ABSTRACT
Nontypeable Haemophilus influenzae (NTHi) is an exclusively human pathobiont that plays a critical role in the course and pathogenesis of chronic obstructive pulmonary disease (COPD). NTHi causes acute exacerbations of COPD and also causes persistent infection of the lower airways. NTHi expresses four IgA protease variants (A1, A2, B1, and B2) that play different roles in virulence. Expression of IgA proteases varies among NTHi strains, but little is known about the frequency and mechanisms by which NTHi modulates IgA protease expression during infection in COPD. To assess expression of IgA protease during natural infection in COPD, we studied IgA protease expression by 101 persistent strains (median duration of persistence, 161 days; range, 2 to 1,422 days) collected longitudinally from patients enrolled in a 20-year study of COPD upon initial acquisition and immediately before clearance from the host. Upon acquisition, 89 (88%) expressed IgA protease. A total of 16 of 101 (16%) strains of NTHi altered expression of IgA protease during persistence. Indels and slipped-strand mispairing of mononucleotide repeats conferred changes in expression of igaA1, igaA2, and igaB1. Strains with igaB2 underwent frequent changes in expression of IgA protease B2 during persistence, mediated by slipped-strand mispairing of a 7-nucleotide repeat, TCAAAAT, within the open reading frame of igaB2. We conclude that changes in iga gene sequences result in changes in expression of IgA proteases by NTHi during persistent infection in the respiratory tract of patients with COPD.
KEYWORDS: IgA protease, nontypeable Haemophilus influenzae, chronic obstructive pulmonary disease, respiratory tract infections
INTRODUCTION
Nontypeable Haemophilus influenzae (NTHi) is a pathobiont of the human upper airways commonly found in the nasopharynx of children and adults (1, 2). NTHi is frequently isolated from patients with chronic obstructive pulmonary disease (COPD) and is the most common bacterial cause of acute exacerbations of COPD, which are associated with substantial morbidity and mortality (3, 4). In addition to causing acute exacerbations, NTHi is present in the lower airways of patients during clinically stable periods (5). This exclusively human pathogen has evolved mechanisms and virulence factors to maintain persistent infection in the hostile environment of the human lower airways for months to years (5, 6). One of these virulence factors is immunoglobulin A (IgA) protease.
IgA proteases cleave the hinge region of human IgA1 and also facilitate bacterial infection by a number of mechanisms, including stimulating proinflammatory cytokines, such as tumor necrosis factor alpha, interleukin-1β (IL-1β), and IL-8, and mediating intracellular persistence in respiratory epithelial cells by cleaving lysosome-associated membrane protein 1 (LAMP1) (7–9). NTHi has two IgA protease genes in separate regions of the genome, igaA and igaB, each with 2 variants, for a total of 4 IgA proteases. All strains of NTHi contain igaA, and approximately 40% of strains isolated from patients with COPD also contain the igaB gene (9). Each variant of IgA protease has a unique cleavage site along the hinge region of human IgA1 (9). In addition, IgA proteases B1 and B2 cleave LAMP1 and mediate intracellular survival in human respiratory epithelial cells (8). NTHi strains with both an igaA gene and an igaB gene are adapted for colonization and infection in COPD (10).
Expression of IgA proteases is variable from strain to strain among NTHi (9). Poole et al. (11) challenged healthy adults by nasal inoculation with a strain of NTHi and demonstrated increased expression of IgA protease B2 in the challenge strain after persistence in the nasopharynx. We demonstrated previously that passage of two strains of NTHi through H292 respiratory epithelial cells selected for the expression of IgA proteases B1 and B2 compared to in vitro passage (12). These studies show that selected strains of NTHi regulate expression of IgA proteases under carefully defined but artificial conditions (nasal inoculation of a laboratory-grown strain and in vitro cell culture). The present study builds on these observations in two ways. First, studying IgA protease expression of strains of NTHi isolated at the time of acquisition by the patient and after persistence in human airways allows conclusions regarding changes in IgA protease expression of NTHi strains acquired by natural infection and during persistence in the airways of COPD patients. Second, our unique collection of 101 prospectively collected strains of NTHi from adults with COPD provides an accurate estimate of the incidence of changes in IgA protease expression during natural infection in humans. Thus, the goal of this study was to examine the extent to which NTHi changes the expression of IgA proteases during persistence in the human respiratory tract by 101 strains isolated from patients with COPD as part of a 20-year longitudinal study to further our understanding of the role of IgA proteases in NTHi infection in the clinical setting of COPD.
RESULTS
Expression of IgA protease during persistence.
To assess the expression of IgA proteases in NTHi strains that persisted in COPD patient airways, IgA protease expression was determined in the first isolate upon acquisition of a strain of NTHi by the patient and the final isolate of the strain just prior to clearance for 101 persistent strains of NTHi (median duration of persistence, 161 days, range, 2 to 1,422 days). In all strains in which no IgA protease expression was observed in the first isolate (n = 12), that pattern of negative expression continued in the final isolate of the same strain of NTHi. Analysis of igaA1 and igaA2 gene sequences in the 12 isolates that did not express the IgA protease upon acquisition of the strain revealed that mutations in the open reading frames caused the gene to be out of frame, resulting in no IgA protease expression.
Figure 1 shows that most strains that expressed IgA protease in the first isolate upon acquisition continued to express the same IgA protease throughout persistence. However, selected strains of each of the 4 IgA protease variants demonstrated a change in expression between the first isolate and the final isolate during persistence.
FIG 1.
