Extraintestinal pathogenic Escherichia coli (ExPEC) is responsible for various infections outside the gastrointestinal tract in humans and other animals. ExPEC strain MT78 is invasive to various nonphagocytic cells and highly virulent in vivo.
KEYWORDS: ExPEC, extraintestinal pathogenic E. coli, MT78, TreA, avian fibroblasts, signature-tagged mutagenesis, trehalase, trehalose, type 1 fimbriae, urinary tract infection
ABSTRACT
Extraintestinal pathogenic Escherichia coli (ExPEC) is responsible for various infections outside the gastrointestinal tract in humans and other animals. ExPEC strain MT78 is invasive to various nonphagocytic cells and highly virulent in vivo. To identify genes required for invasion of nonphagocytic cells by this strain, we applied signature-tagged mutagenesis to generate a library of mutants and tested them for invasion of avian fibroblasts. Mutants showing reduced cellular invasion included those with insertions in the fim operon, encoding type 1 fimbriae. Another attenuated mutant showed a disruption in the treA gene, which encodes a periplasmic trehalase. The substrate of TreA, trehalose, can be metabolized and used as a carbon source or can serve as an osmoprotectant under conditions of osmotic stress in E. coli K-12. We generated and characterized mutant MT78ΔtreA. In contrast to the wild type, MT78ΔtreA was able to grow under osmotic stress caused by 0.6 M urea but not in minimal M9 medium with trehalose as the only carbon source. It presented decreased association and invasion of avian fibroblasts, decreased yeast agglutination titer, and impaired type 1 fimbria production. In a murine model of urinary tract infection, MT78ΔtreA was less able to colonize the bladder. All phenotypes were rescued in the complemented mutant. Our results show that the treA gene is needed for optimal production of type 1 fimbriae in ExPEC strain MT78 and that loss of treA significantly reduces its cell invasion capacity and colonization of the bladder in a murine model of urinary tract infection.
INTRODUCTION
Extraintestinal pathogenic Escherichia coli (ExPEC) is responsible for various infections outside the gastrointestinal tract in humans and other animals, such as dogs, cats, and farm animals (1). Urinary tract infections (UTIs) are among the most common infections in humans, being caused mainly by ExPEC (2). ExPEC is also an important cause of neonatal meningitis among newborns with very low body weight (3, 4). In the poultry industry, the various forms of colibacillosis, comprising either localized or systemic infections caused by ExPEC, are of great economic importance worldwide (5). Importantly, certain phylogenetic or clonal groups of ExPEC have also been shown to be commonly associated with infections in poultry and in humans, suggesting that poultry may provide a potential reservoir for ExPEC strains in humans (6, 7).
ExPEC strain MT78 (O2:H+:K1, sequence type 95 [ST]95), also known as BEN2908, was isolated from the trachea of a diseased chicken in France in 1982 (8). Since then, this strain has been well characterized: MT78 is an efficient colonizer of the chicken intestine (9, 10), is highly virulent in vivo (11, 12), strongly interacts with and resists killing by avian macrophages and heterophils (12, 13), and causes apoptosis in macrophages in vitro (12). The most remarkable capacity of this strain is that it is highly invasive to nonphagocytic cells, such as avian fibroblasts (CEC-32) (14) and hepatocytes (LHM), human type II pneumocytes (A549) (15), and brain microvascular endothelial cells (16). Other ExPEC strains can be as virulent as MT78, but they are less able to invade eukaryotic cells (12, 14). The specific mechanisms underlying the invasion capacity of strain MT78, however, have not been fully elucidated.
In order to identify genes required for invasion of nonphagocytic cells by ExPEC MT78, we applied a signature-tagged mutagenesis (STM) approach to generate a library of mutants and tested them in vitro for invasion of avian fibroblasts. One of the mutants attenuated for cellular interaction and invasion contained a transposon insertion in treA that encodes a periplasmic trehalase, TreA, involved in catabolism of trehalose in the periplasm (17). Trehalose is an α(1→1)-dimer of glucose that plays a dual role in E. coli and many other bacterial species. It can be metabolized and used directly as a carbon source, or, under conditions of osmotic stress, it can serve as an osmoprotective agent (18). Although the role of TreA in trehalose metabolism and osmoprotection was demonstrated in the 1990s, to the best of our knowledge, there is no reported role for TreA in the virulence or fitness of E. coli.
The aim of this work was to characterize the treA mutant of the ExPEC strain MT78 in order to better understand the role of the periplasmic trehalase in the virulence and fitness of this strain. To achieve that, we tested the mutant for its growth under different conditions, its capacity of interaction with eukaryotic cells, its production of type 1 fimbriae, and its virulence in a mouse urinary tract infection model.
RESULTS
Individual mutants from the STM library showed reduced adhesion and invasion of avian fibroblasts.
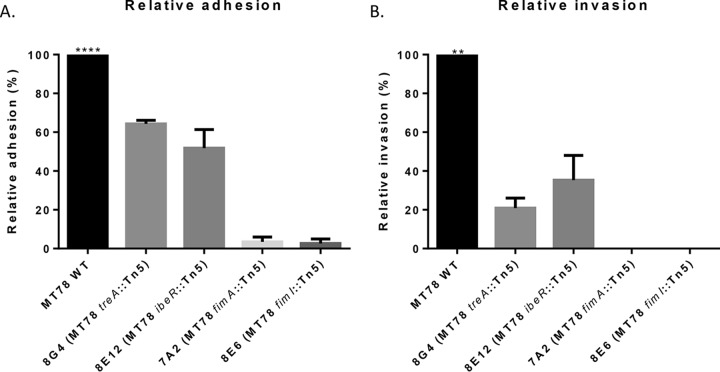
Nineteen pools of 90 STM mutants each were tested for invasion in CEC-32 fibroblasts, and 68 mutants showed a reduced hybridization signal when recovered from inside the cells. Since only a small amount of the inoculum actually invades the fibroblasts, it is possible that not all 90 mutants within the pool may invade cells, regardless of whether or not the transposon insertion disrupted a gene involved in adherence/invasion. Some of these signal-attenuated mutants were therefore tested individually for adhesion to and invasion of avian fibroblasts. Figure 1 shows the results for four mutants, 8G4, 8E12, 7A2, and 8C6, tested individually. The transposon insertion regions of these mutants were determined. The two nonadherent and noninvasive mutants, 7A2 and 8C6, had transposon insertions within the fim operon, in fimA and fimI, respectively. Type 1 fimbriae, encoded by the fim operon, are the major fimbrial structures in many E. coli strains and play an important role in ExPEC adhesion and cellular invasion (19, 20). As such, it was expected that disruption of the fim operon could reduce the capacity of these mutants to adhere to and invade eukaryotic cells. Mutant 8E12, however, was able to adhere to and invade cells but to a lesser extent than wild-type (WT) strain MT78. 8E12 was found to contain a transposon insertion within ibeR, a regulator of the ibeA gene, which contributes to adhesion/invasion in some E. coli strains by promoting or regulating the expression of type 1 fimbriae (20). These results validated the STM screening procedure. Mutant 8G4 displayed decreased cell adhesion and invasion. This mutant contained an insertion in the treA gene, which codes for a periplasmic trehalase. TreA has not previously been identified to play a role in E. coli adhesion or invasion of host cells.
