Abstract
This study analyzed the genetic diversity by MIRU-VNTR of Mycobacterium leprae isolates from nasal cavities and related to epidemiological and clinical data. The sample consisted of 48 newly diagnosed leprosy cases that tested positive for M. leprae PCR in nasal secretion (NS) attending to the National Reference Center of Dermatology Dona Libania (CDERM), Fortaleza, Brazil. Total DNA was extracted from NS of each patient and used for amplification of four M. leprae VNTR loci. Four clusters of M. leprae isolates were formed with identical genotypes. In the spatial analysis, 12 leprosy cases presented similar genotypes organized into 4 clusters. The most common genotypes in the current study was AC8b: 8, AC9: 7, AC8a: 8, GTA9: 10, which may represent a genotype of circulating strains most often in Ceará. A minimum set of four MIRU-VNTR loci was demonstrated to study the genetic diversity of M. leprae isolates from NS.
Keywords: Leprosy, Mycobacterium leprae, Brazil, nasal cavity, genotype, genetic marker, VNTR loci, cluster analysis
Introduction
Despite global efforts to eradicate leprosy, Brazil is still considered as and endemic country, with 213,899 new cases reported globally in 2014 [1]. Brazil, India and Indonesia accounts for 81% of all reported new cases. In Brazil, during the period from 2012 to 2014, the areas of greatest risk for leprosy were concentrated in pockets of the Central region and its neighborhood in the North and Northeast regions. These three areas concentrate only 14% of the population of the Brazilian country and accounts for 44% (13,597/31,044) of the new cases diagnosed in 2014 [1]. In the same year, the State of Ceará, Northeast Brazil, reported 2.069 new cases, with a coefficient of case detection rate (CDR) of 24/100.000. It is also observed that 148 (80.5%) municipalities in Ceará diagnosed new leprosy cases in 2014, 34 (18.4%) of them with more than 10 new cases per 100,000 inhabitants and 23 (12.5%) municipalities were reported as hyper-endemic with a CDR of more than 40/100,000 inhabitants [2].
The conventional epidemiology of leprosy has be improved by strain genotyping tools. The isolates differentiation approach using molecular markers are useful to distinguish different strains of the leprosy bacilli. Variable number tandem repeat (VNTR) provides data about the pattern of variation in the Mycobacterium leprae genome [3–5]. The VNTR typing tool is based on the number of repetitive sequences in polymorphic micro- and mini-satellite regions of the bacteria [6]. Some polymorphic loci are suitable for identifying genotypes according to the discriminatory capacity, stability, and reproducibility. There is considerably more variation in repeat numbers at VNTR locus than in non-repetitive DNA sequences, because length-altering mutations due to slipped-strand mispairing occur at a much higher rate than the inherent DNA substitution or mutation frequency of DNA polymerase [7,8].
Extensive research on different loci has been done on strain typing of the leprosy bacilli and it was achieved the standardization of 17 loci optimized with M. leprae DNA from the strain NHDP63 obtained from infected armadillo tissue [9]. Since M. leprae cannot be cultivated in axenic media, there are several challenges to its molecular typing, including the difficult to obtain sufficient amounts of genomic DNA from clinical samples. In addition, samples from leprosy patients are often of poor quality, and often contaminated with other cultivable agents [10].
Molecular typing of M. leprae through VNTR can establish leprosy transmission chains within populations and allow the evaluation of genomic markers for differentiating bacillus strains. Nasal route is considered as the main entrance and exit of the leprosy bacilli and collection of nasal secretion are not invasive and painless. In this way, genotyping of the bacteria harbored in nasal cavity can provide insights about the circulating strains in the community and active transmission can be correlated. Thus, this study analyzed the genetic diversity by MIRU-VNTR of M. leprae isolates from nasal cavities and related to epidemiological and clinical data.
Materials and methods
Subjects
A cross-sectional study was conducted in Reference Center on Dermatology Dona Libania (CDERM), Fortaleza, capital of Ceará state, northeastern region of Brazil. Nasal samples (NS) from 48 new leprosy cases were collected from June 2009 to December 2010. Cases were diagnosed by clinical evaluation, slit skin smear and biopsy samples. They were classified according to Ridley-Jopling criteria based on histological study and bacilloscopic index (BI) [11]. A well structured questionnaire in combination with a review of medical records were used to collect socio-demographic data.
Ethical considerations
Written consent was obtained from the participants and authorized the collection of samples. The study was approved by the CDERM Ethics Committee and guidelines of the National Ethical Committee were followed to conduct the research.
