SUMMARY
Bloodstream infections are associated with considerable morbidity and health care costs. Molecular rapid diagnostic tests (mRDTs) are a promising complement to conventional laboratory methods for the diagnosis of bloodstream infections and may reduce the time to effective therapy among patients with bloodstream infections. The concurrent implementation of antimicrobial stewardship programs (ASPs) may reinforce these benefits. The aim of this study was to evaluate the cost-effectivenesses of competing strategies for the diagnosis of bloodstream infection alone or combined with an ASP. To this effect, we constructed a decision-analytic model comparing 12 strategies for the diagnosis of bloodstream infection. The main arms compared the use of mRDT and conventional laboratory methods with or without an ASP. The baseline strategy used as the standard was the use of conventional laboratory methods without an ASP, and our decision-analytic model assessed the cost-effectivenesses of 5 principal strategies: mRDT (with and without an ASP), mRDT with an ASP, mRDT without an ASP, conventional laboratory methods with an ASP, and conventional laboratory methods without an ASP. Furthermore, based on the availability of data in the literature, we assessed the cost-effectivenesses of 7 mRDT subcategories, as follows: PCR with an ASP, matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) analysis with an ASP, peptide nucleic acid fluorescent in situ hybridization (PNA-FISH) with an ASP, a blood culture nanotechnology microarray system for Gram-negative bacteria (BC-GP) with an ASP, a blood culture nanotechnology microarray system for Gram-positive bacteria (BC-GN) with an ASP, PCR without an ASP, and PNA-FISH without an ASP. Our patient population consisted of adult inpatients in U.S. hospitals with suspected bloodstream infection. The time horizon of the model was the projected life expectancy of the patients. In a base-case analysis, cost-effectiveness was determined by calculating the numbers of bloodstream infection deaths averted, the numbers of quality-adjusted life years gained, and incremental cost-effectiveness ratios (ICERs). In a probabilistic analysis, uncertainty was addressed by plotting cost-effectiveness planes and acceptability curves for various willingness-to-pay thresholds. In the base-case analysis, MALDI-TOF analysis with an ASP was the most cost-effective strategy, resulting in savings of $29,205 per quality-adjusted life year and preventing 1 death per 14 patients with suspected bloodstream infection tested compared to conventional laboratory methods without an ASP (ICER, −$29,205/quality-adjusted life year). BC-GN with an ASP (ICER, −$23,587/quality-adjusted life year), PCR with an ASP (ICER, −$19,833/quality-adjusted life year), and PCR without an ASP (ICER, −$21,039/quality-adjusted life year) were other cost-effective options. In the probabilistic analysis, mRDT was dominant and cost-effective in 85.1% of simulations. Importantly, mRDT with an ASP had an 80.0% chance of being cost-effective, while mRDT without an ASP had only a 41.1% chance. In conclusion, our findings suggest that mRDTs are cost-effective for the diagnosis of patients with suspected bloodstream infection and can reduce health care expenditures. Notably, the combination of mRDT and an ASP can result in substantial health care savings.
KEYWORDS: bloodstream infections, cost-effectiveness, diagnostics, rapid tests
INTRODUCTION
Bloodstream infections are the leading cause of mortality due to infection in the United States (1) and have been associated with prolonged hospital lengths of stay (2) and increased health care expenditures (3). There are over 60,000 bloodstream infection episodes in the United States per year (1), and each bloodstream infection is associated with a cost of at least $10,000 to $20,000 per patient (3), in addition to the general costs of hospitalization. In addition, there has been an increase in the number of bloodstream infections caused by antibiotic-resistant organisms (4).
The timely administration of appropriate antimicrobial therapy can reduce morbidity and mortality in patients with bloodstream infections (5–7) as well as prevent the development of antibiotic resistance (8). However, reliance on blood cultures and conventional laboratory methods for bacterial identification can result in a delay in the timely administration of effective therapy, as these methods require up to 5 days for diagnostic results (9). The rapid identification of microbial pathogens can allow a prompt switch from broad-spectrum antimicrobial agents to targeted therapy, potentially reducing drug toxicity, antimicrobial drug resistance, and health care costs (10).
The replacement of conventional laboratory methods with molecular rapid diagnostic tests (mRDTs) that require ≤24 h for organism identification can reduce the time to effective therapy and improve outcomes among patients with bloodstream infections, benefits which may be reinforced by the concurrent implementation of antimicrobial stewardship programs (ASPs) (11). The expert guidance of an ASP allows the effective incorporation of mRDT results in clinical practice, improving patient outcomes and conserving health care resources (12).
The Infectious Diseases Society of America (IDSA) supports the use of mRDTs and ASPs as a means of optimizing antimicrobial therapy (13). In spite of these recommendations, mRDTs have yet to be widely implemented, due to high initial costs (14) and uncertainty with respect to the magnitude of the long-term savings that may result from the use of mRDT (15). As the establishment of novel diagnostic methods necessitates evidence of both clinical effectiveness and economic efficiency, it is essential to identify cost-effective strategies for the diagnosis of bloodstream infections. The current emphasis of clinical decision-makers and of the Department of Health and Human Services on achieving high-value, affordable care and implementing value-based policies by linking fee-for-service payments to the quality of an intervention (16) further underlines the need for information on the cost-effectivenesses of different diagnostic strategies.
Data from several single-center studies suggest that the use of mRDT is associated with significant health care cost savings and improved outcomes (17–25). However, a policy paper by the IDSA (14) along with recently reported reviews (10, 12, 15) have underlined the lack of studies on the cost-effectiveness of mRDT (10). In addition, those reports underlined the need for cost-effectiveness studies that take into consideration the length of stay and the effect of mRDTs on mortality. The aim of this study is to address this gap by determining the cost-effectiveness of mRDT alone or combined with an ASP, according to the recommendations provided by the Second Panel on Cost-Effectiveness Analysis (26).
COST-EFFECTIVENESS GUIDELINES AND MODEL STRUCTURE
The cost-effectiveness guidelines reported in 2017 by the Second Panel on Cost-Effectiveness Analysis, which served as an update to the guidelines provided by the First Panel on Cost-Effectiveness, were followed in conducting this study (26).
Model Structure and Inputs
Model structure.
A decision-analytic model was constructed to compare the cost-effectivenesses of different strategies used for the identification of bacterial organisms and/or the presence of antibiotic resistance among patients with a suspected bloodstream infection. To retrieve pertinent data for our model, we searched the published literature for full-length studies and conference abstracts. The source of each piece of data is noted below. Most of the data for the model were obtained from recent reviews (27–29) and a meta-analysis (11) that compared the rates of survival of patients with bloodstream infections who underwent bacterial species identification with either mRDT or conventional laboratory methods, with or without a concurrent ASP.
The analysis was performed from the perspective of a U.S.-based hospital. An impact inventory, presented in Table 1, was used to identify the health and economic consequences to be quantified in the model. Consequently, we incorporated information from studies conducted in the United States and relied on U.S. sources to assign diagnostic and treatment costs (30–32). Outcomes and costs were calculated from the time when a blood culture was ordered for a suspected bloodstream infection to patient discharge or death and information on mortality for the first 30 days after patient admission. Quality-adjusted life years (QALYs) were calculated for the lifetime of a patient. The patient population consisted of adult, hospital inpatients with suspected bacteremia for whom blood cultures were ordered. The analytical model was developed by using TreeAge Pro 2017 modeling software (TreeAge, Williamstown, MA).
TABLE 1.
Impact inventory
| Sector and type(s) of impact | Included in this analysis from the perspective of the health care sector? | Note on sources of evidence |
|---|---|---|
| Formal health care sector | ||
| Health | ||
| Health outcomes (effects) | ||
| Longevity effects (yr) | ✔ | See “Model Structure and Inputs” |
| Health-related quality-of-life effects (QALYs) | ✔ | See “Model Structure and Inputs” |
| Mortality for 30 days after admission | ✔ | See “Model Structure and Inputs” |
| Medical costs | ||
| Paid for by third-party payers | ✔ | |
| Paid for by patients out of pocket | ✔ | |
| Future related medical costs (payers and patients | ✘ | No data available |
| Future unrelated medical costs (payers and patients) | ✘ | No data available |
| Informal healthcare sector | ||
| Health | ||
| Patient time costs | ✘ | |
| Unpaid caregiver time costs | ✘ | |
| Transportation costs | ✘ |
(i) Diagnostic strategies.
mRDT was defined as any molecular method capable of providing a diagnosis in ≤24 h (11) after a positive blood culture. Specifically, our mRDT definition included methods such as PCR, matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) analysis, peptide nucleic acid fluorescent in situ hybridization (PNA-FISH), a blood culture nanotechnology microarray system for Gram-negative bacteria (BC-GN), and a blood culture nanotechnology microarray system for Gram-positive bacteria (BC-GP). On the other hand, conventional laboratory methods were defined as methods that rely on biochemical reactions and organism-specific characteristics, such as growth, pH changes, enzymatic reactions, and metabolic activity, to provide organism identification (33). In this study, we also took into consideration whether the diagnostic technique was combined with an ASP.
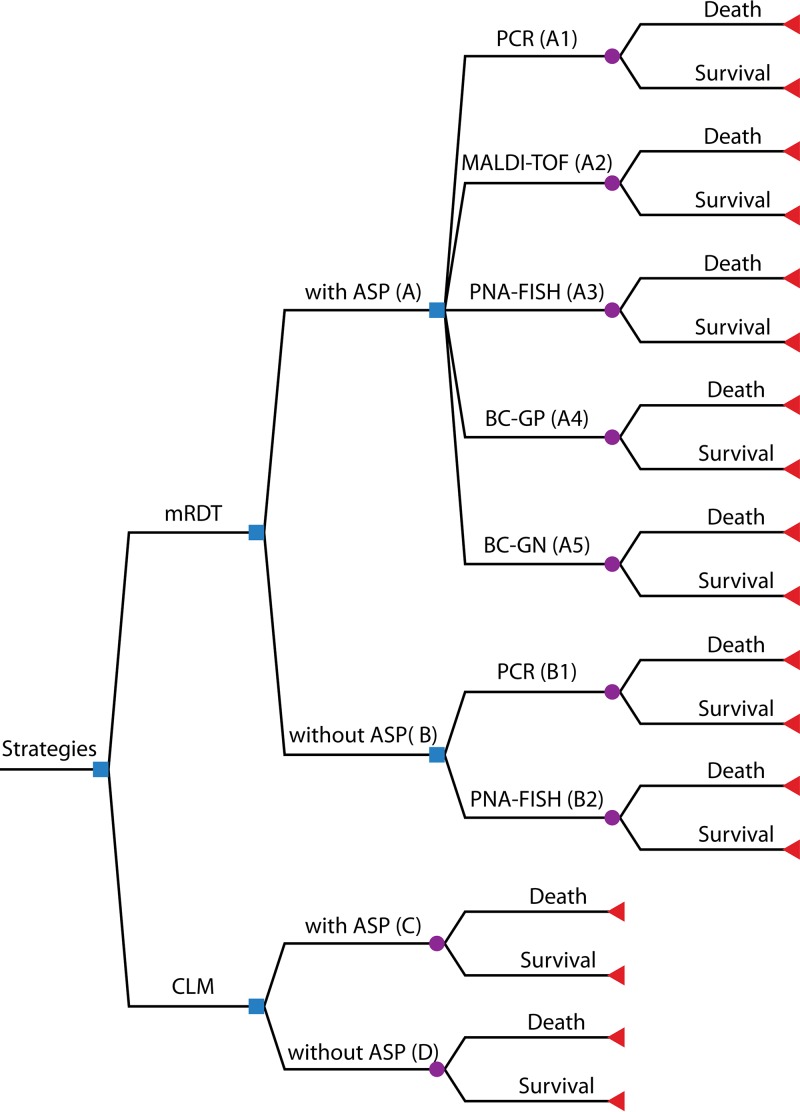
Our decision-analytic model (Fig. 1) assessed the cost-effectivenesses of the following 5 principal strategies: an mRDT (with or without an ASP), an mRDT with an ASP (strategy A), an mRDT without an ASP (strategy B), conventional laboratory methods with an ASP (strategy C), and conventional laboratory methods without an ASP (strategy D). Furthermore, we also assessed the cost-effectivenesses of 7 mRDT subcategories with or without an ASP depending on the availability of relevant data, as follows: PCR with an ASP (strategy A1), MALDI-TOF analysis with an ASP (strategy A2), PNA-FISH with an ASP (strategy A3), BC-GP with an ASP (strategy A4), BC-GN with an ASP (strategy A5), PCR without an ASP (strategy B1), and PNA-FISH without an ASP (strategy B2). BC-GN without an ASP, BC-GP without an ASP, MALDI-TOF analysis without an ASP, and diagnostic tests that do not rely on blood cultures were not included in our analysis, as there is a paucity of data on clinical outcomes.
