Klebsiella pneumoniae is an opportunistic pathogen, and its hypervirulent variants cause serious invasive community-acquired infections. A genomic view of K. pneumoniae NTUH-2044 for the carbohydrate phosphotransferase system (PTS) found a putative fructose PTS, namely, the Frw PTS gene cluster.
KEYWORDS: Klebsiella pneumoniae, CRP, PTS, virulence
ABSTRACT
Klebsiella pneumoniae is an opportunistic pathogen, and its hypervirulent variants cause serious invasive community-acquired infections. A genomic view of K. pneumoniae NTUH-2044 for the carbohydrate phosphotransferase system (PTS) found a putative fructose PTS, namely, the Frw PTS gene cluster. The deletion mutant and the complemented mutant of frwC (KP1_1992), which encodes the putative fructose-specific enzyme IIC, were constructed, and the phenotypes were characterized. This transmembrane PTS protein is responsible for fructose utilization. frwC deletion can enhance biofilm formation and capsular polysaccharide (CPS) biosynthesis but decreases the growth rate and lethality in mice. frwC expression was repressed in the cyclic AMP receptor protein (CRP) mutant. Electrophoretic mobility shift assay showed that CRP can directly bind to the promoter of frwC. These results indicated that frwC expression is controlled by CRP directly and that such regulation contributes to bacterial growth, CPS synthesis, and the virulence of the Δcrp strain. The findings help elucidate fructose metabolism and the CRP regulatory mechanism in K. pneumoniae.
INTRODUCTION
Klebsiella pneumoniae is an opportunistic Gram-negative bacterium that mainly causes urinary tract infection, nosocomial pneumonia, and septicemia in immunocompromised individuals (1). In recent years, the hypervirulent variant of K. pneumoniae, which can cause community-acquired pyogenic liver abscess even in healthy people, has been reported worldwide, especially in Asia (2, 3). The main virulence factors responsible for the hypervirulent phenotype include the capsular polysaccharide (CPS), the lipopolysaccharide O antigen, pili or fimbrial adherence factors, aerobactin siderophores, and biofilm formation (4–8).
Our previous research has proven the association of in vitro growth, CPS production, biofilm formation, and lethality in mice with the cyclic AMP receptor protein (CRP), a well-studied global regulatory protein (9). Electrophoretic mobility shift assay (EMSA) also implied that CRP can directly repress the transcription of wzi and manC and indirectly repress the transcription of galF via rcsA to affect CPS biosynthesis (10). In Escherichia coli, more than 260 CRP binding sites have been identified (11), and the pseudopalindromic consensus of the CRP binding box is TGTGA-N(6)-TCACA (12–15). Through bioinformatics analysis, we found a putative CRP binding box (ATTGCGATCCACCTCAAATC) located at positions −49 to −30 relative to the translation start site of frwC, a gene encoding the putative fructose-specific phosphotransferase system (PTS) component EIIC.
The PTS is a major transport system for carbohydrate transport and involves biofilm formation and virulence in many pathogens (16–19). The system consists of two cytoplasmic energy-coupling proteins (enzyme I and HPr) and a range of carbohydrate-specific enzyme II (EII) types, which catalyze concomitant carbohydrate translocation and phosphorylation. Each EII type is composed of two cytosolic components (EIIA and -B); an integral membrane domain (EIIC); and, in some cases, a fourth component (EIID) (16). In K. pneumoniae, a putative Frw fructose PTS gene cluster from KP1_1987 to KP1_1993 was identified through a genomic view of the PTS. This fructose PTS cluster achieved four distinct PTS protein-encoding genes: one for the phosphoenolpyruvate protein phosphotransferase (PtsA), two encoding EIIB-like proteins (FrwB and FrwD), and one encoding an EIIC-like protein (FrwC).
In the present study, we investigated whether CRP binds directly to the predicted binding site of frwC, which in turn influenced the virulence, growth, and biofilm formation of K. pneumoniae NTUH-2044. Our data revealed that the global transcriptional factor CRP directly positively regulated frwC and that the regulation of frwC was involved in the in vitro growth, biofilm formation, virulence, and hypermucoviscous phenotype of K. pneumoniae.
RESULTS
Mutation and complementation.
The majority of the frwC coding region was in-frame deleted from the wild type (WT) by allelic exchange to generate the ΔfrwC mutant, after which the cis-trans-complemented C-frwC mutant was constructed from the ΔfrwC mutant. To validate the mutation and complementation, reverse transcription (RT)-PCR experiments were performed to detect the frwC transcripts in the WT, ΔfrwC, and C-frwC strains. As expected, the frwC transcript was lacking in the ΔfrwC mutant but restored in the C-frwC strain (Fig. 1).
FIG 1.
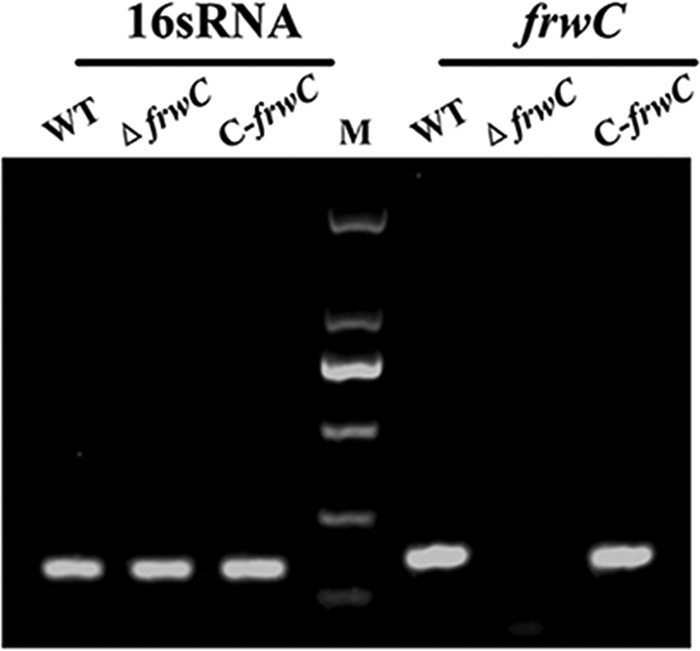
RT-PCR. The expression levels of 16S rRNA and frwC in K. pneumoniae strains were monitored by RT-PCR. The frwC transcript was lacking in the ΔfrwC mutant but present in the WT and C-frwC strains. M, DL 2000 DNA marker.