Sampling collection and DNA extraction
NS were collected from all participants using sterile cotton perinasal swabs pre-moistened with Tris-EDTA buffer (pH 8.0). Nasal specimens were obtained by rotating in each nostril a single swab over the lateral anterior conchae. Swabs were then placed into a sterile and labeled tube and stored at −20 °C until processing. DNA extraction procedure was conducted as previously described [12].
Detection of M. leprae DNA
DNA from NS of participants was searched for a 238-pb fragment of RLEP2 (GenBank accession NC002677) in a semi-nested PCR reaction following the protocol previously described [12]. The PCR products were submitted to the ABI Prism 3730 automated DNA sequencer using the ABI PRISM BigDye Terminator v 3.0 sequencing kit (Applied Biosystems, Foster City, CA). The sequences were identified using SecScape software v.2.7 (Applied Biosystems, Foster City, CA).
MIRU-VNTR genotyping
To assess the genetic diversity of M. leprae among individuals, DNA obtained from NS were analyzed by VNTR analysis targeting 4 loci: AC8b, AC9, AC8a, and GTA9. To determine how many short tandem repeat segments are present in each sample allele, we made a comparison with the positive control NHDP63 strain of M. leprae. The NHDP63 VNTR and amplicon size were verified through gene sequencing. The amplicon sizes at each locus for the positive control were 384 bp to AC8b, 207 bp to AC9, 125 bp to AC8a and 307 bp to GTA9 [4,9].
PCR amplifications were performed using using the HotStartTaq Master Mix (Qiagen, Hilden, Germany), 0.2 μM of each primer pair and 2 μl of template DNA. Thermocycling conditions used were: initial denaturation at 95 °C for 15 min followed by 40 cycles of 94 °C for 30s, 60 °C for 90s, 72 °C for 90s followed by a final elongation step f 72 °C for 10 m [4,9].
Analysis of fragments
Fluorescently-labeled primers were used in each reaction, being amplified in a total of four reactions. PCR products were diluted 30–60 times in water and 1 μl was mixed with 0.3 μl of GeneScan™ 500 LIZ® Size Standard molecular weight marker (Applied Biosystems, Foster City, CA) and 8.7 μl deionized formamide was added. Allele copy number was determined by denaturation of amplicons and capillary gel electrophoresis on the sequencer ABI 3130 Genetic Analyzer (Applied Biosystems, Foster City, CA). Capillary electrophoresis (length 50 cm, polymer POP-7) was conducted at 15 kV over 60 °C with a running time of 45 min [4,9]. Copy number definition of each of the four allele were defined using the Peak Scanner software (Applied Biosystems, Foster City, CA).
Calculation of allelic diversity and discriminatory power
The Hunter – Gaston discrimination index (HGDI) was used as a numeric parameter VNTR discriminatory power. The HGDI was calculated as recommended by Hunter and Gaston [13] by using the formula: 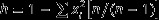 where xi is the frequency of the allele i at locus, n is the number of isolates). The sample is considered to be highly discriminatory polymorphic if h ≥ 0.6; moderately discriminatory if 0.3 ≤ h ≤ 0.6 and weakly discriminatory if h ≤ 0.3 [13].
where xi is the frequency of the allele i at locus, n is the number of isolates). The sample is considered to be highly discriminatory polymorphic if h ≥ 0.6; moderately discriminatory if 0.3 ≤ h ≤ 0.6 and weakly discriminatory if h ≤ 0.3 [13].
Cluster definition and genotype comparison
Definition of clustering was based on comparison of the copy number of the VNTRs, considering identical genotypes those that presented identical copy number for all four alleles.
Spatial analysis
The home addresses of the new leprosy cases were georeferenced using Google Earth (https://www.google.com/earth/) to define latitude and longitude. The spatial database was visualized using the QuantumGis Geographic Information System 18.1.0® licensed by General Public License) (http://www.qgisbrasil.org). The cartographic bases used were obtained from the Brazilian Institute of Geography and Statistics [14]. Data were used to generate graphics showing the distribution of the georeferenced individuals in Fortaleza by VNTR typing.
Statistical analysis
For evaluation of the association of the demographic, clinical and environmental/behavior variables and having a clustered or a unique M. leprae genotype, we used squared and the Fisher exact two-tailed test. A P value of ≤ 0.05 was considered statistically significant. Mann-Whitney test was used to evaluate the differences between a single characteristic between individuals with clustered genotypes or unique patterns. STATA version 12.0 (Stata Corp., College Station, TX, USA) was also used to analyze the data.