FIG 1.
Adherence and invasion assays of individual transposon mutants obtained from the STM screen. Cells were infected at an MOI of 200 CFU/cell. Bars represent normalized percentages + standard error of recovered bacteria at 1 h after infection (A) and at 4 h after infection, the first hour without antibiotic, followed by washes, and the last 3 h in the presence of gentamicin 50 μg/ml (B). (Adhesion of the WT ranged from 22% to 35% of the inoculum; invasion ranged from 0.8% to 1.5%.) **, P < 0.01; ****, P < 0.001 (Kruskal-Wallis test).
The treA mutant is able to grow on LB agar with 0.6 M urea.
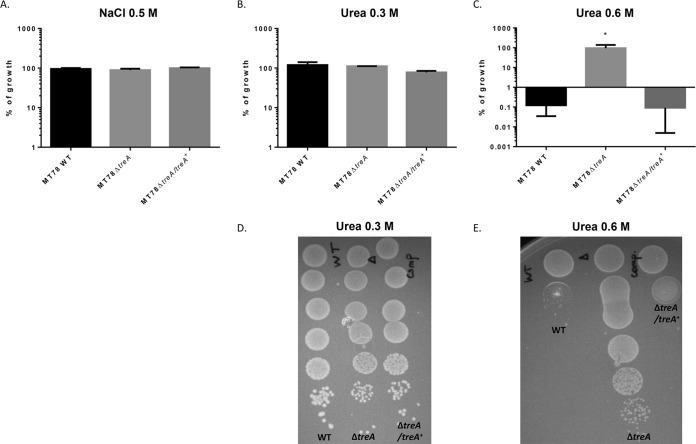
Trehalose metabolism is one of the systems that respond to osmotic stress and has been characterized in E. coli laboratory strain K-12 (17, 18, 21). We therefore tested the growth of the wild type, mutant MT78ΔtreA, and a complemented mutant on modified LB agar containing different concentrations of NaCl or urea. All three strains were able to grow on LB agar with 0.5 M NaCl (Fig. 2A) and with 0.3 M urea (Fig. 2B and D), and no strain could grow on LB agar with 1 M NaCl (results not shown). In contrast, on LB agar with 0.6 M urea, while the WT and the complemented mutant were virtually unable to grow, the mutant MT78ΔtreA grew as well as it did in the absence of urea (Fig. 2C and E). This result suggests that an accumulation of trehalose in the periplasm, due to the absence of the enzyme TreA, could possibly protect the MT78ΔtreA mutant against the osmotic stress caused by the addition of 0.6 M urea on LB agar.
FIG 2.
Growth under conditions of osmotic stress. Strains were grown with shaking in LB medium until mid-log phase (OD600, 0.6) and plated on LB agar (taken as 100% growth) and LB agar with 0.5 M NaCl (A), LB agar with 0.3 M urea (B), and LB agar with 0.6 M urea (C). Graphs show the mean growth relative to that in LB with no additions, plus standard error. Assays were performed three times in duplicate. *, P < 0.05 (Kruskal-Wallis test). Plates showing growth on LB agar with 0.3 M (D) and 0.6 M (E) urea. Each drop represents a 1/10 dilution, from the most concentrated (top) to the most diluted (bottom).
The treA mutant did not grow in M9 minimal medium with trehalose as the only carbon source.
We also tested the growth of MT78ΔtreA in M9 minimal medium with trehalose as the only carbon source, under the same conditions as described for growth in LB. While the WT strain was able to grow, in agreement with previous data (22), the treA mutant did not grow, suggesting that the periplasmic TreA enzyme is required for catabolism of trehalose in MT78, in contrast to what has been described for E. coli K-12 (17).
The treA mutant is less adherent and invasive for avian cells.
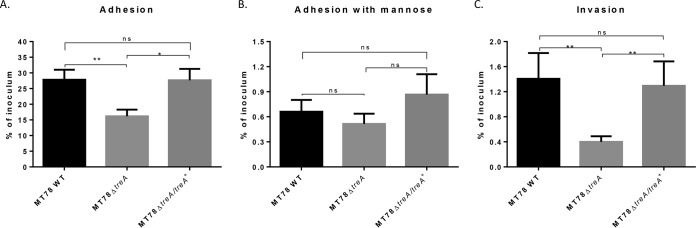
ExPEC strain MT78 is a highly adherent and invasive strain in different cell models (14–16). The adhesion and invasion capacities of the treA mutant were tested using the avian fibroblast CEC-32 cell line. The results are presented in Fig. 3. Whereas 28% of the initial inoculum from the wild-type strain was recovered after 1 h of interaction with avian fibroblasts, only 16% of the treA mutant inoculum was recovered. The adhesive capacity of the treA mutant was successfully rescued by complementation through the introduction of a single copy of the treA gene (Fig. 3A). Inclusion of d-mannose or d-mannopyranose (3%) in cultures to block the effect of type 1 fimbriae greatly reduced cell association of all strains tested in the cell association assay (Fig. 3B). The effect of the treA deletion was even more pronounced in the capacity of the mutant to invade avian fibroblasts: compared to the WT parent, the level of invasion by the treA mutant decreased by 70%, from a mean invasion of 1.4% of the initial inoculum to only 0.4%. Further, the invasive capacity of the treA mutant was regained in the complemented mutant (Fig. 3C).
FIG 3.
Adhesion to and invasion of CEC-32 avian fibroblasts. Cells were infected at an MOI of 10 CFU/cell, as described in Materials and Methods. Bars represent the mean percentage + standard error of recovered CFU compared to that of inocula 1 h after infection (A), 1 h after infection in medium with 3% d-mannose or d-mannopyranose (note that the scale y axis is reduced) (B), and 4 h after infection, the first hour without gentamicin, followed by washes and reincubation in the presence of gentamicin at 50 μg/ml for a further 3 h (C). Assays were performed at least four times in triplicate for each condition. ns, statistically nonsignificant; *, P < 0.05; **, P < 0.01 (Mann-Whitney test).
Deletion of treA reduces the yeast agglutination titer.