CRP directly positively regulates frwC transcription.
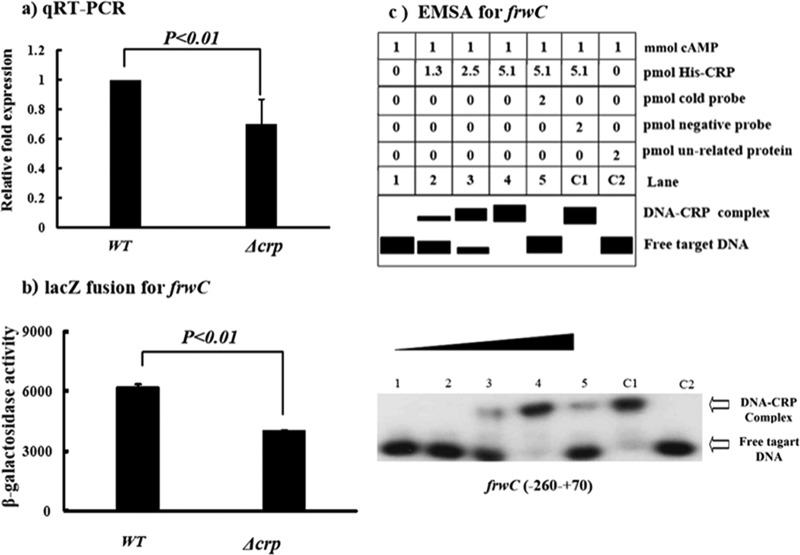
The relative mRNA levels of frwC were determined by quantitative RT (qRT)-PCR to be lower in the Δcrp mutant than in the WT. This observation was further confirmed by determining the activities of the promoter of frwC (PfrwC) by LacZ fusion, which suggested that CRP positively regulated frwC expression (Fig. 2a and b). To determine the interaction between CRP and the promoter of frwC, the CRP protein was cloned, and EMSA was conducted. Figure 2c shows that the CRP protein can bind to the promoter-proximal region of frwC in a dose-dependent manner. DNA-protein binding complexes were observed after incubating 2.5 pmol purified His6-CRP with PfrwC. CRP can regulate frwC transcription by binding directly to the predicted CRP binding site of frwC, which was located at the −49 to −30 positions upstream.
FIG 2.
CRP directly affects frwC transcription. (a) qRT-PCR. The mRNA levels of frwC were compared between the Δcrp and WT strains. A standard curve was prepared for each RNA preparation with the 16S rRNA gene. The relative fold expression of frwC in the Δcrp mutant was determined by the 2−ΔΔCt method and compared with that in the WT. The error bars indicate standard deviations. (b) lacZ fusion. A promoter-proximal region of frwC was cloned into the lacZ transcriptional-fusion vector pHRP309. The frwC-lacZ fusion plasmid was transformed into the WT and Δcrp strains, and frwC promoter activities were determined. (c) EMSAs for binding of CRP with the promoter of frwC. The radioactively labeled PfrwC DNA fragment was incubated with increasing amounts of purified His6-CRP (lanes 1 to 4: 0, 1.3, 2.5, and 5.1 pmol, respectively) and subjected to native 4% polyacrylamide gel electrophoresis. The band of free DNA disappeared with increasing amounts of His6-CRP, resulting in a retarded DNA band with decreased mobility, which presumably represented the DNA-CRP complex.
Growth of different sugars in different media.
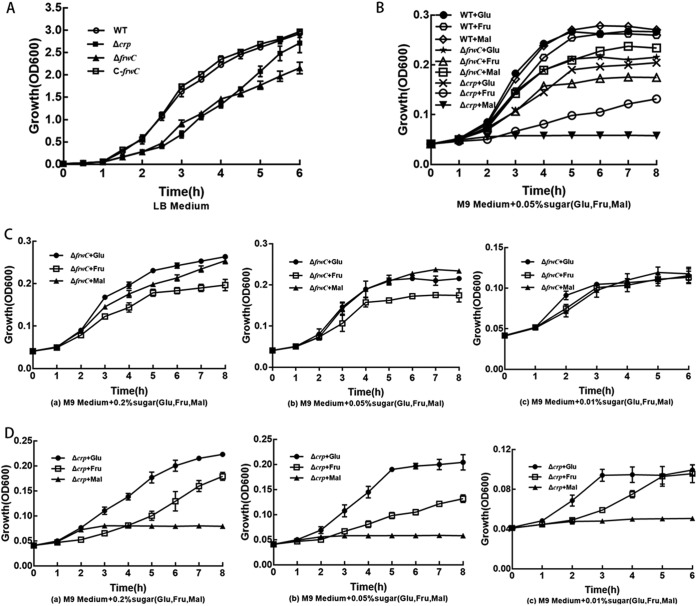
The growth rates of the Δcrp, ΔfrwC, and C-frwC strains were examined in Luria-Bertani (LB) medium. The Δcrp and ΔfrwC strains grew more slowly than did the wild-type strain (Fig. 3A) (P < 0.001). Complementation with frwC restored growth to a level comparable to that of the wild-type parent (P = 0.196). This result suggests that the crp and frwC gene products are required for K. pneumoniae growth.
FIG 3.