FIG 1.
Decision tree model. Shown is a summary of the 5 principal and 7 specific mRDT strategies used for the diagnosis of bloodstream infection. Squares indicate decision nodes, circles indicate chance nodes, and triangles indicate endpoints. CLM, conventional laboratory methods.
(ii) Model inputs.
Model inputs, including mortality probabilities, length-of-stay estimates, and costs, were obtained from the published literature. Previous reviews have summarized studies assessing the use of mRDTs for the diagnosis of bloodstream infections (11, 27–29). A recently published meta-analysis (11) was used to identify studies estimating clinical outcomes among patients who underwent a diagnosis of bloodstream infection with either mRDT or conventional laboratory methods (11). The effectiveness of each diagnostic strategy was assessed in terms of mortality risk. In turn, mortality was defined as either all-cause 30-day mortality (17, 23, 34–42) or in-hospital mortality (9, 18, 22, 24, 25, 43).
All studies that provided available mortality data (9, 17–25, 34, 36–45) were included in our analysis, while studies conducted outside the United States (46–51) or studies that included pediatric patients (35) were excluded. Overall mortality estimates, which were then used to yield probabilities of survival, were obtained by pooling the mortality rates of the included studies with the use of a random-effects meta-analysis (Der Simonian and Laird) (52, 53).
Furthermore, the Freeman-Tukey double-arcsine transformation (54) was performed to facilitate the statistical weighting of extreme mortality values (close to zero or unity). A random-effects meta-analysis was also used to estimate the average length of stay associated with each strategy by pooling the respective results of individual studies (17–19, 22, 23, 34, 37, 38, 40, 55). This method, as opposed to a fixed-effect model, was chosen as it accounts for the considerable interstudy differences and heterogeneity among the data from the included studies by using a simple noniterative method to estimate the interstudy effect variance (53).
To estimate QALYs for the population of our study, we assumed a cohort population with characteristics similar to those of the average for our included studies. Specifically, using the median values across data from the included studies in the analysis, we assumed a population with a size of 195 patients (sample index) and a median age of 58 years that is composed of 53% males and 47% females. The pertinent life expectancy data were obtained from U.S. life expectancy tables (56). We extracted the quality of life for the general U.S. population from the European Quality of Life-5 Dimensions (EQ-5D) index population norm data (57). The EQ-5D index was chosen as it is recommended by guidelines (58, 59). For the first 5 years after the initial bloodstream infection episode, patients were expected to accumulate QALYs poorly, as suggested by data in the literature (60–62), and were assigned a quality-of-life utility score of 0.68 (61). This was obtained from a study by Cuthbertson et al. that estimated the EQ-5D score 5 years after a severe sepsis episode. The quality-of-life value was discounted by 3.0% per year, as suggested by guidelines (26). The mean discounted QALY value that we obtained and that was used as the basis of our analysis was 13.47 (standard deviation, 1.49).
All costs obtained from the literature were adjusted to 2016 dollars by using the personal health care (PHC) expenditure deflator from the Agency of Healthcare Research and Quality (63) and then adjusted to 2017 dollars (fourth quarter) by using the personal consumption expenditure (PCE) price index (64), as recommended by guidelines (59). The gold standard of bloodstream infection diagnosis is the collection of blood cultures. For our study, the estimated base cost of a single blood culture was set at $118 ($178.05 adjusted) and was obtained from data reported by Shapiro et al. (65). This baseline cost was multiplied by 3 for each patient, to better reflect the widely accepted approach that 2 to 3 blood cultures should be ordered in cases of suspected bacteremia (66). Subsequently, the costs of different methods used for pathogen identification and antimicrobial susceptibility testing, after a positive blood culture, were also estimated (29).
In particular, semiautomated systems, such as Phoenix (Becton, Dickinson), Vitek 2 (bioMérieux), and MicroScan WalkAway (Siemens), are conventionally used for this purpose (67) and fall under our definition of conventional laboratory methods. Nonetheless, the average costs of conventional laboratory methods per pathogen identified vary widely, depending on hospital identification protocols and the specific sequence of reactions needed to identify each organism. As such, we approximated the average adjusted cost for conventional laboratory methods to be $3.97 ($5.09 adjusted) per patient, by employing data from a study by Tan et al. (conducted at The Johns Hopkins Hospital, Baltimore, MD) that reported the overall cost for the identification of the 55 most common pathogens causing bloodstream infections over a 1-year period (30). The cost of mRDT was similarly obtained from data in the literature. The base-case cost values used were $45 ($67.90 adjusted), $80 ($99.61 adjusted), $99 ($105.89 adjusted), $82.72 ($139.05 adjusted), and $43 ($45.99 adjusted) for PCR (68), BC-GP (69), BC-GN (42), PNA-FISH (70), and MALDI-TOF analysis (71), respectively. For the cost of ASPs, we used the cost estimate provided by Scheetz et al., namely, $125 ($179.29 adjusted) per patient (31).
The patient hospitalization cost for each strategy was estimated by multiplying the average pooled length of stay for that strategy by the cost of hospitalization per day for general-medicine patients. Specifically, we used the most recent estimate (2015) of hospitalization costs in Rhode Island, provided by the Kaiser Family Foundation, which is $2,759 ($3,080.80 adjusted) per day (32). This value was chosen both because it lies in the middle of the hospitalization cost range for various U.S. states and because Rhode Island is the geographic base of our group. Notably, this estimate accounts for all inpatient expenses, including the cost of antimicrobial agents (32). Model inputs, including probability values, length-of-stay estimates, and costs, are summarized in Table 2. In the text, costs, probabilities, and QALYs have been rounded to two decimals, and incremental cost-effectiveness ratio (ICER) values have been rounded to zero decimals.
TABLE 2.
Model inputs and baseline estimates for probabilities, lengths of stay, and costs
| Input | Mean base-case value (range) | Type of distribution (range or mean [SD]) | Reference(s) |
|---|---|---|---|
| Mortality probabilities | |||
| mRDT | 0.11 (0.08–0.14) | Uniform (0.08–0.14) | 9, 17–25, 34, 36–45 |
| Conventional laboratory methods | 0.16 (0.12–0.19) | Uniform (0.12–0.19) | 9, 17–25, 34, 36–45 |
| mRDT with ASP (A) | 0.11 (0.08–0.15) | Uniform (0.08–0.15) | 9, 17–23, 25, 34, 36, 37, 39–42, 44, 45 |
| mRDT without ASP (B) | 0.10 (0.07–0.14) | Uniform (0.07–0.14) | 24, 38, 43 |
| Conventional laboratory methods with ASP (C) | 0.16 (0.12–0.20) | Uniform (0.12–0.20) | 9, 17–23, 25, 34, 36, 37, 39–42, 44, 45 |
| Conventional laboratory methods without ASP (D) | 0.15 (0.11–0.19) | Uniform (0.11–0.19) | 24, 38, 43 |
| PCR with ASP (A1) | 0.12 (0.05–0.22) | Uniform (0.05–0.22) | 9, 18, 25, 44 |
| MALDI-TOF analysis with ASP (A2) | 0.08 (0.04–0.12) | Uniform (0.04–0.12) | 17, 23, 37, 39 |
| PNA-FISH with ASP (A3) | 0.15 (0.03–0.33) | Uniform (0.03–0.33) | 20-22, 36 |
| BC-GP with ASP (A4) | 0.16 (0.04–0.33) | Uniform (0.04–0.33) | 19, 40, 45 |
| BC-GN with ASP (A5) | 0.08 (0.04–0.12) | Uniform (0.04–0.12) | 34, 41, 42 |
| PCR without ASP (B1) | 0.12 (0.07–0.18) | Uniform (0.07–0.18) | 38, 43 |
| PNA-FISH without ASP (B2) | 0.08 (0.03–0.15) | Uniform (0.03–0.15) | 24 |
| Length of stay (days) | |||
| mRDT | 11.58 (9.44–13.72) | Gamma (11.58 [0.71]) | 17–19, 22, 23, 34, 37, 38, 40, 55 |
| Conventional laboratory methods | 14.02 (11.23–16.80) | Gamma (14.02 [0.93]) | 17–19, 22, 23, 34, 37, 38, 40, 55 |
| mRDT with ASP (A) | 9.89 (8.04–11.74) | Gamma (9.89 [0.62]) | 17–19, 22, 23, 34, 37, 40 |
| mRDT without ASP (B) | 18.37 (13.79–22.97) | Gamma (18.37 [1.53]) | 38, 55 |
| Conventional laboratory methods with ASP (C) | 13.31 (10.42–16.19) | Gamma (13.31 [0.96]) | 17–19, 22, 23, 34, 37, 40 |
| Conventional laboratory methods without ASP (D) | 17.98 (8.99–35.96) | Gamma (17.89 [4.50]) | 38, 55 |
| PCR with ASP (A1) | 15.30 (12.25–18.35) | Gamma (15.30 [1.02]) | 18 |
| MALDI-TOF analysis with ASP (A2) | 8.97 (5.84–12.09) | Gamma (8.97 [1.04]) | 23, 28, 37 |
| PNA-FISH with ASP (A3) | 17.00 (9.86–24.14) | Gamma (17.00 [2.38]) | 22 |
| BC-GP with ASP (A4) | 7.82 (6.90–8.75) | Gamma (7.82 [0.31]) | 19, 40 |
| BC-GN with ASP (A5) | 10.67 (7.46–13.88) | Gamma (10.67 [1.07]) | 34 |
| PCR without ASP (B) | 15.20 (8.83–21.57) | Gamma (15.20 [2.12]) | 38 |
| PNA-FISH without ASP (B2) | 18.70 (15.47–21.93) | Gamma (18.70 [1.0]) | 55 |
| Costs (US$) | |||
| Blood culture | 178.05 (89.03–356.10) | Gamma (178.05 [44.51]) | 65 |
| PCR | 67.90 (33.95–135.80) | Gamma (67.90 [16.98]) | 68 |
| BC-GP | 99.61 (49.81–199.22) | Gamma (99.61 [24.90]) | 69 |
| BC-GN | 105.89 (52.95–211.78) | Gamma (105.89 [26.47]) | 42 |
| PNA-FISH | 139.05 (69.53–278.10) | Gamma (139.05 [34.76]) | 70 |
| MALDI-TOF analysis | 45.99 (23.00–91.98) | Gamma (45.99 [11.50]) | 71 |
| Conventional laboratory methods | 5.09 (2.55–10.18) | Gamma (5.09 [1.27]) | 30 |
| mRDT | 91.69 (45.85–183.38) | Gamma (91.69 [22.92]) | 42, 68–71 |
| ASP per patient | 179.29 (89.65–358.58) | Gamma (179.29 [44.82]) | 31 |
| In-patient hospitalization per day for Rhode Island, 2015 | 3,080.80 (1,540.40–6,161.60) | Gamma (3,080.80 [770.20]) | 32 |
Outcome measure and analysis.
The outcome for the base-case analysis was the ICER, defined as the excess cost of a strategy over the cost of the baseline strategy (conventional laboratory methods without an ASP) divided by the incremental difference in effectivenesses between the strategy in question and the baseline strategy. In turn, the incremental difference in effectivenesses was defined in terms of both the number of QALYs gained and the number of deaths averted. Incremental costs were determined by calculating the differences in the attributed costs of each strategy. Cost-effectiveness was estimated for various cost-effectiveness thresholds.
To account for random variation in the included cost and outcome variables and to evaluate the robustness of our results, we used both deterministic (one-way sensitivity analysis) and probabilistic (Monte Carlo analysis) methods. The deterministic one-way sensitivity analysis (72) was performed for all cost and outcome variables, which allowed each variable to vary within a range of values, as summarized in Table 2. Beta distributions were used to account for variations in the probabilities of patient death/survival, and gamma distributions were used for costs, as recommended by guidelines (73). Furthermore, length of stay was also modeled by using a gamma distribution to reflect the skewed distribution of this parameter (74).