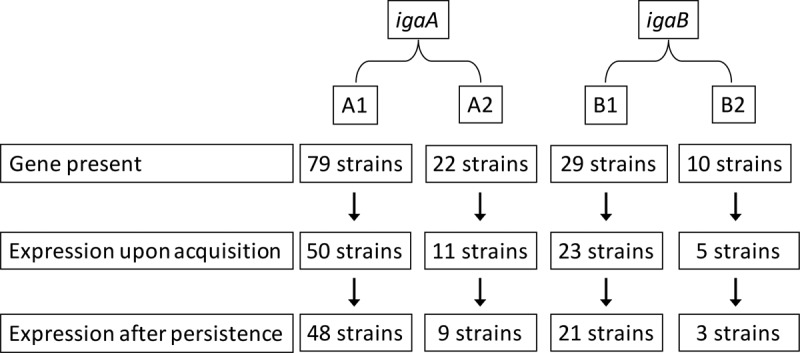
Distribution of iga genes and IgA protease activity upon initial acquisition and final isolation in 101 persistent strains of nontypeable Haemophilus influenzae isolated from sputum samples from patients with chronic obstructive pulmonary disease.
Mechanism of changes in expression: IgA protease A.
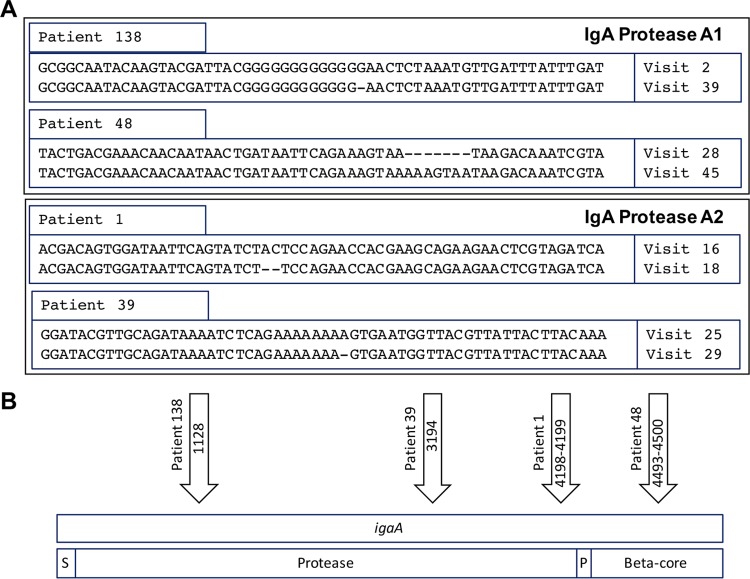
Of the 50 strains that expressed IgA protease A1 at the time of acquisition by the patient, 48 strains showed continued expression following persistence (median, 161 days; range, 2 to 1,420 days), while 2 strains no longer expressed IgA protease A1 in the final isolate (Fig. 2). To explore the mechanism that accounts for changes in igaA1 protease expression, the sequences of the igaA1 genes in isolates of the persistent strains that ceased expression were aligned and compared. A single base deletion in a single nucleotide repeat of 12 G residues was present in the last (nonexpressing) isolate of the strain obtained from patient 138 compared to the sequence of the first isolate, causing the gene to be out of frame. The strain from patient 48 acquired a 7-bp insertion, causing a frameshift in igaA1, accounting for the loss of expression of IgA protease A1 during persistence (Fig. 3A). To begin to assess the diversity of the population of these isolates at the time that they were cultured, individual colonies isolated from the original sputum sample were each grown independently and assayed for IgA protease expression. In each case, all colonies from a sputum sample showed the same expression as the others in the same sample. The number of colonies examined for each isolate is noted in Fig. 2.
FIG 2.
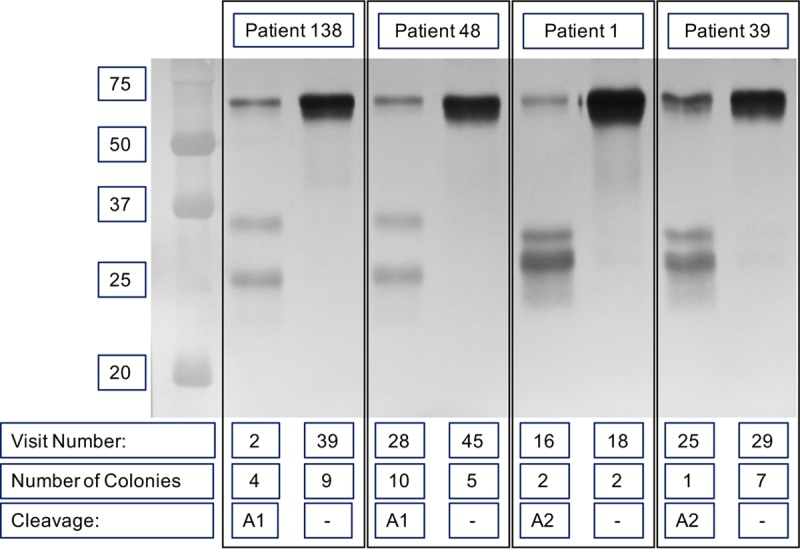
Immunoblot probed with peroxidase-conjugated anti-human IgA showing the IgA protease activity of strains isolated from four patients with COPD (noted at the top) upon acquisition (the lower visit number noted at the bottom) and just before clearance from the host (the higher visit number noted at the bottom). Number of colonies, the number of individually collected colonies that were tested for IgA protease expression upon isolation; Cleavage, IgA protease activity. Molecular mass markers (in kilodaltons) are shown on the left.
FIG 3.
(A) Genome fragments of igaA that contain the change between the first and final isolates of persistent NTHi strains during persistent infection in COPD. The visit numbers on the right indicate the monthly clinic visit at which the isolate was recovered from each patient. (B) Schematic representing the igaA gene (top) and functional domains (bottom). Arrows, the location in the open reading frame of the genomic changes shown in panel A; S, 25-amino-acid signal sequence; Protease, secreted IgA1 protease domain; P, short conserved peptide; Beta-core, beta-barrel component of the type V secretion system.