Growth assays. Cultures were inoculated in LB medium overnight and 100-fold diluted in 30 ml fresh LB broth or M9 minimal medium supplemented with various concentrations of glucose, fructose, or maltose (0.2%, 0.05%, and 0.01%). Culture growth was monitored by measuring the optical density at 600 nm hourly or every half hour. The optical density values are the mean values for data from three independent experiments. The error bars represent standard deviations. (A) Growth of the wild-type, ΔfrwC, Δcrp, and C-frwC strains in LB broth. (B) Growth of the wild-type strain and the ΔfrwC or the Δcrp strain in M9 minimal medium supplemented with 0.05% glucose, 0.05% fructose, or 0.05% maltose. (C) Growth of the ΔfrwC strain in M9 medium supplemented with 0.2% (a), 0.05% (b), or 0.01% (c) glucose, fructose, or maltose. (D) Growth of the Δcrp strain in M9 medium supplemented with 0.2% (a), 0.05% (b), or 0.01% (c) glucose, fructose, or maltose.
The growth rates of the Δcrp and ΔfrwC strains in M9 medium supplemented with individual sugars (glucose, fructose, or maltose) at different concentrations (0.2%, 0.05%, or 0.01%) were examined. The growth curves of the Δcrp and ΔfrwC mutants rose more slowly than did that of the WT in all the minimal media supplemented with different sugars (Fig. 3B) (P < 0.05), suggesting crp and frwC may not be associated only with fructose utilization, but also with that of glucose and maltose. Compared with the ΔfrwC mutant, the Δcrp mutant grew more slowly, especially in minimal medium supplemented with maltose (Fig. 3B) (P < 0.05). The growth rate of the ΔfrwC mutant rose with increasing sugar concentrations. The growth rate of the ΔfrwC strain did not differ significantly in minimal medium supplemented with glucose, fructose, or maltose at 0.01% (Fig. 3C) (P > 0.3). However, at higher sugar concentrations (0.05% and 0.2%), the ΔfrwC strain grew more slowly in minimal medium supplemented with fructose than in medium supplemented with glucose or maltose (Fig. 3C) (P < 0.05). These results suggest that the frwC gene product is required for K. pneumoniae uptake of sugars, especially fructose, at high concentrations. The growth trends of the strain with CRP deleted in glucose and fructose were similar to those of the ΔfrwC strain. At higher sugar concentrations (0.05% and 0.2%), the Δcrp mutant grew more slowly in medium containing fructose than in medium with glucose (Fig. 3D) (P < 0.05). However, the Δcrp mutant did not grow in minimal medium with maltose. These results suggest that CRP can regulate fructose utilization through the transcription of frwC and CRP can regulate maltose utilization through an unknown mechanism.
Deletion of frwC increases biofilm formation.
To investigate the influence of frwC and crp deletions on biofilm formation, we analyzed the biofilm-forming capacity in LB medium of each of the mutants through pellicle formation in glass culture tubes and biofilm formation in 48-well microtiter plates in a crystal violet staining assay. The Δcrp strain formed thin pellicles in static liquid cultures (Fig. 4A), unlike the wild-type and ΔfrwC strains, which formed thick, robust pellicles. A microtiter plate assay was also used to examine biofilm formation. At least nine replicate wells were prepared to study biofilm formation for each strain. A significant increase in biofilm biomass was noted for the ΔfrwC mutant after 2 days (Fig. 4B). This result indicated that frwC negatively regulated biofilm formation in K. pneumoniae. However, CRP helped positively regulate bacterial biofilm formation. This finding indicates that the decreased pellicle- and biofilm-forming capacities of the crp mutant were due to regulation of the other genes in the cell.
FIG 4.
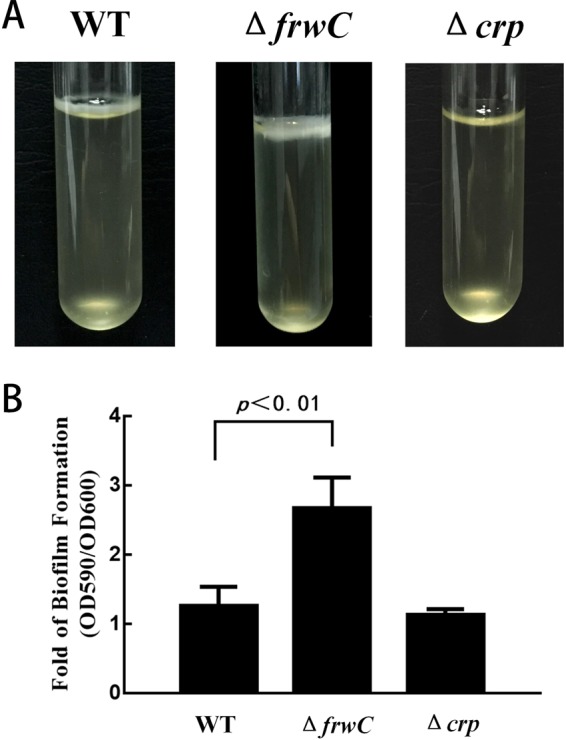
Biofilm formation in LB medium of K. pneumoniae NTUH-K2044 ΔfrwC and Δcrp mutants. (A) Biofilm development by K. pneumoniae ΔfrwC and Δcrp mutants in static liquid cultures after 2 days at 37°C. The effect of pellicle formation showed that the ΔfrwC strain formed the thickest, most robust pellicles, whereas the Δcrp strain generated thin pellicles. (B) Biofilm development by K. pneumoniae ΔfrwC and Δcrp mutants in 48-well microtiter plates in a crystal violet staining assay. The bound dye was dissolved using 95% ethanol, and the optical density at 590 nm was determined. OD600 values were used for normalization to eliminate the effects of the growth rate and cell density. The error bars represent the standard errors of three biological replicates. The P value for the frwC mutant was calculated by ANOVA.