For variables without an available range, a range of possible values was approximated by allowing the variable to vary between 50% below the base-case value and 200% above the base-case value (75). This approximation was also used to determine the range for the length of stay with conventional laboratory methods without an ASP, as there were only two studies available (38, 55). In cases where the standard deviation was not available, the standard deviation was approximated with the aid of an appropriate estimation formula (76), according to which sample variance for data that are not normally distributed can be estimated by dividing the range by 6.
As noted above, we also performed a probabilistic Monte Carlo sensitivity analysis (77) using 10,000 simulations (75). The model was run 10,000 times, and each time, a value, from the predetermined distributions (Table 2), was randomly selected for each variable. Subsequently, the results of each simulation were plotted on a cost-effectiveness plane (x, y), with x representing incremental effectiveness and y representing incremental cost. Points (x, y) that were located within the southeast (dominant) quadrant of the resulting cost-effectiveness plane graph were considered to be cost-effective (78). The percentage of simulations falling within this quadrant was estimated. Furthermore, the outputs obtained from the probabilistic analysis were used in order to obtain cost-effectiveness acceptability curves (79). These curves evaluate the cost-effectiveness of a particular strategy compared to the baseline strategy for a range of different cost-effectiveness thresholds.
CONVENTIONAL LABORATORY METHODS
Conventional Laboratory Methods without an ASP
Conventional laboratory methods without an ASP served as our baseline diagnostic strategy. All other strategies were compared against this standard with respect to the dual outcomes of clinical effectiveness (measured in terms of the probability of survival) and cost. This baseline strategy provided the reference cost and reference effectiveness values used in ICER calculations. In the base-case analysis, the cost of conventional laboratory methods without an ASP was determined to be $55,932.02 per patient after accounting for the cost of blood culture (65), the cost of conventional laboratory methods (30), the length of stay (38, 55), and the cost of hospitalization (32). In turn, the mean effectiveness, measured as the pooled probability of survival that was provided by the data from the included studies (24, 38, 43), was estimated to be 0.85. The mean number of calculated QALYs was 11.45. The calculations for the base-case analysis can be found in the appendix. In the probabilistic analysis, after running the model for 10,000 simulations to estimate the effect of variability, the mean cost was estimated to be $56,095.83 (95% confidence interval [CI], $55,707.78 to $56,483.88), while the mean number of QALYs was estimated to be 11.44 (95% CI, 11.42 to 11.47).
Conventional Laboratory Methods with an ASP
In the base-case analysis, the cost of conventional laboratory methods with an ASP, which accounted for the cost of blood culture (65), the cost of conventional laboratory methods (30), the length of stay (17–19, 22, 23, 34, 37, 40), the cost of hospitalization (32), and the cost of an ASP (31), was calculated to be $41,723.98 (see the appendix). Similarly, the probability of survival was estimated to be 0.84 (9, 17–23, 25, 34, 36, 37, 39–42, 44, 45). The mean calculated number of QALYs was 11.31. Even though the use of conventional laboratory methods with an ASP was less costly than the baseline strategy ($41,723.98 [17–19, 22, 23, 31, 32, 34, 37, 40, 65] versus $55,932.02 [30, 32, 38, 55, 65]), it was also marginally less effective (probability of survival of 0.84 [9, 17–23, 25, 34, 36, 37, 39–42, 44, 45] versus 0.85 [24, 38, 43]), rendering it a suboptimal, non-cost-effective alternative to the baseline strategy.
However, based on the results from the sensitivity analysis, which allows us to vary each model variable within the limits specified in Table 2, conventional laboratory methods with an ASP become cost-effective when the probability of mortality for conventional laboratory methods with an ASP is <0.15 or when the probability of mortality with the baseline strategy is >0.16. Given that these threshold values are very close to the base-case values, the cost-effectiveness outcomes are very sensitive to changes in the mortality values. In the probabilistic analysis, the mean cost for conventional laboratory methods with an ASP was $41,707.71 (95% CI, $41,498.99 to $41,916.44), which is in agreement with the cost obtained from the base-case analysis. The mean number of QALYs was 11.30 (95% CI, 11.27 to 11.32).
RAPID DIAGNOSTIC TESTING
Overall, mRDT (when all mRDT strategies with or without an ASP are combined) was associated with an average cost of $36,301.50 (see the appendix) when accounting for the cost of blood culture (65), the average cost of mRDT (42, 68–71), and the cost of hospitalization (17–19, 22, 23, 32, 34, 37, 38, 40, 55). In turn, the mean probability of survival was estimated to be 0.89 (9, 17–25, 34, 36–45). The difference in effectivenesses between overall mRDT and the baseline strategy, measured as the probability of survival (0.89 for overall mRDT [9, 17–25, 34, 36–45] versus 0.85 for the baseline strategy [24, 38, 43]), was 0.04. In turn, the base-case ICER value, determined as the ratio of the incremental cost (−$19,630.52) (see the appendix) over the incremental difference in the numbers of QALYs (0.54) between mRDT with an ASP and the baseline strategy, was −$36,434/QALY gained (ICER, −$36,434/QALY gained) (see the appendix), rendering this strategy a cost-effective option.
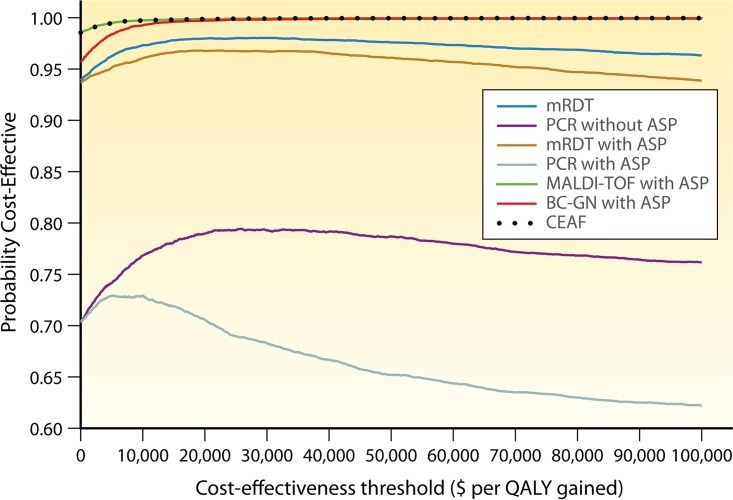
In the sensitivity analysis, no thresholds were identified within the ranges specified in Table 2, suggesting that our findings were robust with respect to this strategy. In the probabilistic analysis, which uses the output information from the Monte Carlo simulation to estimate variability, the mean cost for mRDT was $36,397.47 (95% CI, $36,217.71 to $36,577.23), while the mean number of QALYs was 11.99 (95% CI, 11.96 to 12.01). With respect to the cost-effectiveness planes, the strategy was localized in the cost-effective quadrant in 85.1% of simulations, and as can be seen from the cost-effectiveness acceptability curves (Fig. 2), it was cost-effective in 93.9% to 96.33% of simulations, for a cost-effectiveness threshold ranging between $0 and $100,000.
FIG 2.
Cost-effectiveness acceptability curves and cost-effectiveness acceptability frontier (CEAF). The cost-effectiveness acceptability curves show the probability that a strategy is cost-effective compared to the baseline strategy for a range of different cost-effectiveness thresholds.
Rapid Diagnostic Testing without an ASP
mRDT without an ASP was associated with an average cost of $57,220.14 (see the appendix), after accounting for the cost of blood culture (65), the average cost of mRDT (42, 68–71), and the cost of hospitalization (23, 28, 37, 38, 55). Despite the fact that mRDT without an ASP was associated with a higher probability of survival than the baseline strategy (probability of 0.90 [24, 38, 43] versus 0.85 [24, 38, 43] and 12.12 QALYs versus 11.45 QALYs), it was also more costly ($57,220.14 [38, 55, 65] versus $55,932.02 [30, 32, 38, 55, 65]). Importantly, with an additional cost of $1,913 per QALY gained (ICER, $1,913/QALY gained) (see the appendix), mRDT without an ASP was not shown to be a cost-effective strategy.
In the sensitivity analysis, mRDT without an ASP becomes cost-effective when the length of stay for mRDT without an ASP is less than 18.0 days or when the length of stay for the baseline strategy is more than 18.4 days. This suggests that our finding is not robust with respect to the length of stay and that for patients with a length of stay of <18.0 days, even mRDT without an ASP is cost-effective. In the probabilistic analysis, which aims to further incorporate uncertainty into the analysis, the mean cost for mRDT without an ASP was $57,313.74 (95% CI, $57,018.24 to $57,609.24), while the mean number of QALYs was 12.05 (95% CI, 12.03 to 12.08). Below, we examine specific strategies that fall within the category of mRDT without an ASP (based on available data).
PNA-FISH without an ASP.
The average cost for PNA-FISH without an ASP, including the cost of blood culture (65), the cost of PNA-FISH (70), and the cost of hospitalization (32, 55), was $58,284.16 (see the appendix), while the mean effectiveness was 0.92 (24, 38, 43). The mean number of QALYs was 12.39. PNA-FISH without an ASP was associated with a higher effectiveness than the baseline strategy (probability of survival of 0.92 [24] versus 0.85 [24, 38, 43]). However, higher effectiveness came at the expense of a higher cost ($58,284.16 [32, 55, 65, 70] versus $55,932.02 [30, 32, 38, 55, 65]) (ICER, $2,495/QALY gained) (see the appendix). As a result, PNA-FISH was not a cost-effective alternative, as it resulted in additional costs per QALY gained. This could be due to the fact that PNA-FISH is labor-intensive, requires skilled technicians to interpret the results, and currently provides a limited panel for pathogen detection (24, 29, 80).
In the one-way sensitivity analysis, PNA-FISH without an ASP becomes cost-effective when the length of stay for PNA-FISH without an ASP is less than 18.0 days or when the length of stay for the baseline strategy is more than 18.7 days, suggesting that small variations in hospitalization costs might render this strategy cost-effective. In the probabilistic analysis, the mean cost for PNA-FISH without an ASP was $58,396.27 (95% CI, $58,104.83 to $58,687.72), while the mean number of QALYs was 12.29 (95% CI, 12.26 to 12.31).
PCR without an ASP.
The mean cost of PCR without an ASP, which included the cost of blood culture (65), the cost of PCR (68), and the cost of hospitalization (32, 38), was $47,430.21 (see the appendix), while the mean probability of survival was 0.88. PCR without an ASP was a cost-effective strategy that resulted in savings of $21,039 per QALY gained (ICER, −$21,039/QALY gained) (see the appendix). This finding could potentially be due to the high sensitivity and specificity of PCR (29).
In the sensitivity analysis, PCR without an ASP is no longer cost-effective when the probability of mortality for PCR without an ASP is more than 0.15, when the probability of mortality for the baseline strategy is less than 0.12, when the length of stay for PCR without an ASP is more than 18.0 days, or when the length of stay for the baseline strategy is less than 15 days. This suggests that the cost-effectiveness of this strategy is valid only for values very close to the base-case estimates that we used. In cases where the probability of mortality associated with the use of PCR without an ASP is >0.15 or the length of stay is >18.0 days, PCR without an ASP might not be a cost-effective option.
In the probabilistic analysis, which aimed to further address uncertainty, the mean cost for PCR without an ASP was $47,292.62 (95% CI, $47,027.26 to $47,557.98), which is in agreement with the cost that we obtained in the base-case analysis. The mean number of QALYs was 11.79 (95% CI, 11.76 to 11.81). With respect to the cost-effectiveness planes, this strategy was localized in the cost-effective quadrant in 50.4% of simulations, and as can be seen from the cost-effectiveness acceptability curves (Fig. 2), it was cost-effective in 70.3% to 76.2% of simulations, for a cost-effectiveness threshold ranging between $0 and $100,000.
Rapid Diagnostic Testing with an ASP
The cost of mRDT with an ASP accounted for the cost of blood culture (65), the cost of an ASP (31), the average cost of mRDT (42, 68–71), and the cost of hospitalization (17–19, 22, 23, 32, 34, 37, 40). When comparing mRDT with an ASP to conventional laboratory methods without an ASP, the difference in total costs per patient between the two strategies was $24,657.78 ($31,274.24 for mRDT with an ASP versus $55,932.02 for the baseline strategy [30, 32, 38, 55, 65]) (see the appendix). The difference in effectivenesses between the two strategies measured as the probability of survival (0.89 for mRDT with an ASP [9, 17–23, 25, 34, 36, 37, 39–42, 44, 45] versus 0.85 for the baseline strategy [24, 38, 43]) was 0.04. In turn, the base-case ICER value, calculated as the ratio of the incremental cost (−$24,657.78) over the incremental effectiveness in QALYs (0.54) of mRDT with an ASP versus the baseline strategy was −$45,764/QALY gained (ICER, −$45,764/QALY gained) (see the appendix).
