncoding RNAs, DNA, and proteins [4]. Exosomes are secreted by several cell types and captured by receptor cells in target organs, where they regulate several normal and pathological physiological processes. They play an active role in the metastatic process by modifying the surrounding stroma in order to prepare the tissue microenvironment for the anchoring of metastatic cells [5], [6], [7].
The majority of studies to identify circulating biomarkers of metastases are performed in blood obtained from the peripheral vein (PV) of the forearm. However, by the time the blood reaches the PV, the biomarkers associated with metastases may well have been retained in the target organ and will thus be present at lower levels in the PV blood sample, which may skew study results. Blood from the colon flows through the mesenteric veins (MVs) into the vena porta, carrying the tumor cells and exosomes associated with CC. For this reason, metastases associated with CC are primarily locoregional (in the liver or peritoneum) and less frequently distant (in the lung or bone) [8]. It is thus logical to suppose that obtaining blood from the MV will allow us to identify CC tumor biomarkers before they reach the target organs [9], [10].
We have examined exosomes and their protein cargo by proteomic analysis in MV blood samples from relapsed and relapse-free CC patients who were initially diagnosed with stage I-III disease. The aim of the study was to identify exosomal proteins that can reliably predict the development of metastases in CC patients.
Material and Methods
Patients
From August 2009 to August 2013, samples were obtained from 31 patients initially diagnosed with stage I-III CC who underwent surgical resection at the Municipal Hospital of Badalona (Badalona, Spain). At the time of performing the present study, we selected 16 patients who had not relapsed and 15 who had. Four of the relapsed patients had liver metastases, seven had peritoneal metastases, and four had lung metastases (Table 1). All 31 patients had undergone a complete history and physical examination prior to surgery. In addition, we obtained blood samples from 10 healthy controls.
Table 1.
Patient Characteristics and Univariate P Values for TTR in 31 Surgical CC Patients
| Characteristics | N (%) | P |
|---|---|---|
| Sex | .42 | |
| Male | 18 (58) | |
| Female | 13 (42) | |
| Median age, years | 72 | .92 |
| CEA levels | .01 | |
| <=5 | 24 (77) | |
| >5 | 7 (23) | |
| C 19.9 levels | .80 | |
| <=37 | 27 (87) | |
| >37 | 4 (13) | |
| Tumor location | .99 | |
| Left colon | 16 (52) | |
| Right colon | 15 (48) | |
| Tumor size (cm) | .24 | |
| <=5 | 28 (90) | |
| >5 | 3 (10) | |
| Histological type | .03 | |
| Well differentiated | 28 (90) | |
| Poorly differentiated | 3 (10) | |
| Preexistent polyp | .65 | |
| Absent | 24 (77) | |
| Present | 7 (23) | |
| TNM stage | .003 | |
| I-II | 18 (58) | |
| III | 13 (42) | |
| Lymph nodes examined | .13 | |
| <12 | 8(26) | |
| >12 | 23(74) | |
| Relapsed | ||
| Yes | 15 (48) | |
| No | 16 (52) | |
| Metastatic site | ||
| Liver | 4 (27) | |
| Peritoneum | 7 (46) | |
| Lung | 4 (27) |
CEA, carcinoma embryonic antigen; TNM, tumor, nodule, metastasis.
Approval for the study was obtained from the institutional review board of the hospital, and signed informed consent was obtained from all patients in accordance with the Declaration of Helsinki.
Blood Samples
Paired MV and PV blood was obtained from all 31 patients as previously described [9]. On the day of surgery, 5 ml of blood was drawn from the PV and stored in heparinized tubes. During surgery, with vascular ligation before tumor resection, an additional 5 ml of blood was drawn from either the superior or the inferior MV, according to the anatomic location of the tumor. Healthy control PV blood samples were obtained from the Hospital Clinic Blood Bank.
Sample Processing and Exosome Isolation
Plasma from all blood samples was obtained by centrifugation of the whole blood at 5000g during 10 minutes and saved frozen at −80°C until further use.
Exosomes were isolated from 50 μl of plasma samples by ultracentrifugation as previously described [11]. Briefly, samples were sequentially centrifuged at 4°C at 300g for 5 minutes, followed by 2500g for 20 minutes and finally 10,000g for 30 minutes, followed by ultracentrifugation at 100,000g for 2 hours. Then the pellet was washed with DPBS and ultracentrifuged again at 100,000g for 1 hour in a Sorvall MX Plus Micro-Ultracentrifuge with S140AT Rotor and Polycarbonate Tubes (Thermo Scientific).