Deletion of frwC enhances mucoid capsule production.
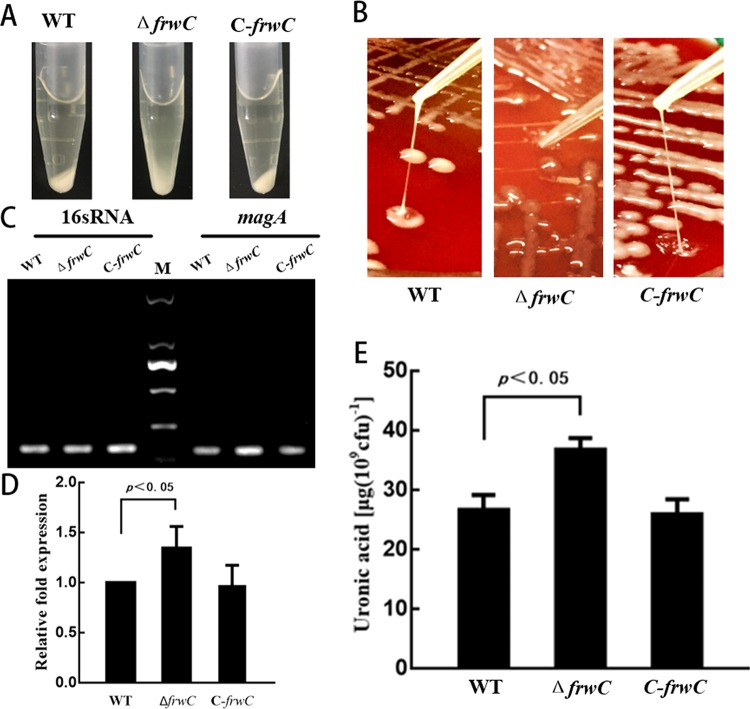
The precipitation of 1 ml mid-logarithmic-phase cell cultures of the WT, ΔfrwC, and C-frwC strains was carried out by centrifugation at 10,000 × g for 5 min to measure the effect of frwC deletion on CPS production. Given that bacteria with thicker and more mucoid capsules were pelleted less readily, the WT and C-frwC strains formed pellets denser than those of the ΔfrwC mutant (Fig. 5A). String tests of the WT and C-frwC strains were considered positive with the formation of a ≥5-mm viscous filament. However, the ΔfrwC colony stretched as a whole, but not as a string. This result suggests that the viscidity between the bacteria was very sticky (Fig. 5B). The expression of mucoviscosity-associated gene A (magA) (20), identified as the gene responsible for the hypermucoviscous phenotype, was increased in the ΔfrwC strain (Fig. 5C and D). The amount of capsule, as a function of uronic acid content, was quantified. There was a statistical difference between the amounts of uronic acid detected in overnight cultures of the wild-type and frwC mutant strains (Fig. 5E), indicating that frwC decreased capsule production.
FIG 5.
Capsular polysaccharide biosynthesis test. (A) A precipitation test was performed with centrifugation at 10,000 × g for 5 min, and the resultant pellet was checked for mucoid appearance. The WT and C-frwC strains were found to have denser pellets than the ΔfrwC mutant. M, DL 2000 DNA marker. (B) String test. The strains were grown on 5% sheep blood plates at 37°C for 16 h, and the WT and C-frwC strains displayed viscous filaments stretched by a tip, but the ΔfrwC mutant colony was stretched as a whole. (C) RT-PCR. The expression levels of 16S rRNA and magA (a gene identified as responsible for the hypermucoviscous phenotype) of K. pneumoniae strains were monitored by RT-PCR. The magA transcript level was increased in the ΔfrwC strain. (D) magA expression. The magA transcript level was quantified with Image Lab software. (E) CPS quantification. CPS was isolated and expressed as the amount of glucuronic acid per 109 CFU of bacterial cells. The error bars represent the standard errors of three biological replicates. The P value for the frwC mutant was calculated by ANOVA.
FrwC is essential for mouse virulence.
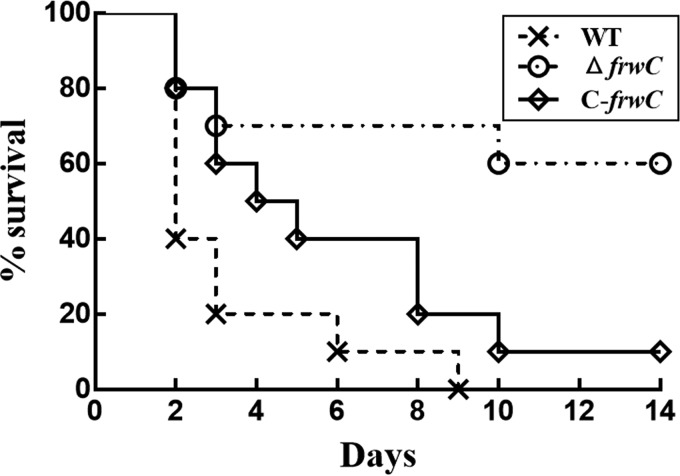
We investigated whether the deletion of frwC would affect the virulence of K. pneumoniae NTUH-K2044. Ten mice were infected intraperitoneally with 104 CFU each of the WT, ΔfrwC, and C-frwC strains, and the corresponding survival curves for 14 days were calculated. All of the mice inoculated with the wild-type strain died 9 days after infection. In contrast, 60% of the mice infected with the ΔfrwC mutant survived for 14 days (Fig. 6). The survival rate of the mice infected with the complementation strain decreased to 10%. This result suggests that the complementation restored the attenuated virulence of the ΔfrwC mutant. At a dose of 105 CFU, all the mice infected intraperitoneally with the WT, ΔfrwC, and C-frwC strains died within 3 days after infection. In contrast, 50% of the mice inoculated with the Δcrp mutant survived 14 days postinfection (data from reference 9). These findings indicate the strong role of CRP in positively modulating bacterial virulence, not only through regulating the role of the Frw fructose PTS.
