ABSTRACT
The type VI secretion system (T6SS) is a macromolecular machine that delivers protein effectors into host cells and/or competing bacteria. The effectors may be delivered as noncovalently bound cargo of T6SS needle proteins (VgrG/Hcp/PAAR) or as C-terminal extensions of these proteins. Many Acinetobacter baumannii strains produce a T6SS, but little is known about the specific effectors or how they are delivered. In this study, we show that A. baumannii AB307-0294 encodes three vgrG loci, each containing a vgrG gene, a T6SS toxic effector gene, and an antitoxin/immunity gene. Each of the T6SS toxic effectors could kill Escherichia coli when produced in trans unless the cognate immunity protein was coproduced. To determine the role of each VgrG in effector delivery, we performed interbacterial competitive killing assays using A. baumannii AB307-0294 vgrG mutants, together with Acinetobacter baylyi prey cells expressing pairs of immunity genes that protected against two toxic effectors but not a third. Using this approach, we showed that AB307-0294 produces only three T6SS toxic effectors capable of killing A. baylyi and that each VgrG protein is specific for the carriage of one effector. Finally, we analyzed a number of A. baumannii genomes and identified significant diversity in the range of encoded T6SS VgrG and effector proteins, with correlations between effector types and A. baumannii global clone lineages.
KEYWORDS: Acinetobacter baumannii, type VI secretion system, antibacterial toxins, effectors, effector functions
INTRODUCTION
The Acinetobacter genus encompasses a diverse group of pathogenic and environmental Gram-negative bacilli. Among this genus, the species Acinetobacter baumannii has risen to prominence as a major, globally distributed, hospital-acquired pathogen. This species is responsible for a range of nosocomial infections, primarily pneumonia but also wound and burn infections, urinary tract infections, meningitis, and sepsis. Increasingly, multidrug-resistant A. baumannii strains are being isolated and some strains display resistance to all clinically relevant antibiotics (1, 2). The rapid evolution of A. baumannii has led to the emergence of a large number of strains; however, the species is currently dominated by two globally distributed clonal lineages, international clone I (IC1) and international clone II (IC2) (3).
Recently, a number of groups have described the activity of a type VI secretion system (T6SS) in A. baumannii and other Acinetobacter species (4–9). The T6SS is a recently discovered Gram-negative secretion system that is widespread among the proteobacteria (10). The T6SS apparatus is functionally analogous to the T4 bacteriophage tail spike and is typically comprised of 13 highly conserved core proteins. The T6SS apparatus consists of a baseplate (comprised of TssAEFGK), a membrane-linked stabilizing structure (TssMJL), a contractile sheath (TssBC), a cytoplasmic sheath recycling protein (TssH), and an injectable needle (Hcp/TssD) that is capped by a puncturing tip (VgrG/TssI) (11–13). Each bacterial species/strain may also produce its own set of accessory proteins; these may have chaperone, structural, or regulatory roles. Importantly, each species/strain produces its own arsenal of secreted T6SS effectors that have anti-host or, more commonly, antibacterial properties (14–21).
The antibacterial T6SS effectors of a number of species have been identified and include a range of proteins with diverse subcellular targets. Identified T6SS effectors include DNases, RNases, peptidoglycan hydrolases, lipases, and phospholipases (22). Regardless of the target, all currently identified antibacterial T6SS effectors are coexpressed together with a cognate immunity protein, the function of which is to prevent self-intoxication or sibling cell intoxication (16). Some effectors are translocated into target cells in the form of a translational fusion with T6SS VgrG/Hcp/PAAR needle proteins (specialized effectors) while others are delivered as noncovalently bound cargo (cargo effectors) (22). Several studies have also shown an association between the Hcp needle protein and specific effector proteins (23–25). In Pseudomonas aeruginosa, several effectors are protected from proteolytic degradation by Hcp, including Tse2, which localizes to the inner surface of the Hcp needle, suggesting a chaperone role for Hcp in effector translocation (25). A common delivery method for cargo effectors is via attachment to a specific VgrG protein present at the tip of the needle apparatus. In a number of species this intimate effector/VgrG association has resulted in “evolved” VgrG proteins that contain the effector in the form of a domain at the C-terminal end (26). Stand-alone effector proteins are often encoded on the chromosome immediately adjacent to a gene that encodes a VgrG protein that is required for the secretion of that specific effector (6, 27–29). The loading of effectors onto the T6SS apparatus is dependent upon a variety of chaperones or linker proteins. Attachment of effector proteins to VgrG proteins is often facilitated by the presence of a PAAR motif that may be found on an independent protein or as a domain within the effector protein itself (6, 24, 29–31). An additional conserved motif, termed MIX (marker for type six effectors) has recently been identified in a class of T6SS effectors and is hypothesized to be involved in the attachment of effectors to the secretion apparatus (32). Effector proteins containing the MIX domain are often found downstream of genes encoding proteins with the conserved domain DUF4123 (33, 34). In Serratia marcescens, the effector-associated protein EagR1 is essential for the antibacterial activity of the effector protein Rhs1, and the gene encoding this protein is immediately upstream of rhs1. However, the specific function of EagR1 is unknown (19).
The T6SSs of A. baumannii and of the related species Acinetobacter baylyi and Acinetobacter nosocomialis have been demonstrated to possess antibacterial activity against laboratory Escherichia coli strains (4, 5, 7–9, 35). Furthermore, A. baumannii is capable of using its T6SS to give a competitive advantage against other A. baumannii strains as well as against Klebsiella pneumoniae and P. aeruginosa, species with which it shares the hospital environment and forms mixed infections (4, 5, 7, 27). Recently, it was shown that A. baylyi contains five effector and immunity pairs although only four of these effectors caused death of E. coli cells during competition (8). Currently, only one A. baumannii T6SS effector has been described, called Tse3AB, which is encoded by strain ATCC 17978 and is a homolog of the A. baylyi T6SS effector Tse2 (8, 9). Therefore, much remains to be understood regarding the different A. baumannii T6SS effectors that give a competitive advantage against rival bacteria, as well as the secretion mechanisms used by different effectors. In this study, we analyzed the ability of the A. baumannii IC1 isolate AB307-0294 to outcompete E. coli, A. baylyi, and other A. baumannii strains in a T6SS-dependent manner. Furthermore, we describe novel A. baumannii T6SS toxic effectors and demonstrate the importance of specific VgrG proteins for effector secretion.
RESULTS
Identification of T6SS-encoding genes in A. baumannii strain AB307-0294.
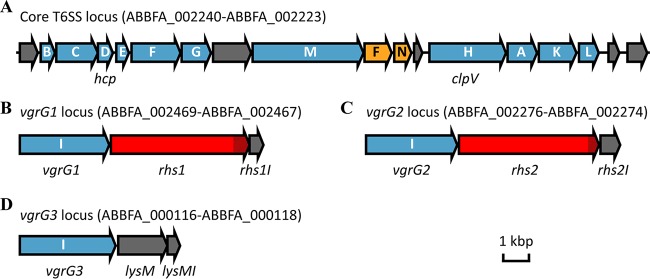
In most bacterial species that produce a functional T6SS, the genes encoding the 13 core proteins required for T6SS apparatus assembly are located within a single locus. Also within the T6SS locus are genes encoding proteins that are predicted to have T6SS accessory, regulatory, or effector roles. In most A. baumannii strains, including strain AB307-0294, the T6SS locus contains 18 genes (Fig. 1A; ABBFA_002240 to ABBFA_002223) (35, 36). These genes encode homologs of the conserved core components TssB, TssC, Hcp (also known as TssD), TssE, TssF, TssG, TssM, TssH (also known as ClpV), TssA, TssK, and TssL. Notably, the A. baumannii T6SS locus lacks a gene encoding a homolog of the conserved TssJ, an outer membrane lipoprotein that is considered a T6SS core protein in other bacteria (35, 37). Also present in the A. baumannii T6SS locus are two T6SS-associated genes (tagF and tagN) and five genes encoding proteins with no known homologs outside the Acinetobacter genus (ABFFA_002240, ABFFA_002233, ABFFA_002229, ABFFA_002224, and ABFFA_002223).
FIG 1.
Schematic representation of the genetic organization of the T6SS locus (A) and the T6SS-associated vgrG loci (B, C, and D) in A. baumannii AB307-0294. Each gene is labeled with its single-letter identifier, and the tss or tag prefixes have been omitted for clarity. Locus tags for each region are also shown. The tss genes (in blue) encode T6SS conserved components. These include three tssI genes, each encoding a VgrG protein, that we have named vgrG1, vgrG2, and vgrG3. The tag genes (in yellow) encode nonconserved, T6SS-associated proteins. Genes unique to the Acinetobacter genus are shown in gray and include the genes encoding the effector protein LysM and the three T6SS immunity proteins, LysMI, Rhs1I, and Rhs2I. Genes encoding T6SS effector proteins with shared identity to known Rhs family T6SS effector proteins in other bacteria are shown in red with the cytotoxic C-terminal region hatched.
Bioinformatic analysis of the AB307-0294 genome revealed three genes (ABBFA_002469, ABBFA_002276, and ABBFA_000116) that encoded proteins with high identity to VgrG (TssI) proteins, which form the needle tip of the T6SS in other species (27–29). Accordingly, these genes were named vgrG1 (ABBFA_002469), vgrG2 (ABBFA_002276), and vgrG3 (ABBFA_000116) (Fig. 1B to D). Amino acid sequence alignments of the three AB307-0294 VgrG proteins revealed that VgrG1 and VgrG2 shared the highest identity (76%), followed by VgrG1 and VgrG3 (69%) and VgrG2 and VgrG3 (66%). Each vgrG gene was the first gene within a three-gene locus. The second genes in each of the vgrG1 and vgrG2 loci (annotated as ABBFA_002468 and ABBFA_002275, respectively) encoded proteins with shared identity to Rhs T6SS effector proteins in P. aeruginosa and S. marcescens (19, 31, 38). Accordingly, these genes were named rhs1 and rhs2, respectively (Fig. 1B and C). In contrast, the second gene in the vgrG3 locus encoded a protein with significant identity to the T6SS LysM family of peptidoglycan hydrolases. Accordingly, we named this gene lysM (Fig. 1D). In other bacteria, such as P. aeruginosa, genes encoding the T6SS effectors are immediately followed by genes encoding cognate immunity proteins (39). Based on this knowledge and on the similar arrangement of the vgrG genes in A. baumannii, we predicted that the genes ABBFA_002467, ABBFA_002274, and ABBFA_001118 each encoded an immunity protein and were thus named rhs1I, rhs2I, and lysMI, respectively (Fig. 1B to D).
The T6SS of A. baumannii strain AB307-0294 is required for active secretion of Hcp.
In order to investigate the T6SS of A. baumannii strain AB307-0294, we first generated a T6SS-deficient strain by replacing tssM, which encodes an essential component of the T6SS membrane-stabilizing structure, with a kanamycin resistance marker using allelic exchange. There was no significant difference in the growth rate of the ΔtssM strain compared to that of the parent strain AB307-0294 under normal laboratory growth conditions (data not shown). As the secretion of Hcp is often used as a marker for a functional T6SS (40), we determined if wild-type (WT) AB307-0294 and AB307-0294 ΔtssM were able to secrete Hcp by analyzing cell-free culture supernatants and whole-cell lysates for the presence of Hcp using anti-Hcp antibodies (Fig. 2). The T6SS Hcp protein, approximately 18.7 kDa in size, was detected in the whole-cell lysates and in the culture supernatant generated from the WT culture, indicating that AB307-0294 actively secretes Hcp under normal laboratory growth conditions. Hcp protein was also detected in whole-cell lysates generated from the AB307-0294 ΔtssM culture, proving that this strain still produced Hcp. However, Hcp was not detected in the AB307-0294 ΔtssM culture supernatant, demonstrating that the tssM mutant lacked a functional T6SS. Importantly, secretion of Hcp was restored when the mutant strain was provided with an intact copy of tssM on a plasmid but not when the mutant was provided with vector only (Fig. 2).
FIG 2.

Analysis of Hcp secretion by the AB307-0294 wild-type, tssM mutant, and complemented strains. Western immunoblotting was performed using an Hcp-specific antiserum of cell-free supernatants (top panel) and whole-cell lysates (bottom panel) from monocultures containing the following strains: the AB307-0294 wild type (WT), the ΔtssM mutant, the ΔtssM mutant with empty vector, and the ΔtssM mutant provided with a functional copy of tssM on a plasmid, as indicated. The position of the Hcp protein is shown at the left. The position of the 16-kDa marker from a SeeBlue Plus2 prestained protein standard (Invitrogen) is indicated on the right.
A. baumannii strain AB307-0294 out-competes E. coli in a T6SS-dependent manner.
A number of A. baumannii strains have been shown to use the T6SS to gain a competitive advantage against E. coli (5, 7), and therefore we investigated this phenotype for strain AB307-0294 under standard laboratory growth conditions. E. coli DH10B cells, grown at 37°C to mid-log phase in lysogeny broth (LB), were mixed at a ratio of 1:10 with A. baumannii AB307-0294 WT, AB307-0294 ΔtssM, AB307-0294 ΔtssM provided with an intact copy of tssM, or the AB307-0294 ΔtssM strain with vector only, and the cocultures were incubated on solid LB medium at 37°C. The number of surviving bacteria was then determined by serially diluting the cells recovered from the coculture and plating them onto LB agar supplemented with the appropriate antibiotic for selection of either E. coli or A. baumannii (Fig. 3). Coincubation of E. coli DH10B with the AB307-0294 WT resulted in an approximately 10,000-fold drop in E. coli viability compared to that with incubation in LB medium alone. However, survival of DH10B following coincubation with the AB307-0294 ΔtssM was indistinguishable from survival of DH10B following incubation in LB medium alone. Therefore, A. baumannii can kill E. coli DH10B in a T6SS-dependent manner. Complementation of the tssM mutant with an intact copy of tssM restored its ability to kill E. coli DH10B to levels similar to the level observed following incubation with the WT strain. No difference in recovery of the A. baumannii strains was observed following incubation with E. coli (data not shown).
