Abstract
Medical oncology is in need of a mathematical modeling toolkit that can leverage clinically-available measurements to optimize treatment selection and schedules for patients. Just as the therapeutic choice has been optimized to match tumor genetics, the delivery of those therapeutics should be optimized based on patient-specific pharmacokinetic/pharmacodynamic properties. Under the current approach to treatment response planning and assessment, there does not exist an efficient method to consolidate biomarker changes into a holistic understanding of treatment response. While the majority of research on chemotherapies focus on cellular and genetic mechanisms of resistance, there are numerous patient-specific and tumor-specific measures that contribute to treatment response. New approaches that consolidate multimodal information into actionable data are needed. Mathematical modeling offers a solution to this problem. In this perspective, we first focus on the particular case of breast cancer to highlight how mathematical models have shaped the current approaches to treatment. Then we compare chemotherapy to radiation therapy. Finally, we identify opportunities to improve chemotherapy treatments using the model of radiation therapy. We posit that mathematical models can improve the application of anticancer therapeutics in the era of precision medicine. By highlighting a number of historical examples of the contributions of mathematical models to cancer therapy, we hope that this contribution serves to engage investigators who may not have previously considered how mathematical modeling can provide real insights into breast cancer therapy.
Introduction
On May 25, 1961, President Kennedy proposed to Congress that the United States should commit itself to “landing a man on the moon and returning him safely to earth” by the end of the decade. Similarly, on December 23, 1971 President Nixon signed into law the National Cancer Act and stated it was time for the concentrated effort that resulted in the lunar landings to be turned towards conquering cancer. Of course, Neil Armstrong first set foot on the lunar surface on July 20, 1969, yet 46 years after Nixon’s announcement we have made only modest advances in controlling this disease. This is particularly striking with the renewed lunar-centric announcement of the Cancer Moonshot Initiative by former President Obama in his 2016 State of the Union. A fundamental difference between the planetary and cancer moonshots is that the basic mathematics for gravity were known for nearly three centuries at the time of Kennedy’s speech, while we still do not have a mathematical description of cancer that allows us to compute the spatiotemporal evolution of an individual patient’s tumor. In the current state of oncology, we are tasked with getting to the moon without knowing F = ma.
Precision medicine is the concept of incorporating patient-specific variability into prevention and treatment strategies [1]. The advent of precision medicine has brought significant advances to oncology. The majority of these efforts have focused on the use of genetics to classify and pharmaceutically target cancers [2]. This approach has led to a paradigm in which tumor genotypes are matched to appropriate treatments [3], [4]. For example, the addition of trastuzumab, a monoclonal antibody targeting the human epidermal receptor 2 (HER2) protein, to chemotherapeutic regimens in breast cancer patients with HER2-positive disease has resulted in improved disease-free and overall survival [5]. While the current genetic-centric approach to cancer therapy has great merit in appropriately selecting therapies and identifying new pharmaceutical targets, it can frequently overlook a host of patient-specific measures that influence response to therapy. For example, the microenvironment of the tumor alters response [6], delivery of therapy to tumors is variable as tumor perfusion is limited [7], [8], and patient-specific pharmacokinetic properties vary [9], [10]. Intratumor heterogeneity, at the genetic and epigenetic levels, complicates the use of gene-centric precision medicine approaches. In some tumors, a single dominant clone may be identified [11], [12] and that clone may be targetable by therapy; however, neutral evolution and vast clonal diversity are more common scenarios [13], [14], [15]. For example, a single hepatocellular carcinoma may include more than 100 million different coding region mutations, including multiple sets of potential ‘driver’ mutations [16]. Further, the schedule on which therapy is given may significantly alter response [17], [18], [19]. These issues may be partly responsible for the high attrition rates of proposed cancer therapeutics [20].
The goal of precision medicine is to tailor therapeutic strategies to each patient’s specific biology. More specifically, we define the goal of precision medicine to be the use of the optimal dose of the optimal therapy on the optimal schedule for each patient. Under this interpretation, there is an opportunity to expand precision oncology beyond the tumor-genotype-driven selection of therapy. To achieve this goal, new hypotheses related to optimal dosing and scheduling are needed. Whereas the hypotheses in genetic studies often compare tumor volume changes to a static genetic marker, dosing and scheduling require temporally-resolved hypotheses and concomitant treatment response measures. In particular, such hypotheses would need to specify quantitatively how the tumor microenvironment and/or patient pharmacokinetics influence response to therapy in order to adapt therapeutic approaches to measured responses. Fortunately, the tools to probe cancer from the genetic to tumor scales have rapidly matured over the past decade. While more time is needed to fully understand and contextualize the micro-, meso-, and macro-scale data coming online, several groups have demonstrated the utility of new technologies. For example, advances in imaging technologies, such as diffusion weighted magnetic resonance imaging (DW-MRI) and dynamic contrast enhanced MRI (DCE-MRI), have led to the discovery of clinically-relevant biomarkers that are predictive of response [21]. We (and others [22], [23], [24]) believe that mathematical modeling holds the potential to synthesize available biomarkers to test new hypotheses. These models will not only improve our ability to treat cancer, but it will also allow precision cancer care to enter the dosing and scheduling domains.
A goal of mathematical modeling is to abstract the key features of a physical system to succinctly describe its behavior in a series of mathematical equations. In this way, the system can be simulated in silico to further understand system behavior, generate hypotheses, and guide experimental design. When experimental data is available, model predictions can be compared to those data. The model can then be iteratively refined to account for data-prediction mismatches. Models can also identify high-yield experiments in cases where an exhaustive investigation of experimental conditions is infeasible [25]. Traditionally, cancer models are built off of first order biological and physical principles, such as evolution [26] and diffusion [27]. Part of the recent excitement about applications of mathematical models to cancer is the discovery of higher-order, emergent properties that any one model component does not possess [28]. For example, cancer models have been constructed to investigate the role of tumor cell-matrix interactions in shaping tumor geometry and in enhancing selective pressures [29]. Fundamentally, models built from these first principles are designed to discover new biological behaviors and principles, identify new hypotheses for further investigation, and predict the behavior of cancer systems to perturbations. These models are tuned with any available data and simulated to discover system properties [24]. However, the majority of these models are not structured to leverage currently-available clinical data to make patient-specific predictions [30]. Often, these complex mechanism-based models have been limited to in silico exploration, and their utility in generating patient-specific predictions remains to be investigated. Medical oncology is in need of a mathematical, mechanism-based modeling framework to leverage all available clinical information, spanning from tumor genetic to tumor imaging data, to make impactful changes on patient management [31]. In this way, models can be used to make specific and measurable predictions of the response of an individual patient to an individualized therapeutic regimen. While these models may not explicitly consider all scales of biological interactions, they may be of practical utility by consolidating clinically-available data sources into a coherent understanding of tumor growth and treatment response.
The interaction of matter is governed by weak nuclear, strong nuclear, gravitational, and electromagnetic forces just as the behavior of cells is governed by genetics and genetic expression. However, for macroscopic objects traveling at speeds much less than the speed of light, F = ma is an excellent approximation of the movement of those objects. While the understanding of fundamental physical laws is still being advanced, a complete understanding is not necessary to leverage classical mechanical models to engineer mechanical tools (such as a rocket to lift astronauts to the moon). There is an opportunity in oncology to develop an analogous “classical oncology” toolkit. We posit that a complete understanding of cancer is not necessary to create tools that leverage clinical data to improve the treatment of cancer. This toolkit will likely consist of “simple” models that approximate the behavior and treatment response of tumors. Fortunately, the tools to make analogous force measurements in cancer already exist.
This perspective will highlight the utility of modeling and discuss opportunities for modeling in breast cancer treatment. Our target audience is composed of investigators with expertise in the biological sciences and interest in how mathematical modeling can inform the selection and optimization of therapies for breast cancer. We begin by reviewing the use of mathematical models in clinical oncology, including those used in radiation oncology. We then draw parallels between dose planning in radiation therapy and chemotherapy and propose how mathematical modeling approaches can leverage current technologies to more precisely apply anti-cancer chemotherapies. We then highlight opportunities for investigation in the clinical evaluation of response in the context of patient-specific modeling. To limit the scope of this perspective, we will focus on cytotoxic chemotherapeutics (defined below) in breast cancer. It is the goal of this perspective to provide guidance and highlight opportunities for a classical oncology toolkit.
Models for Clinical Oncology
We now discuss the mathematical theory and models used to define administration schedules and dosing in both medical and radiation oncology. We primarily focus on select dynamic treatment response models that have penetrated clinical practice and those with promising clinical utility. There exists a wide range of applications for mathematical models in oncology. For example, models have provided insight into tumor evolution and the development of intratumor heterogeneity [32]. Mathematical modeling has also been used to explore tumor initiation and development, focusing on tumor vasculature [33] and the tumor microenvironment [34]. We direct the interested reader to a review on mathematical oncology for a broader overview of the applications of mathematical modeling in cancer [35]. Further, while surgical oncology has incorporated mathematical modeling approaches, especially in image-guided surgical approaches, such discussion falls outside of the scope of this perspective. We review these concepts in the context of our definition of precision medicine: the use of the optimal dose of the optimal therapy on the optimal schedule for each patient.
