Coagulase activation of prothrombin by staphylococcus induces the formation of fibrin deposition that facilitates the establishment of infection by Staphylococcus species. Coagulase activity is a key characteristic of Staphylococcus pseudintermedius; however, no coagulase gene or associated protein has been studied to characterize this activity.
KEYWORDS: coagulase, immunoglobulin binding, Staphylococcus, Staphylococcus pseudintermedius, complement C3
ABSTRACT
Coagulase activation of prothrombin by staphylococcus induces the formation of fibrin deposition that facilitates the establishment of infection by Staphylococcus species. Coagulase activity is a key characteristic of Staphylococcus pseudintermedius; however, no coagulase gene or associated protein has been studied to characterize this activity. We report a recombinant protein sharing 40% similarity to Staphylococcus aureus coagulase produced from a putative S. pseudintermedius coagulase gene. Prothrombin activation by the protein was measured with a chromogenic assay using thrombin tripeptide substrate. Stronger interaction with bovine prothrombin than with human prothrombin was observed. The S. pseudintermedius coagulase protein also bound complement C3 and immunoglobulin. Recombinant coagulase facilitated the escape of S. pseudintermedius from phagocytosis, presumably by forming a bridge between opsonizing antibody, complement, and fibrinogen. Evidence from this work suggests that S. pseudintermedius coagulase has multifunctional properties that contribute to immune evasion that likely plays an important role in virulence.
INTRODUCTION
Staphylococcus pseudintermedius is a Gram-positive, coagulase-positive opportunistic bacterial pathogen commonly found on the skin and in the nares, mouth, pharynx, and anus of dogs, cats, and horses (1). Bacterial pyoderma caused by S. pseudintermedius is one of the most common diseases seen in small-animal veterinary practices worldwide, and it occasionally causes other diseases, including otitis and urinary tract infections (2).
The coagulation system prevents dissemination of microbial organisms by fibrin deposition, with subsequent removal by phagocytes (3). However, some bacterial pathogens, such as Staphylococcus aureus, employ a virulence strategy that utilizes coagulation proteins, providing protection from immune defenses and escape from phagocytic killing. Staphylocoagulase activates a host trypsinogen-like enzyme precursor, prothrombin, without proteolytic cleavage to form staphylothrombin and directly initiate blood clotting and subsequent conversion of fibrinogen to fibrinopeptides and self-polymerizing fibrin (4–6). These products mediate adhesion to plasma and extracellular matrix proteins (5, 7). Thus, coagulase embeds bacteria within a network of fibrin, protecting them from immune recognition, and enables staphylococci to multiply as a bacterial community at the center of lesions, forming bacterial microcolonies (8).
Coagulase activity observed in S. aureus, S. pseudintermedius, Staphylococcus intermedius, Staphylococcus delphini, and Staphylococcus schleiferi subsp. coagulans and in a coagulase-variable species, Staphylococcus hyicus subsp. hyicus, is associated with virulence (9–11). Most of these species, including S. pseudintermedius, have been classified as coagulase positive based on plasma clotting tests. However, the protein responsible for this activity is not well defined except in S. aureus. In 2001, Komori et al. isolated a protein with coagulase activity from S. intermedius that differs physically from other coagulases (12). Since that time strains once identified as S. intermedius have been reclassified as S. intermedius and S. pseudintermedius strains (1). The latter species contains a gene often annotated as encoding coagulase protein (13). The predicted protein, however, is not the same as the previously reported S. intermedius protein in size or amino acid composition (12). Preliminary analysis revealed that only one S. pseudintermedius protein contains the domains associated with bacterial coagulase activity and that it is present in all available genomes. This candidate S. pseudintermedius coagulase protein was studied to determine its biological properties and potential role in the virulence of S. pseudintermedius.
RESULTS
Amplification and sequencing of the coa gene from S. pseudintermedius.
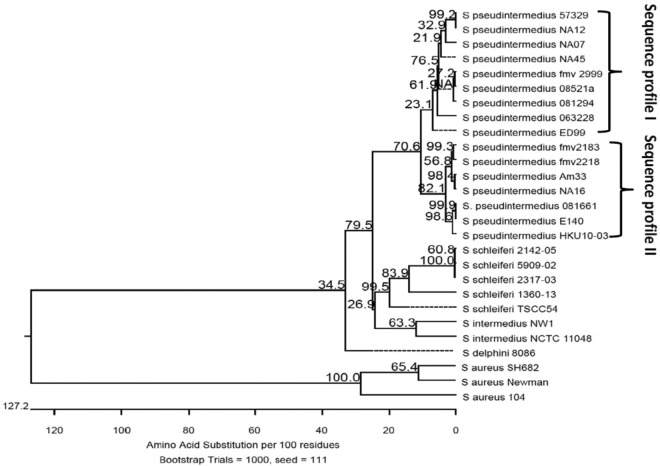
The putative S. pseudintermedius coagulase gene (coa) was amplified by PCR from all 15 strains of S. pseudintermedius tested and yielded a product of the expected size of approximately 1,500 bp. DNA sequencing among the 15 isolates showed that two coagulase sequence profiles were observed; sequence profile I was homologous to that of strain 063228, and sequence profile II was homologous to that of strain 081661, as shown in Fig. 1. The complete coa gene encodes a protein with a predicted molecular mass of 56,490 Da. It corresponds to bases 2612742 to 2614241 in the genome of S. pseudintermedius strain 081661 (GenBank accession number CP016073.1). The predicted protein has a signal sequence in residues 1 to 26 and contains an N-terminal prothrombin binding site in residues 27 to 247, with 34% identity to the D1 and D2 domains of S. aureus coagulase. At residues 280 to 331 in the linker region between the prothrombin binding domain and fibrinogen binding domains, it contains a site similar to the two IgG binding domains of surface immunoglobulin-binding protein (Sbi) of S. aureus (GenBank accession number WP_061740244.1) with 19% and 21% identity, respectively. At fibrinogen binding regions, residues 403 to 430 and residues 431 to 467, it shares 38% and 50% amino acid identity, respectively, to the corresponding regions in S. aureus extracellular fibrinogen binding protein domain A (Efb-A) (GenBank accession number SAZ21881.1). Residues 467 to 491 have 44% identity to complement binding domains of the Sbi protein of S. aureus (GenBank accession number WP_061740244.1) (Fig. 2; see also Fig. S1 to S4 in the supplemental material). Whereas 25% identity is generally considered a good predictor of conserved function, only phenotypic testing can provide definitive results (14).