Exosomes were characterized by two methods: cryo–transmission electron microscopy (cryo-TEM) in a Jeol JEM 2011 transmission electron microscope at the Microscope Facility of the Autonomous University of Barcelona and Western blot analysis using the exosome marker TSG101.
Sample Preparation for Mass Spectrometry
For the analysis of exosome protein cargo by mass spectrometry, the exosome dry pellet was resuspended in 50 μl of DPBS. A training set of 11 different samples was analyzed.
Proteins from exosomes were solubilized in an SDS buffer (0.5% SDS, 50 mM Tris HCL, 5 mM DTT), incubated during 60 minutes at 65°C to denaturate proteins, and alkylated with 55 mM iodoacetamide and digested by trypsin. Following digestion, each sample was labeled with one specifically coded TMT reagent. The multiplex experiment was made by virtue of a specific reporter fragment ion of m/z 126, 127, 128, 129, 130, and 131. In a six-plex experiment, one channel was reserved for a reference sample that was created as a combination of aliquots from each primary sample in the study. The remaining five channels were utilized for samples. Labeled samples were combined into a tub, mixed, and fractionated by strong cation exchange into 10 fractions. Each of the fractions was further separated by HPLC and analyzed online in a LTQ VELOS Orbitrap mass spectrometer (Thermo).
Western Blot and Antibodies
Western blot analysis was performed as previously described [11]. Briefly, exosome dry pellets were mixed with LDS sample buffer (Life Technologies) and sample reducing agent (Life Technologies) according to the manufacturer’s protocol. After incubation for 10 minutes at 97°C, exosome samples were loaded onto a SDS-PAGE (Novex 4%-12% Bis-Tris gel, Life Technologies), transferred to a nitrocellulose membrane by iBlot (Life Technologies), and blocked for 1 hour with 1× TBST buffer (Fisher Scientific) with 5% w/v nonfat dry milk, according to the manufacturer’s instructions. The following primary antibodies were used and incubated in blocking buffer at 4°C overnight: TSG101 (ab83, Abcam) as a marker of exosomes and ECM1 (sc-365946, Santa Cruz Biotechnology) as a marker of the main protein isolated inside the exosomes. The signal was obtained using the Novex ECL Chemiluminescent Substrate Reagent kit (WP 20005, Invitrogen), and the images were developed using the Chemidoc System (Bio-Rad).
Statistical analyses
All 31 patients were evaluable for time to relapse (TTR), calculated from the date of surgery to the date of relapse or last follow-up. The univariate analysis of TTR according to protein expression was performed with the Kaplan-Meier method and compared using the log-rank test. The optimal cutoff point for protein expression was assessed by the Cutoff Finder software (http://molpath.charite.de/cutoff/index.jsp) [12]. Statistical analyses were performed with SPSS 22 (SPSS Inc., Chicago, IL), R 2.6.0 Software (Vienna, Austria), BRB Array Tools version 3.5.0 software (Richard Simon & BRB-ArrayTools Development Team, http://linus.nci.nih.gov/BRB-ArrayTools.html, National Cancer Institute, Bethesda, MD), and TIGR Multiexperiment viewer version 4.0 software (Dana-Farber Cancer Institute, Boston, MA). Statistical significance was set at P < .05.
Results
Patients
Thirty-one patients were included in the study, all of whom were initially diagnosed with stage I-III CC and followed using the standard protocol followed by Hospital Municipal de Badalona. Median age was 72 years, and 18 (58%) were males. Eighteen patients had stage I-II disease. Fifteen patients later relapsed (4 stage II and 11 stage III). Mean follow-up was 45.2 months (range, 26.4-63.8) (Table 1). Approval for the study was obtained from the institutional review board of the hospital, and signed informed consent was obtained from all patients and controls in accordance with the Declaration of Helsinki.
All 31 patients underwent a complete history and physical examination including routine hematological and biochemical analyses, chest radiographs, and computed tomography (CT) of the thorax and abdomen. Target lesions detected by abdominal ultrasound were also assessed by CT or magnetic resonance imaging.
Exosome Isolation and Proteomic Analysis
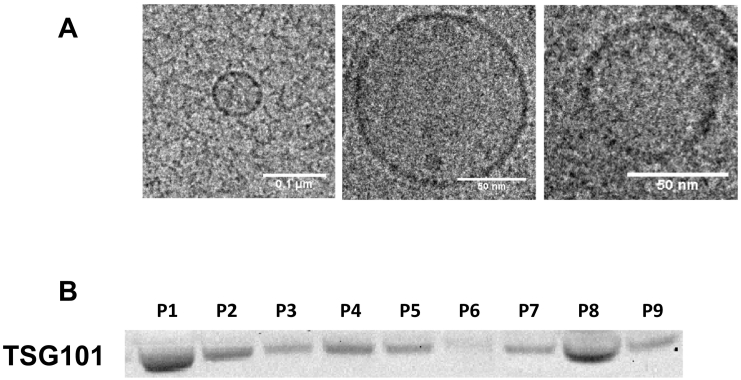
Both MV and PV plasma were obtained from all 31 patients, and exosomes were isolated from all plasma samples. The morphological analysis by cryo-TEM showed electron dense round 30- to 100-nm vesicles with a bilayer membrane [13]. In addition, Western blot analysis confirmed the presence of the exosomal marker TSG101 (Figure 1).