Medical Oncology
Cytotoxic drugs, which are designed to inflict lethal insults on rapidly-dividing cells, were among the first pharmaceuticals used to treat breast cancer (the first clinical trial started in 1958 [36]), and they remain a critical component of current therapeutic regimens. The modern era of chemotherapy was born from the observation that mustard gas induced myelosuppressive states and was effective in treating hematologic malignancies [37]. Dosing schemes with these agents all follow a common pattern: cycles of a high dose nearing the maximum tolerated dose followed by a recovery period. The goal of this strategy is to maximize tumor cell kill, while trying to minimize adverse effects via drug holidays between each cycle. While tumors often respond to these therapies, there is a high rate of tumor recurrence. For example, the 5-year progression free survival rate for triple negative breast cancer (TNBC) patients is 61% [38]. Furthermore, cytotoxic therapies often have lasting effects on survivors, adversely affecting their quality of life. For example, doxorubicin, a standard-of-care therapy for the treatment of TNBC, is associated with cardiomyopathy [39].
When cytotoxic therapy was first applied to cancer, few mathematical principles existed to guide its use [40]. While the cytotoxic properties of these agents had clearly been demonstrated in animal models, the subsequent translation into a human population lagged behind. Skipper first observed the relationship between tumor size and treatment response when he discovered leukemia response to therapy to be proportional to the number of malignant cells [41]. He hypothesized that each dose of treatment kills a fixed percentage of tumor cells. This necessitates repeated dosing strategies to increase the odds of tumor eradication. Despite the relatively simplicity of the model, its practical implication was profound: chemotherapies should be delivered several times, even after the disappearance of macroscopic tumors, to eradicate all tumor cells. This was a departure from the current practice of the time, in which chemotherapeutic agents were given over a short course to treat solid tumors [36]. Skipper’s observation challenged this paradigm, and multi-dose treatment regimens were supported by subsequent clinical trials in the 1970’s [42], [43], forming the basis of modern adjuvant and neoadjuvant chemotherapy approaches. Subsequently, investigators sought to improve response through dose escalation. The dose escalation trials were met with limited success, as several agents demonstrated a saturated response curve at high doses [44], [45].
Investigation into the scheduling of therapeutics was advanced when Norton and Simon hypothesized that tumors grow according to Gompertzian kinetics [46], [47]. Qualitatively, Gompertzian kinetics posit that tumors grow exponentially, with an exponentially decreasing growth rate. Treatment response was assumed proportional to tumor growth rate, with smaller, faster growing tumors responding more robustly to treatment than larger slower-growing tumors. Similar to the log-kill model, the Norton-Simon hypothesis is relatively simple yet impactful. The model indicates that chemotherapy is best delivered to small, fast-growing tumors on a dose-dense schedule, minimizing the time between treatments. This approach limits the regrowth of tumors between treatments, meaning smaller tumors are being treated. Per the model, smaller tumors grow more quickly, rendering them increasingly responsive to treatment thereby maximizing therapeutic effect. This dose-dense approach demonstrated improvement over conventional dosing schedules in a clinical trial [17].
Multi-agent regimens were introduced in order to address tumor heterogeneity, in the hopes of eliminating tumor cells resistant to single agent therapy. Following the Goldie-Coldman hypothesis, which proposes that multi-agent chemotherapies should be delivered in alternating courses (e.g., ABABAB instead of AAABBB) to minimize the probability of developing resistance [48], empiric schedules of administration for these multi-agent regimens were tested in clinical trials [49]. While multi-agent regimens demonstrate improved efficacy relative to single-agent treatments, the scheduling of therapeutics remains an open question. For example, in trials investigating the reordering of treatments to avoid development of resistance according to the Goldie-Coldman hypothesis, the schedules that delivered therapy most quickly (regardless of order) were found to be superior [49]. While different schedules have been hypothesized to significantly impact response [19], [50], [51], empiric schedules remain as a matter of practicality as there exist innumerable combinations of drugs and schedules that cannot be tested clinically.
While several more complicated models of tumor growth and treatment response have been proposed in the literature [35], the models highlighted have been the only to penetrate clinical practice. We suppose these have gained traction because each provided a precise, clinically-testable hypothesis for improved cancer treatment. However, these models are limited to making general predictions for the use of chemotherapy. Further, the above hypotheses were developed to maximize the rate of tumor cell kill, which is assumed to improve long-term, disease-free survival; however, growing evidence suggests this may not be the optimal therapeutic approach [52].
The dosing of chemotherapeutics also has a mathematical basis. Doses of chemotherapeutic agents are often personalized through use of patient body surface area (BSA) [53], [54]. BSA was first proposed as a guide for chemotherapy dosing by Pinkel, noting that the accepted cytotoxic dose for pediatric and adult patients, and the dose used in laboratory animals correlated with BSA across those scales [55]. Several BSA models have been developed over time, primarily differing in the coefficients used in their calculation. While a BSA-based dosing strategy is of great practical utility for calculating doses for each patient, BSA correlates poorly with the underlying physiological processes that affect drug pharmacology (e.g., liver metabolism and glomerular filtration rate) [56]. Specifically, BSA has been found to correlate poorly with patient pharmacokinetic properties for several chemotherapies [57]. For example, in a study of 110 patients receiving doxorubicin therapy, doxorubicin clearance was found to weakly correlate with BSA [9]. Despite the weak relationship between BSA and pharmacokinetics for several therapeutics, BSA remains widely used to guide dosing in the clinic.
Radiation Oncology
Similar to chemotherapy, radiation therapy was once delivered in a single, high dose [58]. Contrary to chemotherapy, in which a theory of treatment response was established prior to changes in therapeutic application, radiation doses were quickly fractionated to account for excessive toxicities in healthy tissue. Briefly, radiation therapy leverages ionizing radiation to damage the DNA of tumor cells [59]. The DNA damage induced by radiation can lead to immediate cell death via apoptosis, senescence, autophagy, or necrosis or a delayed cell death via mitotic catastrophe [60].
The interaction of photons with DNA can be physically modeled as a stochastic process. The probability of the number of photon-tissue interactions can be described using Poisson statistics:
where P(n) is the probability of n interactions, and D is radiation dose in units of Gray (defined to be one joule of energy absorbed per kilogram of matter). If a single interaction is assumed to result in cell death, the probability of survival (n = 0) is simply e-D. For viruses, bacteria, and very sensitive human cells, it is an appropriate model of survival [61]. However, this model fails to describe survival in other human cell types. For these tissues, the linear-quadratic (LQ) model was found to be the most parsimonious model that fit the observed survival curves [61], [62]. The LQ model is expressed as:
where α and β are radiosensitivity parameters, and D is dose. As β approaches zero, the LQ model approaches the Poisson model of cell survival. The LQ model can be used to characterize the radiosensitivity of different tissues with two parameters (α and β). One potential biological interpretation of the linear-quadratic model is offered by the lethal-potentially lethal damage (LPL) model [63]. The LPL model posits that the linear portion of the LQ model are cells that receive non-repairable lethal lesions after a single hit (radiation dose). The quadratic portion is representative of repairable lesions that may eventually die to subsequent lesions or misrepair.
The LQ formalism can be used to explain why fractionated radiotherapy was superior to single doses (there are additional biological rationales for the use of fractionated therapy [64], but these have yet to be formalized into a mathematical modeling framework). Fractionation approaches leverage differential radiosensitivities of tissues (i.e., tumor and healthy tissues) to maximize efficacy while minimizing off-target toxicities. In planning treatment schedules, the effect of therapy on the tumor (generally high α/β ratios) must be balanced with both the acute and long-term toxicities of surrounding, healthy tissue (lower α/β ratios). For a fixed duration of treatment, the isoeffect doses (i.e., doses that have an equivalent biological effect) of different fractionation schedules can be compared [65]:
where Di is the total dose for each fractionation scheme, di is the dose per fraction, and α/β is a measure of tissue-specific radiosensitivity. For late-responding healthy tissues (i.e., for tissue with low α/β), the total isoeffective dose increases more quickly than acutely-responding tissue (i.e., high α/β) when doses are hyperfractionated (i.e., smaller doses with more fractions). This means that fractionation schedules allow for higher isoeffective doses in tumor tissues compared to surrounding healthy tissue. For this reason, radiotherapy is typically given at low doses over several sessions to maximize tumor dose and to minimize damage to healthy tissue. For example, in head and neck cancer with high α/β ratios (>7 Gy) [66], a hyperfractioned schedule has been shown to be superior to conventional schedules with fewer fractions [67]. While patient-specific biology underlies the α/β parameters for tumors and surrounding tissue, interpatient variability in parameters is often not considered in clinical practice, yielding a single schedule for many patients receiving radiotherapy. For example, some tumors demonstrate similar α/β ratios to the surrounding healthy tissue. Specifically, breast cancers have relatively smaller α/β ratios (4 Gy) [66]. In this case, a schedule using higher doses and fewer treatment sessions (hypofractionation) may be superior [68].
In addition to its explicit consideration of off-target toxicities, radiation therapy differs from chemotherapy in dose planning. As noted above, in radiation therapy, dose is defined as the energy absorbed per unit mass. This differs from the use of “dose” in chemotherapy as the amount of drug given to the patient (not necessarily the amount of drug delivered to tissue). Radiation dose planning involves leveraging patient-specific anatomy to maximize dose delivered to the tumor while minimizing off-target effects [69]. As the physics governing tissue irradiation are well-characterized, physical models can be defined to estimate spatially-resolved radiation dose prior to treatment. Several algorithms have been developed to efficiently calculate dose distribution for each patient [70]. Generally, these methods model photon interactions (e.g., photoelectric effect and Compton scattering) to simulate the energy absorbed by tissue. Several of these methods leverage a Monte Carlo approach to estimate spatially-resolved dose estimates, simulating the path of each photon through tissue probabilistically with a random number generator [71]. Briefly, the probability that a photon will travel a distance l without undergoing any interactions can be defined:
where μ is the attenuation coefficient, which is a function of photon energy (E) and the physical properties of the material the photon encounters:
where ρ is the mass density, NA is the Avogadro constant, wi the elemental weight (i.e., fractional composition) of element i in the material, Ai is the atomic mass of element i, and σi is the total cross section for element i (which is a value describing element-photon interactions such as Compton scattering) [72], [73], [74]. By modeling these interactions, spatially-resolved dose maps and the corresponding uncertainty in those estimates can be calculated. Importantly, the uncertainty in radiation dose translates into uncertainty in tumor control probability [75]. While this relationship depends on tumor-specific dose response curves, Boyer and Schulteiss estimated that the cure of early stage patients increases 2% for every 1% improvement in accuracy of dose delivery (i.e., spatially-resolved dose deposition) [76].