FIG 1.
Coagulase phylogenetic relationships. Phylogenetic tree showing the inferred evolutionary relationships among coagulase amino acid sequences from various staphylococcus species. The scale bar represents a genetic distance that is equivalent to the number of amino acid substitutions per 100 residues. Bootstrap probability is indicated at diverging points of branches. Coagulase sequences and their GenBank accession numbers are as follows: S. aureus strain SH682, coagulase type III (ABW83067.1), S. aureus subsp. aureus strain Newman, coagulase type III (WP_000744074), S. aureus strain 104, coagulase type I (BAD98736.1), S. intermedius NW1 (WP_086428747.1), S. intermedius NCTC 11048 (WP_019169028.1), S. schleiferi TSCC54 (WP_060829724.1), S. schleiferi 1360-13 (WP_050345467.1), S. schleiferi 2142-05 (AKS71128.1), S. schleiferi 2317-03 (AKS73300.1), S. schleiferi 5909-02 (WP_050337525.1), S. delphini 8086 (WP_019166910.1), S. pseudintermedius HKU-103 (WP_015728558.1), and S. pseudintermedius ED99 (WP_014614858.1). Brackets indicate the two coagulase sequence profiles; sequence profile I is similar to the coagulase sequence of strain 063228, and sequence profile II is similar to the coagulase sequence of strain 081661.
FIG 2.
Coagulase protein structure. The predicted structure of S. pseudintermedius coagulase protein from S. pseudintermedius strain 081661, including the predicted signal peptide (S), the D1 and D2 domains for prothrombin binding, the linker (L) domain containing an IgG binding site, the fibrinogen binding domain of coagulase, and complement binding domain, is shown. Numbers indicate amino acid positions.
Coagulase tests.
The clotting activity of S. pseudintermedius strains (n = 15) was determined by the plasma tube clotting method, and results showed coagulase-positive reactions in 40% (6/15) of the isolates, with only strain 081661 producing a positive result at 4 h of incubation while strains NA12, 063228, 08521a, 081294, and E140 produced positive results after a 48-h incubation.
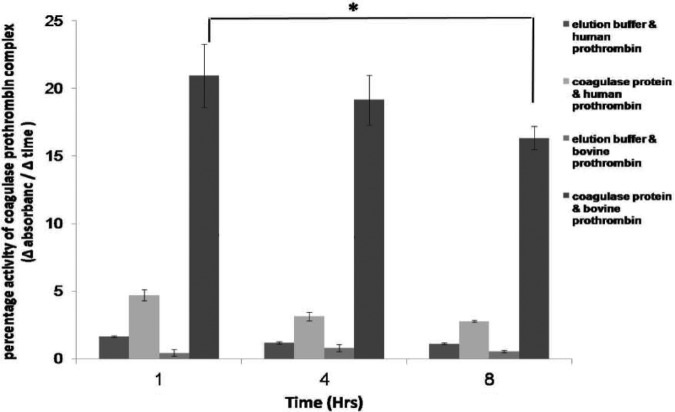
Tested with a chromogenic assay, coagulase activity was detected in supernatants from all S. pseudintermedius strains within 1 h (Fig. 3). Greater hydrolysis of the chromogenic substrate occurred with bovine prothrombin than with human prothrombin. The reactivity of S. pseudintermedius strains with bovine prothrombin was higher than that of the S. aureus positive control. Coagulase activity of isolate E140 was higher than that of all other strains, followed by that of NA45 and 081661, with strain 063228 exhibiting the lowest activity. Significantly higher reactivity occurred with both human and bovine prothrombins among strains with sequence profile II (including all sequence type 71 [ST71] isolates) than in isolates with sequence profile I (including all ST68 isolates) (P < 0.001). S. aureus hydrolysis of the chromogenic substrate was significantly greater than that of all S. pseudintermedius strains in its reactivity with human prothrombin, followed, in order, by the reactivity of S. pseudintermedius strains 081294, NA45, 08521a, and 063228.
FIG 3.
Coagulase activity of S. pseudintermedius. Coagulase activity in concentrated bacterial supernatants was determined using a chromogenic assay. The percentage of coagulase-prothrombin complex activity was inferred by the change in the rate of chromogenic substrate S-2238 hydrolysis (Δ absorbance) over time (Δ time). The parenthetical I and II with the designations refer to coagulase sequence profiles. ST68 isolates (063228, 08521a, and 081294) had significantly lower levels of human and bovine prothrombin activation than ST71 isolates (081661, E140, and NA16) (P < 0.001).
Recombinant coagulase binding to prothrombin, IgG, and complement C3.
Recombinant S. pseudintermedius coagulase protein (prCoa) bound whole IgG and both Fc and Fab fragments of canine IgG, as determined by enzyme-linked immunosorbent assay (ELISA) (Fig. 4A), with the greatest binding to IgG, followed by that to the Fc fragment and less binding to Fab fragments. prCoa bound canine C3 captured from serum in a concentration-dependent manner (Fig. 4B). There was no significant difference in the binding levels of prCoa to bovine and human prothrombins (Fig. 4C).
FIG 4.