Of the 11 strains that expressed IgA protease A2 at the first isolation, 9 strains showed continued expression during persistence (median duration of persistence, 77 days; range, 27 to 1,175 days) and 2 strains stopped expression (Fig. 2). Aligning the sequences of the igaA2 genes from the strain from patient 1 that ceased expression showed a 2-bp deletion resulting in a frameshift in the gene and loss of expression in the final isolate of this strain. Comparison of the gene sequences of igaA2 from the first and final isolates of the persistent strain from patient 39 revealed a loss of a single adenine in a single nucleotide repeat of 8 A residues from the initial acquisition isolate, resulting in a frameshift (Fig. 3A). All available colonies of each isolate were tested independently, and in each case, all colonies from a sputum sample showed the same expression as the others in the same sample. The number of colonies examined for each isolate is noted in Fig. 2.
In addition to the variability of the genetic changes within the open reading frames of IgA proteases A1 and A2 (insertions and deletions of various lengths), the location of the changes in the iga genes of the different strains was distributed throughout the length of the igaA1 and igA2 genes and the changes were not localized to one region or to one domain of the gene. Changes occurred in the functional protease domain and the beta-core required for type V secretion of IgA proteases (13) (Fig. 3B). The observation that the sequence change in igaA1 in the strain from patient 48 that is near the C terminus of the gene results in the total loss of IgA protease expression indicates that the beta-core is required for secretion of the functional protease into the extracellular space.
Mechanism of changes in expression: IgA protease B1.
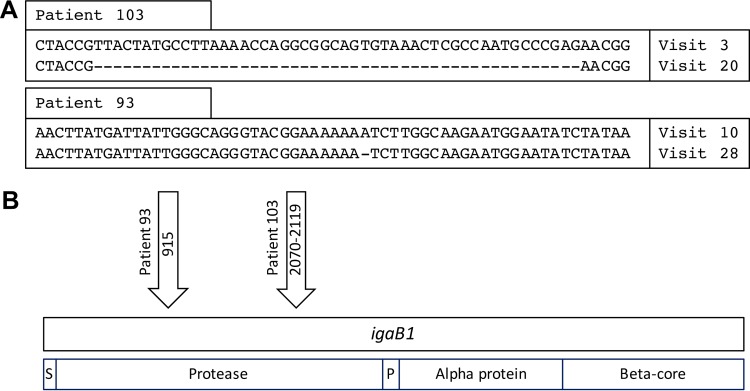
Of the 23 persistent strains that expressed IgA protease B1 upon acquisition by the patient, 21 continued to express IgA protease B1 during persistence (median duration of persistence, 277 days; range, 17 to 1,193 days) (Fig. 4). All 6 persistent strains that had the igaB1 gene and that did not express IgA protease B1 upon acquisition continued to show no expression in the final isolate. Alignment of the sequences of igaB1 of the first and final isolates demonstrated a 49-bp deletion in the strain obtained from patient 103 and a single adenine deletion in a mononucleotide repeat of 7 A residues in the strain obtained from patient 93, causing the final isolates before clearance to be frameshifted and no longer express IgA protease B1 (Fig. 5A). The changes in both of the igaB1 genes were located within the open reading frame near the N terminus, in the structural IgA protease domain of the gene (Fig. 5B). All available colonies of each isolate were tested independently, and in each case, all colonies from a sputum sample showed the same expression as the others in the same sample. The number of colonies examined for each isolate is noted in Fig. 4.
FIG 4.
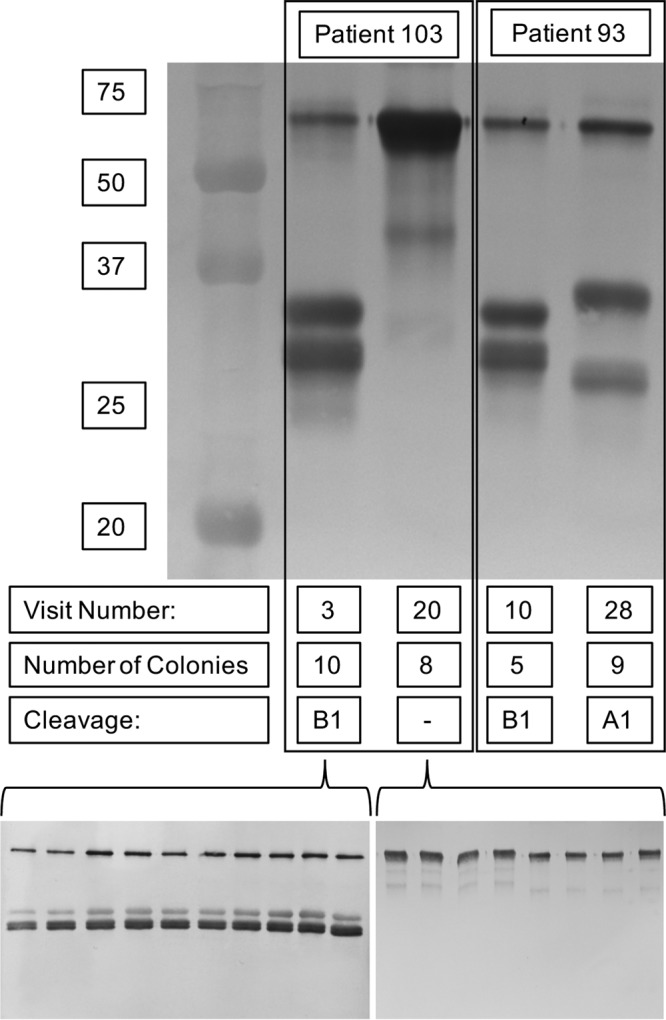
Immunoblot probed with peroxidase-conjugated anti-human IgA showing IgA protease activity of strains isolated from two patients with COPD (noted at the top) upon acquisition (the lower visit number noted at the bottom) and just before clearance from the host (the higher visit number noted at the bottom). Number of colonies, the number of individually collected colonies that were tested for IgA protease expression upon isolation; Cleavage, IgA protease activity. Molecular mass markers (in kilodaltons) are shown on the left. The lower portion demonstrates individual colonies at the time of collection that were assayed for IgA protease activity to show the population of expression.