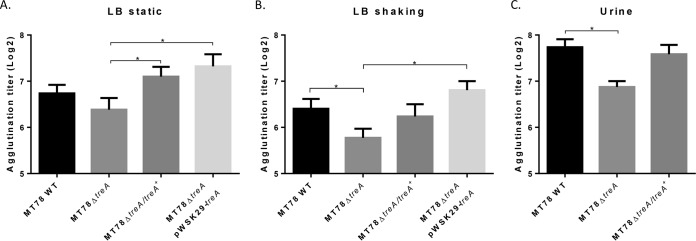
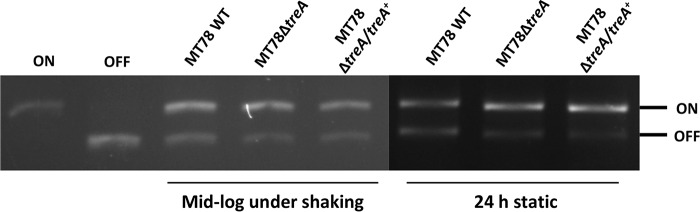
To determine if deletion of treA reduces surface levels of type 1 fimbriae, we performed yeast agglutination experiments with the WT MT78 strain, the MT78ΔtreA mutant, and the complemented mutants grown in different media and under different conditions. Yeast agglutination is a well-established assay to test the production of type 1 fimbriae. After stationary growth in LB, mid-log growth with shaking in LB, and overnight growth in human urine with shaking, MT78ΔtreA agglutination titers were reduced by 22%, 36%, and 45%, respectively, compared to that of the WT strain (Fig. 4). Agglutination titers of both chromosomal and plasmid-complemented mutants regained levels similar to that of the WT strain (Fig. 4). To verify if type 1 fimbriae were reduced in MT78ΔtreA, we performed Western blotting, which confirmed that type 1 fimbriae were indeed reduced in the mutant compared to the wild type and the plasmid-complemented mutant (Fig. 5). To determine if reduction of type 1 fimbriae was due to the position of the invertible promoter, we tested the switch of the fimS promoter region, from the ON to the OFF position. We did not observe any difference between the wild-type strain and the mutant under any of the conditions tested (Fig. 6).
FIG 4.
Yeast agglutination. Yeast agglutination, which correlates with the level of type 1 fimbria functionality, was measured under three different conditions: after static growth overnight in LB (A), after mid-log growth with shaking in LB (B), and after overnight growth with shaking in human urine (C). *, P < 0.05 (Mann-Whitney test).
FIG 5.
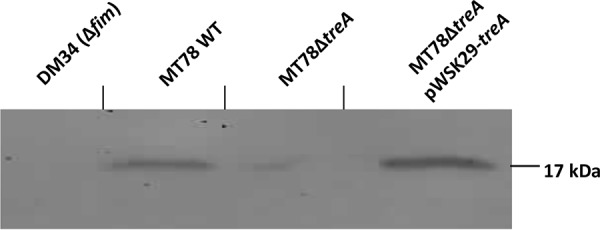
Western blot analysis. Western blotting of fimbrial extracts of strains cultured to the mid-log phase of growth in LB medium.
FIG 6.
Verification of fimS orientation. The fimS region was PCR amplified, and the product was digested with HinfI. Fragments of different sizes indicate the ON or OFF orientation. Only the top half of the gel is shown.
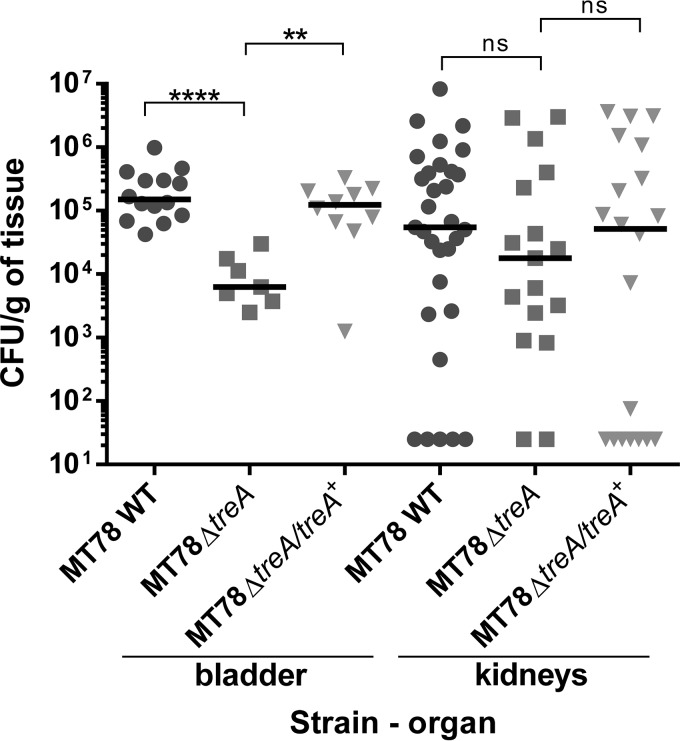
The treA mutant is less able to colonize the bladder in the murine urinary tract infection model.
Type 1 fimbriae are important structures for ExPEC colonization of the bladder in UTIs (1). Since the treA mutant demonstrated decreased yeast agglutination associated with type 1 fimbria production, we tested its capacity to cause urinary tract infection in the CBA/J mouse model. Forty-eight hours after urethral inoculation of 2 × 109 CFU, bladders and kidneys were collected, homogenized, diluted, and plated for bacterial counting. The treA mutant colonized the bladder 10-fold less than the wild-type strain (P < 0.0001) (Fig. 7). In an independent assay, the complemented mutant regained the capacity of colonization (P < 0.01). This reduction in colonization may be due to reduced production of type 1 fimbriae. In contrast to the colonization of the bladder, there was no significant difference in bacterial numbers in the kidneys.
FIG 7.
In vivo urinary tract infection in mice. CBA/J mice were infected transurethrally, the animals were euthanized, and organs were collected 48 h postinfection. Each data point represents a sample from an individual mouse, and horizontal bars indicate the medians. Two independent monoinfections were performed: with MT78 WT and MT78ΔtreA and with MT78 WT and MT78ΔtreA/treA+. Results for MT78 WT were pooled. Each kidney was sampled separately. ns, statistically nonsignificant; **, P < 0.01; ****, P < 0.0001 (Mann-Whitney test).
The FimH adhesin of strain MT78 and that of noninvasive ExPEC are nearly identical.
A possible explanation for the invasive phenotype of MT78 may be the differences in sequence of the FimH fimbrial adhesive tip. We thus aligned the amino acid sequences of FimH from MT78, uropathogenic E. coli (UPEC) CFT073, and phylogenetically similar avian pathogenic E. coli (APEC) IMT5155, since these last two strains are noninvasive in the CEC-32 model (14; Daniel Pavanelo, unpublished results). There are only two amino acid differences among the strains, a G180-to-S180 substitution in MT78, in comparison to the sequences of CFT073 and IMT5155, and an A184-to-V184 substitution compared to the sequence of CFT073. These two differences are conservative substitutions, since glycine and serine are polar and alanine and valine are nonpolar amino acids. It is thus unlikely that the differences observed among these strains concerning interaction with cells could be attributed solely to specific amino acid sequence differences in FimH proteins.