Figure 1.
Exosome characterization in plasma samples from colon cancer patients by (A) cryo-TEM and (B) Western blot using TSG101 marker.
In a training set of 11 of the 31 patients, using mass spectrometry, we profiled the exosome content of the plasma samples. A change of more than one-fold is generally accepted as a significant cutoff value in tandem mass tag technology [14], [15]. We identified a total of 202 proteins in the exosomes of the 11 patients. Of these 202 proteins, an exclusive panel of 49 proteins was detected as upregulated at least one-fold only in exosomes from relapsed patients in comparison with those from relapse-free patients. Twenty of these were considered exosomal proteins as they were annotated in curated databases of extracellular vesicles such as Vesiclepedia (http://www.microvesicles.org) or Exocarta (http://www.exocarta.org), and 14 were specifically identified in colorectal cancer cells (Table 2).
Table 2.
Abundance of the Selected Exosomal Proteins Identified by Mass Spectrometry in a Training Set of 11 CC Patients
| Relapse-Free Patients | Relapsed Patients | Ratio of Relapsed to Relapse-Free Patients | |
|---|---|---|---|
| ITIH4 | 0.67 | 0.71 | 1.06 |
| IGHM | 0.65 | 0.70 | 1.08 |
| CD5L | 0.74 | 0.81 | 1.09 |
| APOA1 | 0.58 | 0.64 | 1.12 |
| APOB | 0.62 | 0.76 | 1.23 |
| ITIH2 | 0.62 | 0.79 | 1.27 |
| C4BPA | 0.66 | 0.90 | 1.36 |
| IGHA1 | 0.56 | 0.88 | 1.57 |
| AMBP | 0.69 | 1.14 | 1.65 |
| C1QC | 0.46 | 0.82 | 1.77 |
| IGHG3 | 0.53 | 0.93 | 1.77 |
| C1QB | 0.48 | 0.92 | 1.92 |
| IGKC | 0.56 | 1.14 | 2.02 |
| ECM1 | 0.06 | 0.88 | 13.55 |
The ratio between relapsed versus relapse-free patients is expressed as a quotient and indicates the relationship in quantity between the two groups of patients.
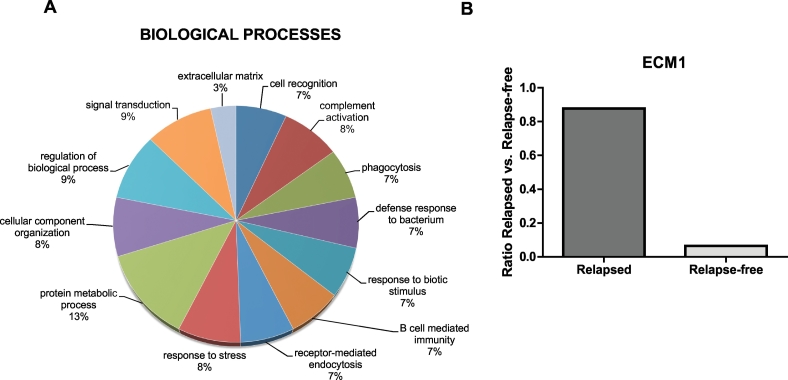
Gene ontology (GO) analysis was performed on these 14 upregulated proteins using Panther (version 9.0; http://www.pantherdb.org) according to the GO terms for molecular function, biological process and cellular component [16]. These 14 upregulated proteins are involved in various biological processes, including cell recognition, complement activation, phagocytosis, defense response, B cell–mediated immunity, receptor-mediated endocytosis, response to stress, protein metabolic process, cellular component organization, regulation of biological process, signal transduction, and extracellular matrix function (Figure 2A).
Figure 2.
(A) The biological processes involving the proteins identified by mass spectrometry as being differentially expressed in relapsed and relapse-free patients. of the mainly differentially expressed proteins in relapsed patients identified by mass spectrometry. GO software was used for the classification analysis, and the information was retrieved from the “protein center” database. (B) ECM1 was identified as the protein most differentially expressed between relapsed and relapse-free patients.