Results
Genotyping
The differentiating power of the four allele for the 48 M. leprae NS isolates from leprosy new cases were analyzed, and moderate discriminatory index was found for the chosen VNTRs loci. Variations in copy number ranged from 6 to 8 for AC8b locus with h = 0.47, from 7 to 9 for AC9 locus with h = 0.51, from 8 to 11 for AC8a locus with h = 0.57, and from 8 to 16 for GT9 locus with h = 0.57. The most common genotypes in the current study were AC8b with 8 or 7 copies, AC9 with 7 or 8 copies, AC8a with 8 or 9 copies and GTA9 with 10 or 9 copies (Table 1).
Table 1.
Allelic diversity of the 4 loci of VNTRs.
| VNTR | Number of allele | Allelic diversity (h) | Conclusion | Total samples | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 6 | 7 | 8 | 9 | 10 | 11 | 12 | 16 | ||||
| AC 8b | 1 | 30 | 17 | 0.47 | Moderately discriminatory | 48 | |||||
| AC 9 | 19 | 19 | 1 | 0.51 | Moderately discriminatory | 39 | |||||
| AC 8a | 20 | 20 | 3 | 1 | 0.57 | Moderately discriminatory | 44 | ||||
| GTA 9 | 2 | 14 | 24 | 2 | 1 | 0.57 | Moderately discriminatory | 43 | |||
Among the four loci, three (AC9, GTA9 and AC8a) showed absence of PCR products for some isolates and were thus subsequently excluded from the final selection. In addition, other isolates have not enough demographic data and were also excluded. Thus, a remaining 24 NS provided enough information for the genotypic comparison and descriptive analysis (Table 2). By using the 24 remaining isolates, four clusters of identical genotypes were formed resulting in a HGDI of 0.73.
Table 2.
VNTRs from 48 nasal secretions.
| Sample ID | AC 8b | AC 9 | AC 8a | GTA 9 |
|---|---|---|---|---|
| 84267* | 8 | 8 | 8 | 10 |
| 85.886 | 7 | 8 | 9 | 9 |
| 86187* | 8 | 7 | 8 | 10 |
| 86.190 | 7 | 8 | 9 | 9 |
| 86.340 | 7 | 7 | 8 | 9 |
| 86.362 | 7 | 8 | 9 | 9 |
| 86.394 | 7 | 7 | 8 | – |
| 86.498 | 7 | 7 | 8 | 16 |
| 86.555 | 7 | 8 | 9 | 10 |
| 86610* | 6 | – | 8 | 9 |
| 86.645 | 7 | – | 9 | – |
| 86776* | 7 | 8 | 8 | 9 |
| 86.829 | 7 | – | 8 | 9 |
| 86.855 | 8 | 7 | 8 | 10 |
| 86962* | 8 | 7 | 8 | 10 |
| 87126* | 8 | 7 | 8 | 10 |
| 87184* | 7 | 8 | 9 | – |
| 87212* | 7 | 8 | 9 | 8 |
| 87.214 | 8 | 7 | 8 | 10 |
| 87330* | 7 | 7 | 10 | 8 |
| 87401* | 8 | 7 | 8 | 10 |
| 87.419 | 7 | – | 9 | – |
| 87.549 | 8 | 7 | 9 | 10 |
| 87.560 | 7 | 8 | 9 | 9 |
| 87914* | 8 | 7 | 8 | 10 |
| 88.276 | 8 | – | – | – |
| 88689* | 8 | 7 | 8 | 10 |
| 88742* | 7 | 8 | 9 | 10 |
| 88935* | 7 | 8 | 9 | 12 |
| 89.115 | 7 | 8 | 9 | 9 |
| 89160* | 7 | 8 | 8 | 10 |
| 89167* | 7 | 8 | 9 | 9 |
| 89.167 | 7 | 8 | 9 | 9 |
| 90148* | 8 | 7 | 9 | 10 |
| 90373* | 7 | 7 | 9 | 10 |
| 90.730 | 8 | – | 10 | – |
| 90.780 | 8 | – | – | 10 |
| 91.371 | 8 | – | – | 10 |
| 91.796 | 7 | 8 | 9 | 9 |
| 91845* | 7 | 8 | 10 | 10 |
| 91.979 | 8 | 7 | 9 | 10 |
| 92008* | 7 | 8 | 9 | 10 |
| 92070* | 7 | 9 | 11 | 10 |
| 92114* | 7 | 7 | 8 | 10 |
| 92.275 | 7 | – | – | 10 |
| 92662* | 7 | 8 | 8 | 9 |
| 92.642* | 7 | 7 | 8 | 10 |
| – | 8 | 7 | 8 | 12 |
24 samples used for descriptive analysis.
