Abstract
Mycobacterium leprae bacilli are mainly transmitted by the dissemination of nasal aerosols from multibacillary (MB) patients to susceptible individuals through inhalation. The upper respiratory tract represents the main entry and exit routes of M. leprae. Therefore, this study aimed to evaluate the sensitivity and specificity of real-time quantitative polymerase chain reaction (qPCR) in detecting M. leprae in nasal secretion (NS) and skin biopsy (SB) samples from MB and paucibacillary (PB) cases. Fifty-four NS samples were obtained from leprosy patients at the Dona Libânia National Reference Centre for Sanitary Dermatology in Ceará, Brazil. Among them, 19 MB cases provided both NS and SB samples. Bacilloscopy index assays were conducted and qPCR amplification was performed using specific primers for M. leprae 16S rRNA gene, generating a 124-bp fragment. Primer specificity was verified by determining the amplicon melting temperature (Tm = 79.5 °C) and detection limit of qPCR was 20 fg of M. leprae DNA. Results were positive for 89.7 and 73.3% of NS samples from MB and PB cases, respectively. SB samples from MB patients were 100% positive. The number of bacilli detected in NS samples were 1.39 × 103–8.02 × 105, and in SB samples from MB patients were 1.87 × 103–1.50 × 106. Therefore, qPCR assays using SYBR Green targeting M. leprae 16S rRNA region can be employed in detecting M. leprae in nasal swabs from leprosy patients, validating this method for epidemiological studies aiming to identify healthy carriers among household contacts or within populations of an endemic area.
Keywords: Mycobacterium leprae, quantitative real-time PCR, nasal cavity, biopsy, paucibacillary leprosy
Introduction
Leprosy is a chronic infectious disease caused by the acid-fast bacilli Mycobacterium leprae. Variations in susceptibility to M. leprae and clinical manifestations of the infection are attributed to the pattern of the host immune response. M. leprae bacilli mainly invade the Schwann cells in the peripheral nerves, leading to nerve damage and development of physical disabilities [1,2]. Even with the implementation of multidrug therapy, leprosy continues to be a public health concern that has not yet been eliminated. The number of new cases reported globally in 2016 was 214,783. Brazil is the second most affected country, where 28,761 new cases were observed in 2015, accounting for 13% of all new cases detected worldwide [3]. Northeast Brazil is considered a highly endemic area for leprosy, and in 2016, leprosy detection rate in the overall population of the state of Ceará was 18.9/100,000 [4].
Leprosy diagnosis is based on clinical examinations, bacilloscopy of slit-skin smears, and histopathology of skin biopsies; however, paucibacillary (PB) forms are not easily detected by the latter two methods [5]. Despite its low sensitivity, detection by bacilloscopy of slit-skin smears is recommended as the ‘gold-standard’ by the Brazilian health authorities, as it is cheap and non-invasive compared with skin biopsies [6]. The direct detection of acid-fast bacilli in slit-skin smears has a high specificity but a low sensitivity, as approximately 50% of all leprosy patients are slit-skin smears negative [5,6]. Moreover, the bacilloscopy index is not sensitive enough for the diagnosis of subclinical infections, including household contacts of leprosy cases [7–9]. Molecular-based approaches using conventional and quantitative PCR (qPCR) have already been demonstrated as having higher sensitivities than the sensitivity observed for bacilloscopy of skin biopsies and slit-skin smears [8,10–12].
The upper respiratory tract of a susceptible person is considered to be the main entry and exit route of M. leprae [13], and individuals with active disease - multibacillary (MB) cases in particular - are the main sources of infection [14]. In addition, several studies based on nucleic acid amplification have demonstrated that nasal cavities are mainly responsible for the transmission of bacilli [13–17]. Thus, nasal secretion (NS) [7,13,18,19] and skin biopsy (SB) [11,20,21] samples have been widely investigated by both conventional and qPCR, the latter proving much more sensitive. The aim of this work was to evaluate the sensitivity and specificity of qPCR for the detection of M. leprae in NS and SB samples from MB and PB cases.
