Abstract
Intratumoral heterogeneity of breast cancer remains a major challenge in successful treatment. Failure of cancer therapies can also be accredited to inability to systemically eradicate cancer stem cells (CSCs). Recent evidence points to the role of epithelial-mesenchymal transition (EMT) in expanding the pool of tumor cells with CSCs features. Thus, we assessed expression level as well as heterogeneity of CSCs markers in primary tumors (PT), lymph node metastasis (LNM), and circulating tumor cells (CTCs)–enriched blood fractions in order to correlate them with signs of EMT activation as well as clinicopathological data of breast cancer patients. Level of CSCs markers (ALDH1, CD44, CD133, OCT-4, NANOG) and EMT markers was quantified in PT (N=107), LNM (N=56), and CTCs-enriched blood fractions (N=85). Heterogeneity of CSCs markers expression within each PT and LNM was assessed by calculating Gini Index. Percentage of ALDH1-positive cells was elevated in PT in comparison to LNM (P = .005). However, heterogeneity of the four CSCs markers: ALDH1 (P = .019), CD133 (P = .009), OCT-4 (P = .027), and CD44 (P < .001) was decreased in LNM. Samples classified as mesenchymal (post-EMT) showed elevated expression of CSCs markers (OCT-4 and CD44 in PT; OCT-4 in LNM; ALDH1, OCT-4, NANOG, CD44 in CTCs). Patients with mesenchymal-like CTCs had worse prognosis than patients with epithelial-like or no CTCs (P = .0025). CSCs markers are enriched in PT, LNM, and CTCs with mesenchymal features, but their heterogeneity is decreased in metastatic lymph nodes. Mesenchymal CTCs phenotype correlates with poor prognosis of the patients.
Introduction
In solid tumors, development of deadly metastases starts when cells of the primary tumor gain the ability to spread to other organs [1]. So far, we are unable to predict which clones within individual primary tumor possess this capability; thus, analysis of cancer cells which passed through the selected stages of metastatic cascade broadens our knowledge of cancer dissemination. Not surprisingly, data suggest that relying on primary tumor characteristics may be insufficient in predicting patients outcome and responsiveness to the therapy [1], [2]. Therefore, not only characterization of primary tumor but also cells which disseminated are necessary to obtain more informative picture of the disease [2]. Distant metastases however are not routinely tested, limiting our understanding of successful metastatic clones. Alternatively, information can be gained for regional lymphatic metastases, which are removed during curative surgery. Additionally, circulating tumor cells (CTCs), which exemplify malignant cells able to detach from the primary tumor PT and enter circulation, are easily obtained from just few milliliters of venous blood [3]. Presence of CTCs is connected with poorer prognosis [4], [5], [6]. Although majority of methods detect cells with epithelial markers [7], new data indicate that CTCs undergoing epithelial to mesenchymal transition (EMT) lose epithelial and gain mesenchymal markers [8], [9], which also confer poorer prognosis of metastatic breast cancer patients [10]. The exact reasons for this increased aggressiveness are not yet clear; one of the explanations is linked with stem cell properties of mesenchymal phenotype. Cancer stem cells (CSCs) possess the ability of self-renewal and to generate heterogeneous lineages of cancer cells and are characterized by multiple markers like CD44+/CD24− [11], ALDH1, CD133, OCT4, and NANOG [12], [13]. CD44 is a hyaluronic acid receptor interacting with tumor stroma [14]; it has been shown to be upregulated in cells with stem cells properties [15], [16], also manifesting EMT phenotype [15]. Another marker found to be upregulated in breast cells with progenitor/stem cells properties is aldehyde dehydrogenase ALDH1 [17], [18] involved in converting retinol to retinoic acid [19]. It is expressed in breast cancers presenting with worse clinicopathological characteristics, and it confers poor prognosis to patients [17]. CD133, also known as prominin 1, is a transmembrane protein of not yet known function, associated with progenitor/stem properties, high tumor grade, and lymph node involvement in breast cancer patients [20]. NANOG is a transcription factor found to promote tumorigenesis and metastasis by enhancing growth and invasion of human breast cancer cells [21]. OCT-4 is essential for maintenance of self-renewal in embryonic stem cells, and it is expressed in human breast cancer stem-like cells [22]. NANOG and OCT-4 induce the expression of each other [23]. In lung adenocarcinoma cell line, double knockdown of NANOG and OCT-4 reverses EMT; in breast cancer, NANOG and OCT-4 promote EMT [24], [25].
Previously, we have shown that loss of E-cadherin in nonlobular primary breast tumors is related to hematogenous and lymphatic dissemination [26]. CTCs seeded from PT with E-cadherin loss showed mesenchymal characteristics more frequently, confirming invasive nature of cancer cells with mesenchymal phenotype. Since loss of E-cadherin (in nonlobular cancers) might point to EMT activation, which in turn might induce stem cell-like profile [15], [27], [28], we asked whether mesenchymal phenotype is also linked with expression of CSCs markers in breast cancer clinical samples comprising different stages of metastatic cascade—primary tumors, lymph node metastases, and circulating tumor cells. We aimed at assessing levels of expression and heterogeneity of CSCs markers in breast cancer CTCs, PT, and LNM, also in matched subset of samples.
Materials and Methods
Primary tumors of nonlobular histological type (N=107), lymph node metastases (N=56), and CTCs-enriched blood samples (n=85) from 107 breast cancer patients (stage I-III) treated in the Medical University Hospital in Gdansk were investigated. Median age of the patients was 61 years (28-86 years); clinical characteristics of the group are presented in Table 1. Informed consent was collected from all participants included in the study, and the agreement to perform the study was given by the Bioethical Committee of the Medical University of Gdansk. General schematics presenting types, total number of samples, and the number of matched samples in each group are depicted in Additional file 1: Figure S1.
Table 1.
Clinicopathological Characteristics of the Study Group
| Variable | Number of cases | % |
|---|---|---|
| Age | ||
| <50 years | 28 | 26.2 |
| ≥50 years | 79 | 73.8 |
| Tumor stage | ||
| T1 | 47 | 43.9 |
| T2 | 54 | 50.5 |
| T3 | 3 | 2.8 |
| T4 | 2 | 1.9 |
| Missing data | 1 | 0.9 |
| Tumor grade | ||
| G1 | 14 | 13.1 |
| G2 | 53 | 49.5 |
| G3 | 40 | 37.4 |
| Missing data | 0 | 0.0 |
| N stage | ||
| N− | 51 | 47.7 |
| N+ | 56 | 52.3 |
| Missing data | 0 | 0.0 |
| ER status | ||
| Positive | 25 | 23.4 |
| Negative | 82 | 76.6 |
| Missing data | 0 | 0.0 |
| PR status | ||
| Positive | 29 | 27.1 |
| Negative | 78 | 72.9 |
| Missing data | 0 | 0.0 |
| HER2 status | ||
| Positive | 76 | 71.0 |
| Negative | 29 | 27.1 |
| Missing data | 2 | 1.9 |
Gene Expression Analysis in CTCs
Peripheral blood samples for CTCs enrichment were collected prior to surgery and only from neoadjuvant chemotherapy-naive patients. Blood samples were enriched for CTCs by density gradient centrifugation and negative immunomagnetic selection with anti-CD45–covered magnetic particles (CD45 Dynabeads, Invitrogen). RNA was isolated from CTCs-enriched blood fractions using TRIzol reagent (Invitrogen). Expression of cytokeratin 19 (CK19), mammaglobin-1 (MGB1), vimentin (VIM), and HER2 was analyzed by qPCR (CFX96 cycler, Bio-Rad) using commercially available TaqMan probes and Universal PCR Mastermix (Applied Biosystems). qPCRs were performed in duplicates on 96-well plates in the following conditions: 2 minutes at 50°C, 10 minutes at 95°C, and 45 cycles of 1 minute at 60°C and 15 seconds at 95°C. Results were analyzed in a relative manner using modified ΔΔCt approach in qBase software (Biogazelle). Samples were classified as CTCs positive when at least one of mammary epithelial transcripts (HER2 or MGB1) and one of the epithelial/mesenchymal markers (CK19 or VIM) were detected [29]. For example, presence of epithelial CTCs was called when the sample was CK19+/VIM− and at least one of MGB1 or HER2 was detected, whereas mesenchymal CTCs were called when sample was CK19−/VIM+ and at least one of MGB1 or HER2 was detected. Samples in which CTCs markers were detected are for short referred to as CTCs. Detailed description of the methods is available in Markiewicz et al. [29].