Critically, X-ray computed tomography images, which generate spatially-resolved μ values, can be used to estimate the tissue parameters needed for Monte Carlo simulation of dose distribution [74]. This modeling framework allows for the use of patient-specific imaging data to design patient-specific dose plans. Indeed, Rockne and colleagues demonstrated how imaging data can be used to estimate radiation response parameters to design treatment schedules that maximize tumor response in glioblastoma [23], [77].
Current Opportunities in Modeling Systemic Therapies
A key step in the evolution of precision cancer therapy will be understanding interpatient variability in drug delivery and drug response and using those differences to personalize drug dosing and administration schedules [78]. Mathematical models can be used to explore these relationships. However, model behavior is reliant on the parameter values used in model evaluation, and many of the variables in proposed models are difficult to measure clinically [25]. This presents a fundamental hurdle in the translation of these approaches into clinical practice. If these models are dependent on un-observable data, the utility of these models in making patient-specific measurements and predictions is greatly reduced.
There is a need to develop methods to measure the biological processes underlying treatment response variability. These measurements can then be used to parameterize predictive mathematical models to optimize treatment plans. Just as the linear-quadratic model can be used to characterize the radio-sensitivity of tissue, models can be applied to clinically-available data to derive measurements of tumor behavior. Below, we reimagine the use of cytotoxic chemotherapies considering this interpatient variability, applying lessons learned from radiation oncology to the technologies available clinically. We again focus specifically on pharmacokinetic and pharmacodynamic models, providing select examples for illustrative purposes.
While the differences in chemotherapy and radiotherapy are apparent, we note fundamental similarities between these modalities. First, several commonly-used chemotherapeutics, such as doxorubicin and cisplatin, are DNA-damaging agents. The response to these therapies can reasonably be compared to the DNA damage of photon therapy. Second, both chemotherapy and radiation therapy share a fractionated dosing schedule. While there exists a formalism for dose fractionation in radiation therapy with the linear-quadratic model, chemotherapies lack a widely-adopted quantitative approach to dose scheduling that balances tumor efficacy with off-target effects.
In our opinion, one of the more prominent discrepancies in these treatment modalities is their respective definitions of dose. There may exist practical reasons for this difference. An external radiation beam can be accurately tuned and targeted, and the physics of photon interactions are well-understood. Alternatively, medical oncologists must leverage patients’ circulatory systems to delivery therapeutics to tumors. While the pharmacokinetic properties of patients can be measured, this delivery method is inherently more imprecise. However, as we highlight below, the technology to estimate patient-specific pharmacokinetic and pharmacodynamic (PK/PD) properties may already be available clinically.
Therapeutic drug monitoring
Therapeutic drug monitoring (TDM) is the concept of adjusting therapeutic doses on a patient-specific basis to maximize drug efficacy. Paci et al. reviewed the relevance of TDM in the use of cytotoxic anticancer drugs [79]. They argue that the use of cytotoxic drugs meets the prerequisites for TDM, specifically: 1) a large variance in inter-patient PK parameters, 2) a defined relationship between PK and PD parameters, and 3) a delay between PD end-point and time of measurement of plasma concentration. For several cytotoxic agents dosed by BSA, pharmacokinetic measurements among patients may vary over an order of magnitude [57]. Given the high variability in PK properties and the narrow therapeutic window (i.e., the range of drug doses that can effectively treat a disease process without having toxic effects) for cytotoxic agents, this variability may be a cause for treatment failures [80], [81]. For example, significantly better outcomes were observed in children with B-lineage acute lymphoblastic leukemia when chemotherapy was dosed to reflect patient-specific clearance rates instead of BSA [82].
The concentration of drug in blood plasma can be measured via a variety of clinical chemistry techniques (e.g., immunoassays or chromatography [83]), and these measurements can be used to parameterize pharmacokinetic models that describe the absorption, distribution, metabolism, and excretion of a therapeutic agent [84]. Compartment models are often employed as pharmacokinetic models. In the context of pharmacokinetics, compartment models separate the body into physiologically-defined volumes (e.g., blood plasma, liver, kidney) that are each assumed to be homogenous with respect to drug concentration. These compartments are defined to communicate with each other with a set of rate constants. Such physiology-based pharmacokinetic models have been leveraged to describe the pharmacokinetics of several anti-cancer agents including doxorubicin [85].
Measurements of plasma drug concentrations offer an alternative to BSA to more precisely account for inter-patient variability in drug pharmacokinetics. For example, Bayesian methods have been employed to leverage limited blood plasma samples to estimate an individual’s pharmacokinetic properties [86]. These a posteriori estimates can be used to guide future dosing of therapeutics. Indeed, some clinical trials have leveraged simple PK/PD models to optimize therapy for patients [87], [88]. Alternatively, a priori dose adjustments can be made leveraging covarying patient properties. For example, carboplatin clearance was found to strongly correlate with kidney function, allowing for an empiric formula based on glomerular filtration rate (a measure of kidney function) to be derived for dosing [89]. Using these approaches to populate pharmacokinetic models will help reduce inter-patient variability and will play a role in the realization of personalized drug treatment schedules [84], [90].
Tumor-specific drug distribution
Inducing and sustaining angiogenesis is a hallmark of cancer [91]. Tumor vasculature is often morphologically and functionally immature. Relative to a healthy vasculature, tumor vasculature is tortuous and leaky with numerous blind endings and arteriovenous shunts. This impairs delivery of nutrients causing local microenvironmental changes that alter the response to therapy [6], [92], [93]. Further, significant heterogeneity in perfusion exists within a tumor, impacting both tumor growth and drug delivery [94]. Differences in treatment response may arise due to variability in tumor perfusion.
Tumor vasculature can be assessed with dynamic contrast enhanced magnetic resonance imaging (DCE-MRI). In DCE-MRI, a series of images are collected before and after a contrast agent is injected into a peripheral vein. Each image represents a snapshot of the tumor in time. Each voxel in the image set gives rise to its own time course which can be analyzed with a pharmacokinetic model to estimate physiological parameters such as the contrast agent transfer rate (Ktrans, related to vessel perfusion and permeability), the extravascular extracellular volume fraction (ve), and the plasma volume (vp) [95].
DCE-MRI parameters have been shown to be predictive of tumor response to therapy [96]. DCE-MRI data have been used in mechanistic models to estimate local nutrient and drug gradients within tumors. For example, in a model of treatment response in breast cancer, increased heterogeneity on DCE-MRI was identified to be a predictor of poor treatment outcomes as increased transport heterogeneity is coupled with increased tumor growth and poor drug response [97]. In theory, DCE-MRI data could be coupled with patient-specific PK measures (i.e., plasma drug concentration timecourses) to create tumor-specific drug distribution maps. Tagami et al. realized the goal of estimating intratumoral drug distribution through a related MRI approach which employed a drug-encapsulated approach with an MRI contrast agent. Changes in MR T1 relaxation time were measured and correlated with distribution of drug within tumors [98]. Coupling measurements of tumor vasculature with mathematical models of drug diffusion through tissue [99] will allow for the modeling the response of tumor cells to therapy to be decoupled from the tumor vasculature, thereby removing a source of variability in patient response.
Tumor-Specific PD Modeling
The efficacy of cytotoxic agents is defined by their ability to induce tumor cell death. Even within a clinically-defined grouping of tumors (e.g., triple negative breast cancer), there exists significant differences in tumor sensitivity to treatment [100]. The assessment of tumor pharmacodynamics is limited to unidimensional tumor changes as defined by the Response Evaluation Criteria in Solid Tumors (RECIST [101]). Briefly, RECIST focuses on changes in the sum of the longest dimension of tumors to assess response to treatment. These changes in tumor size are temporally downstream effects of therapy, limiting the utility of this approach to adapt treatments based on patient-specific tumor measurements. The ability to assess tumor response to treatment in real time is needed to adapt therapy schedules to maximize the odds of treatment success. We now describe three technologies that have been used to monitor treatment response upstream of tumor volume changes: diffusion-weighted magnetic resonance imaging (DW-MRI), fluoro-deoxyglucose positron emission tomography (FDG-PET), and circulating tumor DNA (ctDNA) samples.
Cellular changes within the tumor precede tumor volume changes. In DW-MRI, the diffusion of water molecules through tissue is measured and described by the apparent diffusion coefficient (ADC). This modality relies on the thermally-induced random movement of water molecules (known as Brownian motion). In tissue, this movement is not entirely random as water molecules encounter a number of barriers to diffusion (e.g., cell membranes and extracellular matrix), and the observed diffusion largely depends on the number and separation of barriers that a water molecule encounters. DW-MRI methods have been developed to measure the ADC at the voxel level, and in well-controlled situations the variations in ADC have been shown to correlate inversely with tissue cellularity [102]. Changes in tumor ADC precede tumor volume changes, providing an early biomarker of treatment response [103].