Materials and methods
Collection and processing of clinical material
NS and SB specimens were obtained from patients at the Dona Libânia National Reference Centre for Sanitary Dermatology, Ceará, Brazil. Untreated leprosy cases were included and confirmed by clinical skin examinations, skin smears, and biopsies. They were classified using the Ridley-Jopling [22] criteria based on histology and bacilloscopy indices (BI) and according to the World Health Organization (WHO) [23] as PB or MB cases. Recruitment of cases was random. A total of 54 NS samples from both nostrils were obtained from patients with different clinical forms, 39 MB (20 lepromatous leprosy [LL] and 19 borderline-borderline [BB]) and 15 PB (14 tuberculoid [T] and one indeterminate [I]). In addition, 19 SB specimens (paired with NS samples) were obtained from MB cases (10 BB and 9 LL). Dried slit-skin smear slides were stained by the Ziehl-Neelsen carbol-fuchsin procedure as described previously [6]. Stained slit-skin smears were examined by optical microscopy and the bacilloscopy index (BI) was calculated according to the Ridley scale. MB patients (LL and BB) exhibited BIs ranging from +1 to +6.0, while all PB cases (including T and I forms) had bacilloscopy-negative [6].
During examination and after the confirmation of a new leprosy case, these patients were invited to participate in the study. The research was approved by the Institutional Ethical Committee (protocol number 011/07) and all participants signed an informed consent form and authorized the collection of samples.
NS samples were obtained from all participants by gently rubbing a nasal swab, previously wetted with Tris-EDTA buffer (pH 8.0), in the vestibule on each side of the nose. After collection, each swab was immersed in a labelled sterile tube and stored at −20 °C until processing. DNA was extracted as described by Lima et al. [19] and was later eluted in dH2O and stored at −20 °C until amplification.
Skin biopsies were collected using a 6-mm diameter punch (Kolplast, Brazil) to obtain tissues from new skin lesions of untreated leprosy cases. DNA extraction was performed using the DNeasy® blood and tissue kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions.
qPCR
Primers were designed using the Primer3Plus software [24] based on the nucleotide sequence of the 16S ribosomal RNA of M. leprae (GenBank accession number: X53999.1). A 124-bp region was amplified using the primers 16S2_For rRNA (5′-AGTGGCGAACGGGTGAGTAA-3′) and 16S2_Rev rRNA (5′-CGCAAAAAGCTTTCCACCAC-3′). Both forward and reverse primers had the same melting temperature (Tm) of 62.9 °C. The PCR amplification mixture contained 10 μL of Power SYBR® Green PCR master mix (Applied Biosystems, Foster City, CA), 40 ng μL of sample DNA (template), and 100 nM of each primer in a total volume of 20 μL. For each qPCR assay, a positive control of 20 pg genomic M. leprae DNA was included, as was a negative control without the target DNA. Amplification was performed using a CFX96 Touch™ real-time PCR detection system (Bio-Rad Laboratories, Hercules, CA). PCR cycling conditions consisted of initial denaturation at 95 °C for 7 min, followed by 45 cycles of denaturation at 94 °C for 30 s, annealing at 62 °C for 30 s, and extension at 72 °C for 1 min.
Data were analyzed by the CFX™ Manager software (version 3.0; Bio-Rad Laboratories) to assess the mean quantitative cycle (Cq). Samples were considered negative when there was no increase in fluorescence signals until 45 cycles (Cq = 45). A standard curve ranging from 2 ng to 0.2 fg was generated by serial dilution of the plasmid pIDT16SrRNAMleprae (pIDT Blue; Integrated DNA Technologies, Coalville, IA) containing a 171-bp region of the 16S rRNA gene of M. leprae [25]. To quantify the number of colony-forming units/mg of each sample tested, the mean Cq values obtained (of each sample tested in duplicate for each batch) were interpolated from the constructed standard curve.
Amplification efficiency and limit of detection (LoD) of qPCR
Amplification efficiency curves were determined for three different assays performed on different days. For this purpose, the linearity of each assay was determined using serial 10-fold dilutions of 2 μL of each DNA sample at the following concentrations: 10, 102, 103, 104, 105, and 106 IU/mL. At each concentration, three replicates were tested in a single run. The following data were determined as estimators of the amplification efficiency: slope, coefficient of determination (R2), and efficiency parameters. We determined the qualitative LoD using diluted samples that were no longer showing a high amplification efficiency by repeating the qPCR amplification reaction 10 times. The LoD was defined as the concentration at which amplification was detected before 37 cycles 95% of the time.
Sensitivity and specificity of qPCR
To ensure specificity of the PCR products, we conducted a melting curve analysis, in which the reaction temperature was increased by 0.5 °C every 20 s, beginning at 60 °C and ending at 95 °C. Throughout the curve construction process, the changes in fluorescence were measured, and the data acquired using the iQ™5 Optical System software (version 2.0, Bio-Rad Laboratories) were processed to verify if a single peak was obtained. Subsequently, PCR products were electrophoresed using a 2% agarose gel stained with ethidium bromide was performed and analyzed using an ImageQuant™ 300 biomolecular imager (GE Healthcare, Little Chalfont, UK).