Due to low amount of RNA isolated from CTCs-enriched blood samples, gene expression analysis of stem cell markers was performed after optimized targeted cDNA preamplification. Briefly, cDNA (1 μl) was diluted and mixed with TaqMan PreAmp Master Mix 2× (Applied Biosystems) and pooled TaqMan Assays to preamplify genes of interest, including ALDH1 (Hs00946916_m1), CD44 (Hs01075862_m1), OCT-4 (also known as POU5F1, Hs00999632_g1), NANOG (Hs02387400_g1), CD133 (also known as PROM1, Hs01009250_m1), and two reference genes: GAPDH (Hs99999905_m1) and YWHAZ (Hs03044281_g1). Preamplification PCRs were performed in Mastercycler gradient thermal cycler (Eppendorf) using the following protocol: 95°C 10 minutes and 10 cycles of 15 seconds at 95°C then 4 minutes at 60°C. Lack of preamplification bias was checked by comparing expression of the genes of interest in nonpreamplified and preamplified control cDNA samples. qPCR cycling parameters were the same as for nonpreamplified samples.
Protein Levels in Tissue Microarrays
Tissue microarrays (TMAs) were prepared by sampling up to five nonadjacent tissue cores of 1-mm diameter from each formalin-fixed paraffin-embedded PT and LNM. Serial sections were analyzed by manual immunohistochemical staining with commercially available antibodies against ALDH1 (ALDH1 clone 44, BD Biosciences), CD44 (DF1485, Dako), CD133 (AC133, Miltenyi Biotech), NANOG (NBP104320, Sti), and OCT4 (MRQ-10, Roche). Appropriate secondary antibodies were used for each primary antibody; detection system was based on activity of horseradish peroxidase (Novolink Max-Polymer Detection System, Novocastra). Percentage of stained cells in each tissue core was evaluated by qualified pathologist (J.S, H.M). The staining characteristics were as follows: ALDH1, cytoplasm; CD133, membrane/cytoplasm; CD44, membrane; and NANOG and OCT-4, nucleus/cytoplasm staining. Exemplary staining of CSCs markers in PT and matched LNM is shown in Additional file 2: Figure S2. Heterogeneity of expression within each PT or LNM sample was assessed by calculating Gini Index (GI) [30], taking as input values percentage of positive cells in each of the TMA core. GI describes diversity of marker expression across a sample and ranges from 0 (no diversity) to 1 (maximal possible diversity). Expression of a marker in at least 10% of tumor cells in TMA core was the chosen positivity criterion; if less than 10% of cells were positive in all of the TMA cores, sample was considered to be homogenous for a given marker and assigned GI of 0. Additionally, vimentin (VIM), E-cadherin, and N-cadherin were stained on large sections of PT and LNM in order to capture signs of EMT activation also within tumor margin. PT and LNM samples were classified as displaying epithelial phenotype when E-cadherin was present and VIM and N-cadherin were absent; mesenchymal phenotype was noted when samples had one of the following: E-cadherin loss or VIM expression or N-cadherin expression. E-cadherin loss and VIM/N-cadherin positivity were defined according to previously published criteria on the same set of samples [26].
Statistical Analysis
Patients’ survival was calculated from time of surgery to death or censoring; median follow-up time from the diagnosis was 4.1 years. Kaplan-Meier curves were plotted and compared with the F Cox test for survival in two groups; for three-group analysis, χ2 statistics were used.
For the comparison of the continuous values and clinicopathological data (two groups), nonparametric Mann-Whitney U test was used. P value less than .05 was considered significant. In contingency tables, χ2 test was used, with Fisher’s exact test where appropriate.
All analyses were performed with Statistica version 12 (StatSoft) software.
Results
CSCs Markers in CTCs, PT, and LNM
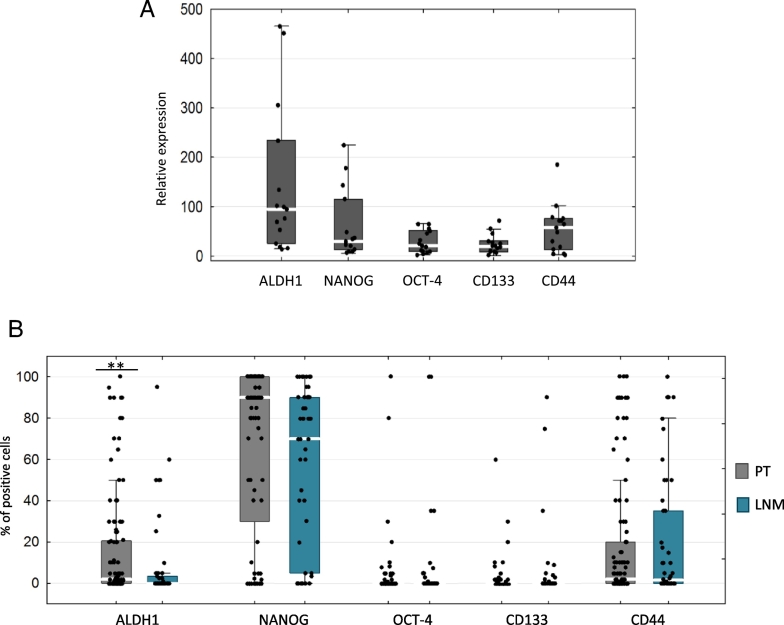
Of the 85 blood samples tested, 23 (27%) had CTCs detected—13/23 (15%) had epithelial phenotype and 10/23 (12%) mesenchymal phenotype. Preamplification was successful for 15/23 (65%) of CTCs samples and allowed for CSCs markers expression analysis. In CTCs, the most highly expressed CSCs markers (analyzed by qPCR) were ALDH1, NANOG, and CD44 (relative median gene expression 94.6, 29 and 57,5, respectively; Figure 1A), whereas median percentage of CSCs markers–positive cells (by IHC) was the highest for NANOG (90%), ALDH1 (2%), and CD44 (2%) in PT and NANOG (70%) and CD44 (1.5%) in LNM. In PT, more ALDH1-positive cells were observed than in LNM (P = .005, Figure 1B). It was also the case when only the subset of matched PT-LNM was considered (N=56)—median percentage of ALDH1-positive cells was 2% in PT vs 0%. Here the in matched LNM (P = .0027; average percentage was 17.6% in PT and 8.8% in LNM) (Additional file 3: Figure S3A). For other CSCs markers, percentage of positive cells was similar between PT and LNM both in all samples analyzed (Figure 1B) and in the matched set (Additional file 3: Figure S3A).
Figure 1.
CSCs markers expression in clinical samples. (A) Relative gene expression level (by qPCR) in CTC-enriched blood fractions. (B) Median percentage of CSCs markers–positive cells (by IHC) in PT and LNM. *P < .05, **P < .005 (Mann-Whitney test). Bars correspond to 25-75 percentile; whiskers cover values of nonoutliers. White gaps in bars represent median value.
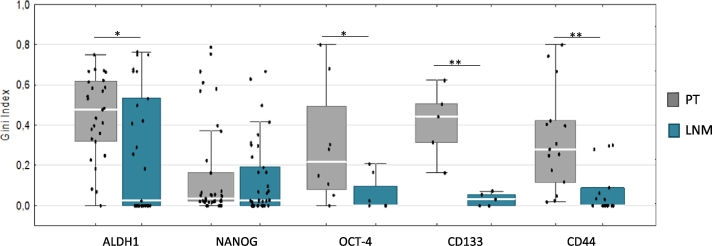
To analyze heterogeneity of CSCs expression within PT and LNM, we selected a group of samples with PT (and their matched LNM) classified as positive for a given marker (at least 10% of positive cells in any of the areas tested on TMA). Here, we observed that PT seeded LNM in which heterogeneity of CSCs markers (measured as GI) was significantly lower—this was the case for CD133, ALDH1, CD44, and OCT-4 (Figure 2). As expression of CD133, NANOG, and CD44 (trend) in PT correlated with increased Ki67 in PT (Additional file 4: Figure S4), we could infer proliferative advantage of CSCs marker–positive cells, which might facilitate new niches (possibly also LN) colonization.
Figure 2.
Heterogeneity of CSCs markers measured by Gini Index in PT and matched LNM. Only positive samples included. In case of ALDH1, OCT-4, CD133, and CD44, heterogeneity was significantly lower in LNM than in PT (P = .019, P = .028, P = .0079, and P = .001, respectively; calculated by Mann-Whitney test). Bars correspond to 25-75 percentile; whiskers cover values of nonoutliers. Median values are represented by white gaps in bars. *P < .05, **P ≤ .008.
Comparing populations of CSCs markers–positive cells between two niches, we need to keep in mind that expression of CSCs markers might not be a stable trait, marking population of phenotypically plastic cells. Such plasticity is also seen in cells undergoing EMT process. We therefore asked if EMT (mesenchymal phenotype) is also linked with expression of CSCs markers in PT, LNM, and CTCs in breast cancer patients.