Among the 24 isolates, 12 had unique genotypes and the remaining 12 isolates form four clusters of genotypes. Cluster 1 to 3 with two isolates each, and cluster 4 with 6 isolates (Table 3).
Table 3.
Groups of samples with identical M. leprae genotype.
| Sample ID | Group | AC 8b | AC 9 | AC 8a | GTA 9 |
|---|---|---|---|---|---|
| 86.776 | 1 | 7 | 8 | 8 | 9 |
| 92.662 | 1 | 7 | 8 | 8 | 9 |
| 92.642 | 2 | 7 | 7 | 8 | 10 |
| 92.114 | 2 | 7 | 7 | 8 | 10 |
| 88.742 | 3 | 7 | 8 | 9 | 10 |
| 92.008 | 3 | 7 | 8 | 9 | 10 |
| 87.401 | 4 | 8 | 7 | 8 | 10 |
| 86.187 | 4 | 8 | 7 | 8 | 10 |
| 87.126 | 4 | 8 | 7 | 8 | 10 |
| 87.914 | 4 | 8 | 7 | 8 | 10 |
| 86.962 | 4 | 8 | 7 | 8 | 10 |
| 88.689 | 4 | 8 | 7 | 8 | 10 |
Spatial analysis and clusters
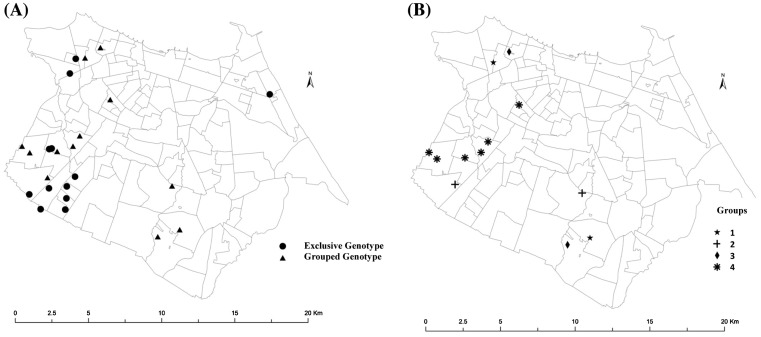
The distribution of the 24 cases was plotted in the map of the city of Fortaleza and spatial analysis according the clustering was performed. Although it was observed that cases were spread over the city, the Fortaleza administrative regions V and I were associated with the clusters. Cluster 4, with five of the six cases distributed in three neighborhoods of the administrative region V, 2 cases in Bonsucesso, 1 in Granja Portugal and 2 in Granja Lisboa (Figure 1).
Figure 1.
Maps of the Municipality of Fortaleza with the dwelling of the individuals with the genotypes of the M. leprae strain analyzed.
Source: (A) Location of patients’ home according to those with strains of exclusive genotypes and strains of grouped genotypes. (B) Location of the patients’ dwelling according to the groups formed from the strains with similar genotypes.
In all four clusters, the 12 leprosy cases reported to have a previous contact with at least one leprosy case. Group 1 was composed of two individuals with lepromatous leprosy, BCG scar, aged 51 and 62, born in the same countryside of Ceará, and living there for 12 years. They were diagnosed in a difference of 17 months (Table 2). Group 2 was composed of two individuals with borderline leprosy, BCG scar, aged 59–60 years, diagnosed in 2010 with a difference in date of diagnosis of 4 months. Despite they were born in different cities of Ceará, they had moved to Fortaleza in 1995 and 1970, respectively. Group 3 was composed of two individuals aged 34 and 32 years and diagnosed within ten months of each other. The first was born and lived in Fortaleza, was diagnosed with borderline leprosy, and presented with a BCG scar. The second, a lepromatous leprosy without a BCG scar, was born in São Paulo, and living in Fortaleza since 1997, and had. Group 4 was composed of six participants with ages ranging from 12 to 71 years, all six cases were diagnosed from February to August 2009. Four cases had borderline leprosy and the two had lepromatous leprosy (Table 4).
Table 4.