DISCUSSION
Several research groups have successfully used STM to identify virulence-associated genes in a series of bacterial pathogens and models (23–27). Our STM library screening resulted in the identification of mutants that were attenuated in cellular invasion of fibroblasts with transposon insertions in genes of the fim operon and in treA, which encodes a periplasmic trehalase. In E. coli K-12, the final cytosolic concentration of trehalose is regulated by a futile cycle: cytosolic enzymes OtsA and OtsB synthesize trehalose from glucose in the cytoplasm (28), which is translocated to the periplasm and degraded by TreA into two molecules of glucose (17). Under osmotic stress, the final concentration of the osmoprotectant trehalose is increased due to higher activity of both cytosolic OtsA and OtsB, despite a concomitant increase in periplasmic TreA activity (21). Therefore, one would expect a treA mutant to produce higher levels of trehalose and be potentially more resistant to osmotic stress. Although there was no difference between the wild type and mutant concerning resistance to osmotic stress caused by NaCl, the treA mutation did confer increased resistance to stress caused by 0.6 M urea, a condition under which growth of the wild-type strain was impaired.
In a recent report, TreA has been shown to be involved in the virulence of Burkholderia pseudomallei (29). Because the MT78ΔtreA mutant displayed an attenuated capacity to invade eukaryotic cells (Fig. 1 and 3), we performed yeast agglutination assays and Western blotting against type 1 fimbriae to investigate any effect on type 1 fimbria production. As shown in Fig. 4 and 5, in comparison to the WT strain, the mutant had a reduced production of type 1 fimbriae and lowered yeast agglutination. This reduction in type 1 fimbriae cannot be explained by the fimS switch, since we did not observe any differences in orientation of the fimS switch in these strains (Fig. 6). Type 1 fimbria expression of ExPEC is regulated by several mechanisms, such as global regulators H-NS, Lrp, IHF, and FNR (30, 31), recombinases FimB and FimE, some virulence-associated genes, such as ibeA and ibeT (20) in certain strains, and a phosphate metabolism operon, pst (19), among others. Since the position of the promoter suggests that the expression of type 1 fimbriae is not reduced, the effect of deletion of treA on type 1 fimbria production is likely a posttranscriptional or posttranslational event.
Our results showed a somewhat more pronounced reduction of yeast agglutination when bacteria were grown under agitation until the mid-log phase instead of when bacteria were grown statically (Fig. 4). In cell assays, the reduction in adhesion and invasion capacities was also more marked when bacteria were grown under agitation until the mid-log phase (results not shown). Similarly, other mutations in genes such as the pst system also resulted in a more marked change in production of type 1 fimbriae during growth to mid-log phase than during overnight static growth (19).
It is not the first time that a gene related to carbohydrate metabolism is reported to be involved in the virulence of ExPEC strain MT78. A selC-associated genomic island, named AGI-3, was found in the MT78 genome, with three genes coding for sugar metabolism: a transcriptional regulator of the LacI family, a hexuronate transporter, and an α-glycosidase. Without these genes, MT78 was impaired in its ability to metabolize carbohydrates, including trehalose, and was less able to cause bacteremia and colonize the liver of 3-week-old white Leghorn chickens 24 h and 48 h postinfection (22). The metabolic operon frz was also found in the MT78 genome, and when this operon was deleted, the strain was less able to interact with lung (A549), liver (LMH), and intestinal (Caco-2) cells, to colonize the intestine of axenic white Leghorn chicks, and, as in our work, to express type 1 fimbriae (32). The mechanism by which a loss of sugar metabolism genes affects the expression of type 1 fimbriae, however, remains to be elucidated.
Some studies relating osmotic stress and type 1 fimbria expression in UPEC strains have shown contradictory results and are so far inconclusive. In 2002, Schwan and colleagues showed that a decrease in type 1 fimbria expression occurred under osmotic stress caused by NaCl and low pH in the UPEC strain NU149 (33). In 2004, Snyder and colleagues performed a transcriptome analysis of the UPEC strain CFT073 during UTI, and type 1 fimbria expression was upregulated together with genes that are regulated by the osmotic stress response (34). In 2013, Withman and colleagues showed, also for CFT073, that type 1 fimbriae were more expressed under osmotic stress caused by an increase in urea but did not respond to an increase in NaCl (35). Finally, in 2015, Greene and colleagues concluded that for UPEC strain UTI89, type 1 fimbriae are less expressed in human urine (36). According to our results, MT78 showed a higher yeast agglutination titer in human urine than in LB (Fig. 4), but urine composition is widely variable, which could explain this difference. In E. coli K-12, treA expression has been involved in the osmotic stress response, whereas we found a reduction in type 1 fimbria production in the MT78ΔtreA mutant. We cannot strictly define the specific roles of osmotic stress or its regulation in type 1 fimbriae in E. coli based on our results and other reports, which have included investigations with a number of different E. coli strains, each of which may have distinct mechanisms of regulation and production of type 1 fimbriae. However, from our data, it is clear that TreA plays a role mediating cell invasion by strain MT78 and its level of production of type 1 fimbriae.
FimH of MT78 is almost identical to the adhesin tip of UPEC CFT073 and APEC IMT5155. Despite these three strains behaving differentially in contact with cells (12, 14), they would all have a high-affinity FimH, because they harbor an aspartate residue at position 188. This particular residue has been shown to increase the affinity of FimH to mannose, and strains with this characteristic tend to be more focally adherent and to spread less throughout the host (37). Mutations at positions 27, 62, 70, 78, and 128 could also explain differences in FimH-mannose affinity (37); however, strains MT78, CFT073, and IMT5155 show the same residues at these sites, which are frequently variable. Since type 1 fimbriae are an important adhesin and the FimH adhesin is highly similar in these three E. coli strains, despite their demonstrating differences in cell adherence/invasion, other differences among these strains or distinct regulation of type 1 fimbriae may contribute to the capacity of strain MT78 to invade a wide variety of eukaryotic cells.
E. coli is a natural colonizer of warm-blooded animals, but some clones can cause disease in a wide variety of niches. ExPEC strains cause diseases in humans and other animals and have a zoonotic potential. Neonatal meningitis E. coli (NMEC) strains have been shown to successfully colonize chickens in a chick colisepticemia model, and APEC strains were able to cause meningitis in a rat neonatal meningitis model (38). In addition, Skyberg and colleagues have shown that the introduction of a plasmid from an APEC strain into a commensal E. coli strain from poultry conferred an increased capacity to colonize the urinary tract of mice (39). In our current report, APEC strain MT78, which belongs to ST95, a common ExPEC sequence type, was shown to cause urinary tract infection in mice at levels comparable to those caused by human UPEC strains such as CFT073. This result also reinforces the zoonotic potential of certain ExPEC isolates.