Expression of CSCs and Proliferation Markers in Relation to EMT Status of CTCs, PT, and LNM
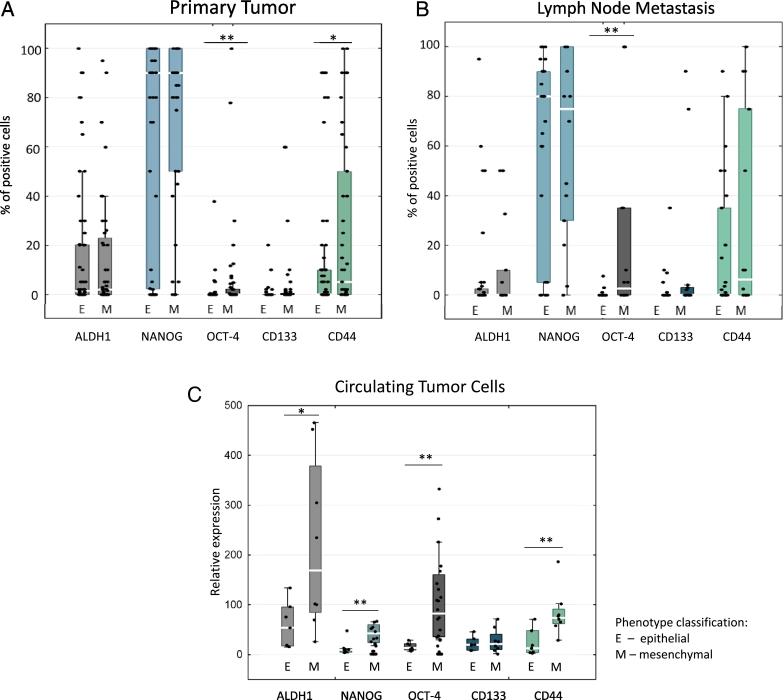
When samples were divided to epithelial- and mesenchymal-like phenotypes (according to criteria mentioned in Methods), we observed significant differences in CSCs markers expression in all three compartments: PT, LNM, and CTCs. However, the expression pattern was not identical among them. In PT, average percentage of OCT-4– and CD44–positive cells was higher in mesenchymal vs epithelial tumors (median percentage 0% in both groups, average percentage 5.99% vs 0.41% for OCT-4, P = .004; for CD44 median percentage 0% vs 5%, average percentage 27.37% vs 12.32%, P = .037; Figure 3A). ALDH1, NANOG, and CD133 showed no differences in expression. In case of LNM, higher levels of OCT-4 were observed in mesenchymal, than in epithelial LNM (median percentages 2% vs 0%, average percentages 20.71% vs 0.36%, P = .001) (Figure 3B). When only the subset of matched PT and LNM was considered, OCT-4 was higher in mesenchymal PT (P = .017) and LNM (P = .001) (Additional file 3: Figure S3, B and C).
Figure 3.
Median percentage of CSCs marker–positive cells in primary tumors (A) and lymph node metastases (B) classified as epithelial (E: expression of E-cadherin and lack of VIM and N-cadherin) or mesenchymal (M: E-cadherin loss or expression of VIM and/or N-cadherin) in IHC evaluation. (C) Relative gene expression level (by qPCR) of CSCs markers in CTC-enriched blood fractions classified as epithelial (E: CK19+/VIM− and MGB1+ and/or HER2+) or mesenchymal (M: CK19−/VIM+ and MGB1+ and/or HER2+). *P < .05, **P ≤ .007 (Mann-Whitney test). Bars correspond to 25-75 percentile; whiskers cover values of nonoutliers. Median values are represented by white gaps in bars.
Similar to PT and LNM, CTCs of mesenchymal-like phenotype expressed higher levels of OCT-4 (relative gene expression level 82 vs 13, P = .001), CD44 (73 vs 12, P = .007), ALDH1 (168 vs 54, P = .032) and NANOG (42 vs 9, P = .007) in comparison to epithelial-like CTCs (Figure 3C). Data including healthy donors, in which no CTCs markers expression were detected, are presented in Additional file 5.
Also, proliferative capacities of PT and LNM were different when subdivided to epithelial and mesenchymal types. Mesenchymal phenotype was characterized by twice as high Ki67 labeling index in comparison to epithelial phenotype (20% vs 10% for PT, P = .0019 and 12% vs 6% for LNM, P = .04; Additional file 6: Figure S6, A and B). Such observation would support more general proliferation-promoting function of EMT (via induction of CSCs phenotype and thus increased proliferation) rather than exclusively associating epithelial phenotype with rapid cycling.
Clinical Significance of EMT and CSCs Markers
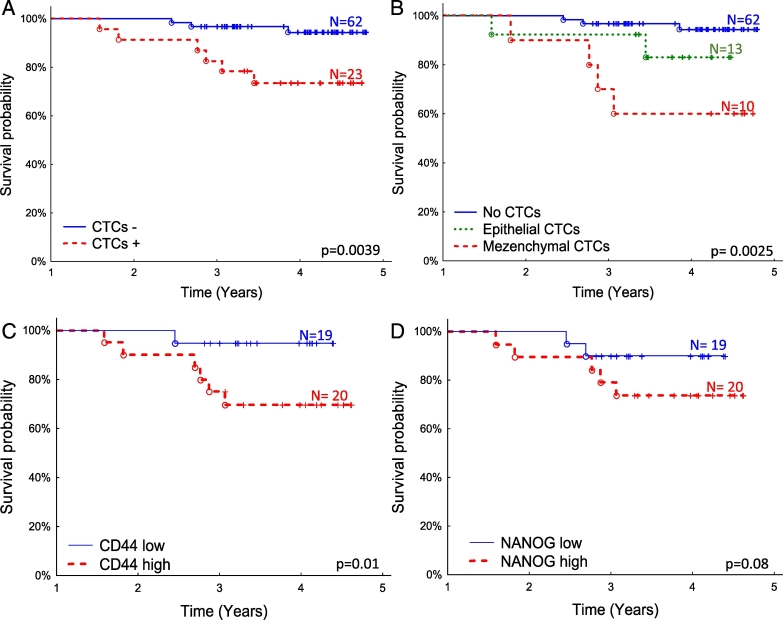
Presence of the CTCs in blood of breast cancer patients is described as an unfavorable independent prognostic and predictive factor [6]. We observed not only that the presence of CTCs is correlated with patients’ poor prognosis (Figure 4A) but also that the phenotype of CTCs could be an important risk factor (Figure 4B). In our study, patients with mesenchymal-like CTCs had significantly worse prognosis than patients with epithelial-like CTCs, with 5-year survival rates of 60% and 82%, respectively. For patients without detected CTCs, survival rate was 94% (P = .0025) (Figure 4, A and B).
Figure 4.
Overall survival probability of breast cancer patients (A) with and without CTCs detected, (B) further subdivided into patients with no CTCs, epithelial or mesenchymal CTCs detected underlining poor prognosis of patients with mesenchymal CTCs; (C) patients with CD44 high and CD44 low expression (division based on median expression level) in CTCs-enriched blood fraction (including samples in which no CTCs markers were detected), (D) patients with NANOG high and NANOG low (division based on median expression level) expression in CTCs-enriched blood fraction (including samples in which no CTCs markers were detected).
Relatively low number of samples with CTCs detected precluded assessing prognostic significance of CSCs markers in this subgroup. Therefore, we tested the prognostic impact of CSCs markers in all available CTCs-enriched blood fractions (including samples in which no CTCs markers were detected). High expression (higher than median) of CD44 was a poor prognostic factor (P = .01), with 70% vs 94% survival rates at the end of the follow-up for positive and negative samples, respectively (Figure 4C). Similar trend was observed for NANOG (P = .08) with 74% vs 90% survival rates for positive and negative samples, respectively (Figure 4D).
In CTCs, a trend was observed towards high expression of NANOG (P = .06) and CD44 (P = .08) in patients with higher T stage (Table 2). Interestingly, increased expression of CSCs markers in all CTCs-enriched blood fractions (including samples negative for CTCs markers) correlated with worse clinical status. High expression of NANOG (P = .02) and CD44 (P = .02) in CTCs-enriched blood fractions was observed in patients with lymph node involvement. Moreover, NANOG (P = .004), OCT-4 (P = .01), and CD44 (P = .002) were higher in tumors of more advanced T stage (Additional file 7: Table S1).
Table 2.