FIG 5.
(A) Genome fragments of igaB1 that contain the change between the first and final isolates of persistent NTHi strains during persistent infection in COPD patients. (B) Schematic representing the igaB1 gene (top) and functional domains (bottom). Arrows, the location in the open reading frame of the genomic changes shown in panel A; S, 25-amino-acid signal sequence; Protease, secreted IgA1 protease domain; P, short conserved peptide; Alpha protein, alpha protein; Beta-core, beta-barrel component of the type V secretion system.
Mechanism of changes in expression: IgA protease B2.
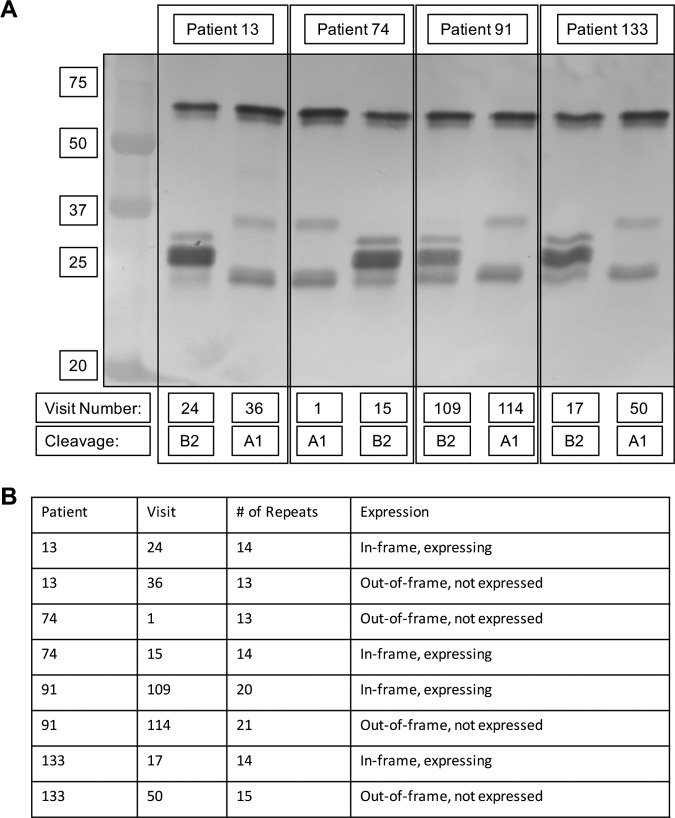
Inspection of the expression patterns of the first and final isolates of the 10 persistent strains with the igaB2 gene showed a variety of patterns, including (i) expression of IgA protease B2 by 6 of 10 strains initially, (ii) loss of expression by 3 of these 6 strains, and (iii) a transition from no expression to expression by 1 of 4 strains that were not expressing IgA protease B2 upon acquisition. In contrast to strains with the igaA1, igaA2, and igaB1 genes, analysis of the igaB2 genes of strains that changed expression during persistence did not reveal insertions or deletions within the open reading frame causing a frameshift. Instead, a simple sequence repeat 7 bp in length, TCAAAAT, was present immediately downstream of the ATG start codon. When the number of TCAAAAT repeats caused the gene to be out of frame, IgA protease B2 was not expressed. Conversely, when the number of TCAAAAT repeats caused the igaB2 gene to be in frame, IgA protease B2 expression was observed. Figure 6A shows immunoblots for the three strains that expressed IgA protease B2 upon acquisition and the loss of expression in the final isolate (patients 13, 91, and 133) and one strain that showed no expression upon acquisition but expressed IgA protease B2 in the final isolate after 421 days of persistence (patient 74). In each case, expression was determined by the number of 7-bp repeats placing the gene in frame or out of frame. The change in repeat number fluctuated by only one between the first and final isolates in each strain (Fig. 6B).
FIG 6.
(A) Immunoblot probed with peroxidase-conjugated anti-human IgA showing the IgA protease activity of strains isolated from four patients with COPD (noted at the top) upon acquisition (the lower visit number noted at the bottom) and just before clearance from the host (the higher visit number noted at that bottom). Cleavage, IgA protease activity. Molecular mass markers (in kilodaltons) are shown on the left. (B) Table showing the relationship between the number of simple sequence repeats within the igaB2 gene (number of repeats) and IgA protease B2 expression.
Figures 4 and 6A show that the IgA protease A1 cleavage pattern is observed in isolates when igaB2 is out of frame. We have shown previously that transcription of the igaB genes is approximately 100-fold greater than that of the igaA genes in quantitative real-time PCR, accounting for this observation (9).
To analyze more rigorously the relationship between the number of TCAAAAT repeats and expression of IgA protease B2, a fragment spanning the repeat region was amplified by PCR and the sequence was determined in 66 isolates. IgA protease expression was assayed in the same 66 isolates. In each isolate, IgA protease B2 was expressed when the number of repeats placed the igaB2 gene in frame. Similarly, IgA protease B2 was not expressed when the number of repeats placed the igaB2 gene out of frame (Table 1).
TABLE 1.