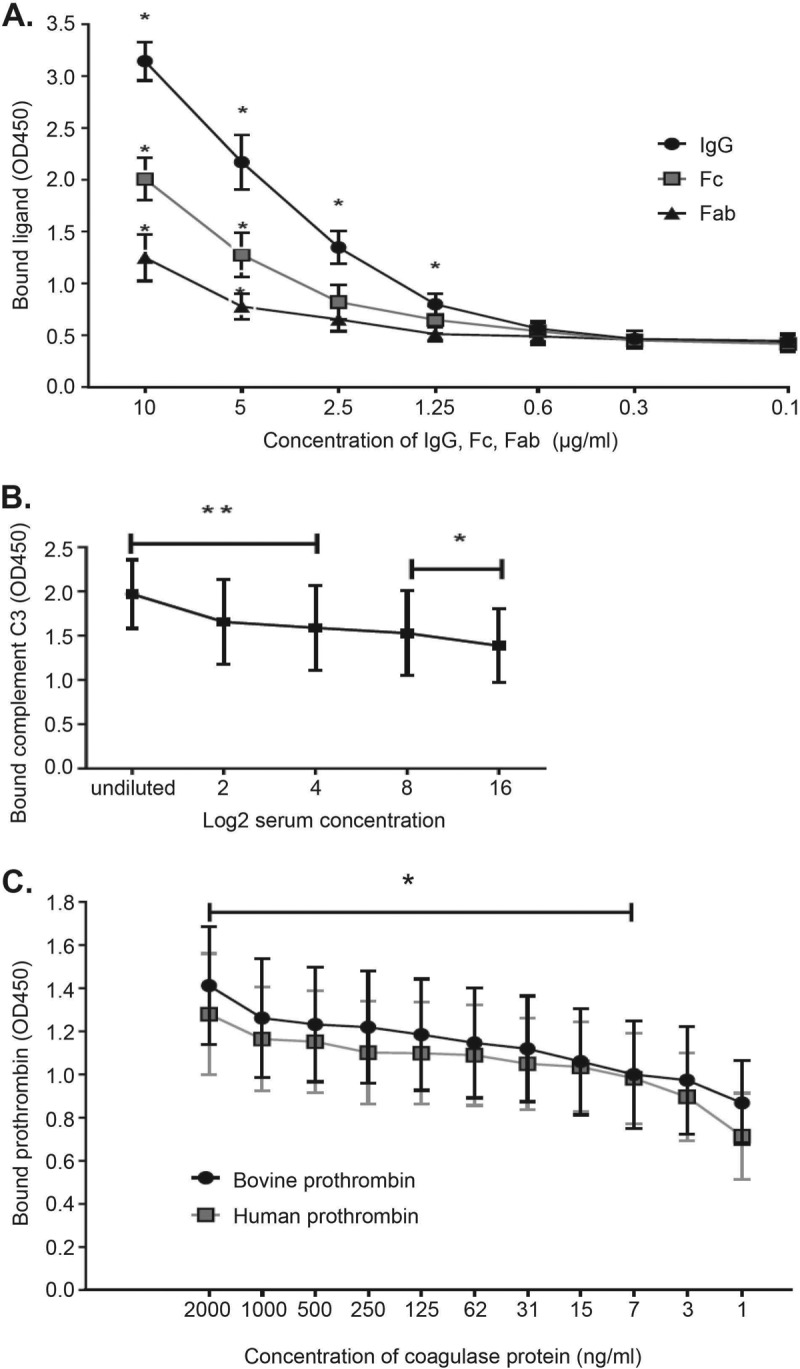
Full-length coagulase protein binding to prothrombin, canine IgG, and complement C3. Data were obtained from three independent experiments. (A) ELISA results showing coagulase binding of canine IgG, Fc, and Fab. There was a significant interaction of treatment (IgG, Fc, and Fab) with the binding of coagulase protein (P = 0.010). All three treatments had significant differences between results at different concentrations (P < 0.001). For IgG concentrations of 10, 5, 2.5, and 1.25 μg/ml, binding was significantly different from that of the negative control (*, P < 0.001). For Fc and Fab concentrations of 5 and 10 μg/ml, binding was significantly different from that of the negative control (*, P < 0.001). The coagulase protein bound more IgG and Fc than Fab. At 5 and 10 μg/ml, IgG binding was significantly higher than that of Fab (P = 0.008), and binding of IgG and Fc was marginally different (P = 0.054). (B) Binding of coagulase to immobilized complement C3. Results for all concentrations were significantly different from the result for the negative control. The first three concentrations differed at a P value of <0.01 (**) (0.000, 0.001, and 0.003, respectively), and the last two concentrations differed at a P value of <0.05 (*) (0.011 and 0.045). (C) Binding of recombinant coagulase protein to immobilized human and bovine prothrombins. There was no significant difference (P = 0.785) in coagulase protein binding levels to human and bovine prothrombin. Binding at all concentrations significantly differed from that of the negative control (*, P < 0.01) except at coagulase concentrations of 3.9 ng/ml (P = 0.053) and 1.9 ng/ml (P = 0.739). OD450, optical density at 450 nm.
Recombinant coagulase activity.
prCoa (50 and 100 μg/ml) yielded positive coagulase activity within 5 h after inoculation by the plasma tube clotting method and remained positive at 24 h. It produced detectable hydrolysis by 1 h in the chromogenic coagulase assay, and this hydrolysis was greater with bovine prothrombin than with human prothrombin (Fig. 5).
FIG 5.
Activity of Staphylococcus pseudintermedius coagulase recombinant protein by chromogenic assay. The percentage of coagulase-prothrombin complex activity was inferred by the change in the rate of chromogen S-2238 hydrolysis (Δ absorbance) over time (Δ time). The coagulase-bovine prothrombin complex percentage activity was significantly higher than the activities of the coagulase-human prothrombin complex and of elution buffer with bovine prothrombin (*, P < 0.001). There was no significant difference between coagulase and elution buffer interactions with human prothrombin (P = 0.522). The percentage of activity did not significantly decrease over time.
Coagulase-mediated binding of fibrinogen to S. pseudintermedius and Staphylococcus epidermidis.