Higher expression of exosomal ECM1 protein in MV plasma from relapsed patients
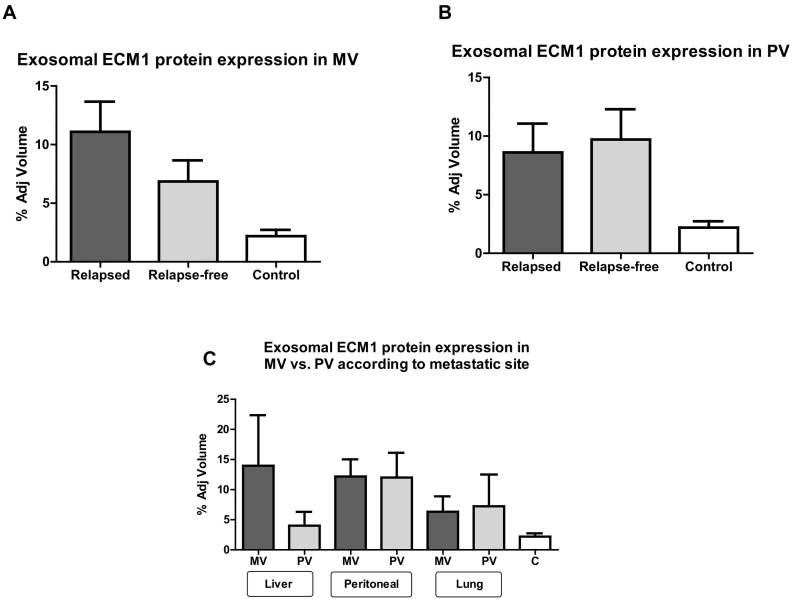
Of the 14 exosomal proteins specifically related to colorectal cancer identified by mass spectrometry in the training set, only extracellular matrix protein 1 (ECM1) was differentially expressed in MV plasma from relapsed versus relapse-free patients. ECM1 levels were 13.55 times higher in relapsed than in relapse-free patients (Figure 2B). We then validated this finding in the entire cohort of 31 patients by Western blot analysis, which confirmed a higher ECM1 expression in relapsed patients (Figure 3A).
Figure 3.
ECM1 protein expression levels (fold change) in exosomes isolated from plasma from the (A) MV and (B) paired PV of relapsed and relapse-free patients. (C) Exosomal ECM1 protein expression levels in MV and PV from relapsed patients classified according to the site of metastasis.
In contrast, in PV plasma, the expression levels of ECM1 were similar in relapsed and relapse-free patients (Figure 3B).
Exosomal ECM1 levels were then analyzed in MV and PV plasma from the 15 relapsed patients, classified according to metastatic site. In patients with liver metastases, ECM1 levels were much higher in MV than in PV plasma, while levels in patients with peritoneal or lung metastases were similar in MV and PV (Figure 3C).
Exosomal ECM1 Protein in MV Plasma Associated with TTR
We then examined the potential prognostic impact of ECM1 expression in all 31 patients and found that high MV expression of exosomal ECM1 was associated with shorter TTR. Mean TTR was 40.2 months (95% CI 33.5-46.9 months) for the 17 patients with low ECM1 levels compared to 31.3 months (95% CI 21.4-41.2 months) for the 14 patients with high levels (P = .04) (Figure 4).
Figure 4.
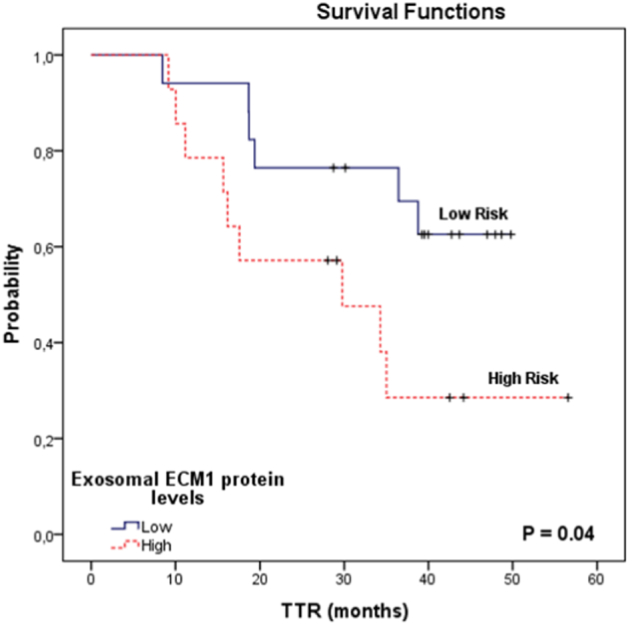
TTR in 31 colon cancer patients according to exosomal ECM1 protein expression levels in plasma from the MV.