Overall, mRDT with an ASP prevented 1 death per 25 patients tested compared to the baseline strategy and was the most cost-effective strategy compared to the baseline strategy (Table 2). As mRDT with an ASP is both less costly ($31,274.24 [17–19, 22, 23, 31, 34, 37, 38, 40, 55, 65] versus $55,932.02 [30, 32, 38, 55, 65]) and more effective (probability of survival of 0.89 [17–19, 22, 23, 34, 37, 40] versus 0.85 [24, 38, 43]) than the baseline strategy, it is a cost-effective alternative for the diagnosis of bloodstream infections.
Sensitivity analysis revealed that when the probability of mortality for mRDT with an ASP is 0.15, the ICER becomes zero, and when the length of stay for the baseline strategy is <10 days, mRDT with an ASP is no longer a cost-effective strategy. The mortality threshold for mRDT with an ASP coincides with the upper limit of the range that we used for the sensitivity analysis (Table 2), which means that mRDT with an ASP would be cost-effective for most clinical settings. The threshold with respect to the length of stay suggests that the strategy is not cost-effective for large lengths of stay (such as those in long-term-care facilities and other subacute settings). In the probabilistic analysis, the mean cost was $31,513.02 (95% CI, $31,272.20 to $31,753.85), while the mean number of QALYs was 11.91 (95% CI, 11.89 to 11.94). With respect to the cost-effectiveness planes (see Fig. S1 in the supplemental material), the strategy was localized in the cost-effective quadrant in 80.0% of simulations, and as can be seen from cost-effectiveness acceptability curves (Fig. 2), mRDT with an ASP was cost-effective in 93.7% to 93.9% of simulations, for a cost-effectiveness threshold ranging between $0 and $100,000.
PCR with an ASP.
When including the cost of blood culture (65), the cost of an ASP (31), the average cost of PCR (68), and the cost of hospitalization (18, 32), the average calculated cost for PCR with an ASP was $47,917.58 (18, 31, 68) (see the appendix), while the mean probability of survival was 0.88 (9, 18, 25, 44). The calculated ICER value suggested that PCR with an ASP resulted in savings of $19,833 per QALY gained (ICER, −$19,833/QALY gained) (see the appendix), suggesting that PCR with an ASP was a cost-effective strategy.
Results from the sensitivity analysis indicate that PCR with an ASP is no longer cost-effective when the probability of mortality with PCR with an ASP is >0.15, when the probability of mortality with the baseline strategy is <0.12, when the length of stay for PCR with ASP is >17.9 days, or when the length of stay for the baseline strategy is <15.4 days. These findings suggest that PCR with an ASP would not be cost-effective for settings that approach the outer limits of the ranges specified in Table 2 but would be cost-effective for most other cases.
In the probabilistic analysis, the mean cost, as determined by Monte Carlo analysis, was $47,950.72 (95% CI, $47,710.99 to $48,190.44), while the mean number of QALYs was 11.67 (95% CI, 11.65 to 11.70). With respect to cost-effectiveness planes, the strategy was in the cost-effective quadrant in 40.9% of simulations, and as can be seen from cost-effectiveness acceptability curves (Fig. 2), PCR with an ASP was cost-effective in 70.4% to 62.2% of simulations, for a cost-effectiveness threshold ranging between $0 and $100,000.
MALDI-TOF analysis with an ASP.
When comparing MALDI-TOF analysis with an ASP with conventional laboratory methods without an ASP, the difference in costs between the two strategies was $27,537.81 ($28,394.21 for MALDI-TOF analysis [23, 28, 32, 37, 38, 55] with an ASP versus $55,932.02 [30, 32, 38, 55, 65] for the baseline strategy) (see the appendix). The cost of MALDI-TOF analysis with an ASP accounted for the cost of blood culture (65), the cost of an ASP (31), the cost of MALDI-TOF analysis (71), and the cost of hospitalization (23, 28, 32, 37).
The difference in effectivenesses between the two strategies, measured as the probability of survival (0.92 for MALDI-TOF analysis [17, 23, 37, 39] versus 0.85 for the baseline strategy [24, 38, 43]), was 0.07. The difference in the number of QALYs was 0.94 (12.39 QALYs versus 11.45 QALYs). In turn, the base-case ICER value, determined as the ratio of the incremental cost ($27,537.81) over the incremental difference in the number of QALYs (0.94) between MALDI-TOF analysis with an ASP and the baseline strategy, was −$29,205/QALY gained (ICER, −$29,205/QALY gained) (see the appendix). MALDI-TOF analysis with an ASP prevented 1 death per 14 patients tested compared to the baseline strategy and was the most cost-effective strategy. In fact, our results suggest that among individual strategies, MALDI-TOF analysis with an ASP was associated with the highest savings per QALY gained (Fig. 3 and 4). This could be due to the fact that MALDI-TOF analysis has a quick turnaround time and can detect a very broad range of pathogens (27).
FIG 3.
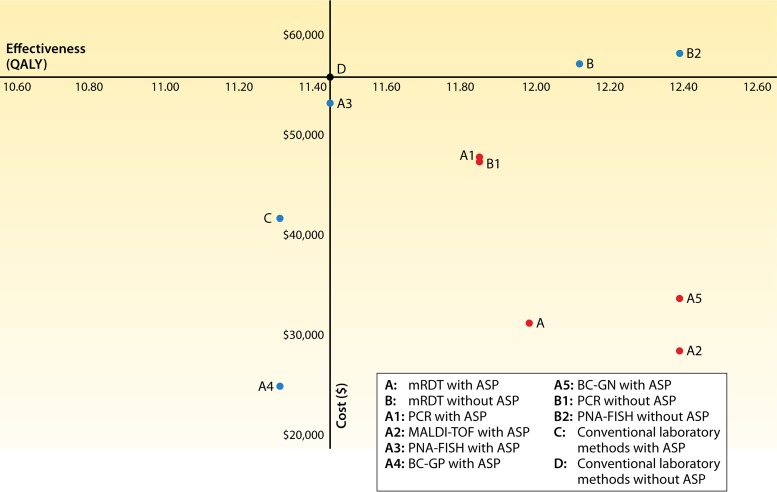
Cost-effectiveness plane for all of the strategies. The y axis represents the average cost of a strategy, while the x axis represents the average effectiveness of a strategy. Cost-effective strategies are depicted with red markers, the baseline strategy is depicted with a black marker, and the remaining strategies that are suboptimal or not cost-effective are indicated with blue markers.
FIG 4.
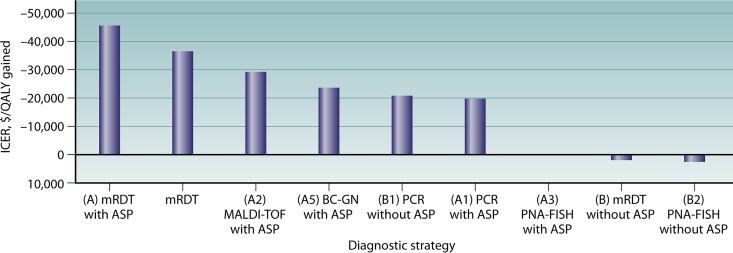
ICER per strategy. mRDT with an ASP resulted in the highest savings per death averted. mRDT (combines all individual mRDT strategies with and without an ASP), MALDI-TOF analysis with an ASP, BC-GN with an ASP, PCR without an ASP, and PCR with an ASP were other cost-effective strategies. mRDT without an ASP and PNA-FISH without an ASP were not cost-effective. PNA-FISH was equivalent to the baseline strategy. BC-GP with an ASP and conventional laboratory methods with an ASP were not included in the graph, as they were of suboptimal effectiveness.
In the sensitivity analysis, no thresholds were identified, suggesting that our findings are robust for this strategy. In the probabilistic analysis, the mean cost was $28,358.74 (95% CI, $28,209.9 to $28,507.58), and the mean number of QALYs was 12.39 (95% CI, 12.36 to 12.41). With respect to the cost-effectiveness planes, the strategy was in the cost-effective quadrant in 97.8% of simulations, and as can be seen from cost-effectiveness acceptability curves (Fig. 2), it was cost-effective in 98.6% to 99.9% of simulations, for a cost-effectiveness threshold ranging between $0 and $100,000.
PNA-FISH with an ASP.
The mean cost for PNA-FISH with an ASP, which included the cost of blood culture (65), the cost of an ASP (31), the cost of PNA-FISH (70), and the cost of hospitalization (22, 32), was $53,226.09 (see the appendix), while the mean effectiveness was 0.85 (20–22, 36), and the mean number of QALYs was 11.45. As PNA-FISH with an ASP had the same effectiveness as the baseline strategy (probability of survival of 0.85 [20–22, 36] versus 0.85 [24, 38, 43]) and a similar cost ($53,226.09 [22, 31, 32, 65, 70] versus $55,932.02 [30, 32, 38, 55, 65]), it resulted in no net savings or extra costs per death averted.
In the one-way sensitivity analysis, PNA-FISH with an ASP becomes cost-effective when the probability of mortality for the baseline strategy is >0.15 or when the probability of mortality for PNA-FISH with an ASP is <0.15. This finding, along with the large range of the probabilities of mortality for PNA-FISH with an ASP, suggests that small variations in mortality would be decisive in determining whether this strategy is cost-effective or not. In the probabilistic analysis, the mean cost for PNA-FISH with an ASP was $52,987.4 (95% CI, $52,692.31 to $53,282.49), while the mean number of QALYs was 11.03 (95% CI, 11 to 11.07).
BC-GP with an ASP.
BC-GP with an ASP was associated with an average cost of $24,904.91 when accounting for the cost of blood culture (65), the cost of an ASP (31), the cost of BC-GP (69), and the cost of hospitalization (19, 32, 40), which accounted for inpatient costs only. More specifically, the use of BC-GP with an ASP was associated with lower costs than with the baseline strategy ($24,904.91 [19, 31, 40, 65, 69] versus $55,932.02 [30, 32, 38, 55, 65]) (see the appendix) but at the expense of decreased effectiveness (probability of survival of 0.84 [19, 40, 45] versus 0.85 [24, 38, 43]; 11.31 QALYs). This decreased effectiveness could be due to the fact that Gram-positive bacteria are associated with a higher percentage of sepsis hospital admissions (81) and a higher rate of mortality than Gram-negative bacteria (82). BC-GP with an ASP was not a cost-effective alternative compared to the baseline strategy, as its use resulted in additional costs per death averted.
In the sensitivity analysis, BC-GP with an ASP becomes cost-effective when the probability of mortality for the baseline strategy is above 0.16 or when the probability of mortality for BC-GP with an ASP is below 0.15. These values are close to the base-case estimates, suggesting that even small variations in mortality could alter the cost-effectiveness findings. In the probabilistic analysis, after running 10,000 Monte Carlo simulations, the mean cost for BC-GP with an ASP was $24,934.62 (95% CI, $24,812.99 to $25,056.25), while the mean number of QALYs was 10.94 (95% CI, 10.91 to 10.97).
BC-GN with an ASP.
The calculated cost for BC-GN, which accounted for the cost of blood culture (65), the cost of an ASP (31), the cost of BC-GN (42), and the cost of hospitalization (32, 34), was $33,691.47 (see the appendix), and the respective survival probability was 0.92 (34, 41, 42). The estimated number of QALYs was 12.39. The calculated ICER value suggested that BC-GN with an ASP results in savings of $23,587 per QALY gained (ICER, −$23,587/QALY gained) (see the appendix), suggesting that BC-GN with an ASP is a cost-effective strategy. This finding is probably explained by the fact that BC-GN reduces the time needed for the detection of bacteremia due to Gram-negative bacteria, which is extremely costly with conventional methods (42, 83).
In the sensitivity analysis, no thresholds were identified, indicating that our findings on the cost-effectiveness of BC-GN with an ASP are robust within the ranges specified in Table 2. In the probabilistic analysis, the mean cost for BC-GN with an ASP was $33,609.38 (95% CI, $33,436.75 to $33,782.02), while the mean number of QALYs was 12.4 (95% CI, 12.38 to 12.43). With respect to the cost-effectiveness planes, the strategy was in the cost-effective quadrant in 95.1% of simulations, and as can be seen from the cost-effectiveness acceptability curves (Fig. 2), it was cost-effective in 95.7% to 99.9% of simulations, for a cost-effectiveness threshold ranging between $0 and $100,000.