FIG 3.
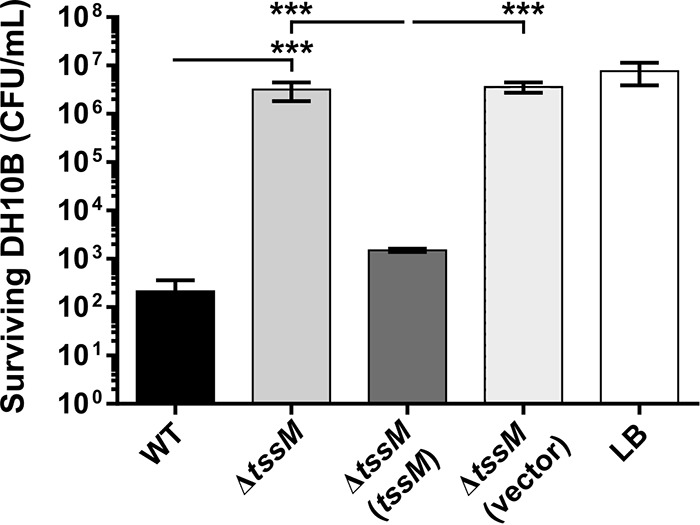
Interbacterial competitive growth assays show that a functional T6SS is essential for interbacterial killing. Values represent surviving E. coli DH10B (CFU/ml) following 4 h of coincubation with A. baumannii AB307-0294 (WT), the ΔtssM mutant, the ΔtssM mutant provided with a functional copy of tssM on a plasmid, the ΔtssM strain containing empty vector, or medium alone (LB), as indicated. Error bars represent standard errors of the means of three independent experiments performed in triplicate. ***, P < 0.001.
The T6SS of A. baumannii AB307-0294 provides a competitive advantage against some Acinetobacter strains but not others.
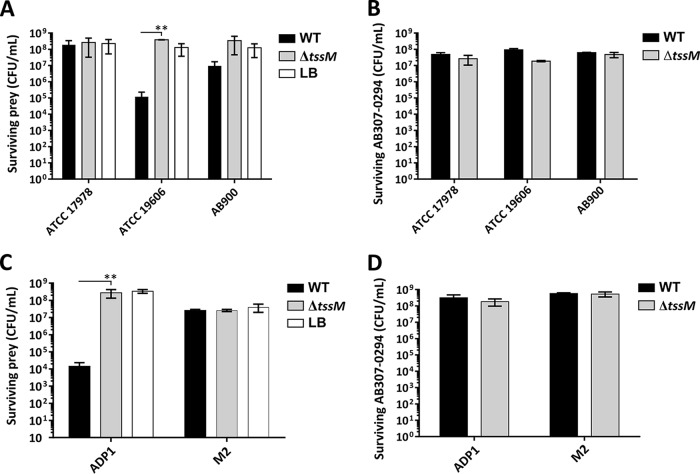
Interbacterial competitive growth assays were extended to examine the ability of A. baumannii AB307-0294 WT or AB307-0294 ΔtssM to outcompete other A. baumannii strains (ATCC 17978, ATCC 19606, and AB900) or the related species A. nosocomialis (strain M2) and A. baylyi (strain ADP1). The wild-type A. baumannii strain AB307-0294 could strongly out-compete A. baumannii strain ATCC 19606 and A. baylyi ADP1 (Fig. 4A and C). In contrast, coincubation of A. baumannii strain ATCC 19606 or A. baylyi ADP1 with AB307-0294 ΔtssM did not affect the survival of either strain, clearly proving that the ability to outcompete these species was dependent on a functional T6SS. However, no significant difference was observed in the ability of A. baumannii strains AB900 and ATCC 17978 or A. nosocomialis strain M2 to survive following competitive growth with WT AB307-0294 or AB307-0294 ΔtssM. It is known that ATCC 17978 harbors a large conjugative resistance plasmid, pAB3, which encodes T6SS-regulating genes which suppress T6SS activity (7). To determine if the presence of this plasmid influenced AB307-0294 T6SS-mediated killing of ATCC 17978, we performed interbacterial killing assays using ATCC 17978, with and without pAB3, as prey. However, AB307-0294 showed no killing activity against ATCC 17978 regardless of the presence or absence of the pAB3 plasmid (data not shown). In all of the assays, none of the strains used had an impact on the survival of the A. baumannii AB307-0294 WT or AB307-0294 ΔtssM strain (Fig. 4B and D).
FIG 4.
The role of the AB307-0294 T6SS in intraspecies and interspecies competitive growth. (A) Surviving numbers of A. baumannii prey strains (ATCC 17978, ATCC 19606, and AB900) following 24 h of coincubation with the predator A. baumannii strain AB307-0294 (WT), the AB307-0294 ΔtssM mutant, or LB medium alone, as indicated. (B) Surviving numbers of the predator strain AB307-0294 WT or tssM mutant (ΔtssM) following 24 h of coincubation with the prey strain A. baumannii ATCC 17978, ATCC 19606, or AB900. (C) Surviving numbers of the prey strain A. baylyi ADP1 or A. nosocomialis M2 following 24 h of coincubation with the predator AB307-0294 wild-type strain (WT), the ΔtssM mutant, or LB medium alone. (D) Surviving numbers of the predator strain AB307-0294 WT or ΔtssM mutant following 24 h of coincubation with the prey strain A. baylyi ADP1 or A. nosocomialis M2. Error bars represent standard errors of the means of three independent experiments performed in triplicate. **, P < 0.01.
Identification of the A. baumannii strain AB307-0294 T6SS effector proteins.
To determine if the predicted T6SS VgrG, effector, and immunity proteins were actively secreted by the A. baumannii strain AB307-0294 T6SS, quantitative high-performance liquid chromatography-tandem mass spectrometry (HPLC-MS/MS) was performed on cell-free supernatant samples obtained from the WT AB307-0294 strain and AB307-0294 ΔtssM. A total of 576 secreted proteins were identified in both of the culture supernatants obtained from the two strains (see Table S1 in the supplemental material). Fourteen proteins showed significantly decreased abundance in the AB307-0294 ΔtssM culture supernatant compared to that in the AB307-0294 WT culture supernatant (>4-fold change in secretion [less than −2.0 log2]; false discovery rate [FDR], <0.01) (Table 1), suggestive of these being directly secreted by the T6SS. These included a number of known T6SS proteins, including the T6SS structural proteins Hcp and TssM, the three predicted T6SS VgrG tip proteins VgrG1, VgrG2, and VgrG3 (ABBFA_002469, ABBFA_002276, and ABBFA_000116, respectively), and two PAAR domain-containing proteins, one encoded within the main T6SS locus (ABBFA_002229) and one encoded elsewhere on the genome (ABBFA_002520). Importantly, all three predicted toxic effector proteins, Rhs1 (ABBFA_002468), Rhs2 (ABBFA_002275), and LysM (ABBFA_000117), were significantly less abundant (233-fold, 84-fold, and 5.9-fold decreased production, respectively) in the supernatant derived from AB307-0294 ΔtssM, indicating that these three proteins are bona fide T6SS-secreted proteins. Five proteins showed significantly increased abundance in the AB307-0294 ΔtssM culture supernatant compared to levels in the AB307-0294 WT culture supernatant (Table S1).
TABLE 1.
Proteins identified as differentially secreted between AB307-0294 WT and AB307-0294 ΔtssM
| AB307-0294 locus tag | Protein name | Predicted functiona | Secretion ratio (log2)b | FDRc | No. of unique peptidesd | % Sequence coveraged |
|---|---|---|---|---|---|---|
| ABBFA_000116 | VgrG3 | T6SS VgrG3 tip protein | −8.76 | 2.4E−05 | 47 | 63.2 |
| ABBFA_000117 | LysM | T6SS LysM effector | −2.56 | 5.7E−03 | 3 | 7.9 |
| ABBFA_001718 | Outer membrane usher protein mrkC precursor | −7.79 | 1.7E−04 | 33 | 56.7 | |
| ABBFA_002011 | Tautomerase enzyme family protein | −7.52 | 2.1E−06 | 7 | 60.5 | |
| ABBFA_002229 | T6SS PAAR protein | −5.26 | 8.3E−05 | 2 | 62.1 | |
| ABBFA_002232 | TssM | T6SS structural protein | −4.65 | 1.5E−03 | 3 | 2.7 |
| ABBFA_002237 | Hcp | T6SS tube protein | −9.34 | 3.4E−07 | 21 | 94.6 |
| ABBFA_002275 | Rhs2 | T6SS Rhs effector | −6.39 | 7.4E−06 | 13 | 7.4 |
| ABBFA_002276 | VgrG2 | T6SS VgrG2 tip protein | −11.61 | 3.8E−06 | 43 | 52.1 |
| ABBFA_002468 | Rhs1 | T6SS Rhs effector | −7.87 | 7.0E−07 | 18 | 15.6 |
| ABBFA_002469 | VgrG1 | T6SS VgrG1 tip protein | −13.19 | 3.1E−05 | 47 | 66.6 |
| ABBFA_002520 | PAAR domain protein | −5.58 | 7.4E−06 | 7 | 36.9 | |
| ABBFA_003026 | GrxC | Glutaredoxin 3 | −5.32 | 4.3E−05 | 4 | 71.8 |
| ABBFA_003460 | Wzb | Protein tyrosine phosphatase, ptp | −5.51 | 3.1E−05 | 10 | 83.1 |
The predicted function of each protein was based on the results of a pBLAST search available at The National Center for Biotechnology Information.
Ratio of secretion in the ΔtssM strain to that in the WT.
FDR, false discovery rate.
The number of unique peptides and sequence coverage were determined using MaxQuant proteomics software (61).
The Rhs1/Rhs1I, Rhs2/Rhs2I, and LysM/LysMI proteins are functional toxic effector/immunity pairs.
To determine if the predicted T6SS effectors Rhs1, Rhs2, and LysM had toxic activity and if this activity could be neutralized by the presence of the predicted cognate immunity proteins (Rhs1I, Rhs2I, and LysMI, respectively), each effector gene, or a specific region of each effector gene, was cloned and expressed in trans in E. coli, either alone or together with the gene encoding the predicted cognate immunity protein. The Rhs family of proteins are characterized by a PXXXXDPXGL motif that demarcates the variable C-terminal (CT) region harboring the catalytic motif from the remainder of the protein (41). Heterologous expression of only the CT region from a number of Rhs proteins has previously demonstrated that these truncated proteins display antibacterial activity (31, 42). Therefore, we cloned only the CT region (Fig. 1, hatched regions) of each rhs gene, rhs1-CT and rhs2-CT, into pBAD30 and under the control of the PBAD promoter to produce plasmids pAL1234 and pAL1237 (Table 2) and then used these plasmids to separately transform E. coli BL21(DE3). Recombinant expression was induced by addition of arabinose to the medium and repressed by the addition of glucose. The cognate immunity genes rhs1I and rhs2I were then cloned into pET28a and under the control of an isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible T7 promoter to generate the recombinant plasmids pAL1282 and pAL1283 (Table 2), respectively, that were used to transform the appropriate E. coli strain above. The expression of rhs1-CT and rhs2-CT was then induced in the E. coli strains harboring the dual expression plasmids to assess toxicity, with or without induction of expression of the cognate immunity gene.
TABLE 2.