Correlations of CSCs Markers Expression in CTCs and Clinicopathological data of the Patients
| Media Expression (25-75 Percentile) |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Variable | ALDH1 | P | NANOG | P | Oct-4 | P | CD133 | P | CD44 | P |
| Grading | P = .95 | P = .27 | P = .69 | P = .11 | P = .86 | |||||
| G1-2 | 130 (82.99-178.87) | 22.92 (15.51-30.33) | 50.41 (31.14-69.68) | 17.08 (12.79-21.37) | 45.04 (35.00-55.08) | |||||
| G3 | 163.20 (92.28-234.12) | 36.03 (26.71-45.35) | 76.48 (40.60-112.35) | 34.55 (24.88-44.22) | 69.67 (41.72-97.63) | |||||
| T stage | P = .34 | P = .06 | P = .14 | P = 1.00 | P = .08 | |||||
| T1 | 65.68 (36.69-94.67) | 8.33 (6.74-9.93) | 17.84 (13.51-22.17) | 22.77 (14.77-30.76) | 20.21 (10.56-29.86) | |||||
| T2-4 | 172.26 (122.11-222.41) | 35.38 (28.63-42.12) | 76.47 (53.61-99.33) | 24.54 (18.18-30.91) | 67.51 (52.37-82.64) | |||||
| N stage | P = .93 | P = .93 | P = 1.00 | P = .80 | P = 1.00 | |||||
| N− | 126.42 (18.34-234.51) | 30.11 (8.81-51.42) | 63.80 (12.57-115.02) | 15.53 (11.67-19.40) | 44.27 (12.34-76.20) | |||||
| N+ | 146.52 (102.95-190.08) | 27.86 (21.53-34.20) | 60.38 (40.36-80.40) | 25.38 (19.70-31.06) | 56.53 (42.53-70.53) | |||||
| ER status | P = 1.00 | P = .93 | P = .69 | P = .57 | P = .17 | |||||
| Negative | 85.32 (68.79-133.19) | 24.95 (17.05-32.85) | 35.21 (33.57-36.86) | 13.55 (1.00-26.11) | 86.11 (70.46-101.77) | |||||
| Positive | 152.84 (108.14-197.54) | 28-66 (21.95-35.37) | 64.78 (44.17-85.39) | 25.69 (20.22-31.15) | 50.09 (36.19-64.00) | |||||
| PR status | P = .93 | P = .48 | P = 1.00 | P = .23 | P = .00 | |||||
| Negative | 101.00 (68.79-133.19) | 12.91 (8.77-17.05) | 29.14 (21.42-36.86) | 35.66 (26.11-45.21) | 52.53 (3.30-101.77) | |||||
| Positive | 150.43 (105.58-195.28) | 30.51 (24.00-37.02) | 65.71 (45.25-86.18) | 22.29 (16.78-27.80) | 55.26 (41.85-68.66) | |||||
| HER2 | P = .95 | P = .37 | P = .68 | P = .77 | P = .86 | |||||
| Negative | 159.71 (105.64-213.77) | 24.46 (17.13-31.80) | 68.22 (43.72-92.72) | 24.99 (18.81-31.18) | 50.20 (39.07-60.73) | |||||
| Positive | 112.10 (62.38-161.82) | 35.57 (25.75-45.38) | 46.07 (21.44-70.70) | 22.22 (12.83-31.60) | 64.29 (30.94-97.64) | |||||
In PT and LNM, high levels of CD133 and CD44 were significantly correlated with lack of ER and PR (P < .01 for all). Furthermore, CD133 in PT and LNM was associated with higher tumor grade (P = .007 and P = .035, respectively) (Table 3, Table 4).
Table 3.
Correlations of CSCs Markers Expression in PT and Clinicopathological Data
| Average Expression (± Standard Error) |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Variable | ALDH1 | P | NANOG | P | Oct-4 | P | CD133 | P | CD44 | P |
| Grading | P = .80 | P = .26 | P = .32 | P < .001 | P = .06 | |||||
| G1-2 | 18.12 (14.5-21.73) | 62.88 (57.54-68.23) | 1.37 (0.79-1.96) | 0.13 (0.04-0.22) | 14.09 (10.89-17.30) | |||||
| G3 | 13.52 (9.85-17.20) | 74.87 (69.33-80.42) | 5.1 (1.95-8.25) | 5.26 (3.04-7.48) | 26.36 (20.58-32.14) | |||||
| T stage | P = .84 | P = .46 | P = .10 | P = .78 | P = .96 | |||||
| T1 | 17.69 (13.21-22.17) | 68.75 (62.66-74.94) | 0.98 (0.30-1.66) | 2.56 (0.97-4.14) | 18.59 (13.72-23.46) | |||||
| T2-4 | 15.63 (12.39-18-87) | 66.35 (61.11-71.58) | 4.23 (2.06-6.40) | 1.60 (0.52-2.67) | 19.22 (15.35-23.09) | |||||
| N stage | P = .35 | P = .43 | P = .71 | P = .45 | P = .20 | |||||
| N− | 14.72 (11.01-18.44) | 66.63 (60.62-72.64) | 1.03 (0.57-1.49) | 2.41 (0.93-3.4) | 19.69 (15.4-23.99) | |||||
| N+ | 17.8 (14.06-21.54) | 68.26 (63.02-73.51) | 4.34 (2.02-6.66) | 2 (0.84-3.16) | 17.96 (13.73-22.18) | |||||
| ER status | P = .29 | P = .38 | P = .24 | P < .001 | P < .001 | |||||
| Negative | 21.8 (15.6-28) | 77.6 (71-84.2) | 3.76 (0.57-6.95) | 8.54 (5.13-11.95) | 40.06 (32.64-47.48) | |||||
| Positive | 14.63 (11.77-17.48) | 64.3 (59.58-69.02) | 2.47 (1.16-3.79) | 0.07 (0.03-0.11) | 12.11 (9.28-14.94) | |||||
| PR status | P = .06 | P = .15 | P = .16 | P < .001 | P = .01 | |||||
| Negative | 21.45 (15.65-27.24) | 80 (74.07-85.99) | 4.28 (1.38-7.17) | 7.36 (4.38-10.35) | 28.84 (22.23-35.46) | |||||
| Positive | 14.47 (11.58-17.37) | 62.89 (58.05-67.73) | 2.21 (0.88-3.55) | 0.07 (0.03-0.11) | 14.92 (11.71-18.14) | |||||
| HER2 | P = .07 | P = .76 | P = .35 | P = .53 | P = .65 | |||||
| Negative | 15.06 (12.05-18.07) | 66.48 (61.77-71.19) | 3.7 (1.95-5.45) | 2.95 (1.63-4.27) | 20.28 (16.47-24.11) | |||||
| Positive | 20.21 (14.52-25.90) | 71.43 (64.09-78.77) | 0.59 (0.2-0.97) | 0.52 (0.22-0.82) | ||||||
| CTCs | P = .27 | P = .58 | P = .96 | P = .91 | P = .27 | |||||
| Negative | 17.71 (14.16-21.25) | 68.99 (63.74-74.24) | 2.45 (0.8-4.1) | 1.05 (0.6-1.5) | 16.07 (12.46-19.67) | |||||
| Positive | 12.04 (7.31-16.78) | 63.26 (54.34-72.18) | 5.13 (1.48-8.79) | 2.84 (0.12-5.56) | 13.57 (7.73-19.40) | |||||
Table 4.
Correlations of CSCs Markers Expression in LNM and Clinicopathological Data
| Average Expression (± Standard Error) |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Variable | ALDH1 | P | NANOG | P | Oct-4 | P | CD133 | P | CD44 | P |
| Grading | P = .85 | P = .051 | P = .007 | P = .035 | P = .11 | |||||
| G1-2 | 10.16 (5.8-14.51) | 45 (36.55-53.45) | 1.48 (0.18-2.78) | 0.43 (0.1-0.76) | 9.59 (6.34-12.84) | |||||
| G3 | 7.07 (3.58-10.55) | 72.55 (65.99-79.11) | 12.11 (5.83-18.39) | 10.57 (5.06-16.08) | 32.74 (24.23-41.24) | |||||
| T stage | P = .60 | P = .14 | P = .76 | P = .60 | P = .049 | |||||
| T1 | 4.36 (0.81-7.90) | 70.71 (60.95-80.48) | 4.73 (1.56-7.91) | 5.33 (0.35-10.32) | 10.39 (3.32-17.47) | |||||
| T2-4 | 10.72 (6.96-14.48) | 50.58 (43.39-57.77) | 7.14 (2.94-11.33) | 3.61 (0.87-6.34) | 23.21 (17.61-28.82) | |||||
| ER status | P = .16 | P = .04 | P = .32 | P < .001 | P = .003 | |||||
| Negative | 2.88 (0.39-5.38) | 77.38 (68.59-86.16) | 11.54 (3.69-19.39) | 17.83 (8.63-27.04) | 42.71 (31.57-53.84) | |||||
| Positive | 10.78 (7.1-14.45) | 50.43 (43.53-57.33) | 4.35 (1.44-7.26) | 0.54 (0.18-0.90) | 12.1 (8.14-15.97) | |||||
| PR status | P = .27 | P = .64 | P = .15 | P = .003 | P = .009 | |||||
| Negative | 8.28 (2.15-14.41) | 61.23 (50.57-71.90) | 12.33 (5.33-19.33) | 14.2 (6.65-21.75) | 34.97 (25.19-44.74) | |||||
| Positive | 8.99 (5.87-12.10) | 55.47 (48.43-62.51) | 3.57 (0.66-6.51) | 0.62 (0.23-1.01) | 12.79 (8.55-17.03) | |||||
| HER2 | P = .11 | P = .87 | P = .78 | P = .83 | P = .43 | |||||
| Negative | 4.99 (2.70-7.27) | 59.31 (52.39-66.23) | 7.78 (3.47-12.04) | 4.73 (1.84-7.62) | 17.33 (12.31-22.35) | |||||
| Positive | 14.22 (7.13-21.3) | 57 (46-68) | 3.37 (1.03-5.70) | 5.2 (0.21-10.19) | 26.13 (16.90-35.37) | |||||
| CTCs | P = .65 | P = .032 | P = .74 | P = .80 | P = .79 | |||||
| Negative | 9 (4.29-13.71) | 49.13 (40.71-57.55) | 6.29 (1.96-10.62) | 5.33 (1.96-8.71) | 16.5 (10.53-22.47) | |||||
| Positive | 6.57 (2.41-10.72) | 75.36 (65.7-85.01) | 10.39 (3.06-17.72) | 6.8 (0.82-12.78) | 23.67 (14.4-32.93) | |||||
In the forward stepwise selection, T stage, lymph node involvement, and CTCs phenotype were included in the multivariate Cox regression analysis. CTCs phenotype (where no CTCs or epithelial CTCs were assigned a hazard ratio of 1) was the only significant variable, with presence of mesenchymal CTCs being related to 5.63 risk of death (CI 1.23-25.86, P = .026; Table 5).