VALUE OF ASPs
The usefulness of mRDT without an ASP has been questioned, as significant reductions in mortality have been observed only when mRDT is used in combination with an ASP (11). Indeed, one of the main findings of our analysis was that mRDT with an ASP has a much higher probability of being cost-effective and dominant than mRDT without an ASP (80.0% versus 41.1%). This finding highlights the superiority of using mRDT with an ASP as a strategy for the diagnosis of bloodstream infection. It underlines how that the addition of an ASP to mRDT results in not only improved patient outcomes but also considerable health care savings.
Interestingly, among the individual strategies, PCR was the only one that resulted in savings that were similar irrespective of whether it was combined with an ASP or not, potentially due to the lack of uniformity between the different PCR assays and kits used (9, 18, 25, 38, 43, 44) and the de facto linkage of “homemade” PCR assays with clinical teams.
The findings on the cost-effectiveness of ASPs are in line with data from previous studies that associated the implementation of ASPs with both improved outcomes and significant cost reductions (84). More precisely, the combination of mRDT with an active ASP improves outcomes, as the expert guidance offered by specialists, combined with the earlier availability of results, allows the early administration of effective treatment (28). Moreover, as the number of new diagnostic methods increases, it becomes increasingly complex for laboratories to determine the most appropriate mRDT for each patient (12, 85).
INFLUENCE OF MODEL VARIABLES
The results of the probabilistic analysis were in agreement with the results of the base-case analysis (summarized in Fig. 3 and 4 and Table 3), underlining the consistency of our findings. In addition, single-center studies have reported reductions in costs associated with the use of specific mRDTs, which are consistent with the estimates that we obtained (9, 18, 19, 21, 22, 25, 45). For instance, we estimated that the use of MALDI-TOF analysis with an ASP is associated with an average cost of $28,394.21 per patient with suspected bacteremia tested. This is similar to the mean cost value estimated by Perez et al., who reported that the use of MALDI-TOF analysis decreased hospital costs from $45,709 to $26,162 per patient (17). Likewise, in our study, the estimated mean cost for the use of PCR with an ASP was $47,917.58, which is in agreement with the cost of $48,350 per patient reported by Bauer et al. in a study that compared rapid PCR (rPCR) for methicillin-resistant Staphylococcus aureus (MRSA) with conventional blood culture (18).
TABLE 3.
Base-case analysis results for competing strategies
| Strategy | Base-case estimate |
||||
|---|---|---|---|---|---|
| Cost ($) | Effect |
ICER |
|||
| Probability of survival | QALY value | $/death averted | $/QALY gained | ||
| mRDT (with and without ASP) | 36,301.50 | 0.89 | 11.9883 | −490,763 | −36,434 |
| mRDT with ASP (A) | 31,274.24 | 0.89 | 11.9883 | −616,445 | −45,764 |
| mRDT without ASP (B) | 57,220.14 | 0.90 | 12.1230 | 25,762 | 1,913 |
| PCR with ASP (A1) | 47,917.58 | 0.88 | 11.8536 | −267,148 | −19,833 |
| MALDI-TOF analysis with ASP (A2) | 28,394.21 | 0.92 | 12.3924 | −393,397 | −29,205 |
| PNA-FISH with ASP (A3) | 53,226.09 | 0.85 | 11.4495 | 0 | 0 |
| BC-GP with ASP (A4) | 24,904.91 | 0.84 | 11.3148 | Dominated | Dominated |
| BC-GN with ASP (A5) | 33,691.47 | 0.92 | 12.3924 | −317,722 | −23,587 |
| PCR without ASP (B1) | 47,430.21 | 0.88 | 11.8536 | −283,394 | −21,039 |
| PNA-FISH without ASP (B2) | 58,284.16 | 0.92 | 12.3924 | 33,602 | 2,495 |
| Conventional laboratory methods with ASP (C) | 41,723.98 | 0.84 | 11.3148 | Dominated | Dominated |
| Conventional laboratory methods without ASP (D) | 55,932.02 | 0.85 | 11.4495 | Baseline | Baseline |
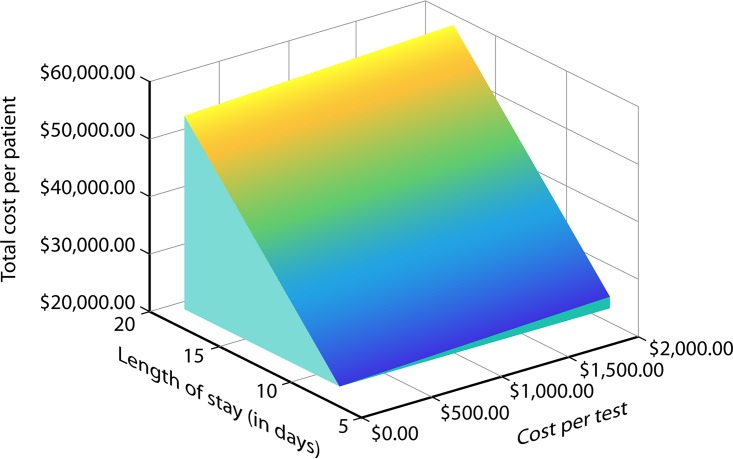
Nevertheless, the analysis suggests that the total cost of hospitalization (see the appendix) was the decisive factor in determining the cost-effectiveness outcomes. Tornado diagrams (see Fig. S2 in the supplemental material), which are a graphical depiction of how variations in each model variable affect the cost-effectiveness output, show that among the parameters that influence cost, the cost of hospitalization per day and the length of stay are the most influential variables in determining the cost-effectiveness results. This is even more evident in Fig. 5, which is a three-dimensional plane surface that shows how the total cost per patient varies as a function of the length of stay and the cost per test. Figure 5 shows that small decreases in the length of hospital stay result in large decreases in the total cost per patient. As such, a hypothetical new diagnostic test that considerably decreases the length of stay would be cost-effective even if the cost for the diagnostic test itself was high.
FIG 5.
Three-dimensional plane showing the relationship between length of stay (x axis) (ranging from 6 to 18 days), total cost per patient (y axis) (ranging from $22,322.04 to $58,167.84), and cost per test per patient (z axis) (ranging from $43 to $2,000).
LIMITATIONS
The present cost-effectiveness analysis is a comprehensive synthesis of data from the relevant literature, but certain limitations should nonetheless be considered when evaluating these findings. First, our findings are more representative of the clinical and epidemiologic reality of a U.S. hospital. Cost values may differ from the ones that we have used due to interstate and interinstitutional differences. Also, to obtain estimates for mortality and length of stay for the competing strategies, we pooled data from available studies. As such, MALDI-TOF analysis without an ASP and BC-GN without an ASP were not included in our study due to a lack of pertinent data, and the results for other strategies with limited clinical information, such as PCR without an ASP and PNA-FISH without an ASP, should be interpreted with caution. Our data have not been stratified by age and other comorbidities. In addition, our model includes outcomes only for 30 days after the blood cultures were ordered, and we have not accounted for readmission costs or the costs associated with antibiotic discontinuation, as such data were not available. Importantly, we did not account for the initial cost of acquiring the mRDT devices, as hospital pricing may reflect acquisition costs. Despite these limitations, our results were robust in the probabilistic and sensitivity analyses that aimed to address uncertainty regarding our assumptions. Of note is that even though the available data do not allow us to account for antibiotic-related adverse events, they are partly reflected in the length-of-stay estimates, and fully accounting for them would result in even further savings. Finally, it should be noted that we did not perform an analysis from the perspective of the society, due to the lack of relevant data. As such, our findings are of interest primarily to the health care industry, which is the principal driver of investment in diagnostic technologies in the United States. While the combination of improved survival and reduced hospitalization costs in a primarily working-age population (the average age of the study population was 58 years, compared to an average retirement age of 66 years in the United States for 2016 [86]) might be expected to yield wider benefits for society as well, prospective, long-term follow-up studies of these patients are needed to confirm these hypotheses and assess the potential for “hidden” societal costs (e.g., reduced hospitalization may be associated with a need for more unpaid care by family members) that could not be considered in the present study. When these data become available, revisiting our results from a societal perspective will likely prove worthwhile.
CONCLUSIONS
In recent years, health care improvement efforts in the United States have focused on achieving high-quality value-based care (87, 88), which has been closely tied with efforts to widely establish the use of value-based payments (16). In turn, cost-effectiveness studies, which integrate information about health outcomes and health care expenditures, are tools commonly employed for assessing the value of clinical interventions (26, 89) and can help inform value-based clinical decision-making (90, 91).
mRDTs that provide quick results, allowing the timely administration of appropriate therapy (9) and reducing mortality and the length of hospital stay (5–7), could help achieve economic efficiency in the clinical setting. Our study, which aimed to evaluate the cost-effectivenesses of competing strategies for the diagnosis of bloodstream infections among patients with suspected bacteremia, found that the use of mRDTs was a cost-effective strategy that was associated with high therapeutic effectiveness and health care cost savings.
In cost-effectiveness planes, mRDT had an 85.1% chance of being cost-effective compared to conventional laboratory methods without an ASP. Importantly, the use of mRDTs constitutes a cost-effective alternative to conventional laboratory methods for the diagnosis of bloodstream infections, whether it is used in combination with an ASP or not. The probabilities of cost-effectiveness in cost-effectiveness planes were 80.0% for mRDT with an ASP and 41.1% for mRDT without an ASP. Interestingly, even though mRDT-based strategies appear to be less cost-effective in the absence of an ASP, they still remain more cost-effective than conventional laboratory methods without an ASP. It should be noted that in our study, we did not distinguish between various ASP strategies, such as prospective audit and feedback, real-time decision support, or electronic decision support, due to a paucity of data on such outcomes (92, 93). Future cost-effectiveness modeling studies could distinguish between different types of ASPs, with the aim of determining whether a particular ASP strategy is more cost-effective than others.
Our model structure allowed us to combine strategies in one decision tree, thus facilitating comparisons between strategies. More precisely, under baseline assumptions, the most cost-effective strategy was MALDI-TOF analysis with an ASP (ICER, −$29,205/QALY gained), while BC-GN with an ASP (ICER, −$23,587/QALY gained), PCR with an ASP (ICER, −$19,833/QALY gained), and PCR without an ASP (ICER, −$21,039/QALY gained) were other cost-effective options. Nonetheless, as noted above, the results of these comparisons should be interpreted with caution due to limited data.
In the future, cost-effectiveness studies incorporating more strategy-specific data, as they become available, could help us distinguish whether there are differences within each of the 7 mRDT subcategories that we have examined. For example, our data on the effectiveness of MALDI-TOF analysis were derived from both studies that performed MALDI-TOF analysis directly on broth from positive blood culture bottles (17, 23) and studies that performed MALDI-TOF analysis on the growth found on a solid culture medium plate following inoculation from a positive blood culture bottle (37, 39). It would be interesting to examine whether this difference would affect cost-effectiveness outcomes. In addition, it should be noted that even though we evaluated mRDTs as separate entities in our study, microbiology laboratories often employ combinations of the individual mRDTs based on specific circumstances. Nevertheless, our results on the use of individual strategies, such as the finding that BC-GN with an ASP results in considerable health care savings, could assist laboratories in identifying situations in which processing of samples could be more cost-effective.
In summary, our results on the cost-effectivenesses of both the combined and the individual testing strategies provide data that can be used to inform cost-efficient clinical decision-making. Given that decisions about the appropriateness of a test should be based on both the test's performance for various targets and economic considerations (12), our findings could be used to better inform the selection of diagnostic methods. The ASP team can play a critical role in this respect and may be instrumental in increasing diagnostic accuracy and treatment efficacy while reducing costs. Specifically, the ASP team is particularly well placed to ensure that diagnostic tests are tailored to the clinical problem at hand, mRDT results are interpreted correctly, and antimicrobial agents are prescribed appropriately, thus limiting the use of unnecessary empirical therapy (12).
APPENDIX
The calculations that were performed in the base-case analysis, in order to obtain the average cost and ICER associated with the use of each strategy, are as follows.