Plasmids used in this study
| Plasmid | Description | Reference or source |
|---|---|---|
| pAL1211 | pWH1266 containing full-length tssM gene, PCR amplified from AB307-0294 using primers tssMFwd and tssMRev and cloned into PvuI/PstI sites; Tetr | This study |
| pAL1234 | pBAD30 containing cytotoxic domain of rhs1 (residues 1396 to 1591), PCR amplified from AB307-0294 using primers rhs1-CTFwd/rhs1-CTRev and cloned into EcoRI/HindIII sites; Ampr | This study |
| pAL1237 | pBAD30 containing cytotoxic domain of rhs2 (residues 1496 to 1624), PCR amplified from AB307-0294 using primers rhs2-CTFwd/rhs2-CTRev, and cloned into EcoRI/HindIII sites; Ampr | This study |
| pAL1249 | pWH1266 containing full length rhs1I, PCR amplified from AB307-0294 using primers rhs1IAbFwd/rhs1IAbRevI, cloned into PvuI/PstI sites, and introduced at a 3′ SacI site; Tetr | This study |
| pAL1250 | pWH1266 containing full length rhs1I, PCR amplified from AB307-0294 using primers rhs1IAbFwd/rhs1IAbRevII, cloned into PvuI/PstI sites, and introduced at a 3′ XmaI site; Tetr | This study |
| pAL1251 | pWH1266 containing full length rhs2I, PCR amplified from AB307-0294 using primers rhs2IAbFwd/rhs2IAbRev, cloned into PvuI/PstI sites, and introduced at a 3′ XmaI site; Tetr | This study |
| pAL1252 | pWH1266 containing full length lysMI, PCR amplified from AB307-0294 using primers lysMIAbFwd/lysMIAbRev, cloned into PvuI/PstI sites, and introduced at a 5′ XmaI site; Tetr | This study |
| pAL1263 (rhs1I, rhs2I) | pWH1266 containing rhs1I and rhs2I, generated by PCR amplification of rhs2I from AB307-0294 using primers rhs2IAbFwd/rhs2IAbRev; the product was then cloned into SacI (introduced at the 3′ end of the rhs1I fragment) and PstI sites of pAL1249; TetR | This study |
| pAL1264 (rhs1I, lysMI) | pWH1266 containing rhs1I and lysMI, generated by PCR amplification of lysMI from AB307-0294 using primers lysMIAbFwd/lysMIAbRev; the product was then cloned into XmaI (introduced at the 3′ end of the rhs1I fragment) and PstI sites of pAL1250; Tetr | This study |
| pAL1265 (rhs2I, lysMI) | pWH1266 containing rhs2I and lysMI, generated by PCR amplification of lysMI from AB307-0294 using primers lysMIAbFwd/lysMIAbRev; the product was then cloned into XmaI (introduced at the 3′ end of the rhs2I fragment) and PstI sites of pAL1251; Tetr | This study |
| pAL1275 | pBAD30 containing full-length lysM with additional pelB leader sequence, PCR amplified using primers pelBFwd and lysMRev with template pAL1330, and cloned into EcoRI/HindIII sites; Ampr | This study |
| pAL1282 | pET28a containing full length rhs1I, PCR amplified from AB307-0294 using primers rhs1IFwd/rhs1IRev, and cloned into NcoI/HindIII sites; Kanr | This study |
| pAL1283 | pET28a containing full length rhs2I, PCR amplified from AB307-0294 using primers rhs2IFwd/rhs2IRev, and cloned into NcoI/HindIII sites; Kanr | This study |
| pAL1319 (rhs1I, rhs2I, lysMI) | pWH1266 containing rhs1I, rhs2I and lysMI, generated by PCR amplifying lysMI from AB307-0294 using primers lysMIFwd/lysMIRev; the product was then cloned into XmaI and PstI sites of pAL1263; TetR | This study |
| pAL1330 | pET22b containing full-length lysM, PCR amplified from AB307-0294 using primers lysMFwd and lysMRev, and cloned into NcoI and HindIII sites; the vector encodes PelB leader sequence upstream of insertion site | This study |
| pAL1331 | pET28a containing full-length lysMI, PCR amplified from AB307-0294 using primers lysMIFwd and lysMIRev, and cloned into NcoI/HindIII sites; Kanr | This study |
| pAL1351 | Plasmid containing a ColE1 origin of replication, PCR amplified from pBR322 using primers colE1Fwd and colE1Rev, an ampicillin resistance gene, PCR amplified from pUC19 using primers blaFwd and blaRev, with fragments ligated together using NotI/XhoI sites; Ampr | This study |
| pAL1352 | pAL1351 with Acinetobacter origin of replication, PCR amplified from pWH1266 using primers oriAbFwd and oriAbRev, and cloned into XhoI and EcoRI sites; Ampr | This study |
| pAL1353 | pAL1352 with a multiple-cloning site, PCR amplified from pUC19 using primers MCSFwd and MCSRev, and cloned into NcoI and XhoI sites; Ampr | This study |
| pAL1415 | pBASE with vgrG1, PCR amplified from AB307-0294 using primers vgrG1Fwd and vgrG1Rev, and cloned into KpnI and SacI sites; Ampr | This study |
| pAL1416 | pBASE with vgrG2, PCR amplified from AB307-0294 using primers vgrG2Fwd and vgrG2Rev, and cloned into KpnI and SacI sites; Ampr | This study |
| pAL1446 | pBASE containing the 5′ end (nucleotides −22 to 1609) of vgrG3, amplified from AB307-0294 using primers BAP8501 and BAP8502, and cloned into XbaI and SmaI sites of pBASE | This study |
| pAL1447 | Plasmid containing full-length AB307-0294 vgrG3 with the 3′ end of vgrG3 (3′ nucleotides 1610 to 3307) amplified using primers BAP8521 and BAP8522 and cloned into SmaI and SacI sites of pAL1446 | This study |
| pBAD30 | Arabinose-inducible recombinant protein expression vector; Ampr | 70 |
| pBASE | A. baumannii/E. coli shuttle and protein expression vector; pAL1353 with tac promoter, PCR amplified from pGEX-4T-3 using primers PTACFwd and PTACRev and cloned into NcoI and HindIII sites; Ampr | This study |
| pCR-BluntII-TOPO | Suicide vector for A. baumannii; Kanr Zeor | Invitrogen |
| pET22b | IPTG-inducible recombinant protein expression vector; Ampr | Novagen |
| pET28a | IPTG-inducible recombinant protein expression vector; Kanr | Novagen |
| pWH1266 | E. coli/Acinetobacter sp. shuttle vector; Ampr Tetr | 43 |
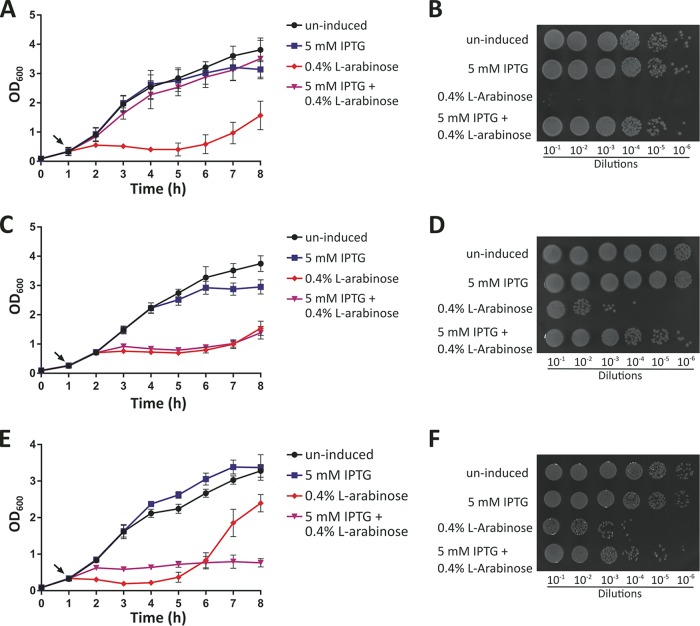
Expression of rhs1-CT alone within the cytoplasm of E. coli (following arabinose induction) resulted in the arrest of E. coli growth within 1 h following induction of expression (Fig. 5A and B). E. coli expressing both rhs1-CT and the predicted cognate immunity gene, rhs1I, grew at normal WT levels, confirming that Rhs1 and Rhs1I are a genuine effector-immunity pair. Similarly, expression of rhs2-CT alone in E. coli resulted in rapid arrest of E. coli growth (Fig. 5C and D). Coexpression of rhs2-CT with the predicted cognate immunity gene rhs2I did not result in growth at WT levels, but viable-cell counts performed 2 h after the induction of effector and immunity expression revealed approximately 1,000-fold more viable cells than when the effector gene rhs2-CT was expressed alone (Fig. 5D). Thus, coexpression of rhs2-CT with the predicted cognate immunity gene rhs2I resulted in a high degree of protection of E. coli against the activity of Rhs2-CT, indicating that Rhs2 and Rh2I are an effector-immunity pair.
FIG 5.
Growth of E. coli BL21(DE3) cells containing relevant pairs of toxic effector and immunity proteins. A growth curve (A, C, and E) and semiquantitative analysis of surviving bacteria at 2 h postinduction (B, D, and F) are shown for each set of toxic effector/immunity strain expression experiments. (A and B) E. coli BL21(DE3) containing the rhs1-CT–rhs1I T6SS effector/immunity cognate pair (pAL1234+pAL1282). (C and D) E. coli BL21(DE3) cells containing the rhs2-CT–rhs2I T6SS effector/immunity cognate pair (pAL1237+pAL1283). (E and F) E. coli BL21(DE3) cells containing the lysM-lysMI T6SS effector/immunity cognate pair (pAL1275+pAL1331). Cultures were grown for 1 h prior to the addition (designated by arrows in panels A, C, and E) of buffer (uninduced), 5 mM IPTG, 0.4% l-arabinose, or 5 mM IPTG and 0.4% l-arabinose and then grown for a further 7 h with optical density measurements taken every 1 h. Error bars represent standard errors of the means of three independent experiments performed in triplicate. Images are representative of three independent experiments.
Recombinant production of LysM, a predicted peptidoglycan hydrolase, within the cytoplasm of E. coli cells did not result in any cell toxicity (data not shown). In contrast, when lysM was cloned in such a way that the gene was fused with the sequence encoding an N-terminal PelB leader sequence (plasmid pAL1275) (Table 2), allowing the fusion protein to be exported to the E. coli periplasm, there was a measurable decrease in the optical density of the E. coli cultures, indicative of cell lysis (Fig. 5E). Coproduction of the PelB-LysM fusion protein with the predicted cognate immunity protein LysMI resulted in no observable lysis of the cell culture, but WT levels of growth were not restored. However, viable cell counts conducted postinduction showed that cell viability was approximately 100-fold higher when the cognate immunity protein was coexpressed compared to cell viability following PelB-LysM expression alone (Fig. 5F). Together, these results show that Rhs1, Rhs2, and LysM are directly toxic to E. coli and support the prediction that Rhs1/Rhs1I, Rhs2/Rhs2I, and LysM/LysMI are effector-immunity pairs.
Rhs2-CT functions as a nuclease.
Bioinformatic analysis revealed that both Rhs1 and Rhs2 shared amino acid identity with several previously characterized T6SS Rhs family nuclease effectors (19, 30). Furthermore, Rhs2 contained an AHH (alanine-histidine-histidine) motif within the C-terminal region that is characteristic of AHH superfamily nucleases. No known motifs were identified in the predicted Rhs1 catalytic domain. To determine if any of the three toxic effectors had nuclease activity, crude cell lysates of the E. coli BL21(DE3) strains harboring the rhs1-CT, rhs2-CT, and the lysM expression plasmids (pAL1234, pAL1237, and pAL1275, respectively) (Table 2) were separately incubated with purified genomic DNA from the known prey strain A. baylyi ADP1 (Fig. 6). Lysates were generated from cells grown with the appropriate antibiotic selection to maintain the plasmid and in the presence or absence of arabinose, which induced the expression of the recombinant protein. As controls, buffer or cell lysates of E. coli BL21(DE3) harboring vector only were also separately incubated with A. baylyi genomic DNA. The A. baylyi genomic DNA was completely degraded in the presence of E. coli lysate containing Rhs2-CT but was not degraded by E. coli lysates containing Rhs1-CT or LysM or when recombinant rhs2-CT expression was not induced (Fig. 6). Therefore, we predict that Rhs2 is the only T6SS effector produced by A. baumannii strain AB307-0294 that acts as a DNA nuclease.
FIG 6.
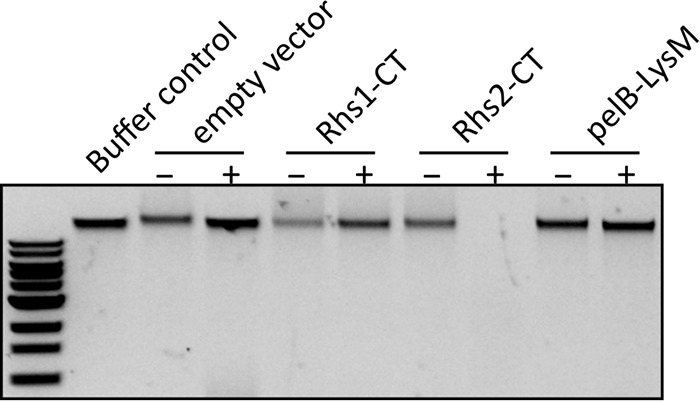
Rhs2-CT is a nuclease. An agarose gel shows the integrity of A. baylyi ADP1 genomic DNA following incubation for 2 h with crude lysates of E. coli cells carrying pBAD30 (empty vector) or pBAD30 derivatives expressing the Rhs1 C-terminal domain (Rhs1-CT; pAL1234), Rhs2 C-terminal domain (Rhs2-CT; pAL1237), or LysM with a PelB leader sequence (pelB-LysM; pAL1275), either with (+) or without (−) prior l-arabinose induction. A. baylyi genomic DNA incubated in buffer only was used as a control. A standard New England Biolab 1-kb DNA ladder is shown at the left.
A. baumannii strain AB307-0294 secretes all three effector proteins while in competition against A. baylyi.
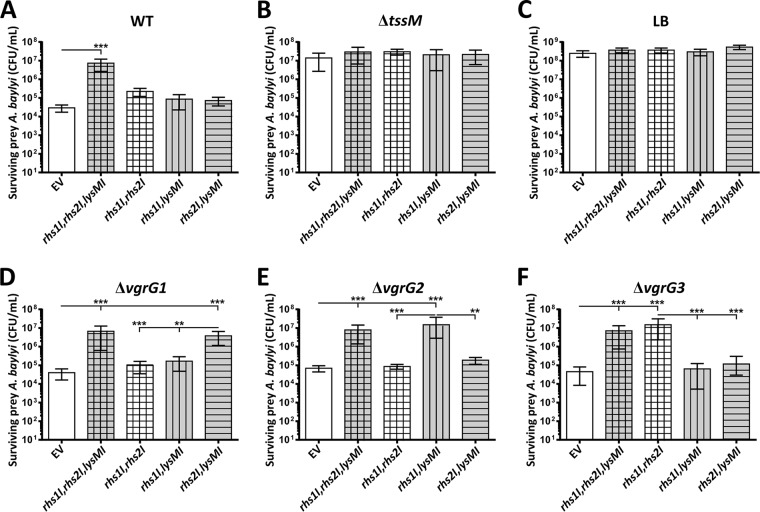
To determine if A. baumannii strain AB307-0294 delivered all three toxic effectors into susceptible competitor cells, we engineered a series of plasmids that allowed the constitutive expression of AB307-0294 immunity genes, in various combinations, in the otherwise susceptible A. baylyi ADP1 strain. This strain was chosen as prey in interbacterial competitive growth assays because of its sensitivity to the A. baumannii AB307-0294 T6SS and limited antibiotic resistance profile and because of our desire to express the immunity proteins in a more natural context in a related Acinetobacter species. We used the Acinetobacter/E. coli shuttle vector pWH1266 (43) to construct four recombinant plasmids, one containing all three immunity genes (rhs1I, rhs2I, and lysMI in pAL1319) (Table 2) and three others that each contained two of the three AB307-0294 immunity genes (rhs1I and rhs2I in pAL1263, rhs1I and lysMI in pAL1264, and rhs2I and lysMI in pAL1265) (Table 2). Interbacterial competitive growth assays were then performed with each assay containing one predator strain, A. baumannii AB307-0294 WT or A. baumannii AB307-0294 ΔtssM, and one prey strain, A. baylyi ADP1 with vector only or A. baylyi ADP1 with a recombinant plasmid expressing rhs1I, rhs2I, and lysMI or rhs1I and rhs2I, rhs1I and lysMI, or rhs2I and lysMI (Fig. 7). None of the A. baylyi strains affected the growth of the AB307-0294 WT or AB307-0294 ΔtssM (data not shown), and AB307-0294 ΔtssM, that is unable to assemble a functional T6SS, did not kill any of the A. baylyi prey strains (Fig. 7B). In contrast, the AB307-0294 WT strain was able to kill the A. baylyi strain containing the empty vector and all of the strains producing only two of the three recombinant immunity proteins derived from AB307-0294 (Fig. 7A), confirming that any one of the three AB307-0294 effectors can act alone and kill A. baylyi. Importantly, the A. baylyi strain producing all three A. baumannii AB307-0294 immunity proteins (Rhs1I, Rhs2I, and LysMI) was completely protected from killing by the AB307-0294 WT strain (Fig. 7A), confirming that each of the three immunity proteins could neutralize their cognate effector protein. These results indicate that Rhs1, Rhs2, and LysM are the only three antibacterial effector proteins secreted by the A. baumannii strain AB307-0294 T6SS that are active against A. baylyi strain ADP1.