Discussion
Metastases are the primary cause of death in cancer patients. Although the exact mechanism whereby metastases occur is unknown, recent findings have shown that cells from the primary tumor release extracellular vesicles, such as exosomes, which play an important role in the metastatic process [17]. When exosomes reach their target organ, they release their cargo of miRNAs and proteins, thus modifying the function of the receptor cells, in order to prepare the premetastatic niche for the anchoring and growth of circulating tumor cells [18], [19], [20]. The exosomal cargo thus acts directly on the target organ to initiate the metastatic process.
Several studies have found that the detection of exosomal biomarkers in blood can help predict the formation of metastases [21], [22], [23]. The majority of these studies have focused on exosomal miRNAs [22], [24], [25], [26], [27], while few have examined the role of exosomal proteins [28], [29], [30]. Moreover, these studies of exosomal cargo have primarily analyzed blood drawn from the PV, although the exosomal molecules involved in metastasis will already have been retained in the target organ before reaching the PV. This phenomenon may explain the conflicting findings on the impact of exosomal cargo in colorectal cancer [31], [32]. However, a recent study comparing exosomal miRNAs in blood from the MV versus the PV found that exosomal miR-328 from the MV was associated with metastases, while there was no correlation between miRNAs from the PV and metastases [10]. This finding highlights the importance in CC of obtaining tumor biomarkers from the MV before they reach the liver.
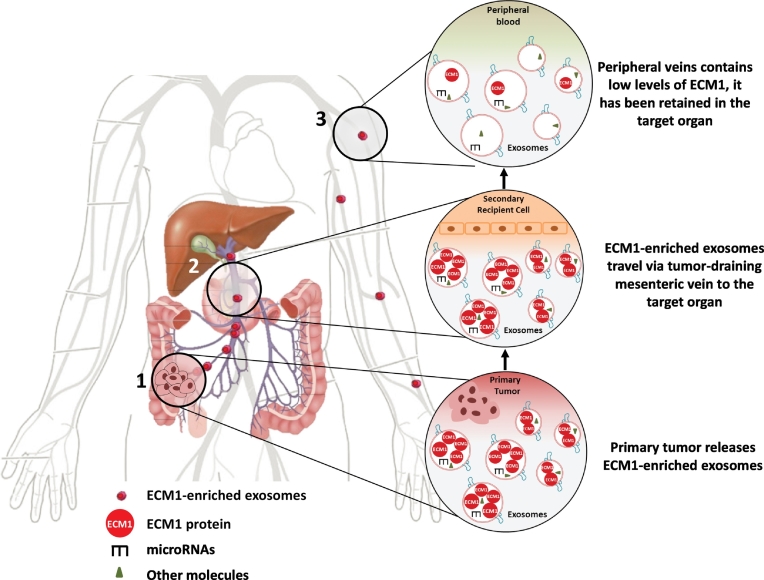
In the present study, we have examined whether the exosomal proteins in plasma obtained from the MV and paired PV are differentially expressed between relapsed and relapse-free CC patients. We have found that although exosomal proteins are present in both MV and PV plasma, which was to be expected since exosomes are released by both normal and tumor cells [33], they are more highly expressed in MV than in PV plasma. Moreover, while the levels of most proteins were quite similar in relapsed and relapse-free patients, relapsed patients had much higher levels of ECM1 than those who were relapse-free, suggesting that exosomal ECM1 could play a role in the formation of metastases (Figure 5).
Figure 5.
Anatomic route of exosomes released by the primary colon tumor. Venous return of the colon occurs through the MVs. Both MVs flow into the hepatic portal vein, which carries blood to the target organ, the liver, before reaching the peripheral veins. This anatomic distribution could explain the higher frequency of liver metastases associated with primary tumors located in the colon.
ECM1 is a 85-kDa secreted glycoprotein that participates in several normal physiological functions, such as the maintenance of skin integrity and homeostasis [34]. However, it is overexpressed in many epithelial tumors, including breast cancer, squamous cell carcinoma of the esophagus, gastric cancer, and CC [35]. Interestingly, the exosomal ECM1 protein has recently been detected in 82% of breast cancer patients with nontuberculous mycobacterial disease [36].
In line with these findings, we have also observed high levels of exosomal ECM1 in MV plasma from CC patients. Furthermore, relapsed patients had even higher levels than those who were relapse-free. We can, therefore, speculate that high exosomal ECM1 levels in plasma obtained from the MV, before the exosomes arrive at the target organ, can help identify patients more likely to relapse. Moreover, an association between ECM1 and metastasis has been reported in several studies [37], [38].