Conventional Laboratory Methods without an ASP (Baseline)
Total cost = [3 × cost of blood culture (65)] + cost of conventional laboratory methods (30) + [cost of hospitalization per day (32) × length of stay for conventional laboratory methods without ASP (38, 55)].
Conventional Laboratory Methods with an ASP
Total cost = [3 × cost of blood culture (65)] + cost of ASP (31) + cost of conventional laboratory methods (30) + [cost of hospitalization per day (32) × length of stay for conventional laboratory methods with ASP (17–19, 22, 23, 34, 37, 40)].
ICER = (total cost of conventional laboratory methods with ASP − total cost of baseline)/(QALY value for conventional laboratory methods with ASP − QALY value for baseline strategy).
mRDT
Total cost = [3 × cost of blood culture (65)] + cost of mRDT (42, 68–71) + [cost of hospitalization per day (32) × length of stay for mRDT (17–19, 22, 23, 34, 37, 38, 40, 55)].
ICER = (total cost of mRDT − total cost of baseline strategy)/(QALY value with mRDT − QALY value for baseline strategy).
mRDT without an ASP
Total cost = [3 × cost of blood culture (65)] + cost of mRDT (42, 68–71) + [cost of hospitalization per day (32) × length of stay for mRDT without ASP (38, 55)].
ICER = (total cost of mRDT without ASP − total cost of baseline strategy)/(number of QALYs for mRDT without ASP − QALY value for baseline strategy).
PCR without an ASP
Total cost = [3 × cost of blood culture (65)] + cost of PCR (68) + [cost of hospitalization per day (32) × length of stay for PCR without ASP (38)].
ICER = (total cost of PCR without ASP − total cost of baseline strategy)/(QALY value for PCR without ASP − QALY value for baseline strategy).
PNA-FISH without an ASP
Total cost = [3 × cost of blood culture (65)] + cost of PNA-FISH (70) + [cost of hospitalization per day (32) × length of stay for PNA-FISH without ASP (55)].
ICER = (total cost of PNA-FISH without ASP − total cost of baseline strategy)/(number of QALYs for PNA-FISH without ASP − QALY value for baseline strategy).
mRDT with an ASP
Total cost = [3 × cost of blood culture (65)] + cost of ASP (31) + cost of mRDT (42, 68–71) + [cost of hospitalization per day (32) × length of stay for mRDT with ASP (17–19, 22, 23, 34, 37, 40)].
ICER = (total cost of mRDT with ASP − total cost of baseline strategy)/(QALY value for mRDT with ASP − QALY value for baseline strategy).
PCR with an ASP
Total cost = [3 × cost of blood culture (65)] + cost of ASP (31) + cost of PCR (68) + [cost of hospitalization per day (32) × length of stay for PCR with ASP (18)].
ICER = (total cost of PCR with ASP − total cost of baseline strategy)/(QALY value for PCR with ASP − QALY value for baseline strategy).
MALDI-TOF Analysis with an ASP
Total cost = [3 × cost of blood culture (65)] + cost of ASP (31) + cost of MALDI-TOF analysis (71) + [cost of hospitalization per day (32) × length of stay for MALDI-TOF analysis with ASP (23, 28, 37)].
ICER = (total cost of MALDI-TOF analysis with ASP − total cost of baseline strategy)/(QALY value for MALDI-TOF analysis with ASP − QALY value for baseline strategy).
PNA-FISH with an ASP
Total cost = [3 × cost of blood culture (65)] + cost of ASP (31) + cost of PNA-FISH (70) + [cost of hospitalization per day (32) × length of stay for PNA-FISH with ASP (22)].
ICER = (total cost of PNA-FISH with ASP − total cost of baseline strategy)/(QALY value for PNA-FISH with ASP − QALY value for baseline strategy).
BC-GP with an ASP
Total cost = [3 × cost of blood culture (65)] + cost of ASP (31) + cost of BC-GP (69) + [cost of hospitalization per day (32) × length of stay for BC-GP with ASP (19, 40)].
ICER = (total cost of BC-GP with ASP − total cost of baseline strategy)/(QALY value for BC-GP with ASP − QALY value for baseline strategy).
BC-GN with an ASP
Total cost = [3 × cost of blood culture (65)] + cost of ASP (31) + cost of BC-GN (42) + [cost of hospitalization per day (32) × length of stay for BC-GN with ASP (34)].
ICER = (total cost of BC-GP with ASP − total cost of baseline strategy)/(QALY value for BC-GP with ASP − QALY value for baseline strategy).
Supplementary Material
ACKNOWLEDGMENTS
E.M. has received grant support from T2 Biosystems and Astellas Pharma. All other authors report no potential conflicts.
Biographies

Elina Eleftheria Pliakos received an Sc.B. in Biology with Honors from Brown University and completed a second concentration in Science and Society. As a student at Brown, she received an Undergraduate Research and Teaching award and was elected to be a member of Brown's Chapter of Sigma Xi, The Scientific Research Society. She is currently a Clinical Research Assistant for the Division of Infectious Diseases at Rhode Island Hospital. Her research interests include health economics, outcomes research, and epidemiology.

Nikolaos Andreatos, M.D., completed his medical studies at the University of Athens, Greece, graduating in 2015. After completing a clinical research fellowship at Johns Hopkins University, Dr. Andreatos moved to Providence, RI, to take up a position as a study coordinator at the Warren Alpert Medical School of Brown University. In this capacity, he has been involved in the design and conduct of 2 clinical trials, in addition to clinical and outcomes research. His research currently focuses on the epidemiology and treatment of infections among vulnerable patient populations, such as cancer patients, as well as on antimicrobial stewardship and public health. His published work includes meta-analyses, county-level analyses, and retrospective-cohort studies. He has authored 40 peer-reviewed publications in the fields of hematology/oncology, infectious disease, and epidemiology.

Fadi Shehadeh received his M.Eng. in Electrical and Computer Engineering from the National Technical University of Athens. His master's thesis focused on machine learning and spatial statistics on spatiotemporal big data. He is currently a Senior Research Assistant at Brown University and Rhode Island Hospital, working in the Division of Infectious Diseases. His research interests include the use of artificial intelligence, big data, and real-time Internet data mining for infectious disease risk assessment and outbreak prediction. He is also interested in the development of new diagnostic techniques and disease surveillance systems.

Panayiotis D. Ziakas, M.D., completed his medical residency training and Hematology training at the University of Athens, Greece, in 2006. He also received a master's degree in Biostatistics at Athens Medical School and Department of Mathematics, University of Athens, in 2006. In 2008, he received his Ph.D. at Athens Medical School and worked as a Postdoctoral Clinical and Research Fellow at Athens Medical School until 2013. He joined the Alpert Medical School of Brown University and the Infectious Disease Division in 2013 as a research associate to pursue his research. His research focuses on nosocomial infections and the appropriate use of antimicrobial and antifungal agents among high risk-populations, namely, immunocompromised individuals and patients with hematologic malignancy. His recent work also includes meta-analyses, decision analyses, as well as genetic association studies. He has authored more than 90 peer-reviewed publications in the fields of infections, hematology/oncology, autoimmunity, and epidemiology.

Eleftherios Mylonakis completed medical training at the University of Athens and completed his medical residency training at Brown University and his Infectious Disease Fellowship at Massachusetts General Hospital. He is the Charles C. J. Carpenter Professor of Infectious Disease and Director Chief of the Infectious Diseases at the Warren Alpert Medical School of Brown University. He has authored over 300 peer-reviewed publications and has received a number of awards, and his research interests include microbial pathogenesis and immune responses in invertebrate model hosts. He is also actively involved in clinical research on fungal diagnostics and treatment, and his research is supported by the NIH and private foundations.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/CMR.00095-17.
REFERENCES
- 1.Goto M, Al-Hasan MN. 2013. Overall burden of bloodstream infection and nosocomial bloodstream infection in North America and Europe. Clin Microbiol Infect 19:501–509. doi: 10.1111/1469-0691.12195. [DOI] [PubMed] [Google Scholar]
- 2.Kaye KS, Marchaim D, Chen TY, Baures T, Anderson DJ, Choi Y, Sloane R, Schmader KE. 2014. Effect of nosocomial bloodstream infections on mortality, length of stay, and hospital costs in older adults. J Am Geriatr Soc 62:306–311. doi: 10.1111/jgs.12634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kilgore M, Brossette S. 2008. Cost of bloodstream infections. Am J Infect Control 36:S172.e1–S172.e3. doi: 10.1016/j.ajic.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 4.Wisplinghoff H, Bischoff T, Tallent SM, Seifert H, Wenzel RP, Edmond MB. 2004. Nosocomial bloodstream infections in US hospitals: analysis of 24,179 cases from a prospective nationwide surveillance study. Clin Infect Dis 39:309–317. doi: 10.1086/421946. [DOI] [PubMed] [Google Scholar]
- 5.Leibovici L, Shraga I, Drucker M, Konigsberger H, Samra Z, Pitlik SD. 1998. The benefit of appropriate empirical antibiotic treatment in patients with bloodstream infection. J Intern Med 244:379–386. doi: 10.1046/j.1365-2796.1998.00379.x. [DOI] [PubMed] [Google Scholar]
- 6.Lodise TP, McKinnon PS, Swiderski L, Rybak MJ. 2003. Outcomes analysis of delayed antibiotic treatment for hospital-acquired Staphylococcus aureus bacteremia. Clin Infect Dis 36:1418–1423. doi: 10.1086/375057. [DOI] [PubMed] [Google Scholar]
- 7.Fraser A, Paul M, Almanasreh N, Tacconelli E, Frank U, Cauda R, Borok S, Cohen M, Andreassen S, Nielsen AD, Leibovici L, TREAT Study Group. 2006. Benefit of appropriate empirical antibiotic treatment: thirty-day mortality and duration of hospital stay. Am J Med 119:970–976. doi: 10.1016/j.amjmed.2006.03.034. [DOI] [PubMed] [Google Scholar]
- 8.Kollef M. 2003. Appropriate empirical antibacterial therapy for nosocomial infections: getting it right the first time. Drugs 63:2157–2168. doi: 10.2165/00003495-200363200-00001. [DOI] [PubMed] [Google Scholar]
- 9.Pardo J, Klinker KP, Borgert SJ, Butler BM, Giglio PG, Rand KH. 2016. Clinical and economic impact of antimicrobial stewardship interventions with the FilmArray blood culture identification panel. Diagn Microbiol Infect Dis 84:159–164. doi: 10.1016/j.diagmicrobio.2015.10.023. [DOI] [PubMed] [Google Scholar]
- 10.Buehler SS, Madison B, Snyder SR, Derzon JH, Cornish NE, Saubolle MA, Weissfeld AS, Weinstein MP, Liebow EB, Wolk DM. 2016. Effectiveness of practices to increase timeliness of providing targeted therapy for inpatients with bloodstream infections: a laboratory medicine best practices systematic review and meta-analysis. Clin Microbiol Rev 29:59–103. doi: 10.1128/CMR.00053-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Timbrook TT, Morton JB, McConeghy KW, Caffrey AR, Mylonakis E, LaPlante KL. 2017. The effect of molecular rapid diagnostic testing on clinical outcomes in bloodstream infections: a systematic review and meta-analysis. Clin Infect Dis 64:15–23. doi: 10.1093/cid/ciw649. [DOI] [PubMed] [Google Scholar]