In summary, we generated a mutant library using signature-tagged mutagenesis to better understand the invasive phenotype of MT78 in host cells. Attenuated mutants showed transposon insertions in genes from the fim operon or in genes that affect type 1 fimbria expression. Our data strongly suggest a critical role of type 1 fimbriae in MT78 invasion of eukaryotic cells. Specifically, our results demonstrate that the treA gene is needed for the optimal production of type 1 fimbriae in this particular ExPEC strain and that the loss of treA significantly reduces cell invasion and colonization of the bladder in a murine urinary tract infection model.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
All strains and plasmids used in this study are listed in Table 1. Strains and mutants were grown in LB, LB with different amounts of NaCl or urea, M9 with trehalose as the sole carbon source, human urine, or brain heart infusion (BHI) medium. Stock cultures were maintained in glycerol stocks at −80°C.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Characteristic(s) | Resistance | Reference or source |
|---|---|---|---|
| Strains | |||
| MT78 (O2:H+:K1) | WT ExPEC strain | Nalidixic acid | 8, 14 |
| S17-1 λpir | Conjugative strain used for creation of STM library | None | 25 |
| DH5α | Vector strain | None | Bethesda Laboratories |
| MT78ΔtreA | treA::Km | Nalidixic acid, kanamycin | This study |
| MT78ΔtreA/treA+ | treA::Km, treA+cat | Nalidixic acid, kanamycin, chloramphenicol | This study |
| MT78ΔtreA/pWSK29-treA | treA::Km, pWSK29-treA | Nalidixic acid, kanamycin, ampicillin | This study |
| 7A2 (STM mutant) | MT78 fimA::Tn5 | Nalidixic acid, kanamycin | This study |
| 8C6 (STM mutant) | MT78 fimI::Tn5 | Nalidixic acid, kanamycin | This study |
| 8E12 (STM mutant) | MT78 ibeR::Tn5 | Nalidixic acid, kanamycin | This study |
| 8G4 (STM mutant) | MT78 treA::Tn5 | Nalidixic acid, kanamycin | This study |
| DM34 | MT78Δfim | Nalidixic acid | 40 |
| Plasmids | |||
| pUT-mini-Tn5km2 | Plasmid used for creation of STM library (90 different tags within the transposon) | Ampicillin, kanamycin | 23, 25, 41 |
| pKD46 | Plasmid used for nonpolar mutation | Ampicillin | 42 |
| pKD4 | Plasmid used as template for kanamycin cassette amplification | Kanamycin | 42 |
| pGPTn7-Cm | Plasmid used for complementation | Ampicillin, chloramphenicol | 43 |
| pWSK29 | Plasmid used for complementation | Ampicillin | 44 |
Construction and analysis of the STM library.
Random mutants were generated as described previously (25). A total of 19 pools with 90 mutants each (n = 1,710) were screened in cell invasion assays (described below). Identification of mutants with reduced invasion was determined as described previously, using input and output pools from the cell assays (25). Attenuated mutants had the transposon insertion region amplified by nested PCR, and unique bands were selected, gel purified, and sequenced by Sanger sequencing (25) at ATCGene (Porto Alegre, Brazil) or the Genome Canada facility (McGill University, Montreal, Canada). Primers used are listed in Table 2.
TABLE 2.
Oligonucleotides used in this study
| Primer | Sequence (5′–3′) | Function | Reference |
|---|---|---|---|
| P6 | CCT AGG CGG CCA GAT CTG AT | Amplification of transposon insertion region | 25 |
| P9 | CGC AGG GCT TTA TTG ATT C | Amplification of transposon insertion region | 25 |
| Arbi 1 | GGC CAC GCG TCG ACT AGT ACN NNN NNN NNN GAT AT | Amplification of transposon insertion region | 25 |
| Arbi 2 | GGC CAC GCG TCG ACT AGT AC | Amplification of transposon insertion region | 25 |
| Arbi 3 | GGC CAC GCG TCG ACT AGT ACN NNN NNN NNN TGA CG | Amplification of transposon insertion region | 25 |
| Arbi 4 | GGC CAC GCG TCG ACT AGT ACN NNN NNN NNN ACG CC | Amplification of transposon insertion region | 25 |
| Arbi 5 | GGC CAC GCG TCG ACT AGT ACN NNN NNN NNN TAC NG | Amplification of transposon insertion region | 25 |
| treA_F | CGT AAT CGG CAT ACC AGC CT | Verification of treA presence | This study |
| treA_R | TGT TGA AAA CCT GCA AGC CG | Verification of treA presence and of treA chromosomal complementation | This study |
| treAKO_F | ATG AAA TCC CCC GCA CCT TCT CGC CCG CAA AAA ATG GCG TTA ATT CCA GCG TGT AGG CTG GAG C TG CTT C | Mutation of treA | This study |
| treAKO_R | GGA CGC GTC GCC GGA ACA TTG TCA CAC GGT TGC TCT TTC GGG CAG ATC AAA TGG GAA TTA GCC ATG GTC C | Mutation of treA | This study |
| QFtreApWSK29compl_F | GTA CCG GGC CCC CCC TCG AGT CCG GCA ATT TAC TCT GCA CT | Cloning of treA into pWSK29 plasmid | This study |
| QFtreApWSK29compl_R | TCC CCC GGG CTG CAG GAA TTC AGG GAG AAT GGG GAG TGG GGG | Cloning of treA into pWSK29 plasmid | This study |
| QFtreAcompl_F | CCG GGC CCA AGC TTC TCG AGT CCG GCA ATT TAC TCT GCA CT | Cloning of treA into pGPTn7Cm plasmid and amplification of treA-cat fragment for complementation | This study |
| QFtreATn7compl_R | CCC CGG GCT GCA GGA ATT CAG GGA GAA TGG GGA GTG GGG G | Cloning of treA into pGPTn7Cm plasmid | This study |
| Dat_treA_compl_R | CGG CGA AAT AGA GTG ATA AAA TAA CAT CTG TTT ATT AGT CAG CCA GCG ACA AGC TTT CAG AAC GCT CGG TTG CCG C | Amplification of treA-cat fragment for chromosomal complementation | This study |
| screening_treAKO_F | ACC GTT CGG ATG GCA TCA TT | Checking of treA mutation | This study |
| K1 | CAG TCA TAG CCG AAT AGC CT | Checking of treA mutation | This study |
| Pri3 | TCG TTT TGC CGG ATT ATG GG | Verification of fimS orientation | 19 |
| Pri4 | AGT GAA CGG TCC CAC CAT TAA CC | Verification of fimS orientation | 19 |
Construction of specific mutants and the complemented strain.
Specific mutants were generated by the procedure described by Datsenko and Wanner, using plasmid pKD4 as the template for the kanamycin resistance cassette (42). Primers used are listed in Table 2.
The treA mutant was complemented with a low-copy-number plasmid and with a single chromosomal gene copy. For the plasmid complementation, MT78 treA was PCR amplified with primers QFtreApWSK29compl_F and QFtreApWSK29compl_R (Table 2) and cloned into a pWSK29 low-copy-number plasmid (44). The plasmid pWSK29-treA was used to transform, by electroporation, the mutant MT78ΔtreA.
For the chromosomal complementation, the treA gene with its promoter was cloned into a pGPTn7-Cm plasmid upstream of a chloramphenicol cassette (43). The entire region comprising treA plus cassette was PCR amplified and used to complement the treA mutant by the same method with which the mutation was generated. Primers used in this reaction are listed in Table 2. The original treA promoter region was replaced by this treA allele by homologous recombination. The complemented strain contains both chloramphenicol and kanamycin cassettes inserted downstream of the treA gene.