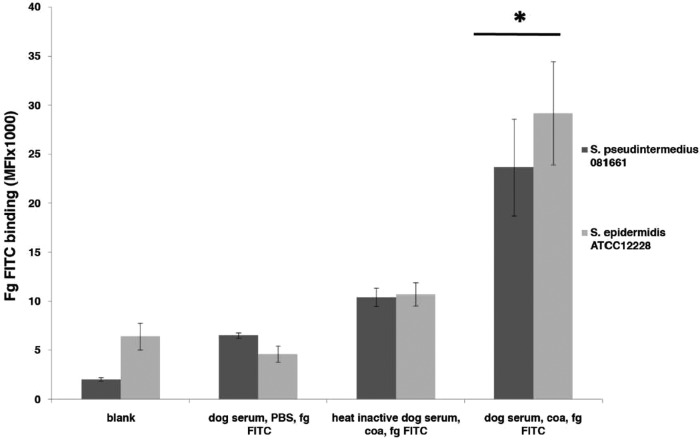
Using flow cytometry it was observed that coagulase facilitated fibrinogen binding to opsonized bacteria (Fig. 6 and S5). To confirm the function of the putative coagulase, we analyzed the ability of purified prCoa to facilitate fibrinogen deposition on S. pseudintermedius and the coagulase-negative, non-biofilm producer S. epidermidis ATCC 12228. The amount of fibrinogen deposited on S. epidermidis was equivalent to that found on S. pseudintermedius.
FIG 6.
Coagulase-mediated fibrinogen binding. Binding of FITC-labeled fibrinogen (Fg; 50 μg/ml) to serum-opsonized S. pseudintermedius and S. epidermidis in the presence of coagulase (coa) was determined. The graph represents the mean and standard error of three independent experiments. The difference in FITC-fibrinogen deposition levels between bacterial strains (S. pseudintermedius 081661 and S. epidermidis ATCC 12228) were not significant (P = 0.287). When the result for each serum type was compared to that of the negative control (blank), only the combination of dog serum, coagulase, and fibrinogen-FITC had a significantly high mean fluorescence intensity (MFI) (*, P < 0.001). Values for the combinations of dog serum and fibrinogen-FITC (P = 1.000) and of heat-inactivated dog serum, coagulase, and FITC-fibrinogen (P = 0.091) did not significantly differ from the value for the negative control.
Coagulase inhibition of phagocytosis in the presence of rabbit plasma.
prCoa reduced phagocytosis when incubated with rabbit plasma and dog blood. The reduction of phagocytosis was dependent on the concentration of coagulase (Fig. 7 and S6).
FIG 7.
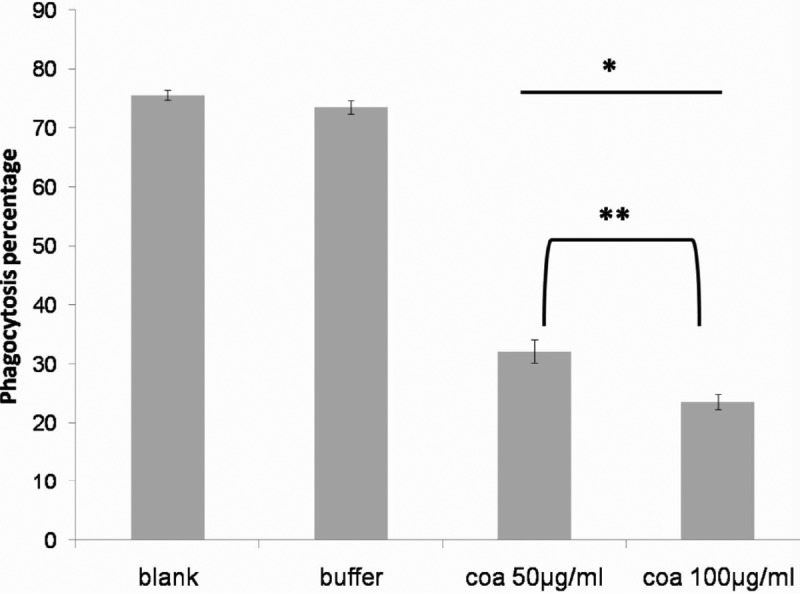
Recombinant coagulase inhibits phagocytosis of S. pseudintermedius. Phagocytosis of fluorescein-labeled bacteria incubated in the presence of rabbit plasma, whole dog blood, and coagulase (coa; 50 and 100 μg/ml) is shown. The graph represents the means and standard errors of three independent experiments, and there was a significant treatment difference (P < 0.001). Comparing the phagocyte-associated fluorescence for each treatment, the result for the blank did not differ from that for the elution buffer control (P = 1.000), and both showed significant increases in phagocyte-associated bacteria (P = 0.015 and P = 0.002, respectively). Values for the blank and buffer significantly differed from those for both coagulase treatments (*, P < 0.001). The two coagulase treatments showed significant decreases in phagocyte-associated bacteria (P = 0.003 for 50 μg/ml coagulase and P = 0.001 for 100 μg/ml coagulase). There was a small but significant difference between results for 50 μg/ml and 100 μg/ml coagulase (**, P = 0.012).
DISCUSSION
The goal of this study was to identify the S. pseudintermedius coagulase gene and to characterize the protein it encodes. Coagulase induces fibrin deposition and enables the establishment of staphylococcal disease (8). S. pseudintermedius is a major etiological agent of skin and soft tissue infections in canines (1, 2), and coagulase activity is considered a key virulence factor for this organism. The protein responsible for this activity and its associated gene have not been previously definitively identified and characterized. A protein with coagulase activity was isolated from S. intermedius in 2001 (12) prior to the delineation of S. pseudintermedius as a separate species in 2005 (1). Neither the sequence of the S. intermedius protein nor the gene encoding it was determined, and, therefore, a direct comparison with our findings is not possible. However, the S. intermedius coagulase protein differs in size and amino acid composition from the one described in the current study. The S. intermedius protein has a molecular weight of 64,500 Da and consists of 615 amino acids. The predicted mature S. pseudintermedius coagulase is considerably smaller, consisting of 499 amino acids with a molecular mass of 53,760 Da without its signal sequence and a mass of 56,490 Da with the signal sequence (varying slightly between strains). It migrates on SDS-PAGE gels at approximately 62 kDa (see Fig. S7 in the supplemental material). Thus, these appear to be two distinctly different proteins or variants.
S. aureus produces a family of well-characterized proteins with overlapping functions called zymogen activator and adhesion proteins (ZAAPs) that serve as a basis for comparison with other species of staphylococci (7). These proteins include staphylocoagulase, von Willebrand factor binding protein (vWBP), and fibrinogen binding protein. The S. pseudintermedius coagulase in the current study has a domain structure similar to the structures of ZAAPs and is 40% similar to the closest corresponding S. aureus coagulase protein. It differs from vWBP in that it lacks a 26-amino-acid motif associated with vWBP activity (15).