FIG 7.
Survival of A. baylyi cells expressing combinations of T6SS immunity proteins and showing that the inactivation of vgrG1, vgrG2, or vgrG3 prevents translocation of the effector protein Rhs1, Rhs2, or LysM, respectively. Graphs show quantitative assessments of the number of the surviving prey bacteria, A. baylyi strain ADP1, following coincubation with the predator strain A. baumannii AB307-0294 WT (A), AB307-0294 tssM (B), AB307-0294 ΔvgrG1 (D), AB307-0294 ΔvgrG2 (E), or AB307-0294 ΔvgrG3 (F) or with LB medium only (C). For each graph the survival of the A. baylyi prey strains containing empty pWH1266 vector (EV) or a recombinant plasmid expressing two or three of the recombinant immunity genes cloned from AB307-0294, as indicated below each bar, is shown. Expression of lysMI in A. baylyi is represented by gray fill, rhs1I expression is represented by vertical stripes, and rhs2I expression is represented by horizontal stripes. Error bars represent standard errors of the means of three independent experiments performed in triplicate. **, P < 0.01; ***, P < 0.0001.
VgrG proteins are predicted to form the tip of the T6SS needle and in several species have been demonstrated to be essential for the secretion of specific effectors (24, 27, 31). Given that each vgrG gene (vgrG1, vgrG2, and vgrG3) was located within a three-gene locus that contained a T6SS effector gene (rhs1, rhs2, and lysM, respectively) and a cognate immunity gene (rhs1I, rhs2I, and lysMI, respectively), we predicted that the three VgrG proteins were likely to have a similar specificities for their cognate effectors (Fig. 1). In order to investigate this relationship, we constructed three AB307-0294 vgrG mutant strains by allelic exchange; these were designated the ΔvgrG1, ΔvgrG2, and ΔvgrG3 strains (Table 3). All three AB307-0294 vgrG mutants had the same growth rate as the parent strain AB307-0294 under normal laboratory growth conditions (data not shown). Western blot analysis showed that each vgrG mutant retained the ability to secrete wild-type levels of the essential T6SS structural protein, Hcp, indicating that the loss of a single VgrG does not affect the overall function of the T6SS in AB307-0294 (Fig. 8). To determine if each VgrG protein was required for the secretion of the effector protein encoded within the same locus, we utilized interbacterial competitive growth assays. Each assay contained one predator strain, A. baumannii AB307-0294 WT, AB307-0294 ΔvgrG1, AB307-0294 ΔvgrG2, or AB307-0294 ΔvgrG3, coincubated with one prey strain, A. baylyi ADP1 with vector only or A. baylyi ADP1 with a recombinant plasmid expressing rhs1I, rhs2I, and lysMI or rhs1I and rhs2I, rhs1I and lysMI, or rhs2I and lysMI (Fig. 7D, E, and F). Each individual vgrG mutant retained the ability to kill A. baylyi containing empty vector, confirming that the T6SS was active in each mutant and that the secretion of some antibacterial effector proteins was occurring. As expected, none of the vgrG mutants was able to kill the A. baylyi recombinant strain expressing all three cognate immunity proteins (rhs1I, rhs2I, and lysMI). The ΔvgrG1 strain was able to kill the A. baylyi strain producing the Rhs1I and Rhs2I immunity proteins (via the action of LysM) and the A. baylyi strain producing the Rhs1I and LysMI immunity proteins (via the action of Rhs2), but it was unable to kill the A. baylyi strain producing Rhs2I and LysMI (Fig. 7D). These results indicate that AB307-0294 ΔvgrG1 is unable to deliver Rhs1, which is encoded within the same locus as vgrG1. Complementation of the AB307-0294 ΔvgrG1 with an intact copy of vgrG1 restored the ability of the strain to kill A. baylyi expressing Rhs2I and LysMI, confirming that VgrG1 is required for the secretion of Rhs1 (Fig. 9). Similar results were observed for coincubation experiments using the AB307-0294 ΔvgrG2 and AB307-0294 ΔvgrG3 strains. AB307-0294 ΔvgrG2 was unable to deliver Rhs2, as shown by its inability to kill A. baylyi expressing Rhs1I and LysMI, but it could still kill A. baylyi expressing Rhs1I/Rhs2I (via the action of LysM) or Rhs2I/LysMI (via the action of Rhs1) (Fig. 7E). Complementation restored the ability of the AB307-0294 ΔvgrG2 strain to kill via the action of Rhs2 (Fig. 9). AB307-0294 ΔvgrG3 was unable to deliver LysM, as demonstrated by its inability to kill the A. baylyi strain expressing Rhs1I and Rhs2I, but retained the ability to kill the A. baylyi strains that lacked one of the two Rhs immunity proteins (Fig. 7F). Complementation restored the ability of the AB307-0294 ΔvgrG3 strain to kill via the action of LysM (Fig. 9). Together, these results show that each VgrG protein is indispensable for delivery of the specific effector protein that is encoded within the same vgrG locus.
TABLE 3.
Bacterial strains used in this study
| Strain | Description | Reference or source |
|---|---|---|
| E. coli strains | ||
| BL21(DE3) | E. coli strain used for recombinant protein expression; F− ompT hsdSB(rB− mB−) galdcm (DE3) | Novagen |
| DH5α | E. coli general laboratory strain; l− f80dlacZDM15 D(lacZYA-argF)U169 recA1 endA hsdR17(rK− mK−) supE44 thi-1 gyrA relA1 | Bethesda Research Laboratories |
| DH10B | E. coli strain used for interbacterial competitive growth assays; F− araD139 Δ(ara leu)7697 ΔlacX74 galUgalKmcrA Δ(mrr-hsdRMS-mcrBC) rpsLdeoR ϕ80dlacZΔM15 recA1endA1 nupG | 67 |
| Acinetobacter strains | ||
| AB307-0294 | A. baumannii AB307-0294 wild type, clinical isolate | 58 |
| AB900 | A. baumannii strain AB900 wild type, clinical isolate | 58 |
| ADP1 | A. baylyi ADP1 wild type | 68 |
| AL2730 ΔtssM | AB307-0294 derivative with tssM replaced by allelic exchange with a kanamycin resistance cassette; Kanr | This study |
| AL2734 ΔtssM (vector only) | AL2730 with pWH1266, complementation control; Kanr Tetr | This study |
| AL2735 ΔtssM tssM | AL2730 complemented with pAL1211; Kanr Tetr | This study |
| AL2751 ΔvgrG1 | AB307-0294 derivative with vgrG1 replaced by allelic exchange with kanamycin resistance cassette; Kanr | This study |
| AL2752 ΔvgrG2 | AB307-0294 derivative with vgrG2 replaced by allelic exchange with kanamycin resistance cassette; Kanr | This study |
| AL2753 ΔvgrG3 | AB307-0294 derivative with vgrG3 replaced by allelic exchange with kanamycin resistance cassette; Kanr | This study |
| AL3261 | AL2751 harboring pBASE as a complementation control; Ampr | This study |
| AL3262 | AL2752 harboring pBASE as a complementation control; Ampr | This study |
| AL3263 | AL2753 harboring pBASE as a complementation control; Ampr | This study |
| AL3264 | AL2751 complemented with pAL1415; Ampr | This study |
| AL3265 | AL2752 complemented with pAL1416; Ampr | This study |
| AL3352 | AL2753 complemented with pAL1447 | This study |
| ATCC 17978 | A. baumannii strain ATCC 17978 wild type, clinical isolate | American Type Culture Collection |
| ATCC 19606 | A. baumannii strain ATCC 19606 wild type, clinical isolate | American Type Culture Collection |
| M2 | A. nosocomialis M2 wild type, clinical isolate | 69 |
FIG 8.
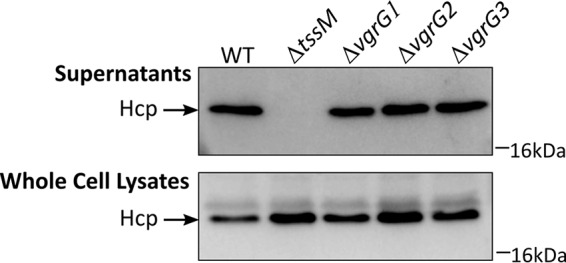
Hcp secretion by A. baumannii strain AB307-0294 is unaffected by the loss of an individual VgrG protein. Samples of cell-free culture supernatants and whole-cell lysates derived from AB307-0294 wild type (WT), AB307-0294 ΔtssM, AB307-0294 ΔvgrG1, AB307-0294 ΔvgrG2, and AB307-0294 ΔvgrG3 were separated by SDS-PAGE and transferred to a PVDF membrane for Western immunoblotting using Hcp-specific antiserum. The position of the Hcp protein is shown at the left. The position of the 16-kDa marker from a SeeBlue Plus2 prestained protein standard (Invitrogen) is indicated on the right.
FIG 9.

A. baylyi and A. baumannii interbacterial competitive growth assays showing that the complementation of vgrG1, vgrG2, or vgrG3 restores the translocation of the effector proteins Rhs1, Rhs2, and LysM, respectively. Graphs show quantitative assessments of the number of surviving prey bacteria, A. baylyi strain ADP1, following coincubation with the predator ΔvgrG1 mutant provided with empty vector (EV) or a functional vgrG1 (C), the ΔvgrG2 mutant provided with empty vector (EV) or a functional vgrG2 (C), or the ΔvgrG3 mutant provided with empty vector (EV) or a functional vgrG3 (C). The prey bacteria contained the appropriate dual-immunity expression plasmids as shown at the top right: rhs2I and lysMI for the ΔvgrG1 strain complementation experiments, rhs1I and lysMI for the ΔvgrG2 strain complementation experiments, and rhs1I and rhs2I for the ΔvgrG3 strain complementation experiments. Expression of lysMI in A. baylyi is represented by gray fill, rhs1I expression is represented by vertical stripes, and rhs2I expression is represented by horizontal stripes. Error bars represent standard errors of the means of three biological replicate experiments. ***, P < 0.0001.
A. baumannii vgrG genes colocalize with putative effector and immunity pairs across many strains.
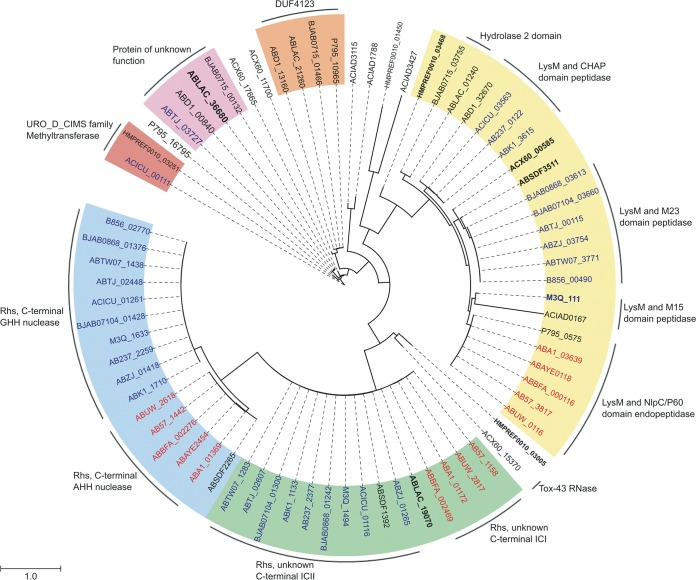
Our results demonstrate that in A. baumannii strain AB307-0294, three T6SS antibacterial toxic effectors, Rhs1, Rhs2, and LysM, and their cognate immunity proteins, Rhs1I, Rhs2I, and LysMI, are each encoded downstream of a VgrG-encoding gene (vgrG1, vgrG2, and vgrG3, respectively). Given this gene arrangement in AB307-0294, we analyzed the 22 publicly available genomes of other A. baumannii strains to determine the range of vgrG and effector/immunity gene sets present in the species. These genomes represented both historical and modern isolates from a range of geographical locations and included 4 strains belonging to IC1, 11 strains belonging to IC2, and 7 non-IC sequence type isolates. Also included in these analyses was the A. baylyi strain ADP1, which was used in our interbacterial competitive growth assays with A. baumannii strain AB307-0294. We identified 73 VgrG proteins across the 23 genomes analyzed. The majority of vgrG genes in each A. baumannii genome were the first gene within a three-gene locus, similar to the arrangement identified in the A. baumannii strain AB307-0294 VgrG loci. We therefore predict that these genes in such loci are likely to encode cognate effector and immunity proteins. Preliminary comparative analyses revealed significant diversity between the putative A. baumannii VgrG proteins, primarily in the C-terminal region that is known to be the most variable part of these proteins. All identified VgrG proteins contained the characteristic canonical VgrG domains, including the phage GPD (pfam05954), phage baseplate V (pfam04717), and T6SS_Vgr (pfam13296) domains (44). Furthermore, each of the VgrG proteins contained an amino acid region with at least a partial match to the 2345 domain of unknown function (DUF2345). To delineate the relationship between each of the VgrG proteins, we constructed a phylogenetic tree using the amino acid sequence of each VgrG protein (Fig. 10). The phylogenetic analysis revealed that the VgrG proteins could be divided into at least six distinct classes. Interestingly, closely related VgrG proteins were always found upstream of similar effector-encoding genes (Fig. 10, outer ring). Analysis of the putative effector proteins, encoded downstream of the vgrG genes, identified potentially novel peptidoglycan hydrolases, Rhs family toxins (with C-terminal nuclease domains or with C-terminal domains of unknown function), RNases, methyltransferases, and numerous proteins of unknown function (Fig. 10). Preceding one class of VgrG proteins was a protein containing a DUF4123 domain, which in Vibrio cholerae and Aeromonas hydrophila is involved in effector protein attachment to VgrG (34, 45). Although the identified repertoire of potential effector proteins is diverse, it is interesting that almost all examined IC1 lineage strains shared identical predicted effector and immunity pairs. Similarly, the IC2 lineage strains also generally shared the same predicted effector and immunity pairs. Together, strains within the IC1 and IC2 lineages were the predominant strains that encoded predicted Rhs family effectors.
