Human innate immunity employs cellular and humoral mechanisms to facilitate rapid killing of invading bacteria. The direct killing of bacteria by human serum is attributed mainly to the activity of the complement system, which forms pores in Gram-negative bacteria.
KEYWORDS: Enterococcus faecium, commensals, group IIA secreted phospholipase A2, innate immune system, pathogens
ABSTRACT
Human innate immunity employs cellular and humoral mechanisms to facilitate rapid killing of invading bacteria. The direct killing of bacteria by human serum is attributed mainly to the activity of the complement system, which forms pores in Gram-negative bacteria. Although Gram-positive bacteria are considered resistant to killing by serum, we uncover here that normal human serum effectively kills Enterococcus faecium. Comparison of a well-characterized collection of commensal and clinical E. faecium isolates revealed that human serum specifically kills commensal E. faecium strains isolated from normal gut microbiota but not clinical isolates. Inhibitor studies show that the human group IIA secreted phospholipase A2 (hGIIA), but not complement, is responsible for killing of commensal E. faecium strains in human normal serum. This is remarkable since the hGIIA concentration in “noninflamed” serum was considered too low to be bactericidal against Gram-positive bacteria. Mechanistic studies showed that serum hGIIA specifically causes permeabilization of commensal E. faecium membranes. Altogether, we find that a normal concentration of hGIIA in serum effectively kills commensal E. faecium and that resistance of clinical E. faecium to hGIIA could have contributed to the ability of these strains to become opportunistic pathogens in hospitalized patients.
INTRODUCTION
The human immune system is essential to protect us against invading bacterial infections. The first line of immune defense is comprised of cellular and humoral factors that together fight bacteria in the first minutes to hours of an infection. Phagocytic cells, such as neutrophils, are able to engulf invading bacteria by bacterial recognition, which is enhanced by bacterial labeling with serum factors such as antibodies and complement activation products (1). However, human serum also harbors various antimicrobial proteins and peptides that can directly lyse bacteria without the help of immune cells (2). This serum bactericidal activity is effective mainly against Gram-negative bacteria that are sensitive to the pore-forming membrane attack complex (MAC) of the human complement system (3). For long, it has been known that Gram-positive bacteria are resistant to this “serum bactericidal activity.” It is generally assumed that all Gram-positive bacteria (both pathogenic and nonpathogenic) protect themselves against complement-induced pore formation via a thick layer of peptidoglycan that surrounds the bacterial membrane (4). We found here that normal human serum selectively kills commensal Enterococcus faecium strains whereas disease-associated E. faecium strains remain viable.
E. faecium is a common inhabitant of the gut of mammals, birds, and insects (5, 6). While E. faecium colonization is harmless in healthy individuals, this bacterium can cause serious infections such as bacteremia and endocarditis in immunocompromised patients (7). In fact, multidrug-resistant E. faecium, most notably vancomycin-resistant E. faecium (VRE), has emerged as an important cause of nosocomial infections worldwide (8). Recent work showed that clinical E. faecium isolates are genetically distinct from commensal strains (9). Whole-genome comparison and functional assays identified various virulence factors and carbohydrate metabolism gene clusters that were enriched in clinical isolates and that might contribute to successful niche adaptation in hospitals (9–12). Hospitalized patients suffering from clinical E. faecium infections frequently have a severely compromised cellular immunity. Yet, innate humoral immunity could still aid in E. faecium infection control (13). To what extent this is relevant in this infection type (by clinical E. faecium strains) is currently not completely understood (14–19).
Screening of a collection of commensal and multidrug-resistant clinical E. faecium strains revealed that normal human serum specifically kills commensal strains. We found that the group IIA secreted phospholipase A2 (hGIIA) is key to the bactericidal activity of serum and that resistance to serum hGIIA could have contributed to the adaptation of clinical E. faecium isolates.
RESULTS
Human serum kills commensal but not clinical Enterococcus faecium strains.
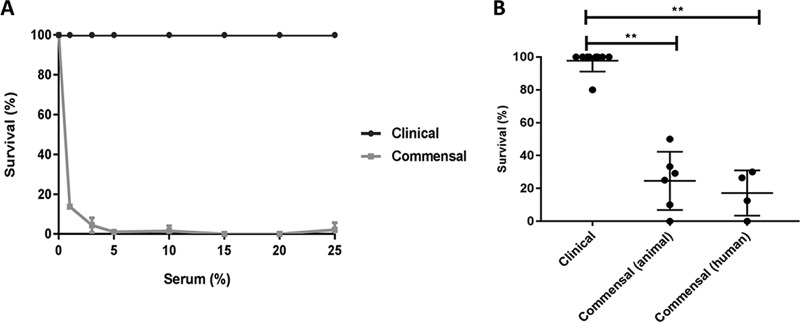
While human serum effectively kills Gram-negative bacteria (20), it is generally accepted that serum does not kill pathogenic and nonpathogenic Gram-positive bacteria (21). Serendipitously, however, we discovered that a commensal strain of E. faecium (E1007, isolated from feces of a healthy individual) was effectively killed by human serum. We incubated 105 exponential-phase E. faecium E1007 cells with normal human serum (pooled from 20 healthy volunteers) and quantified bacterial survival via colony enumeration on agar plates (Fig. 1A). Within 30 min, 10% human serum completely killed 105 E1007 bacteria. In contrast, a clinical E. faecium strain (E1162, isolated from blood of a hospitalized patient) was fully resistant to killing by serum in a similar assay. Based on these results, we extended these analyses to a broader panel of genomically well-characterized clinical and commensal E. faecium isolates. We selected clinical E. faecium strains isolated from hospitalized patients and compared their serum susceptibility to that of a selection of commensal E. faecium strains isolated from healthy individuals or animals. Exposure of all 19 strains (described in Table 1; see also the phylogenetic tree in Fig. S1 in the supplemental material) to 25% human serum revealed that human serum specifically kills commensal strains derived from animals (76% killing) and humans (83% killing), respectively, but does not kill clinical E. faecium strains (2% killing) (Fig. 1B).