Changes in tumor metabolism can precede tumor morphology changes and may be predictive of treatment response in breast cancer [104]. FDG-PET provides a measure of glucose metabolism in tumors. In FDG-PET, 18F-FDG is injected into a peripheral vein. As it circulates, the FDG is transported into cells and phosphorylated, trapping the FDG within cells. As 18F-FDG decays, it emits positrons, which annihilate with nearby electrons. Each annihilation yields two (nearly) antiparallel 511 kEV photons, which are detected and used to map FDG distribution. FDG-PET data are summarized by the standardized uptake value (SUV), which normalizes for patient weight and injected dose [105].
Tumors continually shed DNA into the bloodstream during the course of tumor development. These circulating tumor DNA (ctDNA) potentially may serve as “liquid biopsies,” providing measurements on the mutational status of breast cancers, assessment of treatment response, and guidance for therapy selection [106], [107], [108]. Notably, these data have been shown to be an early predictor of relapse in breast cancer patients [109].
Taken together, these measurements of tumor pharmacodynamics can be leveraged to parameterize models to describe the tumor response to treatment. For example, since ADC changes following treatment are predictive of ultimate treatment response [110], our group has demonstrated how ADC values can be used to estimate response rates of tumors:
where N(x̅, t) is the number of tumor cells at position x̅ and time t and k is the spatially-dependent growth rate [27]. This measure of tumor response can be combined with the assessment of off-target hematologic toxicities, providing a pathway to personalize chemotherapy schedules through PK/PD optimization [111]. Similarly, Liu and colleagues have incorporated SUV measurements derived from FDG-PET imaging into a predictive tumor growth model [112]. ctDNA data can be used to track tumor genetic changes and populate evolutionary dynamics models to predict treatment response [24]. The above technologies present independent means to assess tumor response to therapy. With appropriate mathematical models incorporating the data from these modalities, real-time adjustment of therapeutic schedules in response to tumor changes may be possible.
Vision for Systemic Chemotherapy
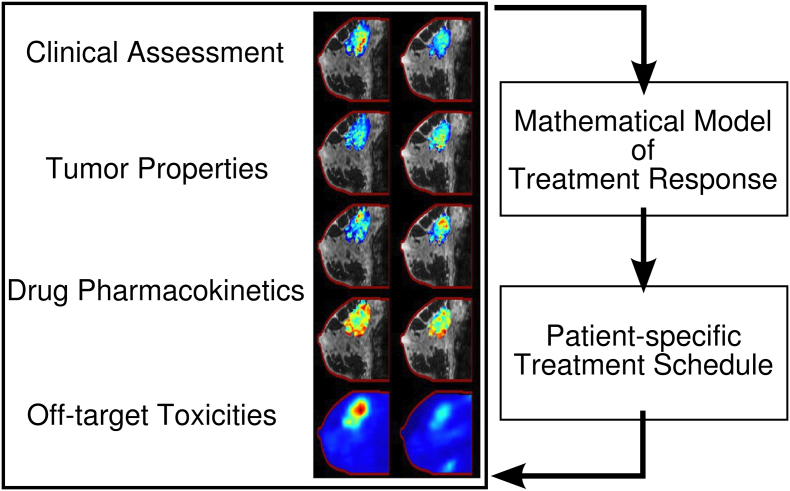
Given the goal of delivering the optimal therapy on the optimal schedule for each patient, we highlighted some potential tools for realizing that goal in section 3. As noted above, overly complex models, which require several parameters to be estimated for each patient, present a difficult task in translation to a clinical population. Radiation oncology relies on a relatively simplistic approximation of dose response to develop treatment schedules. Potentially, such simple models may improve the use of chemotherapy by integrating currently available measurements of treatment response. It is our vision that a classical oncology toolkit be available to clinicians, to leverage measurable patient data to not only select appropriate treatments but also optimize the schedule on which those therapies are given (Figure 1).
Figure 1.
Vision for systemic chemotherapy. Following diagnosis and staging of a cancer, a patient is evaluated clinically with a panel of imaging tests and bloodwork. These data are used to quantify various tumor properties, drug pharmacokinetics, and off-target toxicities to parameterize a mathematical model of treatment response. The model is leveraged to identify optimal treatment plans. This process is repeated throughout the course of treatment to yield treatment plans that co-evolve with the patient’s tumor.
Following diagnosis and staging of tumors, the patient would be evaluated with a panel of imaging tests and bloodwork. Following an initial round of therapy (and, on occasion, through the course of therapy), the testing is repeated, providing data to initialize and constrain predictive models of treatment response. A pre-defined objective function that balances tumor efficacy with off-target toxicities is parameterized with the patient-specific data and is optimized to identify a patient-specific treatment schedule. Simply, the objective of cancer therapy is to maximize survival while minimizing morbidity. Formally, we define:
where x is the therapy schedule, f is the functional relationship between tumor behavior and survival, and g is the functional relationship between off-target toxicities and survival. Fortunately, the toxicity limits of various tissues have been defined, and clinical assays have been developed to monitor those toxicities. For example, hematologic toxicities can be measured through blood sampling. Cardiotoxicity can be assessed through electro- and echocardiography. Thus, the function g can be defined. However, Tumor(x), how a tumor responds to treatment plan x, and f, the relationship between survival and tumor behavior, must be defined. If these functions can be defined, a robust literature for optimization problems already exists [113]. Thus, the question becomes, “How can we use (for example) the technologies highlighted above to define and parameterize these functions?”
Next Steps
Medical oncology is in need of a mathematical modeling toolkit that can leverage clinically-available measurements to optimize treatment selection and schedules in the same way radiation oncologists use clinically-available imaging data for treatment planning. Just as the therapeutic choice has been optimized to match tumor genetics, the delivery of those therapeutics can be optimized based on patient-specific PK/PD properties.
Under the current approach to breast cancer therapy, treating clinicians are tasked with integrating multi-modal biomarker changes into a holistic understanding of treatment response. They are challenged to intervene based on those patient-specific measures. In this context, treatment decisions become increasingly complex with the advent of new technologies and treatments. Mathematical modeling offers a means to build a structured, theoretical understanding which summarizes this complexity, providing clinicians assistance in developing treatment plans. For preclinical investigators, modeling similarly can expedite experimental investigations. Simulations are inexpensive relative to in vitro and in vivo experiments. Further, modeling provides opportunities for discovery when model predictions do not match experimental data. In this way, mathematical modeling has the potential to expedite translation of medical discoveries into patient care.
The Cancer Moonshot Initiative [114] highlights the opportunity that exists by adopting screening and treatment plans known to work on a wide-scale basis. There is a need for such implementation science in the development and deployment of cancer therapeutics. While tumor genotype most likely plays an outsized role in determining response, other measurable factors such as tumor microenviroment and patient pharmacokinetics also influence response. The extensive characterization of tumor genetics has yielded an arsenal of therapeutics that can more precisely target cancer cells. An equally focused approach to the science of deploying these therapeutics on an optimal schedule is now needed. In the dosing and scheduling domains, we are in a similar position to cancer therapy prior to the advent of genotyping technologies. Advances in clinical chemistry and imaging sciences offer platforms to develop biologically-driven, treatment response models while maintaining the ability to translate those models to a clinical population. These tools will provide the measurements needed to test various dose and scheduling hypotheses. Revisiting our earlier analogy, what is the F = ma for cancer? We have the means to measure tumor “mass” and “acceleration” (i.e., the multifactorial response of a tumor to therapy). Further, we can measure treatment “force” (i.e., drug pharmacokinetics). A modeling framework that relates these variables would offer the opportunity to adjust and optimize treatment regimens to maximize response. Mathematical models will form the foundation of this approach, and they will hasten the implementation and maximize the benefit of current (and future) therapeutics.
Acknowledgements
This work was supported by the National Institutes of Health (grant numbers R01CA186193, U01CA174706, and F30CA203220), and the Cancer Prevention and Research Institute of Texas RR160005.
Footnotes
Conflicts of Interest: The authors declare no potential conflicts of interest.