FIG 10.
Acinetobacter VgrG proteins are separated into distinct phylogenetic classes. A maximum likelihood phylogenetic tree was generated from 73 aligned VgrG protein sequences belonging to 22 A. baumannii strains and 1 A. baylyi strain. Phylogenetic classes containing more than one intact member are indicated by a colored background. Class 1 (green), class 2 (blue), and class 3 (yellow) have been numbered as such because they contain VgrG1, VgrG2, and VgrG3 of AB307-0294, respectively. Class 4 (orange), class 5 (lilac), and class 6 (red) were arbitrarily numbered. The outer ring denotes proteins encoded downstream of the respective vgrG genes predicted to belong to the same effector/accessory protein families. VgrG proteins produced by strains belonging to international clonal lineage IC1 are indicated in red text, VgrG proteins produced by strains belonging to the IC2 lineage are indicated in blue text, and non-IC lineages are indicated in black text. Boldface type denotes annotated vgrG sequences containing frameshifts or mobile element disruptions. A. baumannii strains (locus tag identifiers) are as follows: AB307-0294 (ABBFA), ATCC 17978 (ACX60), AB0057 (AB57), ACICU (ACICU), ATCC 19606 (HMPREF0010), AYE (ABAYE), SDF (ABSDF), 1656-2 (ABK1), BJAB0715 (BJAB0715), BJAB0714 (BJAB0714), BJAB0868 (BJAB0868), D1279779 (ABD1), LAC-4 (ABLAC), MDR-TJ (ABTJ), MDR-ZJ06 (ABZJ), NCGM 237 (AB237), TCDC-0715 (ABTW07), TYTH-1 (M3Q), ZW85-1 (P795), AC30 (B856), A1 (ABA1), and AB5075-UW (ABUW). The A. baylyi strain is ADP1 (ACIAD).
DISCUSSION
In this study, we analyzed the T6SS of A. baumannii strain AB307-0294 and determined that this system plays a major role in interbacterial competition. We then identified and characterized the three T6SS effector proteins, their cognate immunity proteins, and the associated VgrG tip proteins that mediate this interaction. It is clear that the T6SS is a weapon deployed by A. baumannii to fight rival bacteria at both the intra- and interspecies levels. Interstrain T6SS-mediated killing has been documented for other bacteria, including S. marcescens, Proteus mirabilis, and V. cholerae (19, 46, 47). This interstrain killing may in part explain the rise of certain dominant clonal lineages globally and within a geographic region. This is particularly relevant to A. baumannii: despite a large number of identified clonal lineages, the species is dominated by strains belonging to the globally distributed IC1 and IC2 lineages. Several groups of investigators have described the capacity of A. baumannii and other Acinetobacter species to kill E. coli in a T6SS-dependent manner (4, 5, 7, 27). Similarly, in this study we showed that A. baumannii strain AB307-0294 is able to kill E. coli strain DH10B and that this activity is dependent upon a functional T6SS apparatus. A. baumannii AB307-0294 was also able to kill the A. baylyi strain ADP1 and the A. baumannii strain ATCC 19606 in a T6SS-dependent manner. However, it was not able to kill A. nosocomialis M2 nor the A. baumannii strain AB900. Moreover, A. baumannii AB307-0294 was unable to kill A. baumannii strain ATCC 17978, with or without the T6SS-suppressing plasmid pAB3 (7), even though a previous study showed that a different A. baumannii strain, DSM3001, was able to outcompete A. baumannii strain ATCC 17978 (5).
The inability of A. baumannii AB307-0294 to kill ATCC 17978 and AB900 was unexpected as analysis of the genomes of these strains did not identify encoded immunity proteins with significant identity to the AB307-0294 immunity proteins, and therefore it is unlikely that they are immune to the AB307-0294 toxic effectors. Indeed, expression of the AB307-0294 Rhs1 and Rhs2 toxic effectors in ATCC 17978 led to rapid death of the ATCC 17978 strain (data not shown), supporting the proposal that the inability of AB307-0294 to kill ATCC 17978 is not due to cross-immunity. We are currently exploring why AB307-0294 is unable to target some A. baumannii strains, including ATCC 17978.
The outcome of competition between rival bacteria is often dependent upon the secretion of T6SS effector proteins that are lethal to the target bacterium. Three effector and immunity pairs, Rhs1/Rhs1I, Rhs2/Rhs2I, and LysM/LysMI, were identified in A. baumannii strain AB307-0294. When each effector gene was separately expressed in E. coli, the viability of the recombinant strain was severely inhibited unless the gene was coexpressed with the cognate immunity protein, confirming that these proteins form genuine effector-immunity pairs. While full viability of E. coli producing Rhs2 and LysM was not completely restored when the Rhs1I and LysMI proteins were coexpressed in the appropriate strains, we predict that this is likely due to insufficient production of the recombinant immunity proteins. Supporting this proposal, recombinant expression of the rhs2I and lysMI immunity genes in E. coli strongly protected against Rhs2 and LysM T6SS-dependent killing by the AB307-0294 ΔvgrG1 strain (1,000-fold increased viability compared to E. coli containing vector only). Similarly, heterologous expression of all three immunity genes in the normally susceptible bacterium A. baylyi gave A. baylyi full protection against Rhs1-, Rhs2-, and LysM-mediated killing by AB307-0294, indicating that these three effectors are the only antibacterial effectors delivered by this strain that are active against A. baylyi and E. coli. Finally, our interbacterial competitive growth assays using A. baylyi strains coexpressing two of the three immunity genes demonstrated that any one of the three effectors delivered by AB307-0294 can kill WT A. baylyi.
The C-terminal domain of Rhs2 is toxic when produced in the cytoplasm of E. coli and was confirmed to function as a DNase. The targeting of nucleic acid components of the cell appears to be a common characteristic of antibacterial Rhs effectors. The Dickeya dadantii Rhs effector proteins have C-terminal domains with DNase activity (31), and an enterotoxigenic E. coli strain produces an Rhs T6SS effector protein that harbors a C-terminal restriction endonuclease-3 domain (14). Moreover, S. marcescens produces one Rhs protein that is a known DNase and a second Rhs protein whose function is unknown but is lethal to E. coli when expressed in the cytoplasm (19). E. coli strains produce Rhs proteins with C-terminal domains with predicted metallopeptidase, DNase, RNase, deamidase, and pore-forming activities although these have yet to be experimentally verified (14). The Rhs1-CT fragment derived from the Rhs1 effector produced by A. baumannii strain AB307-0294 was shown in this study to be toxic to E. coli when produced within the cytoplasm although the precise subcellular target is unknown. Although Rhs1-CT shares no sequence identity with any known domains, it is likely that it represents a novel toxin that belongs to one of these broad functional classes.
In contrast to Rhs1 and Rhs2, the predicted peptidoglycan hydrolase, LysM, is lethal only when targeted to the periplasm of the Gram-negative bacterial cell. A large number of T6SS-secreted peptidoglycan hydrolases, including both amidases and glycosidases, have been described in a number of species (39, 48–51). Although LysM shares only low levels of identity with members of the currently recognized amidase families of T6SS bacteriolytic effectors, it shares 35% amino acid sequence identity with a predicted peptidoglycan targeting effector, Tae1, from A. baylyi (8). Together, these observations suggest that LysM represents a new class of T6SS effector peptidoglycan hydrolase that is unique to A. baumannii.
The delivery of three distinct T6SS effectors by A. baumannii strain AB307-0294 has likely evolved as a strategy to circumvent the emergence of any resistance against an individual effector or as a means by which strain AB307-0294 can gain a competitive advantage over closely related strains with similar effector/immunity pairs. Although all three effector proteins are delivered, it is unknown if all three are delivered in a single secretion event or are utilized in a regulated manner. The secretion of each effector was absolutely dependent upon the expression of the vgrG gene with which the effector gene is colocated on the chromosome. Such dependency of a T6SS effector upon a specific VgrG protein for secretion has been documented in a number of species (19, 24, 27). The specificity of a particular effector protein for one VgrG protein is primarily dependent on the amino acid sequence at the VgrG C-terminal domain although the specific interactions between the two proteins are poorly defined (28). Supporting this was our bioinformatic analysis of more than 70 A. baumannii VgrG proteins that revealed that the greatest variation between the VgrG proteins was within the C-terminal domain. Thus, it is likely that VgrG proteins produced by a range of A. baumannii strains are highly specialized proteins that define the needle tip and are also required for the secretion of a specific effector protein.
Bioinformatic analysis identified a large number of putative T6SS effector proteins genetically linked to vgrG genes in other A. baumannii strains and A. baylyi, indicating that significant diversity exists among the arsenal of potential effector proteins within the species. Interestingly, Rhs effectors were identified almost exclusively in strains belonging to the clonal lineages IC1 and IC2, with a range of predicted toxin domains at the C-terminal end, including GHH- and AHH-type amino acid motifs characteristic of the HNH-endonuclease VII (HNH-EndoVII) superfamily of nucleases as well as C-terminal domains of unknown function (52). The reasons why certain A. baumannii strains/lineages are more dominant globally than others have not been determined; increased acquisition of antibiotic resistance genes, virulence, and biofilm-forming ability have all been assessed, but a clear correlation has not been identified (53–56). Given our results, it is possible that certain A. baumannii strains dominate globally due to their ability to outcompete rival strains via the use of the T6SS apparatus to deliver antibacterial effector proteins. For example, the almost exclusive production of Rhs T6SS effectors and their cognate Rhs immunity proteins in strains belonging to the IC1 and IC2 clonal lineages could allow killing of vulnerable rival/competing bacteria and, at the same time, protect against Rhs-mediated attack from others. The production of these Rhs T6SS effectors could allow the IC1 and IC2 strains to dominate mixed bacterial niches, including niches within the hospital setting where these strains are most prevalent. It has also been shown in A. baylyi that T6SS-mediated lysis of prey cells can enhance horizontal gene transfer, allowing the acquisition of novel antibiotic resistance genes and suggesting that T6SS-mediated killing may play a role in the spread of antibiotic resistance among A. baumannii strains (8, 57). All A. baumannii strains examined in this study contained putative peptidoglycan-targeting effectors, theoretically capable of inducing cell lysis in prey strains and therefore allowing uptake of prey DNA. However, the secretion of DNases, such as Rhs2 in AB307-0294, would seemingly reduce the rate of gene transfer. Thus, further investigation is required to fully understand the role of the T6SS in the spread of antibiotic resistance among A. baumannii strains.
In conclusion, we have shown that the T6SS of A. baumannii strain AB307-0294 secretes three antibacterial effectors and produces three cognate immunity proteins to stop self and sibling intoxication. Each of the identified effector proteins was able to individually kill rival bacterial cells, and one of these toxic effectors, Rhs2, was identified as a DNA nuclease. Furthermore, each effector was dependent upon a specific VgrG protein for secretion, and each set of cognate vgrG, effector, and immunity genes was colocalized on the A. baumannii chromosome. Homologs of the three effector proteins, Rhs1, Rhs2, and LysM, produced by A. baumannii strain AB307-0294 were encoded in a large number of strains belonging to the IC1 clonal lineage, and Rhs1 and Rhs2 effectors were also present in most IC2 strains. In contrast, a diverse repertoire of putative effector proteins was identified in strains belonging to groups outside these lineages. The diversity of the A. baumannii effector proteins will likely play an important role in determining the survival of A. baumannii in its environment when it interacts with other bacteria and may in part explain the success of this organism in the hospital environment.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains used in this study are listed in Table 3. E. coli DH5α was used for all cloning experiments and DNA manipulations. A. baumannii strain AB307-0294 was isolated from a human patient with bacteremia (58). Bacteria were grown in lysogeny broth (LB) (Oxoid) or M9 minimal medium with shaking at 200 rpm at 37°C. For solid medium, 1.0 to 1.5% (wt/vol) agar was added to LB. Medium was supplemented with the following antibiotics as required: 100 μg/ml ampicillin or 50 μg/ml carbenicillin (A. baumannii strain AB307-0294), 50 μg/ml kanamycin, and 10 μg/ml tetracycline.
Generation of Hcp antiserum.
The full-length hcp gene from AB307-0294 was amplified by PCR using the primers hcpFwd and hcpRev (see Table S2 in the supplemental material) and then cloned into the expression vector pENTR/D/SD-TOPO using the Gateway cloning system as described previously (59). The section of the plasmid containing the hcp gene was then transferred to the protein expression vector pBAD-DEST49BA (59), such that the N-terminal end of the recombinant Hcp protein was fused to an NusA solubility tag and hexahistidine tag, allowing recombinant protein purification as described previously (59). Chicken polyclonal antiserum was generated as described previously (59).
Detection of the T6SS protein Hcp.