Secreted coagulase protein does not interact directly with the bacterial surface but can induce the formation of a fibrinogen shield at some distance from it (16). Coagulase has the ability to associate directly with fibrinogen without the aid of prothrombin (17, 18). This interaction allows coagulase to localize at the cell surface by binding to fibrinogen retained by cell wall-anchored proteins such as clumping factor A (ClfA). The similarity of the fibrinogen binding domains in extracellular fibrinogen binding protein and S. pseudintermedius coagulase shown in this study suggests that coagulase may bind fibrinogen to induce a protective fibrinogen-containing barrier. In this study, using flow cytometry, we identified the coagulase-mediated attachment of soluble fibrinogen to the opsonized bacterial surface, creating a potentially protective shield. S. pseudintermedius recombinant coagulase-mediated fibrinogen deposition occurred not only with S. pseudintermedius but also on non-coagulase-producing S. epidermidis. This observation supports the hypothesis that S. pseudintermedius coagulase may facilitate the coating of coagulase-negative bacteria with a protective layer of fibrinogen. This finding warrants further investigation.
Phagocytosis by neutrophils is strongly enhanced by opsonization of bacteria with plasma factors such as antibodies and complement activation products (C3b and iC3b) (8). S. pseudintermedius coagulase has a novel property in that it binds canine IgG, Fc, and Fab. Based on the similarity of the IgG binding domain of S. aureus Sbi and linker domain of S. pseudintermedius coagulase, this domain is the most likely region of S. pseudintermedius that binds IgG. Moreover, this protein is able to bind canine C3, as shown in binding assays performed in this study. The most likely region for binding with C3 is the coagulase segment of residues 467 to 491 as it has 44% identity to the complement binding domain of the Sbi protein of S. aureus, and both contain the same R and N amino acids with 7 linker amino acids in between.
All of the S. pseudintermedius coagulase sequences in this study contain the same functional motifs. The predicted S. pseudintermedius coagulase proteins diverge into two major groups that correlate with multilocus sequence types (MLSTs). The clinical relevance of higher reactivity among isolates sharing the 081661 genotype, which included all of the ST71 isolates, is of interest. Coagulase tests are routinely used to identify S. aureus and other pathogenic staphylococci. Although less than half of the S. pseudintermedius isolates in this study tested positive with the rabbit plasma tube coagulation test, the coa gene was present in all samples, as determined using PCR and sequencing. Furthermore, all of the isolates tested positive with a chromogenic coagulase assay. The discrepancies between the tests may result from generally weak sensitivity of the rabbit plasma tube coagulase test to S. pseudintermedius. Whereas S. aureus isolates are typically coagulase positive at 4 h, S. pseudintermedius may have a delayed tube coagulase result of over 72 h (19).
In this study, we demonstrated that S. pseudintermedius coagulase effectively reduced phagocytosis of bacteria and decreased phagocytosis in a coagulase dose-dependent manner in a reaction that depends on the interaction of coa with both prothrombin and fibrinogen to form a protective fibrin shield around bacteria.
In summary, the gene examined in this study encodes a novel S. pseudintermedius protein that displays coagulase activity, binds prothrombin, immunoglobulin, and complement C3, facilitates the deposition of fibrinogen on the bacterial surface, and provides protection from phagocytosis.
MATERIALS AND METHODS
Bacterial strains.
A total of 15 clinical isolates obtained from the University of Tennessee, College of Veterinary Medicine Clinical Bacteriology Laboratory, as well as from European and North American collaborators through previous studies, as noted in Table 1, were used (20, 21). They were identified as S. pseudintermedius and represent nine MLST lineages associated with methicillin resistance (21). Methods for bacterial isolation, identification, and antimicrobial susceptibility testing were those routinely used in the laboratory (22). Coagulase-positive S. aureus ATCC 23529 and coagulase-negative S. epidermidis ATCC 12228 were used as assay controls.
TABLE 1.
Origins and sequence types of S. pseudintermedius strains used in this study
| S. pseudintermedius strain | Country of isolation | Multilocus sequence type |
|---|---|---|
| NA45 | USA | ST84 |
| NA12 | USA | ST64 |
| NA07 | USA | ST124 |
| NA16 | USA | ST71 |
| 081661 | USA | ST71 |
| 063228 | USA | ST68 |
| 0821a | USA | ST68 |
| 081294 | USA | ST68 |
| FMV2999-10 | Portugal | ST199 |
| FMV2218-10 | Portugal | ST198 |
| FMV2183-10 | Portugal | ST197 |
| Am33 | Thailand | ST111 |
| 57395 | Israel | ST45 |
| E140 (DK729) | Denmark | ST71 |
| E141 (ED99) | Scotland | ST25 |
DNA extraction and PCR amplification.
Bacterial strains were grown on Trypticase soy agar plates with 5% sheep blood overnight at 37°C. Bacteria derived from a single colony were suspended in 5 ml of Trypticase soy broth (TSB) (Becton, Dickinson, and Co., Sparks, MD) and incubated in a rotary shaker at 225 rpm at 37°C. Bacteria were harvested from 1.8 ml of microbial culture, and DNA was extracted using an UltraClean Microbial DNA isolation kit (Qiagen, Carlsbad, CA).
PCR primers were designed, using the IDT SciTools application (Integrated DNA Technologies, Coralville, IA), for amplification and sequencing of the S. pseudintermedius putative coagulase gene based on genomic data obtained for strains NA45, 063228, and 081661 (13) with GenBank accession numbers CP016072.1, CP015626.1, and CP016073.1, respectively. PCR amplification of full-length coagulase genes was carried out using the oligonucleotide primers pseud coagulase F (5′-TTTGGCCATGGATGAAAAAGAAATTGCTT-3′) and pseud coagulase R (5′-TTTGGGGATCCTGACCGTTGTAAGCTTTAT-3′) containing restriction enzyme sites for NcoI and BamH1 (underlined). The reaction mixtures consisted of a 25-μl total volume containing 2.5 μl of genomic DNA, 20 pmol of each primer (1 μl), 12.5 μl of rTaq polymerase enzyme, and 8 μl of nuclease-free water. Amplification conditions consisted of an initial denaturation (94°C for 1.5 min), followed by 35 cycles of denaturation (94°C for 60s), annealing (55°C for 2 min), and extension (72°C for 60 s), with a single final extension (72°C for 5 min). PCR products were resolved and visualized on 1.4% agarose gels.