References
- 1.National Research Council (US) Committee on A Framework for Developing a New Framework for Developing a New Taxonomy of Disease, Taxonomy of Toward Precision Medicine [Internet] Towar. Precis. Med. Build. a Knowl. Netw. Biomed. Res. a New Taxon. Dis. National Academies Press (US); 2011. [[cited 2016 Dec 20]. Available from http://www.ncbi.nlm.nih.gov/pubmed/22536618] [Google Scholar]
- 2.Garraway LA, Verweij J, Ballman KV. Precision oncology: an overview. J Clin Oncol. 2013;31:1803–1805. doi: 10.1200/JCO.2013.49.4799. [[cited 2016 Dec 9] Available from http://jco.ascopubs.org/cgi/doi/10.1200/JCO.2013.49.4799] [DOI] [PubMed] [Google Scholar]
- 3.Garraway LA. Genomics-driven oncology: framework for an emerging paradigm. J Clin Oncol. 2013;31:1806–1814. doi: 10.1200/JCO.2012.46.8934. [[cited 2016 Dec 9] Available from http://jco.ascopubs.org/cgi/doi/10.1200/JCO.2012.46.8934] [DOI] [PubMed] [Google Scholar]
- 4.Olopade OI, Grushko TA, Nanda R, Huo D. Advances in breast cancer: Pathways to personalized medicine. Clin Cancer Res. 2008;14:7988–7999. doi: 10.1158/1078-0432.CCR-08-1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Romond EH, Perez EA, Bryant J, Suman VJ, Geyer CE, Davidson NE, Tan-Chiu E, Martino S, Paik S, Kaufman PA. Trastuzumab plus Adjuvant Chemotherapy for Operable HER2-Positive Breast Cancer. N Engl J Med. 2005;353:1673–1684. doi: 10.1056/NEJMoa052122. [[cited 2017 Jan 3] Available from: http://www.nejm.org/doi/abs/10.1056/NEJMoa052122] [DOI] [PubMed] [Google Scholar]
- 6.Trédan O, Galmarini CM, Patel K, Tannock IF. Drug resistance and the solid tumor microenvironment. J Natl Cancer Inst. 2007;99:1441–1454. doi: 10.1093/jnci/djm135. [Available from http://www.ncbi.nlm.nih.gov/pubmed/17895480] [DOI] [PubMed] [Google Scholar]
- 7.Jain RK. Normalizing tumor microenvironment to treat cancer: bench to bedside to biomarkers. J Clin Oncol. 2013;31:2205–2218. doi: 10.1200/JCO.2012.46.3653. [[cited 2014 Dec 22]Available from http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3731977&tool=pmcentrez&rendertype=abstract] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jain RK. Normalization of tumor vasculature: an emerging concept in antiangiogenic therapy. Science. 2005;307:58–62. doi: 10.1126/science.1104819. [DOI] [PubMed] [Google Scholar]
- 9.Rudek M, Sparreboom A, Garrett-Mayer E, Armstrong D, Wolff A, Verweij J, Baker SD. Factors affecting pharmacokinetic variability following doxorubicin and docetaxel-based therapy. Eur J Cancer. 2004;40:1170–1178. doi: 10.1016/j.ejca.2003.12.026. [DOI] [PubMed] [Google Scholar]
- 10.Weinshilboum RM, Wang L. Pharmacogenetics and pharmacogenomics: development, science, and translation. Annu Rev Genomics Hum Genet. 2006;7:223–245. doi: 10.1146/annurev.genom.6.080604.162315. [Available from http://www.annualreviews.org/doi/10.1146/annurev.genom.6.080604.162315] [DOI] [PubMed] [Google Scholar]
- 11.Notta F, Mullighan CG, Wang JCY, Poeppl A, Doulatov S, Phillips LA, Ma J, Minden MD, Downing JR, Dick JE. Evolution of human BCR–ABL1 lymphoblastic leukaemia-initiating cells. Nature. 2011;469:362–367. doi: 10.1038/nature09733. [cited 2017 Dec 21 Available from http://www.ncbi.nlm.nih.gov/pubmed/21248843] [DOI] [PubMed] [Google Scholar]
- 12.Navin N, Krasnitz A, Rodgers L, Cook K, Meth J, Kendall J, Riggs M, Eberling Y, Troge J, Grubor V. Vol. 20. Cold Spring Harbor Laboratory Press; 2010. Inferring tumor progression from genomic heterogeneity; pp. 68–80. (Genome Res.). [cited 2017 Dec 21 Available from http://www.ncbi.nlm.nih.gov/pubmed/19903760] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gerlinger M, Horswell S, Larkin J, Rowan AJ, Salm MP, Varela I, Fisher R, McGranahan N, Matthews N, Santos CR. Genomic architecture and evolution of clear cell renal cell carcinomas defined by multiregion sequencing. Nat Genet. 2014;46:225–233. doi: 10.1038/ng.2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Anderson K, Lutz C, Van Delft FW, Bateman CM, Guo Y, Colman SM, Kempski H, Moorman AV, Titley I, Swansbury J. Genetic variegation of clonal architecture and propagating cells in leukaemia. Nature. 2011;469:356–361. doi: 10.1038/nature09650. [DOI] [PubMed] [Google Scholar]
- 15.Williams MJ, Werner B, Barnes CP, Graham TA, Sottoriva A. Identification of neutral tumor evolution across cancer types. Nat Genet. 2016;48:238–244. doi: 10.1038/ng.3489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ling S, Hu Z, Yang Z, Yang F, Li Y, Lin P, Chen K, Dong L, Cao L, Tao Y. Extremely high genetic diversity in a single tumor points to prevalence of non-Darwinian cell evolution. Proc Natl Acad Sci. 2015;112:E6496–505. doi: 10.1073/pnas.1519556112. Available from: http://www.pnas.org/lookup/doi/10.1073/pnas.1519556112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Citron ML, Berry DA, Cirrincione C, Hudis C, Winer EP, Gradishar WJ, Davidson NE, Martino S, Livingston R, Ingle JN. Randomized trial of dose-dense versus conventionally scheduled and sequential versus concurrent combination chemotherapy as postoperative adjuvant treatment of node-positive primary breast cancer: First report of Intergroup Trial C9741/Cancer and Leukemia. J Clin Oncol. 2003;21:1431–1439. doi: 10.1200/JCO.2003.09.081. [DOI] [PubMed] [Google Scholar]
- 18.Montagna E, Cancello G, Dellapasqua S, Munzone E, Colleoni M. Metronomic therapy and breast cancer: A systematic review. Cancer Treat Rev. 2014;40:942–950. doi: 10.1016/j.ctrv.2014.06.002. [DOI] [PubMed] [Google Scholar]
- 19.Sorace AG, Quarles CC, Whisenant JG, Hanker AB, McIntyre JO, Sanchez VM, Yankeelov TE. Trastuzumab improves tumor perfusion and vascular delivery of cytotoxic therapy in a murine model of HER2+ breast cancer: preliminary results. Breast Cancer Res Treat. 2016;155:273–284. doi: 10.1007/s10549-016-3680-8. Available from: http://link.springer.com/10.1007/s10549-016-3680-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moreno L, Pearson AD. How can attrition rates be reduced in cancer drug discovery? Expert Opin Drug Discov. 2013;8:363–368. doi: 10.1517/17460441.2013.768984. [Available from http://www.tandfonline.com/doi/full/10.1517/17460441.2013.768984] [DOI] [PubMed] [Google Scholar]
- 21.Li X, Abramson RG, Arlinghaus LR, Kang H, Chakravarthy AB, Abramson VG, Farley J, Mayer IA, Kelley MC, Meszoely IM. Multiparametric magnetic resonance imaging for predicting pathological response after the first cycle of neoadjuvant chemotherapy in breast cancer. Investig Radiol. 2015;50:195–204. doi: 10.1097/RLI.0000000000000100. Available from: http://www.ncbi.nlm.nih.gov/pubmed/25360603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gillies RJ, Verduzco D, Gatenby RA. Evolutionary dynamics of carcinogenesis and why targeted therapy does not work. Nat Rev Cancer. 2012;12:487–493. doi: 10.1038/nrc3298. [Available from http://www.nature.com/doifinder/10.1038/nrc3298] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rockne R, Alvord EC, Jr., Rockhill JK, Swanson KR. A mathematical model for brain tumor response to radiation therapy. J Math Biol. 2009;58:561–578. doi: 10.1007/s00285-008-0219-6. http://www.ncbi.nlm.nih.gov/pubmed/18815786%5Cnhttp://www.ncbi.nlm.nih.gov/pmc/articles/PMC3784027/pdf/nihms511107.pdf Available from. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Altrock PM, Liu LL, Michor F. The mathematics of cancer: integrating quantitative models. Nat Rev Cancer. 2015;15:730–745. doi: 10.1038/nrc4029. [Available from http://www.nature.com/doifinder/10.1038/nrc4029] [DOI] [PubMed] [Google Scholar]
- 25.Yankeelov TE, Atuegwu N, Hormuth D, Weis JA, Barnes SL, Miga MI, Reircha EC, Quaranta V. Clinically relevant modeling of tumor growth and treatment response. Sci Transl Med. 2013;5 doi: 10.1126/scitranslmed.3005686. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3938952&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gatenby RA, Silva AS, Gillies RJ, Frieden BR. Adaptive therapy. Cancer Res. 2009;69:4894–4903. doi: 10.1158/0008-5472.CAN-08-3658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Atuegwu NC, Arlinghaus LR, Li X, Welch EB, Chakravarthy BA, Gore JC, Yankeelov TE. Integration of diffusion-weighted MRI data and a simple mathematical model to predict breast tumor cellularity during neoadjuvant chemotherapy. Magn Reson Med. 2011;66:1689–1696. doi: 10.1002/mrm.23203. Available from: http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=21956404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Anderson AR, Quaranta V. Integrative mathematical oncology. Nat Rev Cancer. 2008;8:227–234. doi: 10.1038/nrc2329. [DOI] [PubMed] [Google Scholar]