For the detection of Hcp in A. baumannii whole-cell lysates and culture supernatants by Western immunoblotting, whole-cell protein fractions were prepared and analyzed as follows. A. baumannii strains were grown to stationary phase (16 h) in LB medium with or without antibiotics as required. To prepare culture supernatants, 1 ml of bacterial culture was subjected to centrifugation at 10,000 × g for 5 min. The supernatant was removed and filtered using a 0.22-μm-pore-size syringe filter and then mixed at 1:1 volume ratio with 2× SDS-PAGE sample buffer. The cell pellet was washed twice with phosphate-buffered saline (PBS; pH 7.4), resuspended in 100 μl of PBS, and then mixed at 1:1 volume ratio with 2× SDS-PAGE sample buffer. Samples were boiled for 5 min and then centrifuged for 5 min (13,000 × g). A sample volume representing proteins derived from approximately 1 × 109 cells (pelleted material) and 1 × 108 cells (supernatant) was loaded into each lane and separated using SDS-PAGE on 15% acrylamide gels and then transferred to polyvinylidene fluoride (PVDF) (Merck Millipore, USA) membranes via electroblotting. Hcp was detected using polyclonal chicken antiserum raised against recombinant Hcp (1/10,000), followed by the secondary antibody, donkey anti-chicken horseradish peroxidase (HRP) conjugate. Antibody binding was analyzed with Amersham ECL Western blotting detection reagent (GE Healthcare, NSW, Australia), and visualized by exposure to X-ray film (Kodak, NY, USA) or a Fujifilm LAS-3000 image reader (chemiluminescence filter).
In vitro interbacterial competitive growth assays.
Assays to determine the ability of A. baumannii strains to out-compete rival bacteria were performed as described previously (35, 48) with minor modifications. Each of the strains required for the assay was grown in LB to mid-logarithmic growth phase, equivalent to an optical density at 600 nm (OD600) of 0.5 to 0.6 at 37°C with shaking. Culture density was normalized to an OD600 of 0.4 by the addition of LB medium. Predator (A. baumannii) and prey strains were mixed at a ratio of 10:1 when E. coli was used as prey and at a ratio of 1:1 when an Acinetobacter species were used as prey. A 10-μl aliquot of each coculture was spotted onto LB agar prewarmed to 37°C, allowed to dry, and then incubated at 37°C for 4 h (E. coli prey strain) or 24 h (Acinetobacter prey strains). Following the incubation period, an agar plug containing each coculture was excised from the solid medium, and the bacterial cells were resuspended in 1 ml of PBS by vortexing. To determine the number of predator and prey cells in the coculture following incubation, 10-fold serial dilutions were performed in LB medium, and then 100-μl aliquots of selected dilutions were plated onto LB agar supplemented with the appropriate antibiotic for selection of either the predator or the prey strain. The number of CFU per milliliter of surviving predator and prey cells was then enumerated. To allow identification and enumeration of the Acinetobacter prey strain following coculture, each strain was freshly transformed with pWH1266 or a pWH1266 derivative plasmid containing a tetracycline antibiotic resistance gene.
Nano-HPLC-MS/MS analysis of A. baumannii culture supernatants.
For each sample, 1 ml of an A. baumannii overnight culture (grown in LB at 37°C with shaking) was pelleted by centrifugation (10,000 × g, 1 min), washed in an equal volume of M9 minimal medium supplemented with 20 mM sodium butyrate and 1% Casamino Acids ([M9Sup] for optimal A. baumannii growth), and then diluted 1/500 into 100 ml of fresh M9Sup medium and grown with shaking (200 rpm) at 37°C to an OD600 of 0.8. The culture supernatant was collected following centrifugation (4,000 × g, 4°C, 10 min) and then filtered through a 0.22-μm-pore-size vacuum filter (Corning, USA) to remove any remaining cellular material. The filtered supernatant was then concentrated approximately 200-fold (to 500 μl), and the culture medium was exchanged to rebuffer solution (50 mM Tris, pH 8.0, 150 mM NaCl), using a 3-kDa Amicon Ultra-15 centrifugal filter unit (Merck Millipore, USA) in a swinging basket rotor, according to the manufacturer's instructions, and stored at −80°C until required. The protein concentration in each concentrated supernatant was determined using a NanoDrop 1000 spectrophotometer (Thermo Fisher Scientific, CA, USA), and then a volume equivalent to a total protein concentration of 50 μg was diluted in rebuffer solution to give a final volume of 220 μl. Following this, the proteins were denatured via the addition of 1,4-dithiothreitol (DTT) to a final concentration of 10 mM and incubated at 65°C for 30 min; then proteins were alkylated via the addition of 40 mM chloroacetamide (CAA) (Sigma) and incubation at room temperature for 20 min in the absence of light. Trypsin Gold (2 μg/ml; Promega, USA) was then added at a ratio of 1:100 (vol/vol), and the sample was incubated overnight at 37°C. To stop the reaction, the sample was acidified (to approximately pH 3.0) by the addition of reagent-grade formic acid to a final concentration of 1%. Samples were concentrated to a final volume of 50 μl by vacuum drying, desalted using P-10 ZipTip columns (OMIX-Mini Bed 96 C18; Agilent), vacuumed dried, and then reconstituted in 20 μl of 0.1% formic acid prior to mass spectrometry.
Peptide identification using a Dionex UltiMate 3000 RSLCnano system equipped with a Dionex 732 UltiMate 3000 RS autosampler was performed as described previously (60) with the exception that that peptides were analyzed using an Orbitrap Fusion Tribrid mass spectrometer (Thermo Scientific). Acquired raw files were identified and quantified with MaxQuant (61) software. Statistical analyses were performed using Limma within the R studio, where the FDR is derived from the Benjamini-Hochberg procedure. Proteins with a >4-fold (2-log2) change and an FDR of ≤0.01 were considered differentially expressed.
General DNA manipulations.
A NucleoSpin plasmid kit (Macherey-Nagel GmbH & Co. KG) and a Genomic DNA Extraction kit (RBC Bioscience Corp.) were used to extract plasmid and genomic DNA, respectively, according to the manufacturer's instructions. Restriction endonucleases and ligase were obtained from New England BioLabs and Roche, respectively, and used according to the manufacturer's instructions. For PCRs, the DNA polymerases Taq and the high-fidelity Phusion were obtained from Roche, and the oligonucleotides (primers) were manufactured by Sigma-Aldrich. PCR products were purified using a NucleoSpin Gel and PCR Clean Up kit (Macherey-Nagel). Sanger sequencing was performed using BigDye Terminator, version 3.1 (Applied Biosystems), with plasmid DNA or PCR products as templates. The DNA sequence was determined on a capillary platform Genetic Analyzer (Applied Biosystems 3730) and analyzed using Vector NTI Advance 11 (Invitrogen). All plasmids and primers used in this study are listed in Tables 2 and S2, respectively. Primers were designed using publicly available genome sequences.
Construction of A. baumannii/E. coli shuttle expression vector, pBASE.
For the construction of the A. baumannii/E. coli shuttle expression vector, pBASE, a DNA fragment containing the ColE1 origin of replication was PCR amplified from pBR322 using primers colE1Fwd/colE1Rev (Table S2), digested with NotI/XhoI, and ligated to an NotI/XhoI-digested DNA fragment containing the β-lactamase resistance gene bla (PCR-amplified from pUC19 using primers blaFwd/blaRev). The interim plasmid, pAL1351, was then digested with XhoI and EcoRI and ligated to a similarly digested PCR product containing the Acinetobacter origin of replication from pWH1266 (amplified using primers oriAbFwd/oriAbRev) to generate the interim plasmid pAL1352. To introduce a multiple-cloning site, the appropriate region of pUC19 was PCR amplified using primers MCSFwd/MCSRev, digested with NcoI and XhoI, and cloned into similarly digested pAL1352 to generate the plasmid pAL1353. Finally, the Ptac promoter was introduced upstream of the multiple-cloning site; the appropriate region from pGEX-4T-3 was PCR amplified using primers PTACFwd/PTACRev, digested with NcoI and HindIII, and cloned into similarly digested pAL1353 to generate the final vector plasmid, which we called pBASE.
Construction of the A. baumannii mutant strains and complementation plasmids.
A. baumannii mutants were constructed as previously described (62). Briefly, two 1.5-kb DNA fragments that represented the region upstream and downstream of the gene of interest ([gene]) were amplified by PCR using the primer pair [gene]UpFwd with [gene]UpRev and the pair [gene]DwnFwd with [gene]DwnRev, respectively. A third PCR fragment (900 bp in size) was also generated that contained the kanamycin resistance gene, neo, from pCR-BluntII-TOPO using primers kanFwd and kanRev. The three DNA fragments were column purified and then combined at equimolar concentrations in a splice overlap extension (SOE) PCR using primers [gene]NestFwd and [gene]NestRev. The SOE PCR product used for each gene inactivation was introduced into A. baumannii strain AB307-0294 by electroporation as previously described (63), and putative mutants were selected on LB agar supplemented with 50 μg/ml kanamycin. Genomic DNA was isolated from kanamycin-resistant colonies, and the replacement of the gene of interest with the kanamycin cassette was confirmed by PCR amplification using primers specific for the region flanking the insertion ([gene]UpFwd and [gene]UpRev), followed by nucleotide sequencing of the PCR product to check that the region upstream and downstream of the mutation site was 100% identical at nucleotide level to the corresponding sequence in the WT AB307-0294 genome. For complementation of AB307-0294 ΔtssM (AL2730), the full-length tssM gene of A. baumannii strain AB307-0294 was amplified using the oligonucleotides tssMFwd and tssMRev that contained PvuI and PstI sites, respectively. The purified PCR fragment generated was digested with PstI and PvuI, column purified, and then ligated to pWH1266 digested with PstI and PvuI. The ligated products were then used to transform AB307-0294 ΔtssM by electroporation. One transformant, AL2735, was selected that contained the correct plasmid, pAL1211, as determined by sequencing. To generate the control strain, AL2734, the empty pWH1266 plasmid was separately used to transform AB307-0294 ΔtssM.
For complementation of the ΔvgrG1 and ΔvgrG2 strains, the full-length vgrG1 and vgrG2 genes from A. baumannii strain AB307-0294 were amplified using the primer pairs vgrG1Fwd/vgrG1Rev and vgrG2Fwd/vgrG2Rev, respectively. The purified PCR products generated were digested with KpnI and SacI and ligated to similarly digested pBASE to generate the plasmids pAL1415 (containing vgrG1) and pAL1416 (containing vgrG2) that were then used to separately transform (via electroporation) the strains AB307-0294 ΔvgrG1 and AB307-0294 ΔvrgG2, respectively. A single ΔvgrG1 transformant containing pAL1415 (AL3264) and a single ΔvgrG2 transformant containing pAL1416 (AL3265) were selected (Table 3). We were unable to generate a plasmid containing a functional vgrG3 using this cloning strategy. Instead, vgrG3 was cloned in a two-step process. Briefly, the DNA section (nucleotides −22 to 1609) encoding the N-terminal region of VgrG3 was PCR amplified using the primer vgrG3NFwd (containing an XbaI site) and vgrG3NRev, containing a silent, single-nucleotide change at the 5′ end to generate half of an SmaI site (and thus would generate a full SmaI site when ligated together with a DNA fragment cut with SmaI). The amplified product was then digested with XbaI and ligated to XbaI/SmaI-digested pBASE to generate the vgrG3 subclone pAL1446 (Table 3). To clone the remainder of the vgrG3 gene, primers vgrG3CFwd and vgrG3CRev were used to amplify nucleotides 1610 to 3307 of the gene. The generated product was digested with SacI and then ligated to SmaI- and SacI-digested pAL1446. The plasmid with the correct sequence, pAL1447, was then introduced via electroporation into strain AB307-0294 ΔvgrG3 to generate the complementation strain AL3352 (Table 3). To generate the control strains for the vgrG complementation studies, the pBASE vector was separately used to transform the strains AB307-0294 ΔvgrG1, AB307-0294 ΔvrgG2, and AB307-0294 ΔvrgG3 to generate strains AL3261, AL3262, and AL3263, respectively (Table 3).
Plasmid construction for heterologous protein expression in E. coli.
For cloning of rhs1-CT and rhs2-CT, the DNA regions encoding the putative C-terminal toxic domain of each protein (residues 1396 to 1591 and 1496 to 1624, respectively) were PCR amplified using AB307-0294 genomic DNA as the template with the appropriate primers (rhs1-CTFwd/rhs1-CTRev and rhs2-CTFwd/rhs2-CTRev, respectively [see Table S2 in the supplemental material]). The purified PCR products containing rhs1-CT and rhs2-CT were then digested with EcoRI and HindIII and separately cloned into EcoRI- and HindIII-digested vector pBAD30, generating the plasmids pAL1234 and pAL1237, respectively (Table 2). To generate LysM containing a PelB leader sequence, a PCR product containing the entire lysM open reading frame from AB307-0294 was generated using primers lysMFwd and lysMRev. The purified PCR product was digested with NcoI and HindIII and ligated to NcoI- and HindIII-digested vector pET22b, which encodes a PelB leader sequence upstream of the multiple-cloning site, to generate the plasmid pAL1330 (Table 2). A PCR product containing lysM with the pelB leader sequence was then generated using pAL1330 as the template with the primers pelBFwd and lysMRev (Table S2). The purified PCR product was digested with EcoRI and HindIII and then ligated to similarly digested pBAD30 to generate the plasmid pAL1275 (Table 2). For construction of the plasmids used for heterologous expression of rhs1I, rhs2I, and lysMI in E. coli, each gene was amplified from AB307-0294 genomic DNA using the appropriate primer pairs (rhs1IFwd/rhs1IRev, rhs2IFwd/rhs2IRev, and lysMIFwd/lysMIRev, respectively [Table S2]), digested with NcoI and HindIII, and cloned into similarly digested pET28a to generate the plasmids pAL1282, pAL1283, and pAL1331, respectively (Table 2).
Construction of plasmids for immunity protein expression in A. baylyi.