Relationship of expression of IgA protease B2 to the number of TCAAAAT repeats in the igaB2 gene
| No. of repeats | No. of isolates | Frame | No. of isolates expressing IgA protease B2 | % of isolates expressing IgA protease B2 |
|---|---|---|---|---|
| 1 | 3 | Out | 0 | 0 |
| 10 | 1 | Out | 0 | 0 |
| 11 | 10 | In | 10 | 100 |
| 12 | 24 | Out | 0 | 0 |
| 13 | 11 | Out | 0 | 0 |
| 14 | 9 | In | 9 | 100 |
| 15 | 1 | Out | 0 | 0 |
| 16 | 2 | Out | 0 | 0 |
| 17 | 2 | In | 2 | 100 |
| 19 | 0 | Out | 0 | 0 |
| 20 | 1 | In | 1 | 100 |
| 21 | 2 | Out | 0 | 0 |
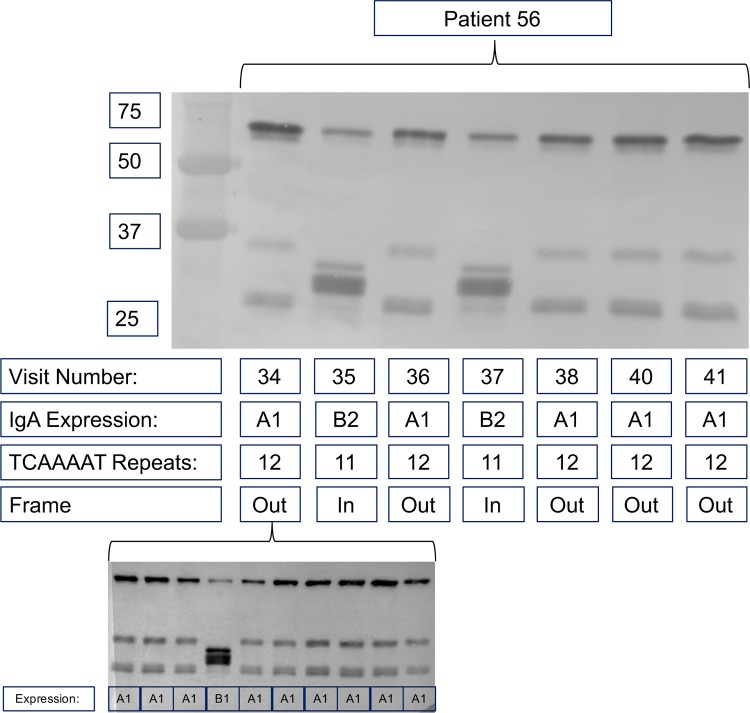
Analysis of sequentially collected intervening isolates of selected strains of NTHi containing the igaB2 gene revealed that expression of IgA protease B2 varies throughout the duration of persistence. For example, Fig. 7 shows a timeline of 7 sequentially collected isolates of a single strain over 7 monthly clinic visits of patient 56. The observation that the initial isolate at visit number 34 and the final isolate before clearance at visit number 41 both demonstrated a lack of IgA protease B2 expression while two isolates at visits 35 and 37 expressed IgA protease B2 supports the conclusion that frequent changes in expression due to slipped-strand mispairing occur during persistent infection in strains of NTHi that contain the igaB2 gene.
FIG 7.
Immunoblot probed with peroxidase-conjugated anti-human immunoglobulin IgA showing IgA protease expression of sequential isolates of a persistent NTHi strain from patient 56 over the course of 217 days. Visit number, the monthly clinic visit; IgA expression, the IgA protease variant expressed; TCAAAAT repeats, the number of repeats in the igaB2 gene; Frame, whether the number of repeats places the igaB2 gene in frame or out of frame. Molecular mass markers (in kilodaltons) are shown on the left. The lower blot shows IgA protease expression from cultures grown from individual colonies obtained from the original culture of sputum collected at visit 34.
Cultures grown from individual colonies from each visit were examined for IgA protease expression. We observed expression of different IgA protease variants from the population of isolates collected from the same culture in the case of IgA protease B2 (Fig. 7), in contrast to that observed in the case of IgA proteases A1, A2, and B1. The individual colonies were confirmed to be the same strain by molecular typing. We conclude that expression of IgA protease B2 is highly variable during persistence in COPD airways and is mediated by a slipped-strand mispairing.
Changes in the number of TCAAAAT repeats in the igaB2 gene alter the frame of the gene and therefore change IgA protease B2 expression during persistence in the human respiratory tract. We conclude that expression of IgA protease B2 during persistence in COPD is dynamic and that the frequent changes in expression are regulated by slipped-strand mispairing of the igaB2 gene. NTHi changes expression in different microenvironments in the airways.
DISCUSSION
The focus of this study was the expression of IgA proteases, important virulence factors of NTHi, during persistence in the airways of adults with COPD. Our observations are based on analysis of strains of NTHi that persisted in the airways of adults with COPD for months to years and that were isolated as part of a 20-year longitudinal study (5, 14). The present study shows that some strains altered their expression of IgA proteases and further shows that changes in the genome during persistence account for changes in expression, advancing understanding of the mechanisms by which NTHi alters expression of each of the four IgA protease variants. IgA proteases B1 and B2 cleave lysosome-associated membrane protein 1 (LAMP1) and mediate intracellular survival in human respiratory epithelial cells, facilitating the persistence of NTHi (8, 12). IgA proteases A1 and A2 do not cleave LAMP1, nor do they enhance intracellular survival in human respiratory epithelial cells compared to strains that lack IgA protease expression (8, 12).