The sensitivity and specificity of the 16SrRNA assays were assessed by testing M. tuberculosis ATCC 697-7, Mycobacterium sp., and Streptococcus pneumoniae ATCC 49,619. The number of genome equivalents (GEs) was calculated according to the following equation, number of GEs = Avogadro constant (GEs g−1 mol−1) × DNA concentration (C) (ng/μL)/Genome length in bp × molecular mass of 1 bp (g mol−1 bp−1) × 1.0 × 109 (ng). Assays were determined by testing serial decimal dilutions of genomic DNA in triplicate ranging from 1.0 × 108 (approximately 2.0 × 107 GEs) to 1.0 × 101 fg (approximately 2 GEs).
Statistical analysis
A standard linear regression analysis of DNA from M. leprae or the number of copies of the M. leprae curve vs. Cq values was calculated automatically by the CFX Manager™. Differences in the Cq mean values between the MB and PB groups were analyzed by the Mann-Whitney U test using Prism software (version 6.01; GraphPad Software Inc., La Jolla, CA). In addition, the correlation between the Cq values in paired NS and SB samples from MB cases was analyzed using Excel 2013 (Microsoft, Redmond, WA). For all tests, p-values < 0.05 were considered significant.
Results
To validate the sensitivity of the primer set used for qPCR analysis, we analyzed DNA samples extracted from NS and SB samples from patients with leprosy. The primer pair amplified a specific gene fragment from the 16S ribosomal RNA of M. leprae. Ten-fold serial dilutions of the DNA samples ranging from 2 ng to 0.2 fg were prepared and tested in triplicate. The same melting temperature (Tm = 79.5 °C) was observed for all dilutions tested. The assay showed a LoD of less than 20 fg of M. leprae DNA with a Cq value ≤ 37 cycles. All negative control samples were negative for M. leprae DNA detection.
The specificity of each probe-primer set was evaluated by testing DNA samples from other bacteria. The 16S rRNA assays correctly detected all M. leprae sample cases. The assay also amplified the DNA of M. tuberculosis ATCC 697-7 and Mycobacterium sp., but the Tm values of their PCR products differed from that of M. leprae DNA.
Of the 54 M. leprae NS samples, 46 tested positive by the qPCR assay (85.2%), while all SB samples tested positive. Of the 15 nasal samples from PB and 39 from MB patients, 11 (73.3%) and 35 (89.7%) tested positive, respectively (Table 1). As expected, all PB cases had negative BI while the average BI for MB was 3.6 (± 1.5). A higher BI was observed in LL cases (4.5 ± 0.7) compared with borderline cases (2.6 ± 1.5). Among the M. leprae NS samples, the number of bacilli ranged from 5.6 × 102 to 8.0 × 105 and the Cq values ranged from 27.5 to 38.2 cycles (mean of 35.5 cycles). As expected, low Cq values were found for MB cases (mean of 35.5 cycles) compared with PB cases (mean of 35.7 cycles). The tuberculoid cases had the highest Cq (mean of 35.7 cycles) and the lowest bacilli load mean (1.1 × 104 copies). As shown in table 2, only one NS sample from the 19 MB paired samples was not found positive by the qPCR assay.
Table 1.
Bacilloscopy index, frequency of qPCR positivity, and mean number of M. leprae DNA copies and Cq values in nasal samples of leprosy cases, according to the operational and Ridley-Jopling classifications.
| Operational classification | BI | Positive 16s qPCR, N (%) | Mean of Cq values | Mean of bacilli copy number (range) |
|---|---|---|---|---|
| Mean (±SD) | ||||
| PB | 0 | 11 (73.3) | 35.7 | 1.1 × 104 (1.4 × 103 to 6.1 × 104) |
| N = 15 | ||||
| Tuberculoid | 0 | 10 (71.4) | 35.7 | 1.1 × 104 (1.4 × 103 to 6.1 × 104) |
| N = 14 | ||||
| Indeterminate | 0 | 1 | 34.7 | 1.8 × 104 |
| N = 1 | ||||
| MB | 3.6 (±1.5) | 35 (89.7) | 35.5 | 3.7 × 104 (5.6 × 102 to 8.0 × 105) |
| N = 39 | ||||
| Borderline | 2.6 (±1.5) | 17 (89.4) | 35.6 | 2.0 × 104 (9.0 × 102 to 1.7 × 105) |
| N = 19 | ||||
| Lepromatous | 4.5 (±0.7) | 18 (90) | 35.4 | 5.2 × 104 (5.6 × 102 to 8.0 × 105) |
| N = 20 | ||||
| Total | 46 (85.2) | 35.5 | 2.9 × 104 (5.6 × 102 to 8.0 × 105) | |
| N = 54 |
BI, bacilloscopy index; SD, standard deviation; Cq, quantification cycles; MB, mutibacillary; PB, paucibacillary.