For coexpression experiments using rhs1I, rhs2I, and lysMI in A. baylyi strain ADP1, each gene was PCR amplified from AB307-0294 genomic DNA using the following primers: rhs1I primer pair rhs1IAbFwd/rhs1IAbRevI or rhs1IAbFwd/rhs1IAbRevII, rhs2I primer pair rhs2IAbFwd/rhs2IAbRev, and lysM primer pair lysMIAbFwd/lysMIAbRev (Table S2). Each of the purified PCR products was digested with PstI and PvuI and separately cloned into similarly digested pWH1266, producing the plasmids pAL1249 (containing a SacI site upstream of rhs1I), pAL1250 (containing an XmaI site upstream of rhs1I), pAL1251 (containing an XmaI site upstream of rhs2I), and pAL1252 (containing lysMI). To generate the plasmids expressing the gene pair rhs1I and lysMI or the gene pair rhs2I and lysMI, the primer pair lysMIAbFwd/lysMIAbRev was used to amplify lysMI from AB307-0294, and the fragment was digested with XmaI and PstI and then cloned into similarly digested pAL1250 or pAL1251 to produce the plasmid pAL1264 or pAL1265, respectively. To generate the plasmid expressing the gene pair rhs1I and rhs2I, pAL1263, the primer pair rhs2IAbFwd/rhs2IAbRev containing an XmaI site at the 5′ end was used to amplify rhs2I from AB307-0294 by PCR, and this fragment was digested with SacI and PstI and cloned into similarly digested pAL1249. To generate the triple-expression plasmid expressing rhs1I, rhs2I, and lysMI, the primer pair lysMIAbFwd/lysMIAbRev was used to amplify lysMI, which was then digested with XmaI and PstI, and the fragment was cloned into similarly digested pAL1263 to produce the plasmid pAL1319.
Overexpression of effector/immunity pairs in E. coli.
E. coli BL21(DE3) cells containing a pBAD30 effector expression plasmid and a pET28a immunity expression plasmid were grown overnight at 37°C in LB medium supplemented with 100 μg/ml ampicillin, 50 μg/ml kanamycin, and 0.4% glucose. Overnight cultures were diluted 1/100 into fresh LB medium (containing the appropriate antibiotics for plasmid maintenance) and grown to an OD600 of 0.1. Each culture was divided into four separate flasks and incubated for a further 1 h at 37°C before the OD600 was measured, and one of the following solutions was added to each culture. To induce expression of the recombinant effector protein, 10% l-arabinose was added to give a final concentration of 0.1% (vol/vol). To induce the expression of the recombinant immunity protein, 1 M isopropyl β-d-1-thiogalactopyranoside (IPTG) (Sigma-Aldrich, USA) was added to give a final concentration of 5 mM. To induce the expression of the effector and immunity proteins, 10% l-arabinose and 1 M IPTG (0.1% and 5 mM final concentrations, respectively) were added. As a control, the final culture received the equivalent amount of sterile distilled H2O (dH2O). The cultures were incubated at 37°C with shaking at 200 rpm for a further 7 h, and OD600 measurements were recorded each hour. In addition, at 2 h postinduction, a sample of each culture was taken for determination of the number of viable CFU in each culture by serial dilution and plating on LB medium.
Statistical analysis.
To determine if any of the differences in the ability of the A. baumannii strains (wild type, mutants, and complemented mutants) to kill various bacterial strains were statistically significant, two-way analysis of variance (ANOVA) was employed (GraphPad Prism, version 7) to analyze the data generated in the interbacterial competitive growth assays. Posttesting for all pairs was calculated automatically within GraphPad using Tukey's multiple-comparison test. A P value of <0.05 was accepted as statistically significant.
DNA nuclease assay.
E. coli strains harboring appropriate expression plasmids were grown to late exponential growth phase in LB medium at 37°C with shaking in the presence of the appropriate antibiotic for plasmid maintenance. Each 20-ml culture was then divided in half; one aliquot received l-arabinose (0.4% final concentration) to induce recombinant protein expression, and the other aliquot received d-glucose (0.4% final concentration) to repress recombinant protein expression. Cultures were incubated for a further 1 h at 37°C with shaking, and the optical density was measured to determine cell density. A volume of each culture equivalent to approximately 1 × 109 CFU was collected and pelleted by centrifugation (12, 000 × g, 1 min), and then the pellet was washed three times with sterile PBS. Excess liquid was removed from the tube, and the wet weight of the cell pellet was measured to ensure an equal cell mass for each sample. Each cell pellet was resuspended in 1 ml of filter-sterilized sonication buffer (phosphate-buffered saline containing 0.5% glycerol, 2 mM phenylmethylsulfonyl fluoride, 1 mM EDTA, and 10 mM β-mercaptoethanol), flash frozen in an ethanol (EtOH) dry-ice bath, and then stored at −80°C until lysis by sonication using a Branson Sonifier 450. Cell debris was removed by centrifugation at 16,000 × g for 30 min at 4°C, and the clarified lysates were stored at −80°C until required. Samples of the crude E. coli cell extracts were diluted 1/5 in sterile DNase activity buffer (20 mM Tris-Cl, pH 7.5, 50 mM NaCl, 10 mM MgSO4, 1 mM β-mercaptoethanol); then a 5-μl aliquot was added to 1 μg of purified A. baylyi genomic DNA, and the reaction volume was made up to 25 μl with DNase activity buffer. Reaction mixtures were incubated for 2 h at 37°C. The integrity of the A. baylyi genomic DNA was assessed by electrophoresis in a 0.8% agarose Tris-acetate-EDTA (TAE) gel, followed by visualization using a Fujifilm LAS300 image reader.
Phylogenetic tree construction.
Genes predicted to encode VgrG homologs were identified by tBLASTn searches using the three VgrG proteins present in AB307-0294 as sequential queries and the publically available genomes of one A. baylyi and 21 A. baumannii strains as the subjects. All genomes are available from the NCBI website. A. baumannii strains and sequence identifiers are as follows (locus tag identifiers): AB307-0294 (ABBFA), ATCC 17978 (ACX60), AB0057 (AB57), ACICU (ACICU), ATCC 19606 (HMPREF0010), AYE (ABAYE), SDF (ABSDF), 1656-2 (ABK1), BJAB0715 (BJAB0715), BJAB07104 (BJAB07104), BJAB0868 (BJAB0868), D1279779 (ABD1), LAC-4 (ABLAC), MDR-TJ (ABTJ), MDR-ZJ06 (ABZJ), NCGM 237 (AB237), TCDC-0715 (ABTW07), TYTH-1 (M3Q), ZW85-1 (P795), AC30 (B856), A1 (ABA1), and AB5075-UW (ABUW). For A. baylyi, the strain ADP1 (ACIAD) was used. A total of 73 unique vgrG gene sequences were identified, and the amino acid sequence of each VgrG was aligned using the multiple-sequence alignment program MAFFT (version 7) using the e-ins-i option (multiple conserved domains and long gaps), BLosum62 scoring matrix, and the remaining features set to default (64). A maximum likelihood phylogenetic tree was calculated using phyML with the LG substitution model, no invariable sites, nearest-neighbor interchange tree improvement, topology and branch optimization, and aBayes branch support calculation (65). Trees were visualized with iTOL (interactive tree of life) (66). Each of these programs is publically available online: MAFFT is available at http://mafft.cbrc.jp/alignment/server/index.html, phyML is available at http://www.atgc-montpellier.fr/phyml, and iTOL is available at http://itol.embl.de/itol.cgi.
Accession number(s).
The mass spectrometry proteomics data have been deposited in the ProteomeXchange Consortium via the PRIDE partner repository with the data set identifier PXD009528.
Supplementary Material
ACKNOWLEDGMENT
We gratefully acknowledge the excellent technical assistance of Haseena Mohideen, who initially cloned the rhs1 and rhs1I genes.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/IAI.00297-18.
REFERENCES
- 1.Gottig S, Gruber TM, Higgins PG, Wachsmuth M, Seifert H, Kempf VA. 2014. Detection of pan drug-resistant Acinetobacter baumannii in Germany. J Antimicrob Chemother 69:2578–2579. doi: 10.1093/jac/dku170. [DOI] [PubMed] [Google Scholar]
- 2.Rodriguez-Hernandez MJ, Saugar J, Docobo-Perez F, de la Torre BG, Pachon-Ibanez ME, Garcia-Curiel A, Fernandez-Cuenca F, Andreu D, Rivas L, Pachon J. 2006. Studies on the antimicrobial activity of cecropin A-melittin hybrid peptides in colistin-resistant clinical isolates of Acinetobacter baumannii. J Antimicrob Chemother 58:95–100. doi: 10.1093/jac/dkl145. [DOI] [PubMed] [Google Scholar]
- 3.Zarrilli R, Pournaras S, Giannouli M, Tsakris A. 2013. Global evolution of multidrug-resistant Acinetobacter baumannii clonal lineages. Int J Antimicrob Agents 41:11–19. doi: 10.1016/j.ijantimicag.2012.09.008. [DOI] [PubMed] [Google Scholar]
- 4.Carruthers MD, Nicholson PA, Tracy EN, Munson RS Jr. 2013. Acinetobacter baumannii utilizes a type VI secretion system for bacterial competition. PLoS One 8:e59388. doi: 10.1371/journal.pone.0059388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Repizo GD, Gagne S, Foucault-Grunenwald ML, Borges V, Charpentier X, Limansky AS, Gomes JP, Viale AM, Salcedo SP. 2015. Differential role of the T6SS in Acinetobacter baumannii virulence. PLoS One 10:e0138265. doi: 10.1371/journal.pone.0138265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shneider MM, Buth SA, Ho BT, Basler M, Mekalanos JJ, Leiman PG. 2013. PAAR-repeat proteins sharpen and diversify the type VI secretion system spike. Nature 500:350–353. doi: 10.1038/nature12453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weber BS, Ly PM, Irwin JN, Pukatzki S, Feldman MF. 2015. A multidrug resistance plasmid contains the molecular switch for type VI secretion in Acinetobacter baumannii. Proc Natl Acad Sci U S A 112:9442–9447. doi: 10.1073/pnas.1502966112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ringel PD, Hu D, Basler M. 2017. The role of type VI secretion system effectors in target cell lysis and subsequent horizontal gene transfer. Cell Rep 21:3927–3940. doi: 10.1016/j.celrep.2017.12.020. [DOI] [PubMed] [Google Scholar]
- 9.Weber BS, Hennon SW, Wright MS, Scott NE, de Berardinis V, Foster LJ, Ayala JA, Adams MD, Feldman MF. 2016. Genetic dissection of the type VI secretion system in Acinetobacter and identification of a novel peptidoglycan hydrolase, TagX, required for its biogenesis. mBio 7:e01253-16. doi: 10.1128/mBio.01253-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bingle LE, Bailey CM, Pallen MJ. 2008. Type VI secretion: a beginner's guide. Curr Opin Microbiol 11:3–8. doi: 10.1016/j.mib.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 11.Brunet YR, Zoued A, Boyer F, Douzi B, Cascales E. 2015. The type VI secretion TssEFGK-VgrG phage-like baseplate is recruited to the TssJLM membrane complex via multiple contacts and serves as assembly platform for tail tube/sheath polymerization. PLoS Genet 11:e1005545. doi: 10.1371/journal.pgen.1005545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lossi NS, Manoli E, Forster A, Dajani R, Pape T, Freemont P, Filloux A. 2013. The HsiB1C1 (TssB-TssC) complex of the Pseudomonas aeruginosa type VI secretion system forms a bacteriophage tail sheathlike structure. J Biol Chem 288:7536–7548. doi: 10.1074/jbc.M112.439273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bonemann G, Pietrosiuk A, Diemand A, Zentgraf H, Mogk A. 2009. Remodelling of VipA/VipB tubules by ClpV-mediated threading is crucial for type VI protein secretion. EMBO J 28:315–325. doi: 10.1038/emboj.2008.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ma J, Sun M, Dong W, Pan Z, Lu C, Yao H. 2017. PAAR-Rhs proteins harbor various C-terminal toxins to diversify the antibacterial pathways of type VI secretion systems. Environ Microbiol 19:345–360. doi: 10.1111/1462-2920.13621. [DOI] [PubMed] [Google Scholar]
- 15.Cianfanelli FR, Monlezun L, Coulthurst SJ. 2016. Aim, load, fire: the type VI secretion system, a bacterial nanoweapon. Trends Microbiol 24:51–62. doi: 10.1016/j.tim.2015.10.005. [DOI] [PubMed] [Google Scholar]
- 16.Jamet A, Nassif X. 2015. New players in the toxin field: polymorphic toxin systems in bacteria. mBio 6:e00285-15. doi: 10.1128/mBio.00285-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Basler M. 2015. Type VI secretion system: secretion by a contractile nanomachine. Philos Trans R Soc Lond B Biol Sci 370:20150021. doi: 10.1098/rstb.2015.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alcoforado Diniz J, Liu YC, Coulthurst SJ. 2015. Molecular weaponry: diverse effectors delivered by the Type VI secretion system. Cell Microbiol 17:1742–1751. doi: 10.1111/cmi.12532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alcoforado Diniz J, Coulthurst SJ. 2015. Intraspecies competition in Serratia marcescens is mediated by type VI-secreted Rhs effectors and a conserved effector-associated accessory protein. J Bacteriol 197:2350–2360. doi: 10.1128/JB.00199-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Burtnick MN, Brett PJ, Harding SV, Ngugi SA, Ribot WJ, Chantratita N, Scorpio A, Milne TS, Dean RE, Fritz DL, Peacock SJ, Prior JL, Atkins TP, Deshazer D. 2011. The cluster 1 type VI secretion system is a major virulence determinant in Burkholderia pseudomallei. Infect Immun 79:1512–1525. doi: 10.1128/IAI.01218-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Coulthurst SJ. 2013. The type VI secretion system-a widespread and versatile cell targeting system. Res Microbiol 164:640–654. doi: 10.1016/j.resmic.2013.03.017. [DOI] [PubMed] [Google Scholar]
- 22.Durand E, Cambillau C, Cascales E, Journet L. 2014. VgrG, Tae, Tle, and beyond: the versatile arsenal of Type VI secretion effectors. Trends Microbiol 22:498–507. doi: 10.1016/j.tim.2014.06.004. [DOI] [PubMed] [Google Scholar]