Strains of NTHi that did not express IgA protease upon acquisition by the patient continued that pattern of negative expression throughout persistence. Two strains each of those initially expressing IgA proteases A1, A2, and B1 stopped expression during persistence due to insertions or deletions ranging in size from 1 to 49 bp in the open reading frame of the gene, causing the gene to be out of frame. The changes in three of these six strains occurred in mononucleotide repeats in a guanine or adenine residue repeat region. A single base change in a mononucleotide repeat region could be interpreted either as an indel or, alternatively, as a change in a simple sequence repeat, consistent with slipped-strand mispairing. Mononucleotide simple sequence repeats mediate phase variation of genes in many species, including virulence-associated genes, such as pilin genes, hemagglutinins, and glycosyltransferases in H. influenzae (34). These previously described mononucleotide repeats associated with frameshift mutations are generally located upstream of the gene or near the 5′ regions of the open reading frame. An interesting observation in this study was that changes in mononuclear repeats during persistence were located throughout the open reading frame of IgA proteases A1, A2, and B1 (Fig. 3B and 5B). Slipped-strand mispairing and indels resulting in changes in expression occurred in the functional protease domain and the beta-core region within the iga genes. The beta-core is associated with formation of the beta-barrel for type V secretion of IgA proteases from the periplasmic space to the extracellular space (13).
IgA protease B2 expression is regulated by slipped-strand mispairing mediated by a change in the number of a 7-bp repeat, TCAAAAT, following the ATG start codon of the igaB2 gene (9, 11). Poole et al. (11) observed phase variation of igaB2 in a laboratory strain of NTHi following experimental human nasopharyngeal challenge. The igaB2 gene was phase-off upon challenge and switched to phase on at rates ranging from 3.8% of isolates at the time of administration of the inoculum to 12.5% of isolates by day 6 (11). Our study of clinical isolates of 23 naturally acquired strains of NTHi showed substantial variability in expression of IgA protease B2, with individual strains turning expression on and off during persistence.
Previously, we showed that passage of the same strains through H292 respiratory epithelial cells resulted in changes in the genome that switched from no expression to expression of IgA proteases B1 and B2, suggesting that intracellular conditions select for expression of IgA proteases B1 and B2. Strains that lost expression of IgA proteases show reduced intracellular survival (12). The results of the present study indicate that genome changes alter IgA protease expression in the natural ecological niche of NTHi, the human respiratory tract. We cannot draw a definitive conclusion that the conditions of the airway selected for these changes. However, given the results of in vitro studies and those obtained with the human NTHi challenge model of Poole et al. (11), we speculate that the microenvironment of the human airways selects for changes in IgA protease expression.
It is curious that passage through respiratory epithelial cells and nasopharyngeal colonization following challenge of healthy adults selected for expression of IgA protease, whereas the strains in the present study that changed expression went from expression to no expression, suggesting selection against expression. Our interpretation is that NTHi adapts to microenvironments in the human airways. When NTHi colonizes the human respiratory tract, it may be present in the lumen, being bound to mucin or adhering to epithelial cells, and it is also present in epithelial cells, macrophages, and intercellular spaces (15–22). We speculate that NTHi alters expression of IgA proteases, depending on its immediate environment, to adapt to the changing conditions of the human airways. We have shown recently that the genomes of NTHi strains that persist in COPD patient airways undergo changes in simple sequence repeats in numerous genes, indicating that the organism uses slipped-strand mispairing of multiple genes simultaneously to survive in the airways (23). The present work advances our understanding of how NTHi adapts to human airways by elucidating changes in IgA protease expression that occur during natural infection in the clinical setting of COPD.
MATERIALS AND METHODS
Prospective study of COPD.
Strains of NTHi were isolated from patients with COPD as a part of a 20-year prospective study in Buffalo, NY, as described previously (14). The institutional review boards of the University at Buffalo and the Veterans Affairs Western New York Healthcare System approved this study; study participants provided written informed consent before enrollment. In brief, patients were seen monthly and at the time of suspected exacerbations. Expectorated sputum samples were collected at each visit and were subjected to bacterial culture.
Bacterial strains.
H. influenzae strains were identified using standard techniques and were distinguished from Haemophilus haemolyticus using monoclonal antibody 7F3, which recognizes an epitope on the P6 protein (24). Strains of NTHi were characterized by multilocus sequence typing (MLST) (25). Strains that were isolated at 2 or more monthly visits and that belonged to the same MLST were defined as “persistent” strains. We studied the first isolate upon acquisition of the strain by the patient and the last isolate before clearance of each strain, referred to here as the “first” and “final” isolates. These observations regarding the identity of strains were confirmed by whole-genome sequencing of the first and final isolates (23). A total of 101 persistent strains of NTHi were isolated during the prospective study, and these strains are the subject of the current study.
IgA protease immunoblot assay.
The expression of IgA proteases (by strains passaged a total of three times from the strain originally isolated from patient expectorated sputum) was determined by immunoblot assay as described previously (9, 26, 27). In brief, strains of NTHi were grown to late log phase in brain heart infusion (BHI) supplemented with hemin and β-NAD to a final concentration of 10 μg/ml each to an optical density at 600 nm of 0.8. Cells were removed by centrifugation at 13,000 × g for 5 min at room temperature, and culture supernatants were incubated with 20-μg/ml human IgA1 myeloma (Calbiochem, Temecula, CA) at 37°C overnight. The supernatants were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to nitrocellulose. Membranes were probed overnight at room temperature with goat anti-human IgA-horseradish peroxidase (1:1,000) conjugate (KPL, Gaithersburg, MD) diluted in buffer A (0.01 M Tris, 0.15 M NaCl, pH 7.4) and developed with H2O2 and color developer. Immunoblots were inspected for IgA protease cleavage patterns.
Sequence analysis methods.