Main characteristics of individuals belonging to the groups formed.
| Sample ID | Group | Date of diagnosis | Clinical form | City of birth | Birth date | Age | BCG Scar |
|---|---|---|---|---|---|---|---|
| 86776 | 1 | 11/03/2009 | Lepromatous | Aracati/CE | 29/11/1946 | 62 | Yes |
| 92662 | 1 | 19/08/2010 | Lepromatous | Aracati/CE | 08/10/1958 | 51 | Yes |
| 92642 | 2 | 18/08/2010 | Borderline | Missão Velha/CE | 18/08/1950 | 60 | Yes |
| 92114 | 2 | 29/04/2010 | Borderline | Morada Nova/CE | 16/04/1951 | 59 | Yes |
| 88742 | 3 | 09/09/2009 | Borderline | Fortaleza/CE | 16/06/1975 | 34 | No |
| 92008 | 3 | 21/06/2010 | Lepromatous | Sao Paulo/SP | 12/12/1977 | 32 | No |
| 87401 | 4 | 25/04/2009 | Lepromatous | Uruburetama/CE | 23/05/1964 | 45 | Yes |
| 86187 | 4 | 01/02/2009 | Borderline | Canide/CE | 01/08/1937 | 71 | No |
| 87126 | 4 | 01/04/2009 | Lepromatous | Capistrano de Abreu/CE | 29/10/1955 | 53 | No |
| 87914 | 4 | 01/07/2009 | Borderline | Fortaleza/CE | 19/03/1966 | 43 | No |
| 86962 | 4 | 01/03/2009 | Borderline | Fortaleza/CE | 22/05/1995 | 13 | Yes |
| 88689 | 4 | 28/08/2009 | Borderline | Sao Joao da Se/PI | 01/07/1997 | 12 | Yes |
The distribution of cases according to belonging to a cluster of genotype and not belonging to any cluster were related to age, to time in months between symptoms and diagnosis and to bacilloscopic index was analyzed. The age of non clustered cases ranged from 17 to 34 years, while to the clustered ranged from 26 to 64 years old, being the overall mean age of the non clustered (25.5 years), much lower than the clustered (44.6 years; p < 0.0110) ones. Despite not significant, the mean time between beginning of symptoms and leprosy diagnosis among the clustered isolates (20.3 months) was lower than those with unique genotype (22.6 months). No differences were seen among bacilloscopic index of the groups of isolates (Table 5).
Table 5.
Comparison of characteristics of patients with samples exclusive genotypes and grouped genotypes by the Mann-Whitney test.
| Samples | N | Mean | sd | Median | p | |
|---|---|---|---|---|---|---|
| Age | Exclusive Genotype | 12 | 25.500 | 8.163 | 23.5 | 0.0110 |
| Grouped genotypes | 12 | 44.583 | 18.807 | 48.0 | ||
| Time (months) between symptoms and diagnosis | Exclusive Genotype | 12 | 22.625 | 17.391 | 24.8 | 0.7728 |
| Grouped genotypes | 12 | 20.304 | 17.505 | 13.1 | ||
| Bacilloscopic index | Exclusive Genotype | 9 | 4.239 | 0.302 | 4.0 | 0.9670 |
| Grouped genotypes | 10 | 4.100 | 1.312 | 4.3 |
sd, Standard Deviation.
Discussion
Genotyping of M. leprae bacilli has been reported in several countries, such as India [15], Indonesia [16], China [17] and Brazil [18]. However, most of the studies are based on M. leprae isolates from biopsy samples. A recent study conducted by our group had demonstrated that NS also can be used to genotype approaches [8].
The difficulty in analyzing the VNTRs from NS in the current research was also observed in Hyderabad, India [19], in which only three loci from samples of different tissues from the same patient were studied. A previous study also conducted in Ceará, Brazil, discuss lack of amplification in NS compared to biopsy samples [8]. A study conducted in Rio de Janeiro, Brazil, with skin biopsy samples had used four short tandem repeats (GAA GTA9, AT17 and TA18), had also demonstrated failure to amplify all loci [20]. M. leprae DNA extracted from NS is often of poor quality, in insufficient amounts, and demonstrates higher contamination with microorganisms derived from the community and from the environment.
The chosen four genetic markers (AC8b, AC9, AC8a, GTA9) used in this study to analyze the NS have been well studied for their stability potential and polymorphic diversity [8,9,19,21,22]. Genetic epidemiological analysis are considered appropriate when demonstrating a discrimination index of at least 90% [14]. In the present study, we report an HGDI of 73% for four VNTRs loci, but for this calculation, only 24 samples were used. In the current study, AC8b presented an h index of 0.47, in contrast to another study conducted in Brazil with samples from the states of São Paulo and Rio de Janeiro, with an HGDI of 0.05 [18]. In our study, this locus showed 6–8 repetitions, 7 being the most common, similar to the study conducted in São Paulo and Rio de Janeiro [18]. In studies conducted in Thailand, Indonesia, Korea and Japan, AC8b were often present with 8 repetitions [6,23].