Table 5.
The Risk of Death of Breast Cancer Patients in Uni- and Multivariate Analysis
| Variable |
Univariate Analysis |
Multivariate Analysis |
||||
|---|---|---|---|---|---|---|
| Hazard Ratio | 95% CI | P Value | Hazard Ratio | 95% CI | P Value | |
| Grade | 1.401 | 0.427-4.591 | .578 | not included | ||
| T stage | 2.183 | 0.579-8.231 | .249 | 0.77 | 0.14-4.10 | .76 |
| N stage | 2.456 | 0.651-9.257 | .185 | 2.83 | 0.52-15.3 | .23 |
| ER status | 0.809 | 0.214-3.048 | .753 | not included | ||
| PR status | 1.722 | 0.372-7.969 | .487 | not included | ||
| HER2 status | 0.969 | 0.257-3.652 | .962 | not included | ||
| CTCs phenotype | 6.489 | 1.741-24.188 | .005 | 5.63 | 1.23-25.86 | .026 |
Discussion
The consensus view on CSCs portrays them as drivers of tumor growth and regeneration [31], [32]. Less acknowledged is the origin of CSCs; whether they are descendants of normal residual CSCs or they arise de novo from transformed cancer cells or from non-CSCs following the instructions sent from the microenvironment (CSCs plasticity model) [33], [34]. Analysis of CSCs often concentrates on PT, from which CSCs need to disseminate first to initiate metastases. But it is not well known how the profile of CSCs markers changes during disease progression. In the current work, we asked whether differences can be observed in the profile of CSCs markers of tumor cells present in different environments at different stages of metastatic process—in the primary site, metastatic LN, and blood, where CTCs are found. Since microenvironment can influence the population of CSCs, via e.g. induction of EMT, we asked if there is a link between EMT activation and the population of CSCs markers–positive cells.
We observed that PT and LNM showing features of EMT (E-cadherin loss and/or acquisition of vimentin or N-cadherin) have increased proliferation rate. Results of other studies are not clear on that matter. Pro-proliferative effect of EMT inducer TGFβ has been described for cancers like gliomas [35] and osteosarcomas [36]; in epithelial cells, EMT is thought to exert antiproliferative effect [37], [38], but not influencing metastases initiation potential [39] due to occurrence of mesenchymal-epithelial transition at the metastatic site, which restores proliferative phenotype [40]. Technical limitation of our research does not allow to test whether 1) it is the mesenchymal phenotype itself which is related to higher proliferation rate or 2) it is the EMT-linked process, like generation of cells with the stem cell properties, which fuels growth of the tumor. However, the latter explanation is supported by our own results showing enrichment of CSCs markers–expressing cells in samples of mesenchymal phenotype and also by data from other studies revealing that cells after EMT are enriched in tumor-forming abilities [15], [41], [42]. Thus, even transient activation of EMT can increase overall proliferative index of the tumor, by temporarily expanding population of cells with cancer stem cell properties.
Analysis of CSCs markers revealed that they do not present with a universal pattern of expression in cancer cells at different stages of metastatic cascade, which concerned both the expression level and the heterogeneity of expression. In case of ALDH1, LNM showed reduced expression in comparison to PT, whereas in CTCs, ALDH1 was the highest of all tested CSCs markers. Decreased ALDH1 levels in regional and distant metastases were previously observed in renal cell carcinoma [43] and colorectal cancer [44]. In breast cancers, ALDH1 was not shown to be different between PT and LNM [45]. Since population of ALDH1+ breast CSCs was shown to be controlled by microenvironment [46], we could speculate that the differences in composition of PT and metastatic LN niches could be one of the factors contributing to the observed difference. Additionally, ALDH1 was the most heterogeneous marker in our analyses (GI for all PT: 0.464). Percentage of ALDH1+ cells within one tumor ranged from 0 to 100% depending on the analyzed area. Therefore, the comparison of ALDH1 levels in PT and its matched LNM is largely dependent on the number of tumor sections/areas analyzed; such prominent heterogeneity should be considered in future studies addressing CSCs markers in breast cancer.
By analyzing EMT markers in the collected samples, we could show that mesenchymal features of PT, LNM, and CTCs correlate with CSCs markers expression, which corroborate the role of EMT in conferring extended malignant phenotype to cancer cells also in nonmetastatic patients [25], [47], [48]. These differences were the most ubiquitous in CTCs, where four CSCs markers (ALDH1, NANOG, OCT4, and CD44) were elevated in mesenchymal samples. Induction of CD44 and concomitant CSCs phenotype in cells undergoing EMT was previously reported in cell lines, in breast cancer mouse models, and in patient samples [15], [28], [49]. High CD44 expression itself in CTCs-enriched blood fractions was a poor prognostic factor related to decreased overall survival of the patients. However, expression of CD44 was also observed in healthy controls, which may be a consequence of sample contamination with immune cells [50]. Therefore, expression of CD44 in CTCs requires further investigation due to its potential prognostics values. Interestingly, no difference in survival was observed when CD44 was measured in PT or LNM. However, others showed that CD44 expression in patients with positive lymph nodes or large tumors correlates with decreased disease free survival [16]. Another CSCs marker, OCT-4, was consistently elevated in all sample types classified as mesenchymal—in PT, LNM, and CTCs. Since correlative results do not allow for inferring causality of events (one of the limitations of our study), we can only presume that OCT-4 and other CSCs markers tested might be linked with EMT, which on mechanistic level was shown by others. In lung carcinoma cells, OCT-4 together with NANOG was inducing EMT, and their double knockdown reverted EMT and blocked the tumorigenic and metastatic ability [24], [51]. Beltran et al. described generation of tumor-initiating cells with EMT gene signature by transduction of normal breast primary cultures with OCT-4, which generated cells capable to form breast carcinomas in nude mice [51]. Moreover, OCT-4 expression in hormone-positive breast cancer correlated with poor clinical outcome, aggressive features, and tamoxifen resistance [52]. In the current work, OCT-4 correlated with more aggressive tumor characteristics when measured in LNM and CTCs-enriched blood fractions but not in PT. Also, mesenchymal phenotype of CTCs was linked with poor prognosis in multivariate analysis, with 5.63 risk of death, in comparison to patients with no CTCs or epithelial CTCs. Thus, overall, prognostic significance of the mesenchymal and CSCs phenotype in the current work was only observed for cancer cells outside of the PT. A similar picture emerges from our previous study in breast cancer patients, where we concluded LNM to be a surrogate marker of the PT metastatic potential [53]. In the current work we noted that molecular profile of CTCs is clinically more informative than the analysis of PT. Heterogeneity of the tumor mass, which we showed is higher in PT than LNM, might render identification of small malignant clones more challenging in the PT than in LNM or CTCs, which already contain a preselected population of cancer cells, having been capable of passing through selected stages of metastatic cascade. Interestingly, CSCs markers were prognostic when analyzed in CTCs-enriched blood fractions (encompassing also samples negative for CTCs markers). Further studies are required to determine if additional CTCs markers (other than the ones tested in the current work) can be detected in samples with high expression of CSCs markers.