Table 2.
Frequency of qPCR positivity in detecting the 124-bp region of the 16S rRNA gene of M. leprae in 19 paired multibacillary (MB) leprosy patient skin biopsies (SB) and nasal samples (NS).
| Operational classification | Positive 16S rRNA qPCR | Total of cases | |
|---|---|---|---|
| NS | SB | ||
| MB | 18 | 19 | 19 |
| Borderline | 9 | 10 | 10 |
| Lepromatous | 9 | 9 | 9 |
NS, nasal secretion; SB, skin biopsy; MB, multibacillary.
To obtain a standard curve for the absolute quantitation of M. leprae DNA, 10-fold dilutions of the plasmid pIDT16SrRNAMleprae were used as template. The standard curve that was generated showed a linear relationship from 103 to 106 DNA copies (Supplementary figure). Linear regression analysis yielded an R2 of 0.98. The slope value for the plasmid was −3.460.
The bacterial load of the 19 SB samples ranged from 30 to 1.5 × 106 copies and the Cq ranged from 26.5 to 42.6 cycles. NS from the same patients ranged from 2.3 × 102 to 8.8 × 106 bacilli and the Cq values ranged from 27.5 to 39.6 cycles (Table 3).
Table 3.
Bacilli copy number and Cq values in nasal and skin biopsy samples of 19 multibacillary paired cases.
| Sample ID | BI | Skin biopsy | Nasal secretiona | ||
|---|---|---|---|---|---|
| Cq | M. leprae copy number | Cq | M. leprae copy number | ||
| 336 | 4 | 31.0 | 7.5 × 104 | 36.3 | 2.1 × 103 |
| 372 | 1.8 | 33.0 | 2.0 × 104 | 37.6 | 9.0 × 102 |
| 381 | 0 | 36.5 | 1.9 × 103 | 36.6 | 1.7 × 103 |
| 386 | 5 | 29.2 | 2.7 × 105 | 35.5 | 3.8 × 103 |
| 387 | 5.5 | 26.5 | 1.5 × 106 | 32.9 | 2.0 × 104 |
| 389 | 4 | 33.5 | 1.4 × 104 | 27.5 | 8.8 × 106 |
| 393 | 1 | 37.4 | 9.4 × 102 | 35.4 | 3.8 × 103 |
| 398 | 4.5 | 39.4 | 2.6 × 102 | 36.5 | 1.8 × 103 |
| 400 | 2.8 | 30.6 | 9.4 × 104 | 35.6 | 3.3 × 103 |
| 401 | 1.8 | 33.9 | 3.1 × 104 | 32.5 | 7.6 × 104 |
| 403 | 4.8 | 39.2 | 2.9 × 102 | 29.8 | 1.7 × 105 |
| 422 | 2.8 | 28.3 | 4.9 × 105 | 37.3 | 1.1 × 103 |
| 423 | 5 | 28.5 | 4.1 × 105 | 36.5 | 1.9 × 103 |
| 429 | 4.8 | 28.4 | 4.5 × 105 | 36.6 | 1.7 × 103 |
| 431 | 3.7 | 37.4 | 1.0 × 103 | 37.2 | 1.2 × 103 |
| 437 | 3.5 | 42.6 | 3.0 × 101 | 39.6 | 2.3 × 102 |
| 441 | 1.6 | 37.2 | 1.2 × 103 | 0.0 | – |
| 443 | 3.8 | 34.3 | 8.0 × 103 | 38.2 | 5.6 × 102 |
| 463 | 3.6 | 34.9 | 5.5 × 103 | 38.7 | 4.3 × 102 |
| Mean | 3.4 | 33.8 | 1.8 × 105 | 35.6 | 5.1 × 105 |
| Range | 0 to 5.5 | 26.5 to 42.6 | 30 to 1.5 × 106 | 27.5 to 39.6 | 2.3 × 102 to 8.8 × 106 |
The sample ID 441 was not considered for the mean and range calculation.
BI, bacilloscopy index; Cq, quantification cycles; SD, standard deviation.