Reference-free whole-genome multiple-sequence alignment-based comparative analyses of diverse subsets of the NTHi genomes were performed using the CloVR-Comparative pipeline (28, 29). The pipeline also generates the Sybil interactive web interface (30, 31) for interrogation of the comparative genomics data. An independent set of clusters of orthologs, Jaccard-filtered clusters of orthologs (JOCs) based on reciprocal best matches by analysis with BLAST software, was generated by performing all-versus-all searches of all the genes predicted in our NTHi genomes. The IgA protease A and IgA protease B gene sequences were obtained by a cluster search of the clusters of orthologous groups on the Sybil interface, using the terms “immunoglobulin A1 protease autotransporter precursor” and “IgA-specific serine endopeptidase autotransporter precursor,” respectively. We also performed comprehensive searches of all subtypes with the BLAST program to ensure identification of all genes, as some frameshifted or fragmented genes would not have been included in the JOC clusters of the Sybil interface. The iga genes in the genomes of the first and last isolates of 101 persistent strains were analyzed using the MacVector (version 14.5.3) application (32, 33). Differences in sequence were observed after using the ClustalW program to align the sequences.
ACKNOWLEDGMENTS
This work was supported by NIH grant R01 AI19641 (to T.F.M., M.M.P., and H.T.) and by the National Center for Advancing Translational Sciences award UL1 TR001412 to the University at Buffalo. R.S.B. and S.E. were supported by training grant T35 AI089693.
REFERENCES
- 1.Clementi CF, Murphy TF. 2011. Non-typeable Haemophilus influenzae invasion and persistence in the human respiratory tract. Front Cell Infect Microbiol 1:1. doi: 10.3389/fcimb.2011.00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Finney LJ, Ritchie A, Pollard E, Johnston SL, Mallia P. 2014. Lower airway colonization and inflammatory response in COPD: a focus on Haemophilus influenzae. Int J Chron Obstruct Pulmon Dis 9:1119–1132. doi: 10.2147/COPD.S54477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sethi S, Murphy TF. 2008. Infection in the pathogenesis and course of chronic obstructive pulmonary disease. N Engl J Med 359:2355–2365. doi: 10.1056/NEJMra0800353. [DOI] [PubMed] [Google Scholar]
- 4.Eldika N, Sethi S. 2006. Role of nontypeable Haemophilus influenzae in exacerbations and progression of chronic obstructive pulmonary disease. Curr Opin Pulm Med 12:118–124. doi: 10.1097/01.mcp.0000208451.50231.8f. [DOI] [PubMed] [Google Scholar]
- 5.Murphy TF, Brauer AL, Schiffmacher AT, Sethi S. 2004. Persistent colonization by Haemophilus influenzae in chronic obstructive pulmonary disease. Am J Respir Crit Care Med 170:266–272. doi: 10.1164/rccm.200403-354OC. [DOI] [PubMed] [Google Scholar]
- 6.Ahearn CP, Gallo MC, Murphy TF. 2017. Insights on persistent airway infection by non-typeable Haemophilus influenzae in chronic obstructive pulmonary disease. Pathog Dis 75:ftx042. doi: 10.1093/femspd/ftx042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lorenzen DR, Dux F, Wolk U, Tsirpouchtsidis A, Haas G, Meyer TF. 1999. Immunoglobulin A1 protease, an exoenzyme of pathogenic Neisseriae, is a potent inducer of proinflammatory cytokines. J Exp Med 190:1049–1058. doi: 10.1084/jem.190.8.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clementi CF, Hakansson AP, Murphy TF. 2014. Internalization and trafficking of nontypeable Haemophilus influenzae in human respiratory epithelial cells and roles of IgA1 proteases for optimal invasion and persistence. Infect Immun 82:433–444. doi: 10.1128/IAI.00864-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Murphy TF, Kirkham C, Jones MM, Sethi S, Kong Y, Pettigrew MM. 2015. Expression of IgA proteases by Haemophilus influenzae in the respiratory tract of adults with chronic obstructive pulmonary disease. J Infect Dis 212:1798–1805. doi: 10.1093/infdis/jiv299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Murphy TF, Lesse AJ, Kirkham C, Zhong H, Sethi S, Munson RS Jr. 2011. A clonal group of nontypeable Haemophilus influenzae with two IgA proteases is adapted to infection in chronic obstructive pulmonary disease. PLoS One 6:e25923. doi: 10.1371/journal.pone.0025923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Poole J, Foster E, Chaloner K, Hunt J, Jennings MP, Bair T, Knudtson K, Christensen E, Munson RS Jr, Winokur PL, Apicella MA. 2013. Analysis of nontypeable Haemophilus influenzae phase-variable genes during experimental human nasopharyngeal colonization. J Infect Dis 208:720–727. doi: 10.1093/infdis/jit240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Murphy TF, Kirkham C, Gallo MC, Yang Y, Wilding GE, Pettigrew MM. 2017. IgA protease variants facilitate intracellular survival in epithelial cells by nontypeable Haemophilus influenzae that persist in the human respiratory tract in COPD. J Infect Dis 216:1295–1302. doi: 10.1093/infdis/jix471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leo JC, Grin I, Linke D. 2012. Type V secretion: mechanism(s) of autotransport through the bacterial outer membrane. Philos Trans R Soc Lond B Biol Sci 367:1088–1101. doi: 10.1098/rstb.2011.0208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sethi S, Evans N, Grant BJ, Murphy TF. 2002. New strains of bacteria and exacerbations of chronic obstructive pulmonary disease. N Engl J Med 347:465–471. doi: 10.1056/NEJMoa012561. [DOI] [PubMed] [Google Scholar]