Coagulase sequence analysis.
PCR products were enzymatically treated to destroy single-stranded DNA (ExoSap-IT; USB Corp., Cleveland, OH) and submitted to the University of Tennessee Genomics Core Facility for DNA sequencing using the dideoxy chain-termination method. PCR primers were used for direct DNA sequencing of PCR amplification products. The BLAST sequence alignment tool (http://www.ncbi.nlm.nih.gov/blast/) and Geneious software (Biomatters, Auckland, New Zealand) were used to determine nucleotide sequence similarities between S. pseudintermedius isolates. Protein sequences were predicted from each DNA sequence using an online tool (Expasy translate [http://web.expasy.org/translate/]), and a phylogenetic tree was generated using Lasergene MegAlign (version 14.1.0 using Clustal W) comparing coagulase proteins from S. aureus, S. intermedius, S. schleiferi, and S. delphini with sequences obtained from GenBank.
Cloning, expression, and purification of coagulase protein.
Cloning of coa was carried out using the pETBlue-2 vector (MilliporeSigma, Burlington, MA). PCR amplification of full-length coagulase was done with oligonucleotide primers containing NcoI and BamHI restriction sites. PCR products from S. pseudintermedius strain 081661 were digested with NcoI and BamHI and purified (Wizard SV Gel and PCR Clean-Up System; Promega, Madison, WI). They were ligated into the vector, transformed into DH5α Escherichia Coli, and plated on LB agar containing 50 μg/ml ampicillin, 0.1 μM isopropyl-β-d-thiogalactopyranoside (IPTG), and 40 μg/ml 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal). The presence of the cloned fragment was confirmed by PCR.
The coa gene construct was expressed in E. coli Tuner(DE3)/pLacI competent cells (MilliporeSigma). They were plated on LB agar containing 50 μg/ml ampicillin and 34 μg/ml chloramphenicol (LB amp-cam). Stationary-phase broth cultures were grown from freshly transformed E. coli for approximately 16 h. Overnight starter cultures were inoculated into fresh medium at a 1:100 dilution and grown to an optical density (at 600 nm) of 0.6. IPTG (MilliporeSigma) was added to a final concentration of 1 mM, and the cells were incubated at 30°C for an additional 4 h in a shaking incubator at 225 rpm for induction of protein expression.
For the purification of recombinant protein, 100 ml of induced bacterial cultures was centrifuged at 10,000 × g for 15 min. Bacteria were suspended in 5 ml of solubilization buffer (BugBuster master mix; MilliporeSigma) containing 20 μl of protease inhibitor (Cocktail Set III, EDTA-free; MilliporeSigma) and incubated for 30 min at 37°C in a shaking incubator at 225 rpm. Samples were centrifuged at 16,000 × g for 45 min at 4°C to remove insoluble cell debris. Proteins were applied to an immobilized metal ion affinity chromatography column (PrepEase Ni-TED column; Thermo Fisher Scientific, Waltham, MA) and eluted with successively higher concentrations of imidazole up to 250 mM (His-Select elution buffer; MilliporeSigma). Protein concentrations were determined with a bicinchoninic acid (BCA) assay (Thermo Fisher Scientific).
Chromogenic coagulase assay.
A chromogenic assay was used to measure the activity of S. pseudintermedius coagulase. Single colonies of bacterial isolates were cultured overnight in 2 ml of TSB at 37°C. Bacteria were centrifuged at 12,000 × g for 2 min, and then supernatants were concentrated as described above. Tests were performed in flat-bottom microtiter plates. The molar concentrations of coagulase and prothrombin were calculated using molecular mass estimates of 62,000 Da for coagulase and 72,000 Da for human and bovine prothrombin.
An equimolar amount of purified prCoa was mixed with 1 × 10−16 M human prothrombin or bovine prothrombin, or an 18-μl aliquot of concentrated supernatant was mixed with 1 × 10−16 M human prothrombin or bovine prothrombin and incubated for 30 min at 37°C. Chromogenic thrombin tripeptide substrate H-d-Phe-Pip-Arg-pNA (Molecular Innovations, Novi, MI) was added to a final concentration of 1 mM in a total reaction buffer volume of 100 μl of phosphate-buffered saline (PBS) per well. After an initial reading the reaction was allowed to proceed by incubation in the dark for 1, 4, or 8 h at 37°C. The absorbance was measured at 405 nm. Reagent controls (prothrombin alone without substrate), the S. aureus ATCC 25923 positive control, and the S. epidermidis ATCC 12228 negative control were included with each batch. The change in absorbance (A) was plotted, and the slope of the curve (dA/dt) was interpreted to be the rate of substrate hydrolysis measured as the increase in the absorbance and thus reflective of enzymatic function (percent coagulase prothrombin complex activity) (5).
Tube coagulase test.
prCoa at final concentrations of 50 and 100 μg/ml or a single overnight bacterial colony was inoculated into 0.25 ml of EDTA-treated pooled rabbit coagulase plasma (Difco Laboratories, East Molesey, United Kingdom) in a sterile plastic tube, mixed, and then incubated at 37°C. After 4, 24, and 48 h, the mixtures were examined for clotting by tipping the tubes to a 45° angle. Any detectable clotting was interpreted as a positive reaction. S. aureus (ATCC 25923) and S. epidermidis (ATCC 12228) were included as positive and negative controls, respectively. EDTA-treated plasma alone with buffer was included as an additional negative control.
Phagocytosis assay.