Growth under conditions of osmotic stress.
Strains were tested for the capacity to grow under conditions of osmotic stress caused by NaCl or urea. Strains were inoculated 1:100 from an overnight preinoculum in LB medium and grown until mid-log phase with shaking. They were serially diluted and plated on LB agar alone and LB agar with 0.5 M NaCl, 1 M NaCl, 0.3 M urea, and 0.6 M urea. Colonies were enumerated, and growth under each condition was compared to growth on LB agar.
Cell association and invasion assays.
The eukaryotic cell assays were performed with CEC-32 avian fibroblasts (45) as described previously (14), with some modifications. Briefly, 5 × 104 cells/well were distributed in 96-well plates, or 2 × 105 cells/well were cultured in 24-well plates, in high-glucose Dulbecco's modified Eagle's medium (DMEM) with 10% fetal bovine serum (infection medium). Immediately before infection, cultures were washed once with phosphate-buffered saline (PBS) to remove dead cells, returned to the infection medium, and infected at an estimated multiplicity of infection (MOI) of 10, 20, or 200 CFU per cell. Bacterial strains were grown in LB medium with shaking (240 rpm) until mid-log phase. For STM library screening, a set of 90 mutants were pooled from overnight cultures (10 μl each) and inoculated in 50 ml of LB with shaking until mid-log phase. Aliquots of the inocula used for infections were diluted and plated to confirm the MOI, in single infections. For STM screening, samples were collected and used as input pools. After 1 h of incubation at 37°C, the medium was removed, and the cells were washed three times with PBS. For the association assays, the cells were then lysed with 1% (vol/vol) Triton X-100 in PBS for 5 min. The lysates were serially diluted and plated on LB agar for CFU counting. For the invasion assays, the cells were further incubated in culture medium supplemented with 50 μg/ml gentamicin for 3 h, after which they were lysed, and the lysates were diluted and plated. For STM screening, only invasion assays were performed. Recovered bacteria were used as the output pools.
To verify type 1 fimbria-specific adherence, all assays were performed in the presence or absence of 3% α-d-mannose or d-mannopyranose.
Yeast agglutination.
In order to detect differences in type 1 fimbria production, strains were tested for yeast agglutination as described previously (19, 46). Bacterial strains were incubated in LB medium or human urine at 37°C until mid-log or stationary phase with or without agitation. A suspension of approximately 2 ×109 bacterial cells in PBS was serially diluted 1:2 to a dilution of 2−10 bacterial cells in microtiter plates. In each well, an equal volume of a 1.5% commercially available yeast suspension was added. After 30 min of incubation on ice, agglutination was visually monitored and the agglutination titer was determined using the most diluted well in which agglutination was observed.
Preparation of fimbrial extracts and Western blotting.
Preparation of fimbrial extracts was performed as described previously (47), with slight modifications. Briefly, bacteria were grown in 100 ml LB medium with shaking; after reaching mid-log phase (optical density at 600 nm [OD600], 0.6), cells were collected by centrifugation at 1,000 × g for 15 min and resuspended in 1 ml of 150 mM NaCl–50 mM Tris-HCl, pH 7.8. Cell suspensions were incubated at 56°C for 60 min and vortexed at maximum speed for 2 min before being pelleted by centrifugation (maximum speed) in a microcentrifuge for 2 min. Proteins were precipitated with 100% trichloroacetic acid (TCA) at a final ratio of 1:10. The tube was incubated for 20 min on ice, followed by centrifugation at 20,000 × g for 15 min at 4°C. The protein pellet was washed twice with 50 mM Tris-EDTA (pH 12.0) and resuspended in 50 mM Tris-EDTA, pH 8.0, at 1/10 of the supernatant's initial volume. Western blotting was performed as described previously (46).
Detection of the ON/OFF state of the fimS region.
Detection of the orientation of the fimS region was performed as described previously (19). Strains were cultured statically in LB for 24 h to enhance the ON orientation in the bacterial population and with shaking until mid-log phase to observe the orientation at the time point of cell assays and yeast agglutination. As a control, the wild-type strain was statically grown for 96 h to select for increased switching toward the ON orientation and repetitively on agar plates to select for an increased switching toward the OFF orientation.
Experimental UTIs in CBA/J mice.
Experimental urinary tract infections in CBA/J mice were performed as previously described (48). Briefly, 5-week-old female mice were infected with 20 μl (2 × 109 CFU) of bacteria through a catheter inserted in the urethra. At 48 h postinfection, the mice were euthanized. Bladders and kidneys were then collected, homogenized, diluted, and plated on MacConkey agar plates to enumerate bacterial CFU.
Alignment of FimH amino acid sequences.
Amino acid sequences of the FimH protein from CFT073 (GenBank accession no. AE014075.1), IMT5155 (GenBank accession no. CP005930.1), and MT78 (8, 14; sequence not public) were aligned using the Clustal Omega Multiple Sequence Alignment server (49).
Statistical analysis.
The Mann-Whitney test was used to compare the samples by pairs, and the Kruskal-Wallis test was used to compare groups, since the distribution of results was nonparametric. Tests were performed using GraphPad Prism version 6.00 for Windows (GraphPad Software, La Jolla, CA, USA).
Ethics statement.
The experimental protocol for handling animals was approved by the Ethical Committee of INRS (protocol number 1206-03).
ACKNOWLEDGMENTS
This work was supported by CAPES, Ministry of Education, Brazil (PVE A093/2013, number 23038.009613/2013-11) and NSERC Canada Discovery grant 2014-06622 (C.M.D.). D.B.P. was the recipient of a CAPES Ph.D. studentship (CAPES PROEX and SFS-PVE-S 99999.009562/2014-01).