FIG 6.
Mouse lethality assay. Ten mice were infected intraperitoneally with 104 CFU each of the WT, ΔfrwC, and C-frwC strains, and the corresponding survival curves for 14 days were calculated. The survival percentages showed significant differences between the WT and mutant strains (P < 0.001; Kaplan-Meier test), but the WT and complemented strains exhibited no difference (P = 0.114; Kaplan-Meier test).
DISCUSSION
CRP is a global regulator of gene expression and was originally identified as a major regulator of catabolite repression (21, 22). In K. pneumoniae, CRP acts as a negative regulator of CPS biosynthesis. The crp mutant produces increased CPS levels and decreased biofilm formation, growth rate, and virulence (9). When the regulatory mechanism of CRP was further elucidated, CRP was directly bound to the predictive CRP binding sites and repressed the transcription of wzi, manC, and rcsA, which regulate CPS production (10).
Besides CPS, various PTSs have been linked to biofilm formation and infection. The mutant of the cellobiose-specific PTS component IIC gene celB of K. pneumoniae reduces biofilm production and virulence (23). Fructose PTS is another such PTS. In K. pneumoniae, fructose PTS includes the Fru operon, Frv operon, and Frw gene cluster (24). The putative Frw fructose PTS gene clusters from KP1_1987 to KP1_1993 produce proteins similar to those produced by the Fru operon in E. coli. This Frw PTS cluster includes the following four distinct PTS protein-encoding genes: one for the phosphoenolpyruvate protein phosphotransferase (ptsA), two encoding IIB-like proteins (frwB and frwD), and one encoding a IIC-like protein (frwC). qRT-PCR and a lacZ fusion assay revealed that frwC expression decreased in the crp deletion mutant in our study and thereby suggested that CRP can positively regulate the Frw PTS in K. pneumoniae. To investigate whether CRP binds directly to the frwC promoter region, we performed EMSA. DNA-protein-binding complexes were observed after incubation of 2.5 pmol purified His6-CRP with PfrwC (Fig. 2c). Therefore, we suggest that CRP binds directly to the CRP binding site in PfrwC to control frwC transcription. FrwC is highly homologous to the EIIC domain of FruI of the fructose PTS in Streptococcus gordonii, and nonpolar inactivation of fruI results in a biofilm-defective phenotype and reduced virulence in S. gordonii (17). These findings suggest that CRP may also influence biofilm formation and virulence by regulating the Frw fructose PTS in K. pneumoniae.
The underlying mechanisms of Frw PTS in sugar utility, biofilm formation, and virulence of K. pneumoniae were studied. Growth assays and carbon utilization profiles revealed that frwC was required for growth in fructose. When grown in minimal medium supplemented with 0.01% glucose, maltose, or fructose, the growth rates of the ΔfrwC strain were similar in different sugars. However, at higher fructose concentrations (0.05% and 0.2%), the ΔfrwC mutant grew much more slowly in fructose than in the corresponding glucose and maltose. In addition to the Frw PTS, there are several other fructose PTSs, such as the Fru PTS, hypothesized to be involved in the utilization of fructose. Fructose can be fully utilized by other PTSs at low concentrations. At high concentrations, the Frw fructose system is needed to fully transport fructose. This could also explain why there was still a significant level of growth of the frwC mutant in minimal medium with fructose. The growth trend of the Δcrp strain in M9 medium supplemented with fructose was similar to that of the ΔfrwC strain, which indicates that CRP can influence fructose utilization by regulating frwC expression. The growth rate of the ΔfrwC mutant was lower than that of the WT but faster than that of the Δcrp strain. These results suggest that, in addition to frwC, there are other genes that take part in the utilization of fructose that are regulated by CRP. Interestingly, the Δcrp strain did not grow in M9 medium with maltose as the sole carbon source. The underlying mechanisms of the effect of CRP on maltose utilization in K. pneumoniae are under investigation in our laboratory.
Although the growth rate of the ΔfrwC mutant was lower than that of the WT, the ΔfrwC mutant formed a biofilm thicker than that of the WT. The FrwC protein can increase biofilm formation by regulating CPS biosynthesis. The centrifugation test and CPS quantification proved that the number of mucoid capsules of the ΔfrwC mutant increased relative to that in the WT strain. Although the string test of the ΔfrwC mutant was negative, not all mucoid colonies of K. pneumoniae showed a positive string test (25). The ΔfrwC colony attached as a whole to the tip. This phenomenon highlights a distinct difference between mucoid capsular strains and the hypermucoviscous variants. Mucoviscosity-associated gene A (magA) encodes a polymerase essential for capsule synthesis. Deletion of the magA gene prevents the production of the polysaccharide capsule (26). RT-PCR results showed that transcription of magA increased in the ΔfrwC strains. Such increases then augmented the production of CPS and the mucoviscosity of K. pneumoniae. Interestingly, CRP positively regulates frwC expression directly and FrwC increases biofilm formation, and the Δcrp strain formed thinner biofilm than the WT. The reason was deduced to be as follows: CRP is a global regulatory protein that regulates many genes, such as the CPS biosynthesis genes except for frwC, and the decrease of biofilm formation by the CRP mutant may be the comprehensive result of regulation.
Although K. pneumoniae ΔfrwC produced more CPS and biofilm than the WT, lethality in mice was attenuated in the former strain. This observation may be related to the decreased bacterial growth during infection. Although 60% of the mice infected with 104 CFU of the ΔfrwC mutant survived, the mice infected with 105 CFU of the ΔfrwC mutant died at the same time as the WT.