Furthermore, in the present study, we have observed a higher level of exosomal ECM1 in MV than PV plasma among patients with liver metastases, while levels in MV and PV plasma were similar among patients with peritoneal or lung metastases. This finding indicates that the exosomes released by the primary tumor may be organotropic and can be retained specifically by liver tissue. In fact, proteomic studies have shown that the exosomal αvβ5 integrin was associated with liver metastases, while the α6β4 and α6β1 integrins were associated with lung metastases [5].
We have also found that patients with high levels of exosomal ECM1 have a shorter TTR than those with low levels. Although the exact role of ECM1 in metastasis formation is not clear, findings in breast cancer indicate that ECM1 helps to increase cancer stem cells by stabilizing β-catenin [38]. In addition, ECM1 protein overexpression induces tumor progression in liver cancer through epithelial-mesenchymal transition [37].
In summary, our findings indicate that the analysis of exosomal ECM1 in blood drawn from the MV is a simple surgical method that can help identify patients with a higher risk of relapse by detecting tumor markers before they reach the target organ. Taken together with previous findings on exosomal miRNAs [10] and integrins [5], our findings lead us to suggest that ECM1, ideally in combination with other known metastasis-associated molecules, may well prove to be a sensitive marker for diagnosis and prognosis in CC patients. Moreover, we recommend the use of MV blood over PV whenever possible in future biomarker studies. In addition, future studies, ideally with a larger number of patients, should focus on the organotropism of exosomes to examine their association with specific metastatic sites, as well as on the functional mechanism of ECM1 whereby it impacts metastasis.
Conflict of Interest Statement
The authors declare no conflicts of interest.
Acknowledgements
The authors thank Renée Grupp for assistance in drafting the manuscript.
Footnotes
Funding: This work was supported by a grant from Servei de Donació del Cos a la Ciència (Body Donation Service) of the University of Barcelona. S. S. and J. J. C. are APIF fellows of the University of Barcelona. Neither of these funding bodies had a role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript; or decision to submit the manuscript for publication.
Contributor Information
Sandra Santasusagna, Email: sandra.santasusagna@ub.edu.
Isabel Moreno, Email: imoreno@bsa.cat.
Alfons Navarro, Email: anavarroponz@ub.edu.
Joan J. Castellano, Email: joan.castellano@ub.edu.
Francisco Martinez, Email: fmartinez@bsa.cat, fmrodenas@gmail.com.
Raquel Hernández, Email: rahernandez@bsa.cat.
Carmen Muñoz, Email: carmen.munoz@ub.edu.
Mariano Monzo, Email: mmonzo@ub.edu.
References
- 1.Miller KD, Siegel RL, Lin CC, Mariotto AB, Kramer JL, Rowland JH. Cancer treatment and survivorship statistics, 2016. CA Cancer J Clin. 2016;66(4):271–289. doi: 10.3322/caac.21349. [DOI] [PubMed] [Google Scholar]
- 2.Benson AB, III, Schrag D, Somerfield MR, Cohen AM, Figueredo AT, Flynn PJ. American Society of Clinical Oncology recommendations on adjuvant chemotherapy for stage II colon cancer. J Clin Oncol. 2004;22(16):3408–3419. doi: 10.1200/JCO.2004.05.063. [DOI] [PubMed] [Google Scholar]
- 3.van der Stok EP, Spaander MCW, Grunhagen DJ, Verhoef C, Kuipers EJ. Surveillance after curative treatment for colorectal cancer. Nat Rev Clin Oncol. 2017;14(5):297–315. doi: 10.1038/nrclinonc.2016.199. [DOI] [PubMed] [Google Scholar]
- 4.Hamam R, Ali AM, Alsaleh KA, Kassem M, Alfayez M, Aldahmash A. microRNA expression profiling on individual breast cancer patients identifies novel panel of circulating microRNA for early detection. Sci Rep. 2016;6:25997. doi: 10.1038/srep25997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hoshino A, Costa-Silva B, Shen T-L, Rodrigues G, Hashimoto A, Mark MT. Tumour exosome integrins determine organotropic metastasis. Nature. 2015;527(7578):329. doi: 10.1038/nature15756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Milane L, Singh A, Mattheolabakis G, Suresh M, Amiji MM. Exosome mediated communication within the tumor microenvironment. J Control Release. 2015;219:278–294. doi: 10.1016/j.jconrel.2015.06.029. [DOI] [PubMed] [Google Scholar]