FIG 1.
Human serum kills commensal but not clinical E. faecium isolates. (A) Comparison of bacterial survival of clinical E. faecium strain E1162 (clinical) and commensal E. faecium strain E1007 (commensal) at different concentrations of pooled normal human serum. (B) Killing of E. faecium isolates originating from hospitalized patients (clinical), healthy animals (commensal animal), and healthy humans (commensal humans) in 25% pooled human serum. Killing was quantified by comparison of total CFU after 30 min of incubation in the negative-control buffer (RPMI) and in 25% human serum. Each data point represents one E. faecium strain; means ± standard deviations (SD) are indicated. Statistical significance was determined using the Kruskal-Wallis test followed by Dunn's multiple-comparison test (**, P ≤ 0.01). Epidemiological details of E. faecium strains are listed in Table 1.
TABLE 1.
Ecological origin and clade assignment of 19 E. faecium strainsa
| Strain name | Alternate strain name | Category | Cladeb | MLST ST | Country | Yr | Source | Isolation site | Reference |
|---|---|---|---|---|---|---|---|---|---|
| E0333 | EnGen0013 | Clinical | A1 | 80 | ISR | 1997 | Hospitalized patient | Blood | 9 |
| E0745 | E0745 | Clinical | A1 | 16 | NL | 2000 | Hospitalized patient | Feces | 53 |
| E1162 | E1162 | Clinical | A1 | 17 | FR | 1997 | Hospitalized patient | Blood | 54 |
| E1321 | EnGen0054 | Clinical | A1 | 78 | IT | 1999 | Hospitalized patient | Catheter | 9 |
| E1392 | EnGen0016 | Clinical | A1 | 64 | GBR | 2000 | Hospitalized patient | ND | 9 |
| E1644 | EnGen0051 | Clinical | A1 | 78 | GER | 2002 | Hospitalized patient | ND | 9 |
| E1731 | EnGen0036 | Clinical | A1 | 18 | TZA | 2002 | Hospitalized patient | Blood | 9 |
| E2297 | EnGen0034 | Clinical | A1 | 117 | USA | 2001 | Hospitalized patient | Urine | 9 |
| E2560 | EnGen0046 | Clinical | A1 | 78 | NL | 2006 | Hospitalized patient | Blood | 9 |
| E0045 | EnGen0005 | Commensal animal | A2 | 9 | GBR | 1992 | Healthy poultry | Feces | 9 |
| E0164 | EnGen0010 | Commensal animal | A2 | 26 | NL | 1996 | Healthy poultry | Feces | 9 |
| E1573 | EnGen0009 | Commensal animal | A1 | 21 | BE | 1994 | Healthy bison | Rumen | 9 |
| E1604 | EnGen0028 | Commensal animal | B | 75 | NO | 1956 | Healthy cow | Cheese | 9 |
| E2134 | EnGen0043 | Commensal animal | A2 | 12 | NL | 2004 | Healthy poultry | ND | 9 |
| E4215 | EnGen0048 | Commensal animal | A2 | 310 | SWE | 2004 | Healthy poultry | ND | 9 |
| E1007 | EnGen0015 | Commensal human | B | 61 | NL | 1998 | Healthy human | Feces | 9 |
| E1050 | EnGen0017 | Commensal human | A1 | 92 | NL | 1998 | Healthy human | Feces | 9 |
| E1590 | EnGen0003 | Commensal human | B | 163 | IRL | 2001 | Healthy human | Feces | 9 |
| E3548 | EnGen0047 | Commensal human | B | 328 | NL | 2004 | Healthy human | Blood | 9 |
Abbreviations: MLST ST, sequence type by multilocus sequence typing; NL, Netherlands; ISR, Israel; FR, France; IT, Italy; GBR, Great Britain; GER, Germany; TZA, Tanzania; USA, United States; BE, Belgium; NOR, Norway; SWE, Sweden; IRL, Ireland; ND, not determined.
Clade structure was based on the classification described by Lebreton et al. (9).
Complement does not contribute to serum-mediated killing of commensal E. faecium.
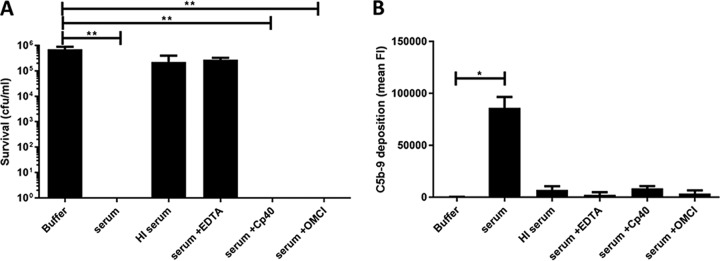
To investigate how human serum kills commensal E. faecium strains, we first used several approaches to inactivate the complement system. Bactericidal activity of the complement system is mediated by the membrane attack complex (MAC), a ring-structured pore consisting of proteins C5b, C6, C7, and C8 and multiple copies of C9 (C5b-9). The formation of pores in bacterial membranes occurs rapidly following an enzymatic chain reaction on the cell surface. Initial experiments seemed to suggest that complement indeed played a role in killing of E. faecium. For instance, we found that E. faecium killing could be blocked by exposing the serum to 56°C (heat-inactivated [HI] serum), a method commonly used to inactivate certain heat-labile complement components (22) (Fig. 2A and B). Furthermore, addition of the chelating agent EDTA, known to block the complement reaction (23), also interfered with serum-mediated killing (Fig. 2A and B). However, when we used more-specific inhibitors to block the complement reaction, we found that these inhibitors did not affect serum-mediated killing of E. faecium (Fig. 2A). For instance, application of C3 inhibitor Cp40 (24) and C5 inhibitor OmCI (25) did not block bactericidal activity (Fig. 2A), although the two inhibitors effectively blocked MAC deposition on the bacterial surface (Fig. 2B). The resistance of the clinical E. faecium strain E1162 to serum was not affected by any of the serum treatments (see Fig. S2 in the supplemental material). From these data, we concluded that killing of commensal E. faecium in human serum is not mediated by complement but by another heat-sensitive and divalent cation-dependent factor (26).