- 15.St Geme JW III, Falkow S. 1990. Haemophilus influenzae adheres to and enters cultured human epithelial cells. Infect Immun 58:4036–4044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Forsgren J, Samuelson A, Ahlin A, Jonasson J, Rynnel-Dagoo B, Lindberg A. 1994. Haemophilus influenzae resides and multiplies intracellularly in human adenoid tissue as demonstrated by in situ hybridization and bacterial viability assay. Infect Immun 62:673–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Forsgren J, Samuelson A, Borrelli S, Christensson B, Jonasson J, Lindberg AA. 1996. Persistence of nontypeable Haemophilus influenzae in adenoid macrophages: a putative colonization mechanism. Acta Otolaryngol 116:766–773. doi: 10.3109/00016489609137922. [DOI] [PubMed] [Google Scholar]
- 18.Ketterer MR, Shao JQ, Hornick DB, Buscher B, Bandi VK, Apicella MA. 1999. Infection of primary human bronchial epithelial cells by Haemophilus influenzae: macropinocytosis as a mechanism of airway epithelial cell entry. Infect Immun 67:4161–4170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martí-Lliteras P, Regueiro V, Morey P, Hood DW, Saus C, Sauleda J, Agustí AGN, Bengoechea JA, Garmendia J. 2009. Nontypeable Haemophilus influenzae clearance by alveolar macrophages is impaired by exposure to cigarette smoke. Infect Immun 77:4232–4242. doi: 10.1128/IAI.00305-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morey P, Cano V, Marti-Lliteras P, Lopez-Gomez A, Regueiro V, Saus C, Bengoechea JA, Garmendia J. 2011. Evidence for a non-replicative intracellular stage of nontypable Haemophilus influenzae in epithelial cells. Microbiology 157:234–250. doi: 10.1099/mic.0.040451-0. [DOI] [PubMed] [Google Scholar]
- 21.Swords WE, Buscher BA, Ver Steeg K II, Preston A, Nichols WA, Weiser JN, Gibson BW, Apicella MA. 2000. Non-typeable Haemophilus influenzae adhere to and invade human bronchial epithelial cells via an interaction of lipooligosaccharide with the PAF receptor. Mol Microbiol 37:13–27. doi: 10.1046/j.1365-2958.2000.01952.x. [DOI] [PubMed] [Google Scholar]
- 22.van Schilfgaarde M, Eijk P, Regelink A, van Ulsen P, Everts V, Dankert J, van Alphen L. 1999. Haemophilus influenzae localized in epithelial cell layers is shielded from antibiotics and antibody-mediated bactericidal activity. Microb Pathog 26:249–262. doi: 10.1006/mpat.1998.0269. [DOI] [PubMed] [Google Scholar]
- 23.Pettigrew MM, Ahearn CP, Gent JF, Kong Y, Gallo MC, Munro JB, D'Mello A, Sethi S, Tettelin H, Murphy TF. 2018. Haemophilus influenzae genome evolution during persistence in the human airways in chronic obstructive pulmonary disease. Proc Natl Acad Sci U S A 115:E3256–E3265. doi: 10.1073/pnas.1719654115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Murphy TF, Brauer AL, Sethi S, Kilian M, Cai X, Lesse AJ. 2007. Haemophilus haemolyticus: a human respiratory tract commensal to be distinguished from Haemophilus influenzae. J Infect Dis 195:81–89. doi: 10.1086/509824. [DOI] [PubMed] [Google Scholar]
- 25.Meats E, Feil EJ, Stringer S, Cody AJ, Goldstein R, Kroll JS, Popovic T, Spratt BG. 2003. Characterization of encapsulated and noncapsulated Haemophilus influenzae and determination of phylogenetic relationships by multilocus sequence typing. J Clin Microbiol 41:1623–1636. doi: 10.1128/JCM.41.4.1623-1636.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fernaays MM, Lesse AJ, Cai X, Murphy TF. 2006. Characterization of igaB, a second immunoglobulin A1 protease gene in nontypeable Haemophilus influenzae. Infect Immun 74:5860–5870. doi: 10.1128/IAI.00796-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reinholdt J, Kilian M. 1997. Comparative analysis of immunoglobulin A1 protease activity among bacteria representing different genera, species, and strains. Infect Immun 65:4452–4459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Angiuoli SV, Matalka M, Gussman A, Galens K, Vangala M, Riley DR, Arze C, White JR, White O, Fricke WF. 2011. CloVR: a virtual machine for automated and portable sequence analysis from the desktop using cloud computing. BMC Bioinformatics 12:356. doi: 10.1186/1471-2105-12-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Agrawal S, Arze C, Adkins RS, Crabtree J, Riley D, Vangala M, Galens K, Fraser CM, Tettelin H, White O, Angiuoli SV, Mahurkar A, Fricke WF. 2017. CloVR-Comparative: automated, cloud-enabled comparative microbial genome sequence analysis pipeline. BMC Genomics 18:332. doi: 10.1186/s12864-017-3717-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Riley DR, Angiuoli SV, Crabtree J, Dunning Hotopp JC, Tettelin H. 2012. Using Sybil for interactive comparative genomics of microbes on the web. Bioinformatics 28:160–166. doi: 10.1093/bioinformatics/btr652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Crabtree J, Angiuoli SV, Wortman JR, White OR. 2007. Sybil: methods and software for multiple genome comparison and visualization. Methods Mol Biol 408:93–108. doi: 10.1007/978-1-59745-547-3_6. [DOI] [PubMed] [Google Scholar]
- 32.Olson SA. 1994. MacVector: aligning sequences. Methods Mol Biol 25:203–214. [DOI] [PubMed] [Google Scholar]
- 33.Olson SA. 1994. MacVector: an integrated sequence analysis program for the Macintosh. Methods Mol Biol 25:195–201. [DOI] [PubMed] [Google Scholar]
- 34.Power PM, Sweetman WA, Gallacher NJ, Woodhall MR, Kumar GA, Moxon ER, Hood DW. 2009. Simple sequence repeats in Haemophilus influenzae. Infect Genet Evol 9:216–228. doi: 10.1016/j.meegid.2008.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]