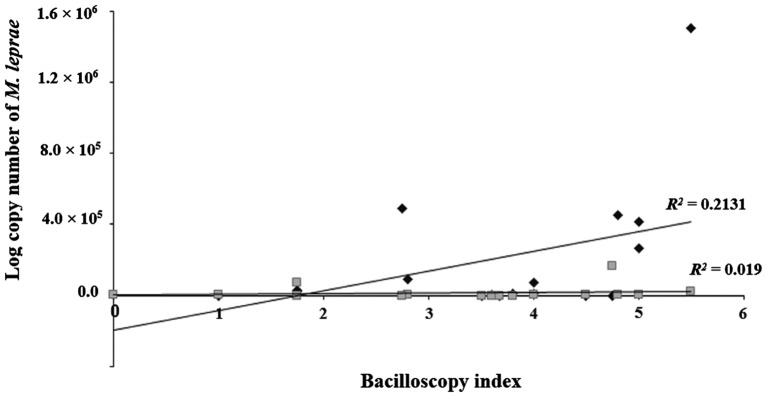
Furthermore, a significant association between the bacilloscopy index values of slit-skin smear samples and the number of M. leprae genomic copies was found while analyzing paired SB and NS samples from MB patients (Figure 1). Higher logarithmic copy numbers of M. leprae bacilli were associated with high bacilloscopy index values in SB samples (R2 = 0.2131; p = 0.001). However, no correlation was observed between the log copy number and bacilloscopy index values of NS samples (R2 = 0.019; p > 0.005).
Figure 1.
Linear range quantification of M. leprae DNA of 18 paired multibacillary (MB) leprosy patients from skin biopsies and nasal samples. The bacilloscopy index (BI) of skin biopsies (♦) and nasal samples (■) was plotted against the logarithmic count number of M. leprae bacilli.
Discussion
In the present study, we identified and quantified M. leprae in nasal secretion samples and biopsies of leprosy cases. Although several studies have reported the detection of M. leprae by real-time PCR in samples from skin biopsies [11,26–28], this is the third study published in the literature that quantified M. leprae in nasal secretion samples [13,18]. The first study investigated 31 nasal secretion samples of leprosy patients without quantifying the number of bacilli [18]. The second study employed qPCR for DNA detection of M. leprae using nasal swabs, nasal turbinate biopsy, and peripheral blood of 113 leprosy cases and 104 household contacts [13]. The same study obtained a 66.4% positive rate of M. leprae DNA detection using TaqMan qPCR [13], whereas our study used the SYBR Green assay and revealed 85.2% positivity of M. leprae DNA in nasal swabs. In addition, our qPCR assay exhibited a higher positivity for PB (11/15, 73.3%) and MB (35/39, 89.7%) compared with the previous report [13] (PB 14/32, 43.8% and MB 61/81, 75.3%) and we found a lower mean of bacilli number per reaction in PB cases (1.1 × 104). In trials where M. leprae DNA was detected by conventional PCR, detection rates of the disease in the general population varied between 9.1 to 84.9% [8,12,14,17,29,30].
Compared with the detection of M. leprae using conventional PCR in leprosy cases, the rate of qPCR positive detection of the 16S rRNA region in nasal secretions (in accordance with the operational classification of positive results) was 73.3% for PB cases, which is far superior to other values reported that included 5.3, 13.3, and 36.4% [8,29,31]. With regards to detection rates of MB cases, we also obtained high results (89.7%) similar to what was found in Minas Gerais (91.9%), Southeastern Brazil [8]. Compared to a study conducted in São Paulo [12], which used qPCR on different biopsy samples, our study had a higher rate of positive results for PB cases and lower rates for MB cases.
Analytical sensitivity refers to the minimum number of PCR amplicons of a sample that can be measured accurately in one assay, whereas clinical sensitivity is the percentage of individuals with a particular disease that the assay identifies as positive for that condition [32]. Our qPCR assay accurately detected 20 fg of M. leprae DNA, equivalent to four bacilli. However, the highest dilution of the standard curve where quantitative analysis of the samples was possible corresponded to Cq ≤ 37. More than this Cq, precise quantification is not possible, and the samples can only be analyzed qualitatively (positive or negative). These findings are similar to those in studies performed with biopsies of patients from Khon Kaen Province, Thailand and from Rio de Janeiro, Brazil [11,20]. However, our study revealed a clinical sensitivity of 73.3% for PB cases, individuals whose diagnosis is more difficult because of the reduced number of bacilli.
The Cq values of the two groups, MB and PB, were not significantly different (p > 0.05). This is probably because the number of PB cases in the study was low. Among the 11 positive cases, only five were Cq ≤ 37 and only these could be analyzed. Although PB cases had fewer bacilli (1.39 × 103–1.42 × 104 copies) in nasal secretions than in MB cases (1.42 × 103–8.02 × 105 copies), it is important to note that PB patients are carriers of the bacillus bacteria and are potential transmitters to susceptible individuals.