FIG 2.
Complement does not contribute to serum-mediated killing of commensal E. faecium. (A) Killing of commensal E. faecium strain E1007 was measured by bacterial viability on blood agar plates after 30 min of incubation under the indicated conditions. (B) Complement system activity in human serum was measured by C5b-9 deposition on the bacterial surface of clinical isolate E1162. FI, fluorescence intensity. OmCI, Ornithodoros moubata C5 inhibitor. Each bar represents the means ± SD from three independent experiments. Statistical significance was determined using the Kruskal-Wallis test followed by Dunn's multiple-comparison test (**, P ≤ 0.01; *, P ≤ 0.05).
hGIIA is essential for serum killing of commensal E. faecium.
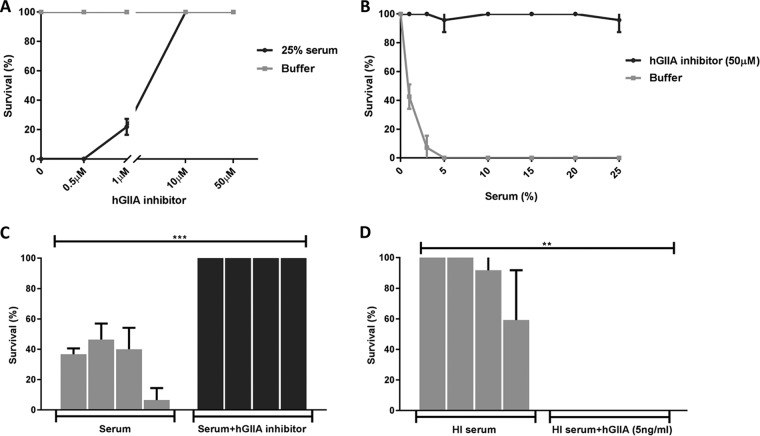
Next, we decided to test the role of hGIIA (14 kDa), the catalytic function of which is calcium dependent (27). hGIIA belongs to the secreted phospholipases A2 (PLA2) family of enzymes present in various genomes, from humans to snakes, including invertebrates, plants, fungi, and even bacteria, that hydrolyze membrane phospholipids at the sn-2 position (28). Although immune cell-derived hGIIA was previously identified as a bactericidal component against several Gram-positive bacteria (29), hGIIA concentrations in normal serum are much lower than the reported bactericidal concentrations for most Gram-positive bacteria (30–33). In our pooled human serum, we observed a mean concentration of hGIIA of 3.8 ng/ml (0.3 nM), similar to concentrations described previously (31, 34) (see Fig. S3 in the supplemental material). Nevertheless, we tested whether a specific hGIIA inhibitor (LY311727; Sigma-Aldrich) could block killing of commensal E. faecium in human serum. We observed that the inhibitor blocks the killing activity in a dose-dependent manner (Fig. 3A) and at different concentrations of human serum (Fig. 3B). Moreover, the hGIIA inhibitor blocked killing of all tested commensal E. faecium strains (Fig. 3C). Conversely, we were able to restore killing of commensal E. faecium in heat-inactivated serum by reconstituting serum with pure recombinant human hGIIA (Fig. 3D). Although purified hGIIA alone in buffer (RPMI) also kills E1007, we observed somewhat better activity in the presence of heat-inactivated human serum (see Fig. S4A in the supplemental material). The MIC of hGIIA for E1007 is around 3 ng/ml, which is equivalent to the hGIIA range in normal serum (1 to 5 ng/ml). At these concentrations, we do not see killing of E1162. At the hGIIA concentration seen in inflamed serum (>100 ng/ml), killing of E1162 was observed, albeit still at a lower efficiency than for E1007 (see Fig. S4B in the supplemental material). In summary, we found that hGIIA is the key player in killing of commensal E. faecium strains by normal human serum.
FIG 3.
hGIIA is essential for serum killing of commensal E. faecium. (A) Killing of commensal E. faecium strain E1007 in 25% pooled human serum is inhibited by hGIIA inhibitor (LY311727) in a dose-dependent manner. (B) Killing of commensal E. faecium strain E1007 in different concentrations of human serum incubated with or without hGIIA inhibitor (LY311727). (C) Killing of E. faecium commensal strains E1050, 1590, E3548, and E1007 in 25% pooled human serum with or without 50 μM hGIIA inhibitor (LY311727). (D) Killing of E. faecium commensal strains E1050, 1590, E3548, and E1007 in 25% pooled heat-inactivated (HI) human serum without or with recombinant hGIIA. Killing was quantified by bacterial viability on blood agar plates after 30 min of incubation under the different conditions. Each bar represents the mean ± SD from three independent experiments. Statistical significance was determined using multiple t tests (***, P ≤ 0.0005; **, P ≤ 0.01).
Serum hGIIA causes membrane destabilization in commensal E. faecium.