- 23.Zheng J, Leung KY. 2007. Dissection of a type VI secretion system in Edwardsiella tarda. Mol Microbiol 66:1192–1206. doi: 10.1111/j.1365-2958.2007.05993.x. [DOI] [PubMed] [Google Scholar]
- 24.Whitney JC, Beck CM, Goo YA, Russell AB, Harding BN, De Leon JA, Cunningham DA, Tran BQ, Low DA, Goodlett DR, Hayes CS, Mougous JD. 2014. Genetically distinct pathways guide effector export through the type VI secretion system. Mol Microbiol 92:529–542. doi: 10.1111/mmi.12571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Silverman JM, Agnello DM, Zheng H, Andrews BT, Li M, Catalano CE, Gonen T, Mougous JD. 2013. Haemolysin coregulated protein is an exported receptor and chaperone of type VI secretion substrates. Mol Cell 51:584–593. doi: 10.1016/j.molcel.2013.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brooks TM, Unterweger D, Bachmann V, Kostiuk B, Pukatzki S. 2013. Lytic activity of the Vibrio cholerae type VI secretion toxin VgrG-3 is inhibited by the antitoxin TsaB. J Biol Chem 288:7618–7625. doi: 10.1074/jbc.M112.436725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hachani A, Allsopp LP, Oduko Y, Filloux A. 2014. The VgrG proteins are “à la carte” delivery systems for bacterial type VI effectors. J Biol Chem 289:17872–17884. doi: 10.1074/jbc.M114.563429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bondage DD, Lin JS, Ma LS, Kuo CH, Lai EM. 2016. VgrG C terminus confers the type VI effector transport specificity and is required for binding with PAAR and adaptor-effector complex. Proc Natl Acad Sci U S A 113:e3931–e3940. doi: 10.1073/pnas.1600428113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cianfanelli FR, Alcoforado Diniz J, Guo M, De Cesare V, Trost M, Coulthurst SJ. 2016. VgrG and PAAR proteins define distinct versions of a functional type VI secretion system. PLoS Pathog 12:e1005735. doi: 10.1371/journal.ppat.1005735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ma LS, Hachani A, Lin JS, Filloux A, Lai EM. 2014. Agrobacterium tumefaciens deploys a superfamily of type VI secretion DNase effectors as weapons for interbacterial competition in planta. Cell Host Microbe 16:94–104. doi: 10.1016/j.chom.2014.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koskiniemi S, Lamoureux JG, Nikolakakis KC, t'Kint de Roodenbeke C, Kaplan MD, Low DA, Hayes CS. 2013. Rhs proteins from diverse bacteria mediate intercellular competition. Proc Natl Acad Sci U S A 110:7032–7037. doi: 10.1073/pnas.1300627110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Salomon D, Kinch LN, Trudgian DC, Guo X, Klimko JA, Grishin NV, Mirzaei H, Orth K. 2014. Marker for type VI secretion system effectors. Proc Natl Acad Sci U S A 111:9271–9276. doi: 10.1073/pnas.1406110111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Unterweger D, Miyata ST, Bachmann V, Brooks TM, Mullins T, Kostiuk B, Provenzano D, Pukatzki S. 2014. The Vibrio cholerae type VI secretion system employs diverse effector modules for intraspecific competition. Nat Commun 5:3549. doi: 10.1038/ncomms4549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liang X, Moore R, Wilton M, Wong MJ, Lam L, Dong TG. 2015. Identification of divergent type VI secretion effectors using a conserved chaperone domain. Proc Natl Acad Sci U S A 112:9106–9111. doi: 10.1073/pnas.1505317112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weber BS, Miyata ST, Iwashkiw JA, Mortensen BL, Skaar EP, Pukatzki S, Feldman MF. 2013. Genomic and functional analysis of the type VI secretion system in Acinetobacter. PLoS One 8:e55142. doi: 10.1371/journal.pone.0055142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Henry R, Vithanage N, Harrison P, Seemann T, Coutts S, Moffatt JH, Nation RL, Li J, Harper M, Adler B, Boyce JD. 2012. Colistin-resistant, lipopolysaccharide-deficient Acinetobacter baumannii responds to lipopolysaccharide loss through increased expression of genes involved in the synthesis and transport of lipoproteins, phospholipids, and poly-beta-1,6-N-acetylglucosamine. Antimicrob Agents Chemother 56:59–69. doi: 10.1128/AAC.05191-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aschtgen MS, Bernard CS, De Bentzmann S, Lloubes R, Cascales E. 2008. SciN is an outer membrane lipoprotein required for type VI secretion in enteroaggregative Escherichia coli. J Bacteriol 190:7523–7531. doi: 10.1128/JB.00945-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jones C, Hachani A, Manoli E, Filloux A. 2014. An rhs gene linked to the second type VI secretion cluster is a feature of the Pseudomonas aeruginosa strain PA14. J Bacteriol 196:800–810. doi: 10.1128/JB.00863-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Russell AB, Hood RD, Bui NK, LeRoux M, Vollmer W, Mougous JD. 2011. Type VI secretion delivers bacteriolytic effectors to target cells. Nature 475:343–347. doi: 10.1038/nature10244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pukatzki S, Ma AT, Sturtevant D, Krastins B, Sarracino D, Nelson WC, Heidelberg JF, Mekalanos JJ. 2006. Identification of a conserved bacterial protein secretion system in Vibrio cholerae using the Dictyostelium host model system. Proc Natl Acad Sci U S A 103:1528–1533. doi: 10.1073/pnas.0510322103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jackson AP, Thomas GH, Parkhill J, Thomson NR. 2009. Evolutionary diversification of an ancient gene family (rhs) through C-terminal displacement. BMC Genomics 10:584. doi: 10.1186/1471-2164-10-584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Poole SJ, Diner EJ, Aoki SK, Braaten BA, t'Kint de Roodenbeke C, Low DA, Hayes CS. 2011. Identification of functional toxin/immunity genes linked to contact-dependent growth inhibition (CDI) and rearrangement hotspot (Rhs) systems. PLoS Genet 7:e1002217. doi: 10.1371/journal.pgen.1002217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hunger M, Schmucker R, Kishan V, Hillen W. 1990. Analysis and nucleotide sequence of an origin of DNA replication in Acinetobacter calcoaceticus and its use for Escherichia coli shuttle plasmids. Gene 87:45–51. doi: 10.1016/0378-1119(90)90494-C. [DOI] [PubMed] [Google Scholar]
- 44.Pukatzki S, Ma AT, Revel AT, Sturtevant D, Mekalanos JJ. 2007. Type VI secretion system translocates a phage tail spike-like protein into target cells where it cross-links actin. Proc Natl Acad Sci U S A 104:15508–15513. doi: 10.1073/pnas.0706532104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Unterweger D, Kostiuk B, Otjengerdes R, Wilton A, Diaz-Satizabal L, Pukatzki S. 2015. Chimeric adaptor proteins translocate diverse type VI secretion system effectors in Vibrio cholerae. EMBO J 34:2198–2210. doi: 10.15252/embj.201591163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Alteri CJ, Himpsl SD, Pickens SR, Lindner JR, Zora JS, Miller JE, Arno PD, Straight SW, Mobley HL. 2013. Multicellular bacteria deploy the type VI secretion system to preemptively strike neighboring cells. PLoS Pathog 9:e1003608. doi: 10.1371/journal.ppat.1003608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Unterweger D, Kitaoka M, Miyata ST, Bachmann V, Brooks TM, Moloney J, Sosa O, Silva D, Duran-Gonzalez J, Provenzano D, Pukatzki S. 2012. Constitutive type VI secretion system expression gives Vibrio cholerae intra- and interspecific competitive advantages. PLoS One 7:e48320. doi: 10.1371/journal.pone.0048320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hood RD, Singh P, Hsu F, Guvener T, Carl MA, Trinidad RR, Silverman JM, Ohlson BB, Hicks KG, Plemel RL, Li M, Schwarz S, Wang WY, Merz AJ, Goodlett DR, Mougous JD. 2010. A type VI secretion system of Pseudomonas aeruginosa targets a toxin to bacteria. Cell Host Microbe 7:25–37. doi: 10.1016/j.chom.2009.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Russell AB, Singh P, Brittnacher M, Bui NK, Hood RD, Carl MA, Agnello DM, Schwarz S, Goodlett DR, Vollmer W, Mougous JD. 2012. A widespread bacterial type VI secretion effector superfamily identified using a heuristic approach. Cell Host Microbe 11:538–549. doi: 10.1016/j.chom.2012.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.English G, Trunk K, Rao VA, Srikannathasan V, Hunter WN, Coulthurst SJ. 2012. New secreted toxins and immunity proteins encoded within the type VI secretion system gene cluster of Serratia marcescens. Mol Microbiol 86:921–936. doi: 10.1111/mmi.12028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Whitney JC, Chou S, Russell AB, Biboy J, Gardiner TE, Ferrin MA, Brittnacher M, Vollmer W, Mougous JD. 2013. Identification, structure, and function of a novel type VI secretion peptidoglycan glycoside hydrolase effector-immunity pair. J Biol Chem 288:26616–26624. doi: 10.1074/jbc.M113.488320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang D, de Souza RF, Anantharaman V, Iyer LM, Aravind L. 2012. Polymorphic toxin systems: Comprehensive characterization of trafficking modes, processing, mechanisms of action, immunity and ecology using comparative genomics. Biol Direct 7:18. doi: 10.1186/1745-6150-7-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Antunes LC, Imperi F, Carattoli A, Visca P. 2011. Deciphering the multifactorial nature of Acinetobacter baumannii pathogenicity. PLoS One 6:e22674. doi: 10.1371/journal.pone.0022674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.de Breij A, Dijkshoorn L, Lagendijk E, van der Meer J, Koster A, Bloemberg G, Wolterbeek R, van den Broek P, Nibbering P. 2010. Do biofilm formation and interactions with human cells explain the clinical success of Acinetobacter baumannii? PLoS One 5:e10732. doi: 10.1371/journal.pone.0010732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Giannouli M, Antunes LC, Marchetti V, Triassi M, Visca P, Zarrilli R. 2013. Virulence-related traits of epidemic Acinetobacter baumannii strains belonging to the international clonal lineages I-III and to the emerging genotypes ST25 and ST78. BMC Infect Dis 13:282. doi: 10.1186/1471-2334-13-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sahl JW, Gillece JD, Schupp JM, Waddell VG, Driebe EM, Engelthaler DM, Keim P. 2013. Evolution of a pathogen: a comparative genomics analysis identifies a genetic pathway to pathogenesis in Acinetobacter. PLoS One 8:e54287. doi: 10.1371/journal.pone.0054287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cooper RM, Tsimring L, Hasty J. 2017. Inter-species population dynamics enhance microbial horizontal gene transfer and spread of antibiotic resistance. Elife 6:e25950. doi: 10.7554/eLife.25950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Adams MD, Goglin K, Molyneaux N, Hujer KM, Lavender H, Jamison JJ, MacDonald IJ, Martin KM, Russo T, Campagnari AA, Hujer AM, Bonomo RA, Gill SR. 2008. Comparative genome sequence analysis of multidrug-resistant Acinetobacter baumannii. J Bacteriol 190:8053–8064. doi: 10.1128/JB.00834-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hatfaludi T, Al-Hasani K, Gong L, Boyce JD, Ford M, Wilkie IW, Quinsey N, Dunstone MA, Hoke DE, Adler B. 2012. Screening of 71 P. multocida proteins for protective efficacy in a fowl cholera infection model and characterization of the protective antigen PlpE. PLoS One 7:e39973. doi: 10.1371/journal.pone.0039973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gulliver EL, Wright A, Deveson Lucas D, Megroz M, Kleifeld O, Schittenhelm R, Powell D, Seemann T, Bulitta J, Harper M, Boyce JD. 2018. Determination of the small RNA GcvB regulon in the Gram-negative bacterial pathogen Pasteurella multocida and identification of the GcvB seed binding region. RNA 24:704–720. doi: 10.1261/rna.063248.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cox J, Mann M. 2008. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol 26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 62.Aranda J, Poza M, Pardo BG, Rumbo S, Rumbo C, Parreira JR, Rodriguez-Velo P, Bou G. 2010. A rapid and simple method for constructing stable mutants of Acinetobacter baumannii. BMC Microbiol 10:279. doi: 10.1186/1471-2180-10-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Choi KH, Kumar A, Schweizer HP. 2006. A 10-min method for preparation of highly electrocompetent Pseudomonas aeruginosa cells: application for DNA fragment transfer between chromosomes and plasmid transformation. J Microbiol Methods 64:391–397. doi: 10.1016/j.mimet.2005.06.001. [DOI] [PubMed] [Google Scholar]
- 64.Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 66.Letunic I, Bork P. 2011. Interactive Tree Of Life v2: online annotation and display of phylogenetic trees made easy. Nucleic Acids Res 39:W475–8. doi: 10.1093/nar/gkr201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Grant SG, Jessee J, Bloom FR, Hanahan D. 1990. Differential plasmid rescue from transgenic mouse DNAs into Escherichia coli methylation-restriction mutants. Proc Natl Acad Sci U S A 87:4645–4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Barbe V, Vallenet D, Fonknechten N, Kreimeyer A, Oztas S, Labarre L, Cruveiller S, Robert C, Duprat S, Wincker P, Ornston LN, Weissenbach J, Marliere P, Cohen GN, Medigue C. 2004. Unique features revealed by the genome sequence of Acinetobacter sp. ADP1, a versatile and naturally transformation competent bacterium. Nucleic Acids Res 32:5766–5779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Niu C, Clemmer KM, Bonomo RA, Rather PN. 2008. Isolation and characterization of an autoinducer synthase from Acinetobacter baumannii. J Bacteriol 190:3386–3392. doi: 10.1128/JB.01929-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Guzman LM, Belin D, Carson MJ, Beckwith J. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol 177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.