In conclusion, our study proved from a genomic perspective that the putative fructose PTS actually plays a role in carbon utilization. The gene deletion mutant of frwC, which is a gene encoding an EIIC-like protein of the PTS, can increase biofilm formation and CPS production and decrease the growth rate and lethality in mice. Notably, frwC expression was controlled by CRP directly. Such regulation can contribute to bacterial growth, CPS synthesis, and Δcrp strain virulence. This study helps elucidate the regulatory mechanism of the global regulator CRP in K. pneumoniae.
MATERIALS AND METHODS
Bacterial strains, plasmids, and primers.
The bacterial strains, plasmids, and primers used in this study are listed in Table 1. K. pneumoniae NTUH-2044 serotype K1 with the hypermucoviscosity phenotype was isolated from a Taiwanese liver abscess case (27). K. pneumoniae and E. coli were grown in LB medium or M9 minimal medium with the appropriate sugar at 37°C. For biochemical and phenotypic assays, strains were precultivated overnight in LB broth at 37°C and diluted 100-fold in fresh medium.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Description |
|---|---|
| Strains | |
| K. pneumoniae | |
| NTUH K-2044 (WT) | Wild-type strain; K1 serotype; Apr |
| ΔfrwC | Deletion of frwC from WT; Apr |
| C-frwC | Complemented frwC mutant; Apr Kmr |
| Δcrp | Deletion of crp from WT; Apr |
| E. coli | |
| DH5α | λ− φ80dlacZΔM15 Δ(lacZYA-argF)U169 recA1 endA1 hsdR17(rK− mK−) supE44 thi-1 gyrA relA1 |
| Plasmids | |
| pKO3-Km | pKO3-derived plasmid with insertion of a Km resistance cassette from pUC4K into the AccI site |
| pGEM-T-easy | pGEM-T easy with insertion of a Km cassette from pUC4K into the NdeI site for trans-complementation |
| pHRP309 | lacZ transcriptional fusion vector based on the IncQ plasmid RSF1010 |
Construction of deletion mutants and complementation strains of K. pneumoniae.
The frwC gene in-frame deletion was introduced into K. pneumoniae NTUH-K2044 by using an allelic-exchange strategy as previously described (23). The flanking regions of the frwC gene were cloned into the temperature-sensitive suicide plasmid pKO3-Km, which contains a kanamycin resistance (Kmr) cassette and a sacB gene for positive and negative selection for chromosomal integration and excision, respectively. The resulting plasmid was transformed into NTUH-K2044 (WT strain) and plated on LB agar with Km at the nonpermissive temperature (43°C) to force integration of the plasmid into the bacterial chromosome by a single crossover. Several positive colonies were spread onto an LB agar plate in the presence of sucrose and absence of Km. These colonies were then grown at the permissive temperature (30°C) to select for plasmid excision and loss and to acquire the deletion mutant of frwC (ΔfrwC).
To facilitate the production of a complementary strain, we amplified the DNA fragment containing the 1,077-bp frwC open reading frame; 514-bp upstream and 278-bp downstream regions were amplified and cloned into the km-pGEM-T-easy vector to generate a frwC complementary plasmid. The resulting plasmid was then transformed into the frwC deletion mutant or the CRP deletion mutant to acquire the complementation strains designated C-frwC and Δcrp+frwC. The Δcrp and C-crp strains were previously constructed in our laboratory (9).
RT-PCR experiments were performed to detect the frwC mRNA in the WT, ΔfrwC, and C-frwC strains.
RNA isolation and qRT-PCR analysis.
The total RNAs of the WT, Δcrp, and ΔfrwC strains were extracted using the TRIzol reagent (Invitrogen). The RNA quality was then monitored by agarose gel electrophoresis, and the RNA quantity was determined by spectrophotometry. The contaminated DNA was removed from the RNA samples using Ambion's DNA-free kit. cDNAs were generated by using 5 μg of RNA and 3 μg of random-hexamer primers. qRT-PCR was performed through a StepOnePlus real-time PCR system (ABI), along with SYBR green master mix. The relative fold expression of frwC in the Δcrp mutant were compared with those in the WT and determined by the 2−ΔΔCT method.
lacZ fusion and β-galactosidase.
The promoter-proximal DNA region of frwC was cloned into the low-copy-number transcriptional fusion vector pHRP309 harboring a promoterless lacZ reporter gene. The K. pneumoniae WT and Δcrp strains transformed with the recombinant plasmid were grown to measure β-galactosidase activity in cellular extracts using a β-galactosidase enzyme assay system.
EMSA.
The His6-CRP protein was cloned into pET28a, expressed in E. coli, and then purified using a Ni-nitrilotriacetic acid (NTA) agarose column. The DNA fragment of the putative promoter region of frwC was PCR amplified with 5′-32P-labeled primers. The radioactively labeled DNA fragment was incubated with increasing amounts of purified His6-CRP (0, 1.3, 2.5, and 5.1 pmol). Following incubation at room temperature for 30 min, the mixtures were analyzed on 4% native polyacrylamide gels. Radioactive species were detected by autoradiography.
Growth rate and sugar specificity.
The K. pneumoniae NTUH-K2044 (WT), ΔfrwC, Δcrp, and C-frwC strains were grown in LB medium overnight and 100-fold diluted in 30 ml fresh LB broth or M9 minimal medium supplemented with various concentrations of glucose, fructose, or maltose (0.2%, 0.05%, or 0.01%). Culture growth was monitored by measuring the optical density at 600 nm (OD600) hourly or every half hour.
Biofilm formation assay.