- 12.Messacar K, Parker SK, Todd JK, Dominguez SR. 2017. Implementation of rapid molecular infectious disease diagnostics: the role of diagnostic and antimicrobial stewardship. J Clin Microbiol 55:715–723. doi: 10.1128/JCM.02264-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barlam TF, Cosgrove SE, Abbo LM, MacDougall C, Schuetz AN, Septimus EJ, Srinivasan A, Dellit TH, Falck-Ytter YT, Fishman NO, Hamilton CW, Jenkins TC, Lipsett PA, Malani PN, May LS, Moran GJ, Neuhauser MM, Newland JG, Ohl CA, Samore MH, Seo SK, Trivedi KK. 2016. Implementing an antibiotic stewardship program: guidelines by the Infectious Diseases Society of America and the Society for Healthcare Epidemiology of America. Clin Infect Dis 62:e51–e77. doi: 10.1093/cid/ciw118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Caliendo AM, Gilbert DN, Ginocchio CC, Hanson KE, May L, Quinn TC, Tenover FC, Alland D, Blaschke AJ, Bonomo RA, Carroll KC, Ferraro MJ, Hirschhorn LR, Joseph WP, Karchmer T, MacIntyre AT, Reller LB, Jackson AF, Infectious Diseases Society of America. 2013. Better tests, better care: improved diagnostics for infectious diseases. Clin Infect Dis 57(Suppl 3):S139–S170. doi: 10.1093/cid/cit578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McElvania TeKippe E. 2017. The added cost of rapid diagnostic testing and active antimicrobial stewardship: is it worth it? J Clin Microbiol 55:20–23. doi: 10.1128/JCM.02061-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Burwell SM. 2015. Setting value-based payment goals—HHS efforts to improve U.S. health care. N Engl J Med 372:897–899. doi: 10.1056/NEJMp1500445. [DOI] [PubMed] [Google Scholar]
- 17.Perez KK, Olsen RJ, Musick WL, Cernoch PL, Davis JR, Land GA, Peterson LE, Musser JM. 2013. Integrating rapid pathogen identification and antimicrobial stewardship significantly decreases hospital costs. Arch Pathol Lab Med 137:1247–1254. doi: 10.5858/arpa.2012-0651-OA. [DOI] [PubMed] [Google Scholar]
- 18.Bauer KA, West JE, Balada-Llasat JM, Pancholi P, Stevenson KB, Goff DA. 2010. An antimicrobial stewardship program's impact with rapid polymerase chain reaction methicillin-resistant Staphylococcus aureus/S. aureus blood culture test in patients with S. aureus bacteremia. Clin Infect Dis 51:1074–1080. doi: 10.1086/656623. [DOI] [PubMed] [Google Scholar]
- 19.Box MJ, Sullivan EL, Ortwine KN, Parmenter MA, Quigley MM, Aguilar-Higgins LM, MacIntosh CL, Goerke KF, Lim RA. 2015. Outcomes of rapid identification for gram-positive bacteremia in combination with antibiotic stewardship at a community-based hospital system. Pharmacotherapy 35:269–276. doi: 10.1002/phar.1557. [DOI] [PubMed] [Google Scholar]
- 20.Forrest GN, Mankes K, Jabra-Rizk MA, Weekes E, Johnson JK, Lincalis DP, Venezia RA. 2006. Peptide nucleic acid fluorescence in situ hybridization-based identification of Candida albicans and its impact on mortality and antifungal therapy costs. J Clin Microbiol 44:3381–3383. doi: 10.1128/JCM.00751-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Forrest GN, Mehta S, Weekes E, Lincalis DP, Johnson JK, Venezia RA. 2006. Impact of rapid in situ hybridization testing on coagulase-negative staphylococci positive blood cultures. J Antimicrob Chemother 58:154–158. doi: 10.1093/jac/dkl146. [DOI] [PubMed] [Google Scholar]
- 22.Heil EL, Daniels LM, Long DM, Rodino KG, Weber DJ, Miller MB. 2012. Impact of a rapid peptide nucleic acid fluorescence in situ hybridization assay on treatment of Candida infections. Am J Health Syst Pharm 69:1910–1914. doi: 10.2146/ajhp110604. [DOI] [PubMed] [Google Scholar]
- 23.Lockwood AM, Perez KK, Musick WL, Ikwuagwu JO, Attia E, Fasoranti OO, Cernoch PL, Olsen RJ, Musser JM. 2016. Integrating rapid diagnostics and antimicrobial stewardship in two community hospitals improved process measures and antibiotic adjustment time. Infect Control Hosp Epidemiol 37:425–432. doi: 10.1017/ice.2015.313. [DOI] [PubMed] [Google Scholar]
- 24.Ly T, Gulia J, Pyrgos V, Waga M, Shoham S. 2008. Impact upon clinical outcomes of translation of PNA FISH-generated laboratory data from the clinical microbiology bench to bedside in real time. Ther Clin Risk Manag 4:637–640. doi: 10.2147/TCRM.S2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.MacVane SH, Hurst JM, Boger MS, Gnann JW Jr. 2016. Impact of a rapid multiplex polymerase chain reaction blood culture identification technology on outcomes in patients with vancomycin-resistant enterococcal bacteremia. Infect Dis (Lond) 48:732–737. doi: 10.1080/23744235.2016.1185533. [DOI] [PubMed] [Google Scholar]
- 26.Neumann PJ, Sanders GD. 2017. Cost-effectiveness analysis 2.0. N Engl J Med 376:203–205. doi: 10.1056/NEJMp1612619. [DOI] [PubMed] [Google Scholar]
- 27.Kothari A, Morgan M, Haake DA. 2014. Emerging technologies for rapid identification of bloodstream pathogens. Clin Infect Dis 59:272–278. doi: 10.1093/cid/ciu292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bauer KA, Perez KK, Forrest GN, Goff DA. 2014. Review of rapid diagnostic tests used by antimicrobial stewardship programs. Clin Infect Dis 59(Suppl 3):S134–S145. doi: 10.1093/cid/ciu547. [DOI] [PubMed] [Google Scholar]
- 29.Afshari A, Schrenzel J, Ieven M, Harbarth S. 2012. Bench-to-bedside review: rapid molecular diagnostics for bloodstream infection—a new frontier? Crit Care 16:222. doi: 10.1186/cc11202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tan KE, Ellis BC, Lee R, Stamper PD, Zhang SX, Carroll KC. 2012. Prospective evaluation of a matrix-assisted laser desorption ionization–time of flight mass spectrometry system in a hospital clinical microbiology laboratory for identification of bacteria and yeasts: a bench-by-bench study for assessing the impact on time to identification and cost-effectiveness. J Clin Microbiol 50:3301–3308. doi: 10.1128/JCM.01405-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Scheetz MH, Bolon MK, Postelnick M, Noskin GA, Lee TA. 2009. Cost-effectiveness analysis of an antimicrobial stewardship team on bloodstream infections: a probabilistic analysis. J Antimicrob Chemother 63:816–825. doi: 10.1093/jac/dkp004. [DOI] [PubMed] [Google Scholar]
- 32.Kaiser Family Foundation. 2015. Hospital adjusted expenses per inpatient day. Data source: 1999-2015 AHA annual survey. Kaiser Family Foundation, Washington, DC: http://kff.org/health-costs/state-indicator/expenses-per-inpatient-day/?currentTimeframe=0&sortModel=%7B%22colId%22:%22Location%22,%22sort%22:%22asc%22%7D Accessed 3 May 2017. [Google Scholar]
- 33.Carroll KC, Weinstein MP. 2007. Manual and automated systems for detection and identification of microorganisms, p 192–217. In Murray PR, Baron EJ, Jorgensen JH, Landry ML, Pfaller MA (ed), Manual of clinical microbiology, 9th ed, vol 1 ASM Press, Washington, DC. [Google Scholar]
- 34.Bias T, Jain A, Beil E, Ruffner R, Borodin V, Bruno C, Emery C. 2015. Use of the Nanosphere Verigene gram-negative blood culture (BC-GN) system for more rapid bacterial identification and antimicrobial optimization in patients with gram-negative rod bacteraemia, abstr EP033. Abstr 25th Eur Congr Clin Microbiol Infect Dis, Copenhagen, Denmark. [Google Scholar]
- 35.Felsenstein S, Bender JM, Sposto R, Gentry M, Takemoto C, Bard JD. 2016. Impact of a rapid blood culture assay for Gram-positive identification and detection of resistance markers in a pediatric hospital. Arch Pathol Lab Med 140:267–275. doi: 10.5858/arpa.2015-0119-OA. [DOI] [PubMed] [Google Scholar]
- 36.Forrest GN, Roghmann MC, Toombs LS, Johnson JK, Weekes E, Lincalis DP, Venezia RA. 2008. Peptide nucleic acid fluorescent in situ hybridization for hospital-acquired enterococcal bacteremia: delivering earlier effective antimicrobial therapy. Antimicrob Agents Chemother 52:3558–3563. doi: 10.1128/AAC.00283-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang AM, Newton D, Kunapuli A, Gandhi TN, Washer LL, Isip J, Collins CD, Nagel JL. 2013. Impact of rapid organism identification via matrix-assisted laser desorption/ionization time-of-flight combined with antimicrobial stewardship team intervention in adult patients with bacteremia and candidemia. Clin Infect Dis 57:1237–1245. doi: 10.1093/cid/cit498. [DOI] [PubMed] [Google Scholar]
- 38.Maslonka M, Freifeld AG, Rupp ME, Greer D, Peetz R, Lyden E, Fey PD, Van Schooneveld TC. 2014. Impact of rapid bloodstream pathogen identification in hospitalized patients, abstr P171. Abstr IDWeek 2014, Philadelphia, PA. [Google Scholar]
- 39.Nagel JL, Huang AM, Kunapuli A, Gandhi TN, Washer LL, Lassiter J, Patel T, Newton DW. 2014. Impact of antimicrobial stewardship intervention on coagulase-negative Staphylococcus blood cultures in conjunction with rapid diagnostic testing. J Clin Microbiol 52:2849–2854. doi: 10.1128/JCM.00682-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Revolinski S, Huang A, Peppard W, Ledeboer N. 2015. Outcomes utilizing rapid diagnostic technology coupled with pharmacist intervention in patients with gram-positive bloodstream infections, abstr P0429. Abstr 25th Eur Congr Clin Microbiol Infect Dis, Copenhagen, Denmark. [Google Scholar]
- 41.Sothoron C, Ferreira J, Guzman N, Aldridge P, McCarter YS, Jankowski CA. 2015. A stewardship approach to optimize antimicrobial therapy through use of a rapid microarray assay on blood cultures positive for Gram-negative bacteria. J Clin Microbiol 53:3627–3629. doi: 10.1128/JCM.02161-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Walker T, Dumadag S, Lee CJ, Lee SH, Bender JM, Cupo Abbott J, She RC. 2016. Clinical impact of laboratory implementation of Verigene BC-GN microarray-based assay for detection of Gram-negative bacteria in positive blood cultures. J Clin Microbiol 54:1789–1796. doi: 10.1128/JCM.00376-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Frye AM, Baker CA, Rustvold DL, Heath KA, Hunt J, Leggett JE, Oethinger M. 2012. Clinical impact of a real-time PCR assay for rapid identification of staphylococcal bacteremia. J Clin Microbiol 50:127–133. doi: 10.1128/JCM.06169-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Macvane SH, Nolte FS. 2015. Clinical impact of adding a rapid PCR-based blood culture identification panel (BCID) to an established antimicrobial stewardship intervention (ASI) program for patients with Gram-negative bacteremia (GNB). Abstr 55th Intersci Conf Antimicrob Agents Chemother, San Diego, CA. [Google Scholar]
- 45.Sango A, McCarter YS, Johnson D, Ferreira J, Guzman N, Jankowski CA. 2013. Stewardship approach for optimizing antimicrobial therapy through use of a rapid microarray assay on blood cultures positive for Enterococcus species. J Clin Microbiol 51:4008–4011. doi: 10.1128/JCM.01951-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Beuving J, Wolffs PF, Hansen WL, Stobberingh EE, Bruggeman CA, Kessels A, Verbon A. 2015. Impact of same-day antibiotic susceptibility testing on time to appropriate antibiotic treatment of patients with bacteraemia: a randomised controlled trial. Eur J Clin Microbiol Infect Dis 34:831–838. doi: 10.1007/s10096-014-2299-0. [DOI] [PubMed] [Google Scholar]
- 47.Cattoir V, Merabet L, Djibo N, Rioux C, Legrand P, Girou E, Lesprit P. 2011. Clinical impact of a real-time PCR assay for rapid identification of Staphylococcus aureus and determination of methicillin resistance from positive blood cultures. Clin Microbiol Infect 17:425–431. doi: 10.1111/j.1469-0691.2010.03233.x. [DOI] [PubMed] [Google Scholar]
- 48.Na SH, Kim CJ, Kim M, Park JS, Song KH, Choe PG, Park WB, Bang JH, Kim ES, Park SW, Park KU, Kim NJ, Oh MD, Kim HB. 2016. Impact of the multiplex polymerase chain reaction in culture-positive samples on appropriate antibiotic use in patients with staphylococcal bacteremia. Diagn Microbiol Infect Dis 84:353–357. doi: 10.1016/j.diagmicrobio.2015.12.007. [DOI] [PubMed] [Google Scholar]