In our study, AC9 presented an h index of 0.51, higher than reported in previous studies conducted in Brazil (h = 0.28 on average) [18]. Regarding the number of alleles for this genetic marker, it was observed in our study of 7 and 8 alleles as the most frequent. Although with small sample sizes, other studies found 10 alleles at this locus [6,23,24]. Ten AC9 repetitions are quite rare and not yet reported in samples from Brazil.
Thus, despite the small number of samples analyzed in this study, AC9 and AC8b loci showed greater diversity in samples from Fortaleza compared to isolates from states of São Paulo and Rio de Janeiro, but with a similar number of alleles [18].
In this study, AC8a presented an h index of 0.57, with 8 to 11 alleles, but in several reports, this locus showed more variability in the number of alleles [6,18,23–25]. Similarly, GTA9 showed an h index of 0.57, with 8 to 16 alleles. Other published studies showed more variability in this allele number, from 7 to 45 [18,23–25]. Contrary to these studies, our isolates report 10 alleles for this genetic marker.
The high genetic diversity of isolates in Brazil is demonstrated in the present study with only 24 leprosy cases, and only 4 clusters. This genetic diversity is similar to that reported in India [26]. Brazil and India alone account for 69% of all new leprosy cases detected worldwide. Thus, this high genetic variability of M. leprae would be proportional to the number of cases in these two countries, therefore suggesting the existence of different M. leprae strains in Fortaleza [27].
Our spatial analysis on genotype distribution based on these four loci demonstrated a distribution of clustering similar from disease distribution in Fortaleza in general. A previous study already had demonstrated the concentration of leprosy cases in areas with low socioeconomic status (SES) in Fortaleza [12]. The administrative regions V and I are hyperendemics (40–300/10.000) for leprosy in general population and in children less than 15 years of age (>10–55/10.000) [2]. Since the main route of infection of the bacilli is via nose, the M. leprae isolates from NS represent the truly genetic diversity of the active transmission of the disease in endemic community, such as Fortaleza.
The exact transmission source between individuals could not be established in the clusters with M. leprae isolates with similar genotypes, however transmission insights can be addressed. Given that our samples are derived from a single city, variations in a few VNTR loci are quite significant for the formation of clusters, as also demonstrated in villages of North Maluku, Indonesia [16]. Other studies have also studied the molecular epidemiology of leprosy with few loci of VNTRs [20,21,28].
Primers specific for the RLEP2 region displayed 100% specificity for the only M. leprae species with a single PCR product. Despite M. leprae and M. lepromatosis are highly related mycobacterial species, sharing 93% of nucleotide sequence identity and now referred to as the Leprosy Complex [29], our primers did not show homology to M. lepromatosis. These two species share 75–90% sequence identity in segments of the four families of repetitive DNA [29].
Several studies discuss the ideal number of genetic markers and which loci fulfill the criteria of sufficient variability with required stability and robustness [4,30]. In addition, it was raised the concern that some markers did not yield consistent outcomes for different samples that originated from the same patient [31]. Thus, choosing appropriate markers is of outmost importance for the reliability of genotyping results. In the same way, there are a few information about the genetic diversity among M. leprae isolates from Brazil, in particular in the Northeast region, which is still endemic for the disease. Despite the cure rate in Brazil in 2016 was 81.8% [32], the distribution of leprosy is unevenly, with highly endemic regions in the North, Northeast and Central-West. SNP analysis in previous work revealed large variability in genotypes in Brasil, such as the prevalence of SNP genotype 3 in São Paulo, Rio de Janeiro and Rondônia, and a high prevalence of SNP genotype 4 in Ceará and Pernambuco in states of the Northeast Brazil [3].
In conclusion, we have demonstrated with a minimum set of four MIRU-VNTR loci, the genetic diversity of M. leprae isolates from NS. Since M. leprae isolates from nasal cavity are representative of the transmission within the local neighborhood, more studies are needed to provide insights about the leprosy dynamic. Furthermore, studies using a higher number of VNTR loci are encouraged, including the addition of other reliable and high-resolution molecular markers for evaluating M. leprae transmission.
Funding
This work was supported by the Conselho Nacional de Desenvolvimento Científico e Tecnológico, Brazil under [grant number CNPq–410573/2006-0].