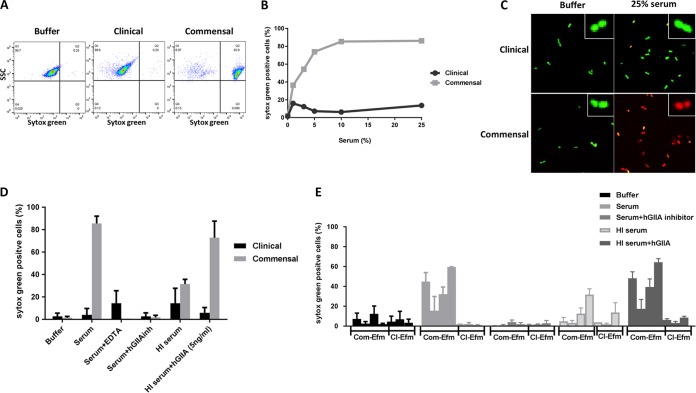
Finally, we studied how hGIIA in serum causes bacterial killing. hGIIA acts by hydrolyzing membrane phospholipids in bacterial membranes, whereas host cells are highly resistant to its activity at normal physiological concentrations (30). Here, we used a flow-cytometric approach to study whether serum hGIIA can induce membrane damage to commensal E. faecium strains. Bacteria were incubated with human serum in the presence of the DNA dye Sytox green, which binds DNA and RNA only when bacterial membranes are damaged (35). While untreated E. faecium cells remained Sytox negative, an average of 86.2% of the commensal E. faecium population became Sytox positive after 30 min of incubation with serum concentrations equal to or greater than 10% (Fig. 4B). No increase in Sytox intensity was observed in the clinical strain E1162 (Fig. 4A and B). Flow-cytometric quantification of Sytox green influx showed a concentration-dependent effect of serum in inducing membrane damage of commensal but not clinical E. faecium strains (Fig. 4B). Confocal microscopy confirmed membrane damage of the commensal E. faecium strain E1007 but not clinical strain E1162 by human serum (Fig. 4C) with different DNA dyes (Syto9 and propidium iodide). Furthermore, we found that the serum-induced membrane permeabilization of strain E1007 was mediated by hGIIA, since EDTA and hGIIA inhibition blocked the observed Sytox green influx (Fig. 4D). Finally, we found that all tested commensal but not clinical E. faecium isolates were sensitive to membrane permeabilization via hGIIA in human serum (Fig. 4E).
FIG 4.
Serum hGIIA causes membrane permeabilization in commensal E. faecium. (A) Representative flow cytometry plots of Sytox green influx (switch to the right) of commensal E. faecium isolate E1007 incubated with RPMI (buffer) and clinical (E1162) and commensal (E1007) strains incubated with 25% pooled human serum. (B) Sytox green influx in E1007 and E1162 in different concentrations of human serum. (C) Confocal pictures of clinical (E1162) and commensal (E1007) E. faecium strains incubated with RPMI (buffer) or 25% human serum in the presence of Syto9 (in green, representing live cells) and propidium iodide (in red, representing damaged bacterial membrane). E. faecium membrane permeabilization of clinical strain E1162 and commensal strain E1007 was measured by Sytox green influx under the described conditions. (D) Sytox green influx upon incubation in serum in the absence or presence of hGIIA inhibitors and upon restoration in heat-inactivated (HI) human serum with recombinant hGIIA. (E) Sytox green influx in E. faecium clinical (Cl) strains E2560, E0745, E1162, and commensal (Com) strains E1050, E1590, E3548, and E1007 in (from left to right) RPMI (buffer), 25% pooled human serum without or with 50 μM hGIIA inhibitor (LY311727), or heat-inactivated (HI) human serum without or with 5 ng/ml recombinant hGIIA. Each bar represents the mean ± SD from three independent experiments. Com, commensal; Cl, clinical; Efm, E. faecium.
Altogether, we conclude that hGIIA is the principal component of human serum that effectively kills commensal E. faecium at low concentrations (in the subnanomolar range) by destabilization of the bacterial membrane.
DISCUSSION
Novel anti-infective strategies are pivotal to curtailing the emergence of multidrug-resistant pathogens. Besides development of direct antibacterial compounds, drugs acting at the level of interaction between microbes and the immune system might be promising. Previous studies revealed that E. faecium can have distinct lifestyles (commensal and pathogenic), which are represented by two distinct genomic clades that differ in genetic polymorphisms and gene repertoire (9, 10). Here we identify that these two clades display distinct susceptibilities to killing by normal human serum. Given the genetic variances in genes that are conserved in commensal and clinical strains as well as the differences in gene repertoire between clinical and commensal E. faecium strains, it is likely that gene expression and/or differences in gene content (a result of gene gain and loss) explain this difference between commensal and clinical strains in resistance to hGIIA. Further investigation is needed to identify the bacterial factors that contribute to E. faecium resistance or susceptibility to serum, but genes related to changes in bacterial surface charge and involved in lipid synthesis in bacteria (36, 37) would be good candidates to be investigated, since hGIIA binding is directly influenced by the negative charge of the bacterial surface and phospholipid modifications, as demonstrated in Staphylococcus aureus (30, 38). We hypothesize that the development of resistance to human serum may have contributed to the decisive step for E. faecium to evolve into a relevant pathogen. On the other hand, the ability of hGIIA to kill commensal Gram-positives can be an evolutionary strategy of the innate immune system to contain commensal bacteria in their specific niches and preclude invasion of sterile tissues. Furthermore, we believe that our findings could also be relevant for E. faecium survival in the intestine, since several lines of evidence support the idea that hGIIA (and other phospholipases) is expressed in the intestine (39). Interestingly, hGIIA concentrations were found to be higher in the small intestine than in the large intestine, the latter being the niche in which E. faecium is found in high abundance after antibiotic treatment.