- 7.O’Driscoll L. Expanding on exosomes and ectosomes in cancer. N Engl J Med. 2015;372(24):2359–2362. doi: 10.1056/NEJMcibr1503100. [DOI] [PubMed] [Google Scholar]
- 8.Labianca R, Beretta GD, Kildani B, Milesi L, Merlin F, Mosconi S. Colon cancer. Crit Rev Oncol Hematol. 2010;74(2):106–133. doi: 10.1016/j.critrevonc.2010.01.010. [DOI] [PubMed] [Google Scholar]
- 9.Monzo M, Martínez-Rodenas F, Moreno I, Navarro A, Santasusagna S, Macias I. Differential MIR-21 expression in plasma from mesenteric versus peripheral veins: an observational study of disease-free survival in surgically resected colon cancer patients. Medicine. 2015;94(1) doi: 10.1097/MD.0000000000000145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Monzo M, Santasusagna S, Moreno I, Martinez F, Hernáez R, Muñoz C. Exosomal microRNAs isolated from plasma of mesenteric veins linked to liver metastases in resected patients with colon cancer. Oncotarget. 2017;8(19):30859. doi: 10.18632/oncotarget.16103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ruiz-Martinez M, Navarro A, Marrades RM, Viñolas N, Santasusagna S, Muñoz C. YKT6 expression, exosome release, and survival in non-small cell lung cancer. Oncotarget. 2016;7(32):51515. doi: 10.18632/oncotarget.9862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Budczies J, Klauschen F, Sinn BV, Győrffy B, Schmitt WD, Darb-Esfahani S. Cutoff Finder: a comprehensive and straightforward Web application enabling rapid biomarker cutoff optimization. PLoS One. 2012;7(12):e51862. doi: 10.1371/journal.pone.0051862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ha D, Yang N, Nadithe V. Exosomes as therapeutic drug carriers and delivery vehicles across biological membranes: current perspectives and future challenges. Acta Pharm Sin B. 2016;6(4):287–296. doi: 10.1016/j.apsb.2016.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zeng X, Yang P, Chen B, Jin X, Liu Y, Zhao X. Quantitative secretome analysis reveals the interactions between epithelia and tumor cells by in vitro modulating colon cancer microenvironment. J Proteomics. 2013;89:51–70. doi: 10.1016/j.jprot.2013.05.032. [DOI] [PubMed] [Google Scholar]
- 15.Fang Z, Yao W, Xiong Y, Zhang J, Liu L, Li J. Functional elucidation and methylation-mediated downregulation of ITGA5 gene in breast cancer cell line MDA-MB-468. J Cell Biochem. 2010;110(5):1130–1141. doi: 10.1002/jcb.22626. [DOI] [PubMed] [Google Scholar]
- 16.Zhang H, Xu Y, Filipovic A, Lit L, Koo C, Stebbing J. SILAC-based phosphoproteomics reveals an inhibitory role of KSR1 in p53 transcriptional activity via modulation of DBC1. Br J Cancer. 2013;109(10):2675. doi: 10.1038/bjc.2013.628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tkach M, Théry C. Communication by extracellular vesicles: where we are and where we need to go. Cell. 2016;164(6):1226–1232. doi: 10.1016/j.cell.2016.01.043. [DOI] [PubMed] [Google Scholar]
- 18.Peinado H, Alečković M, Lavotshkin S, Matei I, Costa-Silva B, Moreno-Bueno G. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat Med. 2012;18(6):883–891. doi: 10.1038/nm.2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Costa-Silva B, Aiello NM, Ocean AJ, Singh S, Zhang H, Thakur BK. Pancreatic cancer exosomes initiate pre-metastatic niche formation in the liver. Nat Cell Biol. 2015;17(6):816–826. doi: 10.1038/ncb3169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.de la Fuente A, Alonso-Alconada L, Costa C, Cueva J, Garcia-Caballero T, Lopez-Lopez R. M-trap: exosome-based capture of tumor cells as a new technology in peritoneal metastasis. J Natl Cancer Inst. 2015;107(9) doi: 10.1093/jnci/djv184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Skog J, Würdinger T, Van Rijn S, Meijer DH, Gainche L, Curry WT. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol. 2008;10(12):1470–1476. doi: 10.1038/ncb1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schwarzenbach H. Clinical relevance of circulating, cell-free and exosomal microRNAs in plasma and serum of breast cancer patients. Oncol Res Treat. 2017;40(7-8):423–429. doi: 10.1159/000478019. [DOI] [PubMed] [Google Scholar]