In the IHC analysis of tumor samples, we have observed different staining patterns of the stem cell factors. OCT4 and NANOG transcription factors showed both nuclear and cytoplasmic staining. Though OCT4 is a transcription factor, it can be found in the cytoplasm as a result of nuclear export. Even transient localization of OCT4 in the nucleus can trigger expression of downstream effectors genes [54]. It was indicated by Plachta et al. that OCT4 kinetics of shifting between nucleus and cytoplasm might be more informative [55]; however, such dynamic changes cannot be assessed in immunohistochemical analysis. In case of NANOG, cellular localization is still a matter of concern. Some studies indicate nuclear localization [56]; others find only cytoplasmic staining [57], [58] or both nuclear and cytoplasmic [59]. Cytoplasmic localization of NANOG was also described as a more significant early cancer risk factor than nuclear NANOG in patients with laryngeal precancerous lesions [60].
For CD133, which is a transmembrane protein, we observed both membrane and cytoplasmic staining. The role of CD133 has not been fully elucidated. Interestingly, it is considered that signaling from cytoplasmic CD133 potentiates prosurvival signals under stress conditions [61]. Cytoplasmic localization of CD133 was observed in other studies evaluating breast cancer [62], [63] and other cancers [64], [65].
Due to methodological limitations, we cannot directly compare levels of CSCs markers in PT/LNM and CTCs—CSCs markers in PT and LNM were analyzed using IHC (where proteins in cells of stromal component are not evaluated); in CTCs, analyses were done with qPCR (measuring gene expression in complete sample). Additionally, analyses of gene expression were performed on CTCs-enriched blood fractions, which might be contaminated with components of the blood not removed during sample preparation. However, cutoffs for detection of CTCs markers were based on the expression of these markers in samples from healthy volunteers (described in [66]).
To summarize, our work underlines the importance of assessing heterogeneity of the CSCs markers since percentage of CSCs markers–positive cells can be vastly different in the same tumor and can change between PT and matched LNM. These differences also point to the significance of molecular characterization of cancer cells outside of the PT, as they might bring additional clinically important information. At last, we showed that CSCs markers expressing cells are enriched in samples with mesenchymal features, possibly explaining the observed poor prognosis of patients with mesenchymal CTCs.
The following are the supplementary data related to this article.
List of samples available from breast cancer patients included into the study. (A) Schematic depicting number of primary tumors (PT), blood samples (for CTCs isolation), lymph node metastases, and their classification into epithelial or mesenchymal phenotype. (B) Total number of lymph node metastases (LNM) available for the immunohistochemical analysis of CSCs markers, as well as the number of matched pairs of LNM and PT.
Representative images of CSCs markers immunohistochemical staining in PT (A-E), matched LNM (F-J), and negative PT (K-O).
Analysis for the subset of matched pairs of PT and LNM. (A) Box plots of median percentage of CSCs markers–positive cells in PT and LNM matched pairs. Percent of ALDH1-positive cells was significantly higher in PT (P = .0027 by Mann-Whitney test). Box plots of median percentage of CSCs markers–positive cells in PT (B) and LNM matched pairs (C) classified as epithelial (E: expression of E-cadherin and lack of VIM and N-cadherin) or mesenchymal (M: E-cadherin loss or expression of VIM and/or N-cadherin) in IHC evaluation. *P < .05, **P ≤ .003 (Mann-Whitney test). Bars correspond to 25-75 percentile; whiskers cover values of nonoutliers. White gaps in bars represent median value.
Box plots of raw data presenting median percentage of Ki67-positive cells in PT classified as positive and negative cells (using 10% cutoff) for CSCs markers. NANOG- and CD133-positive group had higher median percentage of Ki67-positive cells (P = .02 and P = .001, respectively, by Mann-Whitney test). Bars correspond to 25-75 percentile; whiskers cover values of nonoutliers. Median values are represented by white gaps in bars.
Box plots of raw data representing stem cell markers expression in mesenchymal and epithelial CTCs samples as well as in healthy controls with no detected CTCs. Bars correspond to 25-75 percentile; whiskers cover values of nonoutliers. White gaps in bars represent median value. P values for differences in expression between samples are listed in table.
Box plots of raw data presenting median percentage of Ki67-positive cells in PT and LNM classified as mesenchymal and epithelial. In both cases, Ki67 median percentage of positive cells was higher in mesenchymal samples (P = .0019 for PT and P = .04 for LNM by Mann-Whitney test). Bars correspond to 25-75 percentile; whiskers cover values of nonoutliers. Median values are represented by white gaps in bars.
Correlations between CSCs Markers Expression in CTCs-Enriched Blood Fractions and Clinicopathological Data of the Patients.
Acknowledgements
The authors declare that they have no competing interests.
The study was approved by the Ethical Committee of the Medical University of Gdansk, and all patients included in the study have given their informed consent.
Research was funded by a grant no. 2016/22/E/NZ4/00664 and 2016/21/D/NZ3/02629 from the National Science Centre. Anna Nagel was supported by University of Gdansk, 538-M000-B497-17. Aleksandra Markiewicz was supported by the Foundation for Polish Science (FNP).
Contributor Information
Aleksandra Markiewicz, Email: aleksandra.markiewicz@biotech.ug.edu.pl.
Anna Nagel, Email: anna.nagel@biotech.ug.edu.pl.
Jolanta Szade, Email: jszade@gumed.edu.pl.
Hanna Majewska, Email: hania.majewska@gumed.edu.pl.
Jaroslaw Skokowski, Email: jskokowski@gumed.edu.pl.
Barbara Seroczynska, Email: bastrzel@gumed.edu.pl.
Tomasz Stokowy, Email: Tomasz.Stokowy@uib.no.
Marzena Welnicka-Jaskiewicz, Email: mwelj@gumed.edu.pl.
Anna J Zaczek, Email: azaczek@gumed.edu.pl.
References
- 1.Chaffer CL, Weinberg RA. A perspective on cancer cell metastasis. Science. 2011;331(6024):1559–1564. doi: 10.1126/science.1203543. [DOI] [PubMed] [Google Scholar]
- 2.Braun S, Naume B. Circulating and disseminated tumor cells. J Clin Oncol. 2005;23(8):1623–1626. doi: 10.1200/JCO.2005.10.073. [DOI] [PubMed] [Google Scholar]
- 3.Pantel K, Hayes DF. Disseminated breast tumour cells: biological and clinical meaning. Nat Rev Clin Oncol. 2017;15:129–131. doi: 10.1038/nrclinonc.2017.174. [DOI] [PubMed] [Google Scholar]
- 4.Xenidis N, Ignatiadis M, Apostolaki S, Perraki M, Kalbakis K, Agelaki S. Cytokeratin-19 mRNA-positive circulating tumor cells after adjuvant chemotherapy in patients with early breast cancer. J Clin Oncol. 2009;27(13):2177–2184. doi: 10.1200/JCO.2008.18.0497. [DOI] [PubMed] [Google Scholar]
- 5.Rack B, Andergassen U, Neugebauer J, Salmen J, Hepp P, Sommer H. The German SUCCESS C Study—the first European lifestyle study on breast cancer. Breast Care (Basel) 2010;5(6):395–400. doi: 10.1159/000322677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Janni WJ, Rack B, Terstappen LW, Pierga JY, Taran FA, Fehm T. Pooled analysis of the prognostic relevance of circulating tumor cells in primary breast cancer. Clin Cancer Res. 2016;22(10):2583–2593. doi: 10.1158/1078-0432.CCR-15-1603. [DOI] [PubMed] [Google Scholar]
- 7.Micalizzi DS, Haber DA, Maheswaran S. Cancer metastasis through the prism of epithelial-to-mesenchymal transition in circulating tumor cells. Mol Oncol. 2017;11(7):770–780. doi: 10.1002/1878-0261.12081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hyun KA, Koo GB, Han H, Sohn J, Choi W, Kim SI. Epithelial-to-mesenchymal transition leads to loss of EpCAM and different physical properties in circulating tumor cells from metastatic breast cancer. Oncotarget. 2016;7(17):24677–24687. doi: 10.18632/oncotarget.8250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu M, Bardia A, Wittner BS, Stott SL, Smas ME, Ting DT. Circulating breast tumor cells exhibit dynamic changes in epithelial and mesenchymal composition. Science. 2013;339(6119):580–584. doi: 10.1126/science.1228522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bulfoni M, Gerratana L, Del Ben F, Marzinotto S, Sorrentino M, Turetta M. In patients with metastatic breast cancer the identification of circulating tumor cells in epithelial-to-mesenchymal transition is associated with a poor prognosis. Breast Cancer Res. 2016;18(1):30. doi: 10.1186/s13058-016-0687-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A. 2003;100(7):3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clarke MF, Dick JE, Dirks PB, Eaves CJ, Jamieson CH, Jones DL. Cancer stem cells--perspectives on current status and future directions: AACR Workshop on cancer stem cells. Cancer Res. 2006;66(19):9339–9344. doi: 10.1158/0008-5472.CAN-06-3126. [DOI] [PubMed] [Google Scholar]