Our study also highlights an important role for hGIIA in the humoral immune response against E. faecium. While other groups reported that high concentrations of hGIIA (produced locally in lungs and tears [40, 41] or in serum under septic shock conditions [42, 43]) could disrupt certain Gram-positive bacteria, its importance in normal serum or plasma has not been recognized in physiological concentrations, besides its activity against Listeria monocytogenes (34). We observed restoration of killing of commensal E. faecium when recombinant hGIIA was added in physiological concentrations in heat-inactivated human serum. Although our data indicate that hGIIA is the major factor in serum responsible for the killing phenotype observed against commensal E. faecium strains, further studies are necessary to identify the specific conditions in serum that facilitate hGIIA action.
hGIIA is known for its ability to hydrolyze bacterial membrane phospholipids, which is a major structural component of the bacterial cell wall (27). Its relevance during infections stems from in vivo studies, in which mice overexpressing human hGIIA control infections by group B Streptococcus, S. aureus, and Bacillus anthracis better than their control littermates do (43–46). Furthermore, the fact that pathogenic Gram-positive bacteria developed resistance against hGIIA is an indication for its relevance in vivo (46, 47). Future work is needed to unravel whether serum sensitivity to hGIIA-mediated killing is specific for commensal E. faecium strains or whether other nonpathogenic Gram positives may be killed by normal human serum as well. Thus far, nonpathogenic Gram positives appear to be resistant to killing by serum (21). Only B. anthracis was previously shown to be sensitive to normal serum; however, this was fully dependent on growth inhibition by transferrin-mediated iron deprivation (48). hGIIA is an acute-phase reactant protein, whose concentration in serum upon bacterial infection can increase up to 100-fold, which is high enough to kill some Gram-positive pathogens (31). Since hGIIA is able to kill commensal variants of a clinically relevant multiresistant pathogen, and since hGIIA resistance can be an important mechanism of bacterial escape, strategies based on the use of components of the innate immune system, such as hGIIA and even its variants, may be developed to fight against multidrug-resistant bacteria. In conclusion, the findings presented here not only provide fundamental knowledge about how the innate immune system kills bacterial cells but also open up new therapeutic routes to boost immune clearance of bacteria through conversion of resistance to serum.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The 19 E. faecium strains used in this study are from our laboratory collection (Department of Medical Microbiology, UMC Utrecht) and were previously characterized bacteria isolated from healthy and hospitalized humans or animals (Table 1) (9, 49). A phylogenetic tree of the selected strains was constructed based on 1,423 core genes as described by de Been et al. (50). E. faecium was grown at 37°C for 24 h in Trypticase soy agar II (TSA) plates supplemented with 5% sheep blood (BD Biosciences) and tryptic soy broth media (TSB; Oxoid) when indicated.
Serum, plasma, and inhibitors.
Normal human serum (HS) was generated at the Department of Medical Microbiology, UMC Utrecht. As previously described (21), whole blood was drawn via venous puncture from 20 healthy volunteers, who provided written informed consent in accordance with the Declaration of Helsinki; approval was obtained from the medical ethics committee of the UMC Utrecht. Following collection via venous puncture, blood was clotted for 15 min at room temperature. Blood was centrifuged (10 min at 2,700 × g at 4°C), and serum (supernatant) was collected, pooled, and frozen in small aliquots before storage at −80°C. Heat-inactivated (HI) serum was prepared by incubating serum at 56°C for 30 min. When indicated, EDTA (10 mM), C3 inhibitor Cp40 (24) (10 μg/ml), C5 inhibitor OmCI (25) (10 μg/ml), or group IIA secreted phospholipase A2 (hGIIA) inhibitor LY311727 (50 μM; Sigma-Aldrich Ltd.) was used. Pure recombinant hGIIA was prepared in Escherichia coli, without a tag, and full activity and purity were confirmed as described previously (46, 51). hGIIA concentration in human serum was quantified using the human sPLA2-IIA enzyme-linked immunosorbent assay (ELISA) kit from Cayman Chemicals (Ann Arbor, MI).
Serum bactericidal assays.
E. faecium strains were grown in 4 ml TSB to an optical density at 660 nm (OD660) of 0.4 from an overnight culture in TSA plates supplemented with 5% sheep blood (BD Biosciences). Bacteria were centrifuged and resuspended in sterile RPMI (Gibco). For each specific condition, 105 bacteria were incubated with human serum (at the indicated concentrations, with or without inhibitors) in sterile round-bottom 96-well plates (Greiner) under shaking conditions for 30 min at 37°C. Killing was evaluated by serial dilution of samples in RPMI and subsequent plating onto TSA plates supplemented with 5% sheep blood. Following overnight incubation at 37°C, surviving bacteria were quantified by counting the CFU. Killing was measured by comparison of total CFU after 30 min of incubation in the control (RPMI) in relation to the CFU in 25% human serum or other described conditions. Bactericidal assays were performed at least in duplicate for each condition. When indicated, the statistical difference between results under different conditions was calculated using the Kruskal-Wallis test followed by Dunn's multiple-comparison test (Fig. 1B and 2) or multiple t tests (Fig. 3C and D). Statistical analysis was performed in Prism GraphPad.
Bacterial membrane permeabilization.
Bacterial membrane permeability was analyzed by flow cytometry and confocal microscopy. Following incubation of E. faecium with serum, the membrane-impermeable nucleic acid dye Sytox green (1:5,000, vol/vol; Life Technology) was added to the samples. Sytox green staining was quantified by flow cytometry on a BD FACSVerse (488-nm laser; Becton Dickinson, San Jose, CA, USA). Bacteria were gated based on forward and side scatter properties, and the fluorescence of 10,000 gated bacterial cells was quantified. Results were analyzed with FlowJo (version v10). Based on the negative-control buffer condition (RPMI), a threshold was set to determine the increase of Sytox green signal in the population. The increase of Sytox signal was represented as the percentage of the total population that was stained with Sytox green (see results in Fig. 4A for an example of flow cytometry plots). For confocal microscopy, bacteria were incubated with 25% serum as described above in the presence of Syto9 (1.5:1,000, vol/vol) and propidium iodide (1.5:1,000, vol/vol) (both from the BacLight bacteria viability kit; Life Technologies). After 10 min of incubation at room temperature, bacteria were mounted in a glass slide with ProLong antifade mountant (Life Technologies), followed by fluorescence analysis in the confocal microscope (Leica SP5). Syto9 and propidium iodide were excited at 488 nm, and emissions were measured at 500 nm and 636 nm, respectively. Pictures were taken at a magnification of ×40 and an optical zoom of 3×.