REFERENCES
- 1.Kaper JB, Nataro JP, Mobley HLT. 2004. Pathogenic Escherichia coli. Nat Rev Microbiol 2:123–140. doi: 10.1038/nrmicro818. [DOI] [PubMed] [Google Scholar]
- 2.Foxman B. 2014. Urinary tract infection syndromes: occurrence, recurrence, bacteriology, risk factors, and disease burden. Infect Dis Clin North Am 28:1–13. doi: 10.1016/j.idc.2013.09.003. [DOI] [PubMed] [Google Scholar]
- 3.Ku LC, Boggess KA, Cohen-Wolkowiez M. 2015. Bacterial meningitis in infants. Clin Perinatol 42:29–45, vii–viii. doi: 10.1016/j.clp.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stoll BJ, Hansen N, Fanaroff AA, Wright LL, Carlo WA, Ehrenkranz RA, Lemons JA, Donovan EF, Stark AR, Tyson JE, Oh W, Bauer CR, Korones SB, Shankaran S, Laptook AR, Stevenson DK, Papile LA, Poole WK. 2002. Changes in pathogens causing early-onset sepsis in very-low-birth-weight infants. N Engl J Med 347:240–247. doi: 10.1056/NEJMoa012657. [DOI] [PubMed] [Google Scholar]
- 5.Ferreira AJ, Knöbl T. 2000. Colibacilose aviária, p 197–205. In Berchieri A, Macari M (ed), Doença das aves. FACTA, Campinas, Brazil. [Google Scholar]
- 6.Mora A, Viso S, Lopez C, Alonso MP, Garcia-Garrote F, Dabhi G, Mamani R, Herrera A, Marzoa J, Blanco M, Blanco JE, Moulin-Schouleur M, Schouler C, Blanco J. 2013. Poultry as reservoir for extraintestinal pathogenic Escherichia coli O45:K1:H7-B2-ST95 in humans. Vet Microbiol 167:506–512. doi: 10.1016/j.vetmic.2013.08.007. [DOI] [PubMed] [Google Scholar]
- 7.Mitchell NM, Johnson JR, Johnston B, Curtiss R III, Mellata M. 2015. Zoonotic potential of Escherichia coli isolates from retail chicken meat products and eggs. Appl Environ Microbiol 81:1177–1187. doi: 10.1128/AEM.03524-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dho M, Lafont JP. 1982. Escherichia coli colonization of the trachea in poultry—comparison of virulent and avirulent strains in gnotoxenic chickens. Avian Dis 26:787–797. doi: 10.2307/1589865. [DOI] [PubMed] [Google Scholar]
- 9.Schouler C, Taki A, Chouikha I, Moulin-Schouleur M, Gilot P. 2009. A genomic island of an extraintestinal pathogenic Escherichia coli strain enables the metabolism of fructooligosaccharides, which improves intestinal colonization. J Bacteriol 191:388–393. doi: 10.1128/JB.01052-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Porcheron G, Chanteloup NK, Trotereau A, Bree A, Schouler C. 2012. Effect of fructooligosaccharide metabolism on chicken colonization by an extra-intestinal pathogenic Escherichia coli strain. PLoS One 7:e35475. doi: 10.1371/journal.pone.0035475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pourbakhsh SA, Boulianne M, Martineau-Doize B, Fairbrother JM. 1997. Virulence mechanisms of avian fimbriated Escherichia coli in experimentally inoculated chickens. Vet Microbiol 58:195–213. doi: 10.1016/S0378-1135(97)00163-6. [DOI] [PubMed] [Google Scholar]
- 12.Horn F, Correa AMR, Barbieri NL, Glodde S, Weyrauch KD, Kaspers B, Driemeier D, Ewers C, Wieler LH. 2012. Infections with avian pathogenic and fecal Escherichia coli strains display similar lung histopathology and macrophage apoptosis. PLoS One 7:e41031. doi: 10.1371/journal.pone.0041031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mellata M, Dho-Moulin M, Dozois CM, Curtiss R, Lehoux B, Fairbrother JM. 2003. Role of avian pathogenic Escherichia coli virulence factors in bacterial interaction with chicken heterophils and macrophages. Infect Immun 71:494–503. doi: 10.1128/IAI.71.1.494-503.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Matter LB, Barbieri NL, Nordhoff M, Ewers C, Horn F. 2011. Avian pathogenic Escherichia coli MT78 invades chicken fibroblasts. Vet Microbiol 148:51–59. doi: 10.1016/j.vetmic.2010.08.006. [DOI] [PubMed] [Google Scholar]
- 15.Chanteloup NK, Porcheron G, Delaleu B, Germon P, Schouler C, Moulin-Schouleur M, Gilot P. 2011. The extra-intestinal avian pathogenic Escherichia coli strain BEN2908 invades avian and human epithelial cells and survives intracellularly. Vet Microbiol 147:435–439. doi: 10.1016/j.vetmic.2010.07.013. [DOI] [PubMed] [Google Scholar]
- 16.Germon P, Chen YH, He L, Blanco JE, Bree A, Schouler C, Huang SH, Moulin-Schouleur M. 2005. ibeA, a virulence factor of avian pathogenic Escherichia coli. Microbiology 151:1179–1186. doi: 10.1099/mic.0.27809-0. [DOI] [PubMed] [Google Scholar]
- 17.Boos W, Ehmann U, Bremer E, Middendorf A, Postma P. 1987. Trehalase of Escherichia coli. Mapping and cloning of its structural gene and identification of the enzyme as a periplasmic protein induced under high osmolarity growth conditions. J Biol Chem 262:13212–13218. [PubMed] [Google Scholar]
- 18.Styrvold OB, Strom AR. 1991. Synthesis, accumulation, and excretion of trehalose in osmotically stressed Escherichia coli K-12 strains: influence of amber suppressors and function of the periplasmic trehalase. J Bacteriol 173:1187–1192. doi: 10.1128/jb.173.3.1187-1192.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Crepin S, Houle S, Charbonneau ME, Mourez M, Harel J, Dozois CM. 2012. Decreased expression of type 1 fimbriae by a pst mutant of uropathogenic Escherichia coli reduces urinary tract infection. Infect Immun 80:2802–2815. doi: 10.1128/IAI.00162-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cortes MAM, Gibon J, Chanteloup NK, Moulin-Schouleur M, Gilot P, Germon P. 2008. Inactivation of ibeA and ibeT results in decreased expression of type 1 fimbriae in extraintestinal pathogenic Escherichia coli strain BEN2908. Infect Immun 76:4129–4136. doi: 10.1128/IAI.00334-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Strom AR, Kaasen I. 1993. Trehalose metabolism in Escherichia coli: stress protection and stress regulation of gene expression. Mol Microbiol 8:205–210. doi: 10.1111/j.1365-2958.1993.tb01564.x. [DOI] [PubMed] [Google Scholar]
- 22.Chouikha I, Germon P, Bree A, Gilot P, Moulin-Schouleur M, Schouler C. 2006. A selC-associated genomic island of the extraintestinal avian pathogenic Escherichia coli strain BEN2908 is involved in carbohydrate uptake and virulence. J Bacteriol 188:977–987. doi: 10.1128/JB.188.3.977-987.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hensel M, Shea JE, Gleeson C, Jones MD, Dalton E, Holden DW. 1995. Simultaneous identification of bacterial virulence genes by negative selection. Science 269:400–403. doi: 10.1126/science.7618105. [DOI] [PubMed] [Google Scholar]