- 13.Tang DG. Understanding cancer stem cell heterogeneity and plasticity. Cell Res. 2012;22(3):457–472. doi: 10.1038/cr.2012.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zöller M. CD44: can a cancer-initiating cell profit from an abundantly expressed molecule? Nat Rev Cancer. 2011;11(4):254–267. doi: 10.1038/nrc3023. [DOI] [PubMed] [Google Scholar]
- 15.Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133(4):704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McFarlane S, Coulter JA, Tibbits P, O'Grady A, McFarlane C, Montgomery N. CD44 increases the efficiency of distant metastasis of breast cancer. Oncotarget. 2015;6(13):11465–11476. doi: 10.18632/oncotarget.3410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ginestier C, Hur MH, Charafe-Jauffret E, Monville F, Dutcher J, Brown M. ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell. 2007;1(5):555–567. doi: 10.1016/j.stem.2007.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Deng S, Yang X, Lassus H, Liang S, Kaur S, Ye Q. Distinct expression levels and patterns of stem cell marker, aldehyde dehydrogenase isoform 1 (ALDH1), in human epithelial cancers. PLoS One. 2010;5(4) doi: 10.1371/journal.pone.0010277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chute JP, Muramoto GG, Whitesides J, Colvin M, Safi R, Chao NJ. Inhibition of aldehyde dehydrogenase and retinoid signaling induces the expansion of human hematopoietic stem cells. Proc Natl Acad Sci U S A. 2006;103(31):11707–11712. doi: 10.1073/pnas.0603806103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu TJ, Sun BC, Zhao XL, Zhao XM, Sun T, Gu Q. CD133+ cells with cancer stem cell characteristics associates with vasculogenic mimicry in triple-negative breast cancer. Oncogene. 2013;32(5):544–553. doi: 10.1038/onc.2012.85. [DOI] [PubMed] [Google Scholar]
- 21.Lu X, Mazur SJ, Lin T, Appella E, Xu Y. The pluripotency factor nanog promotes breast cancer tumorigenesis and metastasis. Oncogene. 2014;33(20):2655–2664. doi: 10.1038/onc.2013.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ponti D, Costa A, Zaffaroni N, Pratesi G, Petrangolini G, Coradini D. Isolation and in vitro propagation of tumorigenic breast cancer cells with stem/progenitor cell properties. Cancer Res. 2005;65(13):5506–5511. doi: 10.1158/0008-5472.CAN-05-0626. [DOI] [PubMed] [Google Scholar]
- 23.Chambers I, Colby D, Robertson M, Nichols J, Lee S, Tweedie S. Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell. 2003;113(5):643–655. doi: 10.1016/s0092-8674(03)00392-1. [DOI] [PubMed] [Google Scholar]
- 24.Chiou SH, Wang ML, Chou YT, Chen CJ, Hong CF, Hsieh WJ. Coexpression of Oct4 and Nanog enhances malignancy in lung adenocarcinoma by inducing cancer stem cell-like properties and epithelial-mesenchymal transdifferentiation. Cancer Res. 2010;70(24):10433–10444. doi: 10.1158/0008-5472.CAN-10-2638. [DOI] [PubMed] [Google Scholar]
- 25.Wang D, Lu P, Zhang H, Luo M, Zhang X, Wei X. Oct-4 and Nanog promote the epithelial-mesenchymal transition of breast cancer stem cells and are associated with poor prognosis in breast cancer patients. Oncotarget. 2014;5(21):10803–10815. doi: 10.18632/oncotarget.2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Markiewicz A, Wełnicka-Jaśkiewicz M, Seroczyńska B, Skokowski J, Majewska H, Szade J. Epithelial-mesenchymal transition markers in lymph node metastases and primary breast tumors—relation to dissemination and proliferation. Am J Transl Res. 2014;6(6):793–808. [PMC free article] [PubMed] [Google Scholar]
- 27.Morel AP, Lièvre M, Thomas C, Hinkal G, Ansieau S, Puisieux A. Generation of breast cancer stem cells through epithelial-mesenchymal transition. PLoS One. 2008;3(8) doi: 10.1371/journal.pone.0002888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chaffer CL, Marjanovic ND, Lee T, Bell G, Kleer CG, Reinhardt F. Poised chromatin at the ZEB1 promoter enables breast cancer cell plasticity and enhances tumorigenicity. Cell. 2013;154(1):61–74. doi: 10.1016/j.cell.2013.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Markiewicz A, Książkiewicz M, Wełnicka-Jaśkiewicz M, Seroczyńska B, Skokowski J, Szade J. Mesenchymal phenotype of CTC-enriched blood fraction and lymph node metastasis formation potential. PLoS One. 2014;9(4) doi: 10.1371/journal.pone.0093901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Natowicz R, Jiang T, Shi W, Qi Y, Delpech Y, Symmans WF. Correlation of intratumor gene expression heterogeneity with chemotherapy sensitivity in breast cancer. J Clin Oncol. 2013;31(15_suppl):1013. [Google Scholar]
- 31.Beck B, Blanpain C. Unravelling cancer stem cell potential. Nat Rev Cancer. 2013;13(10):727–738. doi: 10.1038/nrc3597. [DOI] [PubMed] [Google Scholar]
- 32.Apostolou P, Toloudi M, Chatziioannou M, Ioannou E, Papasotiriou I. Cancer stem cells stemness transcription factors expression correlates with breast cancer disease stage. Curr Stem Cell Res Ther. 2012;7(6):415–419. doi: 10.2174/157488812804484639. [DOI] [PubMed] [Google Scholar]
- 33.Cabrera MC, Hollingsworth RE, Hurt EM. Cancer stem cell plasticity and tumor hierarchy. World J Stem Cells. 2015;7(1):27–36. doi: 10.4252/wjsc.v7.i1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Plaks V, Kong N, Werb Z. The cancer stem cell niche: how essential is the niche in regulating stemness of tumor cells? Cell Stem Cell. 2015;16(3):225–238. doi: 10.1016/j.stem.2015.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bruna A, Darken RS, Rojo F, Ocaña A, Peñuelas S, Arias A. High TGFbeta-Smad activity confers poor prognosis in glioma patients and promotes cell proliferation depending on the methylation of the PDGF-B gene. Cancer Cell. 2007;11(2):147–160. doi: 10.1016/j.ccr.2006.11.023. [DOI] [PubMed] [Google Scholar]
- 36.Matsuyama S, Iwadate M, Kondo M, Saitoh M, Hanyu A, Shimizu K. SB-431542 and Gleevec inhibit transforming growth factor-beta–induced proliferation of human osteosarcoma cells. Cancer Res. 2003;63(22):7791–7798. [PubMed] [Google Scholar]
- 37.Chanrion M, Kuperstein I, Barrière C, El Marjou F, Cohen D, Vignjevic D. Concomitant Notch activation and p53 deletion trigger epithelial-to-mesenchymal transition and metastasis in mouse gut. Nat Commun. 2014;5:5005. doi: 10.1038/ncomms6005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tsai JH, Donaher JL, Murphy DA, Chau S, Yang J. Spatiotemporal regulation of epithelial-mesenchymal transition is essential for squamous cell carcinoma metastasis. Cancer Cell. 2012;22(6):725–736. doi: 10.1016/j.ccr.2012.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Beerling E, Seinstra D, de Wit E, Kester L, van der Velden D, Maynard C. Plasticity between Epithelial and mesenchymal states unlinks EMT from metastasis-enhancing stem cell capacity. Cell Rep. 2016;14(10):2281–2288. doi: 10.1016/j.celrep.2016.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139(5):871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 41.Morata-Tarifa C, Jiménez G, García MA, Entrena JM, Griñán-Lisón C, Aguilera M. Low adherent cancer cell subpopulations are enriched in tumorigenic and metastatic epithelial-to-mesenchymal transition-induced cancer stem-like cells. Sci Rep. 2016;6:18772. doi: 10.1038/srep18772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mitra A, Mishra L, Li S. EMT, CTCs and CSCs in tumor relapse and drug-resistance. Oncotarget. 2015;6(13):10697–10711. doi: 10.18632/oncotarget.4037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Abourbih S, Sircar K, Tanguay S, Kassouf W, Aprikian A, Mansure J. Aldehyde dehydrogenase 1 expression in primary and metastatic renal cell carcinoma: an immunohistochemistry study. World J Surg Oncol. 2013;11:298. doi: 10.1186/1477-7819-11-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hessman CJ, Bubbers EJ, Billingsley KG, Herzig DO, Wong MH. Loss of expression of the cancer stem cell marker aldehyde dehydrogenase 1 correlates with advanced-stage colorectal cancer. Am J Surg. 2012;203(5):649–653. doi: 10.1016/j.amjsurg.2012.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ito M, Shien T, Omori M, Mizoo T, Iwamoto T, Nogami T. Evaluation of aldehyde dehydrogenase 1 and transcription factors in both primary breast cancer and axillary lymph node metastases as a prognostic factor. Breast Cancer. 2016;23(3):437–444. doi: 10.1007/s12282-015-0583-1. [DOI] [PubMed] [Google Scholar]