- 29.Anderson ARA. A hybrid mathematical model of solid tumour invasion: the importance of cell adhesion. Math Med Biol. 2005;22:163–186. doi: 10.1093/imammb/dqi005. [Available from https://academic.oup.com/imammb/article-lookup/doi/10.1093/imammb/dqi005] [DOI] [PubMed] [Google Scholar]
- 30.Yankeelov TE, Quaranta V, Evans KJ, Rericha EC. Toward a science of tumor forecasting for clinical oncology. Cancer Res. 2015;75:918–923. doi: 10.1158/0008-5472.CAN-14-2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yankeelov TE, An G, Saut O, Luebeck EG, Popel AS, Ribba B, Vicini P, Zhou X, Weis JA, Ye K. Multi-scale Modeling in Clinical Oncology: Opportunities and Barriers to Success. Ann Biomed Eng. 2016;44:2626–2641. doi: 10.1007/s10439-016-1691-6. [cited 2016 Dec 20 Available from: http://link.springer.com/10.1007/s10439-016-1691-6] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marusyk A, Polyak K. Tumor heterogeneity: Causes and consequences. Biochim Biophys Acta. 2010;1805:105–117. doi: 10.1016/j.bbcan.2009.11.002. [[cited 2017 Jul 6] Available from http://linkinghub.elsevier.com/retrieve/pii/S0304419X09000742] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Anderson A, Chaplain MAJ. Continuous and discrete mathematical models of tumor-induced angiogenesis. Bull Math Biol. 1998;60:857–899. doi: 10.1006/bulm.1998.0042. [Springer-Verlag; [cited 2018 Mar 19] Available from http://link.springer.com/10.1006/bulm.1998.0042] [DOI] [PubMed] [Google Scholar]
- 34.Tang L, van de Ven AL, Guo D, Andasari V, Cristini V, Li KC, Zhou X. Computational modeling of 3D tumor growth and angiogenesis for chemotherapy evaluation. PLoS ONE. 2014;9 doi: 10.1371/journal.pone.0083962. [cited 2015 Mar 31 Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3880288&tool=pmcentrez&rendertype=abstract] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Byrne HM. Dissecting cancer through mathematics : from the cell to the animal model. Nat Rev Cancer. 2010;10:221–230. doi: 10.1038/nrc2808. [DOI] [PubMed] [Google Scholar]
- 36.Fisher B, Slack N, Katrych D, Wolmark N. Ten year follow-up results of patients with carcinoma of the breast in a co-operative clinical trial evaluating surgical adjuvant chemotherapy. Surg Gynecol Obstet. 1975;140:528–534. [[cited 2017 Jan 17] Available from http://www.ncbi.nlm.nih.gov/pubmed/805475] [PubMed] [Google Scholar]
- 37.Goodman LS, Wintrobe MM, Dameshek W, Goodman MJ, Gilman A, McLennan MT. Vol. 132. American Medical Association; 1946. Nitrogen Mustard Therapy; pp. 126–132. (J Am Med Assoc.). [cited 2017 Jan 17 Available from: http://jama.jamanetwork.com/article.aspx?doi=10.1001/jama.1946.02870380008004] [DOI] [PubMed] [Google Scholar]
- 38.Liedtke C, Mazouni C, Hess KR, André F, Tordai A, Mejia JA, Symmans WF, Gonzalez-Angulo AM, Hennessy B, Green M. Response to neoadjuvant therapy and long-term survival in patients with triple-negative breast cancer. J Clin Oncol. 2008;26:1275–1281. doi: 10.1200/JCO.2007.14.4147. [cited 2014 Aug 26 Available from: http://www.ncbi.nlm.nih.gov/pubmed/18250347] [DOI] [PubMed] [Google Scholar]
- 39.Chatterjee K, Zhang J, Honbo N, Karliner JS. Doxorubicin cardiomyopathy. Cardiology. 2010;115:155–162. doi: 10.1159/000265166. [Available from http://www.ncbi.nlm.nih.gov/pubmed/20016174] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fisher B. From Halsted to prevention and beyond: advances in the management of breast cancer during the twentieth century. Eur J Cancer. 1999;35:1963–1973. doi: 10.1016/s0959-8049(99)00217-8. [Available from http://www.ncbi.nlm.nih.gov/pubmed/10711239] [DOI] [PubMed] [Google Scholar]
- 41.Skipper HE. Kinetics of mammary tumor cell growth and implications for therapy. Cancer. 1971;28:1479–1499. doi: 10.1002/1097-0142(197112)28:6<1479::aid-cncr2820280622>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 42.Fisher B, Carbone P, Economou SG, Frelick R, Glass A, Lerner H, Redmond C, Zelen M, Band P, Katrych DL. L-Phenylalanine Mustard (L-PAM) in the Management of Primary Breast Cancer. N Engl J Med. 1975;292:117–122. doi: 10.1056/NEJM197501162920301. [cited 2017 Jan 17 Available from: http://www.nejm.org/doi/abs/10.1056/NEJM197501162920301] [DOI] [PubMed] [Google Scholar]
- 43.Bonadonna G, Brusamolino E, Valagussa P, Rossi A, Brugnatelli L, Brambilla C, De Lena M, Tancini G, Bajetta E, Musumeci R. Combination Chemotherapy as an Adjuvant Treatment in Operable Breast Cancer. N Engl J Med. 1976;294:405–410. doi: 10.1056/NEJM197602192940801. [cited 2017 Jan 17 Available from: http://www.nejm.org/doi/abs/10.1056/NEJM197602192940801] [DOI] [PubMed] [Google Scholar]
- 44.Hortobagyi GN. High-dose chemotherapy for primary breast cancer: facts versus anecdotes. J Clin Oncol. 1999;17:25–29. [American Society of Clinical Oncology; [cited 2016 Aug 24]. Available from http://www.ncbi.nlm.nih.gov/pubmed/10630258] [PubMed] [Google Scholar]
- 45.Stadtmauer EA, O’Neill A, Goldstein LJ, Crilley PA, Mangan KF, Ingle JN, Brodsky I, Martino S, Lazarus HM, Erban JK. Conventional-dose chemotherapy compared with high-dose chemotherapy plus autologous hematopoietic stem-cell transplantation for metastatic breast cancer. Philadelphia Bone Marrow Transplant Group. N Engl J Med. 2000;342:1069–1076. doi: 10.1056/NEJM200004133421501. [cited 2016 Aug 24 Available from: http://www.nejm.org/doi/abs/10.1056/NEJM200004133421501] [DOI] [PubMed] [Google Scholar]
- 46.Norton L, Simon R. Growth curve of an experimental solid tumor following radiotherapy. J Natl Cancer Inst. 1977;58:1735–1741. doi: 10.1093/jnci/58.6.1735. [Available from http://www.ncbi.nlm.nih.gov/pubmed/194044] [DOI] [PubMed] [Google Scholar]
- 47.Norton L. A Gompertzian model of human breast cancer growth. Cancer Res. 1988;48:7067–7071. [PubMed] [Google Scholar]
- 48.Goldie JH, Coldman AJ. A mathematic model for relating the drug sensitivity of tumors to their spontaneous mutation rate. Cancer Treat Rep. 1979;63:1727–1733. [Available from http://www.ncbi.nlm.nih.gov/pubmed/526911] [PubMed] [Google Scholar]
- 49.Bonadonna G, Zambetti M, Moliterni A, Gianni L, Valagussa P. Clinical relevance of different sequencing of doxorubicin and cyclophosphamide, methotrexate, and Fluorouracil in operable breast cancer. J Clin Oncol. 2004;22:1614–1620. doi: 10.1200/JCO.2004.07.190. [[cited 2017 Jan 19] Available from http://jco.ascopubs.org/cgi/doi/10.1200/JCO.2004.07.190] [DOI] [PubMed] [Google Scholar]
- 50.Browder T, Butterfield CE, Kräling BM, Shi B, Marshall B, O’Reilly MS, Folkman J. Antiangiogenic scheduling of chemotherapy improves efficacy against experimental drug-resistant cancer. Cancer Res. 2000;60:1878–1886. [PubMed] [Google Scholar]
- 51.Foo J, Michor F. Evolution of resistance to anti-cancer therapy during general dosing schedules. J Theor Biol. 2010;263:179–188. doi: 10.1016/j.jtbi.2009.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gatenby RA. A change of strategy in the war on cancer. Nature. 2009;459:508–509. doi: 10.1038/459508a. [[cited 2017 Feb 16] Available from http://www.nature.com/doifinder/10.1038/459508a] [DOI] [PubMed] [Google Scholar]
- 53.Miller AA. Body surface area in dosing anticancer agents: scratch the surface! J Natl Cancer Inst. 2002;94:1822–1831. doi: 10.1093/jnci/94.24.1822. [[cited 2017 Jul 5]. Available from https://academic.oup.com/jnci/article-lookup/doi/10.1093/jnci/94.24.1822] [DOI] [PubMed] [Google Scholar]
- 54.Redlarski G, Palkowski A, Krawczuk M. Body surface area formulae: an alarming ambiguity. Sci Rep. 2016;6:27966. doi: 10.1038/srep27966. [cited 2017 Jan 20 Available from: http://www.nature.com/articles/srep27966] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pinkel D. The use of body surface area as a criterion of drug dosage in cancer chemotherapy. Cancer Res. 1958;18:853–856. [PubMed] [Google Scholar]
- 56.Sawyer M, Ratain MJ. Body surface area as a determinant of pharmacokinetics and drug dosing. Invest New Drugs. 2001;19:171–177. doi: 10.1023/a:1010639201787. [[cited 2017 Jan 17]. Available from http://link.springer.com/10.1023/A:1010639201787] [DOI] [PubMed] [Google Scholar]
- 57.Gurney H. Dose calculation of anticancer drugs: a review of the current practice and introduction of an alternative. J Clin Oncol. 1996;14:2590–2611. doi: 10.1200/JCO.1996.14.9.2590. [[cited 2016 Dec 1] Available from http://www.ncbi.nlm.nih.gov/pubmed/8823340] [DOI] [PubMed] [Google Scholar]