In the 19 cases of paired biopsies and nasal secretion samples from leprosy patients, the positive results obtained by the bacilloscopy index were confirmed positive by real-time PCR. Biopsies from patients with higher BI values were deemed positive for bacteria earlier in the amplification cycle, as seen by the lower Cq values and high copy numbers of bacilli. However, the correlation between higher copy number of M. leprae and higher BI values of NS samples were not significantly different (p > 0.05), whereas in biopsy specimens the copy number and BI values differed (p = 0.001). The lack of correlation between the 16S rRNA genomic region detection in NS samples and the BI demonstrate that nasal swabs are not suitable to for leprosy diagnosis. Similar results were also seen in other studies [13,27,29].
Primers specific for the 16S rRNA region displayed 100% specificity for M. leprae with a single PCR product at a melting temperature of 79.5 °C. However, the amplification of M. tuberculosis DNA ATCC 697-7 and Mycobacterium sp. occurred at a different Tm than that of M. leprae. The amplification of M. leprae and Mycobacterium sp. regions could not be distinguished in assays with the same primers using conventional PCR. A previous comparative study targeting several genomic regions of M. leprae performed with skin biopsies reported that real-time PCR targeting of the 16S rRNA region was more sensitive in determining viable M. leprae [27].
There are several methodologies available for real-time qPCR, of which TaqMan and SYBR Green are the most common. Real-time PCR has been used for the diagnosis of several diseases. In general, the SYBR Green methodology costs less and is relatively easier compared with the TaqMan assay, which requires probe design and synthesis. The specificity of the TaqMan method is based on the design of labelled oligonucleotide probes and the exonuclease activity of the Taq polymerase enzyme [33], whereas the SYBR Green assay is based on the attachment of a fluorophore to the dsDNA – any nonspecific product may bind to the primers and give false positive results [34]. Therefore, primer design is the most important step of a PCR assay. In this study, we used the Primer3Plus software for primer design and produced a single fragment [24]. We also performed a BLAST analysis (https://blast.ncbi.nlm.nih.gov/Blast.cgi) against all microbial nucleotide databases of the 124-bp 16S rRNA fragment generated from M. leprae, and confirmed 100% specificity to the M. leprae genome. Lower similarities were also found for other species of the Mycobacterium genus. Real-time PCR assays using DNA from other bacterial species demonstrated specificity for M. leprae, except for Mycobacterium sp., which generated an amplification product with a different Tm than M. leprae. These optimizations ensured that the SYBR Green methodology had a similar specificity to that of TaqMan assays.
In endemic regions, detection of M. leprae DNA from nasal secretions does not differentiate contacts from cases [16,19,35]; therefore, quantification of M. leprae DNA can be used in epidemiological studies to identify healthy carriers among household contacts or within populations of an endemic area [7,36].
Conclusions
Our results suggest that the qPCR assay employing SYBR Green and the M. leprae 16S rRNA target region can be utilized for the detection of bacillus in nasal secretion samples from leprosy patients. This validates the method for use in epidemiological studies aiming to identify healthy carriers between leprosy household contacts or within populations of an endemic area.
Funding
This work was supported by the Conselho Nacional de Desenvolvimento Científico e Tecnológico, Brazil [grant number CNPq-410573/2006-0].
Disclosure statement
No potential conflict of interest was reported by the authors.
Geolocation information
Available at: https://goo.gl/maps/uLepvoDvdr42.
Supplemental data
Supplemental data for this article can be accessed here.
Supplementary Material
Acknowledgements
We express our gratitude to the head of the Dona Libânia National Reference Centre of Dermatology and to all participants who voluntarily cooperated in the study.