Disclosure statement
The authors declare that they have no competing interests.
Geolocation information
Acknowledgments
We thank the patients and the support of the staff at Reference Center on Dermatology Dona Libania, Ceará.
References
- [1].Acelerating towards a leprosy-free world [Internet]. World Health Organization 2016. Available from: http://www.searo.who.int/entity/global_leprosy_programme/documents/global_leprosy_strategy_2020/en/
- [2].Boletim Epidemiológico de Hanseníase [Internet]. Secretaria de Saúde, Governo do Estado do Ceará 2017. Available from: http://www.saude.ce.gov.br
- [3].Fontes AN, Gomes HM, Araujo MI, et al. . Genotyping of Mycobacterium leprae present on Ziehl-Neelsen-stained microscopic slides and in skin biopsy samples from leprosy patients in different geographic regions of Brazil. Mem Inst Oswaldo Cruz. 2012;107(Suppl 1):143–149. DOI: 10.1590/S0074-02762012000900021. [DOI] [PubMed] [Google Scholar]
- [4].Jensen RW, Rivest J, Li W, et al. . DNA fingerprinting of Mycobacterium leprae strains using variable number tandem repeat (VNTR) – fragment length analysis (FLA). J Vis Exp. 2011;53:e3104 DOI: 10.3791/3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Taylor GM, Donoghue HD. Multiple loci variable number tandem repeat (VNTR) analysis (MLVA) of Mycobacterium leprae isolates amplified from European archaeological human remains with lepromatous leprosy. Microbes Infect. 2011;13(11):923–929. DOI: 10.1016/j.micinf.2011.05.003. [DOI] [PubMed] [Google Scholar]
- [6].Zhang L, Budiawan T, Matsuoka M. Diversity of potential short tandem repeats in Mycobacterium leprae and application for molecular typing. J Clin Microbiol. 2005;43(10):5221–5229. DOI: 10.1128/jcm.43.10.5221-5229.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Chokkakula S, Abdelrawaf SS, Attitalla IH, et al. . Variable number tandem repeats for global strain typing and strain differentiation of Mycobacterium leprae. J Med Microbiol. 2014;3(4):1 DOI: 10.4172/2161-0703.1000156. [DOI] [Google Scholar]
- [8].Lima L, Fontes AN, Li W, et al. . Intrapatient comparison of Mycobacterium leprae by VNTR analysis in nasal secretions and skin biopsy in a Brazilian leprosy endemic region. Lepr Rev. 2016;87(4):486–500. [PubMed] [Google Scholar]
- [9].Kimura M, Sakamuri RM, Groathouse NA, et al. . Rapid variable-number tandem-repeat genotyping for Mycobacterium leprae clinical specimens. J Clin Microbiol. 2009;47(6):1757–1766. DOI: 10.1128/jcm.02019-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Lavania M, Jadhav R, Turankar RP, et al. . Genotyping of Mycobacterium leprae strains from a region of high endemic leprosy prevalence in India. Infect Genet Evol. 2015;36:256–261. DOI: 10.1016/j.meegid.2015.10.001. [DOI] [PubMed] [Google Scholar]
- [11].Ridley DS, Jopling WH. Classification of leprosy according to immunity. A five-group system. Int J Lepr Other Mycobact Dis. 1966;34(3):255–273. [PubMed] [Google Scholar]
- [12].Lima LN, Frota CC, Mota RM, et al. . Widespread nasal carriage of Mycobacterium leprae among a healthy population in a hyperendemic region of northeastern Brazil. Mem Inst Oswaldo Cruz. 2015;110(7):898–905. DOI: 10.1590/0074-02760150178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Sola C, Filliol I, Legrand E, et al. . Genotyping of the Mycobacterium tuberculosis complex using MIRUs: association with VNTR and spoligotyping for molecular epidemiology and evolutionary genetics. Infect Genet Evol. 2003;3(2):125–133. DOI: 10.1016/S1567-1348(03)00011-X. [DOI] [PubMed] [Google Scholar]