The glass tube assay and microtiter plate assay were performed to observe the progression of biofilm formation. Pellicles were assayed by visual inspection of the air-liquid interface of a static liquid culture in LB medium after 2 days at 37°C in a glass tube. Meanwhile, the microtiter plate assay was modified to examine biofilm formation by K. pneumoniae (28). Briefly, 10 μl of an overnight culture was inoculated into 1 ml of fresh LB medium in each well of a 48-well polystyrene plate. After 2 days of incubation at 37°C, the adherent biofilm formation was measured by staining bound cells for 15 min with a 0.1% (wt/vol) aqueous solution of crystal violet. The bound dye was dissolved using 1 ml 95% ethanol, and the optical density at 590 nm was determined. The OD600 of medium with planktonic cells was used for normalization to eliminate the effects of the growth rate and cell density. The relative capacity for biofilm formation was calculated with the following formula: OD590/OD600.
Mucoviscosity assay.
The mucoviscosity levels were determined by centrifugation and the string test (29). K. pneumoniae NTUH-K2044 and the ΔfrwC and C-frwC strains were cultivated at 37°C overnight. Subsequently, 1-ml aliquots of bacteria were pelleted at 10,000 × g for 5 min to check the mucoid appearance. For the string test, the WT, mutant, and complemented strains were inoculated onto 5% sheep blood plates and incubated at 37°C for 16 h. A standard tip was used to stretch a mucoviscous string from the colony (20). Meanwhile, magA gene expression differences between WT and ΔfrwC strains were amplified by RT-PCR and analyzed by 1.2% agarose gel electrophoresis, which was normalized with 16S rRNA. The magA transcript was quantified using Image Lab software. To quantify the extracted CPS, the K. pneumoniae CPS was isolated using the hot phenol-water method (30). The relative CPS concentration was expressed as the amount (in micrograms) of glucuronic acid per 109 CFU of bacterial cells.
Mouse lethality assay.
Overnight cultures of the WT, ΔfrwC, and C-frwC strains were diluted 1:100 in fresh LB broth and incubated at 37°C until the OD600 reached 1.4. The cells were harvested and diluted with PBS to about 105 CFU/ml. The actual concentrations were plated onto LB agar plates to calculate the number of CFU in the plate. Ten 5-week-old BALB/c female mice from each group were injected intraperitoneally with 0.1 ml of the WT, ΔfrwC, or C-frwC strain. The infected mice were then monitored daily for 14 days to measure the illness severity and survival.
All animal experiments were approved by the Animal Care and Use Committee at Hubei University of Medicine and complied with all ethical and husbandry regulations.
Experimental replicates and statistical methods.
Experiments were performed with three independent bacterial cultures, and values were expressed as means and standard deviations. To determine the statistically significant differences, analysis of variance (ANOVA), repeated measures with least-significant difference (LSD), and Kaplan-Meier tests were performed, and P values of <0.05 and <0.01 were considered to indicate statistical significance.
ACKNOWLEDGMENTS
Financial support was provided by the Foundation for Innovative Research Team of the Hubei Provincial Department of Education (T201612) and the Natural Science Foundation of Hubei Province for Distinguished Young Scholars (2018CFA046).
REFERENCES
- 1.Podschun R, Ullmann U. 1998. Klebsiella spp. as nosocomial pathogens: epidemiology, taxonomy, typing methods, and pathogenicity factors. Clin Microbiol Rev 11:589–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tsai FC, Huang YT, Chang LY, Wang JT. 2008. Pyogenic liver abscess as endemic disease, Taiwan. Emerg Infect Dis 14:1592–1600. doi: 10.3201/eid1410.071254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fang FC, Sandler N, Libby SJ. 2005. Liver abscess caused by magA+ Klebsiella pneumoniae in North America. J Clin Microbiol 43:991–992. doi: 10.1128/JCM.43.2.991-992.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cortes G, Borrell N, de Astorza B, Gomez C, Sauleda J, Alberti S. 2002. Molecular analysis of the contribution of the capsular polysaccharide and the lipopolysaccharide O side chain to the virulence of Klebsiella pneumoniae in a murine model of pneumonia. Infect Immun 70:2583–2590. doi: 10.1128/IAI.70.5.2583-2590.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Struve C, Bojer M, Krogfelt KA. 2009. Identification of a conserved chromosomal region encoding Klebsiella pneumoniae type 1 and type 3 fimbriae and assessment of the role of fimbriae in pathogenicity. Infect Immun 77:5016–5024. doi: 10.1128/IAI.00585-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hsieh PF, Lin TL, Lee CZ, Tsai SF, Wang JT. 2008. Serum-induced iron-acquisition systems and TonB contribute to virulence in Klebsiella pneumoniae causing primary pyogenic liver abscess. J Infect Dis 197:1717–1727. doi: 10.1086/588383. [DOI] [PubMed] [Google Scholar]
- 7.Fierer J. 2012. Biofilm formation and Klebsiella pneumoniae liver abscess: true, true and unrelated. Virulence 3:241–242. doi: 10.4161/viru.20588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vuotto C, Longo F, Balice MP, Donelli G, Varaldo PE. 2014. Antibiotic resistance related to biofilm formation in Klebsiella pneumoniae. Pathogens 3:743–758. doi: 10.3390/pathogens3030743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ou Q, Fan J, Duan D, Xu L, Wang J, Zhou D, Yang H, Li B. 2017. Involvement of cAMP receptor protein in biofilm formation, fimbria production, capsular polysaccharide biosynthesis and lethality in mouse of Klebsiella pneumoniae serotype K1 causing pyogenic liver abscess. J Med Microbiol 66:1–7. doi: 10.1099/jmm.0.000391. [DOI] [PubMed] [Google Scholar]