- 24.Badger J, Wass C, Weissman S, Kim K. 2000. Application of signature-tagged mutagenesis for identification of Escherichia coli K1 genes that contribute to invasion of human brain microvascular endothelial cells. Infect Immun 68:5056–5061. doi: 10.1128/IAI.68.9.5056-5061.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li GW, Laturnus C, Ewers C, Wieler LH. 2005. Identification of genes required for avian Escherichia coli septicemia by signature-tagged mutagenesis. Infect Immun 73:2818–2827. doi: 10.1128/IAI.73.5.2818-2827.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dziva F, Hauser H, Connor TR, van Diemen PM, Prescott G, Langridge GC, Eckert S, Chaudhuri RR, Ewers C, Mellata M, Mukhopadhyay S, Curtiss R III, Dougan G, Wieler LH, Thomson NR, Pickard DJ, Stevens MP. 2013. Sequencing and functional annotation of avian pathogenic Escherichia coli serogroup O78 strains reveal the evolution of E. coli lineages pathogenic for poultry via distinct mechanisms. Infect Immun 81:838–849. doi: 10.1128/IAI.00585-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Antão E-M, Ewers C, Guerlebeck D, Preisinger R, Homeier T, Li G, Wieler LH. 2009. Signature-tagged mutagenesis in a chicken infection model leads to the identification of a novel avian pathogenic Escherichia coli fimbrial adhesin. PLoS One 4:e7796. doi: 10.1371/journal.pone.0007796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Giaever HM, Styrvold OB, Kaasen I, Strom AR. 1988. Biochemical and genetic characterization of osmoregulatory trehalose synthesis in Escherichia coli. J Bacteriol 170:2841–2849. doi: 10.1128/jb.170.6.2841-2849.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vanaporn M, Sarkar-Tyson M, Kovacs-Simon A, Ireland PM, Pumirat P, Korbsrisate S, Titball RW, Butt A. 2017. Trehalase plays a role in macrophage colonization and virulence of Burkholderia pseudomallei in insect and mammalian hosts. Virulence 8:30–40. doi: 10.1080/21505594.2016.1199316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barbieri NL, Nicholson B, Hussein A, Cai W, Wannemuehler YM, Dell'Anna G, Logue CM, Horn F, Nolan LK, Li G. 2014. FNR regulates expression of important virulence factors contributing to pathogenicity of uropathogenic Escherichia coli. Infect Immun 82:5086–5098. doi: 10.1128/IAI.02315-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Corcoran CP, Dorman CJ. 2009. DNA relaxation-dependent phase biasing of the fim genetic switch in Escherichia coli depends on the interplay of H-NS, IHF and LRP. Mol Microbiol 74:1071–1082. doi: 10.1111/j.1365-2958.2009.06919.x. [DOI] [PubMed] [Google Scholar]
- 32.Rouquet G, Porcheron G, Barra C, Reperant M, Chanteloup NK, Schouler C, Gilot P. 2009. A metabolic operon in extraintestinal pathogenic Escherichia coli promotes fitness under stressful conditions and invasion of eukaryotic cells. J Bacteriol 191:4427–4440. doi: 10.1128/JB.00103-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schwan WR, Lee JL, Lenard FA, Matthews BT, Beck MT. 2002. Osmolarity and pH growth conditions regulate fim gene transcription and type 1 pilus expression in uropathogenic Escherichia coli. Infect Immun 70:1391–1402. doi: 10.1128/IAI.70.3.1391-1402.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Snyder JA, Haugen BJ, Buckles EL, Lockatell CV, Johnson DE, Donnenberg MS, Welch RA, Mobley HL. 2004. Transcriptome of uropathogenic Escherichia coli during urinary tract infection. Infect Immun 72:6373–6381. doi: 10.1128/IAI.72.11.6373-6381.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Withman B, Gunasekera TS, Beesetty P, Agans R, Paliy O. 2013. Transcriptional responses of uropathogenic Escherichia coli to increased environmental osmolality caused by salt or urea. Infect Immun 81:80–89. doi: 10.1128/IAI.01049-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Greene SE, Hibbing ME, Janetka J, Chen SL, Hultgren SJ. 2015. Human urine decreases function and expression of type 1 pili in uropathogenic Escherichia coli. mBio 6:e00820-15. doi: 10.1128/mBio.00820-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Eris D, Preston RC, Scharenberg M, Hulliger F, Abgottspon D, Pang L, Jiang X, Schwardt O, Ernst B. 2016. The conformational variability of FimH: which conformation represents the therapeutic target? Chembiochem 17:1012–1020. doi: 10.1002/cbic.201600066. [DOI] [PubMed] [Google Scholar]
- 38.Tivendale KA, Logue CM, Kariyawasam S, Jordan D, Hussein A, Li G, Wannemuehler Y, Nolan LK. 2010. Avian pathogenic Escherichia coli strains are similar to neonatal meningitis E. coli strains and are able to cause meningitis in the rat model of human disease. Infect Immun 78:3412–3419. doi: 10.1128/IAI.00347-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Skyberg JA, Johnson TJ, Johnson JR, Clabots C, Logue CA, Nolan LK. 2006. Acquisition of avian pathogenic Escherichia coli plasmids by a commensal E. coli isolate enhances its abilities to kill chicken embryos, grow in human urine, and colonize the murine kidney. Infect Immun 74:6287–6292. doi: 10.1128/IAI.00363-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marc D, Arne P, Bree A, Dho-Moulin M. 1998. Colonization ability and pathogenic properties of a fim− mutant of an avian strain of Escherichia coli. Res Microbiol 149:473–485. doi: 10.1016/S0923-2508(98)80002-8. [DOI] [PubMed] [Google Scholar]
- 41.de Lorenzo V, Herrero M, Jakubzik U, Timmis KN. 1990. Mini-Tn5 transposon derivates for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in Gram-negative eubacteria. J Bacteriol 172:6568–6572. doi: 10.1128/jb.172.11.6568-6572.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Crepin S, Harel J, Dozois CM. 2012. Chromosomal complementation using Tn7 transposon vectors in Enterobacteriaceae. Appl Environ Microbiol 78:6001–6008. doi: 10.1128/AEM.00986-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang RF, Kushner SR. 1991. Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene 100:195–199. doi: 10.1016/0378-1119(91)90366-J. [DOI] [PubMed] [Google Scholar]
- 45.Kaaden OR, Lange S, Stiburek B. 1982. Establishment and characterization of chicken embryo fibroblast clone LSCC-H32. In Vitro 18:827–834. doi: 10.1007/BF02796323. [DOI] [PubMed] [Google Scholar]
- 46.Crepin S, Lamarche MG, Garneau P, Seguin J, Proulx J, Dozois CM, Harel J. 2008. Genome-wide transcriptional response of an avian pathogenic Escherichia coli (APEC) pst mutant. BMC Genomics 9:568. doi: 10.1186/1471-2164-9-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lymberopoulos MH, Houle S, Daigle F, Leveille S, Bree A, Moulin-Schouleur M, Johnson JR, Dozois CM. 2006. Characterization of Stg fimbriae from an avian pathogenic Escherichia coli O78:K80 strain and assessment of their contribution to colonization of the chicken respiratory tract. J Bacteriol 188:6449–6459. doi: 10.1128/JB.00453-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hagberg L, Engberg I, Freter R, Lam J, Olling S, Svanborg Eden C. 1983. Ascending, unobstructed urinary tract infection in mice caused by pyelonephritogenic Escherichia coli of human origin. Infect Immun 40:273–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Soding J, Thompson JD, Higgins DG. 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]