Surface deposition of activated complement products.
Deposition of C3b molecules or C5b-9 complexes on E. faecium surfaces was quantified by flow cytometry as previously described (21). E. faecium was incubated with human serum (with or without inhibitors) for 30 min at 37°C with shaking. After washing, bacteria were incubated with goat fluorescein isothiocyanate (FITC)-conjugated anti-C3 antibody (1 μg/ml; Protos Immunoresearch) or mouse anti-C5b-9 antibody (1 μg/ml; aE11; Santa Cruz) in phosphate-buffered saline (PBS)–1% bovine serum albumin (BSA) for 30 min at 4°C. For C5b-9 detection, a subsequent incubation with FITC-conjugated goat anti-mouse IgG antibody (1 μg/ml; Dako) was performed (30 min at 4°C). Bacteria were washed once more, and the fluorescence of 10,000 gated bacteria was quantified by flow cytometry using a FACSVerse flow cytometer (Becton Dickinson, San Jose, CA, USA) (52).
Supplementary Material
ACKNOWLEDGMENTS
We thank Susan Lea from University of Oxford for kindly providing OmCI and John Lambris, University of Pennsylvania, Philadelphia, for kindly providing Cp40.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/IAI.00180-18.
REFERENCES
- 1.Doorduijn DJ, Rooijakkers SH, van Schaik W, Bardoel BW. 2016. Complement resistance mechanisms of Klebsiella pneumoniae. Immunobiology 221:1102–1109. doi: 10.1016/j.imbio.2016.06.014. [DOI] [PubMed] [Google Scholar]
- 2.Beutler B. 2004. Innate immunity: an overview. Mol Immunol 40:845–859. doi: 10.1016/j.molimm.2003.10.005. [DOI] [PubMed] [Google Scholar]
- 3.Brogden KA. 2005. Antimicrobial peptides: pore formers or metabolic inhibitors in bacteria? Nat Rev Microbiol 3:238–250. doi: 10.1038/nrmicro1098. [DOI] [PubMed] [Google Scholar]
- 4.Brown EJ. 1985. Interaction of gram-positive microorganisms with complement. Curr Top Microbiol Immunol 121:159–187. [DOI] [PubMed] [Google Scholar]
- 5.Mundt JO. 1963. Occurrence of Enterococci in animals in a wild environment. Appl Microbiol 11:136–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martin JD, Mundt JO. 1972. Enterococci in insects. Appl Microbiol 24:575–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.O'Driscoll T, Crank CW. 2015. Vancomycin-resistant enterococcal infections: epidemiology, clinical manifestations, and optimal management. Infect Drug Resist 8:217–230. doi: 10.2147/IDR.S54125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guzman Prieto AM, van Schaik W, Rogers MR, Coque TM, Baquero F, Corander J, Willems RJ. 2016. Global emergence and dissemination of enterococci as nosocomial pathogens: attack of the clones? Front Microbiol 7:788. doi: 10.3389/fmicb.2016.00788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lebreton F, van Schaik W, McGuire AM, Godfrey P, Griggs A, Mazumdar V, Corander J, Cheng L, Saif S, Young S, Zeng Q, Wortman J, Birren B, Willems RJ, Earl AM, Gilmore MS. 2013. Emergence of epidemic multidrug-resistant Enterococcus faecium from animal and commensal strains. mBio 4:e00534-13. doi: 10.1128/mBio.00534-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Willems RJ, Top J, van Schaik W, Leavis H, Bonten M, Siren J, Hanage WP, Corander J. 2012. Restricted gene flow among hospital subpopulations of Enterococcus faecium. mBio 3:e00151-12. doi: 10.1128/mBio.00151-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang X, Top J, de Been M, Bierschenk D, Rogers M, Leendertse M, Bonten MJ, van der Poll T, Willems RJ, van Schaik W. 2013. Identification of a genetic determinant in clinical Enterococcus faecium strains that contributes to intestinal colonization during antibiotic treatment. J Infect Dis 207:1780–1786. doi: 10.1093/infdis/jit076. [DOI] [PubMed] [Google Scholar]
- 12.Paganelli FL, Huebner J, Singh KV, Zhang X, van Schaik W, Wobser D, Braat JC, Murray BE, Bonten MJ, Willems RJ, Leavis HL. 2016. Genome-wide screening identifies phosphotransferase system permease BepA to be involved in Enterococcus faecium endocarditis and biofilm formation. J Infect Dis 214:189–195. doi: 10.1093/infdis/jiw108. [DOI] [PubMed] [Google Scholar]
- 13.Vaishnava S, Yamamoto M, Severson KM, Ruhn KA, Yu X, Koren O, Ley R, Wakeland EK, Hooper LV. 2011. The antibacterial lectin RegIIIgamma promotes the spatial segregation of microbiota and host in the intestine. Science 334:255–258. doi: 10.1126/science.1209791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leendertse M, Willems RJ, Oei GA, Florquin S, Bonten MJ, van der Poll T. 2009. Intestinal Enterococcus faecium colonization improves host defense during polymicrobial peritonitis. J Infect Dis 200:735–744. doi: 10.1086/603542. [DOI] [PubMed] [Google Scholar]
- 15.Leendertse M, Willems RJ, Giebelen IA, Roelofs JJ, Bonten MJ, van der Poll T. 2009. Neutrophils are essential for rapid clearance of Enterococcus faecium in mice. Infect Immun 77:485–491. doi: 10.1128/IAI.00863-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leendertse M, Willems RJ, Giebelen IA, Roelofs JJ, van Rooijen N, Bonten MJ, van der Poll T. 2009. Peritoneal macrophages are important for the early containment of Enterococcus faecium peritonitis in mice. Innate Immun 15:3–12. doi: 10.1177/1753425908100238. [DOI] [PubMed] [Google Scholar]