- [14].Hunter PR, Gaston MA. Numerical index of the discriminatory ability of typing systems: an application of Simpson’s index of diversity. J Clin Microbiol. 1988;26(11):2465–2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Kuruwa S, Vissa V, Mistry N. Distribution of Mycobacterium leprae strains among cases in a rural and urban population of Maharashtra, India. J Clin Microbiol. 2012;50(4):1406–1411. DOI: 10.1128/jcm.05315-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Matsuoka M, Zhang L, Budiawan T, et al. . Genotyping of Mycobacterium leprae on the basis of the polymorphism of TTC repeats for analysis of leprosy transmission. J Clin Microbiol. 2004;42(2):741–745. DOI: 10.1128/JCM.42.2.741-745.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Weng X, Xing Y, Liu J, et al. . Molecular, ethno-spatial epidemiology of leprosy in China: novel insights for tracing leprosy in endemic and non endemic provinces. Infect Genet Evol. 2013;14:361–368. DOI: 10.1016/j.meegid.2012.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Fontes AN, Sakamuri RM, Baptista IM, et al. . Genetic diversity of Mycobacterium leprae isolates from Brazilian leprosy patients. Lepr Rev. 2009;80(3):302–315. [PubMed] [Google Scholar]
- [19].Young SK, Taylor GM, Jain S, et al. . Microsatellite mapping of Mycobacterium leprae populations in infected humans. J Clin Microbiol. 2004;42(11):4931–4936. DOI: 10.1128/JCM.42.11.4931-4936.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].da Silva RA, Dos Santos AAC, Pignataro P, et al. . Genotyping of Mycobacterium leprae from Brazilian leprosy patients suggests the occurrence of reinfection or of bacterial population shift during disease relapse. J Med Microbiol. 2011;60(10):1441–1446. DOI: 10.1099/jmm.0.029389-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Truman R, Fontes AB, de Miranda AB, et al. . Genotypic variation and stability of four variable-number tandem repeats and their suitability for discriminating strains of Mycobacterium leprae. J Clin Microbiol. 2004;42(6):2558–2565. DOI: 10.1128/JCM.42.6.2558-2565.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Groathouse NA, Rivoire B, Kim H, et al. . Multiple polymorphic loci for molecular typing of strains of Mycobacterium leprae. J Clin Microbiol. 2004;42(4):1666–1672. DOI: 10.1128/JCM.42.4.1666-1672.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Sakamuri RM, Kimura M, Li W, et al. . Population-based molecular epidemiology of leprosy in Cebu, Philippines. J Clin Microbiol. 2009;47(9):2844–2854. DOI: 10.1128/JCM.02021-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Srisungnam S, Rudeeaneksin J, Lukebua A, et al. . Molecular epidemiology of leprosy based on VNTR typing in Thailand. Lepr Rev. 2009;80(3):280. [PubMed] [Google Scholar]
- [25].Weng X, Wang Z, Liu J, et al. . Identification and distribution of Mycobacterium leprae genotypes in a region of high leprosy prevalence in China: a 3-year molecular epidemiological study. J Clin Microbiol. 2007;45(6):1728–1734. DOI: 10.1128/JCM.00018-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Shinde V, Newton H, Sakamuri RM, et al. . VNTR typing of Mycobacterium leprae in South Indian leprosy patients. Lepr Rev. 2009;80(3):290. [PubMed] [Google Scholar]
- [27].Clark-Curtiss JE, Walsh G. Conservation of genomic sequences among isolates of Mycobacterium leprae. J Bacteriol. 1989;171(9):4844–4851. 10.1128/jb.171.9.4844-4851.1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Phetsuksiri B, Srisungngam S, Rudeeaneksin J, et al. . SNP genotypes of Mycobacterium leprae isolates in Thailand and their combination with rpoT and TTC genotyping for analysis of leprosy distribution and transmission. Jpn J Infect Dis. 2012;65(1):52–56. [PubMed] [Google Scholar]
- [29].Singh P, Benjak A, Schuenemann VJ, et al. . Insight into the evolution and origin of leprosy bacilli from the genome sequence of Mycobacterium lepromatosis. Proc Natl Acad Sci USA. 2015;112(14):4459–4464. DOI: 10.1073/pnas.1421504112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Sun Z, Li W, Xu S, et al. . The discovery, function and development of the variable number tandem repeats in different Mycobacterium species. Crit Rev Microbiol. 2016;42(5):738–758. DOI: 10.3109/1040841x.2015.1022506. [DOI] [PubMed] [Google Scholar]
- [31].Young SK, Ponnighaus JM, Jain S, et al. . Use of short tandem repeat sequences to study Mycobacterium leprae in leprosy patients in Malawi and India. PLoS Negl Trop Dis. 2008;2(4):e214 DOI: 10.1371/journal.pntd.0000214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Hanseníase [Internet]. Ministerio da Saúde 2016. Available from: http://portalsaude.saude.gov.br/index.php/o-ministerio/principal/secretarias/svs/hanseniase