- 10.Lin CT, Chen YC, Jinn TR, Wu CC, Hong YM, Wu WH. 2013. Role of the cAMP-dependent carbon catabolite repression in capsular polysaccharide biosynthesis in Klebsiella pneumoniae. PLoS One 8:e54430. doi: 10.1371/journal.pone.0054430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Salgado H, Gama-Castro S, Martinez-Antonio A, Diaz-Peredo E, Sanchez-Solano F, Peralta-Gil M, Garcia-Alonso D, Jimenez-Jacinto V, Santos-Zavaleta A, Bonavides-Martinez C, Collado-Vides J. 2004. RegulonDB (version 4.0): transcriptional regulation, operon organization and growth conditions in Escherichia coli K-12. Nucleic Acids Res 32:D303–D306. doi: 10.1093/nar/gkh140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baichoo N, Heyduk T. 1999. Mapping cyclic nucleotide-induced conformational changes in cyclicAMP receptor protein by a protein footprinting technique using different chemical proteases. Protein Sci 8:518–528. doi: 10.1110/ps.8.3.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gunasekera A, Ebright YW, Ebright RH. 1992. DNA sequence determinants for binding of the Escherichia coli catabolite gene activator protein. J Biol Chem 267:14713–14720. [PubMed] [Google Scholar]
- 14.Busby S, Ebright RH. 1999. Transcription activation by catabolite activator protein (CAP). J Mol Biol 293:199–213. doi: 10.1006/jmbi.1999.3161. [DOI] [PubMed] [Google Scholar]
- 15.Kolb A, Spassky A, Chapon C, Blazy B, Buc H. 1983. On the different binding affinities of CRP at the lac, gal and malT promoter regions Nucleic Acids Res 11:7833–7852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kotrba P, Inui M, Yukawa H. 2001. Bacterial phosphotransferase system (PTS) in carbohydrate uptake and control of carbon metabolism. J Biosci Bioeng 92:502–517. doi: 10.1016/S1389-1723(01)80308-X. [DOI] [PubMed] [Google Scholar]
- 17.Loo CY, Mitrakul K, Voss IB, Hughes CV, Ganeshkumar N. 2003. Involvement of an inducible fructose phosphotransferase operon in Streptococcus gordonii biofilm formation. J Bacteriol 185:6241–6254. doi: 10.1128/JB.185.21.6241-6254.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kilic AO, Tao L, Zhang Y, Lei Y, Khammanivong A, Herzberg MC. 2004. Involvement of Streptococcus gordonii beta-glucoside metabolism systems in adhesion, biofilm formation, and in vivo gene expression. J Bacteriol 186:4246–4253. doi: 10.1128/JB.186.13.4246-4253.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Valdes KM, Sundar GS, Vega LA, Belew AT, Islam E, Binet R, El-Sayed NM, Le Breton Y, McIver KS. 2016. The fruRBA operon is necessary for group A streptococcal growth in fructose and for resistance to neutrophil killing during growth in whole human blood. Infect Immun 84:1016–1031. doi: 10.1128/IAI.01296-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fang CT, Chuang YP, Shun CT, Chang SC, Wang JT. 2004. A novel virulence gene in Klebsiella pneumoniae strains causing primary liver abscess and septic metastatic complications. J Exp Med 199:697–705. doi: 10.1084/jem.20030857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cooper TF, Remold SK, Lenski RE, Schneider D. 2008. Expression profiles reveal parallel evolution of epistatic interactions involving the CRP regulon in Escherichia coli. PLoS Genet 4:e35. doi: 10.1371/journal.pgen.0040035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Willias SP, Chauhan S, Lo CC, Chain PS, Motin VL. 2015. CRP-mediated carbon catabolite regulation of Yersinia pestis biofilm formation is enhanced by the carbon storage regulator protein, CsrA. PLoS One 10:e0135481. doi: 10.1371/journal.pone.0135481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu MC, Chen YC, Lin TL, Hsieh PF, Wang JT. 2012. Cellobiose-specific phosphotransferase system of Klebsiella pneumoniae and its importance in biofilm formation and virulence. Infect Immun 80:2464–2472. doi: 10.1128/IAI.06247-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reizer J, Reizer A, Saier MJ. 1995. Novel phosphotransferase system genes revealed by bacterial genome analysis—a gene cluster encoding a unique enzyme I and the proteins of a fructose-like permease system. Microbiology 141:961–971. doi: 10.1099/13500872-141-4-961. [DOI] [PubMed] [Google Scholar]
- 25.Hagiya H, Watanabe N, Maki M, Murase T, Otsuka F. 2015. Clinical utility of string test as a screening method for hypermucoviscosity-phenotype Klebsiella pneumoniae. Acute Med Surg 1:245–246. doi: 10.1002/ams2.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pan YJ, Lin TL, Chen YH, Hsu CR, Hsieh PF, Wu MC, Wang JT. 2013. Capsular types of Klebsiella pneumoniae revisited by wzc sequencing. PLoS One 8:e80670. doi: 10.1371/journal.pone.0080670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu KM, Li LH, Yan JJ, Tsao N, Liao TL, Tsai HC, Fung CP, Chen HJ, Liu YM, Wang JT, Fang CT, Chang SC, Shu HY, Liu TT, Chen YT, Shiau YR, Lauderdale TL, Su IJ, Kirby R, Tsai SF. 2009. Genome sequencing and comparative analysis of Klebsiella pneumoniae NTUH-K2044, a strain causing liver abscess and meningitis. J Bacteriol 191:4492–4501. doi: 10.1128/JB.00315-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.O'Toole GA, Kolter R. 1998. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol Microbiol 28:449–461. doi: 10.1046/j.1365-2958.1998.00797.x. [DOI] [PubMed] [Google Scholar]
- 29.Wu MC, Lin TL, Hsieh PF, Yang HC, Wang JT. 2011. Isolation of genes involved in biofilm formation of a Klebsiella pneumoniae strain causing pyogenic liver abscess. PLoS One 6:e23500. doi: 10.1371/journal.pone.0023500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chuang YP, Fang CT, Lai SY, Chang SC, Wang JT. 2006. Genetic determinants of capsular serotype K1 of Klebsiella pneumoniae causing primary pyogenic liver abscess. J Infect Dis 193:645–654. doi: 10.1086/499968. [DOI] [PubMed] [Google Scholar]