References
- [1].Britton WJ, Lockwood DN. Leprosy. Lancet. 2004;363:1209–1219. DOI: 10.1016/S0140-6736(04)15952-7 [DOI] [PubMed] [Google Scholar]
- [2].Pinheiro RO, de Souza Salles J, Sarno EN, et al. Mycobacterium leprae-host-cell interactions and genetic determinants in leprosy: an overview. Future Microbiol. 2011;6:217–230. DOI: 10.2217/fmb.10.173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Luta contra hanseníase [Internet]. Portal da Saúde – Ministério da Saúde; 2017. Available from: http://portalsaude.saude.gov.br/index.php/o-ministerio/principal/secretarias/svs/noticias-svs/27487-em-10-anos-o-numero-de-novos-casos-de-hanseniase-cai-34 [Google Scholar]
- [4].Boletim Epidemiológico de Hanseníase [Internet]. Secretaria de Saúde, Governo do Estado do Ceará; 2017. Available from: http://www.saude.ce.gov.br [Google Scholar]
- [5].Bhushan P, Sardana K, Koranne RV, et al. Diagnosing multibacillary leprosy: a comparative evaluation of diagnostic accuracy of slit-skin smear, bacterial index of granuloma and WHO operational classification. Indian J Dermatol Venereol Leprol. 2008;74:322–326. 10.4103/0378-6323.42892 [DOI] [PubMed] [Google Scholar]
- [6].Baptista IMFD, Sartori BGC, Trino LM. Guia de conduta para realização do exame baciloscópico. Hansenol Int (Online). 2006;31:39–41. [Google Scholar]
- [7].Lourenço DDS, Campelo TA, Cruz GA, et al. Detection of subclinical Mycobacterium leprae infection in children, contacts of leprosy cases, Fortaleza-Ceara, Brazil. Lepr Rev. 2017;88:184–196. [Google Scholar]
- [8].Patrocinio LG, Goulart IM, Goulart LR, et al. Detection of Mycobacterium leprae in nasal mucosa biopsies by the polymerase chain reaction. FEMS Immunol Med Microbiol. 2005;44:311–316. DOI: 10.1016/j.femsim.2005.01.002; pii:S0928-8244(05)00019-2 [DOI] [PubMed] [Google Scholar]
- [9].Hacker MA, Duppre NC, Nery JA, et al. Characteristics of leprosy diagnosed through the surveillance of contacts: a comparison with index cases in Rio de Janeiro, 1987–2010. Mem Inst Oswaldo Cruz. 2012;107(Suppl 1):49–54. DOI: 10.1590/S0074-02762012000900009 [DOI] [PubMed] [Google Scholar]
- [10].Turankar RP, Pandey S, Lavania M, et al. Comparative evaluation of PCR amplification of RLEP, 16S rRNA, rpoT and SodA gene targets for detection of M. leprae DNA from clinical and environmental samples. Int J Mycobacteriol. 2015;4:54–59. DOI: 10.1016/j.ijmyco.2014.11.062 [DOI] [PubMed] [Google Scholar]
- [11].Martinez AN, Britto CF, Nery JA, et al. Evaluation of real-time and conventional PCR targeting complex 85 genes for detection of Mycobacterium leprae DNA in skin biopsy samples from patients diagnosed with leprosy. J Clin Microbiol. 2006;44:3154–3159. DOI: 10.1128/JCM.02250-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Azevedo MC, Ramuno NM, Fachin LR, et al. qPCR detection of Mycobacterium leprae in biopsies and slit skin smear of different leprosy clinical forms. Braz J Infect Dis. 2017;21:71–78. DOI: 10.1016/j.bjid.2016.09.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Araujo S, Freitas LO, Goulart LR, et al. Molecular evidence for the aerial route of infection of Mycobacterium leprae and the role of asymptomatic carriers in the persistence of leprosy. Clin Infect Dis. 2016;63:1412–1420. DOI: 10.1093/cid/ciw570 [DOI] [PubMed] [Google Scholar]
- [14].Job CK, Jayakumar J, Kearney M, et al. Transmission of leprosy: a study of skin and nasal secretions of household contacts of leprosy patients using PCR. Am J Trop Med Hyg. 2008;78:518–521. [PubMed] [Google Scholar]
- [15].Almeida EC, Martinez AN, Maniero VC, et al. Detection of Mycobacterium leprae DNA by polymerase chain reaction in the blood and nasal secretion of Brazilian household contacts. Mem Inst Oswaldo Cruz. 2004;99:509–511. pii:S0074-02762004000500009. [DOI] [PubMed] [Google Scholar]
- [16].de Wit MY, Douglas JT, McFadden J, et al. Polymerase chain reaction for detection of Mycobacterium leprae in nasal swab specimens. J Clin Microbiol. 1993;31:502–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Klatser PR, Van BS, Madjid B, et al. Detection of Mycobacterium leprae nasal carriers in populations for which leprosy is endemic. J Clin Microbiol. 1993;31:2947–2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Martins AC, Miranda A, Oliveira ML, et al. Nasal mucosa study of leprosy contacts with positive serology for the phenolic glycolipid 1 antigen. Braz J Otorhinolaryngol. 2010;76:579–587. DOI: 10.1590/S1808-86942010000500008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Lima LN, Frota CC, Mota RM, et al. Widespread nasal carriage of Mycobacterium leprae among a healthy population in a hyperendemic region of Northeastern Brazil. Mem Inst Oswaldo Cruz. 2015;110:898–905. DOI: 10.1590/0074-02760150178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Rudeeaneksin J, Srisungngam S, Sawanpanyalert P, et al. LightCycler™ real-time PCR for rapid detection and quantitation of Mycobacterium leprae in skin specimens. FEMS Immunol Med Microbiol. 2008;54:263–270. DOI: 10.1111/j.1574-695X.2008.00472.x [DOI] [PubMed] [Google Scholar]