- 58.Bernier J, Hall EJ, Giaccia A. Timeline: Radiation oncology: a century of achievements. Nat Rev Cancer. 2004;4:737–747. doi: 10.1038/nrc1451. [cited 2017 Jan 23] Available from http://www.nature.com/doifinder/10.1038/nrc1451] [DOI] [PubMed] [Google Scholar]
- 59.Prise KM, Schettino G, Folkard M, Held KD. New insights on cell death from radiation exposure. Lancet Oncol. 2005;6:520–528. doi: 10.1016/S1470-2045(05)70246-1. [DOI] [PubMed] [Google Scholar]
- 60.Wouters BG. Cell death after irradiation: how, when and why cells die. In: Joiner MC, van der Kogel AJ, editors. Basic Clin Radiobiol. 4th ed. CRC Press; 2009. pp. 27–40. [Google Scholar]
- 61.Joiner MC. Quantifying cell kill and cell survival. In: Joiner MC, van der Kogel AJ, editors. Basic Clin Radiobiol. 4th ed. CRC Press; 2009. pp. 41–55. [Google Scholar]
- 62.Douglas BG, Fowler JF. The effect of multiple small doses of x rays on skin reactions in the mouse and a basic interpretation. Radiat Res. 1976;66:401–426. [PubMed] [Google Scholar]
- 63.Curtis SB. Lethal and potentially lethal lesions induced by radiation – a unified repair model. Radiat Res. 1986;106:252–270. [PubMed] [Google Scholar]
- 64.Pajonk F, Vlashi E, McBride WH. Radiation resistance of cancer stem cells: the 4 R’s of radiobiology revisited. Stem Cells. 2010;28:639–648. doi: 10.1002/stem.318. [[cited 2017 Jan 24] Available from http://www.ncbi.nlm.nih.gov/pubmed/20135685] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Withers HR, Thames HD, Peters LJ. A new isoeffect curve for change in dose per fraction. Radiother Oncol. 1983;1:187–191. doi: 10.1016/s0167-8140(83)80021-8. [DOI] [PubMed] [Google Scholar]
- 66.Bentzen SM, Joiner MC, Joiner M. The linear-quadratic approach in clinical practice. In: van der Kogel A, editor. Basic Clin Radiobiol. 4th ed. CRC Press; 2009. pp. 120–134. [Google Scholar]
- 67.Bourhis J, Overgaard J, Audry H, Ang KK, Saunders M, Bernier J, Horiot JC, Le Maître A, Pajak TF, Poulsen MG. Hyperfractionated or accelerated radiotherapy in head and neck cancer: a meta-analysis. Lancet. 2006;368:843–854. doi: 10.1016/S0140-6736(06)69121-6. [cited 2017 Jun 28 Available from: http://linkinghub.elsevier.com/retrieve/pii/S0140673606691216] [DOI] [PubMed] [Google Scholar]
- 68.Whelan TJ, Pignol J-P, Levine MN, Julian JA, MacKenzie R, Parpia S, Shelley W, Grimard L, Bowen J, Lukka H. Long-Term Results of Hypofractionated Radiation Therapy for Breast Cancer. N Engl J Med. 2010;362:513–520. doi: 10.1056/NEJMoa0906260. [cited 2017 Jun 28 Available from: http://www.nejm.org/doi/abs/10.1056/NEJMoa0906260] [DOI] [PubMed] [Google Scholar]
- 69.Verellen D, De Ridder M, Linthout N, Tournel K, Soete G, Storme G. Innovations in image-guided radiotherapy. Nat Rev Cancer. 2007;7:949–960. doi: 10.1038/nrc2288. [[cited 2017 Feb 2] Available from http://www.nature.com/doifinder/10.1038/nrc2288] [DOI] [PubMed] [Google Scholar]
- 70.Oelkfe U, Scholz C. Dose calculation algorithms. N Technol Radiat Oncol. 2006:187–196. [Berlin/Heidelberg [cited 2017 Jan 18] Available from http://link.springer.com/10.1007/3-540-29999-8_15] [Google Scholar]
- 71.Monte Andreo P. Carlo techniques in medical radiation physics. Phys Med Biol. 1991:861–920. doi: 10.1088/0031-9155/36/7/001. 36. [[cited 2017 Jul 23] Available from http://stacks.iop.org/0031-9155/36/i=7/a=001?key=crossref.ba74de27c7524de42c998f4f8548e6d8] [DOI] [PubMed] [Google Scholar]
- 72.Hubbell JH, Seltzer SM. National Inst. of Standards and Technology-PL; 1995. Tables of X-ray mass attenuation coefficients and mass energy-absorption coefficients 1 keV to 20 MeV for elements Z= 1 to 92 and 48 additional substances of dosimetric interest. [Google Scholar]
- 73.Jackson DF, Hawkes DJ. X-ray attenuation coefficients of elements and mixtures. Phys Rep. 1981;70:169–233. [ [cited 2017 Jul 23] Available from http://linkinghub.elsevier.com/retrieve/pii/0370157381900144] [Google Scholar]
- 74.Schneider W, Bortfeld T, Schlegel W. Correlation between CT numbers and tissue parameters needed for Monte Carlo simulations of clinical dose distributions. Phys Med Biol. 2000;45:459–478. doi: 10.1088/0031-9155/45/2/314. [[cited 2017 Jan 23] Available from http://stacks.iop.org/0031-9155/45/i=2/a=314?key=crossref.e78c9ce7980571a8752f2265863c8960] [DOI] [PubMed] [Google Scholar]
- 75.Brahme A. Dosimetric precision requirements in radiation therapy. Acta Radiol Oncol. 1984;23:379–391. doi: 10.3109/02841868409136037. [[cited 2017 Jul 23] Available from http://www.tandfonline.com/doi/full/10.3109/02841868409136037] [DOI] [PubMed] [Google Scholar]
- 76.Boyer AL, Schultheiss T. Effects of dosimetric and clinical uncertainty on complication-free local tumor control. Radiother Oncol. 1988;11:65–71. doi: 10.1016/0167-8140(88)90046-1. [[cited 2017 Jan 23] Available from http://linkinghub.elsevier.com/retrieve/pii/0167814088900461] [DOI] [PubMed] [Google Scholar]
- 77.Rockne R, Rockhill JK, Mrugala M, Spence AM, Kalet I, Hendrickson K, Lai A, Cloughesy T, Alvord EC, Jr., Swanson KR. Predicting the efficacy of radiotherapy in individual glioblastoma patients in vivo: a mathematical modeling approach. Phys Med Biol. 2010;55:3271–3285. doi: 10.1088/0031-9155/55/12/001. [cited 2015 Mar 22 Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3786554&tool=pmcentrez&rendertype=abstract] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Peck RW. The right dose for every patient: a key step for precision medicine. Nat Rev Drug Discov. 2015;15:145–146. doi: 10.1038/nrd.2015.22. [[cited 2017 Feb 3] Available from http://www.nature.com/doifinder/10.1038/nrd.2015.22] [DOI] [PubMed] [Google Scholar]
- 79.Paci A, Veal G, Bardin C, Levêque D, Widmer N, Beijnen J, Astier A, Chatelut E. Review of therapeutic drug monitoring of anticancer drugs part 1 – Cytotoxics. Eur J Cancer. 2014;50:2010–2019. doi: 10.1016/j.ejca.2014.04.014. [DOI] [PubMed] [Google Scholar]
- 80.Undevia SD, Gomez-Abuin G, Ratain MJ. Pharmacokinetic variability of anticancer agents. Nat Rev Cancer. 2005;5:447–458. doi: 10.1038/nrc1629. [[cited 2017 Feb 27]. Available from http://www.nature.com/doifinder/10.1038/nrc1629] [DOI] [PubMed] [Google Scholar]
- 81.Rousseau A, Marquet P. Application of pharmacokinetic modelling to the routine therapeutic drug monitoring of anticancer drugs. Fundam Clin Pharmacol. 2002;16:253–262. doi: 10.1046/j.1472-8206.2002.00086.x. [[cited 2017 Jul 5] Available from http://doi.wiley.com/10.1046/j.1472-8206.2002.00086.x] [DOI] [PubMed] [Google Scholar]
- 82.Evans WE, Relling MV, Rodman JH, Crom WR, Boyett JM, Pui C-H. Conventional compared with individualized chemotherapy for childhood acute lymphoblastic leukemia. N Engl J Med. 1998;338:499–505. doi: 10.1056/NEJM199802193380803. [[cited 2017 Jul 23]. Available from http://www.nejm.org/doi/abs/10.1056/NEJM199802193380803] [DOI] [PubMed] [Google Scholar]
- 83.Dasgupta A, Datta P. Analytical Techniques for Measuring Concentrations of Therapeutic Drugs in Biological Fluids. Handb Drug Monit Methods. 2008:67–86. [Totowa, NJ: Humana Press cited 2017 Jul 23 Available from http://link.springer.com/10.1007/978-1-59745-031-7_3] [Google Scholar]
- 84.Barbolosi D, Ciccolini J, Lacarelle B, Barlési F, André N. Computational oncology – mathematical modelling of drug regimens for precision medicine. Nat Rev Clin Oncol. 2015;13:242–254. doi: 10.1038/nrclinonc.2015.204. [[cited 2017 Jan 24] Available from http://www.nature.com/doifinder/10.1038/nrclinonc.2015.204] [DOI] [PubMed] [Google Scholar]
- 85.Gustafson DL, Rastatter JC, Colombo T, Long ME. Doxorubicin pharmacokinetics: Macromolecule binding, metabolism, and excretion in the context of a physiologic model. J Pharm Sci. 2002;91:1488–1501. doi: 10.1002/jps.10161. [Wiley Subscription Services, Inc., A Wiley Company [cited 2017 Jul 5] Available from http://linkinghub.elsevier.com/retrieve/pii/S0022354916310243] [DOI] [PubMed] [Google Scholar]
- 86.Rousseau A, Marquet P, Debord J, Sabot C, Lachatre G. Adaptive control methods for the dose individualisation of anticancer agents. Clin Pharmacokinet. 2000;38:315–353. doi: 10.2165/00003088-200038040-00003. [[cited 2017 Feb 14] Available from http://link.springer.com/10.2165/00003088-200038040-00003] [DOI] [PubMed] [Google Scholar]
- 87.Barbolosi D, Ciccolini J, Meille C, Elharrar X, Faivre C, Lacarelle B, André N, Barlesi F. Metronomics chemotherapy: Time for computational decision support. Cancer Chemother Pharmacol. 2014;74:647–652. doi: 10.1007/s00280-014-2546-1. [DOI] [PubMed] [Google Scholar]