- 17.Leendertse M, Willems RJ, Giebelen IA, van den Pangaart PS, Wiersinga WJ, de Vos AF, Florquin S, Bonten MJ, van der Poll T. 2008. TLR2-dependent MyD88 signaling contributes to early host defense in murine Enterococcus faecium peritonitis. J Immunol 180:4865–4874. doi: 10.4049/jimmunol.180.7.4865. [DOI] [PubMed] [Google Scholar]
- 18.Zou J, Shankar N. 2016. Surface protein Esp enhances pro-inflammatory cytokine expression through NF-kappaB activation during enterococcal infection. Innate Immun 22:31–39. doi: 10.1177/1753425915611237. [DOI] [PubMed] [Google Scholar]
- 19.Bloes DA, Otto M, Peschel A, Kretschmer D. 2012. Enterococcus faecium stimulates human neutrophils via the formyl-peptide receptor 2. PLoS One 7:e39910. doi: 10.1371/journal.pone.0039910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Berends ET, Mohan S, Miellet WR, Ruyken M, Rooijakkers SH. 2015. Contribution of the complement membrane attack complex to the bactericidal activity of human serum. Mol Immunol 65:328–335. doi: 10.1016/j.molimm.2015.01.020. [DOI] [PubMed] [Google Scholar]
- 21.Berends ET, Dekkers JF, Nijland R, Kuipers A, Soppe JA, van Strijp JA, Rooijakkers SH. 2013. Distinct localization of the complement C5b-9 complex on Gram-positive bacteria. Cell Microbiol 15:1955–1968. doi: 10.1111/cmi.12170. [DOI] [PubMed] [Google Scholar]
- 22.Joisel F, Leroux-Nicollet I, Lebreton JP, Fontaine M. 1983. A hemolytic assay for clinical investigation of human C2. J Immunol Methods 59:229–235. doi: 10.1016/0022-1759(83)90035-2. [DOI] [PubMed] [Google Scholar]
- 23.Muller-Eberhard HJ. 1969. Complement. Annu Rev Biochem 38:389–414. doi: 10.1146/annurev.bi.38.070169.002133. [DOI] [PubMed] [Google Scholar]
- 24.Sahu A, Kay BK, Lambris JD. 1996. Inhibition of human complement by a C3-binding peptide isolated from a phage-displayed random peptide library. J Immunol 157:884–891. [PubMed] [Google Scholar]
- 25.Nunn MA, Sharma A, Paesen GC, Adamson S, Lissina O, Willis AC, Nuttall PA. 2005. Complement inhibitor of C5 activation from the soft tick Ornithodoros moubata. J Immunol 174:2084–2091. doi: 10.4049/jimmunol.174.4.2084. [DOI] [PubMed] [Google Scholar]
- 26.Nevalainen TJ, Cardoso JC, Riikonen PT. 2012. Conserved domains and evolution of secreted phospholipases A(2). FEBS J 279:636–649. doi: 10.1111/j.1742-4658.2011.08453.x. [DOI] [PubMed] [Google Scholar]
- 27.van den Bosch H. 1980. Intracellular phospholipases A. Biochim Biophys Acta 604:191–246. [DOI] [PubMed] [Google Scholar]
- 28.Lambeau G, Gelb MH. 2008. Biochemistry and physiology of mammalian secreted phospholipases A2. Annu Rev Biochem 77:495–520. doi: 10.1146/annurev.biochem.76.062405.154007. [DOI] [PubMed] [Google Scholar]
- 29.Foreman-Wykert AK, Weinrauch Y, Elsbach P, Weiss J. 1999. Cell-wall determinants of the bactericidal action of group IIA phospholipase A2 against Gram-positive bacteria. J Clin Invest 103:715–721. doi: 10.1172/JCI5468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weiss JP. 2015. Molecular determinants of bacterial sensitivity and resistance to mammalian group IIA phospholipase A2. Biochim Biophys Acta 1848:3072–3077. doi: 10.1016/j.bbamem.2015.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nevalainen TJ, Eerola LI, Rintala E, Laine VJ, Lambeau G, Gelb MH. 2005. Time-resolved fluoroimmunoassays of the complete set of secreted phospholipases A2 in human serum. Biochim Biophys Acta 1733:210–223. doi: 10.1016/j.bbalip.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 32.Nevalainen TJ, Graham GG, Scott KF. 2008. Antibacterial actions of secreted phospholipases A2. Rev Biochim Biophys Acta 1781:1–9. doi: 10.1016/j.bbalip.2007.12.001. [DOI] [PubMed] [Google Scholar]
- 33.Wu Y, Raymond B, Goossens PL, Njamkepo E, Guiso N, Paya M, Touqui L. 2010. Type-IIA secreted phospholipase A2 is an endogenous antibiotic-like protein of the host. Biochimie 92:583–587. doi: 10.1016/j.biochi.2010.01.024. [DOI] [PubMed] [Google Scholar]
- 34.Gronroos JO, Laine VJ, Nevalainen TJ. 2002. Bactericidal group IIA phospholipase A2 in serum of patients with bacterial infections. J Infect Dis 185:1767–1772. doi: 10.1086/340821. [DOI] [PubMed] [Google Scholar]
- 35.Lebaron P, Catala P, Parthuisot N. 1998. Effectiveness of SYTOX Green stain for bacterial viability assessment. Appl Environ Microbiol 64:2697–2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lin Y, Bogdanov M, Lu S, Guan Z, Margolin W, Weiss JP, Zheng L. 2018. The phospholipid-repair system LplT/Aas in Gram-negative bacteria protects the bacterial membrane envelope from host phospholipase A2 attack. J Biol Chem 293:3386–3398. doi: 10.1074/jbc.RA117.001231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Arduino RC, Jacques-Palaz K, Murray BE, Rakita RM. 1994. Resistance of Enterococcus faecium to neutrophil-mediated phagocytosis. Infect Immun 62:5587–5594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Koprivnjak T, Peschel A, Gelb MH, Liang NS, Weiss JP. 2002. Role of charge properties of bacterial envelope in bactericidal action of human group IIA phospholipase A2 against Staphylococcus aureus. J Biol Chem 277:47636–47644. doi: 10.1074/jbc.M205104200. [DOI] [PubMed] [Google Scholar]