- 49.Neuberger A, Oren I, Sprecher H. 2008. Clinical impact of a PCR assay for rapid identification of Klebsiella pneumoniae in blood cultures. J Clin Microbiol 46:377–379. doi: 10.1128/JCM.00568-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Suzuki H, Hitomi S, Yaguchi Y, Tamai K, Ueda A, Kamata K, Tokuda Y, Koganemaru H, Kurihara Y, Ishikawa H, Yanagisawa H, Yanagihara K. 2015. Prospective intervention study with a microarray-based, multiplexed, automated molecular diagnosis instrument (Verigene system) for the rapid diagnosis of bloodstream infections, and its impact on the clinical outcomes. J Infect Chemother 21:849–856. doi: 10.1016/j.jiac.2015.08.019. [DOI] [PubMed] [Google Scholar]
- 51.Wang B, Jessamine P, Desjardins M, Toye B, Ramotar K. 2013. Direct mecA polymerase chain reaction testing of blood culture bottles growing Gram-positive cocci and the clinical potential in optimizing antibiotic therapy for staphylococcal bacteremia. Diagn Microbiol Infect Dis 75:37–41. doi: 10.1016/j.diagmicrobio.2012.09.014. [DOI] [PubMed] [Google Scholar]
- 52.DerSimonian R, Laird N. 1986. Meta-analysis in clinical trials. Control Clin Trials 7:177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 53.DerSimonian R, Kacker R. 2007. Random-effects model for meta-analysis of clinical trials: an update. Contemp Clin Trials 28:105–114. doi: 10.1016/j.cct.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 54.Nyaga VN, Arbyn M, Aerts M. 2014. Metaprop: a Stata command to perform meta-analysis of binomial data. Arch Public Health 72:39. doi: 10.1186/2049-3258-72-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Holtzman C, Whitney D, Barlam T, Miller NS. 2011. Assessment of impact of peptide nucleic acid fluorescence in situ hybridization for rapid identification of coagulase-negative staphylococci in the absence of antimicrobial stewardship intervention. J Clin Microbiol 49:1581–1582. doi: 10.1128/JCM.02461-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Arias E, Heron M, Xu J. 2017. United States life tables, 2013. Natl Vital Stat Rep 66:1–64. [PubMed] [Google Scholar]
- 57.Szende A. 2014. Self-reported population health an international perspective based on EQ-5D. Springer, Dordrecht, Netherlands. [PubMed] [Google Scholar]
- 58.National Institute for Health and Care Excellence. 2013. NICE process and methods guides, guide to the methods of technology appraisal 2013. National Institute for Health and Care Excellence, London, United Kingdom. [Google Scholar]
- 59.Neumann PJ, Ganiats TG, Russell LB, Sanders GD, Siegel JE. 2016. Cost-effectiveness in health and medicine, 2nd ed Oxford University Press, Oxford, United Kingdom. [Google Scholar]
- 60.Karlsson S, Ruokonen E, Varpula T, Ala-Kokko TI, Pettila V, Finnsepsis Study Group. 2009. Long-term outcome and quality-adjusted life years after severe sepsis. Crit Care Med 37:1268–1274. doi: 10.1097/CCM.0b013e31819c13ac. [DOI] [PubMed] [Google Scholar]
- 61.Cuthbertson BH, Elders A, Hall S, Taylor J, MacLennan G, Mackirdy F, Mackenzie SJ, Scottish Critical Care Trials Group, Scottish Intensive Care Society Audit Group. 2013. Mortality and quality of life in the five years after severe sepsis. Crit Care 17:R70. doi: 10.1186/cc12616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Drabinski AWG, Formica C. 2001. Observational evaluation of health state utilities among a cohort of sepsis patients. Value Health 4:128–129. [Google Scholar]
- 63.Agency for Healthcare Research and Quality. 2016. Using appropriate price indices for analysis of health care expenditures or income across multiple years: personal health care expenditure price index. Agency for Healthcare Research and Quality, Rockville, MD: https://meps.ahrq.gov/about_meps/Price_Index.shtml Accessed 13 February 2018. [Google Scholar]
- 64.Bureau of Economic Analysis. 2017. National data: personal consumption expenditures (PCE). Bureau of Economic Analysis, Washington, DC: https://www.bea.gov/iTable/iTable.cfm?reqid=19&step=2#reqid=19&step=3&isuri=1&1921=survey&1903=67 Accessed 13 February 2018. [Google Scholar]
- 65.Shapiro NI, Wolfe RE, Wright SB, Moore R, Bates DW. 2008. Who needs a blood culture? A prospectively derived and validated prediction rule. J Emerg Med 35:255–264. doi: 10.1016/j.jemermed.2008.04.001. [DOI] [PubMed] [Google Scholar]
- 66.Lee A, Mirrett S, Reller LB, Weinstein MP. 2007. Detection of bloodstream infections in adults: how many blood cultures are needed? J Clin Microbiol 45:3546–3548. doi: 10.1128/JCM.01555-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.O'Hara CM. 2005. Manual and automated instrumentation for identification of Enterobacteriaceae and other aerobic gram-negative bacilli. Clin Microbiol Rev 18:147–162. doi: 10.1128/CMR.18.1.147-162.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Brown J, Paladino JA. 2010. Impact of rapid methicillin-resistant Staphylococcus aureus polymerase chain reaction testing on mortality and cost effectiveness in hospitalized patients with bacteraemia: a decision model. Pharmacoeconomics 28:567–575. doi: 10.2165/11533020-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 69.Wojewoda CM, Sercia L, Navas M, Tuohy M, Wilson D, Hall GS, Procop GW, Richter SS. 2013. Evaluation of the Verigene Gram-positive blood culture nucleic acid test for rapid detection of bacteria and resistance determinants. J Clin Microbiol 51:2072–2076. doi: 10.1128/JCM.00831-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Alexander BD, Ashley ED, Reller LB, Reed SD. 2006. Cost savings with implementation of PNA FISH testing for identification of Candida albicans in blood cultures. Diagn Microbiol Infect Dis 54:277–282. doi: 10.1016/j.diagmicrobio.2005.10.011. [DOI] [PubMed] [Google Scholar]
- 71.Patel TS, Kaakeh R, Nagel JL, Newton DW, Stevenson JG. 2017. Cost analysis of implementing matrix-assisted laser desorption ionization–time of flight mass spectrometry plus real-time antimicrobial stewardship intervention for bloodstream infections. J Clin Microbiol 55:60–67. doi: 10.1128/JCM.01452-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jain R, Grabner M, Onukwugha E. 2011. Sensitivity analysis in cost-effectiveness studies: from guidelines to practice. Pharmacoeconomics 29:297–314. doi: 10.2165/11584630-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 73.Briggs ACK, Sculpher M. 2006. Decision modelling for health economic evaluation. Oxford University Press, Oxford, United Kingdom. [Google Scholar]
- 74.Walker B, Powers-Fletcher MV, Schmidt RL, Hanson KE. 2016. Cost-effectiveness analysis of multiplex PCR with magnetic resonance detection versus empiric or blood culture-directed therapy for management of suspected candidemia. J Clin Microbiol 54:718–726. doi: 10.1128/JCM.02971-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ziakas PD, Zacharioudakis IM, Zervou FN, Mylonakis E. 2015. Methicillin-resistant Staphylococcus aureus prevention strategies in the ICU: a clinical decision analysis. Crit Care Med 43:382–393. doi: 10.1097/CCM.0000000000000711. [DOI] [PubMed] [Google Scholar]
- 76.Hozo SP, Djulbegovic B, Hozo I. 2005. Estimating the mean and variance from the median, range, and the size of a sample. BMC Med Res Methodol 5:13. doi: 10.1186/1471-2288-5-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Doubilet P, Begg CB, Weinstein MC, Braun P, McNeil BJ. 1985. Probabilistic sensitivity analysis using Monte Carlo simulation. A practical approach. Med Decis Making 5:157–177. doi: 10.1177/0272989X8500500205. [DOI] [PubMed] [Google Scholar]
- 78.Fenwick E, O'Brien BJ, Briggs A. 2004. Cost-effectiveness acceptability curves—facts, fallacies and frequently asked questions. Health Econ 13:405–415. doi: 10.1002/hec.903. [DOI] [PubMed] [Google Scholar]
- 79.Fenwick E, Byford S. 2005. A guide to cost-effectiveness acceptability curves. Br J Psychiatry 187:106–108. doi: 10.1192/bjp.187.2.106. [DOI] [PubMed] [Google Scholar]
- 80.Peleg AY, Tilahun Y, Fiandaca MJ, D'Agata EM, Venkataraman L, Moellering RC Jr, Eliopoulos GM. 2009. Utility of peptide nucleic acid fluorescence in situ hybridization for rapid detection of Acinetobacter spp. and Pseudomonas aeruginosa. J Clin Microbiol 47:830–832. doi: 10.1128/JCM.01724-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Martin GS, Mannino DM, Eaton S, Moss M. 2003. The epidemiology of sepsis in the United States from 1979 through 2000. N Engl J Med 348:1546–1554. doi: 10.1056/NEJMoa022139. [DOI] [PubMed] [Google Scholar]
- 82.Geerdes HF, Ziegler D, Lode H, Hund M, Loehr A, Fangmann W, Wagner J. 1992. Septicemia in 980 patients at a university hospital in Berlin: prospective studies during 4 selected years between 1979 and 1989. Clin Infect Dis 15:991–1002. doi: 10.1093/clind/15.6.991. [DOI] [PubMed] [Google Scholar]
- 83.Mauldin PD, Salgado CD, Hansen IS, Durup DT, Bosso JA. 2010. Attributable hospital cost and length of stay associated with health care-associated infections caused by antibiotic-resistant Gram-negative bacteria. Antimicrob Agents Chemother 54:109–115. doi: 10.1128/AAC.01041-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wenzler E, Wong JR, Goff DA, Jankowski CA, Bauer KA. 2016. Controversies in antimicrobial stewardship: focus on new rapid diagnostic technologies and antimicrobials. Antibiotics (Basel) 5:E6. doi: 10.3390/antibiotics5010006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Blaschke AJ, Hersh AL, Beekmann SE, Ince D, Polgreen PM, Hanson KE. 2015. Unmet diagnostic needs in infectious disease. Diagn Microbiol Infect Dis 81:57–59. doi: 10.1016/j.diagmicrobio.2014.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Organisation for Economic Co-operation and Development. 2016. Ageing and employment policies—statistics on average effective age of retirement. Organisation for Economic Co-operation and Development, Paris, France: http://www.oecd.org/els/emp/average-effective-age-of-retirement.htm. Accessed 3 March 2018. [Google Scholar]
- 87.Enthoven AC, Crosson FJ, Shortell SM. 2007. ‘Redefining health care’: medical homes or archipelagos to navigate? Health Aff (Millwood) 26:1366–1372. doi: 10.1377/hlthaff.26.5.1366. [DOI] [PubMed] [Google Scholar]
- 88.Porter ME. 2010. What is value in health care? N Engl J Med 363:2477–2481. doi: 10.1056/NEJMp1011024. [DOI] [PubMed] [Google Scholar]
- 89.Garrison LP Jr, Kamal-Bahl S, Towse A. 2017. Toward a broader concept of value: identifying and defining elements for an expanded cost-effectiveness analysis. Value Health 20:213–216. doi: 10.1016/j.jval.2016.12.005. [DOI] [PubMed] [Google Scholar]
- 90.Kang R, Goodney PP, Wong SL. 2016. Importance of cost-effectiveness and value in cancer care and healthcare policy. J Surg Oncol 114:275–280. doi: 10.1002/jso.24331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Stone PW, Cohen C, Pincus HA. 20 April 2017. Comparative and cost-effectiveness research: competencies, opportunities, and training for nurse scientists. Nurs Outlook doi: 10.1016/j.outlook.2017.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Messacar K, Hurst AL, Child J, Campbell K, Palmer C, Hamilton S, Dowell E, Robinson CC, Parker SK, Dominguez SR. 2017. Clinical impact and provider acceptability of real-time antimicrobial stewardship decision support for rapid diagnostics in children with positive blood culture results. J Pediatr Infect Dis Soc 6:267–274. doi: 10.1093/jpids/piw047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Banerjee R, Teng CB, Cunningham SA, Ihde SM, Steckelberg JM, Moriarty JP, Shah ND, Mandrekar JN, Patel R. 2015. Randomized trial of rapid multiplex polymerase chain reaction-based blood culture identification and susceptibility testing. Clin Infect Dis 61:1071–1080. doi: 10.1093/cid/civ447. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.