- [21].Yan W, Xing Y, Yuan LC, et al. Application of RLEP real-time PCR for detection of M. leprae DNA in paraffin-embedded skin biopsy specimens for diagnosis of paucibacillary leprosy. Am J Trop Med Hyg. 2014;90:524–529. DOI: 10.4269/ajtmh.13-0659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Ridley DS, Jopling WH. Classification of leprosy according to immunity. A five-group system. Int J Lepr Other Mycobact Dis. 1966;34:255–273. [PubMed] [Google Scholar]
- [23].Guide to eliminate leprosy as a public health problem [Internet]. World Health Organization; 2000. Available from: http://www.who.int/lep/resources/who_cds_cpe_cee_2000.14/en/ [PubMed] [Google Scholar]
- [24].Untergasser A, Nijveen H, Rao X, et al. Primer3Plus, an enhanced web interface to Primer3. Nucleic Acids Res. 2007;35:W71–W74. DOI: 10.1093/nar/gkm306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Lavania M, Katoch K, Katoch VM, et al. Detection of viable Mycobacterium leprae in soil samples: insights into possible sources of transmission of leprosy. Infect Genet Evol. 2008;8:627–631. DOI: 10.1016/j.meegid.2008.05.007 [DOI] [PubMed] [Google Scholar]
- [26].Martinez TS, Figueira MM, Costa AV, et al. Oral mucosa as a source of Mycobacterium leprae infection and transmission, and implications of bacterial DNA detection and the immunological status. Clin Microbiol Infect. 2011;17:1653–1658. DOI: 10.1371/journal.pntd.0001354 [DOI] [PubMed] [Google Scholar]
- [27].Martinez AN, Lahiri R, Pittman TL, et al. Molecular determination of Mycobacterium leprae viability by use of real-time PCR. J Clin Microbiol. 2009;47:2124–2130. DOI: 10.1128/JCM.00512-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Sharma R, Lavania M, Katoch K, et al. Development and evaluation of real-time RT-PCR assay for quantitative estimation of viable Mycobacterium leprae in clinical samples. Indian J Lepr. 2008;80:315–321. [PubMed] [Google Scholar]
- [29].Araújo S, Lobato J, Reis Érica de Melo, et al. Unveiling healthy carriers and subclinical infections among household contacts of leprosy patients who play potential roles in the disease chain of transmission. Mem Inst Oswaldo Cruz. 2012;107(Suppl 1):55–59. pii:S0074-02762012000900010. 10.1590/S0074-02762012000900010 [DOI] [PubMed] [Google Scholar]
- [30].Cardona-Castro N, Beltrán-Alzate JC, Manrique-Hernández R. Survey to identify Mycobacterium leprae-infected household contacts of patients from prevalent regions of leprosy in Colombia. Mem Inst Oswaldo Cruz. 2008;103:332–336. DOI: 10.1590/S0074-02762008000400003 [DOI] [PubMed] [Google Scholar]
- [31].Pontes AR, Almeida M, Xavier MB, et al. Detection of Mycobacterium leprae DNA in nasal swab. Rev Bras Enferm. 2008;61:734–737. [DOI] [PubMed] [Google Scholar]
- [32].Bustin SA, Benes V, Garson JA, et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem. 2009;55:611–622. DOI: 10.1373/clinchem.2008.112797 [DOI] [PubMed] [Google Scholar]
- [33].Livak KJ, Flood SJ, Marmaro J, et al. Oligonucleotides with fluorescent dyes at opposite ends provide a quenched probe system useful for detecting PCR product and nucleic acid hybridization. Genome Res. 1995;4:357–362. 10.1101/gr.4.6.357 [DOI] [PubMed] [Google Scholar]
- [34].Mackay IM. Real-time PCR in the microbiology laboratory. Clin Microbiol Infect. 2004;10:190–212. DOI: 10.1111/j.1198-743X.2004.00722.x [DOI] [PubMed] [Google Scholar]
- [35].Pattyn SR, Ursi D, Ieven M, et al. Detection of Mycobacterium leprae by the polymerase chain reaction in nasal swabs of leprosy patients and their contacts. Int J Lepr Other Mycobact Dis. 1993;61:389–393. [PubMed] [Google Scholar]
- [36].Arunagiri K, Sangeetha G, Sugashini PK, et al. Nasal PCR assay for the detection of Mycobacterium leprae pra gene to study subclinical infection in a community. Microb Pathog. 2017;104:336–339. DOI: 10.1016/j.micpath.2017.01.046 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.