For fluorescent labeling of S. pseudintermedius strain 081661, bacteria were suspended in PBS, pH 7.2, and incubated with 1 mM pHrodo green dye (Invitrogen, Mulgrave, Australia) in dimethyl sulfoxide (DMSO) for 45 min at room temperature protected from light. This dye is nonfluorescent outside cells but fluorescent in phagosomes (acidic environment). Bacteria were washed twice with PBS, suspended in PBS containing 10% glycerol, and stored at −80°C. Phagocytosis by leukocytes was measured with 10 μl of pHrodo green-labeled S. pseudintermedius strain 081661 bacteria (109 cells/ml) mixed with 10% rabbit plasma for 30 min at 37°C in the presence of prCoa (50 or 100 μg/ml) or buffer. Then, 200 μl of dog blood freshly collected in EDTA was added and incubated for 30 min at 37°C. The reaction was stopped using red blood cell lysis buffer (Sigma). Cells were washed with PBS and analyzed by flow cytometry (Applied Biosystems Attune flow cytometer; Thermo Fisher Scientific). Neutrophils were gated based on forward and side scatter, and for each sample, fluorescence intensity of 10,000 neutrophils was determined. Phagocytosis was expressed as the percentage of neutrophils that fluoresced compared to that of neutrophils that did not fluoresce.
Fibrinogen deposition on S. pseudintermedius and S. epidermidis.
Bacteria grown to mid-log phase were incubated with 50 μl of dog serum for 30 min at 37°C, washed with PBS, and incubated with prCoa (50 μg/ml) for 1 h at 37°C with shaking. After another washing step, bacteria were incubated with fluorescein isothiocyanate (FITC)-conjugated fibrinogen from pooled human plasma (50 μg/ml; Zedira, Darmstadt, Germany) for 1 h at 37°C with shaking. Controls included heat-inactivated serum with added prCoa, FITC-fibrinogen, or dog serum alone with FITC-fibrinogen. Washed bacteria were analyzed by flow cytometry as described above.
Prothrombin, complement, and IgG binding assays.
To detect the ability of prCoa to bind with complement factor C3, microtiter plates were coated overnight at 4°C with affinity-purified goat anti-dog C3 (Bethyl, Montgomery, TX) (10 μg/ml). After wells were washed with phosphate-buffered saline with Tween 20 (PBST), dog serum (undiluted and 2-fold serially diluted) was added, and the plates were incubated for 1 h at 37°C, washed, and then incubated with prCoa (5 μg/ml) for another hour at 37°C and washed. prCoa binding was detected using a 1:3,000 dilution of horseradish peroxidase (HRP)-conjugated goat anti-His monoclonal antibody (Thermo Fisher Scientific) and visualized using TMB (3,3′,5,5′-tetramethylbenzidine) substrate (Thermo Fisher Scientific).
To detect the binding of IgG, IgG-Fc, and IgG-Fab to prCoa, canine IgG was digested with immobilized papain using a Fab preparation kit (Thermo Fisher Scientific) according to the manufacturer's instructions using a 6-h incubation for 1 mg/ml dog IgG whole molecule (Rockland Antibodies and Assays, Limerick, PA) with papain. Digestion was verified by running the digested IgG in 7.5% polyacrylamide gels using sample buffer without reducing agent (Bio-Rad, Hercules, CA). This was followed by purification and separation of Fab fragments from Fc fragments using a protein A spin column (NAb Protein A Plus; Thermo Fisher Scientific). prCoa (2 μg/ml)-coated plates were incubated with canine whole-molecule IgG, Fc, or Fab fragments (beginning concentration of 10 μg/ml and then serially diluted 2-fold) for 1 h at 37°C. After plates were washed, HRP-conjugated sheep anti-dog IgG heavy chain (1:4,000) (Bethyl) was added, and bound antibody was detected with TMB substrate.
To measure the binding of prCoa to prothrombin, human and bovine prothrombins (2 μg/ml) were adsorbed to a microtiter plate well at 4°C for 16 h. After the wells were washed with PBST, prCoa (beginning at 2 μg/ml and then serially diluted 2-fold) was added, and the plates were incubated for 1 h. Following incubation, HRP-conjugated mouse anti-His monoclonal antibody (Thermo Fisher Scientific), diluted 1:3,000, was added and incubated for 1 h. Plates were washed, and TMB substrate was added. Bound proteins were quantified by measuring the absorbance at 450 nm in a microplate reader.
Statistical analysis.
Repeated-measures mixed-effects analyses of variance were used to test within and between subject effects for binding of prCoa to human and bovine prothrombins, complement C3, and IgG and the reactivity of bacterial supernatant and coagulase recombinant protein to prothrombin in the chromogenic and phagocytosis assays. A 2-by-4 analysis of variance was used to test between-subject effects for fibrinogen-FITC deposition. Post hoc analysis included simple contrasts in comparison to a control or pairwise comparisons. All post hoc tests used a Bonferroni adjustment. All analyses were run in SPSS, version 24.
Supplementary Material
ACKNOWLEDGMENTS
We thank the University of Tennessee Institute of Agriculture Center of Excellence in Livestock Diseases and Human Health and the Egyptian Cultural and Educational Bureau for funding and support.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/IAI.00027-18.