- 39.Surrel F, Jemel I, Boilard E, Bollinger JG, Payre C, Mounier CM, Talvinen KA, Laine VJ, Nevalainen TJ, Gelb MH, Lambeau G. 2009. Group X phospholipase A2 stimulates the proliferation of colon cancer cells by producing various lipid mediators. Mol Pharmacol 76:778–790. doi: 10.1124/mol.108.053371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Qu XD, Lehrer RI. 1998. Secretory phospholipase A2 is the principal bactericide for Staphylococci and other gram-positive bacteria in human tears. Infect Immun 66:2791–2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Saari KM, Aho V, Paavilainen V, Nevalainen TJ. 2001. Group II PLA(2) content of tears in normal subjects. Invest Ophthalmol Vis Sci 42:318–320. [PubMed] [Google Scholar]
- 42.Pernet E, Brunet J, Guillemot L, Chignard M, Touqui L, Wu Y. 2015. Staphylococcus aureus adenosine inhibits sPLA2-IIA-mediated host killing in the airways. J Immunol 194:5312–5319. doi: 10.4049/jimmunol.1402665. [DOI] [PubMed] [Google Scholar]
- 43.Pernet E, Guillemot L, Burgel PR, Martin C, Lambeau G, Sermet-Gaudelus I, Sands D, Leduc D, Morand PC, Jeammet L, Chignard M, Wu Y, Touqui L. 2014. Pseudomonas aeruginosa eradicates Staphylococcus aureus by manipulating the host immunity. Nat Commun 5:5105. doi: 10.1038/ncomms6105. [DOI] [PubMed] [Google Scholar]
- 44.Piris-Gimenez A, Paya M, Lambeau G, Chignard M, Mock M, Touqui L, Goossens PL. 2005. In vivo protective role of human group IIa phospholipase A2 against experimental anthrax. J Immunol 175:6786–6791. doi: 10.4049/jimmunol.175.10.6786. [DOI] [PubMed] [Google Scholar]
- 45.Movert E, Wu Y, Lambeau G, Touqui L, Areschoug T. 2011. A novel bacterial resistance mechanism against human group IIA-secreted phospholipase A2: role of Streptococcus pyogenes sortase A. J Immunol 187:6437–6446. doi: 10.4049/jimmunol.1100499. [DOI] [PubMed] [Google Scholar]
- 46.Movert E, Wu Y, Lambeau G, Kahn F, Touqui L, Areschoug T. 2013. Secreted group IIA phospholipase A2 protects humans against the group B Streptococcus: experimental and clinical evidence. J Infect Dis 208:2025–2035. doi: 10.1093/infdis/jit359. [DOI] [PubMed] [Google Scholar]
- 47.Koprivnjak T, Weidenmaier C, Peschel A, Weiss JP. 2008. Wall teichoic acid deficiency in Staphylococcus aureus confers selective resistance to mammalian group IIA phospholipase A(2) and human beta-defensin 3. Infect Immun 76:2169–2176. doi: 10.1128/IAI.01705-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rooijakkers SH, Rasmussen SL, McGillivray SM, Bartnikas TB, Mason AB, Friedlander AM, Nizet V. 2010. Human transferrin confers serum resistance against Bacillus anthracis. J Biol Chem 285:27609–27613. doi: 10.1074/jbc.M110.154930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Paganelli FL, de Been M, Braat JC, Hoogenboezem T, Vink C, Bayjanov J, Rogers MR, Huebner J, Bonten MJ, Willems RJ, Leavis HL. 2015. Distinct SagA from hospital-associated clade A1 Enterococcus faecium strains contributes to biofilm formation. Appl Environ Microbiol 81:6873–6882. doi: 10.1128/AEM.01716-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.de Been M, Pinholt M, Top J, Bletz S, Mellmann A, van Schaik W, Brouwer E, Rogers M, Kraat Y, Bonten M, Corander J, Westh H, Harmsen D, Willems RJ. 2015. Core genome multilocus sequence typing scheme for high-resolution typing of Enterococcus faecium. J Clin Microbiol 53:3788–3797. doi: 10.1128/JCM.01946-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ghomashchi F, Brglez V, Payre C, Jeammet L, Bezzine S, Gelb MH, Lambeau G. 2017. Preparation of the full set of recombinant mouse- and human-secreted phospholipases A2. Methods Enzymol 583:35–69. doi: 10.1016/bs.mie.2016.10.034. [DOI] [PubMed] [Google Scholar]
- 52.Berends ET, Gorham RD Jr, Ruyken M, Soppe JA, Orhan H, Aerts PC, de Haas CJ, Gros P, Rooijakkers SH. 2015. Molecular insights into the surface-specific arrangement of complement C5 convertase enzymes. BMC Biol 13:93. doi: 10.1186/s12915-015-0203-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang X, de Maat V, Guzman Prieto AM, Prajsnar TK, Bayjanov JR, de Been M, Rogers MRC, Bonten MJM, Mesnage S, Willems RJL, van Schaik W. 2017. RNA-seq and Tn-seq reveal fitness determinants of vancomycin-resistant Enterococcus faecium during growth in human serum. BMC Genomics 18:893. doi: 10.1186/s12864-017-4299-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.van Schaik W, Top J, Riley DR, Boekhorst J, Vrijenhoek JE, Schapendonk CM, Hendrickx AP, Nijman IJ, Bonten MJ, Tettelin H, Willems RJ. 2010. Pyrosequencing-based comparative genome analysis of the nosocomial pathogen Enterococcus faecium and identification of a large transferable pathogenicity island. BMC Genomics 11:239. doi: 10.1186/1471-2164-11-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.