- 46.Liu S, Ginestier C, Ou SJ, Clouthier SG, Patel SH, Monville F. Breast cancer stem cells are regulated by mesenchymal stem cells through cytokine networks. Cancer Res. 2011;71(2):614–624. doi: 10.1158/0008-5472.CAN-10-0538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ye X, Tam WL, Shibue T, Kaygusuz Y, Reinhardt F, Ng Eaton E. Distinct EMT programs control normal mammary stem cells and tumour-initiating cells. Nature. 2015;525(7568):256–260. doi: 10.1038/nature14897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.May CD, Sphyris N, Evans KW, Werden SJ, Guo W, Mani SA. Epithelial-mesenchymal transition and cancer stem cells: a dangerously dynamic duo in breast cancer progression. Breast Cancer Res. 2011;13(1):202. doi: 10.1186/bcr2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu S, Cong Y, Wang D, Sun Y, Deng L, Liu Y. Breast cancer stem cells transition between epithelial and mesenchymal states reflective of their normal counterparts. Stem Cell Reports. 2014;2(1):78–91. doi: 10.1016/j.stemcr.2013.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Borland G, Ross JA, Guy K. Forms and functions of CD44. Immunology. 1998;93(2):139–148. doi: 10.1046/j.1365-2567.1998.00431.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Beltran AS, Rivenbark AG, Richardson BT, Yuan X, Quian H, Hunt JP. Generation of tumor-initiating cells by exogenous delivery of OCT4 transcription factor. Breast Cancer Res. 2011;13(5):R94. doi: 10.1186/bcr3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gwak JM, Kim M, Kim HJ, Jang MH, Park SY. Expression of embryonal stem cell transcription factors in breast cancer: Oct4 as an indicator for poor clinical outcome and tamoxifen resistance. Oncotarget. 2017;8(22):36305–36318. doi: 10.18632/oncotarget.16750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Markiewicz A, Ahrends T, Wełnicka-Jaśkiewicz M, Seroczyńska B, Skokowski J, Jaśkiewicz J. Expression of epithelial to mesenchymal transition-related markers in lymph node metastases as a surrogate for primary tumor metastatic potential in breast cancer. J Transl Med. 2012;10:226. doi: 10.1186/1479-5876-10-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Oka M, Moriyama T, Asally M, Kawakami K, Yoneda Y. Differential role for transcription factor Oct4 nucleocytoplasmic dynamics in somatic cell reprogramming and self-renewal of embryonic stem cells. J Biol Chem. 2013;288(21):15085–15097. doi: 10.1074/jbc.M112.448837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Plachta N, Bollenbach T, Pease S, Fraser SE, Pantazis P. Oct4 kinetics predict cell lineage patterning in the early mammalian embryo. Nat Cell Biol. 2011;13(2):117–123. doi: 10.1038/ncb2154. [DOI] [PubMed] [Google Scholar]
- 56.Ishiguro T, Sato A, Ohata H, Sakai H, Nakagama H, Okamoto K. Differential expression of nanog1 and nanogp8 in colon cancer cells. Biochem Biophys Res Commun. 2012;418(2):199–204. doi: 10.1016/j.bbrc.2011.10.123. [DOI] [PubMed] [Google Scholar]
- 57.Luo W, Li S, Peng B, Ye Y, Deng X, Yao K. Embryonic stem cells markers SOX2, OCT4 and Nanog expression and their correlations with epithelial-mesenchymal transition in nasopharyngeal carcinoma. PLoS One. 2013;8(2) doi: 10.1371/journal.pone.0056324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tamaki T, Shimizu T, Niki M, Shimizu M, Nishizawa T, Nomura S. Immunohistochemical analysis of NANOG expression and epithelial-mesenchymal transition in pulmonary sarcomatoid carcinoma. Oncol Lett. 2017;13(5):3695–3702. doi: 10.3892/ol.2017.5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nagata T, Shimada Y, Sekine S, Hori R, Matsui K, Okumura T. Prognostic significance of NANOG and KLF4 for breast cancer. Breast Cancer. 2014;21(1):96–101. doi: 10.1007/s12282-012-0357-y. [DOI] [PubMed] [Google Scholar]
- 60.Rodrigo JP, Villaronga M, Menéndez ST, Hermida-Prado F, Quer M, Vilaseca I. A novel role for Nanog as an early cancer risk marker in patients with laryngeal precancerous lesions. Sci Rep. 2017;7(1):11110. doi: 10.1038/s41598-017-11709-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jang JW, Song Y, Kim SH, Kim J, Seo HR. Potential mechanisms of CD133 in cancer stem cells. Life Sci. 2017;184:25–29. doi: 10.1016/j.lfs.2017.07.008. [DOI] [PubMed] [Google Scholar]
- 62.Yang F, Cao L, Sun Z, Jin J, Fang H, Zhang W. Evaluation of breast cancer stem cells and intratumor stemness heterogeneity in triple-negative breast cancer as prognostic factors. Int J Biol Sci. 2016;12(12):1568–1577. doi: 10.7150/ijbs.16874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Aomatsu N, Yashiro M, Kashiwagi S, Takashima T, Ishikawa T, Ohsawa M. CD133 is a useful surrogate marker for predicting chemosensitivity to neoadjuvant chemotherapy in breast cancer. PLoS One. 2012;7(9) doi: 10.1371/journal.pone.0045865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Han SA, Jang JH, Won KY, Lim SJ, Song JY. Prognostic value of putative cancer stem cell markers (CD24, CD44, CD133, and ALDH1) in human papillary thyroid carcinoma. Pathol Res Pract. 2017;213(8):956–963. doi: 10.1016/j.prp.2017.05.002. [DOI] [PubMed] [Google Scholar]
- 65.Saeednejad Zanjani L, Madjd Z, Abolhasani M, Andersson Y, Rasti A, Shariftabrizi A. Cytoplasmic expression of CD133 stemness marker is associated with tumor aggressiveness in clear cell renal cell carcinoma. Exp Mol Pathol. 2017;103(2):218–228. doi: 10.1016/j.yexmp.2017.10.001. [DOI] [PubMed] [Google Scholar]
- 66.Markiewicz A, Wełnicka-Jaśkiewicz M, Skokowski J, Jaśkiewicz J, Szade J, Jassem J. Prognostic significance of ESR1 amplification and ESR1 PvuII, CYP2C19*2, UGT2B15*2 polymorphisms in breast cancer patients. PLoS One. 2013;8(8) doi: 10.1371/journal.pone.0072219. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of samples available from breast cancer patients included into the study. (A) Schematic depicting number of primary tumors (PT), blood samples (for CTCs isolation), lymph node metastases, and their classification into epithelial or mesenchymal phenotype. (B) Total number of lymph node metastases (LNM) available for the immunohistochemical analysis of CSCs markers, as well as the number of matched pairs of LNM and PT.
Representative images of CSCs markers immunohistochemical staining in PT (A-E), matched LNM (F-J), and negative PT (K-O).
Analysis for the subset of matched pairs of PT and LNM. (A) Box plots of median percentage of CSCs markers–positive cells in PT and LNM matched pairs. Percent of ALDH1-positive cells was significantly higher in PT (P = .0027 by Mann-Whitney test). Box plots of median percentage of CSCs markers–positive cells in PT (B) and LNM matched pairs (C) classified as epithelial (E: expression of E-cadherin and lack of VIM and N-cadherin) or mesenchymal (M: E-cadherin loss or expression of VIM and/or N-cadherin) in IHC evaluation. *P < .05, **P ≤ .003 (Mann-Whitney test). Bars correspond to 25-75 percentile; whiskers cover values of nonoutliers. White gaps in bars represent median value.
Box plots of raw data presenting median percentage of Ki67-positive cells in PT classified as positive and negative cells (using 10% cutoff) for CSCs markers. NANOG- and CD133-positive group had higher median percentage of Ki67-positive cells (P = .02 and P = .001, respectively, by Mann-Whitney test). Bars correspond to 25-75 percentile; whiskers cover values of nonoutliers. Median values are represented by white gaps in bars.
Box plots of raw data representing stem cell markers expression in mesenchymal and epithelial CTCs samples as well as in healthy controls with no detected CTCs. Bars correspond to 25-75 percentile; whiskers cover values of nonoutliers. White gaps in bars represent median value. P values for differences in expression between samples are listed in table.
Box plots of raw data presenting median percentage of Ki67-positive cells in PT and LNM classified as mesenchymal and epithelial. In both cases, Ki67 median percentage of positive cells was higher in mesenchymal samples (P = .0019 for PT and P = .04 for LNM by Mann-Whitney test). Bars correspond to 25-75 percentile; whiskers cover values of nonoutliers. Median values are represented by white gaps in bars.
Correlations between CSCs Markers Expression in CTCs-Enriched Blood Fractions and Clinicopathological Data of the Patients.