- 23.Erb U, Zhao K, Wang Z, Xiao L, Zöller M. Murine and human pancreatic tumor exosome recovery in mouse serum: diagnostic and prognostic potential and target cell delivery. Cancer Lett. 2017;403:1–12. doi: 10.1016/j.canlet.2017.06.005. [DOI] [PubMed] [Google Scholar]
- 24.Wu K, Feng J, Xing F, Liu Y, Sharma S, Watabe K. AACR; 2017. Exosomal miR-19a: a novel communicator between cancer cell and osteoclast in osteolytic bone metastasis of breast cancer. [Google Scholar]
- 25.Lan F, Qing Q, Pan Q, Hu M, Yu H, Yue X. Serum exosomal miR-301a as a potential diagnostic and prognostic biomarker for human glioma. Cell Oncol. 2017:1–9. doi: 10.1007/s13402-017-0355-3. [DOI] [PubMed] [Google Scholar]
- 26.Teng Y, Ren Y, Hu X, Mu J, Samykutty A, Zhuang X. MVP-mediated exosomal sorting of miR-193a promotes colon cancer progression. Nat Commun. 2017;8:14448. doi: 10.1038/ncomms14448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhao L, Yu J, Wang J, Li H, Che J, Cao B. Isolation and identification of miRNAs in exosomes derived from serum of colon cancer patients. J Cancer. 2017;8(7):1145. doi: 10.7150/jca.18026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Campanella C, Rappa F, Sciumè C, Marino Gammazza A, Barone R, Bucchieri F. Heat shock protein 60 levels in tissue and circulating exosomes in human large bowel cancer before and after ablative surgery. Cancer. 2015;121(18):3230–3239. doi: 10.1002/cncr.29499. [DOI] [PubMed] [Google Scholar]
- 29.Yunusova N, Tamkovich S, Stakheeva M, Afanas’ ev S, Frolova A, Kondakova I, editors. The characterization of exosome from blood plasma of patients with colorectal cancer. AIP Conference Proceedings. AIP Publishing; 2016. [Google Scholar]
- 30.Chen Y, Xie Y, Xu L, Zhan S, Xiao Y, Gao Y. Protein content and functional characteristics of serum-purified exosomes from patients with colorectal cancer revealed by quantitative proteomics. Int J Cancer. 2017;140(4):900–913. doi: 10.1002/ijc.30496. [DOI] [PubMed] [Google Scholar]
- 31.Matsumura T, Sugimachi K, Iinuma H, Takahashi Y, Kurashige J, Sawada G. Exosomal microRNA in serum is a novel biomarker of recurrence in human colorectal cancer. Br J Cancer. 2015;113(2):275–281. doi: 10.1038/bjc.2015.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ogata-Kawata H, Izumiya M, Kurioka D, Honma Y, Yamada Y, Furuta K. Circulating exosomal microRNAs as biomarkers of colon cancer. PLoS One. 2014;9(4):e92921. doi: 10.1371/journal.pone.0092921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Piccin A, Murphy WG, Smith OP. Circulating microparticles: pathophysiology and clinical implications. Blood Rev. 2007;21(3):157–171. doi: 10.1016/j.blre.2006.09.001. [DOI] [PubMed] [Google Scholar]
- 34.Sercu S, Lambeir A, Steenackers E, El Ghalbzouri A, Geentjens K, Sasaki T. ECM1 interacts with fibulin-3 and the beta 3 chain of laminin 332 through its serum albumin subdomain-like 2 domain. Matrix Biol. 2009;28(3):160–169. doi: 10.1016/j.matbio.2009.02.003. [DOI] [PubMed] [Google Scholar]
- 35.Wang L, Yu J, Ni J, Xu X-M, Wang J, Ning H. Extracellular matrix protein 1 (ECM1) is over-expressed in malignant epithelial tumors. Cancer Lett. 2003;200(1):57–67. doi: 10.1016/s0304-3835(03)00350-1. [DOI] [PubMed] [Google Scholar]
- 36.Philley JV, Kannan A, Griffith DE, Devine MS, Benwill JL, Wallace RJ., Jr. Exosome secretome and mediated signaling in breast cancer patients with nontuberculous mycobacterial disease. Oncotarget. 2017;8(11):18070. doi: 10.18632/oncotarget.14964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen H, Jia W, Li J. ECM1 promotes migration and invasion of hepatocellular carcinoma by inducing epithelial-mesenchymal transition. World J Surg Oncol. 2016;14(1):195. doi: 10.1186/s12957-016-0952-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee K, Nam K, Oh S, Lim J, Kim R, Shim D. ECM1 regulates tumor metastasis and CSC-like property through stabilization of β-catenin. Oncogene. 2015;34(50):6055–6065. doi: 10.1038/onc.2015.54. [DOI] [PubMed] [Google Scholar]