REFERENCES
- 1.Devriese LA, Vancanneyt M, Baele M, Vaneechoutte M, De Graef E, Snauwaert C, Cleenwerck I, Dawyndt P, Swings J, Decostere A, Haesebrouck F. 2005. Staphylococcus pseudintermedius sp. nov., a coagulase-positive species from animals. Int J Syst Evol Microbiol 55:1569–1573. doi: 10.1099/ijs.0.63413-0. [DOI] [PubMed] [Google Scholar]
- 2.Hill PB, Lo A, Eden CA, Huntley S, Morey V, Ramsey S, Richardson C, Smith DJ, Sutton C, Taylor MD, Thorpe E, Tidmarsh R, Williams V. 2006. Survey of the prevalence, diagnosis and treatment of dermatological conditions in small animals in general practice. Vet Rec 158:533–539. doi: 10.1136/vr.158.16.533. [DOI] [PubMed] [Google Scholar]
- 3.Levi M, Keller TT, van Gorp E, ten Cate H. 2003. Infection and inflammation and the coagulation system. Cardiovasc Res 60:26–39. doi: 10.1016/S0008-6363(02)00857-X. [DOI] [PubMed] [Google Scholar]
- 4.Heilmann C, Herrmann M, Kehrel BE, Peters G. 2002. Platelet-binding domains in 2 fibrinogen-binding proteins of Staphylococcus aureus identified by phage display. J Infect Dis 186:32–39. doi: 10.1086/341081. [DOI] [PubMed] [Google Scholar]
- 5.Hemker HC, Bas BM, Muller AD. 1975. Activation of a pro-enzyme by a stoichiometric reaction with another protein. The reaction between prothrombin and staphylocoagulase. Biochim Biophys Acta 379:180–188. [DOI] [PubMed] [Google Scholar]
- 6.Zajdel M, Wegrzynowicz Z, Jeljaszewicz J, Pulverer G. 1973. Mechanism of action of staphylocoagulase and clumping factor. Contrib Microbiol Immunol 1:364–375. [PubMed] [Google Scholar]
- 7.Friedrich R, Panizzi P, Fuentes-Prior P, Richter K, Verhamme I, Anderson PJ, Kawabata S, Huber R, Bode W, Bock PE. 2003. Staphylocoagulase is a prototype for the mechanism of cofactor-induced zymogen activation. Nature 425:535–539. doi: 10.1038/nature01962. [DOI] [PubMed] [Google Scholar]
- 8.Cheng AG, Kim HK, Burts ML, Krausz T, Schneewind O, Missiakas DM. 2009. Genetic requirements for Staphylococcus aureus abscess formation and persistence in host tissues. FASEB J 23:3393–3404. doi: 10.1096/fj.09-135467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Capurro A, Concha C, Nilsson L, Ostensson K. 1999. Identification of coagulase-positive staphylococci isolated from bovine milk. Acta Vet Scand 40:315–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pantucek R, Gotz F, Doskar J, Rosypal S. 1996. Genomic variability of Staphylococcus aureus and the other coagulase-positive Staphylococcus species estimated by macrorestriction analysis using pulsed-field gel electrophoresis. Int J Syst Bacteriol 46:216–222. doi: 10.1099/00207713-46-1-216. [DOI] [PubMed] [Google Scholar]
- 11.Vandenesch F, Lebeau C, Bes M, Lina G, Lina B, Greenland T, Benito Y, Brun Y, Fleurette J, Etienne J. 1994. Clotting activity in Staphylococcus schleiferi subspecies from human patients. J Clin Microbiol 32:388–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Komori Y, Iimura N, Yamashita R, Sugihara H, Nikai T. 2001. Characterization of coagulase from Staphylococcus intermedius. J Nat Toxins 10:111–118. [PubMed] [Google Scholar]
- 13.Riley MC, Perreten V, Bemis DA, Kania SA. 2016. Complete genome sequences of three important methicillin-resistant clinical isolates of eStaphylococcus pseudintermedius. Genome Announc 4:e01194-16. doi: 10.1128/genomeA.01194-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chang GS, Hong Y, Ko KD, Bhardwaj G, Holmes EC, Patterson RL, van Rossum DB. 2008. Phylogenetic profiles reveal evolutionary relationships within the “twilight zone” of sequence similarity. Proc Natl Acad Sci U S A 105:13474–13479. doi: 10.1073/pnas.0803860105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bjerketorp J, Nilsson M, Ljungh A, Flock JI, Jacobsson K, Frykberg L. 2002. A novel Willebrand factor binding protein expressed by Staphylococcus aureus. Microbiology 148:2037–2044. doi: 10.1099/00221287-148-7-2037. [DOI] [PubMed] [Google Scholar]
- 16.Cheng AG, McAdow M, Kim HK, Bae T, Missiakas DM, Schneewind O. 2010. Contribution of coagulases towards Staphylococcus aureus disease and protective immunity. PLoS Pathog 6:e1001036. doi: 10.1371/journal.ppat.1001036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McDevitt D, Vaudaux P, Foster TJ. 1992. Genetic evidence that bound coagulase of Staphylococcus aureus is not clumping factor. Infect Immun 60:1514–1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Panizzi P, Nahrendorf M, Figueiredo JL, Panizzi J, Marinelli BB, Iwamoto Y, Keliher E, Maddur AA, Waterman P, Kroh HK, Leuschner F, Aikawa E, Swirski FK, Pittet MJ, Hackeng TM, Fuentes-Prior P, Schneewind O, Bock PE, Weissleder R. 2011. In vivo detection of Staphylococcus aureus endocarditis by targeting pathogen-specific prothrombin activation. Nat Med 17:1142–1146. doi: 10.1038/nm.2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Watanabe S, Ito T, Takeuchi F, Endo M, Okuno E, Hiramatsu K. 2005. Structural comparison of ten serotypes of staphylocoagulases in Staphylococcus aureus. J Bacteriol 187:3698–3707. doi: 10.1128/JB.187.11.3698-3707.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Perreten V, Kadlec K, Schwarz S, Gronlund Andersson U, Finn M, Greko C, Moodley A, Kania SA, Frank LA, Bemis DA, Franco A, Iurescia M, Battisti A, Duim B, Wagenaar JA, van Duijkeren E, Weese JS, Fitzgerald JR, Rossano A, Guardabassi L. 2010. Clonal spread of methicillin-resistant Staphylococcus pseudintermedius in Europe and North America: an international multicentre study. J Antimicrob Chemother 65:1145–1154. doi: 10.1093/jac/dkq078. [DOI] [PubMed] [Google Scholar]
- 21.Videla R, Solyman SM, Brahmbhatt A, Sadeghi L, Bemis DA, Kania SA. 2017. Clonal complexes and antimicrobial susceptibility profiles of Staphylococcus pseudintermedius isolates from dogs in the United States. Microb Drug Resist 24:83–88. doi: 10.1089/mdr.2016.0250. [DOI] [PubMed] [Google Scholar]
- 22.Jones RD, Kania SA, Rohrbach BW, Frank LA, Bemis DA. 2007. Prevalence of oxacillin- and multidrug-resistant staphylococci in clinical samples from dogs: 1,772 samples (2001–2005). J Am Vet Med Assoc 230:221–227. doi: 10.2460/javma.230.2.221. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.