- 88.Joerger M, Kraff S, Huitema ADR, Feiss G, Moritz B, Schellens JHM, Beijnen JH, Jaehde U. Vol. 51. Springer International Publishing; 2012. Evaluation of a Pharmacology-Driven Dosing Algorithm of 3-Weekly Paclitaxel Using Therapeutic Drug Monitoring; pp. 607–617. (Clin Pharmacokinet.). [cited 2017 Feb 3 Available from: http://link.springer.com/10.1007/BF03261934] [DOI] [PubMed] [Google Scholar]
- 89.Calvert AH, Newell DR, Gumbrell LA, O’Reilly S, Burnell M, Boxall FE, Sikkik ZH, Judson IR, Gore ME, Wiltshaw E. Carboplatin dosage: prospective evaluation of a simple formula based on renal function. J Clin Oncol. 1989;7:1748–1756. doi: 10.1200/JCO.1989.7.11.1748. [cited 2017 Feb 28 Available from: http://www.ncbi.nlm.nih.gov/pubmed/2681557] [DOI] [PubMed] [Google Scholar]
- 90.Weinshilboum R, Wang L. Pharmacogenomics: bench to bedside. Nat Rev Drug Discov. 2004;3:739–748. doi: 10.1038/nrd1497. [Available from http://www.nature.com/doifinder/10.1038/nrd1497] [DOI] [PubMed] [Google Scholar]
- 91.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 92.Teicher BA. Hypoxia and drug resistance. Cancer Metastasis Rev. 1994;13:139–168. doi: 10.1007/BF00689633. [Kluwer Academic Publishers; [cited 2017 Feb 24] Available from http://link.springer.com/10.1007/BF00689633] [DOI] [PubMed] [Google Scholar]
- 93.Vaupel P. Tumor microenvironmental physiology and its implications for radiation oncology. Semin Radiat Oncol. 2004;14:198–206. doi: 10.1016/j.semradonc.2004.04.008. [[cited 2017 Jun 29] Available from http://linkinghub.elsevier.com/retrieve/pii/S1053429604000591] [DOI] [PubMed] [Google Scholar]
- 94.Minchinton AI, Tannock IF. Drug penetration in solid tumours. Nat Rev Cancer. 2006;6:583–592. doi: 10.1038/nrc1893. [Nature Publishing Group; [cited 2017 Feb 16] Available from http://www.nature.com/doifinder/10.1038/nrc1893] [DOI] [PubMed] [Google Scholar]
- 95.Li X, Arlinghaus LR, Ayers GD, Chakravarthy AB, Abramson RG, Abramson VG, Atuegwu N, Farley J, Mayer IA, Kelley MC. DCE-MRI analysis methods for predicting the response of breast cancer to neoadjuvant chemotherapy: pilot study findings. Magn Reson Med. 2013;71:1592–1602. doi: 10.1002/mrm.24782. Available from: http://www.ncbi.nlm.nih.gov/pubmed/23661583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Yankeelov TE, Arlinghaus LR, Li X, Gore JC. The role of magnetic resonance imaging biomarkers in clinical trials of treatment response in cancer. Semin Oncol. 2011;38:16–25. doi: 10.1053/j.seminoncol.2010.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Venkatasubramanian R, Arenas R, Henson M, Forbes N. Mechanistic modelling of dynamic MRI data predicts that tumour heterogeneity decreases therapeutic response. Br J Cancer. 2010;103:486–497. doi: 10.1038/sj.bjc.6605773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Tagami T, Foltz WD, Ernsting MJ, Lee CM, Tannock IF, May JP, Li SD. MRI monitoring of intratumoral drug delivery and prediction of the therapeutic effect with a multifunctional thermosensitive liposome. Biomaterials. 2011;32:6570–6578. doi: 10.1016/j.biomaterials.2011.05.029. [DOI] [PubMed] [Google Scholar]
- 99.Kim M, Gillies RJ, Rejniak KA. Current advances in mathematical modeling of anti-cancer drug penetration into tumor tissues. Pharmacol Anti-Cancer Drugs. 2013;3:278. doi: 10.3389/fonc.2013.00278. [Available from http://journal.frontiersin.org/article/10.3389/fonc.2013.00278/abstract] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Lehmann BD, Bauer JA, Chen X, Sanders ME, Chakravarthy AB, Shyr Y, Pietenpol JA. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Invest. 2011;121:2750–2767. doi: 10.1172/JCI45014. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3127435&tool=pmcentrez&rendertype=abstract%5Cnhttp://www.ncbi.nlm.nih.gov/pmc/articles/pmc3127435/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Eisenhauer EA, Therasse P, Bogaerts J, Schwartz LH, Sargent D, Ford R, Dancey J, Arbuck S, Gwyther S, Mooney M. New response evaluation criteria in solid tumours: Revised RECIST guideline (version 1.1) Eur J Cancer. 2009;45:228–247. doi: 10.1016/j.ejca.2008.10.026. [DOI] [PubMed] [Google Scholar]
- 102.Atuegwu NC, Gore JC, Yankeelov TE. The integration of quantitative multi-modality imaging data into mathematical models of tumors. Phys Med Biol. 2010;55:2429–2449. doi: 10.1088/0031-9155/55/9/001. [[cited 2015 Mar 6] Available from http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2897238&tool=pmcentrez&rendertype=abstract] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Pickles MD, Gibbs P, Lowry M, Turnbull LW. Diffusion changes precede size reduction in neoadjuvant treatment of breast cancer. Magn Reson Imaging. 2006;24:843–847. doi: 10.1016/j.mri.2005.11.005. [DOI] [PubMed] [Google Scholar]
- 104.Dose Schwarz J, Bader M, Jenicke L, Hemminger G, Jänicke F, Avril N. Early prediction of response to chemotherapy in metastatic breast cancer using sequential 18F-FDG PET. J Nucl Med. 2005;46(7):1144–1150. [Society of Nuclear Medicine 2005;46:1144–50. Available from http://www.ncbi.nlm.nih.gov/pubmed/16000283] [PubMed] [Google Scholar]
- 105.Kinahan PE, Fletcher JW. Positron emission tomography-computed tomography standardized uptake values in clinical practice and assessing response to therapy. Semin Ultrasound CT MR. 2010;31:496–505. doi: 10.1053/j.sult.2010.10.001. [NIH Public Access; [cited 2017 Apr 6]; Available from http://www.ncbi.nlm.nih.gov/pubmed/21147377] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Dawson S-J, Tsui DWY, Murtaza M, Biggs H, Rueda OM, Chin S-F, Dunning MJ, Gale D, Forshew T, Mahler-Arujo B. Vol. 368. Massachusetts Medical Society; 2013. Analysis of Circulating Tumor DNA to Monitor Metastatic Breast Cancer; pp. 1199–1209. (N Engl J Med.). [cited 2017 Apr 6 Available from: http://www.nejm.org/doi/abs/10.1056/NEJMoa1213261] [DOI] [PubMed] [Google Scholar]
- 107.Thress KS, Paweletz CP, Felip E, Cho BC, Stetson D, Dougherty B, Lai Z, Markovets A, Vivancos A, Kuang Y. Acquired EGFR C797S mutation mediates resistance to AZD9291 in non–small cell lung cancer harboring EGFR T790M. Nat Med. 2015;21:560–562. doi: 10.1038/nm.3854. [cited 2017 Jul 26 Available from http://www.ncbi.nlm.nih.gov/pubmed/25939061] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Siravegna G, Mussolin B, Buscarino M, Corti G, Cassingena A, Crisafulli G, Ponzetti A, Cremolini C, Amatu A, Lauricella C. Vol. 21. NIH Public Access; 2015. Clonal evolution and resistance to EGFR blockade in the blood of colorectal cancer patients; pp. 795–801. (Nat Med). [cited 2017 Jul 26 Available from: http://www.ncbi.nlm.nih.gov/pubmed/26030179] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Garcia-Murillas I, Schiavon G, Weigelt B, Ng C, Hrebien S, Cutts RJ, Cheang M, Osin P, Nerurkar A, Kozarewa I. Mutation tracking in circulating tumor DNA predicts relapse in early breast cancer. Sci Transl Med. 2015;7:302ra133. doi: 10.1126/scitranslmed.aab0021. [cited 2017 Apr 6 Available from: http://stm.sciencemag.org/cgi/doi/10.1126/scitranslmed.aab0021] [DOI] [PubMed] [Google Scholar]
- 110.Li X, Abramson RG, Arlinghaus LR, Kang H, Chakravarthy AB, Abramson VG, Farley J, Mayer IA, Kelley MC, Meszoely IM. Combined DCE-MRI and DW-MRI for Predicting Breast Cancer Pathological Response After the First Cycle of Neoadjuvant Chemotherapy. Investig Radiol. 2015;50:195–204. doi: 10.1097/RLI.0000000000000100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Harrold JM, Parker RS. Clinically relevant cancer chemotherapy dose scheduling via mixed-integer optimization. Comput Chem Eng. 2009;33:2042–2054. [[cited 2015 Mar 31] Available from http://linkinghub.elsevier.com/retrieve/pii/S0098135409001525] [Google Scholar]
- 112.Liu Y, Sadowski SM, Weisbrod AB, Kebebew E, Summers RM, Yao J. Patient specific tumor growth prediction using multimodal images. Med Image Anal. 2014;18:555–566. doi: 10.1016/j.media.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Shi J, Alagoz O, Erenay FS, Su Q. A survey of optimization models on cancer chemotherapy treatment planning. Ann Oper Res. 2011;221:331–356. [[cited 2015 Mar 31] Available from http://link.springer.com/10.1007/s10479-011-0869-4] [Google Scholar]
- 114.Singer DS, Jacks T, Jaffee E. A U.S. “Cancer Moonshot” to accelerate cancer research. Science. 2016;353 doi: 10.1126/science.aai7862. [Available from http://science.sciencemag.org/content/353/6304/1105.full] [DOI] [PubMed] [Google Scholar]